Abstract
Multiple myeloma (MM) is a plasma cell malignancy with an estimated 26,850 new cases and 11,240 deaths in 2015 in the United States. Two main classes of agents are the mainstays of therapy - proteasome inhibitors (PIs) and immunomodulatory drugs (IMiDs). Other new targets are emerging rapidly, including monoclonal antibodies and histone deacetylase (HDAC) inhibitors. These therapeutic options have greatly improved overall survival but currently only 15-20% of patients experience long-term progression-free survival or are cured. Therefore, improvement in treatment options is needed. One potential means of improving clinical options is to target resistance mechanisms for current agents. For example, eliminating the cytoprotective heat shock response that protects myeloma cells from proteasome inhibition may enhance PI-based therapies. The transcription factor Heat Shock Factor 1 (HSF1) is the master regulator of the heat shock response. HSF1 is vital in the proteotoxic stress response and its activation is controlled by post-translational modifications (PTMs). This review details the mechanisms of HSF1 regulation and discusses leveraging that regulation to enhance PI activity.
Keywords: Multiple myeloma, HSF1, proteasome inhibition
Non-Proteasome Inhibitor Myeloma Treatments
From 1971-1996, the overall survival rate for MM patients remained largely unchanged[1]. Despite the use of alkylators, corticosteroids (dexamethasone and prednisone), and autologous bone marrow transplantation, little improvement was noted. Then, in 1999, thalidomide (in combination with dexamethasone) became the first new agent with major activity against MM in 37 years[2]. Thalidomide (Thalomid® - 2006 FDA approval) belongs to a class of structurally similar drugs known as immunomodulatory drugs (IMiDs), along with lenalidomide (Revlimid® - 2006) and pomalidomide (Pomalyst® - 2013). IMiDs have helped to improve patient outcomes in recent years along with another major class of MM agents: proteasome inhibitors[3]. The two FDA-approved PIs are bortezomib (Velcade® - 2003) and carfilzomib (Kyprolis® - 2012).
Proteasome Inhibition
The main effector in the ubiquitin-proteasome system (UPS) is the proteasome, a cytoplasmic protein complex responsible for protein degradation[4]. The 26S proteasome is about 2000 kilodaltons (kDa) in molecular mass and consists of one 20S protein subunit and two 19S regulatory cap subunits. Proteasomal degradation removes denatured, misfolded, damaged, or improperly translated proteins from cells. The UPS plays an essential homeostatic role in regulating intracellular protein concentration, as well as being a regulator involved in many cellular processes including DNA repair, sodium channel function, regulation of immune and inflammatory responses, signal transduction and cell cycle progression[5].
Proteasome-mediated degradation is particularly vital for plasma cell quality control because of its role as a professional secretory cell that produces copious amounts of immunoglobulin in a constitutive manner. Therefore, proteasome inhibition can dramatically alter protein homeostasis leading to stress responses and if not resolved, apoptosis[6].
Bortezomib is a highly selective and reversible PI that has a boron atom which binds the β5 subunit (PSMB5)/chymotrypsin-like activity of the 26S proteasome[7]. The proteasome has an ATP-dependent proteolytic activity, therefore, bortezomib's targeting of β5 results in decrease or loss of proteasome function. Bortezomib was first reported as an anti-inflammatory agent for treating polyarthritis in 1998. Palombella et al., used bortezomib as a means for inhibiting NF-κB activation by preventing proteasome-mediated degradation of IκBα, an NF-κB negative regulator[8]. For cancer, bortezomib was first tested in vitro in by Adams et al., in a 60 tumor cell line NCI screen, and was most potent in the prostate cancer cell line, PC-3[9]. Cytotoxicity was speculated to be due to stabilization and dysregulation of cyclins, CDK inhibitors, tumor suppressor proteins, IκB, and other proteins associated with cell cycle progression. Hideshima et al., published the first report on bortezomib in MM cell lines and freshly isolated patient samples[10]. In addition to the NF-κB mechanism described above, bortezomib was shown to alter cellular interactions and cytokine secretion in the bone marrow milieu to inhibit tumor cell growth, induce apoptosis, and overcome drug resistance. Mitsiades et al., used high-dose bortezomib in the human MM cell line, MM.1S, to probe gene expression changes[11]. These changes included a downregulation of growth/survival signaling pathways, upregulation of molecules implicated in pro-apoptotic cascades, and upregulation of ubiquitin/proteasome pathway members and heat shock proteins (HSPs). HSP27, 40, and 70 upregulation was seen as early as two hours post-treatment. Bortezomib was FDA-approved in 2003 for patient use in large part due to the results of a Phase II study of its use in relapsed/refractory MM[12].
Up to this point, while gene expression profiling had been used to characterize the molecular sequelae of bortezomib treatment, mechanisms mediating anti-MM activity had not yet been defined. Questions remained unanswered including, ‘Through what pathway(s) does PI induce apoptosis?’ and ‘Is there a cellular event specific to plasma cells that can predict its effectiveness?’ Hideshima et al., began to scratch the surface of the bortezomib-cell biology connection by linking bortezomib, p53 phosphorylation, JNK activation, caspase-3 and 8 activation, inhibition of DNA damage repair, and cell death[13].
This study led to further investigation into the cell biology changes caused by bortezomib. However, what had not been looked at up to that point was specifically the plasma cell nature of a myeloma cell. Because of their role as immunoglobulin producers, plasma cells are heavily reliant on the unfolded protein response (UPR) for protein quality control[14]. Lee et al., suggested that UPR inhibition, through IRE1α (a UPR transducer) suppression and splicing impairment of its downstream target, XBP1, plays a role in MM PI-induced death[15]. Our group showed that PIs can lead to an accumulation of misfolded proteins and an induction of terminal components of the UPR including PERK, eIF-2α, ATF4, and its downstream target, CHOP[16]. This was one of the first reports detailing how bortezomib was exploiting plasma cell biology, specifically immunoglobulin accumulation and terminal UPR activation, to induce apoptosis. Meister et al., concluded that bortezomib-induced apoptosis is associated with the buildup of defective ribosomal products (DRiPs) and other unfolded proteins in the ER[17]. Also, Bianchi et al., determined that the balance between proteasome workload and degradative capacity represents a critical determinant of apoptotic sensitivity of MM cell lines to PI[18]. Furthermore, Ling et al., showed that low XBP1 levels predict poor response to bortezomib, both in vitro and in MM patients, and ATF6 (a UPR transducer) expression correlates with bortezomib sensitivity[19]. Leung-Hagesteijn et al., proposed that the existence of PI-insensitive Xbp1s- tumor progenitors within primary MM tumors may produce class-effect PI resistance independent of drug identity[20]. Mechanistically, MM Xbp1s suppression induces bortezomib resistance via decommitment to plasma cell maturation and immunoglobulin production, diminishing ER stress-associated cytotoxicity.
In addition to direct inhibition of the proteasome, PI-induced ER stress can also occur from aggresome formation and autophagy[21-23]. Both are thought to be survival mechanisms used by cancer cells, and a recent study suggests that targeting the integrated networks of aggresome formation, proteasome, and autophagy may potentiate ER stress-mediated cell death pathways[21]. However, one potential counter to PI effectiveness is the development of acquired mutations.
The direct target of bortezomib, PSMB5, is the most well-characterized mutation site[24]. The PSMB5 mutation A49T has been shown to play in role in bortezomib resistance[25, 26]. This mutation reduces bortezomib-induced apoptosis through the prevention of ubiquitinated protein accumulation and fatal ER stress in MM. Despite this concern, no clinical evidence of an acquired proteasome subunit mutation has been published[25].
With the success of bortezomib in the clinic, second generation PIs have been developed that have different activities, bioavailability (oral) and toxicity profiles. These agents have been the subject of intense preclinical and clinical studies. The first of these new inhibitors, Carfilzomib, has now been FDA-approved for the treatment of relapsed/refractory MM. Carfilzomib is an intravenous irreversible PI which binds to β5 with greater selectivity than bortezomib[27]. NPI-0052 (marizomib), ONX 0912 (oprozomib), and MLN9708/2238 (ixazomib) are all involved in clinical trials[7, 27]. Marizomib is being tested intravenously and oprozomib and ixazomib are being tested orally in MM. Marizomib is a β-lactone-γ-lactam inhibitor which irreversibly binds β2 and β5 with high affinity and β1 with low affinity, and was granted “orphan drug” status by the FDA for MM treatment. Phase I combination studies are being conducted using marizomib, pomalidomide, and dexamethasone in subjects with relapsed/refractory MM[28]. Oprozomib is an epoxyketone which irreversibly binds β5 with high affinity and was also recently granted “orphan drug” status by the FDA for MM and Waldenström macroglobulinemia treatment. Ixazomib is a boric acid analog which reversibly binds β5 with high affinity and at higher concentrations is able to inhibit β1 and β2. Two recently published companion reports from Phase I oral ixazomib studies in relapsed/refractory MM patients showed that 15-18% of patients achieved partial response or better with 76% reaching a state of stable disease or better in one of the studies[29, 30].
Continued improvement in current treatments and clinical trials including those for second-generation PIs have led some researchers to state that prolonged disease-free survival and a cure for a majority of patients are on the horizon[31]. Improved disease-free survival can only occur if we can identify and target cellular resistance mechanisms. Resistance mechanisms, including HSP upregulation as part of the heat shock response (HSR), can limit PI effectiveness. Therefore, inhibiting the HSR is a therapeutic opportunity for improving PI efficacy.
The Heat Shock Response and Heat Shock Proteins
As mentioned above, HSP family members were reported amongst genes that were highly upregulated by bortezomib[11]. The HSR is part of a cell's internal repair machinery and maintains homeostasis under stressful conditions including infection, inflammation, exercise, exposure to toxins or pharmacological agents, starvation, or hypoxia[32]. This response is carried out by HSPs, many of which act as chaperones assisting in protein folding and establishment of proper conformation while also preventing undesired protein aggregation. HSPs are categorized into five families: (1) HSP70 superfamily (2) DNAJ (HSP40) family (3) HSPB (small heat shock protein) family (4) HSP90/HSPC family (5) Chaperonins and related genes[33]. While the cytoprotective HSR is desired in healthy cells, it could also protect cancer cells from bortezomib's pro-apoptotic effects and is a potential resistance mechanism as demonstrated in bladder cancer cells[34]. Zhang, et al., have published a detailed review of the connection between bortezomib and HSPs in MM[32].
