Abstract
Purpose
Recent genomic approaches have suggested the existence of multiple distinct subtypes of medulloblastoma. We studied a large cohort of medulloblastomas to determine how many subgroups of the disease exist, how they differ, and the extent of overlap between subgroups.
Methods
We determined gene expression profiles and DNA copy number aberrations for 103 primary medulloblastomas. Bioinformatic tools were used for class discovery of medulloblastoma subgroups based on the most informative genes in the data set. Immunohistochemistry for subgroup-specific signature genes was used to determine subgroup affiliation for 294 nonoverlapping medulloblastomas on two independent tissue microarrays.
Results
Multiple unsupervised analyses of transcriptional profiles identified the following four distinct, nonoverlapping molecular variants: WNT, SHH, group C, and group D. Supervised analysis of these four subgroups revealed significant subgroup-specific demographics, histology, metastatic status, and DNA copy number aberrations. Immunohistochemistry for DKK1 (WNT), SFRP1 (SHH), NPR3 (group C), and KCNA1 (group D) could reliably and uniquely classify formalin-fixed medulloblastomas in approximately 98% of patients. Group C patients (NPR3-positive tumors) exhibited a significantly diminished progression-free and overall survival irrespective of their metastatic status.
Conclusion
Our integrative genomics approach to a large cohort of medulloblastomas has identified four disparate subgroups with distinct demographics, clinical presentation, transcriptional profiles, genetic abnormalities, and clinical outcome. Medulloblastomas can be reliably assigned to subgroups through immunohistochemistry, thereby making medulloblastoma subclassification widely available. Future research on medulloblastoma and the development of clinical trials should take into consideration these four distinct types of medulloblastoma.
INTRODUCTION
Brain tumors are the leading cause of cancer-related death in children, and medulloblastoma is the most common malignant pediatric brain tumor. Although overall survival rates have improved in recent years, the mortality rate remains significant, with survivors often suffering from neurologic, endocrinologic, and social sequelae as a result of current treatment options. Completely resected tumors from patients older than 3 years of age with no leptomeningeal dissemination at diagnosis are classified as standard risk, whereas all others are considered high risk. This stratification scheme does not adequately account for the prognostic variability that exists among patients ascribed to either of these risk groups. Rational molecular-based classification methods for medulloblastoma are vital to permit accurate patient stratification, improved clinical trial design, and the future development of molecularly targeted therapies.
Published molecular markers of prognostic value in medulloblastoma include nuclear β-catenin, ERBB2, TP53, and TRKC immunopositivity.1–3 Cytogenetic and genetic events, including chromosome 17 aberrations, CTNNB1 mutation/monosomy 6, and MYC family amplification, are informative predictors of patient outcome.4–10 Recently, a molecular risk stratification approach based on multicolor fluorescence in situ hybridization (FISH) of MYC and MYCN loci, as well as chromosomal alterations on 17q and 6q, proved more effective than conventional clinical criteria at predicting patient survival.6 Although statistically robust, the generalization of this technique could be limited by the geographic availability of high-quality FISH laboratories.
In the present study, we integrated genome-wide DNA copy number and mRNA expression profiles from a large cohort of primary medulloblastomas to generate a novel molecular classification scheme that reliably predicts patient prognosis. Because this is an immunohistochemistry-based approach, we expect that it will have widespread utility in clinical settings around the world, leading to significantly improved patient stratification.
METHODS
Tumor Specimens and Genomic Data Sets
All tumor specimens were obtained in accordance with the Research Ethics Board at the Hospital for Sick Children (Toronto, Canada), as described,11 and the N.N. Burdenko Neurosurgical Institute (Moscow, Russia).6 Copy number and expression array data were generated and analyzed as described.11,12 Content and construction of the medulloblastoma tissue microarrays (TMAs) have been described previously.6,11,13,14
Biostatistics and Bioinformatics
Medulloblastoma samples were classified into molecular subgroups using TM4 Microarray Software Suite (MeV v4.4; Dana-Farber Cancer Institute, Boston, MA) by unsupervised hierarchical clustering (HCL) using the Pearson correlation metric (average linkage) and bootstrapping analysis of high–standard deviation genes. Subgroup-specific signature genes were identified by a multivariate permutation test restricted on the proportion of false discoveries and one-way analysis of variance. Principal component analysis (PCA) of gene expression data was performed using Partek Genomics Suite (Partek, St Louis, MO). Prediction analysis of microarrays was carried out as described,15 using the current data set as a training data set and the data set reported by Kool et al16 (Gene Expression Omnibus accession No. GSE 10327) as a test data set. Pathway analysis was performed using Ingenuity Pathway Analysis (v7.5; Ingenuity Systems, Redwood City, CA). Gene Set Enrichment Analysis (v2.0; Broad Institute, Cambridge, MA), non-negative matrix factorization (NMF), and subclass mapping (SubMap) were carried out as described.17–20
Immunohistochemistry
Antibodies against the following antigens were used: β-catenin (1:100; BD Transduction Laboratories, Franklin Lakes, NJ), DKK1 (1:100; Abnova, Taipei City, Taiwan), GLI1 (1:5,000; Millipore, Billerica, MA), SFRP1 (1:2,000; Abcam, Cambridge, MA), NPR3 (1:200; Abcam), and KCNA1 (1:2,000; Abcam). TMA staining was performed, evaluated, and scored as published.13
RESULTS
Transcriptional Profiling Identifies Four Subgroups of Medulloblastoma With Distinct Demographics
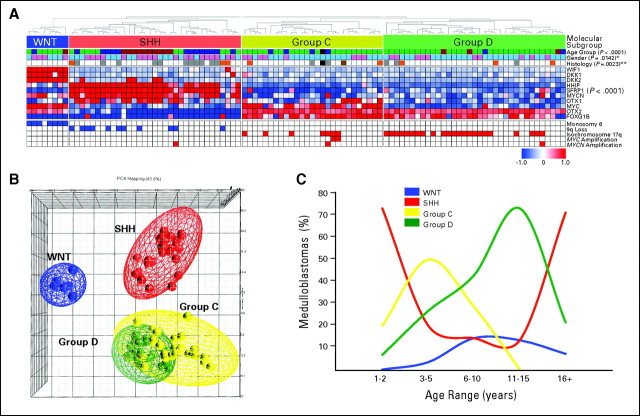
Unsupervised HCL of medulloblastoma expression data identified the following four unique sample clusters: WNT, SHH, group C, and group D (Fig 1A). Support tree analysis of the clustering data reveals high confidence for these four subgroups (≥ 96%; Data Supplement). The degree of separation among the four subgroups was further established by PCA, with WNT and SHH tumors showing clear separation from each other and group C and D tumors (Fig 1B). In contrast, group C and D tumors are more similar to each other and exhibited some proximity in the PCA. Desmoplastic medulloblastomas were found predominantly in the SHH subgroup (P = .0023), but they were also found in groups C and D (Fig 1A; see Data Supplement for all available clinical details).21 Large-cell/anaplastic (LCA) medulloblastomas were found in SHH, group C, and group D. Known targets of the Wnt pathway (WIF1, DKK1, and DKK2) and Shh pathway (HHIP, SFRP1, and MYCN) all show clear, statistically significant differential expression in their respective subgroups (Fig 1A; Data Supplement). Group C and D tumors both express high levels of the known medulloblastoma oncogenes OTX222,23 and FOXG1B.24 Group C and WNT tumors, but not SHH or group D tumors, have high levels of MYC expression (P < .001). SHH tumors, but not group C or group D tumors, have high levels of MYCN expression (P < .001), making group D the only subgroup lacking elevated MYC family expression. As expected, monosomy 6 and 9q deletion were restricted to WNT and SHH tumors, respectively (Fig 1A).4,5,16,25,26 Isochromosome 17q is restricted to groups C and D (P < .001). MYC amplification (two of 103 medulloblastomas) is limited to group C, whereas MYCN amplicons (three of 103 medulloblastomas) are distributed among SHH, group C, and group D.
