Publisher's Note: There is an Inside Blood Commentary on this article in this issue.
Key Points
Type 1 VWD in the United States is highly variable, including patients with very low VWF levels as well as those with mild or minimal VWF deficiency.
The frequency of sequence variants in the VWF gene increases with decreasing VWF level, but BS does not vary by VWF level.
Abstract
von Willebrand disease (VWD) is the most common inherited bleeding disorder, and type 1 VWD is the most common VWD variant. Despite its frequency, diagnosis of type 1 VWD remains the subject of debate. In order to study the spectrum of type 1 VWD in the United States, the Zimmerman Program enrolled 482 subjects with a previous diagnosis of type 1 VWD without stringent laboratory diagnostic criteria. von Willebrand factor (VWF) laboratory testing and full-length VWF gene sequencing was performed for all index cases and healthy control subjects in a central laboratory. Bleeding phenotype was characterized using the International Society on Thrombosis and Haemostasis bleeding assessment tool. At study entry, 64% of subjects had VWF antigen (VWF:Ag) or VWF ristocetin cofactor activity below the lower limit of normal, whereas 36% had normal VWF levels. VWF sequence variations were most frequent in subjects with VWF:Ag <30 IU/dL (82%), whereas subjects with type 1 VWD and VWF:Ag ≥30 IU/dL had an intermediate frequency of variants (44%). Subjects whose VWF testing was normal at study entry had a similar rate of sequence variations as the healthy controls (14%). All subjects with severe type 1 VWD and VWF:Ag ≤5 IU/dL had an abnormal bleeding score (BS), but otherwise BS did not correlate with VWF:Ag. Subjects with a historical diagnosis of type 1 VWD had similar rates of abnormal BS compared with subjects with low VWF levels at study entry. Type 1 VWD in the United States is highly variable, and bleeding symptoms are frequent in this population.
Introduction
von Willebrand disease (VWD) is the most common inherited bleeding disorder, affecting ∼1:1000 individuals.1 The most common variant of VWD in clinical practice is type 1 VWD, which presents with mild to moderate mucosal bleeding symptoms, typically associated with a family history of bleeding and a quantitative reduction in von Willebrand factor (VWF) protein. The true incidence of VWD is difficult to determine. Low levels of VWF are seen in up to 1% of the population, but not all have clinically significant bleeding.2,3 On the other hand, mild bleeding symptoms are not uncommon, such that the coincidental association of low VWF levels and bleeding may lead to an erroneous diagnosis.4,5 Bleeding symptoms are difficult to quantify, but much recent work has been performed adapting bleeding assessment tools (BATs) and assessing their reliability in VWD diagnosis.6
Lack of reliable screening tests for VWD also complicates diagnosis, in that no single screening test can confirm the presence or absence of VWD, and an array of tests is required to diagnose and classify the type of VWD present. VWF antigen (VWF:Ag) is used to measure total VWF protein present in a sample, whereas VWF ristocetin cofactor activity (VWF:RCo) is used as a surrogate measure of VWF platelet binding.7,8 Shear stress initiates VWF platelet interactions in vivo, but no shear-based clinical assays are presently available to allow efficient diagnosis of VWD in the clinical laboratory setting.
The National Heart, Lung, and Blood Institute (NHLBI) published guidelines in 2008 suggesting that laboratory values of VWF:Ag or VWF:RCo <30 IU/dL serve as a cutoff for the diagnosis of type 1 VWD.9 Other groups have suggested 40 IU/dL as a cutoff, although this results in an increased number of VWD patients.10,11 The hematology community has been cautioned about the risk of over diagnosis, including an eloquent plea invoking type 1 VWD as a “diagnosis in search of a disease” by Sadler.4 However, for clinicians faced with patients who bleed and have low VWF levels but an otherwise negative laboratory evaluation, assigning a diagnosis of VWD allows a route to treatment if needed.
Several groups have recently examined cohorts of type 1 VWD patients, including the UK Haemophilia Centre Doctors’ Organisation VWD study,12 the Canadian type 1 VWD study,13 and the European Union Molecular and Clinical Markers for the Diagnosis and Management of Type 1 VWD study.14 We sought to evaluate the spectrum of type 1 VWD in the United States through a large multicenter National Institutes of Health–supported study (The Zimmerman Program for the Molecular and Clinical Biology of VWD, or the Zimmerman Program) that enrolled patients from 31 US hematology centers and also evaluated healthy control subjects for comparison. In order to evaluate the true fidelity of VWD in clinical practice in the United States, inclusion was based on a past diagnosis of VWD and treatment as such by the patients’ physicians without employing additional strict diagnostic criteria. Phenotypic evaluation of bleeding was measured by a BAT; laboratory evaluation of VWF was examined through a series of VWF assays; and genetic evaluation of VWF was performed by Sanger sequencing and comparative genomic hybridization. The results presented here demonstrate the high degree of variability in bleeding symptoms and VWF laboratory testing observed in subjects with a diagnosis of type 1 VWD in the United States.
Methods
Subjects were enrolled in the Zimmerman Program through 8 primary centers and 23 secondary centers across the United States (see supplemental Appendix, available on the Blood Web site). The Institutional Review Board of each study center approved the study, and informed consent was obtained from each subject. A preexisting diagnosis of VWD of any type was required for study entry. Although family members were enrolled, only probands were included in this current analysis. For this analysis, subjects were assigned to the “type 1 VWD” cohort if they had a current laboratory phenotypic diagnosis of type 1 VWD based on either VWF:Ag or VWF:RCo <30 IU/dL as measured by the central laboratory at the time of study entry as per the NHLBI guidelines, or low VWF with VWF:Ag 30 to 49 IU/dL and/or VWF:RCo 30 to 53 IU/dL at the time of study entry to include subjects with levels below the lower limit of normal for each assay. Subjects were assigned to the “historical type 1 VWD” cohort if they were enrolled with a diagnosis of type 1 VWD, but at the time of enrollment, their central laboratory findings did not support the laboratory criteria for diagnosis of VWD.
Phenotypic evaluation
A bleeding questionnaire was administered to each subject, comprised of questions that enabled calculation of a formal bleeding score (BS), as well as Zimmerman Program-specific questions. For the purpose of this study, BS were calculated using the International Society on Thrombosis and Haemostasis (ISTH) BAT.15 The bleeding questionnaire and other questions were asked by a trained study coordinator, nurse, or physician. Race and ethnicity were as self-reported by subject. Subjects were encouraged but not required to answer all questions.
Laboratory evaluation
Blood samples were collected from each subject at the time of study enrollment. Samples collected in 3.2% sodium citrate were centrifuged and plasma frozen at −80°C and sent to the central laboratory (the Hemostasis Reference Laboratory at the BloodCenter of Wisconsin) for further testing. All samples were maintained at −80°C for long-term storage. VWF:Ag was performed by enzyme-linked immunosorbent assay.16 VWF:RCo was performed by automated platelet agglutination.16 Factor VIII activity was performed by a one-stage clotting assay.17 Multimer distribution was assayed by quantitative gel electrophoresis.18 Ligand binding assays were performed as previously described, including VWF binding to type III collagen (VWF:CB3),16 VWF binding to type IV collagen (VWF:CB4),19 and VWF binding to mutant platelet GPIb (VWF:GPIbM).20,21 VWF propeptide (VWFpp) was measured to evaluate for VWF clearance defects.22 Blood type was ascertained by reverse typing.16 When possible, results were compared with historical laboratory results available for the subject. The historical results were performed in a variety of different laboratories, and at a variable number of years prior to study enrollment.