The cytoprotective nature of HSPs has stimulated preclinical testing and clinical trials of HSP90 and HSP70 inhibitors in MM and other cancers. HSP90 inhibitors have been tested either alone or in combination with bortezomib and/or dexamethasone in MM[35, 36]. However, the results of these studies to date have been disappointing and have yet to lead to an FDA-approved HSP90 inhibitor. Usmani et al., have comprehensively reviewed the promise and difficulty of HSP90 inhibition as a therapeutic strategy in MM[37]. Numerous other reports have been published regarding HSP90 inhibitors[32, 38, 39]. HSP70 inhibitors have shown promise in preclinical settings, including MM, both alone and in combination with bortezomib and/or HSP90 inhibitors, but have not progressed to clinical trials[40-43]. Detailed overviews of the role of HSP70 in cancer and the challenges of various HSP70 inhibition strategies have previously been published[44, 45]. For several reasons, both HSP70 and HSP90 inhibitors face a similar challenge: single-target HSP inhibitors may not work in cancer. First, some HSP inhibitors cannot induce apoptosis by themselves at biologically relevant levels[42, 46]. For those inhibitors that do, studies have shown that they induce other chaperones including HSPs as a compensatory mechanism. For example, HSP90 inhibitors induce HSP70 and HSP27, and lead to an increase in HSP90 client proteins[43, 47-53]. In addition, Acquaviva, et al., showed that the treatment of H1975 non-small cell lung cancer or A375 melanoma cells with the HSP90 inhibitor ganetespib leads to an additional compensatory mechanism, nuclear accumulation of the HSR master regulator, Heat Shock Factor 1 (HSF1)[53]. These and other results indicate that individual HSP inhibition only targets a part of the HSR. The combination of compensatory HSP induction and nuclear HSF1 accumulation could lead to increased drug resistance and negate any pro-apoptotic effect of single-target HSP inhibitors. Therefore, to inhibit the entirety of the HSR one would need to inhibit HSF1.
Heat Shock Factor 1
HSF1 is one of four proteins (HSF1-4) involved in stress response and development[54]. It is the factor primarily responsible for HSP gene upregulation when myeloma cells are treated with bortezomib[55]. HSF1 also drives a heat shock-independent tumorigenesis program supporting oncogenic processes such as cell-cycle regulation, signaling, metabolism, adhesion, translation, and reprogramming of neighboring stromal cells to permit a malignant phenotype[56, 57]. HSF2 has a minor role during the stress response[58]. HSF1-HSF2 heterocomplexes form under conditions of cell stress including proteasome inhibition, and HSF2 can modulate inducible HSF1-mediated gene expression[58, 59]. Avian and murine, but not human, HSF3 has been characterized and may have a HSF crosstalk-independent role in activating nonclassical heat-shock genes[58, 60]. HSF4 is involved in the development of different sensory organs in cooperation with HSF1, but has no known role in the HSR[58].
HSF1 is a 57 kDA, cytoplasmic, and inactive protein under non-stress conditions. It forms an inert heterotetramer with HSP40, 70, and 90. HSP90 has been identified as HSF1's major repressor. However, there is evidence that HSF1-HSP70 interactions are also repressive[61, 62]. When activated by stress such as proteasome inhibition, the tetramer dissociates. HSP90 is a cytoplasmic chaperone that binds misfolded proteins while HSP70 and HSP40 can either act as cytoplasmic chaperones or remain associated with HSF1[63, 64]. Upon dissociation, HSF1 trimerizes and translocates into the nucleus. However, there are conflicting views regarding which of these steps occurs first[65, 66]. After trimerization and translocation, HSF1 binds to the heat shock element (human HSE consensus sequence: nTTCnnGAAnnTTCn) in the promoter region of target HSPs. There are multiple HSE within each HSP promoter allowing for binding of multiple HSF1 trimers[67]. In addition, there are interactions between HSF1-HSE and newly recruited activating molecules such as general transcription factors, e.g., ATF1, Mediator complex, elongation factors, the chromatin remodeling complex SWI/SNF, histone modifying proteins, e.g., EP300/CBP, and RNA polymerase II (Pol II)[54, 68]. HSF1 transactivation includes continued binding to HSP70 and/or 40 complexes until shortly after HSF1 binds to HSE[63]. HSP70 and/or 40 associate with HSF1 even when it is bound to DNA, and may continue to repress HSF1 until a secondary stimulus promotes its dissociation.
HSE binding can increase HSP gene transcription by over 100-fold[69]. Transcription attenuation is mediated by a negative feedback loop. The newly translated HSPs themselves, most notably 70 and 40 and potentially 90, bind to the HSF1 transactivation domain (amino acids 440-529, near the HSF1 C-terminus)[63, 70]. Then, HSF1 detaches from the promoter region and leaves the nucleus, mediated in part by members of the 14-3-3 regulatory protein family. Preliminary evidence suggests that the HSF1 trimer is converted back to cytoplasmic monomers, but degradation also remains a possibility. Monomeric HSF1 complexes with HSP40, 70, and 90 to re-form the inactive tetramer.
Regulation of HSF1 by Post-translational Modifications
Since HSF1 is present in an inactive form, activation is mediated through PTMs (Figure 1). These include phosphorylation, sumoylation, and acetylation, in addition to 14-3-3 binding. Table 1 lists kinases and associated phosphorylation sites that have been shown or speculated to be involved with HSF1 dissociation (from the inert cytoplasmic heterotetramer), trimerization, nuclear translocation, HSE binding, transactivation, and HSR attenuation[71].
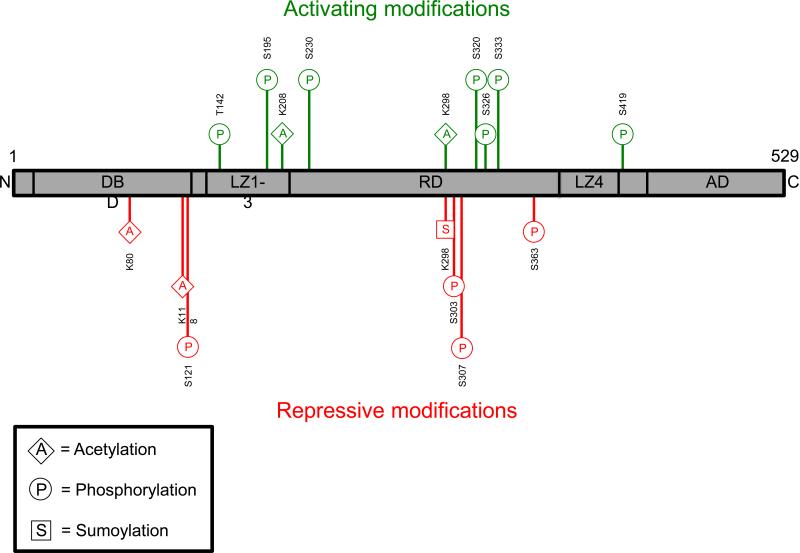
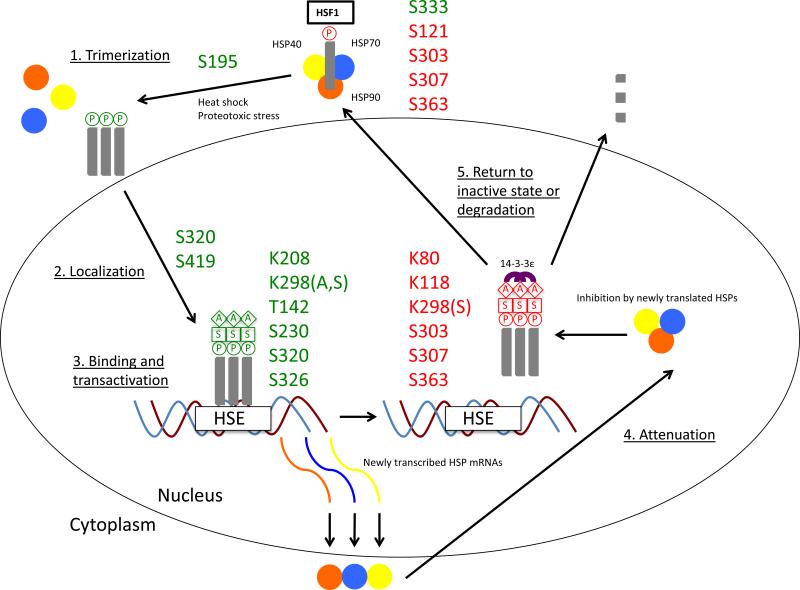
Figure 1. Heat Shock Factor 1 Post-translational Modifications and Activation Lifecycle.
1A: Heat Shock Factor 1 (HSF1) activating (green) and repressive (red) post-translational modifications (PTMs) are shown above. The bottom left box displays a PTM abbreviation key. Amino acids - K, lysine; S, serine; T, threonine. AD, activation domain; C, c-terminus; DBD, DNA-binding domain; LZ, leucine zipper domain; N, n-terminus; RD, regulatory domain.
1B: The Heat Shock Factor 1 (HSF1) activation and attenuation cycle with associated post-translational modifications (PTMs) is shown above. HSF1 forms a constitutively inactive heterotetramer with Heat Shock Protein (HSP) 40, 70, and 90. Serine (S) 121, S303, S307, and S363 phosphorylation aid in heterotetramer maintenance. (1) Upon heat shock or proteotoxic stress, the heterotetramer dissociates and S333 phosphorylation has been linked to dissociation of the repressive HSF1-HSP90 interaction. HSF1 trimerizes and translocates to the nucleus, though which occurs first has not yet been resolved. Here we show trimerization occurring first. S195 phosphorylation occurs concurrently with trimerization but this event effects transactivation downstream and not trimerization. (2) Nuclear localization is positively regulated by S320 and S419 phosphorylation. (3) After trimerization and translocation, HSF1 trimers bind to the Heat Shock Element (HSE) on HSP promoter regions. Binding is followed by transactivation. Binding is positively regulated by T142 and S320 phosphorylation and transactivation is regulated by T142, S230, S320, and S326 phosphorylation, and Lysine (K) 298 sumoylation. In addition, stabilizing acetylation events have been shown at K208 and K298. Notably, stabilizing acetylation is delayed upon transactivation and may proceed even after attenuation has begun. (4) Attenuation is initiated by newly translated HSPs, which bind to HSF1 to block HSE binding and transactivation as part of a regulatory feedback loop. K298 sumoylation and S363 phosphorylation are associated with transactivation repression. Furthermore, K80 and K118 acetylation destabilizes HSE binding. In addition, S303 and S307 phosphorylation are involved in 14-3-3ε binding to HSF1, which helps facilitate its nuclear export. (5) Upon export, HSF1 either returns to its cytoplasmic inactive state or is degraded. A, acetylation; P, phosphorylation; S, sumoylation.