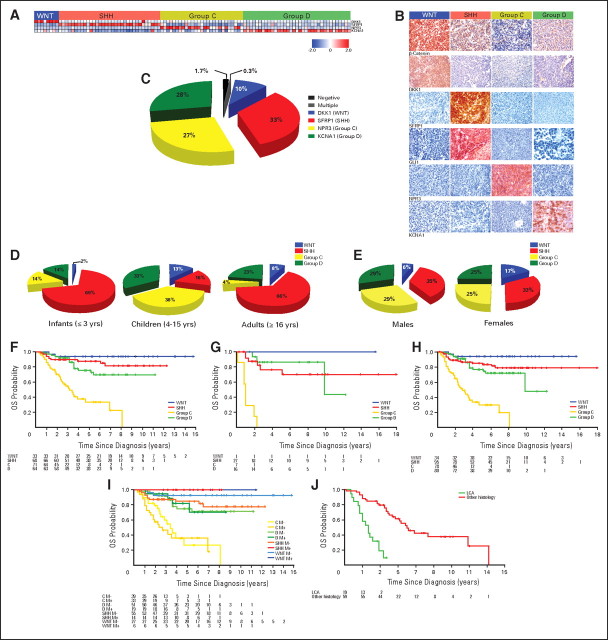
Fig 1.
(A) Unsupervised hierarchical clustering of human 1.0 exon array expression data from 103 primary medulloblastomas using 1,450 high–standard deviation (SD) genes. Clinical features (age group, sex, and histology) for the 103 samples included in the study are shown below the dendrogram (see Data Supplement for summary of all clinical details related to the sample cohort). Age groups include infants (≤ 3 years; blue), children (4 to 15 years; green), adults (≥ 16 years; red), and unknown (black). Sex includes males (blue) and females (pink). Histology includes classic (white), desmoplastic (gray), large-cell/anaplastic (orange), medulloblastoma with extensive nodularity (brown), and unknown (black). Statistical significance for the different clinical features was determined using the χ2 test (age group) and Fisher's exact test (sex and histology). (*) P value determined by comparing sex prevalence in WNT/SHH tumors versus group C/D tumors using Fisher's exact test. (**) P value corresponds to over-representation of desmoplastic tumors in the SHH subgroup as determined using Fisher's exact test. The heatmap below the dendrogram shows the expression profile for 10 genes well characterized in medulloblastoma and demonstrates their significant pattern of differential expression among the four subgroups (see Data Supplement for summary of all differentially expressed genes identified in the four subgroups). Statistical significance of differential gene expression was determined using one-way analysis of variance. Common genomic aberrations known to occur in medulloblastoma are shown below the heatmap. Blue boxes indicate loss/deletion, red boxes indicate gain/amplification, and white boxes denote balanced copy number state for the specified genomic aberration. (B) Principal component analysis (PCA) of the primary medulloblastomas described in (A) using the same 1,450 high-SD genes used in clustering. Individual samples are represented as colored spheres (blue = WNT, red = SHH, yellow = group C, green = group D), and ellipsoids represent two SDs of the data distribution for each subgroup. (C) Age at diagnosis distribution for each of the four medulloblastoma subgroups.
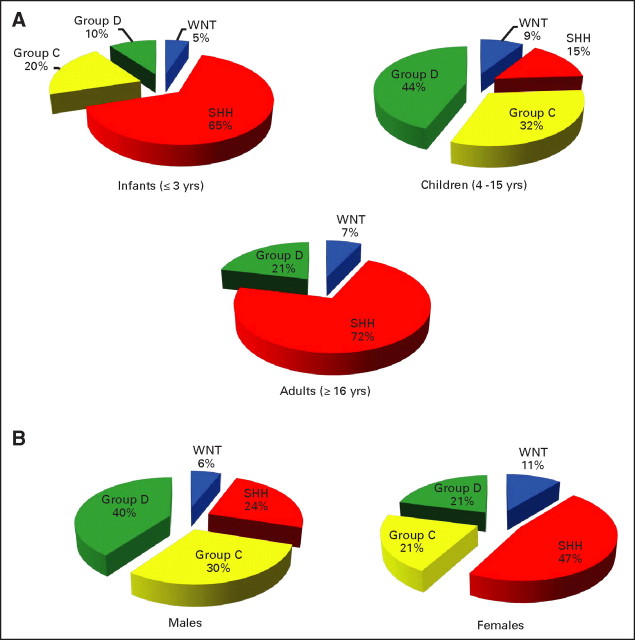
Demographic comparison reveals notable differences between the subgroups. SHH-driven tumors are most common in infants (≤ 3 years old) and adults (≥ 16 years old; P < .001; Fig 1C; Appendix Fig A1A, online only). Group C tumors peak in childhood (age 3 to 10 years) but are completely absent in individuals over the age of 10 years (P = .0184). Group D and WNT tumors show a more distributed age of onset (median age, 9 to 10 years), ranging from infancy to adulthood (Fig 1C and Appendix Fig A1A). The published male-to-female sex ratio for medulloblastoma is approximately 1.5:1; in our cohort, the ratio is 1.55:1. Notably, approximately 70% of males belong to Group C or D compared with only approximately 42% of females (Appendix Fig A1B; P = .0142). Conversely, approximately 47% of females are classified as having SHH tumors versus only approximately 24% of males (P = .0312). The incidence of WNT tumors in female patients (approximately 11%) is nearly double that of males (approximately 6%).