Genetic evaluation
One additional blood sample per subject was collected in EDTA, and the whole blood was shipped at room temperature to the Hemostasis Reference Laboratory at the BloodCenter of Wisconsin. DNA was extracted and subjected to Sanger sequencing of all exons, including intron-exon boundaries and ∼50 to 100 base pairs of intronic sequence at either the Harvard Partners Genome Center or the BloodCenter of Wisconsin using the VWF reference sequence NM_000552.23 For the purpose of this study, sequence variations were stated as such if they were seen in <1% of the Zimmerman Program healthy controls. Any variant present at higher frequencies in healthy control subjects was excluded from this analysis. Comparative genomic hybridization was used to evaluate for large deletions or insertions by analysis of copy number variation.24,25
Statistical analysis
To compare the categorical outcomes, χ2 tests were used and Kruskal–Wallis/Mann–Whitney tests were used to compare the continuous outcomes across the groups. In addition, log transformed outcomes (because the outcomes were fairly skewed) were used for the multivariable model. A generalized linear model was used and a stepwise selection procedure (which included any variable with α ≤ 0.10) was applied to select the best set of predictors for the respective outcomes.
Results
When considering all subjects originally enrolled as type 1 VWD, the median VWF:Ag was 47 IU/dL and the median VWF:RCo was 45 IU/dL. The median BS was 5. We substantiated low VWF levels in 64% of the subjects. These subjects were assigned to the “type 1 VWD” cohort for further analysis. In 36% of subjects, normal VWF levels were found at study entry and were assigned to the “historical type 1 VWD” cohort. The demographic characteristics of each cohort are given in Table 1. The type 1 VWD cohort was further divided into clearance defects (type 1 C), type 1 severe (type 1S, VWF:Ag 1 to 5 IU/dL), or type 1 (supplemental Table 1). A propeptide to antigen ratio >3 was used to define type 1C subjects.22,26 The type 1C cohort had median VWF:Ag of 8 IU/dL and median VWF:RCo of 5 IU/dL. The median VWFpp/VWF:Ag ratio was elevated in type 1C subjects with a median ratio of 5.33. When the entire type 1 cohort with the exception of type 1C subjects was analyzed, the median VWF:Ag was 39 IU/dL and the median VWF:RCo was 38 IU/dL. The historical type 1 VWD cohort had a median VWF:Ag of 76 IU/dL and a median VWF:RCo of 72 IU/dL. Differences in collagen binding were observed as previously reported.19 At the time of initiation of this study, type 2M did not routinely include collagen-binding variants, so these subjects remained in the type 1 cohort for this study.
Table 1.
Characterization of VWD cohorts
| Type 1 VWD cohort | Historical type 1 VWD cohort | Healthy control cohort | |
|---|---|---|---|
| No. of subjects | 310 | 172 | 257 |
| No. Caucasian (%) | 263 (85) | 152 (88) | 139 (54) |
| No. African American (%) | 19 (6) | 7 (4) | 67 (26) |
| No. Hispanic (%) | 35 (11) | 16 (9) | 46 (18) |
| No. female (%) | 204 (66) | 115 (67) | 193 (75) |
| Mean (SD) age at enrollment | 19 (15) | 21 (16) | 38 (11) |
| No. under age 18 (%) | 197 (64) | 100 (58) | 0 |
Subjects self categorized race (Caucasian, African American, Asian, American Indian, Native Hawaiian, and multiple race) and ethnicity (Hispanic and non-Hispanic), and had the option of not answering either question (>95% of each cohort had recorded answers for race and ethnicity). Race and ethnicity were separate questions and therefore the percentages do not always add up to 100%.
SD, standard deviation.
Similar racial and ethnic distributions were observed for each group of type 1 subjects, but increased numbers of African Americans were enrolled as healthy controls, potentially increasing the median VWF:Ag and decreasing the median VWF:RCo (Table 1). Although the numbers of African American subjects with type 1 VWD were small, we did investigate the potential for laboratory differences based on race. No significant difference was seen for VWF:Ag, VWF:RCo, or VWF:RCo/VWF:Ag for African American type 1 subjects as compared with Caucasian type 1 VWD subjects (P = NS), although African American subjects did have a lower mean ratio of 0.87 as compared with a ratio of 1.00 for the Caucasian subjects. When subjects were analyzed based on ethnicity, no difference was seen for Hispanic compared with non-Hispanic subjects for VWF:Ag, VWF:RCo, or VWF:RCo/VWF:Ag (P = NS), although again, VWF:RCo/VWF:Ag ratios trended to be lower in Hispanic subjects (mean ratio, 0.89).
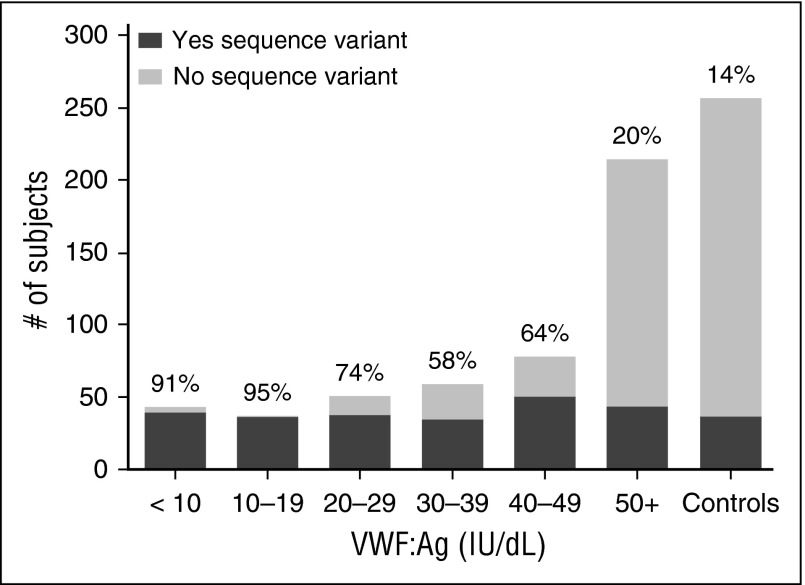
We next examined the genotype of subjects with VWD. Sequence variations, defined as a variant present in <1% of the healthy control population, were found in 45% of subjects enrolled as type 1 VWD (including both type 1 and historical type 1 subjects). In the group with type 1 VWD at study entry, 62% had a sequence variation in the VWF gene (84% of subjects with either VWF:Ag or VWF:RCo <30 IU/dL and 44% of subjects with levels >30 IU/dL). The historical type 1 VWD cohort had only 14% of subjects with sequence variations, similar to the 14% frequency seen in healthy controls. Figure 1 shows the percentage of subjects with sequence variations by VWF:Ag level. Many of the sequence variations found in the healthy control subjects were present in more than 1 subject, and African American healthy controls accounted for many of the sequence variations seen, as previously reported.23 Of the subjects with severe type 1 VWD (VWF:Ag 1 to 5 IU/dL), 100% had a sequence variation, whereas 88% of type 1C subjects had a sequence variation (supplemental Figure 1).