Table 1.
HSF1 kinases, their targets, and functional consequences
| Kinase | Amino acid target | Functional consequences |
|---|---|---|
| AMPKα | S121 | Represses HSE binding and promotes HSF1 binding to HSP90 |
| CAMKII | S230 | Promotes transactivation |
| Casein Kinase II | T142 | Promotes HSE binding and transactivation |
| CDK1 | Meiosis regulation | |
| ERK1/2 | S307,S326,S363 | Represses HSE binding and transactivation and is required for 14-3-3ε binding (S307); promotes transactivation (S326) or may inhibit MEK phosphorylation (S326); may repress HSE binding and transactivation (S363) |
| GSK3α | S303 | Represses trimerization and is required for 14-3-3ε binding |
| JNK | TAD,S307,S320,S363 | Promotes transactivation and prolongs nuclear localization of HSF1 (TAD): represses HSE binding and transactivation (S307); promotes nuclear localization. HSE binding, transactivation, and may reverse nuclear export (S320); may repress HSE binding and transactivation (S363) |
| MAPKAP-K2 | S121 | Represses HSE binding and promotes HSF1 binding to HSP90 |
| MEK | S326 | Promotes transactivation |
| mTOR | S326 | Promotes transactivation |
| P38MAPK | Promotes HSE binding and transactivation | |
| PI3K | S326 | Promotes transactivation |
| PKAcα | S320 | Promotes nuclear localization, HSE binding, transactivation, and may reverse nuclear export |
| PKCα, θ, ζ | S333,S363 | Promotes HSF1 dissociation from HSP90 (S333 [PKCθ only]): may repress HSE binding and transactivation (S363) |
| PLK1 | S216,S419 | Mitosis regulation (S216); promotes nuclear translocation (S419) |
| Rim15 | Yeast only: promotes HSE binding when PKA activity is lowered by glucose deprivation | |
| RSK2 | Represses HSE binding | |
| Slt2/MAPK7 | Represses trimerization | |
| Snf1 | Yeast only; promotes HSE transactivation under conditions of glucose deprivation | |
| Yak1 | Yeast only; promotes HSE binding when PKA activity is lowered by glucose deprivation |
Sourbier, et al., have shown that PKCθ activates HSF1 by S333 phosphorylation in the stress responsive regulatory domain, potentially leading to dissociation of the repressive cytoplasmic HSF1-HSP90 interaction[72]. HSF1-S333A, a mutant HSF1 lacking S333 phosphorylation, associated with endogenous HSP90 to a greater extent than did HSF1-S333E, a mutant HSF1 with constitutively active S333 phosphorylation (phosphomimetic). Also, S333E was twice as efficient at activating HSF1 than S333A.
To date, no published phosphorylation events have been specifically linked to positive regulation of HSF1 trimerization. Kim et al., showed that nuclear translocation is regulated by PLK1-mediated phospho(p)Serine(S)419, but has no role in HSE binding or transactivation[73]. Also, Murshid, et al., demonstrated that shRNA against PKAcα blocked S320 phosphorylation, preventing HSF1 nuclear translocation in addition to disrupting other activation events discussed below[74].
HSE binding and transactivation are distinct activation steps but are regulated by several common phosphorylation events. pS320 is critical for hsp70.1 promoter HSE binding, transactivation, and reversal of HSF1 nuclear export[74]. CKII-mediated pT142 phosphorylation is also vital in HSE binding and transactivation[75]. Soncin et al., showed that a T142A mutant inhibits HSE-binding ready nuclear HSF1 and ultimately, HSP70B gene transcription. In addition, Holmberg et al., observed that the molar ratio between CaMKII-mediated pS230 and repressive PTM sites determines the magnitude of transactivation[76]. However, pS230 is not needed for either stress-induced HSE binding activity or the formation of nuclear stress bodies (the main site of accumulated HSF1, RNA Pol II, and other RNA-binding proteins in stressed cells).
Two related studies demonstrated that an early phosphorylation event, pS195, is critical for breakage of intramolecular interactions between leucine zipper domains (LZ) 2 and 3, an unmasking step required downstream for HSF1 transactivation[77, 78]. In addition, the role of pS326 in transactivation has been widely published on. Guettouche et al., observed in HeLa cervical carcinoma cells that a S326A mutant stimulated HSP70 expression several times worse than wild type HSF1 while having no effect on heat stress-induced DNA binding and nuclear translocation[79]. Li et al., noted that in MDA231 breast cancer cells, direct interaction of mutant p53 with activated pS326 facilitates HSF1 recruitment to HSE and stimulates transactivation under conditions of proteasome inhibition[80].
Chou et al., showed that mTOR is responsible for pS326[81]. Studies have also linked the MAPK/ERK pathway to pS326. However, the role of specific pathway members has not yet been resolved. For example, two studies have shown that MEK directly phosphorylates S326[82, 83]. However, Kim et al., concluded that pS326 is catalyzed by ERK1/2[84].
Sumoylation also positively regulates HSF1 activity. Hong et al., observed K298-dependent HSF1 co-localization with SUMO-1 in nuclear stress bodies[85]. K298 mutation resulted in a significant decrease in stress-induced transactivation in vivo. pS303 has been shown to stimulate K298 sumoylation by causing a conformation change that relieves the inhibitory effect of HSF1's lone C-terminal leucine zipper (LZ4)[86].
Interestingly, Raychaudhuri et al., showed that K298 is acetylated during the stress response in addition to K208. Catalyzed by the acetyltransferase EP300, K298 and K208 stabilize and prevent degradation of the HSE-bound HSF1 trimer[87]. EP300 maintains HSF1 stability in a phosphorylation-independent manner[87]. Ten potentially phosphorylated serines were replaced with alanines, yet HSF1 remained acetylation competent. Notably, HSF1 acetylation kinetics do not match those of transactivation[88]. Stabilizing acetylation is delayed upon onset of HSF1 transactivation and persists when HSF1 activity and DNA binding have attenuated.
PTMs also mediate negative regulation. HSF1 is maintained in an inactive heterotetramer by constitutive phosphorylation at S121, S303, S307, and potentially S363. Liu et al., showed that the linker region enclosing pS121 might be a negative regulator of the monomer to trimer transition[89]. Wang et al., identified MAPKAP-K2 (MKII) as the pS121-specific kinase and noted that pS121 promotes cytoplasmic HSP90 binding to HSF1 to help maintain its inactive state[90]. Another negative regulatory event is ERK1/2-mediated S307 phosphorylation, which has been shown to be a priming event for GSK3β-mediated phosphorylation of S303[91]. pS303 prevents HSF1 trimerization upon stress-induced activation. Thus, the priming requirement by pS307 provides a potential link between the MAPK cascade and HSF1.
However, a contrasting study by Batista-Nascimento et al., showed that when human HSF1 was expressed in yeast, Slt2 (MAPK7) phosphorylated S303 independently of both GSK3β and the pS307 priming event[92]. The authors concluded that differences in HSF1 structure between in vitro and in vivo systems may help to explain why different kinases can mediate S303 phosphorylation under different conditions. Downstream of these phosphorylation events, Wang et al., showed that both GSK3β-mediated pS303 and ERK1-mediated pS307 are prerequisites for HSF1-14-3-3ε binding[93]. HSF1-14-3-3ε binding results in cytoplasmic HSF1 sequestration, specifically of the active, DNA-binding trimers. In addition, Chu et al., demonstrated that pS363 is an early negative regulatory event that ultimately decreases HSP70B promoter activity though exactly where this phosphorylation event occurs is unclear[94]. Contrasting studies suggest S363 is phosphorylated by PKCα/ζ (in vivo and in vitro), JNK (in vitro), or ERK (in vitro)[91, 94, 95].
Post-nuclear translocation negative regulation decreases HSF1 activity through a variety of mechanisms, ultimately leading to HSF1 release from the promoter region of its target gene(s) and export back to the cytoplasm. For example, pS121 can also inhibit HSE binding[90]. In contrast to the positive regulation K298 sumoylation described above, Brunet Simioni et al., have published on a SUMO-2/3 modification at K298 that has been shown to block transactivation capacity[96]. pS303 is also a prerequisite for this modification. Large HSP27 oligomers were shown to act as an E3 factor and serve as a scaffold to strengthen the repressive interaction between the SUMO-E2-conjugating enzyme, Ubc9, and HSF1. Furthermore, Raychaudhuri et al., published on two destabilizing acetylation sites, K80 and K118[87]. K80 and K118 acetylation occurs within the HSF1 DNA binding domain (amino acids 16-123) and these events lead to inhibition of chromatin binding by HSF1. This is a crucial step in the regulated release of HSF1 trimers from DNA, ultimately leading to HSR attenuation. K118 is positively regulated by EP300 like its stabilizing counterparts K208 and K298. (K80 was shown to be EP300-independent.) K118 is negatively regulated by the deacetylase, SIRT1. SIRT1 is regulated by AROS (a deacetylase promoter) and DBC1 (a deacetylase inhibitor). Raynes et al., demonstrated that AROS and DBC1 have an impact on HSF1 acetylation status, HSF1 recruitment to the hsp70 promoter, and hsp70 transcription[97].
In addition to the roles described above, pS303 and pS307 have also been linked to accelerated HSF1 nuclear export through 14-3-3ε[93]. 14-3-3ε binding influences HSF1 interaction with the nuclear export protein CRM1 and leads to enhanced nuclear export. 14-3-3β binding has also been linked to HSF1 nuclear export[98]. Ultimately, a better understanding of positive and negative regulation through HSF1 PTMs may lead to treatments that alter HSF1 activation and help increase the efficacy of PI-based MM therapy.
