Abstract
Trisomy 12 (+12) is detected by fluorescence in situ hybridization (FISH) analysis in up to 20% of patients with chronic lymphocytic leukemia (CLL). Patients with +12 are known to have unique features and to carry an intermediate prognosis. In order to better define this large group, we reviewed the characteristics of 250 untreated patients with +12. When compared to 516 untreated patients negative for +12 by FISH, patients with +12 showed a higher incidence of thrombocytopenia, Richter Transformation (RT) and second malignant neoplasms (SMN), in addition to the expected increased rate of CD38 positivity and atypical immunophenotype. At a median follow-up of 51 months, 57% of patients needed first-line treatment; median time-to-first-treatment was 38 months and on multivariate analysis (MVA) it was shorter in patients with advanced Rai stage, palpable splenomegaly, and deletion 14q by conventional cytogenetic analysis. The overall response rate with first-line treatment was 94%. The median failure-free survival has not been reached, but on MVA it was shorter in patients who achieved a response other than complete remission or with FISH negativity for deletion 13q. The median overall survival for the entire group has not been reached, but on MVA it was shorter in patients with an absolute lymphocyte count >30×109/L or who developed SMN. Eighteen deaths have been observed so far, and RT and SMN were the leading causes of death (3 and 6, respectively). In conclusion, patients with +12 CLL show characteristic clinical and biological features, and may benefit from increased surveillance for second cancers.
Keywords: CLL, trisomy 12, prognosis, second cancers, Richter transformation
Introduction
Chronic lymphocytic leukemia (CLL) is a B-cell malignancy with a variable clinical course for which clonal genomic aberrations play a crucial prognostic role. Trisomy 12 (+12) is the most common abnormality identified by chromosome banding analysis (CBA). It is the third most common abnormality identified by fluorescence in situ hybridization (FISH) analysis using a panel of FISH probes to the common recurrent abnormalities (deletions of 11q, 13q, 17p, and +12), with an incidence of about 16%.1 Trisomy 12 is traditionally associated with an intermediate risk of progression, and a favorable overall survival (OS).2 CLL cells with +12 tend to have atypical morphology, defined as more than 15% of cells with cleaved nuclei and/or lymphoplasmacytoid features, and an atypical immunophenotype, with a modified Matutes score less than 4 (based on expression of CD5, CD23, FMC7, surface immunoglobulin, CD22 and/or CD79b).3–6
In CLL cases with +12 identified by FISH, it is the sole aberration in about 70% of cases. It is associated with deletion 13q (del(13q)), deletion 11q (del(11q)), and deletion 17q (del(17p)) in 18%, 8%, and 4% of cases, respectively.7 By CBA, +12 is identified as the sole abnormality in 30% of cases, but it may also be associated with trisomy 18 (+18, 5% of cases), deletion 14q (del(14q), 3% of cases), t(14;19)(q32;q13), and/or trisomy 19 (+19).1,8,9 Of interest, the incidence of +12 rises from 16 to 36% in cases of small lymphocytic lymphoma (SLL)10, and small case series have reported an incidence of +12 in up to 50–90% of patients with Richter’s Transformation (RT)11–13, though the mechanism remains unclear.14
Recently, interest in +12 CLL has been raised by the discovery of NOTCH1 mutation in up to 24% of CLL patients with +12, particularly in cases with somatically unmutated immunoglobulin heavy chain variable region (IGHV) genes. NOTCH1 mutation is a stable marker, and may be associated with an inferior outcome.7,15,16 However, the impact of NOTCH1 mutation on prognosis may be influenced by other concurrent chromosomal aberrations. For example, NOTCH1 mutation occurs more frequently in patients carrying +12 as the sole abnormality, but a worse outcome is observed among patients with +12 associated with additional chromosomal abnormalities, irrespective of NOTCH1 mutation status.17
Compared to other cytogenetic subtypes, there are few large series in the literature that describe the clinical features of CLL cases with +12;18–21 the largest includes 104 patients.7 Thus, we analyzed and summarized our single-center experience of the clinical and laboratory features of 250 previously-untreated patients with +12 CLL over a period of nine years. Similar to previous reports, we observed an association between +12 and atypical immunophenotype, and a worse outcome in presence of deletion 14q (del14q). However, in contrast to previous reports, we observed an elevated mortality related to the onset of second cancers, suggesting the need for increased surveillance in this genetic subgroup.
Methods
Case selection
We performed a retrospective analysis of 250 treatment-naïve patients with CLL and +12 seen and followed at the University of Texas M.D. Anderson Cancer Center (MDACC) between 2003 (when routine FISH analysis was implemented at MDACC) and 2011. Their baseline characteristics were compared to those of 516 treatment-naïve patients with CLL and negative FISH followed at MDACC in the same time period. The study was approved by and conducted according to the Institutional Review Board of MDACC guidelines and was conducted in accordance with the principles of the Declaration of Helsinki. The clinical and laboratory features were obtained by review of the medical records. Cases were classified using the hierarchical risk model of FISH anomalies.2 Thus, we included cases with del(13q) or diploid cytogenetics in our analysis, but excluded cases with del(11q) or del(17p). The National Cancer Institute-Working Group (NCI-WG) criteria were applied to initiate treatment and to categorize response to treatment and time-to-event endpoints.22,23 We classified front-line therapy as follows: (1) FCR-based regimens, which included fludarabine, cyclophosphamide, and rituximab (FCR), FCR plus mitoxantrone (FCMR), FCR plus granulocyte-macrophage colony-stimulating factor (GM-CSF), FCR plus alemtuzumab (CFAR); (2) rituximab-based regimens, which included rituximab plus high-dose methylprednisone (HDMP), and rituximab plus GM-CSF; (3) investigational drug regimens, which included lenalidomide, idelalisib, and ibrutinib, with or without rituximab.