Figure 1.
Sequence variations in VWD are most common in subjects with VWF:Ag <30 IU/dL. This graph shows the number of subjects with sequence variations (either point mutations, or insertions or deletions) in the VWF coding sequence (dark gray) as compared with those without sequence variations in the VWF coding sequence (light gray) for the entire type 1 VWD cohort by VWF:Ag as compared with the healthy controls. The percent of each group with sequence variations is shown at the top of each column. Sequence variations were most common in those with VWF:Ag <30.
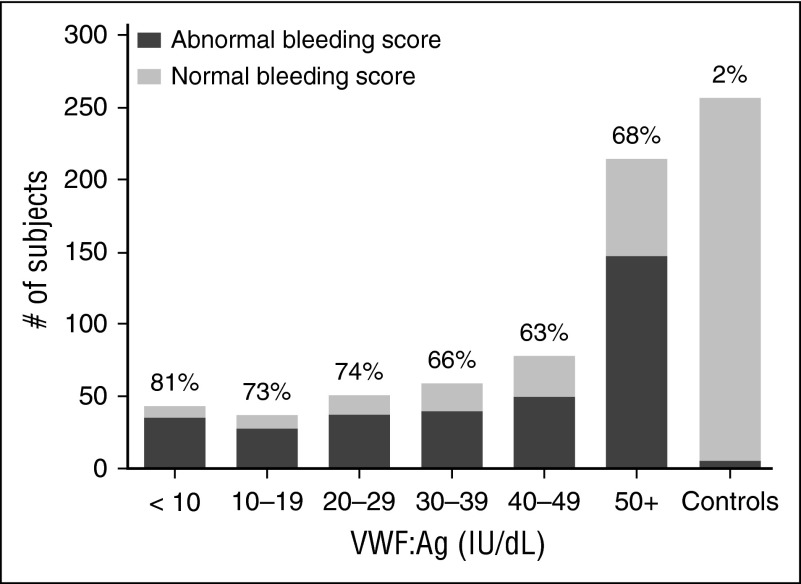
Figure 2 shows the BS by VWF:Ag for all type 1 subjects. When BS as assessed by the ISTH BAT15 was compared, there was no difference in BS between the type 1 cohort and the historical VWD cohort, both with median BS of 5 (P = NS). Both type 1 and historical type 1 subjects had significantly higher BS than the healthy controls (P < .01). When subdivided (supplemental Figure 2), the type 1C subjects had a similar median BS of 6 as compared with the remainder of the type 1 subjects (P = NS), but severe type 1 subjects had a much higher median BS of 15 (P < .001). The Zimmerman Program type 2 VWD subjects as a combined group had a median BS of 8 and type 3 VWD subjects had a median BS of 15.27
Figure 2.
No significant difference in BS for type 1 VWD subjects regardless of VWF:Ag level. This graph shows the number of subjects with abnormal BS (defined as >2 in children <18 years of age, >3 in adult males, and >5 in adult females) in dark gray as compared with those with normal BS (light gray) for the entire type 1 VWD cohort by VWF:Ag. The percent of each group with abnormal BS is shown at the top of each column. BS were similar for type 1 subjects regardless of VWF:Ag.
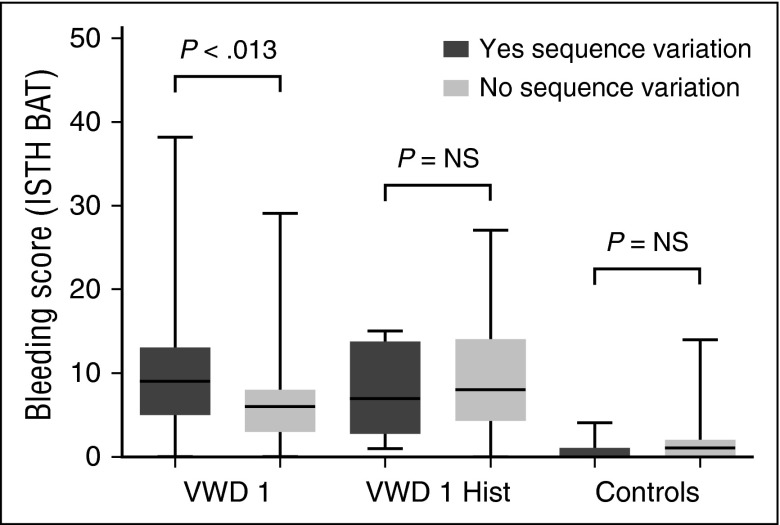
Furthermore, BS within each of the VWD groups varied; 24% of type 1 VWD subjects had BS in the normal range, which is a score of 0 to 3 for adult males, 0 to 5 for adult females, and 0 to 2 for all children <18 years of age.28 This pattern was present in both pediatric and adult subjects ≥18 years of age, suggesting that the variable bleeding phenotype seen in type 1 VWD is not solely a function of age and exposure to hemostatic challenges. There was no difference in BS between males and females in the type 1 cohort, but there was a significant difference between females and males in the historical VWD cohort (P < .001), with higher BS observed in adult female subjects. There was no difference in bleeding phenotype between boys and girls <10 years of age. Comparison of BS for subjects with and without a sequence variation revealed no difference in adult subjects with historical type 1 VWD but a borderline significant difference in those subjects with type 1 VWD (Figure 3).
Figure 3.
Correlation of sequence variations with BS. This box and whisker plot compares BS using the ISTH BAT for adult subjects (≥18 years of age) with type 1 VWD (VWF:Ag and/or VWF:RCo below the lower limit of normal at study entry) in the first pair of columns (“VWD 1”), those with a historical diagnosis of type 1 VWD but normal laboratory findings at study entry in the second pair of columns (“VWD 1 Hist”), and a comparison group of healthy control subjects in the third pair of column (“Controls”). Those subjects with a sequence variation are shown in dark gray, whereas those without a sequence variation are shown in light gray. There was no significant difference in BS between those with and those without a sequence variation for the historical type 1 cohort, and a borderline significant difference for the type 1 VWD cohort. NS, not significant.