HSF1 Inhibition in Cancer Treatment
Targeting HSF1 could be a more effective therapeutic strategy than pursuing individual HSP inhibition. However, developing transcription factor inhibitors is difficult for many reasons. One, transcription factors bind negatively charged DNA and therefore their exposed regions are largely positive. This requires that any inhibitor must be negatively charged, but charged molecules cannot freely diffuse across the cell membrane. Also, the DNA-protein interface is large and developing effective small molecule inhibitors is difficult. To cover the entirety of their binding pockets, a large molecule may have to be developed. Bioavailability may become a concern and promiscuous binding to other targets could cause side effects. Finally, screens for transcription factor inhibitors are less straightforward than those for kinase inhibitors, which are reliant on easier to detect processes such as ATP hydrolysis or phosphate transfer to a substrate. Despite these complexities, multiple HSF1 inhibitor screens have been performed and their various methods are described below.
Whitesell and Lindquist detailed drug-like inhibitors of the HSF1-regulated HSR and concluded that all HSP induction inhibitors suffer from low potency and/or poor specificity[99]. At the time of that publication, those inhibitors included quercetin and its prodrug QC12, NZ28 and its structural analog emunin, KNK437, stresgenin B, and triptolide. Table 2 is an updated HSF1 inhibitor listing and Figure 2 is an illustration of published inhibitor mechanisms. NZ28/emunin and triptolide will be discussed in detail below along with recently published inhibitors, cantharidin, 2,4-bis(4-hydroxybenzyl)phenol, KRIBB11, and Rohinitib (RHT).
Table 2.
HSF1 inhibitors
| Compound | Class |
|---|---|
| 2,4-bis(4-hydroxybenzyl)phenol | Benzyl derivative |
| Cantharidin | Terpenoid |
| Emunin | Emetine derivative |
| KNK437 | Benzylidene lactam |
| KRIBB11 | Diaminopyrimidine |
| NZ28 | Emetine derivative |
| QC12 | Quercetin prodrug |
| Quercetin | Flavonoid |
| Rohinitib | Flavagline derivative |
| Stresgenin B | Streptomyces fermentation product |
| Triptolide | Diterpene triexpoxide |
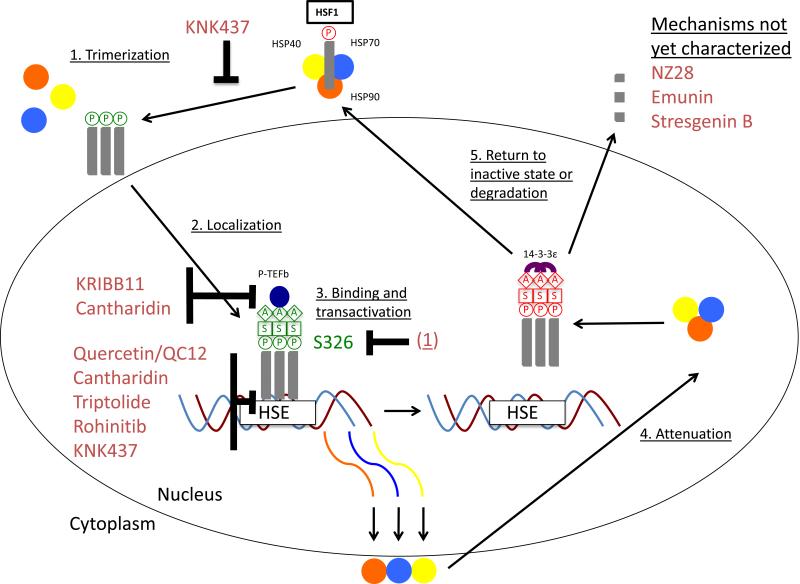
Figure 2. Inhibitors of the Heat Shock Factor 1-dependent Heat Shock Response.
Inhibitors of the Heat Shock Factor 1 (HSF1)-dependent heat shock response are shown above. The mechanism for KRIBB11, Cantharidin, Quercetin/QC12 [a Quercetin prodrug], Triptolide, Rohinitib, and 2,4-Bis(4-hydroxybenzyl)phenol (referred to as (1) above) has been published, while the mechanism for KNK437 has been speculated about. However, the mechanism for NZ28, Emunin, and Stresgenin B remains uncharacterized. (1) KNK437 may repress HSF1 trimerization though no studies have confirmed this hypothesis. (2) To date, no inhibitors have been shown to effect nuclear localization. (3) KRIBB11 and Cantharidin inhibit Positive Transcription Elongation Factor b (P-TEFb) recruitment to HSP promoters. Quercetin/QC12, Cantharidin, and Rohinitib inhibit HSE binding while KNK437 may also inhibit HSE binding. Triptolide inhibits transactivation, and though not shown here, has also been found to decrease HSF1 protein levels in multiple myeloma cell lines.) (1) induces Serine 326 dephosphorylation, leading to a decrease in HSF1 stability and as a result, increased degradation. (4,5) The inhibitor mechanisms presented in (3) accelerate attenuation while no inhibitor to date has been linked to nuclear export. A, acetylation; P, phosphorylation; S, sumoylation.
NZ28/emunin was discovered as the result of a high-throughput screen for small molecules that inhibit HSP induction[100]. The first step was performing a cell-based screen for inhibitors of HSP-mediated refolding of heat-denatured luciferase followed by a counterscreen for toxicity. The second step was direct testing for HSP induction inhibition by immunoblotting against HSP70. Out of 20,000 compounds from several diversity libraries, emunin was found to sensitize PC-3 human prostate cancer cells and MM.1S to proteasome and HSP90 inhibitors without significant toxicity. However, its precise mechanism HSP translation inhibition mechanism is unknown, and may involve events downstream of HSF1, leading to significant concerns over specificity[99].
Triptolide is a diterpenoid epoxide derived from Tripterygium wilfordii, a plant long used in Chinese medicine[101]. Heimberger et al., used triptolide to take advantage of a myeloma cell's sensitivity to proteasome inhibition and subsequent reliance on the cytoprotective HSR[102]. In MM.1S and INA-6 (another human MM cell line), triptolide in combination with bortezomib synergistically induced apoptosis. While this is a promising result, concerns about the specificity of this agent exist. Triptolide interferes with NF-κB, NFAT, AP-1, and p53 activity, and inhibits global gene transcription by inducing RNA Pol II degradation and inhibiting the ATPase activity of the DNA helicase ERCC3[103]. In addition, the in vivo tumor model in mice measuring tumor burden did not extend past 11 days, which raises the question about the durability of triptolide's in vivo effects[102]. While triptolide holds promise as a MM therapeutic, its specific mechanisms must be better understood.
Yoon et al., identified KRIBB11 from a synthetic chemical library screen[104]. A heat shock-dependent luciferase reporter plasmid was used to identify HSF1 inhibitors and KRIBB11 was chosen for further testing from a ~6,230 compound chemical bank. KRIBB11 abolished heat shock-dependent HSP70 induction through HSF1 inhibition in colon carcinoma HCT-116 cells and also inhibited the growth of HCT-116 cells in a nude mouse xenograft regression model. KRIBB11 inhibited PI or HSP90 inhibitor-mediated HSP induction, indicating its potential use in combination therapy. Interestingly, while KRIBB11 does not inhibit heat shock-induced recruitment of HSF1 to the hsp70 promoter, it does inhibit P-TEFb (positive transcription elongation factor, a heterodimer of CDK9 and cyclin T) recruitment. This study was able to show by affinity chromatography and competition assays that KRIBB11 specifically inhibits HSF1. In the competition assay, HSF2, HSP90, and CDK9, common HSF1 binding partners, were not detected, thus further strengthening the argument that KRIBB11 is HSF1-specific. In a separate study, Wiita et al., combined KRIBB11 with low-dose bortezomib in MM.1S and saw an additive apoptotic effect[105]. KRIBB11 shows HSF1 specificity and will be worth monitoring as it progresses through further preclinical studies.
Kim et al., identified the blister beetle-derived compound cantharidin as an HSF1 inhibitor from a similar screen to the one used for KRIBB11[106]. Cantharidin was shown to have inhibitory effects on HSP70 and BAG3 expression in HCT-116 cells. Here, cantharidin blocked HSF1-dependent P-TEFb recruitment to the HSP70 promoter. Cantharidin demonstrated anticancer effects and an additive effect with bortezomib, but its HSF1-specificity is questionable. Cantharidin is known as a PP2A inhibitor[107]. Additionally, it has also been shown to be an activator of serine proteases in epidermal cells[108].
Another natural compound, 2,4-bis(4-hydroxybenzyl)phenol [referred to as (1) in the original publication and here as well], derived from the orchid Gastrodia elata, was identified from a screen using a luciferase reporter under the control of a HSE to find inhibitors of HSF1 activity in NCI-H460 human lung cancer cells[109]. Similar to the previously mentioned studies, data from Yoon et al., indicate that (1) can lead to HSP suppression and an increase in apoptosis. The mechanism proposed is that (1) induces degradation of HSF1 through S326 dephosphorylation. However, HSF1 knockdown with siHSF1 + (1) resulted in increased degradation compared to (1) alone, yet cell death with siHSF1 + (1) is less than that of (1) by itself. Therefore, while this study points to a specific mechanism by which its compound works, more work is needed to confirm that observation.
Santagata et al., used a 300,000+ compound chemical screen to look for HSF1 inhibitors and found that the rocaglate, rocaglamide A, was the most potent and selective hit[110]. Rocaglamide A inhibits translation initiation factor eIF4A, thus providing a link between HSF1 and protein translation flux. Rocaglate specificity for HSE reporter activity inhibition was demonstrated by stably transducing NIH3T3-HGL mouse embryonic fibroblasts with two constructs; one encoding a green fluorescent protein (GFP) driven by HSEs and the other encoding a red fluorescent protein (RFP) driven by a doxycycline-regulated control promoter. Rocaglates suppressed GFP but not RFP activity whereas triptolide, quercetin, and KNK437 (among other previously reported HSF1 inhibitors), suppressed both GFP and RFP. An analog, Rohinitib (RHT, for Rocaglate Heat Shock) was found to be more potent than rocaglamide A while retaining similar selectivity and was used for in vivo mouse studies. An M0-91 mouse acute myeloid leukemia (AML) xenograft model showed that RHT treatment resulted in significantly decreased tumor volume in addition to a dramatic reduction in HSPA8 mRNA. However, rocaglamide derivatives are known to inhibit NF-κB and therefore, RHT HSF1-specificity needs to looked at in further detail[111]. Regardless, investigation of the relationship between the ribosome, translation flux, and HSF1 will provide novel insight into targeting the biology of a cancer cell.