Molecular Classification of Medulloblastoma by Transcriptional Profiling
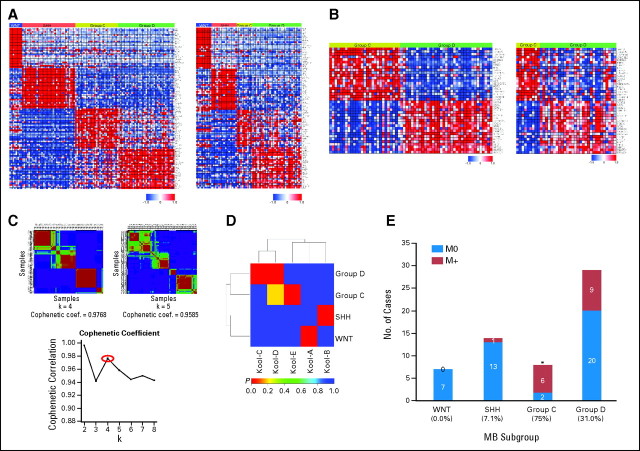
An 84-gene classifier of medulloblastoma subgroup signature genes derived from the current data set using prediction analysis of microarrays15 was applied to a published data set of 62 medulloblastomas reported by Kool et al,16 separating this cohort into the current four subgroups (Appendix Fig A2A, online only; Data Supplement). Comparison of the 20 most highly differentially expressed genes discriminating Groups C and D in both data sets demonstrates that 38 of 40 genes are concordantly differentially expressed (P < .05; Appendix Fig A2B; Data Supplement). Application of consensus NMF,19 an unsupervised bioinformatic tool for determining the number of independent classes within an expression data set, strongly supports the existence of four medulloblastoma subgroups (Appendix Fig A2C; Data Supplement). Direct comparison of clustering results obtained by HCL and NMF confirmed strong concordance between the independent analyses (Rand index, 0.9309; adjusted Rand index, 0.8287).27 Another bioinformatic algorithm, SubMap,20 suggests subgroups C and D in the Kool data set both correspond to our group D (P < .01; Appendix Fig A2D). In the Kool data set, adult tumors (age ≥ 16 years) were devoid of group C cases. Notably, there was a high rate of M+ patients in group C in this data set (75%; P = .0039; Appendix Fig A2E).
Ingenuity Pathway Analysis identified the top canonical signaling pathways over-represented in medulloblastoma subgroups (Table 1). As expected, Wnt signaling was enriched in the WNT group tumors (P < .001), and Shh signaling was enriched in the SHH subgroup (P < .001). However, Wnt pathway genes were also enriched in SHH and group C, but not group D. Both WNT and SHH group tumors had an over-representation of genes involved in axonal guidance. Both group C (phototransduction and glutamate signaling) and group D (semaphorin, cyclic adenosine monophosphate, G protein–coupled receptors, and β-adrenergic signaling) were characterized by an over-representation of pathways involved in neuronal development. Gene Set Enrichment Analysis17 comparison of genes discriminating group C and group D demonstrated that genes upregulated in group C positively correlate with genes associated with medulloblastoma treatment failure identified by Pomeroy et al28 and with genes associated with elevated MYC levels (Data Supplement).
Table 1.
Ingenuity Analysis of Subgroup-Specific Genes in Medulloblastoma
| Ingenuity Canonical Pathways | P | Molecules |
|---|---|---|
| WNT | ||
| Axonal guidance signaling | < .001 | EPHA7, FZD10, BMP4, WNT16, EPHA4, NFATC4, SLIT2, EPHA3, ADAM12, PRKCD, FZD6, ADAM19, PLCB1, SEMA3B |
| WNT/β-catenin signaling | < .001 | FZD10, AXIN2, WIF1, FZD6, DKK4, WNT16, DKK2, LEF1, KREMEN1, DKK1 |
| Factors promoting cardiogenesis in vertebrates | .00339 | FZD10, BMP4, PRKCD, FZD6, LEF1, DKK1 |
| O-glycan biosynthesis | .00417 | GLT8D2, GALNT7, GALNT14, GALNT12, GALNTL2 |
| Basal cell carcinoma signaling | .00776 | FZD10, BMP4, FZD6, WNT16, LEF1 |
| SHH | ||
| SHH signaling | < .001 | DYRK1B, ARRB2, GLI2, GLI3, HHIP, GLI1, PTCH2 |
| Basal cell carcinoma signaling | < .001 | TP53, GLI2, GLI3, LEF1, HHIP, GLI1, BMP5, FZD7, PTCH2 |
| WNT/β-catenin signaling | < .001 | TP53, SFRP4, TCF4, HDAC1, SOX2, SOX9, RARB, TGFB2, SFRP5, PPP2R2C, LEF1, SFRP1, TCF7L2 (includes EG:6934), FZD7 |
| Axonal guidance signaling | < .001 | GLI2, PLXNC1, CXCR4, SEMA6A, GNG3, HHIP, NFATC4, BMP5, ROBO1, PTCH2, PLCB4, GLI3, SRGAP1, SDC2, NTRK3, NGFR, EFNA5, ABLIM3, PDGFD, GLI1, PRKD1, FZD7, UNC5C |
| Human embryonic stem-cell pluripotency | .00295 | SOX2, NTRK3, PDGFRA, SPHK1, TGFB2, LEF1, PDGFD, BMP5, FZD7 |
| Group C | ||
| Phototransduction pathway | < .001 | GNB3, GNGT1, RCVRN, PDE6H |
| WNT/β-catenin signaling | .00933 | MYC, NLK, TGFB1, TGFBR3, PPP2R2B |
| Glutamate receptor signaling | .01122 | GNB3, SLC17A7, SLC1A7 |
| Thyroid cancer signaling | .04074 | MYC, RXRG |
| p38 MAPK signaling | .04266 | MYC, TGFB1, EEF2K |
| Group D | ||
| Semaphorin signaling in neurons | < .001 | ARHGEF12, RND1, RHOT1, DPYSL4, DIRAS3, FNBP1 |
| cAMP-mediated signaling | .00275 | GRK4, GRM8, RGS7, ADCY1, STAT3, PDE4B, AKAP9, CHRM3, PDE1C |
| G protein–coupled receptor signaling | .00490 | HTR2C, GRK4, GPR12, GRM8, RGS7, ADCY1, STAT3, PDE4B, CHRM3, PDE1C |
| p53 signaling | .00603 | KAT2B, CCND2, GADD45G, C12ORF5, HIPK2, TP53BP2 |
| Cardiac β-adrenergic signaling | .00617 | ADCY1, CACNA1C, PDE4B, GNG2, AKAP9, CACNA1A, PDE1C |
Abbreviation: MAPK, mitogen-activated protein kinase; cAMP, cyclic adenosine monophosphate.