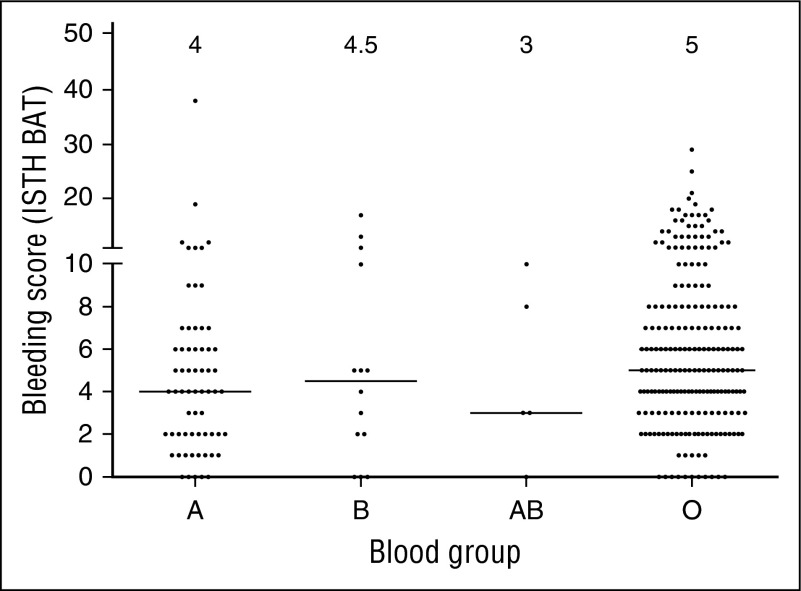
Because blood group O is linked to lower VWF levels, we also investigated each group of subjects by blood type. Although blood group O is present in ∼45% of the general population, subjects with blood group O represented 73% of the type 1 subjects. Group A and B were underrepresented, each present at about half the frequency expected based on population data (21% and 4%, respectively, enrolled in Zimmerman Program vs 40% and 11%, respectively, in the general population). Only a few blood group AB subjects were enrolled, similar to frequencies seen in the normal population. Similar blood group distributions were observed in the historical type 1 VWD cohort. Sequence variations were more frequent in non-group O subjects. Although only 54% of group O subjects had a sequence variation found, 75% of group A, 93% of group B, and 80% of group AB subjects in the type 1 cohort had a sequence variation in the VWF coding sequence. Figure 4 shows BS by blood group.
Figure 4.
BS vary across blood groups in type 1 VWD subjects. This graph shows BS for subjects with blood group A, AB, B, and O. Median BS are shown at the top of the graph. No significant difference was seen between blood group O and blood group B or AB. A borderline significant difference was seen comparing blood group O and blood group A (P < .015).
Alternate assays of VWF function were examined (Table 2), including non–ristocetin-mediated platelet binding (VWF:GPIbM) and collagen binding (VWF:CB3 and VWF:CB4). VWF platelet binding as measured by VWF:GPIbM was similar to the traditional VWF:RCo for type 1 VWD subjects. VWF:CB3/VWF:Ag ratios were normal for subjects with VWF:Ag >10 IU/dL, with the exception of 1 subject with a previously reported p.H1786D sequence variant.29 VWF:CB4/VWF:Ag ratios were normal for most subjects, but 12 subjects (4%) had low VWF:CB4/VWF:Ag ratios, as previously reported.19 The median BS for the subjects with low VWF:CB4 group was 10.5, as compared with a median BS of 5 for the remainder of the type 1 subjects with VWF:CB4/VWF:Ag >0.5 or median BS of 6 when comparison type 1 subjects were matched for comparable VWF:Ag, age, gender, race, and ethnicity. This difference was not statistically significant, but when subjects <18 were excluded (due to having less time to generate significant bleeding symptoms), the median BS was 13.5 for those with low VWF:CB4/VWF:Ag ratios compared with 7 for those with normal ratios (P < .01).
Table 2.
VWF laboratory testing in VWD cohorts
| Type 1 VWD cohort (n = 140) | Historical type 1 VWD cohort (n = 172) | Healthy control cohort (n = 257) | |
|---|---|---|---|
| VWF:Ag (IU/dL) | 36 (17-46) | 76 (64-96) | 104 (85-143) |
| VWF:RCo (IU/dL) | 33 (19-44) | 72 (61-97) | 100 (76-141) |
| VWF:RCo/VWF:Ag ratio | 0.98 (0.84-1.13) | 1.00 (0.87-1.11) | 0.96 (0.84-1.08) |
| No. with normal multimer distribution (%) | 285 (92) | 163 (95) | 255 (99) |
| VWF:GPIbM | 40 (19-54) | 94 (75-121) | 108 (80-142) |
| VWFpp | 53 (39-66) | 74 (66-88) | 88 (75-102) |
| VWFpp/VWF:Ag ratio | 1.50 (1.23-2.28) | 0.98 (0.79-1.15) | 0.80 (0.63-0.99) |
| FVIII:C | 53 (38-71) | 85 (73-101) | 102 (84-125) |
| VWF:CB3 | 39 (20-52) | 83 (72-104) | 121 (3-167) |
| VWF:CB4 | 31 (19-42) | 71 (56-99) | 108 (74-163) |
| No. with sequence variants in VWF (%) | 193 (62) | 24 (14) | 36 (14) |
| BS | 5 (3-8) | 5 (3-9) | 1 (0-2) |
Results are given as median (interquartile range). For VWF:RCo, the lower limit of detection is 10 IU/dL. Therefore levels <10 IU/dL were given an average value of 5 IU/dL for calculation of means, with the underlying assumption that levels below 10 would be normally distributed.
FVIII:C, factor VIII activity.