As noted earlier, the main difficulty of finding small molecule transcription factor inhibitors stems from the size and complexity of the DNA-protein interface. In this regard, RNA aptamer technology may prove useful. RNA aptamers are small oligonucleotides that specifically bind to targets such as small proteins[112]. RNA molecules share some common structural features with DNA, and RNA aptamers have been shown to target the DNA-binding domains of molecules such as NF-κB. Though aptamer technology is in its infancy as a therapeutic strategy, it can currently be used for drug target validation. For example, Salamanca, et al., modified iaRNAHSF1, a Drosophila RNA aptamer, to block HSE binding in HeLa cells and promote apoptosis[113].
In addition to direct HSF1 inhibition, targeting its activation by modulating PTMs is also a potential therapeutic strategy. HSF1 PTMs happen in all stages of activation and attenuation as previously described. The majority of published studies on HSF1 PTMs focus on phosphorylation events and their respective kinases. For example, the aforementioned study describing how HSP90 inhibition leads to nuclear HSF1 accumulation also showed that that accumulation was reduced by mTOR inhibition[53]. Therefore, targeting kinases that activate HSF1 could be a simpler way of modulating targeting this pathway than developing HSF1 inhibitors.
Taken together, the findings described here show that HSF1 is involved in several cancers including MM. HSF1 has drawn interest as a biomarker though there are no known translocation groups or mutations associated with its activity[57, 114, 115]. A broad variety of tumors including carcinomas of the breast, cervix, colon, lung, pancreas and prostate as well as mesenchymal tumors such as meningioma, show increased HSF1 gene copy number, protein expression, or activation compared to their normal counterparts[56, 116]. Dai et al., have shown a therapeutic window between cancer and normal cells by demonstrating that HSF1 depletion minimally impacts normal cell viability, whereas cancer cells are strongly affected by HSF1 depletion[117, 118]. HSF1 inhibitors will likely play a role in treating a diverse range of malignancies including MM because of HSF1's multifaceted role in promoting tumorigenesis[56, 57]. We anticipate that one target MM population will be those who are bortezomib-resistant. An HSF1 inhibitor could help unblock one potential bortezomib resistance mechanism and increase MM apoptosis.
Though there is a demonstrated need for an HSF1 inhibitor, the future of HSF1 drug development will depend in part on the ability for therapeutic agents to be able to effectively and specifically target HSF1. Direct HSF1 inhibition has proven to be an elusive task but the studies presented demonstrate progress. In addition to direct inhibition, new drugs could target HSF1 activation through PTM inhibition; for example, kinase or HDAC inhibition, or anti-SUMO therapies. Regardless of the mechanism, drugs should show the ability to work in tandem with current therapies such as proteasome inhibition because the majority of current induction therapy is based on combination and not single agent treatments.
Conclusions
There is no universal cure for MM but recently developed therapies such as IMiDs and PIs have dramatically increased patient survival. Bortezomib is effective in MM therapy for a variety of reasons including targeting its plasma cell biology. However, MM cells counteract bortezomib treatment by activating the heat shock response. This cytoprotective mechanism is regulated by the master transcription factor, HSF1. Developing a specific and effective HSF1 inhibitor has proven to be a challenge. While that aim is being pursued, a more practical approach is targeting HSF1 regulation. This strategy could have a dramatic impact on patient survival especially when combined with current PI-based therapies, even beyond MM. A genome-wide siRNA screen identified proteasome addiction as a vulnerability of basal-like triple-negative breast cancer (TNBC) cells[119]. MM, TNBC, and bladder cancer are three examples of malignancies whose patients could benefit from a therapeutic strategy of proteasome and HSF1 inhibition.
Acknowledgments
We thank Faith E. Davies for her thoughtful feedback.
Financial support: Support provided by P30 CA138292 and The TJ Martell Foundation. LHB is a Georgia Cancer Coalition Distinguished Cancer Scientist.
Footnotes
Disclosures:
Lonial: Consultancy - Millennium: The Takeda Oncology Company, Celgene, Novartis, Bristol-Myers Squibb, Onyx Pharmaceuticals, Janssen Pharmaceutical Companies: The Pharmaceutical Companies of Johnson & Johnson
References
- 1.Kumar SK, Rajkumar SV, Dispenzieri A, Lacy MQ, Hayman SR, Buadi FK, Zeldenrust SR, Dingli D, Russell SJ, Lust JA, Greipp PR, Kyle RA, Gertz MA. Improved survival in multiple myeloma and the impact of novel therapies. Blood. 2008;111(5):2516–20. doi: 10.1182/blood-2007-10-116129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singhal S, Mehta J, Desikan R, Ayers D, Roberson P, Eddlemon P, Munshi N, Anaissie E, Wilson C, Dhodapkar M, Zeddis J, Barlogie B. Antitumor activity of thalidomide in refractory multiple myeloma. The New England journal of medicine. 1999;341(21):1565–71. doi: 10.1056/NEJM199911183412102. [DOI] [PubMed] [Google Scholar]
- 3.Boise LH, Kaufman JL, Bahlis NJ, Lonial S, Lee KP. The Tao of myeloma. Blood. 2014;124(12):1873–9. doi: 10.1182/blood-2014-05-578732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weathington NM, Mallampalli RK. Emerging therapies targeting the ubiquitin proteasome system in cancer. J Clin Invest. 2014;124(1):6–12. doi: 10.1172/JCI71602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahasrabuddhe AA, Elenitoba-Johnson KSJ. Role of the ubiquitin proteasome system in hematologic malignancies. Immunological Reviews. 2015;263(1):224–39. doi: 10.1111/imr.12236. [DOI] [PubMed] [Google Scholar]
- 6.Hideshima T, Anderson KC. Biologic impact of proteasome inhibition in multiple myeloma cells--from the aspects of preclinical studies. Seminars in hematology. 2012;49(3):223–7. doi: 10.1053/j.seminhematol.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moreau P, Richardson PG, Cavo M, Orlowski RZ, San Miguel JF, Palumbo A, Harousseau JL. Proteasome inhibitors in multiple myeloma: 10 years later. Blood. 2012;120(5):947–59. doi: 10.1182/blood-2012-04-403733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palombella VJ, Conner EM, Fuseler JW, Destree A, Davis JM, Laroux FS, Wolf RE, Huang J, Brand S, Elliott PJ, Lazarus D, McCormack T, Parent L, Stein R, Adams J, Grisham MB. Role of the proteasome and NF-kappaB in streptococcal cell wall-induced polyarthritis. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(26):15671–6. doi: 10.1073/pnas.95.26.15671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams J, Palombella VJ, Sausville EA, Johnson J, Destree A, Lazarus DD, Maas J, Pien CS, Prakash S, Elliott PJ. Proteasome inhibitors: a novel class of potent and effective antitumor agents. Cancer research. 1999;59(11):2615–22. [PubMed] [Google Scholar]
- 10.Hideshima T, Richardson P, Chauhan D, Palombella VJ, Elliott PJ, Adams J, Anderson KC. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer research. 2001;61(7):3071–6. [PubMed] [Google Scholar]
- 11.Mitsiades N, Mitsiades CS, Poulaki V, Chauhan D, Fanourakis G, Gu X, Bailey C, Joseph M, Libermann TA, Treon SP, Munshi NC, Richardson PG, Hideshima T, Anderson KC. Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(22):14374–9. doi: 10.1073/pnas.202445099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richardson PG, Barlogie B, Berenson J, Singhal S, Jagannath S, Irwin D, Rajkumar SV, Srkalovic G, Alsina M, Alexanian R, Siegel D, Orlowski RZ, Kuter D, Limentani SA, Lee S, Hideshima T, Esseltine DL, Kauffman M, Adams J, Schenkein DP, Anderson KC. A phase 2 study of bortezomib in relapsed, refractory myeloma. The New England journal of medicine. 2003;348(26):2609–17. doi: 10.1056/NEJMoa030288. [DOI] [PubMed] [Google Scholar]
- 13.Hideshima T, Mitsiades C, Akiyama M, Hayashi T, Chauhan D, Richardson P, Schlossman R, Podar K, Munshi NC, Mitsiades N, Anderson KC. Molecular mechanisms mediating antimyeloma activity of proteasome inhibitor PS-341. Blood. 2003;101(4):1530–4. doi: 10.1182/blood-2002-08-2543. [DOI] [PubMed] [Google Scholar]
- 14.Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nature immunology. 2003;4(4):321–9. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- 15.Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(17):9946–51. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Obeng EA, Carlson LM, Gutman DM, Harrington WJ, Jr., Lee KP, Boise LH. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood. 2006;107(12):4907–16. doi: 10.1182/blood-2005-08-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meister S, Schubert U, Neubert K, Herrmann K, Burger R, Gramatzki M, Hahn S, Schreiber S, Wilhelm S, Herrmann M, Jack HM, Voll RE. Extensive immunoglobulin production sensitizes myeloma cells for proteasome inhibition. Cancer research. 2007;67(4):1783–92. doi: 10.1158/0008-5472.CAN-06-2258. [DOI] [PubMed] [Google Scholar]
- 18.Bianchi G, Oliva L, Cascio P, Pengo N, Fontana F, Cerruti F, Orsi A, Pasqualetto E, Mezghrani A, Calbi V, Palladini G, Giuliani N, Anderson KC, Sitia R, Cenci S. The proteasome load versus capacity balance determines apoptotic sensitivity of multiple myeloma cells to proteasome inhibition. Blood. 2009;113(13):3040–9. doi: 10.1182/blood-2008-08-172734. [DOI] [PubMed] [Google Scholar]