Subgroup-Specific Genetic Events in the Medulloblastoma Genome
With the exception of i(17)q, which is reported in approximately 30% to 50% of patients,6,11 most of the known chromosomal changes in medulloblastoma occur at low frequency. To determine whether certain genetic aberrations were more prominent when accounting for subgroups, we manually cataloged all chromosomal changes in our data set in a subgroup-specific manner (Appendix Fig A3A, online only; Appendix Table A1, online only; Data Supplement). Not surprisingly, monosomy 6 was found exclusively in WNT tumors,4,12,16,26 and 9q loss was found only in SHH tumors.16
Multiple novel regions of genomic aberration were identified in SHH tumors, including chromosome 9p gain (P < .001), which often co-occurred with 9q loss [ie, i(9)p], as well as gains of 3q (P = .0015), 20q (P = .0331), and 21q (P = .0124). Chromosome 10q loss was primarily limited to SHH and group C tumors (P = .0034). Events significantly over-represented in group C included 1q gain (P < .001), distal 5q loss (P = .0038), and 16q loss (P = .0055). Multiple events were over-represented in both groups C and D compared with WNT/SHH tumors, including gain of chromosomes 17q (P = .0014) and 18 (P = .0136), i(17)q (P < .001), and loss of 11p (P < .001). Despite the commonalities between group C and D, these subgroups could be discriminated by several genetic events that are significantly more common in group C, including 1q gain (P = .0021), loss of distal 5q (P = .0689), and 10q loss (P = .0168; Appendix Fig A3B; Table 2). Isochromosome 17q was significantly more prominent in group D (23 of 35 tumors; 65.7%) than group C (seven of 27 tumors; 25.9%; P = .0024), and loss of the X chromosome occurred more commonly in group D females (P < .001; Appendix Fig A3B; Table 2).
Table 2.
Group C Versus Group D Medulloblastoma
| Genotype/Phenotype | Group C |
Group D |
P | ||
|---|---|---|---|---|---|
| No. | Total No. | No. | Total No. | ||
| Demographics | |||||
| Infant (MB103/TMA) | 10 | 106 | 8 | 114 | NS |
| Child (MB103/TMA) | 94 | 106 | 91 | 114 | NS |
| Adult (MB103/TMA) | 2 | 106 | 15 | 114 | .0018 |
| Gene expression | |||||
| MYC expression (mean signal intensity exon array) | 9.424 | 6.787 | < .001 | ||
| NPR3 expression (mean signal intensity exon array) | 9.195 | 6.280 | < .001 | ||
| KCNA1 expression (mean signal intensity exon array) | 6.812 | 10.702 | < .001 | ||
| NPR3 positive (TMA) | 80 | 80 | 0 | 80 | < .001 |
| KCNA1 positive (TMA) | 0 | 80 | 80 | 80 | < .001 |
| Genetics | |||||
| 1q gain (SNP array) | 12 | 27 | 3 | 35 | .0021 |
| 5q loss (SNP array) | 6 | 27 | 2 | 35 | .0689 |
| 10q loss (SNP array) | 7 | 27 | 1 | 35 | .0168 |
| i(17)q (SNP array) | 7 | 27 | 23 | 35 | .0024 |
| MYC amplification (SNP array/FISH) | 11 | 98 | 0 | 98 | < .001 |
| MYCN amplification (SNP array/FISH) | 8 | 98 | 6 | 98 | NS |
| Histology | |||||
| Classic (TMA-DKFZ) | 54 | 71 | 56 | 63 | .0708 |
| Desmoplastic (TMA-DKFZ) | 1 | 71 | 2 | 63 | NS |
| Large cell/anaplastic (TMA-DKFZ) | 16 | 71 | 5 | 63 | .0306 |
| Outcome | |||||
| M+ (TMA-DKFZ) | 33 | 71 | 19 | 64 | .0527 |
| PFS (TMA-DKFZ), % | 32 | 63 | < .001 | ||
| OS (TMA-DKFZ), % | 33 | 77 | < .001 | ||
Abbreviations: MB, medulloblastoma; TMA, tissue microarray; NS, not significant; SNP, single nucleotide polymorphism; FISH, fluorescent in situ hybridization; DKFZ, German Cancer Research Center; PFS, progression-free survival; OS, overall survival.
The bioinformatic tool Genomic Identification of Significant Targets in Cancer (GISTIC)29 delineated statistically significant, subgroup-specific genetic changes in an unbiased manner. GISTIC confirmed all of the large genomic abnormalities identified by previous manual curation of the data set and revealed several novel subgroup-specific amplifications and deletions (Appendix Figs A3C and A3D). GISTIC identified gain of chromosome 2 (Q, .2412 to .0019) and deletion of chromosome 14 (Q, .7534 to .0017) as significant in the SHH subgroup. Focal MYC amplification on 8q24 was identified exclusively in group C tumors (Q = .0011; Appendix Fig A3C). Loss of chromosome 8p was highly significant in group C (Q, .5086 to .0005), whereas group D tumors exhibited significant loss on both 8p and 8q (Q, .1662 to .0010) Appendix Fig A3D).