Routine laboratory and cytogenetic analyses
Laboratory testing, including evaluation of the IGHV somatic mutation status, and expression of CD38 and ZAP70, were performed as described previously.24,25 CBA was performed on metaphase cells prepared from bone marrow aspirate specimens cultured for 24 hours without mitogens, or for 72 hours with lipopolysaccharide, using standard techniques. Twenty Giemsa-banded metaphases were analyzed, and the results were reported using the International System for Human Cytogenetic Nomenclature. FISH analysis was performed on interphase nuclei prepared from bone marrow samples after culturing cells for 24 hours without mitogens, using the CLL probe panel (Vysis) according to the manufacturer’s recommendations. The panel includes probes specific to TP53 (17p13.1), ATM (11q22.3), D13S319 (13q14.3), LAMP1 (13q34), and the centromeric region of chromosome 12 (12p11.1-q11). Results for 200 analyzed nuclei were reported.
Statistical analysis
Time-to-first-treatment (TTT) and overall survival (OS) were calculated from the date of diagnosis to the date of therapy and death, respectively, or the date of last follow-up. Failure-free survival (FFS) was calculated from the date of first therapy to the date of relapse, death, or last follow-up. Survival distributions were calculated using the method of Kaplan and Meier and were compared using the log-rank test. Categorical and continuous variables were compared using the χ2 or Fisher exact tests, or the Mann-Whitney test, as appropriate. Linear regression and Cox regression were used for multivariable analysis (MVA) of categorical variables and survival, respectively. All P values were two-sided and considered significant if ≤0.05.
Results
Patient characteristics
Two-hundred and fifty treatment-naïve CLL patients with +12 evaluated at MDACC between 2003 and 2011 were included in the study. Of the 250 patients, 191 were evaluated by both CBA and FISH, and 59 by FISH only. Of the 191 patients assessed by both FISH and CBA, 91 were FISH+/CBA+ and 100 were FISH+/CBA− for +12. Trisomy 12 was the only abnormality in 82% of cases by FISH and 21% of cases by CBA (Table I). Baseline patient characteristics at presentation to MDACC are shown in Table II. We compared these patients to a cohort of 516 treatment-naïve CLL patients who lacked the common recurrent abnormalities on FISH analysis (negative for deletions of 11q, 13q, 17p, or +12) evaluated at our institution during the same time period. On univariate analysis (UVA), factors associated with +12 were thrombocytopenia (platelets (PLT) <100×109/L; p <0.001), bone marrow lymphocytosis (BM-L) ≥80% (p=0.05), serum beta-2-microglobulin (B2M) ≥4 mg/L (p=0.02), unmutated IGHV (p=0.02), positivity for CD38 (p<0.001), and Matutes score <4 (p<0.001). Factors not associated with +12 (p>0.05) were male gender, age ≥65years, bulky lymphadenopathy (LN), palpable splenomegaly, palpable hepatomegaly, hemoglobin (HGB) <10 g/dL, absolute lymphocyte count (ALC) >30×109/L, positivity for ZAP70, and use of VH 4-34 or VH 3-23. On multivariate analysis (MVA), factors significantly associated with +12 were PLT <100×109/L (odds ratio (OR) 2.4, p=0.03), positivity for CD38 (OR 2.4, p=0.001), and Matutes score <4 (OR 2.4, p <0.001) (Table II).
Table I.
Patients with +12 by FISH and/or CBA*
| Patients (N=250) | Number (%) |
|---|---|
|
| |
| FISH+/CBA− | 100 (40) |
| FISH+/CBA+ | 91 (36) |
| FISH+ only | 59 (24) |
|
| |
| FISH +12: alone | 206 (82) |
| with del(13q) | 44 (18) |
|
| |
| FISH +12: 7–25% | 56 (22) |
| 26–50% | 86 (34) |
| 51–75% | 94 (38) |
| 76–100% | 14 (6) |
|
| |
| CBA: diploid | 93/191 (47) |
| +12 alone | 40/191 (21) |
| with +19 | 14/191 (7) |
| with del(14q) | 9/191 (5) |
| with +18 | 6/191 (3) |
| others§ | 29/191 (17) |
Cases were classified using the hierarchical risk model of FISH anomalies; thus cases with concomitant del(11q) or del(17p) were not included in the analysis.
Additional chromosomal abnormalities associated with +12 identified by CBA and with a frequency <3% included +8, del13q and t(14;19)(q32;q13).
Abbreviations: FISH, fluorescence in situ hybridization; CBA, chromosome banding analysis.
Table II.