- 19.Ling SC, Lau EK, Al-Shabeeb A, Nikolic A, Catalano A, Iland H, Horvath N, Ho PJ, Harrison S, Fleming S, Joshua DE, Allen JD. Response of myeloma to the proteasome inhibitor bortezomib is correlated with the unfolded protein response regulator XBP-1. Haematologica. 2012;97(1):64–72. doi: 10.3324/haematol.2011.043331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leung-Hagesteijn C, Erdmann N, Cheung G, Keats JJ, Stewart AK, Reece D, Chung KC, Tiedemann RE. Xbp1s-Negative Tumor B Cells and Pre-Plasmablasts Mediate Therapeutic Proteasome Inhibitor Resistance in Multiple Myeloma. Cancer cell. 2013;24(3):289–304. doi: 10.1016/j.ccr.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moriya S, Komatsu S, Yamasaki K, Kawai Y, Kokuba H, Hirota A, Che X-F, Inazu M, Gotoh A, Hiramoto M, Miyazawa K. Targeting the integrated networks of aggresome formation, proteasome, and autophagy potentiates ER stress-mediated cell death in multiple myeloma cells. International journal of oncology. 2015;46(2):474–86. doi: 10.3892/ijo.2014.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selimovic D, Porzig BB, El-Khattouti A, Badura HE, Ahmad M, Ghanjati F, Santourlidis S, Haikel Y, Hassan M. Bortezomib/proteasome inhibitor triggers both apoptosis and autophagy-dependent pathways in melanoma cells. Cellular signalling. 2013;25(1):308–18. doi: 10.1016/j.cellsig.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Catley L, Weisberg E, Kiziltepe T, Tai YT, Hideshima T, Neri P, Tassone P, Atadja P, Chauhan D, Munshi NC, Anderson KC. Aggresome induction by proteasome inhibitor bortezomib and alpha-tubulin hyperacetylation by tubulin deacetylase (TDAC) inhibitor LBH589 are synergistic in myeloma cells. Blood. 2006;108(10):3441–9. doi: 10.1182/blood-2006-04-016055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kale AJ, Moore BS. The molecular mechanisms of acquired proteasome inhibitor resistance. Journal of medicinal chemistry. 2012;55(23):10317–27. doi: 10.1021/jm300434z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ri M, Iida S, Nakashima T, Miyazaki H, Mori F, Ito A, Inagaki A, Kusumoto S, Ishida T, Komatsu H, Shiotsu Y, Ueda R. Bortezomib-resistant myeloma cell lines: a role for mutated PSMB5 in preventing the accumulation of unfolded proteins and fatal ER stress. Leukemia. 2010;24(8):1506–12. doi: 10.1038/leu.2010.137. [DOI] [PubMed] [Google Scholar]
- 26.Oerlemans R, Franke NE, Assaraf YG, Cloos J, van Zantwijk I, Berkers CR, Scheffer GL, Debipersad K, Vojtekova K, Lemos C, van der Heijden JW, Ylstra B, Peters GJ, Kaspers GL, Dijkmans BAC, Scheper RJ, Jansen G. Molecular basis of bortezomib resistance: proteasome subunit β5 (PSMB5) gene mutation and overexpression of PSMB5 protein. Blood. 2008;112(6):2489–99. doi: 10.1182/blood-2007-08-104950. [DOI] [PubMed] [Google Scholar]
- 27.Kubiczkova L, Pour L, Sedlarikova L, Hajek R, Sevcikova S. Proteasome inhibitors -molecular basis and current perspectives in multiple myeloma. Journal of cellular and molecular medicine. 2014;18(6):947–61. doi: 10.1111/jcmm.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Triphase Research and Development Corporation [2014 12/16];Combination Study of Pomalidomide, Marizomib, and Dexamethasone in Relapsed or Refractory Multiple Myeloma ClinicalTrials.gov. 2014 Available from: http://clinicaltrials.gov/ct2/show/NCT02103335.
- 29.Richardson PG, Baz R, Wang M, Jakubowiak AJ, Laubach JP, Harvey RD, Talpaz M, Berg D, Liu G, Yu J, Gupta N, Di Bacco A, Hui AM, Lonial S. Phase 1 study of twice-weekly ixazomib, an oral proteasome inhibitor, in relapsed/refractory multiple myeloma patients. Blood. 2014;124(7):1038–46. doi: 10.1182/blood-2014-01-548826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar SK, Bensinger WI, Zimmerman TM, Reeder CB, Berenson JR, Berg D, Hui AM, Gupta N, Di Bacco A, Yu J, Shou Y, Niesvizky R. Phase 1 study of weekly dosing with the investigational oral proteasome inhibitor ixazomib in relapsed/refractory multiple myeloma. Blood. 2014;124(7):1047–55. doi: 10.1182/blood-2014-01-548941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofmeister C, Lonial S. Is a Cure for Myeloma on the Horizon? Online video clip. Patient Power. 2014 Dec; YouTube Web. Dec 2012. [Google Scholar]
- 32.Zhang L, Fok JH, Davies FE. Heat shock proteins in multiple myeloma. Oncotarget. 2014;5(5):1132–48. doi: 10.18632/oncotarget.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, Bruford EA, Cheetham ME, Chen B, Hightower LE. Guidelines for the nomenclature of the human heat shock proteins. Cell stress & chaperones. 2009;14(1):105–11. doi: 10.1007/s12192-008-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi W, White MC, Choi W, Guo C, Dinney C, McConkey DJ, Siefker-Radtke A. Inhibition of inducible heat shock protein-70 (hsp72) enhances bortezomib-induced cell death in human bladder cancer cells. PloS one. 2013;8(7):e69509. doi: 10.1371/journal.pone.0069509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richardson PG, Mitsiades CS, Laubach JP, Lonial S, Chanan-Khan AA, Anderson KC. Inhibition of heat shock protein 90 (HSP90) as a therapeutic strategy for the treatment of myeloma and other cancers. British journal of haematology. 2011;152(4):367–79. doi: 10.1111/j.1365-2141.2010.08360.x. [DOI] [PubMed] [Google Scholar]
- 36.Ishii T, Seike T, Nakashima T, Juliger S, Maharaj L, Soga S, Akinaga S, Cavenagh J, Joel S, Shiotsu Y. Anti-tumor activity against multiple myeloma by combination of KW-2478, an Hsp90 inhibitor, with bortezomib. Blood Cancer Journal. 2012;2:e68. doi: 10.1038/bcj.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Usmani SZ, Chiosis G. HSP90 inhibitors as therapy for multiple myeloma. Clinical lymphoma, myeloma & leukemia. 2011;11(Suppl 1):S77–81. doi: 10.1016/j.clml.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 38.Jhaveri K, Ochiana SO, Dunphy MP, Gerecitano JF, Corben AD, Peter RI, Janjigian YY, Gomes-DaGama EM, Koren J, 3rd, Modi S, Chiosis G. Heat shock protein 90 inhibitors in the treatment of cancer: current status and future directions. Expert opinion on investigational drugs. 2014;23(5):611–28. doi: 10.1517/13543784.2014.902442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nature reviews Cancer. 2005;5(10):761–72. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 40.Powers MV, Workman P. Inhibitors of the heat shock response: Biology and pharmacology. FEBS letters. 2007;581(19):3758–69. doi: 10.1016/j.febslet.2007.05.040. [DOI] [PubMed] [Google Scholar]
- 41.Schmitt E, Maingret L, Puig PE, Rerole AL, Ghiringhelli F, Hammann A, Solary E, Kroemer G, Garrido C. Heat shock protein 70 neutralization exerts potent antitumor effects in animal models of colon cancer and melanoma. Cancer research. 2006;66(8):4191–7. doi: 10.1158/0008-5472.CAN-05-3778. [DOI] [PubMed] [Google Scholar]
- 42.Braunstein MJ, Scott SS, Scott CM, Behrman S, Walter P, Wipf P, Coplan JD, Chrico W, Joseph D, Brodsky JL, Batuman O. Antimyeloma Effects of the Heat Shock Protein 70 Molecular Chaperone Inhibitor MAL3-101. Journal of oncology. 2011;2011:232037. doi: 10.1155/2011/232037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davenport EL, Zeisig A, Aronson LI, Moore HE, Hockley S, Gonzalez D, Smith EM, Powers MV, Sharp SY, Workman P, Morgan GJ, Davies FE. Targeting heat shock protein 72 enhances Hsp90 inhibitor-induced apoptosis in myeloma. Leukemia. 2010;24(10):1804–7. doi: 10.1038/leu.2010.168. [DOI] [PubMed] [Google Scholar]
- 44.Murphy ME. The HSP70 family and cancer. Carcinogenesis. 2013 doi: 10.1093/carcin/bgt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goloudina AR, Demidov ON, Garrido C. Inhibition of HSP70: A challenging anti-cancer strategy. Cancer letters. 2012;325(2):117–24. doi: 10.1016/j.canlet.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 46.Sauvageot CM, Weatherbee JL, Kesari S, Winters SE, Barnes J, Dellagatta J, Ramakrishna NR, Stiles CD, Kung AL, Kieran MW, Wen PY. Efficacy of the HSP90 inhibitor 17-AAG in human glioma cell lines and tumorigenic glioma stem cells. Neuro-oncology. 2009;11(2):109–21. doi: 10.1215/15228517-2008-060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Erlichman C. Tanespimycin: the opportunities and challenges of targeting heat shock protein 90. Expert opinion on investigational drugs. 2009;18(6):861–8. doi: 10.1517/13543780902953699. [DOI] [PubMed] [Google Scholar]
- 48.McCollum AK, Teneyck CJ, Sauer BM, Toft DO, Erlichman C. Up-regulation of heat shock protein 27 induces resistance to 17-allylamino-demethoxygeldanamycin through a glutathione-mediated mechanism. Cancer research. 2006;66(22):10967–75. doi: 10.1158/0008-5472.CAN-06-1629. [DOI] [PubMed] [Google Scholar]
- 49.Powers MV, Clarke PA, Workman P. Dual targeting of HSC70 and HSP72 inhibits HSP90 function and induces tumor-specific apoptosis. Cancer cell. 2008;14(3):250–62. doi: 10.1016/j.ccr.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 50.Clarke PA, Hostein I, Banerji U, Stefano FD, Maloney A, Walton M, Judson I, Workman P. Gene expression profiling of human colon cancer cells following inhibition of signal transduction by 17-allylamino-17-demethoxygeldanamycin, an inhibitor of the hsp90 molecular chaperone. Oncogene. 2000;19(36):4125–33. doi: 10.1038/sj.onc.1203753. [DOI] [PubMed] [Google Scholar]
- 51.Maloney A, Clarke PA, Naaby-Hansen S, Stein R, Koopman JO, Akpan A, Yang A, Zvelebil M, Cramer R, Stimson L, Aherne W, Banerji U, Judson I, Sharp S, Powers M, deBilly E, Salmons J, Walton M, Burlingame A, Waterfield M, Workman P. Gene and protein expression profiling of human ovarian cancer cells treated with the heat shock protein 90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer research. 2007;67(7):3239–53. doi: 10.1158/0008-5472.CAN-06-2968. [DOI] [PubMed] [Google Scholar]