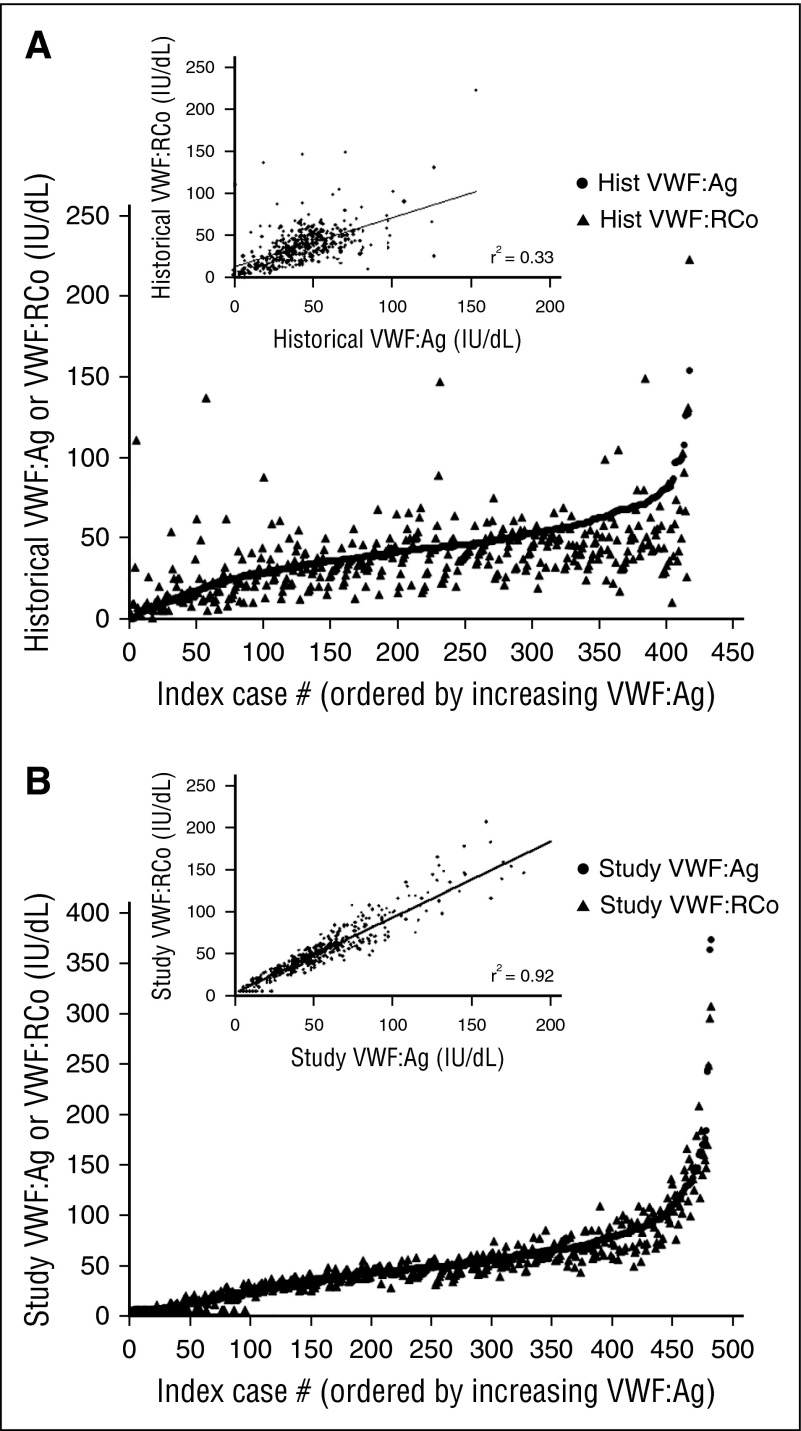
Historical VWF levels were available on 88% of the subjects enrolled initially as type 1 VWD (Figure 5A). VWF:Ag values varied from 0 to 154 IU/dL, whereas VWF:RCo values varied from 0 to 223 IU/dL. These levels were obtained anywhere from 1 to 30 years prior to study enrollment. Recorded levels occasionally might have been obtained following treatment and were not all necessarily the original diagnostic laboratory findings, nor were all VWF:Ag and VWF:RCo necessarily drawn at the same time. Correlation between VWF:RCo and VWF:Ag was improved at study entry (Figure 5B), as both levels were obtained from the same sample and all testing was run in the same laboratory. Of the subjects with historical type 1 VWD and at least 1 additional family member enrolled in the Zimmerman Program, 42% had family members with type 1 VWD (37 subjects) or a diagnosis of historical type 1 VWD (8 subjects), whereas 58% did not have affected family members. However, this data should be interpreted with caution given that not all family members were available for enrollment.
Figure 5.
Variation in historical VWF testing for Zimmerman Program subjects. (A-B) Comparison of the historical (A) and study entry (B) VWF:Ag (circles) and VWF:RCo (triangles) for all subjects enrolled with a diagnosis of type 1 VWD. The insets show the comparison of VWF:Ag on the x-axis and VWF:RCo on the y-axis for historical laboratory values (A) and study entry laboratory values (B). The correlation is much lower for historical values and improved for study entry values, as expected, given that all study testing was performed in the same laboratory and all testing was performed on the same sample for each subject. However, there still remain issues with the lower limit of the ristocetin cofactor assay, as seen by the number of VWF:RCo values at or below the lower limit of detection. hist, historical.
Discussion
Although the initial subject enrollment was performed based on the preexisting diagnoses from the referring center, this current assignment of diagnoses in this study was by phenotypic diagnosis based on careful review of central laboratory results. Discordance between study laboratory findings and the enrollment diagnosis was observed for a significant number of subjects (172 subjects, 36%) with some subjects having historically low VWF levels but normal levels at time of entry. Approximately one-third (36%) of subjects were enrolled with a preexisting diagnosis of type 1 VWD but did not have laboratory evidence of type 1 VWD at the time of study entry. There are a number of possible reasons for this lack of diagnostic fidelity. VWF levels increase with age, such that patients diagnosed many years prior to study entry may have “outgrown” their diagnosis30-32; furthermore, the appropriate reference interval for an older adult is not well defined. Assays for VWF function may not be ideal, resulting in potential false positive or false negative laboratory results. This is particularly true for the VWF:RCo with its high coefficient of variation.33,34 Stress or underlying inflammatory conditions at the time of study entry may have also contributed to increased VWF levels.35 Hormones and pregnancy can elevate VWF levels. Pre-analytical variability may come from specimen handling prior to reaching the laboratory, and laboratory techniques in measuring VWF activity are also subject to variation. It is also plausible that in some cases, the presence of bleeding symptoms and a single low VWF level resulted in a diagnosis of VWD. Almost half of the subjects in the historical type 1 VWD category did have affected family members with current or historical low VWF. Because the subjects enrolled in this study represent only those subjects followed by an adult or pediatric hematologist, typically through a hemophilia treatment center, we suspect that variability in type 1 VWD seen in the community may be even greater than that demonstrated here.
Laboratory findings consistent with a diagnosis of type 1 VWD are generally considered to include decreased but proportional VWF:Ag and activity. Typical VWF activity testing in the United States includes RCo and often multimer distribution, and in some cases, collagen binding with types 1 and/or 3 collagen. Our data showed that subjects with laboratory findings otherwise consistent with type 1 VWD but with low VWF:CB4 had increased bleeding symptoms as measured by BS.19 Although only 12 subjects were affected, this represented 4% of type 1 subjects. This raises the possibility that these subjects may be better classified as type 2M VWD on the basis of a functional defect in the VWF protein, even though the VWF:RCo/VWF:Ag ratio was normal. We elected to include them as type 1 for this analysis, because the collagen testing was performed following study entry as a research test, but they may best fit as type 2M variants.
Genetic analysis of VWF is currently not part of the typical laboratory workup for VWD. We have included as sequence variations any novel or previously reported variant found in <1% of our healthy control population, and excluded variants in 1% or more of the healthy controls for the purpose of this analysis. For example, the p.D1472H sequence variation is found at high frequency in African Americans, and is associated with low VWF:RCo/VWF:Ag ratios but not with an increased risk of bleeding.16 However, there are two potential limitations with this approach. First, not all sequence variations cause disease, and even unique variants may be benign. Therefore, caution should be used in attributing VWD to any specific genetic variant until more careful analysis is performed. Second, even sequence variants occurring at relatively high frequency may result in changes in VWF, which might not be apparent in a healthy control but when inherited or expressed in conjunction with another hemostatic defect, might result in VWD. Further research is needed to clarify both these possibilities and their implications for diagnosis of VWD.
There are now several reported modifier genes not examined in our study that can affect VWF levels, apart from the VWF gene, including CLEC4M and STXBP5.36,37 A chromosome 2 locus affecting VWF levels has also been identified from sibling studies.38 However, our reported frequency of VWF sequence variations of 62% in all subjects with type 1 VWD is similar to that reported in several other studies, including the United Kingdom, Canadian, European Union, and German studies.12-14,39 Four subjects with VWF sequence variations had large deletions that would not have been picked up on conventional sequencing but were picked up via comparative genomic hybridization.