Baseline characteristics and comparison to patients with negative FISH
| Characteristics | +12 (N=250) | neg (N=516) | p-uni | OR [95% CI] | p-multi |
|---|---|---|---|---|---|
| Males | 147 (59%) | 292 (57%) | 0.59 | - | - |
| Age ≥65 years | 92 (37%) | 165 (32%) | 0.19 | - | - |
| Rai stage III–IV | 39 (16%) | 57 (11%) | 0.08 | - | - |
| Bulky (≥5cm) lymph nodes (n) | 5 (2%) | 6 (1%) | 0.35 | - | - |
| Splenomegaly (n) | 40 (16%) | 60 (12%) | 0.11 | - | - |
| Hepatomegaly (n) | 14 (6%) | 32 (6%) | 0.87 | - | - |
| Hemoglobin <11 g/dL | 27 (11%) | 38 (7%) | 0.07 | - | - |
| Platelets <100 X109/L | 37 (15%) | 30 (6%) | <0.001 | 2.4 [1.1–5.3] | 0.03 |
| ALC ≥30 X109/L | 66 (26%) | 105 (20%) | 0.06 | - | - |
| BM-Lymphocytes ≥80% | 43/199 (22%) | 57/385 (15%) | 0.05 | 1.3 | 0.35 |
| B2M ≥4 mg/L | 44/243 (18%) | 60/504 (12%) | 0.02 | 1.6 | 0.18 |
| IGHV unmutated | 103/187 (55%) | 160/362 (44%) | 0.02 | 1.3 | 0.34 |
| CD38 ≥30% | 109/198 (55%) | 109/361 (30%) | <0.001 | 2.4 [1.4–3.9] | 0.001 |
| ZAP70 positive | 88/195 (45%) | 166/342 (48%) | 0.47 | - | - |
| VH4-34 | 21/151 (14%) | 21/250 (8%) | 0.09 | - | - |
| VH3-23 | 17/151 (11%) | 24/250 (10%) | 0.61 | - | - |
| Matutes score <4 | 90/185 (49%) | 96/321 (30%) | <0.001 | 2.4 [1.5–4.1] | <0.001 |
Abbreviations: FISH, fluorescence in situ hybridization; neg, negative FISH; OR, odds ratio; CI, confidence interval; ALC, absolute lymphocyte count; BM, bone marrow; B2M, beta-2-microglobulin
Time-To-first-Treatment
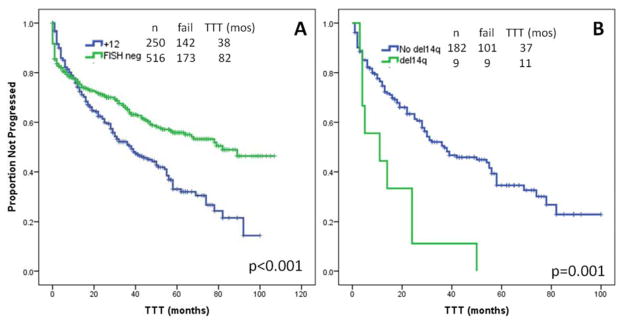
With a median follow-up of 51 months (range, 1–105), 142 patients (57%) required front-line therapy. The median TTT was 38 months (95% confidence interval [CI], 27–48), significantly shorter than FISH-negative patients (38 vs 82 months, p<0.001) (Figure 1A). On UVA, factors significantly associated with shorter TTT were Rai stage III–IV (p<0.001), bulky LN (p=0.02), palpable splenomegaly (p<0.001), HGB <11 g/dL (p<0.001), PLT <100×109/L (p<0.001), ALC ≥30×109/L (p<0.001), BM-L ≥80% (p=0.005), B2M ≥4 mg/L (p<0.001), positivity for ZAP70 (p=0.03), >50% of interphase nuclei positive for +12 by FISH (p=0.04), CBA positivity for +12 (p<0.001), aneuploidy on CBA (p<0.001), and CBA positivity for del(14q) (p=0.001). Factors not associated with shorter TTT on UVA (p>0.05) were male sex, age ≥65 years, palpable hepatomegaly, unmutated IGHV, positivity for CD38, use of VH4-34 or VH3-23, Matutes score <4, FISH positivity for del(13q), CBA positivity for +12 only, for +19, and for +18. The multivariable model for TTT included Rai stage III–IV (hazard ratio [HR] 3.3, p=0.02), palpable splenomegaly (HR 2.3, p=0.007), and CBA positivity for del(14q) (HR 3.5, p=0.004) (Figure 1B) as independently associated with TTT (Table III).
Figure 1.
Time-to-first-treatment (TTT). A. The median TTT was significantly shorter for patients with +12 than for patients with negative FISH (38 vs 82 months, p<0.001). B. On MVA, CBA positivity for del(14q) was associated with a significantly shorter TTT (11 vs 37 months, p=0.001).
Table III.
Multivariable Model for Survival
| TTT (months) | HR | p-multi | FFS (months) | HR | p-multi | 5yrs-OS (%) | HR | p-multi | |
|---|---|---|---|---|---|---|---|---|---|
| Rai stage II–IV | 12 | 3.3 | 0.02 | - | - | - | - | - | - |
| Splenomegaly | 13 | 2.3 | 0.007 | - | - | - | - | - | - |
| CBA+ for del14q | 11 | 3.5 | 0.004 | - | - | - | - | - | - |
| FISH- for del13q | - | - | - | 53 | 6.2 | 0.01 | - | - | - |
| No CR | - | - | - | 41 | 3.3 | 0.003 | - | - | - |
| ALC > 30 X109/L | - | - | - | - | - | - | 78 | 14.5 | 0.04 |
| SMN | - | - | - | - | - | - | 63 | 23.8 | 0.002 |
Abbreviations: TTT, time to first treatment; HR, hazard ratio; FFS, failure-free survival; OS, overall survival; CBA, chromosome banding analysis; FISH, fluorescence in situ hybridization; CR, complete remission; ALC, absolute lymphocyte count; SMN, second malignant neoplasm
Response to First-Line Treatment and Failure-Free Survival
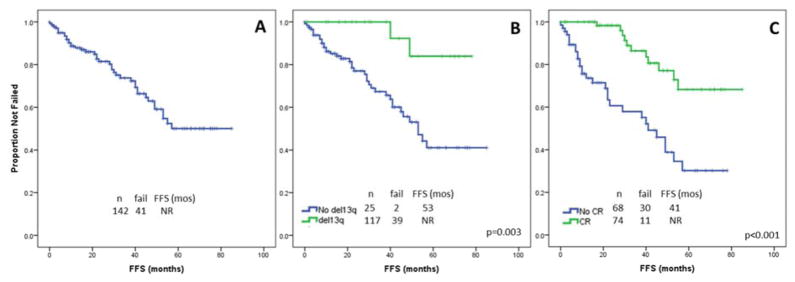