- 52.Yasui H, Hideshima T, Ikeda H, Jin J, Ocio EM, Kiziltepe T, Okawa Y, Vallet S, Podar K, Ishitsuka K, Richardson PG, Pargellis C, Moss N, Raje N, Anderson KC. BIRB 796 enhances cytotoxicity triggered by bortezomib, heat shock protein (Hsp) 90 inhibitor, and dexamethasone via inhibition of p38 mitogen-activated protein kinase/Hsp27 pathway in multiple myeloma cell lines and inhibits paracrine tumour growth. British journal of haematology. 2007;136(3):414–23. doi: 10.1111/j.1365-2141.2006.06443.x. [DOI] [PubMed] [Google Scholar]
- 53.Acquaviva J, He S, Sang J, Smith DL, Sequeira M, Zhang C, Bates RC, Proia DA. mTOR inhibition potentiates HSP90 inhibitor activity via cessation of HSP synthesis. Molecular cancer research : MCR. 2014;12(5):703–13. doi: 10.1158/1541-7786.MCR-13-0605. [DOI] [PubMed] [Google Scholar]
- 54.Vihervaara A, Sistonen L. HSF1 at a glance. Journal of cell science. 2014;127(Pt 2):261–6. doi: 10.1242/jcs.132605. [DOI] [PubMed] [Google Scholar]
- 55.Anckar J, Sistonen L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annual review of biochemistry. 2011;80:1089–115. doi: 10.1146/annurev-biochem-060809-095203. [DOI] [PubMed] [Google Scholar]
- 56.Mendillo ML, Santagata S, Koeva M, Bell GW, Hu R, Tamimi RM, Fraenkel E, Ince TA, Whitesell L, Lindquist S. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150(3):549–62. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scherz-Shouval R, Santagata S, Mendillo ML, Sholl LM, Ben-Aharon I, Beck AH, Dias-Santagata D, Koeva M, Stemmer SM, Whitesell L, Lindquist S. The reprogramming of tumor stroma by HSF1 is a potent enabler of malignancy. Cell. 2014;158(3):564–78. doi: 10.1016/j.cell.2014.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Åkerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11(8):545–55. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rossi A, Riccio A, Coccia M, Trotta E, La Frazia S, Santoro MG. The proteasome inhibitor bortezomib is a potent inducer of zinc finger AN1-type domain 2a gene expression: role of heat shock factor 1 (HSF1)-heat shock factor 2 (HSF2) heterocomplexes. The Journal of biological chemistry. 2014;289(18):12705–15. doi: 10.1074/jbc.M113.513242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Inouye S, Katsuki K, Izu H, Fujimoto M, Sugahara K, Yamada S, Shinkai Y, Oka Y, Katoh Y, Nakai A. Activation of heat shock genes is not necessary for protection by heat shock transcription factor 1 against cell death due to a single exposure to high temperatures. Molecular and cellular biology. 2003;23(16):5882–95. doi: 10.1128/MCB.23.16.5882-5895.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94(4):471–80. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
- 62.Ali A, Bharadwaj S, O'Carroll R, Ovsenek N. HSP90 interacts with and regulates the activity of heat shock factor 1 in Xenopus oocytes. Molecular and cellular biology. 1998;18(9):4949–60. doi: 10.1128/mcb.18.9.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neef DW, Jaeger AM, Thiele DJ. Heat shock transcription factor 1 as a therapeutic target in neurodegenerative diseases. Nature reviews Drug discovery. 2011;10(12):930–44. doi: 10.1038/nrd3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tomanek L, Somero GN. Interspecific-and acclimation-induced variation in levels of heat-shock proteins 70 (hsp70) and 90 (hsp90) and heat-shock transcription factor-1 (HSF1) in congeneric marine snails (genus Tegula): implications for regulation of hsp gene expression. The Journal of experimental biology. 2002;205(Pt 5):677–85. doi: 10.1242/jeb.205.5.677. [DOI] [PubMed] [Google Scholar]
- 65.Verma P, Pfister JA, Mallick S, D'Mello SR. HSF1 protects neurons through a novel trimerization-and HSP-independent mechanism. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34(5):1599–612. doi: 10.1523/JNEUROSCI.3039-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang HY, Fu JC, Lee YC, Lu PJ. Hyperthermia stress activates heat shock protein expression via propyl isomerase 1 regulation with heat shock factor 1. Molecular and cellular biology. 2013;33(24):4889–99. doi: 10.1128/MCB.00475-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Molecular and Physiological Basis of Nematode Survival. CAB International; Oxfordshire, UK: 2011. pp. 238–9. [Google Scholar]
- 68.Takii R, Fujimoto M, Tan K, Takaki E, Hayashida N, Nakato R, Shirahige K, Nakai A. ATF1 Modulates the Heat Shock Response by Regulating the Stress-Inducible Heat Shock Factor 1 Transcription Complex. Molecular and cellular biology. 2015;35(1):11–25. doi: 10.1128/MCB.00754-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gilmour DS, Lis JT. In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster. Molecular and cellular biology. 1985;5(8):2009–18. doi: 10.1128/mcb.5.8.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Taylor DM, Tradewell ML, Minotti S, Durham HD. Characterizing the role of Hsp90 in production of heat shock proteins in motor neurons reveals a suppressive effect of wild-type Hsf1. Cell stress & chaperones. 2007;12(2):151–62. doi: 10.1379/CSC-254R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pockley AG, Calderwood SK, Santoro MG. Prokaryotic and Eukaryotic Heat Shock Proteins in Infectious Disease: Dordrecht. Springer; 2009. [Google Scholar]
- 72.Sourbier C, Scroggins BT, Ratnayake R, Prince TL, Lee S, Lee M-J, Nagy PL, Lee YH, Trepel JB, Beutler JA, Linehan WM, Neckers L. Englerin A stimulates PKCθ to inhibit insulin signaling and simultaneously activate HSF1: An example of pharmacologically induced synthetic lethality. Cancer cell. 2013;23(2):228–37. doi: 10.1016/j.ccr.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim SA, Yoon JH, Lee SH, Ahn SG. Polo-like kinase 1 phosphorylates heat shock transcription factor 1 and mediates its nuclear translocation during heat stress. The Journal of biological chemistry. 2005;280(13):12653–7. doi: 10.1074/jbc.M411908200. [DOI] [PubMed] [Google Scholar]
- 74.Murshid A, Chou SD, Prince T, Zhang Y, Bharti A, Calderwood SK. Protein kinase A binds and activates heat shock factor 1. PloS one. 2010;5(11):e13830. doi: 10.1371/journal.pone.0013830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soncin F, Zhang X, Chu B, Wang X, Asea A, Ann Stevenson M, Sacks DB, Calderwood SK. Transcriptional activity and DNA binding of heat shock factor-1 involve phosphorylation on threonine 142 by CK2. Biochemical and biophysical research communications. 2003;303(2):700–6. doi: 10.1016/s0006-291x(03)00398-x. [DOI] [PubMed] [Google Scholar]
- 76.Holmberg CI, Hietakangas V, Mikhailov A, Rantanen JO, Kallio M, Meinander A, Hellman J, Morrice N, MacKintosh C, Morimoto RI, Eriksson JE, Sistonen L. Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. The EMBO journal. 2001;20(14):3800–10. doi: 10.1093/emboj/20.14.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Calderwood SK, Xie Y, Wang X, Khaleque MA, Chou SD, Murshid A, Prince T, Zhang Y. Signal Transduction Pathways Leading to Heat Shock Transcription. Signal transduction insights. 2010;2:13–24. doi: 10.4137/STI.S3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xie Y, Zhong R, Chen C, Calderwood SK. Heat shock factor 1 contains two functional domains that mediate transcriptional repression of the c-fos and c-fms genes. The Journal of biological chemistry. 2003;278(7):4687–98. doi: 10.1074/jbc.M210189200. [DOI] [PubMed] [Google Scholar]
- 79.Guettouche T, Boellmann F, Lane WS, Voellmy R. Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC biochemistry. 2005;6:4. doi: 10.1186/1471-2091-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li D, Yallowitz A, Ozog L, Marchenko N. A gain-of-function mutant p53-HSF1 feed forward circuit governs adaptation of cancer cells to proteotoxic stress. Cell death & disease. 2014;5:e1194. doi: 10.1038/cddis.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chou SD, Prince T, Gong J, Calderwood SK. mTOR is essential for the proteotoxic stress response, HSF1 activation and heat shock protein synthesis. PloS one. 2012;7(6):e39679. doi: 10.1371/journal.pone.0039679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tang Z, Dai S, He Y, Doty Rosalinda A, Shultz Leonard D, Sampson Stephen B, Dai C. MEK Guards Proteome Stability and Inhibits Tumor-Suppressive Amyloidogenesis via HSF1. Cell. 2015;160(4):729–44. doi: 10.1016/j.cell.2015.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dai C, Santagata S, Tang Z, Shi J, Cao J, Kwon H, Bronson RT, Whitesell L, Lindquist S. Loss of tumor suppressor NF1 activates HSF1 to promote carcinogenesis. The Journal of Clinical Investigation. 2012;122(10):3742–54. doi: 10.1172/JCI62727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim S-Y, Lee H-J, Nam J-W, Seo E-K, Lee Y-S. Coniferyl Aldehyde Reduces Radiation Damage Through Increased Protein Stability of Heat Shock Transcriptional Factor 1 by Phosphorylation. International Journal of Radiation Oncology*Biology*Physics. 2015;91(4):807–16. doi: 10.1016/j.ijrobp.2014.11.031. [DOI] [PubMed] [Google Scholar]
- 85.Hong Y, Rogers R, Matunis MJ, Mayhew CN, Goodson ML, Park-Sarge OK, Sarge KD. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. The Journal of biological chemistry. 2001;276(43):40263–7. doi: 10.1074/jbc.M104714200. [DOI] [PubMed] [Google Scholar]