Validation of Medulloblastoma Subgroups by Immunohistochemistry
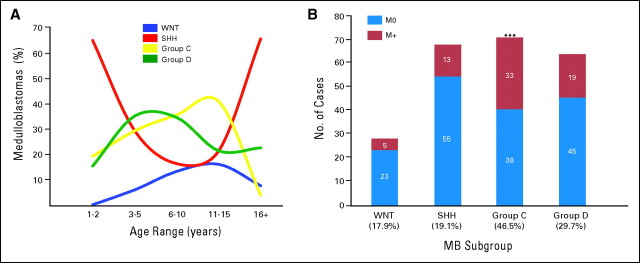
We selected highly expressed, subgroup-specific signature genes based on our microarray expression data set (Appendix Fig A2A; Appendix Fig A4A, online only) and the availability of high-quality commercial antibodies. Staining two separate medulloblastoma TMAs using these antibodies, as well as β-catenin and GLI1, demonstrated robust staining in a subgroup-specific manner (Appendix Fig A4B). Remarkably, 288 (approximately 98%) of 294 tumor samples stained positive for a single marker protein, a failure rate of only approximately 2.1% (P < .001; Appendix Fig A4C). Demographics for the TMA patients validate the results from our microarray data set (Figs 1C and 2A; Appendix Figs A1A, A1B, A4D, and A4E), confirming that SHH tumors occur primarily in infants and adults (P < .001). Group C tumors were largely confined to childhood (P < .001), except for two patients both age 18 years (Fig 2A; Appendix Fig A4D). WNT group tumors were almost three times more common in females than males (17% v 6%, respectively; P = .0046; Appendix Fig A4E). Metastases were significantly over-represented in group C (46.5%; P < .001), followed by group D (29.7%), similar to the Kool data set (Fig 2B; Appendix Fig A2B). MYC amplification was demonstrated by interphase FISH in 11 of 98 group C tumors on the German Cancer Research Center TMA, but not in any SHH, WNT, or group D tumors (Table 2). LCA histology was much more common among group C tumors (23%) compared with group D tumors (8%; P < .001). Overall survival in the German Cancer Research Center cohort (n = 236; Appendix Fig A4F), the Johns Hopkins University cohort (n = 50, a single WNT tumor was excluded; Appendix Fig A4G), and both cohorts combined (n = 287; Appendix Fig A4H) demonstrates that group C tumors have the worst prognosis (log-rank P < .001). Replotting the overall survival curves by dividing each subgroup into M0 and M+ sub-subgroups shows that group C patients exhibit a dismal prognosis regardless of M stage (Appendix Fig A4I). A multivariate analysis evaluating age, M stage, histology, extent of resection, and subgroup (WNT, SHH, group C, or group D) revealed that only LCA histology and group C subgroup were prognostic (Data Supplement). Indeed, group C tumors with LCA histology have the worst prognosis (Appendix Fig A4J).
Fig 2.
(A) Age at diagnosis distribution for each of the four medulloblastoma (MB) subgroups as determined by immunohistochemical staining of the MB tissue microarrays (TMAs). (B) Incidence of metastases by subgroup as determined by TMA staining. Significance was assessed by Fisher's exact test. (***) P < .001.
DISCUSSION
Prior attempts to subgroup medulloblastoma using genomics identified five16 or six26 subgroups. Both studies clearly show the WNT group of tumors clustering alone, whereas SHH-driven tumors did not always segregate as a single subgroup.26 Both the current data set and the data set from Kool et al16 show that although WNT tumors are classic medulloblastomas and most desmoplastic medulloblastomas are SHH tumors, the WNT and SHH subgroups always cluster together, suggesting that they are biologically similar. Although desmoplastic tumors were most commonly seen in the SHH group, they were also found in group C and group D, supporting the known difficulty in making a histologic diagnosis of nodular desmoplastic medulloblastoma.21 We observed LCA tumors in SHH, group C, and group D tumors, and others have described the LCA phenotype in WNT tumors.5 Interestingly, children with a WNT subgroup LCA medulloblastoma have a good prognosis.5
SHH tumors are seen in both infants and adults, whereas WNT and group D tumors are distributed across all age groups. Group C tumors peak in childhood, are rarely seen in older teenagers, and are never seen in adults. The paucity of group C tumors and predominance of group D tumors seen in adolescence may account for the distinct clinical course and pattern of relapse that has been described in adolescents with medulloblastoma.30 The St Jude Medulloblastoma-96 study showed that the male-to-female ratio for high-risk medulloblastoma was 3.9:1, and several reports have suggested that females with medulloblastoma have a better outcome than males.30–32 We believe this is attributable to a higher female incidence of WNT tumors and perhaps SHH tumors. Indeed, the improved survival among females is only seen among older children and adults (in whom WNT tumors are more prevalent), highly supporting a WNT subgroup prevalence as an explanation for the improved outcome in females.32 Prior clinical trials studying medulloblastoma have tended to divide patients by age group, with infants often being studied separately. Our results suggest that infants with medulloblastoma should be broken down by subgroup and that infants with group C/D tumors may account for the children who respond poorly to current therapies.33 This is consistent with identified prognostic factors of desmoplasia (good) and metastatic status (poor), which might be identifying SHH and group C tumors, respectively.33
Analysis of 62 medulloblastomas by Kool et al16 identified three non-SHH/WNT medulloblastoma subgroups, whereas our analysis of 103 medulloblastomas revealed only two non-SHH/WNT subgroups. The statistical support for three non-SHH/WNT subgroups as reported by Kool et al16 was either 21% or 41%, depending on the number of genes used in support tree analysis. In the current study, both HCL and PCA demonstrate that group C is distinct from group D with strong statistical support. NMF supports the existence of four subgroups of medulloblastoma. The initial obvious difference between group C and group D is that although MYC is highly expressed in group C and WNT tumors and MYCN is highly expressed in SHH tumors, neither MYC nor MYCN is highly expressed in group D tumors. Additionally, our data show that group C and group D tumors have different demographics, rate of metastases, genetic profiles, and clinical outcomes (Table 2). Our immunohistochemical analysis of medulloblastoma TMAs revealed that only one of 294 medulloblastomas stained positive for both group C and group D signature genes, demonstrating the lack of overlap between the two groups. NMF and SubMap analysis also support the existence of two non-SHH/WNT groups of medulloblastoma. Taken together, all of these data highly support the existence of two non-SHH/WNT subgroups that we have designated as group C (childhood tumors with high MYC levels and frequent CSF dissemination) and group D.
Our novel four-antibody approach to subgroup medulloblastoma should be broadly applicable across the globe because the technique does not require RNA, FISH, PCR, microarray technology, or any other techniques from molecular biology, but rather uses immunohistochemistry on archival specimens, which is in routine use in most neuropathology laboratories around the world. Furthermore, because marker staining is quite robust, we hope that there will be little interobserver variability.
Analysis of overall survival demonstrates a marked reduction in survival for children with Group C medulloblastoma regardless of metastatic stage. This suggests that some children with average-risk medulloblastoma who subsequently experience recurrence likely belong to group C. Indeed, the failure of metastatic stage to predict prognosis in our multivariate analysis suggests that the published association between poor prognosis and metastatic stage may be attributable to the high rate of metastases in group C tumors.