Front-line treatment consisted of an FCR-based regimen for 91 (64%) patients, an R-based regimen for 29 (20%), and investigational drug regimens for 22 (16%). A similar distribution of treatment regimens was observed in the control cohort. The complete remission (CR) rate was 52% (72%, 3%, and 3%, respectively); the overall response rate (ORR) was 94% (96%, 86% and 95%, respectively). With a median follow-up of 22 months (range, 1–85), the median failure-free-survival (FFS) following front-line treatment has not been reached (Figure 2A); 41 patients (29%) failed first therapy. On UVA, factors significantly associated with shorter FFS following first-line therapy were FISH negativity for del(13q) (p=0.003), therapy other than FCR-based (p<0.001), and response other than CR (p<0.001). Factors not significantly associated with FFS (p>0.05) were male sex, age >65 years, Rai stage III–IV, bulky LN, palpable splenomegaly or hepatomegaly, HGB <11 g/dL, PLT <100 X109/L, ALC ≥30×109/L, BM-L ≥80%, B2M ≥4 mg/L, unmutated IGHV, positivity for CD38 or ZAP70, use of VH4-34 or VH3-23, Matutes score <4, >50% of interphase nuclei positive for +12 by FISH, CBA positive for +12, aneuploidy on CBA, CBA positive for +12 only, CBA positive for +19, del14q or +18, achievement of PR. The multivariable model included FISH negativity for del(13q) (HR 6.2, p=0.01) (Figure 2B) and response other than CR (HR 3.3, p=0.003) (Figure 2C) as independently associated with FFS (Table III). Among the 41 patients who failed first-line therapy, 18 were observed (and 8 of them died), whereas 23 received 2nd line treatment (and 2 of them died). Second line treatment consisted of FCR-based regimen in 11 patients, R-based in 5, investigational in 4, and bendamustine plus rituximab in 3 patients. ORR was 83%.
Figure 2.
Failure-free survival (FFS). A. With a median follow-up of 22 months (range, 1–85), the median FFS following first-line treatment has not been reached. B. On MVA, FISH negativity for del(13q) was associated with a significantly shorter FFS (53 months vs not reached, p=0.003). C. On MVA, response other than CR was associated with a significantly shorter FFS (41 months vs not reached, p<0.001).
Overall survival and causes of death
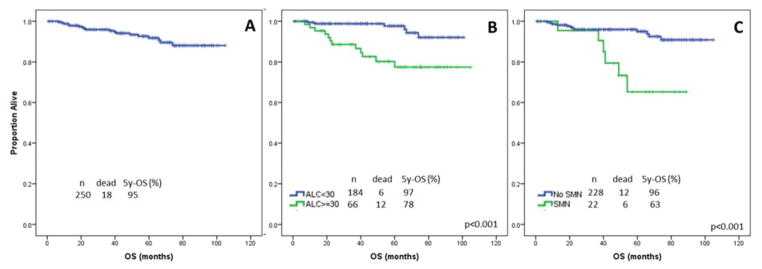
During the time of observation, the overall estimated median survival was not reached and it did not differ significantly between patients with or without +12 by FISH (p=0.22)(Figure 3A). On UVA, factors significantly associated with a shorter OS were age ≥65 years (p=0.02), HGB <11 g/dL (p=0.01), ALC ≥30 X109/L (p<0.001), B2M ≥4 mg/L (p=0.004), use of VH3-23 (p=0.05), aneuploidy on CBA (p=0.03), failure to achieve CR (p=0.03), onset of RS (p<0.001), and development of a second malignant neoplasm (SMN)(p<0.001). Factors not associated with OS on UVA (p>0.05) were male sex, Rai stage III–IV, bulky LN, palpable splenomegaly or hepatomegaly, PLT <100×109/L, BM-L ≥80%, unmutated IGHV, positivity for CD38 or ZAP70, use of VH4-34, Matutes score <4, positivity for del(13q) by FISH, >50% of interphase nuclei positive for +12 by FISH, CBA positivity for +12, CBA positivity for +12 only, +19, del14q, or +18, use of FCR-based therapy, and achievement of PR. In the multivariable model, ALC >300×109/L (HR 14.5, p=0.04) (Figure 3B) and development of a SMN (HR 23.8, p=0.002) (Figure 3C) remained independently associated with OS (Table III).
Figure 3.
Overall survival (OS). A. With a median follow-up of 51 months (range, 1–105), the median OS has not been reached. B. On MVA, ALC ≥30 X109/L was associated with a significantly shorter 5-year OS (78% vs 98%, p<0.001). C. On MVA, onset of SMN was associated with a significantly shorter 5-year OS (63% vs 96%, p<0.001).
Overall, 6 patients (2%) developed histologically-confirmed RT after a median time of 10 months (95% CI, 5–15) from diagnosis. The incidence of RT was significantly higher among treated than untreated patients (4% vs 0%, p= 0.04); it was also higher among patients with +12 FISH-positive than in the control cohort (2 vs 0.4%, p=0.02). Twenty-two patients (9%) developed a SMN other than RT, after a median of 19 months (95% CI, 6–32) from diagnosis. Five SMN were hematologic malignancies (1 acute myeloid leukemia, 1 myelodysplastic syndrome, 1 myeloproliferative neoplasm, and 2 plasma cell myelomas) and 17 were non-hematologic malignancies (1 bladder carcinoma, 1 uterine carcinoma, 1 gastric carcinoma, 1 colon carcinoma, 1 lung carcinoma, 4 prostate carcinomas, 6 invasive squamous cell carcinomas of the skin, 1 sarcoma, 1 thyroid carcinoma). Of the 5 hematologic SMN, 2 occurred after therapy for CLL (after 18 from CFAR and 36 months from R+GMCSF) and, therefore, could be classified as therapy-related. The incidence of SMN did not differ significantly among treated and untreated patients (p=0.12), but it was significantly higher in +12 FISH-positive patients than in the control cohort (9% vs 1%, p<0.001).