- 86.Hietakangas V, Ahlskog JK, Jakobsson AM, Hellesuo M, Sahlberg NM, Holmberg CI, Mikhailov A, Palvimo JJ, Pirkkala L, Sistonen L. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Molecular and cellular biology. 2003;23(8):2953–68. doi: 10.1128/MCB.23.8.2953-2968.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Raychaudhuri S, Loew C, Korner R, Pinkert S, Theis M, Hayer-Hartl M, Buchholz F, Hartl FU. Interplay of acetyltransferase EP300 and the proteasome system in regulating heat shock transcription factor 1. Cell. 2014;156(5):975–85. doi: 10.1016/j.cell.2014.01.055. [DOI] [PubMed] [Google Scholar]
- 88.Westerheide SD, Anckar J, Stevens SM, Jr., Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science (New York, NY) 2009;323(5917):1063–6. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu PC, Thiele DJ. Modulation of human heat shock factor trimerization by the linker domain. The Journal of biological chemistry. 1999;274(24):17219–25. doi: 10.1074/jbc.274.24.17219. [DOI] [PubMed] [Google Scholar]
- 90.Wang X, Khaleque MA, Zhao MJ, Zhong R, Gaestel M, Calderwood SK. Phosphorylation of HSF1 by MAPK-activated protein kinase 2 on serine 121, inhibits transcriptional activity and promotes HSP90 binding. The Journal of biological chemistry. 2006;281(2):782–91. doi: 10.1074/jbc.M505822200. [DOI] [PubMed] [Google Scholar]
- 91.Chu B, Soncin F, Price BD, Stevenson MA, Calderwood SK. Sequential phosphorylation by mitogen-activated protein kinase and glycogen synthase kinase 3 represses transcriptional activation by heat shock factor-1. The Journal of biological chemistry. 1996;271(48):30847–57. doi: 10.1074/jbc.271.48.30847. [DOI] [PubMed] [Google Scholar]
- 92.Batista-Nascimento L, Neef DW, Liu PC, Rodrigues-Pousada C, Thiele DJ. Deciphering human heat shock transcription factor 1 regulation via post-translational modification in yeast. PloS one. 2011;6(1):e15976. doi: 10.1371/journal.pone.0015976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang X, Grammatikakis N, Siganou A, Calderwood SK. Regulation of molecular chaperone gene transcription involves the serine phosphorylation, 14-3-3 epsilon binding, and cytoplasmic sequestration of heat shock factor 1. Molecular and cellular biology. 2003;23(17):6013–26. doi: 10.1128/MCB.23.17.6013-6026.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chu B, Zhong R, Soncin F, Stevenson MA, Calderwood SK. Transcriptional activity of heat shock factor 1 at 37 degrees C is repressed through phosphorylation on two distinct serine residues by glycogen synthase kinase 3 and protein kinases Calpha and Czeta. The Journal of biological chemistry. 1998;273(29):18640–6. doi: 10.1074/jbc.273.29.18640. [DOI] [PubMed] [Google Scholar]
- 95.Dai R, Frejtag W, He B, Zhang Y, Mivechi NF. c-Jun NH2-terminal kinase targeting and phosphorylation of heat shock factor-1 suppress its transcriptional activity. The Journal of biological chemistry. 2000;275(24):18210–8. doi: 10.1074/jbc.M000958200. [DOI] [PubMed] [Google Scholar]
- 96.Brunet Simioni M, De Thonel A, Hammann A, Joly AL, Bossis G, Fourmaux E, Bouchot A, Landry J, Piechaczyk M, Garrido C. Heat shock protein 27 is involved in SUMO-2/3 modification of heat shock factor 1 and thereby modulates the transcription factor activity. Oncogene. 2009;28(37):3332–44. doi: 10.1038/onc.2009.188. [DOI] [PubMed] [Google Scholar]
- 97.Raynes R, Pombier KM, Nguyen K, Brunquell J, Mendez JE, Westerheide SD. The SIRT1 modulators AROS and DBC1 regulate HSF1 activity and the heat shock response. PloS one. 2013;8(1):e54364. doi: 10.1371/journal.pone.0054364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fu Q, Wang J, Boerma M, Berbee M, Qiu X, Fink LM, Hauer-Jensen M. Involvement of heat shock factor 1 in statin-induced transcriptional upregulation of endothelial thrombomodulin. Circulation research. 2008;103(4):369–77. doi: 10.1161/CIRCRESAHA.108.174607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Whitesell L, Lindquist S. Inhibiting the transcription factor HSF1 as an anticancer strategy. Expert opinion on therapeutic targets. 2009;13(4):469–78. doi: 10.1517/14728220902832697. [DOI] [PubMed] [Google Scholar]
- 100.Zaarur N, Gabai VL, Porco JA, Jr., Calderwood S, Sherman MY. Targeting heat shock response to sensitize cancer cells to proteasome and Hsp90 inhibitors. Cancer research. 2006;66(3):1783–91. doi: 10.1158/0008-5472.CAN-05-3692. [DOI] [PubMed] [Google Scholar]
- 101.Kupchan SM, Court WA, Dailey RG, Jr., Gilmore CJ, Bryan RF. Triptolide and tripdiolide, novel antileukemic diterpenoid triepoxides from Tripterygium wilfordii. Journal of the American Chemical Society. 1972;94(20):7194–5. doi: 10.1021/ja00775a078. [DOI] [PubMed] [Google Scholar]
- 102.Heimberger T, Andrulis M, Riedel S, Stuhmer T, Schraud H, Beilhack A, Bumm T, Bogen B, Einsele H, Bargou RC, Chatterjee M. The heat shock transcription factor 1 as a potential new therapeutic target in multiple myeloma. British journal of haematology. 2013;160(4):465–76. doi: 10.1111/bjh.12164. [DOI] [PubMed] [Google Scholar]
- 103.Titov DV, Gilman B, He QL, Bhat S, Low WK, Dang Y, Smeaton M, Demain AL, Miller PS, Kugel JF, Goodrich JA, Liu JO. XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nature chemical biology. 2011;7(3):182–8. doi: 10.1038/nchembio.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yoon YJ, Kim JA, Shin KD, Shin DS, Han YM, Lee YJ, Lee JS, Kwon BM, Han DC. KRIBB11 inhibits HSP70 synthesis through inhibition of heat shock factor 1 function by impairing the recruitment of positive transcription elongation factor b to the hsp70 promoter. The Journal of biological chemistry. 2011;286(3):1737–47. doi: 10.1074/jbc.M110.179440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wiita AP, Ziv E, Wiita PJ, Urisman A, Julien O, Burlingame AL, Weissman JS, Wells JA. Global cellular response to chemotherapy-induced apoptosis. eLife. 2013;2:e01236. doi: 10.7554/eLife.01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim JA, Kim Y, Kwon BM, Han DC. The natural compound cantharidin induces cancer cell death through inhibition of heat shock protein 70 (HSP70) and Bcl-2-associated athanogene domain 3 (BAG3) expression by blocking heat shock factor 1 (HSF1) binding to promoters. The Journal of biological chemistry. 2013;288(40):28713–26. doi: 10.1074/jbc.M113.488346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li W, Xie L, Chen Z, Zhu Y, Sun Y, Miao Y, Xu Z, Han X. Cantharidin, a potent and selective PP2A inhibitor, induces an oxidative stress-independent growth inhibition of pancreatic cancer cells through G2/M cell-cycle arrest and apoptosis. Cancer science. 2010;101(5):1226–33. doi: 10.1111/j.1349-7006.2010.01523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dubertret L, Bertaux B, Fosse M, Touraine R. Psoriasis: a defect in the regulation of epidermal proteases, as shown by serial biopsies after cantharidin application. The British journal of dermatology. 1984;110(4):405–10. doi: 10.1111/j.1365-2133.1984.tb04654.x. [DOI] [PubMed] [Google Scholar]
- 109.Yoon T, Kang GY, Han AR, Seo EK, Lee YS. 2,4-Bis(4-hydroxybenzyl)phenol inhibits heat shock transcription factor 1 and sensitizes lung cancer cells to conventional anticancer modalities. Journal of natural products. 2014;77(5):1123–9. doi: 10.1021/np4009333. [DOI] [PubMed] [Google Scholar]
- 110.Santagata S, Mendillo ML, Tang YC, Subramanian A, Perley CC, Roche SP, Wong B, Narayan R, Kwon H, Koeva M, Amon A, Golub TR, Porco JA, Jr., Whitesell L, Lindquist S. Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science (New York, NY) 2013;341(6143):1238303. doi: 10.1126/science.1238303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Baumann B, Bohnenstengel F, Siegmund D, Wajant H, Weber C, Herr I, Debatin K-M, Proksch P, Wirth T. Rocaglamide Derivatives Are Potent Inhibitors of NF-κB Activation in T-cells. Journal of Biological Chemistry. 2002;277(47):44791–800. doi: 10.1074/jbc.M208003200. [DOI] [PubMed] [Google Scholar]
- 112.Kang KN, Lee YS. RNA aptamers: a review of recent trends and applications. Advances in biochemical engineering/biotechnology. 2013;131:153–69. doi: 10.1007/10_2012_136. [DOI] [PubMed] [Google Scholar]
- 113.Salamanca HH, Antonyak MA, Cerione RA, Shi H, Lis JT. Inhibiting heat shock factor 1 in human cancer cells with a potent RNA aptamer. PloS one. 2014;9(5):e96330. doi: 10.1371/journal.pone.0096330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.de Billy E, Clarke PA, Workman P. HSF1 in Translation. Cancer cell. 2013;24(2):147–9. doi: 10.1016/j.ccr.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 115.Villanueva MT. Microenvironment: HSF1, the troublemaker next door. Nature reviews Cancer. 2014;14(9):579. doi: 10.1038/nrc3807. [DOI] [PubMed] [Google Scholar]
- 116.Dai C, Dai S, Cao J. Proteotoxic stress of cancer: implication of the heat-shock response in oncogenesis. Journal of cellular physiology. 2012;227(8):2982–7. doi: 10.1002/jcp.24017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130(6):1005–18. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Villicana C, Cruz G, Zurita M. The basal transcription machinery as a target for cancer therapy. Cancer cell international. 2014;14(1):18. doi: 10.1186/1475-2867-14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Petrocca F, Altschuler G, Tan Shen M, Mendillo Marc L, Yan H, Jerry DJ, Kung Andrew L, Hide W, Ince Tan A, Lieberman J. A Genome-wide siRNA Screen Identifies Proteasome Addiction as a Vulnerability of Basal-like Triple-Negative Breast Cancer Cells. Cancer cell. 2013;24(2):182–96. doi: 10.1016/j.ccr.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]