Historically, small round blue cell tumors of the cerebellum have been grouped under the rubric of medulloblastoma. Recently, atypical teratoid and rhabdoid tumors were shown to represent a distinct subgroup of blue cell tumors based on different histopathology and, subsequently, a lack of expression of the hSNF5/INI1 tumor suppressor gene.34,35 Our data suggest that the histologic entity of medulloblastoma comprises four distinct molecular variants that are demographically, clinically, transcriptionally, and genetically distinct and can be distinguished through the application of four commercial antibodies on formalin-fixed, paraffin-embedded tumor material. Preliminary analysis of the SHH subgroup tumors in this study suggest the existence of three transcriptionally and genetically distinct clusters (data not shown). We anticipate that analysis of much larger cohorts of medulloblastoma will likely reveal subclusters within the four main subgroups. Future clinical trials should prospectively validate our four-antibody immunohistochemical approach. Most importantly, because of the nonoverlapping character of the four types of medulloblastoma, we suggest that targeted therapies may need to be developed against each subtype individually because they are biologically distinct. Finally, prospective investigation into the cellular origin(s) and specific genetic events found in each subgroup will be necessary to determine whether these molecular variants constitute four distinct diseases.
Supplementary Material
Acknowledgment
We thank Susan Archer for editing the manuscript. We acknowledge David Shih for assistance with statistical analyses and Christian Smith for help with preparation of figures and tables. We thank the patients and families for participating in this research.
Appendix
Fig A1.
(A) Pie charts showing the frequency of the four molecular subgroups among the following three patient age categories: infants (≤ 3 years), children (4 to 15 years), and adults (≥ 16 years). (B) Pie charts showing the frequency of medulloblastoma subgroups by sex.
Fig A2.
(A) Left panel: Supervised analysis of the four medulloblastoma subgroups showing the most differentially expressed signature genes that discriminate the subgroups. The top 25 signature genes for each subgroup are shown. Right panel: Prediction analysis of microarrays (PAM) was used to identify a gene signature that could robustly classify our training data set into the four molecular subgroups and predict the subgroup affiliation of samples in a test data set of 62 medulloblastomas published by Kool et al.16 The expression profile of signature genes identified in the test data set is shown for the predicted subgroups of the Kool data set. (B) Expression heatmap showing 40 significant genes discriminating group C and group D medulloblastomas in our cohort (left panel) and the same 40 genes described in group C and group D tumors from the Kool data set (right panel). (C) Non-negative matrix factorization (NMF) consensus analysis of our medulloblastoma cohort provides strong statistical support for the existence of four medulloblastoma subgroups. Agreement between hierarchical clustering and NMF clustering data was supported by calculation of the Rand index (Rand index, 0.931; adjusted Rand index, 0.829). (D) Subclass mapping (SubMap) analysis comparing the four medulloblastoma subgroups identified in the current study to the five subgroups previously reported by Kool et al.16 SubMap supports the existence of four medulloblastoma subgroups (WNT = Kool-A, SHH = Kool-B, Group C = Kool-E, Group D = Kool-C/Kool-D). (E) Incidence of metastasis in predicted medulloblastoma subgroups of the Kool data set. Subgroup affiliation was predicted for the Kool samples using PAM as described in (A), and patient metastatic status was then plotted for each of the predicted subgroups. Significance was assessed by Fisher's exact test. Coef., coefficient. *P < .01.
Fig A3.
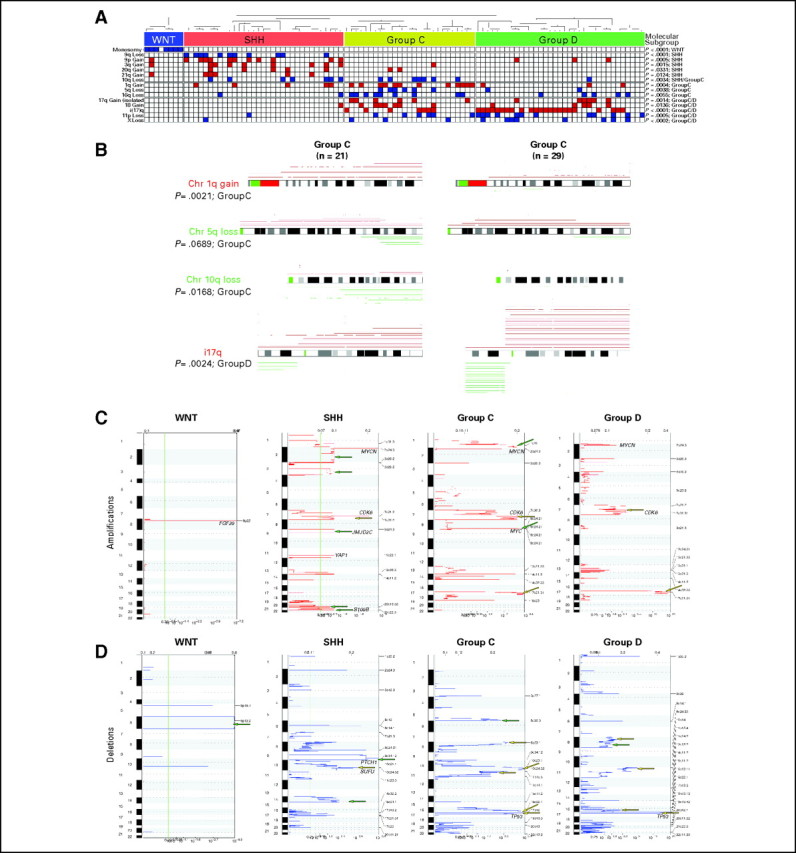
(A) Distribution of statistically significant, subgroup-specific copy number aberrations identified by manual curation of our medulloblastoma series. Significance was determined by Fisher's exact test. Blue boxes indicate loss/deletion, red boxes indicate gain/amplification, and white boxes denote balanced copy number state for the specified genomic aberration. (B) Comparison of statistically significant copy number aberrations over-represented in group C and group D medulloblastomas. Significance was determined by Fisher's exact test. (C, D) Genomic Identification of Significant Targets in Cancer analysis of the four medulloblastoma subgroups represented in our data series. Significance plots show regions of statistically significant (C) gains/amplifications and (D) losses/deletions in the medulloblastoma subgroups. Yellow arrows indicate copy number aberrations restricted to a single subgroup, and green arrows mark regions that are significant in multiple subgroups. Loci marking select candidate genes are shown for reference.