Eighteen patients (7%) died during the time of observation, 15 of whom had received treatment for CLL. Seven patients died of CLL-related (4 patients) or RT-related (3 patients) causes, i.e. severe infections, bleeding, organ infiltration. Six patients died of a SMN (1 acute myeloid leukemia, 1 gastric carcinoma, 1 colon carcinoma, 1 lung carcinoma, 1 bladder carcinoma, 1 sarcoma), and 5 died of unrelated causes. The distribution of the causes of death did not differ significantly among treated and untreated patients (p=0.24).
Discussion
Although the most common cytogenetic abnormalities identified by FISH in CLL are deletions, +12 is the most commonly identified abnormality on conventional karyotypic analysis. Trisomy 12 has been traditionally considered an intermediate-risk prognostic factor in CLL.2 Cases of CLL with +12 comprise up to 16% of all patients assessed by FISH, yet relatively little is known about its pathophysiology compared to other genetic subtypes. In the current study we analyzed 250 treatment-naive patients with +12 CLL, the largest series in the literature.
In our series, +12 was the sole abnormality in 82% of cases assessed by FISH; it was associated with del(13q) in the remaining cases, similar to previous reports.7,18 Of interest, because we used the hierarchical FISH model, we excluded cases with associated del(11q) or del(17p) from our analysis.2 When evaluated by CBA, +12 was the sole abnormality in 21% of cases; the remaining cases were diploid or showed +12 in association with +19, del14q, and +18, or less commonly, with trisomy 8, del(13q), and t(14;19)(q32;q13).
Recent data suggest that +12 associated with additional chromosomal abnormalities portends a poor prognosis, independent of NOTCH1 mutation, which is more common in cases with isolated +12 compared to cases with additional abnormalities.17 New methods to stimulate CLL cells to divide in culture (developed after the period of our current study) have increased the sensitivity to detect chromosomal abnormalities and karyotypic complexity.26 Using these methods, it is unclear whether in cases with +12 karyotypic complexity or NOTCH1 mutation status will have a greater impact on prognosis.
In our study, several clinical and laboratory features distinguished CLL patients with +12 from the cohort with negative FISH. Similar to previous studies, we found an association between +12, atypical immunophenotype (low Matutes score), and positivity for CD38 (that did not correlate with the somatic mutation status of the IGHV genes or positivity for ZAP70).3–5,20,21,27,28 CD38, whose expression is often associated with +12, is a highly conserved transmembrane glycoprotein that plays a critical role in lymphocyte trafficking between blood and lymphoid organs, and in survival and proliferation within the lymphoid organs. It is conceivable that CD38 inhibitors currently under development may have particular efficacy in this cytogenetic subtype.29–32
Unlike other studies, we found that patients with +12 were more likely to present with thrombocytopenia. During the course of disease, cytopenias have been observed in up to 24% of patients with CLL, resulting from bone marrow failure or autoimmune disease.33,34 While the diagnosis of autoimmune hemolytic anemia is based on specific laboratory findings, identifying an autoimmune thrombocytopenia can be problematic.35,36 In our series, bone marrow infiltration was not associated with +12 on MVA, suggesting an autoimmune etiology for the elevated rate of thrombocytopenia that we observed. A previous study showed that patients with +12 had a higher incidence of autoimmune rather than infiltrative cytopenias.33 Taken together, the findings raise the possibility that CLL patient with +12 may be at increased risk to develop autoimmune thrombocytopenia.
Most of the patients in our study (57%) required treatment during the time of observation, with a median TTT of about 3 years. A similar proportion of treated patients (50%) but with a slightly shorter TTT (14–33 months) has been described previously in a smaller series of 47 patients.2,37 In the latter study, baseline characteristics for patients with +12 were not reported, which may account for this slight difference. In our study, a shorter TTT was observed for patients with Rai stage III–IV, palpable splenomegaly, and del(14q) on CBA. Advanced Rai stage and massive splenomegaly are established indications for therapy, so their association with a shorter TTT is not surprising.22 It has been shown recently that del(14q), often associated with +12 as well as with unmutated IGHV and NOTCH1 mutations, portends a poor prognosis.38 The number of patients with del(14q) in our study is small. However, our findings suggest that evaluation for this abnormality, either by conventional cytogenetic analysis and/or FISH analysis, may have prognostic relevance in these patients.
Although many patients required treatment, the ORR to first-line therapy was 94% and the median FFS has not been reached yet. Factors associated with a shorter FFS were response other than CR and FISH negativity for del(13q). The quality of the response to therapy is an independent prognostic factor in CLL, demonstrated by recent studies analyzing the importance of minimal residual disease eradication.39,40 Further studies are needed to shed light on the biological mechanisms favoring a longer FFS when del13q and +12 are concomitant.