Phenotypic assessment of bleeding symptoms is challenging, but the advent of new BATs allows for calculation of a numerical BS for patients. In our study, there was little difference in median BS for subjects with low VWF:Ag as compared with subjects with higher VWF:Ag. The relatively low scores could be due to the inclusion of a large number of children, who have had less time to accumulate bleeding symptoms. It is possible that the BAT may be less sensitive in children with fewer hemostatic challenges, although different normal ranges are used in children.28 In addition, our BAT was performed following diagnosis, such that some patients may have acquired higher scores due to a history of previous treatment of known VWD. Evaluation of BAT at time of diagnosis and following changes in BAT and VWF levels over time may be more predictive. Previous studies have showed that the BAT has excellent negative predictive value but lower positive predictive value when used as a screening tool.40 It may also be very sensitive to mild decreases in VWF level, but in this study did not predict the presence of a sequence variation or low VWF levels. Other investigators have examined the use of BS as a predictor of VWD and found that higher BS, particularly in families where multiple members have low VWF levels, were highly predictive of the presence of VWD.41 However, the spectrum of type 1 VWD includes mild bleeding that may not be easily distinguished from normal by current BATs, and an individualized approach that accounts for the observed bleeding rate in a given person may be more useful in terms of treatment.42
The Zimmerman Program type 1 cohort has several limitations. Historical VWF levels and the timing of those levels were not available to the study investigators for all subjects. Patients were not systematically investigated for non-VWF causes of a possible bleeding disorder, which may have confounded the results, particularly in the historical type 1 cohort. It is possible that mild factor deficiencies or platelet function defects could have been missed, and raises the question of the need for more thorough evaluation, including specific factor activities (factors IX, XI, and XIII) and more extensive platelet testing (aggregation and release). The subjects were recruited from academic medical centers, meaning that these results may not fully represent the community practice in the United States, and the relatively young median age of the subjects in this study may have influenced the BS and reduced the applicability to the general population. Individuals with lower VWF levels, however, are in theory more likely to present at an earlier age due to increased bleeding symptoms. The high numbers of subjects with low VWF levels in the range of 30 to 50 IU/dL and elevated BS suggest that this population merits further study, and consideration of the concept that low VWF may be a contributory risk factor for bleeding, even if it is insufficient to classify a patient as having VWD. Treatment of surgical procedures or bleeding episodes may in fact be indicated in this group based on symptoms.
Our study measured VWF activity using VWF:RCo, although we did also analyze a research laboratory VWF:GPIbM assay looking at direct binding of VWF to mutated GPIb in the absence of ristocetin.20 We did not have available the current commercial VWF:GPIbM assay used in many clinical laboratories, particularly in Europe and Canada.43 In our study, results with VWF:RCo and VWF:GPIbM were similar, however, we did have a number of subjects with normal VWF:Ag included as type 1 VWD because of a low VWF:RCo, and a number of historical type 1 VWD subjects included due to a single low VWF:RCo as well. Unlike the VWF:GPIbM,43 the VWF:RCo is affected by VWF sequence variations that alter ristocetin binding but not VWF function.16
Despite these limitations, this study does demonstrate several key points. First, genetic analysis of VWF in type 1 VWD is not currently sufficient to confirm the diagnosis, although sequence variants are clearly more common in subjects with lower VWF levels. Genetic analysis of the VWF gene in type 1 VWD is not supported by current evidence. Second, VWF levels, and sequence variations, do not always correlate with BS. BS may be more valuable at initial presentation, supported by data showing that BS were more predictive in family members than in the index case.44 Obtaining a BAT at time of diagnosis, and following changes with time and age, may ultimately be more useful than retrospective assessment. Third, there appears to be a subgroup of patients who are potentially misclassified as type 1 VWD because standard assessment does not include evaluation of the interaction of VWF with collagen. Fourth, approximately one-third of subjects who carried a diagnosis of type 1 VWD actually had VWF levels in the normal reference range upon study entry. The fact that these individuals had BS similar to those of subjects with type 1 VWD suggests that this group merits additional study. Assigning a diagnosis based on low VWF at one visit may mean limiting the exploration for alternate bleeding disorders, whereas merely stating that they do not meet the criteria for VWD may be denying these patients needed treatment of the ultimate cause of their bleeding. In addition, some information would suggest that individuals with higher BS are more likely to bleed in the future.45,46
This study highlights several critical areas in VWD diagnosis that require additional investigation. First, improved evaluation of phenotype, either through BATs or careful clinical evaluation, including repeat testing, should help define which patients require additional workup and treatment, the subject of further Zimmerman Program investigations. The ultimate goal is to accurately assess which patients require treatment, while limiting the diagnosis of patients with low VWF levels who lack bleeding symptoms. Second, improved laboratory tests are needed to provide more accurate and efficient diagnosis of VWD. The advent of commercially available VWF:GPIbM assays may help reduce the variability seen with the VWF:RCo, but repeat testing of borderline patients may still be necessary due to the numerous external influences on VWF levels. Third, improved understanding of both VWF genetics and potential modifier genes is required to interpret genotypic variation in type 1 VWD. These efforts will thus guide appropriate diagnosis and ultimately improve care of patients with type 1 VWD.
Acknowledgments
The authors acknowledge everyone involved in this undertaking, including the research coordinators, nurses, physicians, laboratory personnel, and of course the patients, without whom this study would not have been possible.
Funding for this study was provided by a grant from the National Institutes of Health NHLBI for the Zimmerman Program and multiple investigators (HL081588 and HL102260). Additional support was provided by the Midwest Athletes Against Childhood Cancer and the BloodCenter Research Fund.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: V.H.F. and P.A.C. designed the research, analyzed data, and wrote the manuscript; J.C.G. contributed to enrollment, analyzed data, and edited the manuscript; K.D.F. and S.L.H. analyzed data and edited the manuscript; D.B.B., R.A.U., and K.T.M. performed DNA sequencing analysis and edited the manuscript; M.D. and R.G.H. performed the statistical analysis; M.V.R., A.D.S., J.M.L., S.R.L., T.C.A., C.L., W.K.H., M.J.M.-J., R.A.G., and L.N.B. contributed to enrollment and edited the manuscript; A.C.G., P.D.J., D.L., and I.R.P. assisted with study design and edited the manuscript; and R.R.M. conceived the original study, designed the research, analyzed data, and helped write the manuscript.
Conflict-of-interest disclosure: V.H.F. has served as a consultant for Baxter and CSL Behring; J.C.G. has served on advisory boards for Baxalta, Bayer, and CSL Behring; K.D.F. has served as a consultant for CSL Behring, Novo Nordisk, and Werfen, and on the speakers bureau for Alexion; M.V.R. serves on advisory boards for Baxalta, Biogen, BioMarin, Dimensions, Shire, and Tacere Benitec, and has received research funding from Alnylam, Baxalta, Biogen, BioMarin, CSL Behring, Dimension, Ferring, Genentech/Roche, Medscape, Pfizer, SPARK, and Shire; S.R.L. has served as a consultant for Novo Nordisk; T.C.A. serves on the advisory board for CSL Behring; C.L. has served on advisory boards for Baxalta, Bayer, CSL Behring, and Kedrion; and R.R.M. is a consultant or advisor for AstraZeneca, Baxter, Bayer, Biogen Idec, CSL Behring, Grifols, Immucor, and Octapharma. The remaining authors declare no competing financial interests.