Fig A4.
(A) Differential expression of four selected genes (DKK1, SFRP1, NPR3, and KCNA1) in the four medulloblastoma subgroups as determined by exon array expression profiling. (B) Representative immunohistochemistry for β-catenin, DKK1, SFRP1, GLI1, NPR3, and KCNA1 on a medulloblastoma tissue microarray (TMA). (C) Results obtained from immunohistochemical staining of two independent medulloblastoma TMAs (German Cancer Research Center [DKFZ] and Johns Hopkins University [JHU]) consisting of 294 nonoverlapping primary cases. Pie chart demonstrates that 288 (approximately 98%) of 294 of tumors stained positive for a single marker (DKK1, SFRP1, NPR3, or KCNA1), one (approximately 0.3%) of 294 tumors stained positive for multiple markers, and five (approximately 1.7%) of 294 tumors did not stain for any of the four markers. Significance was determined using Fisher's exact test. (D) Pie charts showing the frequency of the four molecular subgroups based on TMA staining among the following three patient age categories: infants (≤ 3 years), children (4 to 15 years), and adults (≥ 16 years). (E) Pie charts showing the frequency of medulloblastoma subgroups by sex based on TMA staining. (F) Kaplan-Meier analysis showing overall survival (OS) probability for patients on the DKFZ medulloblastoma TMA (n = 236) separated by subgroup. (G) Kaplan-Meier analysis showing OS probability for patients on the JHU TMA (n = 50) separated by subgroup. (H) Combined OS probability for both the DKFZ and JHU TMAs (n = 287) separated by subgroup. (I) Combined OS probability for both TMAs showing medulloblastoma subgroups separated by metastatic status. (J) Kaplan-Meier analysis discriminating large-cell/anaplastic (LCA; green) from non-LCA (red) histology in group C medulloblastomas present on the TMAs.
Table A1.
Recurrent Chromosomal Aberrations in Medulloblastoma Subgroups
| Aberration | % of Aberration |
||||
|---|---|---|---|---|---|
| MB | WNT | SHH | Group C | Group D | |
| Monosomy 6 | 6.8 | 87.5 | 0.0 | 0.0 | 0.0 |
| 9q loss | 10.7 | 0.0 | 33.3 | 0.0 | 0.0 |
| 9p gain | 14.6 | 12.5 | 33.3 | 0.0 | 8.6 |
| 3q gain | 9.7 | 12.5 | 24.2 | 3.7 | 0.0 |
| 20q gain | 6.8 | 0.0 | 15.2 | 7.4 | 0.0 |
| 21q gain | 5.8 | 12.5 | 15.2 | 0.0 | 0.0 |
| 10q loss | 14.6 | 0.0 | 21.2 | 25.9 | 2.9 |
| 1q gain | 19.4 | 0.0 | 15.2 | 44.4 | 8.6 |
| 5q loss | 7.8 | 0.0 | 0.0 | 22.2 | 5.7 |
| 16q loss | 15.5 | 0.0 | 6.1 | 33.3 | 14.3 |
| 17q gain | 12.6 | 0.0 | 0.0 | 22.2 | 20.0 |
| 18 gain | 12.6 | 0.0 | 3.0 | 25.9 | 14.3 |
| i17q | 29.1 | 0.0 | 0.0 | 25.9 | 65.7 |
| 11p loss | 18.4 | 0.0 | 3.0 | 22.2 | 34.3 |
| X loss (females) | 30.0 | 0.0 | 6.0 | 38.0 | 80.0 |
Abbreviation: MB, medulloblastoma.
Footnotes
Supported by a Clinician-Scientist Phase II Award (M.D.T.) from the Canadian Institutes of Health Research; a Distinguished Scientist Award (M.D.T.) from the Sontag Foundation; the Canadian Cancer Society; the Pediatric Brain Tumor Foundation; the American Brain Tumor Association; and grants from the Deutsche Kinderkrebsstiftung (S.P.) and the German Ministry of Health and Education (S.P.).
Presented at the Inaugural Pediatric Neuro-Oncology Basic and Translational Research Conference, October 2, 2009, Asheville, NC.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Paul A. Northcott, Andrey Korshunov, Stefan Pfister, Michael D. Taylor
Financial support: Michael D. Taylor
Provision of study materials or patients: Andrey Korshunov, Charles G. Eberhart, Pim French, Stefan Pfister, Michael D. Taylor
Collection and assembly of data: Paul A. Northcott, Andrey Korshunov, Hendrik Witt, Thomas Hielscher, Stephen Mack, Pim French, Stefan Pfister, Michael D. Taylor
Data analysis and interpretation: Paul A. Northcott, Andrey Korshunov, Hendrik Witt, Thomas Hielscher, Charles G. Eberhart, Stephen Mack, Eric Bouffet, Steven C. Clifford, Cynthia E. Hawkins, Pim French, James T. Rutka, Stefan Pfister, Michael D. Taylor
Manuscript writing: Paul A. Northcott, Andrey Korshunov, Eric Bouffet, Steven C. Clifford, Cynthia E. Hawkins, Pim French, James T. Rutka, Stefan Pfister, Michael D. Taylor
Final approval of manuscript: Paul A. Northcott, Andrey Korshunov, Hendrik Witt, Thomas Hielscher, Charles G. Eberhart, Stephen Mack, Eric Bouffet, Steven C. Clifford, Cynthia E. Hawkins, Pim French, James T. Rutka, Stefan Pfister, Michael D. Taylor
REFERENCES
- 1.Ray A, Ho M, Ma J, et al. A clinicobiological model predicting survival in medulloblastoma. Clin Cancer Res. 2004;10:7613–7620. doi: 10.1158/1078-0432.CCR-04-0499. [DOI] [PubMed] [Google Scholar]
- 2.Gilbertson RJ, Perry RH, Kelly PJ, et al. Prognostic significance of HER2 and HER4 coexpression in childhood medulloblastoma. Cancer Res. 1997;57:3272–3280. [PubMed] [Google Scholar]
- 3.Grotzer MA, Janss AJ, Fung K, et al. TrkC expression predicts good clinical outcome in primitive neuroectodermal brain tumors. J Clin Oncol. 2000;18:1027–1035. doi: 10.1200/JCO.2000.18.5.1027. [DOI] [PubMed] [Google Scholar]
- 4.Clifford SC, Lusher ME, Lindsey JC, et al. Wnt/Wingless pathway activation and chromosome 6 loss characterize a distinct molecular sub-group of medulloblastomas associated with a favorable prognosis. Cell Cycle. 2006;5:2666–2670. doi: 10.4161/cc.5.22.3446. [DOI] [PubMed] [Google Scholar]
- 5.Ellison DW, Onilude OE, Lindsey JC, et al. Beta-Catenin status predicts a favorable outcome in childhood medulloblastoma: The United Kingdom Children's Cancer Study Group Brain Tumour Committee. J Clin Oncol. 2005;23:7951–7957. doi: 10.1200/JCO.2005.01.5479. [DOI] [PubMed] [Google Scholar]