Similarly, median OS has not been reached in our analysis and only 18 patients died during the time of observation. Other studies have reported similar results, with a median OS of 133 months despite a significantly shorted median TTT.2,7,41 Surprisingly, the mortality of patients with +12 was only partly related to complications of CLL; the leading causes of death were RT and SMN. In our series, RT was observed in 2% of patients with +12, in line with other large retrospective analyses.42 However, this was significantly higher than in patients with negative FISH (0.4%), as partially reported in other series.13,43 Though some hypothesis have been done, the mechanism linking +12 to RT still remains unclear.14 Similarly, in our study the incidence of SMN among patients with +12 was 9%, significantly higher than in patient with negative FISH (1%). A correlation between CLL and SMN has been reported previously44, though a specific association with +12 is a novel finding. The mechanism linking +12 to the onset of SMN remains unclear. However, given the recent evidence of a worse outcome for SMN in CLL45, increased surveillance of patients in this specific group should be considered.
It is interesting that the MVA for TTT, FFS and OS revealed different factors, and that not a single variable appeared to be associated uniformly with survival. CLL +12 has a higher expression of CD20, probably determining a better response to R-based regimens.46 Factors determining CD20 expression may influence response to treatment, and be totally independent from factors determining disease progression to first therapy (presence of del14q). At the same time, unless the burden of disease is high (elevated ALC), a deeper response to therapy may be achieved, and subsequently OS may be determined by other factors (such as second cancers).
Clinical Practice Point.
Patients with trisomy 12 (+12) CLL have a higher incidence of thrombocytopenia, Richter Transformation (RT) and second cancers, in addition to the expected increased rate of CD38 positivity and atypical immunophenotype
Time to first treatment is shorter in patients with advanced Rai stage, splenomegaly, and deletion 14q by cytogenetic.
Failure-free survival is shorter in patients who don’t achieve complete remission or in absence of deletion 13q by fluorescence in situ hybridization.
OS is shorter in patients with lymphocytosis and who developed SMN
Mortality rate is low (7%) and mostly related to RT and SMN
Increased surveillance for second cancers is needed in this group
Acknowledgments
The authors are grateful to Susan Lerner and Susan Smith for their help with data collection.
Footnotes
Authorship Contributions
M.J.K. and P.S. designed the study, analyzed the data, and wrote the paper; A.F. designed the study and provided supervision and advice in data analysis and manuscript preparation; L.V.A. supervised the conventional cytogenetic and FISH testing and provided advice and oversight in manuscript preparation; W.W. and S.O. contributed to the design of the study, verified the accuracy of patient data, and provided advice on statistical analysis.
Disclosure of Conflict of Interest
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Haferlach C, Dicker F, Schnittger S, Kern W, Haferlach T. Comprehensive genetic characterization of CLL: a study on 506 cases analysed with chromosome banding analysis, interphase FISH, IgV(H) status and immunophenotyping. Leukemia. 2007;21(12):2442–2451. doi: 10.1038/sj.leu.2404935. [DOI] [PubMed] [Google Scholar]
- 2.Dohner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 3.Cro L, Ferrario A, Lionetti M, et al. The clinical and biological features of a series of immunophenotypic variant of B-CLL. European journal of haematology. 2010;85(2):120–129. doi: 10.1111/j.1600-0609.2010.01454.x. Prepublished on 2010/04/23. [DOI] [PubMed] [Google Scholar]
- 4.Matutes E, Oscier D, Garcia-Marco J, et al. Trisomy 12 defines a group of CLL with atypical morphology: correlation between cytogenetic, clinical and laboratory features in 544 patients. British journal of haematology. 1996;92(2):382–388. doi: 10.1046/j.1365-2141.1996.d01-1478.x. Prepublished on 1996/02/01. [DOI] [PubMed] [Google Scholar]
- 5.Schlette E, Medeiros LJ, Keating M, Lai R. CD79b expression in chronic lymphocytic leukemia. Association with trisomy 12 and atypical immunophenotype. Archives of pathology & laboratory medicine. 2003;127(5):561–566. doi: 10.1043/0003-9985(2003)127<0561:CEICLL>2.0.CO;2. Prepublished on 2003/04/24. [DOI] [PubMed] [Google Scholar]
- 6.Quijano S, Lopez A, Rasillo A, et al. Impact of trisomy 12, del(13q), del(17p), and del(11q) on the immunophenotype, DNA ploidy status, and proliferative rate of leukemic B-cells in chronic lymphocytic leukemia. Cytometry B Clin Cytom. 2008;74(3):139–149. doi: 10.1002/cyto.b.20390. [DOI] [PubMed] [Google Scholar]
- 7.Del Giudice I, Rossi D, Chiaretti S, et al. NOTCH1 mutations in +12 chronic lymphocytic leukemia (CLL) confer an unfavorable prognosis, induce a distinctive transcriptional profiling and refine the intermediate prognosis of +12 CLL. Haematologica. 2012;97(3):437–441. doi: 10.3324/haematol.2011.060129. Prepublished on 2011/12/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huh YO, Schweighofer CD, Ketterling RP, et al. Chronic lymphocytic leukemia with t(14;19)(q32;q13) is characterized by atypical morphologic and immunophenotypic features and distinctive genetic features. Am J Clin Pathol. 2011;135(5):686–696. doi: 10.1309/AJCPOEFP3SLX6HXJ. [DOI] [PubMed] [Google Scholar]