Correspondence: Veronica H. Flood, Pediatric Hematology/Oncology, Medical College of Wisconsin, 8701 Watertown Plank Rd, Milwaukee, WI 53226; e-mail: vflood@mcw.edu.
References
- 1.Bowman M, Hopman WM, Rapson D, Lillicrap D, James P. The prevalence of symptomatic von Willebrand disease in primary care practice. J Thromb Haemost. 2010;8(1):213–216. doi: 10.1111/j.1538-7836.2009.03661.x. [DOI] [PubMed] [Google Scholar]
- 2.Rodeghiero F, Castaman G, Dini E. Epidemiological investigation of the prevalence of von Willebrand’s disease. Blood. 1987;69(2):454–459. [PubMed] [Google Scholar]
- 3.Werner EJ, Broxson EH, Tucker EL, Giroux DS, Shults J, Abshire TC. Prevalence of von Willebrand disease in children: a multiethnic study. J Pediatr. 1993;123(6):893–898. doi: 10.1016/s0022-3476(05)80384-1. [DOI] [PubMed] [Google Scholar]
- 4.Sadler JE. Von Willebrand disease type 1: a diagnosis in search of a disease. Blood. 2003;101(6):2089–2093. doi: 10.1182/blood-2002-09-2892. [DOI] [PubMed] [Google Scholar]
- 5.Sadler JE. Slippery criteria for von Willebrand disease type 1. J Thromb Haemost. 2004;2(10):1720–1723. doi: 10.1111/j.1538-7836.2004.00933.x. [DOI] [PubMed] [Google Scholar]
- 6.Rydz N, James PD. The evolution and value of bleeding assessment tools. J Thromb Haemost. 2012;10(11):2223–2229. doi: 10.1111/j.1538-7836.2012.04923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingerslev J. A sensitive ELISA for von Willebrand factor (vWf:Ag). Scand J Clin Lab Invest. 1987;47(2):143–149. [PubMed] [Google Scholar]
- 8.Howard MA, Sawers RJ, Firkin BG. Ristocetin: a means of differentiating von Willebrand’s disease into two groups. Blood. 1973;41(5):687–690. [PubMed] [Google Scholar]
- 9.Nichols WL, Hultin MB, James AH, et al. von Willebrand disease (VWD): evidence-based diagnosis and management guidelines, the National Heart, Lung, and Blood Institute (NHLBI) Expert Panel report (USA). Haemophilia. 2008;14(2):171–232. doi: 10.1111/j.1365-2516.2007.01643.x. [DOI] [PubMed] [Google Scholar]
- 10.Tosetto A, Rodeghiero F, Castaman G, et al. Impact of plasma von Willebrand factor levels in the diagnosis of type 1 von Willebrand disease: results from a multicenter European study (MCMDM-1VWD). J Thromb Haemost. 2007;5(4):715–721. doi: 10.1111/j.1538-7836.2007.02444.x. [DOI] [PubMed] [Google Scholar]
- 11.Quiroga T, Goycoolea M, Belmont S, et al. Quantitative impact of using different criteria for the laboratory diagnosis of type 1 von Willebrand disease. J Thromb Haemost. 2014;12(8):1238–1243. doi: 10.1111/jth.12594. [DOI] [PubMed] [Google Scholar]
- 12.Cumming A, Grundy P, Keeney S, et al. UK Haemophilia Centre Doctors’ Organisation. An investigation of the von Willebrand factor genotype in UK patients diagnosed to have type 1 von Willebrand disease. Thromb Haemost. 2006;96(5):630–641. [PubMed] [Google Scholar]
- 13.James PD, Notley C, Hegadorn C, et al. The mutational spectrum of type 1 von Willebrand disease: results from a Canadian cohort study. Blood. 2007;109(1):145–154. doi: 10.1182/blood-2006-05-021105.. [DOI] [PubMed] [Google Scholar]
- 14.Goodeve A, Eikenboom J, Castaman G, et al. Phenotype and genotype of a cohort of families historically diagnosed with type 1 von Willebrand disease in the European study, Molecular and Clinical Markers for the Diagnosis and Management of Type 1 von Willebrand Disease (MCMDM-1VWD). Blood. 2007;109(1):112–121. doi: 10.1182/blood-2006-05-020784. [DOI] [PubMed] [Google Scholar]
- 15.Rodeghiero F, Tosetto A, Abshire T, et al. ISTH/SSC Joint VWF and Perinatal/Pediatric Hemostasis Subcommittees Working Group. ISTH/SSC bleeding assessment tool: a standardized questionnaire and a proposal for a new bleeding score for inherited bleeding disorders. J Thromb Haemost. 2010;8(9):2063–2065. doi: 10.1111/j.1538-7836.2010.03975.x. [DOI] [PubMed] [Google Scholar]
- 16.Flood VH, Gill JC, Morateck PA, et al. Common VWF exon 28 polymorphisms in African Americans affecting the VWF activity assay by ristocetin cofactor. Blood. 2010;116(2):280–286. doi: 10.1182/blood-2009-10-249102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simone JV, Vanderheiden J, Abildgaard CF. A semiautomatic one-stage factor 8 assay with a commercially prepared standard. J Lab Clin Med. 1967;69(4):706–712. [PubMed] [Google Scholar]
- 18.Jacobi PM, Gill JC, Flood VH, Jakab DA, Friedman KD, Haberichter SL. Intersection of mechanisms of type 2A VWD through defects in VWF multimerization, secretion, ADAMTS-13 susceptibility, and regulated storage. Blood. 2012;119(19):4543–4553. doi: 10.1182/blood-2011-06-360875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flood VH, Schlauderaff AC, Haberichter SL, et al. Zimmerman Program Investigators. Crucial role for the VWF A1 domain in binding to type IV collagen. Blood. 2015;125(14):2297–2304. doi: 10.1182/blood-2014-11-610824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flood VH, Gill JC, Morateck PA, et al. Gain-of-function GPIb ELISA assay for VWF activity in the Zimmerman program for the molecular and clinical biology of VWD. Blood. 2011;117(6):e67–e74. doi: 10.1182/blood-2010-08-299016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bodó I, Eikenboom J, Montgomery R, Patzke J, Schneppenheim R, Di Paola J Subcommittee on von Willebrand factor. Platelet-dependent von Willebrand factor activity. Nomenclature and methodology: communication from the SSC of the ISTH. J Thromb Haemost. 2015;13(7):1345–1350. doi: 10.1111/jth.12964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haberichter SL, Balistreri M, Christopherson P, et al. Assay of the von Willebrand factor (VWF) propeptide to identify patients with type 1 von Willebrand disease with decreased VWF survival. Blood. 2006;108(10):3344–3351. doi: 10.1182/blood-2006-04-015065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bellissimo DB, Christopherson PA, Flood VH, et al. VWF mutations and new sequence variations identified in healthy controls are more frequent in the African-American population. Blood. 2012;119(9):2135–2140. doi: 10.1182/blood-2011-10-384610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Askree SH, Chin EL, Bean LH, Coffee B, Tanner A, Hegde M. Detection limit of intragenic deletions with targeted array comparative genomic hybridization. BMC Genet. 2013;14:116. doi: 10.1186/1471-2156-14-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue Y, Ankala A, Wilcox WR, Hegde MR. Solving the molecular diagnostic testing conundrum for Mendelian disorders in the era of next-generation sequencing: single-gene, gene panel, or exome/genome sequencing. Genet Med. 2015;17(6):444–451. doi: 10.1038/gim.2014.122. [DOI] [PubMed] [Google Scholar]