- 6.Pfister S, Remke M, Benner A, et al. Outcome prediction in pediatric medulloblastoma based on DNA copy-number aberrations of chromosomes 6q and 17q and the MYC and MYCN loci. J Clin Oncol. 2009;27:1627–1636. doi: 10.1200/JCO.2008.17.9432. [DOI] [PubMed] [Google Scholar]
- 7.Grotzer MA, Hogarty MD, Janss AJ, et al. MYC messenger RNA expression predicts survival outcome in childhood primitive neuroectodermal tumor/medulloblastoma. Clin Cancer Res. 2001;7:2425–2433. [PubMed] [Google Scholar]
- 8.Gajjar A, Hernan R, Kocak M, et al. Clinical, histopathologic, and molecular markers of prognosis: Toward a new disease risk stratification system for medulloblastoma. J Clin Oncol. 2004;22:984–993. doi: 10.1200/JCO.2004.06.032. [DOI] [PubMed] [Google Scholar]
- 9.Gilbertson R, Wickramasinghe C, Hernan R, et al. Clinical and molecular stratification of disease risk in medulloblastoma. Br J Cancer. 2001;85:705–712. doi: 10.1054/bjoc.2001.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan E, Pellarin M, Holmes E, et al. Isochromosome 17q is a negative prognostic factor in poor-risk childhood medulloblastoma patients. Clin Cancer Res. 2005;11:4733–4740. doi: 10.1158/1078-0432.CCR-04-0465. [DOI] [PubMed] [Google Scholar]
- 11.Northcott PA, Nakahara Y, Wu X, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Northcott PA, Fernandez LA, Hagan JP, et al. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009;69:3249–3255. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neben K, Korshunov A, Benner A, et al. Microarray-based screening for molecular markers in medulloblastoma revealed STK15 as independent predictor for survival. Cancer Res. 2004;64:3103–3111. doi: 10.1158/0008-5472.can-03-3968. [DOI] [PubMed] [Google Scholar]
- 14.Eberhart CG, Chaudhry A, Daniel RW, et al. Increased p53 immunopositivity in anaplastic medulloblastoma and supratentorial PNET is not caused by JC virus. BMC Cancer. 2005;5:19. doi: 10.1186/1471-2407-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tibshirani R, Hastie T, Narasimhan B, et al. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kool M, Koster J, Bunt J, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One. 2008;3:e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 19.Brunet JP, Tamayo P, Golub TR, et al. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci U S A. 2004;101:4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoshida Y, Brunet JP, Tamayo P, et al. Subclass mapping: Identifying common subtypes in independent disease data sets. PLoS One. 2007;2:e1195. doi: 10.1371/journal.pone.0001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McManamy CS, Pears J, Weston CL, et al. Nodule formation and desmoplasia in medulloblastomas-defining the nodular/desmoplastic variant and its biological behavior. Brain Pathol. 2007;17:151–164. doi: 10.1111/j.1750-3639.2007.00058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boon K, Eberhart CG, Riggins GJ. Genomic amplification of orthodenticle homologue 2 in medulloblastomas. Cancer Res. 2005;65:703–707. [PubMed] [Google Scholar]
- 23.Di C, Liao S, Adamson DC, et al. Identification of OTX2 as a medulloblastoma oncogene whose product can be targeted by all-trans retinoic acid. Cancer Res. 2005;65:919–924. [PubMed] [Google Scholar]
- 24.Adesina AM, Nguyen Y, Mehta V, et al. FOXG1 dysregulation is a frequent event in medulloblastoma. J Neurooncol. 2007;85:111–122. doi: 10.1007/s11060-007-9394-3. [DOI] [PubMed] [Google Scholar]
- 25.Fattet S, Haberler C, Legoix P, et al. Beta-catenin status in paediatric medulloblastomas: Correlation of immunohistochemical expression with mutational status, genetic profiles, and clinical characteristics. J Pathol. 2009;218:86–94. doi: 10.1002/path.2514. [DOI] [PubMed] [Google Scholar]
- 26.Thompson MC, Fuller C, Hogg TL, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 27.Hubert L, Arabie P. Comparing partitions. J Classif. 1985;2:193–218. [Google Scholar]
- 28.Pomeroy SL, Tamayo P, Gaasenbeek M, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 29.Beroukhim R, Getz G, Nghiemphu L, et al. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabori U, Sung L, Hukin J, et al. Distinctive clinical course and pattern of relapse in adolescents with medulloblastoma. Int J Radiat Oncol Biol Phys. 2006;64:402–407. doi: 10.1016/j.ijrobp.2005.07.962. [DOI] [PubMed] [Google Scholar]
- 31.Gajjar A, Chintagumpala M, Ashley D, et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): Long-term results from a prospective, multicentre trial. Lancet Oncol. 2006;7:813–820. doi: 10.1016/S1470-2045(06)70867-1. [DOI] [PubMed] [Google Scholar]
- 32.Curran EK, Sainani KL, Le GM, et al. Gender affects survival for medulloblastoma only in older children and adults: A study from the Surveillance Epidemiology and End Results Registry. Pediatr Blood Cancer. 2009;52:60–64. doi: 10.1002/pbc.21832. [DOI] [PubMed] [Google Scholar]
- 33.Rutkowski S, Gerber NU, von Hoff K, et al. Treatment of early childhood medulloblastoma by postoperative chemotherapy and deferred radiotherapy. Neuro Oncol. 2009;11:201–210. doi: 10.1215/15228517-2008-084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Judkins AR, Mauger J, Ht A, et al. Immunohistochemical analysis of hSNF5/INI1 in pediatric CNS neoplasms. Am J Surg Pathol. 2004;28:644–650. doi: 10.1097/00000478-200405000-00013. [DOI] [PubMed] [Google Scholar]
- 35.Biegel JA, Zhou JY, Rorke LB, et al. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59:74–79. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.