- 9.Sellmann L, Gesk S, Walter C, et al. Trisomy 19 is associated with trisomy 12 and mutated IGHV genes in B-chronic lymphocytic leukaemia. Br J Haematol. 2007;138(2):217–220. doi: 10.1111/j.1365-2141.2007.06636.x. [DOI] [PubMed] [Google Scholar]
- 10.Daudignon A, Poulain S, Morel P, et al. Increased trisomy 12 frequency and a biased IgVH 3-21 gene usage characterize small lymphocytic lymphoma. Leuk Res. 2010;34(5):580–584. doi: 10.1016/j.leukres.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Tsimberidou AM, Keating MJ, Wierda WG. Richter’s transformation in chronic lymphocytic leukemia. Curr Hematol Malig Rep. 2007;2(4):265–271. doi: 10.1007/s11899-007-0036-9. [DOI] [PubMed] [Google Scholar]
- 12.Rossi D, Cerri M, Capello D, et al. Biological and clinical risk factors of chronic lymphocytic leukaemia transformation to Richter syndrome. British journal of haematology. 2008;142(2):202–215. doi: 10.1111/j.1365-2141.2008.07166.x. Prepublished on 2008/05/22. [DOI] [PubMed] [Google Scholar]
- 13.Chigrinova E, Rinaldi A, Kwee I, et al. Two main genetic pathways lead to the transformation of chronic lymphocytic leukemia to Richter syndrome. Blood. 2013;122(15):2673–2682. doi: 10.1182/blood-2013-03-489518. Prepublished on 2013/09/06. [DOI] [PubMed] [Google Scholar]
- 14.Falisi E, Novella E, Visco C, et al. B-cell receptor configuration and mutational analysis of patients with chronic lymphocytic leukaemia and trisomy 12 reveal recurrent molecular abnormalities. Hematological oncology. 2014;32(1):22–30. doi: 10.1002/hon.2086. Prepublished on 2013/07/19. [DOI] [PubMed] [Google Scholar]
- 15.Balatti V, Bottoni A, Palamarchuk A, et al. NOTCH1 mutations in CLL associated with trisomy 12. Blood. 2012;119(2):329–331. doi: 10.1182/blood-2011-10-386144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puente XS, Pinyol M, Quesada V, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475(7354):101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez C, Delgado J, Costa D, et al. Different distribution of NOTCH1 mutations in chronic lymphocytic leukemia with isolated trisomy 12 or associated with other chromosomal alterations. Genes Chromosomes Cancer. 2012;51(9):881–889. doi: 10.1002/gcc.21972. [DOI] [PubMed] [Google Scholar]
- 18.Escudier SM, Pereira-Leahy JM, Drach JW, et al. Fluorescent in situ hybridization and cytogenetic studies of trisomy 12 in chronic lymphocytic leukemia. Blood. 1993;81(10):2702–2707. Prepublished on 1993/05/15. [PubMed] [Google Scholar]
- 19.Athanasiadou A, Stamatopoulos K, Tsompanakou A, et al. Clinical, immunophenotypic, and molecular profiling of trisomy 12 in chronic lymphocytic leukemia and comparison with other karyotypic subgroups defined by cytogenetic analysis. Cancer Genet Cytogenet. 2006;168(2):109–119. doi: 10.1016/j.cancergencyto.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Que TH, Marco JG, Ellis J, et al. Trisomy 12 in chronic lymphocytic leukemia detected by fluorescence in situ hybridization: analysis by stage, immunophenotype, and morphology. Blood. 1993;82(2):571–575. Prepublished on 1993/07/15. [PubMed] [Google Scholar]
- 21.Tabernero MD, San Miguel JF, Garcia JL, et al. Clinical, biological, and immunophenotypical characteristics of B-cell chronic lymphocytic leukemia with trisomy 12 by fluorescence in situ hybridization. Cytometry. 1995;22(3):217–222. doi: 10.1002/cyto.990220309. Prepublished on 1995/09/15. [DOI] [PubMed] [Google Scholar]
- 22.Hallek M, Cheson BD, Catovsky D, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111(12):5446–5456. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheson BD, Bennett JM, Grever M, et al. National Cancer Institute-sponsored Working Group guidelines for chronic lymphocytic leukemia: revised guidelines for diagnosis and treatment. Blood. 1996;87(12):4990–4997. Prepublished on 1996/06/15. [PubMed] [Google Scholar]
- 24.Shanafelt TD, Witzig TE, Fink SR, et al. Prospective evaluation of clonal evolution during long-term follow-up of patients with untreated early-stage chronic lymphocytic leukemia. J Clin Oncol. 2006;24(28):4634–4641. doi: 10.1200/JCO.2006.06.9492. [DOI] [PubMed] [Google Scholar]
- 25.Rassenti LZ, Huynh L, Toy TL, et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N Engl J Med. 2004;351(9):893–901. doi: 10.1056/NEJMoa040857. [DOI] [PubMed] [Google Scholar]
- 26.Muthusamy N, Breidenbach H, Andritsos L, et al. Enhanced detection of chromosomal abnormalities in chronic lymphocytic leukemia by conventional cytogenetics using CpG oligonucleotide in combination with pokeweed mitogen and phorbol myristate acetate. Cancer genetics. 2011;204(2):77–83. doi: 10.1016/j.cancergen.2010.12.006. Prepublished on 2011/04/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Criel A, Wlodarska I, Meeus P, et al. Trisomy 12 is uncommon in typical chronic lymphocytic leukaemias. British journal of haematology. 1994;87(3):523–528. doi: 10.1111/j.1365-2141.1994.tb08307.x. Prepublished on 1994/07/01. [DOI] [PubMed] [Google Scholar]