- 26.Haberichter SL, Castaman G, Budde U, et al. Identification of type 1 von Willebrand disease patients with reduced von Willebrand factor survival by assay of the VWF propeptide in the European study: molecular and clinical markers for the diagnosis and management of type 1 VWD (MCMDM-1VWD). Blood. 2008;111(10):4979–4985. doi: 10.1182/blood-2007-09-110940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flood VH, Christopherson PA, Bellissimo DB, et al. Spectrum of type 2 von Willebrand disease in the Zimmerman program [abstract]. Blood. 2014;124(21) Abstract 472. [Google Scholar]
- 28.Elbatarny M, Mollah S, Grabell J, et al. Zimmerman Program Investigators. Normal range of bleeding scores for the ISTH-BAT: adult and pediatric data from the merging project. Haemophilia. 2014;20(6):831–835. doi: 10.1111/hae.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flood VH, Lederman CA, Wren JS, et al. Absent collagen binding in a VWF A3 domain mutant: utility of the VWF:CB in diagnosis of VWD. J Thromb Haemost. 2010;8(6):1431–1433. doi: 10.1111/j.1538-7836.2010.03869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gill JC, Endres-Brooks J, Bauer PJ, Marks WJ, Jr, Montgomery RR. The effect of ABO blood group on the diagnosis of von Willebrand disease. Blood. 1987;69(6):1691–1695. [PubMed] [Google Scholar]
- 31.Sanders YV, Giezenaar MA, Laros-van Gorkom BA, et al. WiN study group. von Willebrand disease and aging: an evolving phenotype. J Thromb Haemost. 2014;12(7):1066–1075. doi: 10.1111/jth.12586. [DOI] [PubMed] [Google Scholar]
- 32.Rydz N, Grabell J, Lillicrap D, James PD. Changes in von Willebrand factor level and von Willebrand activity with age in type 1 von Willebrand disease. Haemophilia. 2015;21(5):636–641. doi: 10.1111/hae.12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitchen S, Jennings I, Woods TA, Kitchen DP, Walker ID, Preston FE. Laboratory tests for measurement of von Willebrand factor show poor agreement among different centers: results from the United Kingdom National External Quality Assessment Scheme for Blood Coagulation. Semin Thromb Hemost. 2006;32(5):492–498. doi: 10.1055/s-2006-947863. [DOI] [PubMed] [Google Scholar]
- 34.Meijer P, Haverkate F. An external quality assessment program for von Willebrand factor laboratory analysis: an overview from the European concerted action on thrombosis and disabilities foundation. Semin Thromb Hemost. 2006;32(5):485–491. doi: 10.1055/s-2006-947862. [DOI] [PubMed] [Google Scholar]
- 35.Pottinger BE, Read RC, Paleolog EM, Higgins PG, Pearson JD. von Willebrand factor is an acute phase reactant in man. Thromb Res. 1989;53(4):387–394. doi: 10.1016/0049-3848(89)90317-4. [DOI] [PubMed] [Google Scholar]
- 36.Rydz N, Swystun LL, Notley C, et al. The C-type lectin receptor CLEC4M binds, internalizes, and clears von Willebrand factor and contributes to the variation in plasma von Willebrand factor levels. Blood. 2013;121(26):5228–5237. doi: 10.1182/blood-2012-10-457507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanders YV, van der Bom JG, Isaacs A, et al. WiN Study Group. CLEC4M and STXBP5 gene variations contribute to von Willebrand factor level variation in von Willebrand disease. J Thromb Haemost. 2015;13(6):956–966. doi: 10.1111/jth.12927. [DOI] [PubMed] [Google Scholar]
- 38.Desch KC, Ozel AB, Siemieniak D, et al. Linkage analysis identifies a locus for plasma von Willebrand factor undetected by genome-wide association. Proc Natl Acad Sci USA. 2013;110(2):588–593. doi: 10.1073/pnas.1219885110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yadegari H, Driesen J, Pavlova A, Biswas A, Hertfelder HJ, Oldenburg J. Mutation distribution in the von Willebrand factor gene related to the different von Willebrand disease (VWD) types in a cohort of VWD patients. Thromb Haemost. 2012;108(4):662–671. doi: 10.1160/TH12-02-0089. [DOI] [PubMed] [Google Scholar]
- 40.Bowman M, Mundell G, Grabell J, et al. Generation and validation of the condensed MCMDM-1VWD bleeding questionnaire for von Willebrand disease. J Thromb Haemost. 2008;6(12):2062–2066. doi: 10.1111/j.1538-7836.2008.03182.x. [DOI] [PubMed] [Google Scholar]
- 41.Tosetto A, Castaman G, Rodeghiero F. Evidence-based diagnosis of type 1 von Willebrand disease: a Bayes theorem approach. Blood. 2008;111(8):3998–4003. doi: 10.1182/blood-2007-08-105940. [DOI] [PubMed] [Google Scholar]
- 42.Tosetto A, Castaman G, Rodeghiero F. Bleeders, bleeding rates, and bleeding score. J Thromb Haemost. 2013;11(suppl 1):142–150. doi: 10.1111/jth.12248. [DOI] [PubMed] [Google Scholar]
- 43.Graf L, Moffat KA, Carlino SA, et al. Evaluation of an automated method for measuring von Willebrand factor activity in clinical samples without ristocetin. Int J Lab Hematol. 2014;36(3):341–351. doi: 10.1111/ijlh.12218. [DOI] [PubMed] [Google Scholar]
- 44.Christopherson PA, Flood VH, Friedman KD, Gill JC, Haberichter SL, Montgomery RR. Improved diagnosis of VWD in affected family members using the ISTH bleeding score. J Thromb Haemost. 2014;12(suppl 1):95. [Google Scholar]
- 45.Tosetto A, Rodeghiero F, Castaman G, et al. A quantitative analysis of bleeding symptoms in type 1 von Willebrand disease: results from a multicenter European study (MCMDM-1 VWD). J Thromb Haemost. 2006;4(4):766–773. doi: 10.1111/j.1538-7836.2006.01847.x. [DOI] [PubMed] [Google Scholar]
- 46.Federici AB, Bucciarelli P, Castaman G, et al. The bleeding score predicts clinical outcomes and replacement therapy in adults with von Willebrand disease. Blood. 2014;123(26):4037–4044. doi: 10.1182/blood-2014-02-557264. [DOI] [PubMed] [Google Scholar]