- 28.Criel A, Verhoef G, Vlietinck R, et al. Further characterization of morphologically defined typical and atypical CLL: a clinical, immunophenotypic, cytogenetic and prognostic study on 390 cases. British journal of haematology. 1997;97(2):383–391. doi: 10.1046/j.1365-2141.1997.402686.x. Prepublished on 1997/05/01. [DOI] [PubMed] [Google Scholar]
- 29.D’Arena G, Musto P, Cascavilla N, et al. CD38 expression correlates with adverse biological features and predicts poor clinical outcome in B-cell chronic lymphocytic leukemia. Leukemia & lymphoma. 2001;42(1–2):109–114. doi: 10.3109/10428190109097682. Prepublished on 2001/11/09. [DOI] [PubMed] [Google Scholar]
- 30.Riches JC, O’Donovan CJ, Kingdon SJ, et al. Trisomy 12 chronic lymphocytic leukemia cells exhibit upregulation of integrin signaling that is modulated by NOTCH1 mutations. Blood. 2014;123(26):4101–4110. doi: 10.1182/blood-2014-01-552307. Prepublished on 2014/05/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brachtl G, Pinon Hofbauer J, Greil R, Hartmann TN. The pathogenic relevance of the prognostic markers CD38 and CD49d in chronic lymphocytic leukemia. Annals of hematology. 2014;93(3):361–374. doi: 10.1007/s00277-013-1967-y. Prepublished on 2013/11/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreau C, Liu Q, Graeff R, et al. CD38 Structure-Based Inhibitor Design Using the 1-Cyclic Inosine 5’-Diphosphate Ribose Template. PloS one. 2013;8(6):e66247. doi: 10.1371/journal.pone.0066247. Prepublished on 2013/07/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zent CS, Ding W, Schwager SM, et al. The prognostic significance of cytopenia in chronic lymphocytic leukaemia/small lymphocytic lymphoma. Br J Haematol. 2008;141(5):615–621. doi: 10.1111/j.1365-2141.2008.07086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strati P, Caligaris-Cappio F. A matter of debate in chronic lymphocytic leukemia: is the occurrence of autoimmune disorders an indicator of chronic lymphocytic leukemia therapy? Curr Opin Oncol. 2011;23(5):455–460. doi: 10.1097/CCO.0b013e328348c683. [DOI] [PubMed] [Google Scholar]
- 35.Bass GF, Tuscano ET, Tuscano JM. Diagnosis and classification of autoimmune hemolytic anemia. Autoimmun Rev. 2014;13(4–5):560–564. doi: 10.1016/j.autrev.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 36.Lo E, Deane S. Diagnosis and classification of immune-mediated thrombocytopenia. Autoimmun Rev. 2014;13(4–5):577–583. doi: 10.1016/j.autrev.2014.01.026. [DOI] [PubMed] [Google Scholar]
- 37.Ripolles L, Ortega M, Ortuno F, et al. Genetic abnormalities and clinical outcome in chronic lymphocytic leukemia. Cancer Genet Cytogenet. 2006;171(1):57–64. doi: 10.1016/j.cancergencyto.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 38.Cosson A, Chapiro E, Belhouachi N, et al. 14q deletions are associated with trisomy 12, NOTCH1 mutations and unmutated IGHV genes in chronic lymphocytic leukemia and small lymphocytic lymphoma. Genes Chromosomes Cancer. 2014 doi: 10.1002/gcc.22176. [DOI] [PubMed] [Google Scholar]
- 39.Bottcher S, Ritgen M, Fischer K, et al. Minimal residual disease quantification is an independent predictor of progression-free and overall survival in chronic lymphocytic leukemia: a multivariate analysis from the randomized GCLLSG CLL8 trial. J Clin Oncol. 2012;30(9):980–988. doi: 10.1200/JCO.2011.36.9348. [DOI] [PubMed] [Google Scholar]
- 40.Strati P, Keating MJ, O’Brien SM, et al. Eradication of bone marrow minimal residual disease with first-line chemoimmunotherapy may prompt early treatment discontinuation in chronic lymphocytic leukemia. Blood. 2014 doi: 10.1182/blood-2013-11-538116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giertlova M, Hajikova M, Vaskova J, et al. Cytogenetic abnormalities predict treatment-free interval and response to therapy in previously untreated chronic lymphocytic leukemia patients. Neoplasma. 2011;58(1):82–88. doi: 10.4149/neo_2011_01_82. [DOI] [PubMed] [Google Scholar]
- 42.Parikh SA, Rabe KG, Call TG, et al. Diffuse large B-cell lymphoma (Richter syndrome) in patients with chronic lymphocytic leukaemia (CLL): a cohort study of newly diagnosed patients. British journal of haematology. 2013;162(6):774–782. doi: 10.1111/bjh.12458. Prepublished on 2013/07/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parikh SA, Rabe KG, Call TG, et al. Diffuse large B-cell lymphoma (Richter syndrome) in patients with chronic lymphocytic leukaemia (CLL): a cohort study of newly diagnosed patients. Br J Haematol. 2013;162(6):774–782. doi: 10.1111/bjh.12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsimberidou AM, Wen S, McLaughlin P, et al. Other malignancies in chronic lymphocytic leukemia/small lymphocytic lymphoma. J Clin Oncol. 2009;27(6):904–910. doi: 10.1200/JCO.2008.17.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solomon BM, Rabe KG, Slager SL, Brewer JD, Cerhan JR, Shanafelt TD. Overall and cancer-specific survival of patients with breast, colon, kidney, and lung cancers with and without chronic lymphocytic leukemia: a SEER population-based study. J Clin Oncol. 2013;31(7):930–937. doi: 10.1200/JCO.2012.43.4449. [DOI] [PubMed] [Google Scholar]
- 46.Tam CS, Otero-Palacios J, Abruzzo LV, et al. Chronic lymphocytic leukaemia CD20 expression is dependent on the genetic subtype: a study of quantitative flow cytometry and fluorescent in-situ hybridization in 510 patients. British journal of haematology. 2008;141(1):36–40. doi: 10.1111/j.1365-2141.2008.07012.x. Prepublished on 2008/03/08. [DOI] [PubMed] [Google Scholar]