Abstract
B-cell chronic lymphocytic leukemia (CLL) is the most common adult human leukemia. Although, the molecular alterations leading to CLL onset and progression are still under investigation (specifically, the interplay and exact role of oncogenes and tumor suppressors in CLL pathogenesis). MicroRNAs are small non-coding RNAs that regulate gene expression and are expressed in a tissue specific manner. Deregulation of microRNAs can alter expression levels of genes involved in the development and/or progression of tumors. In CLL, microRNAs can function as oncogenes or tumor suppressors. Here, we review the most recent findings on the role of microRNAs in the onset/progression of CLL, and how this knowledge can be used to identify new biomarkers and targets to treat this leukemia.
Keywords: CLL, microRNA, miR-15/16
Characteristics and outcomes of chronic lymphocytic leukemia
Chronic lymphocytic leukemia (CLL) is the most frequent human leukemia occurring as an aggressive or indolent disease, both characterized by the accumulation of CD5+ B lymphocytes [1]. Patients with indolent CLL usually do not require treatment for several years (or even decades) while most of aggressive CLL patients require immediate treatment. Moreover, indolent CLL can progress to the aggressive form, thus it is essential to find new markers for a more specific and early diagnosis of CLL onset and staging [2]. Several prognostic markers have been already identified such as the expression of non-mutated immunoglobulin heavy variable genes (UM-IgVH) and high level of 70 kD zeta-associated protein (ZAP70), both associated with unfavorable prognosis [3, 4]. Also, chromosomal alterations are detected in >80% of all CLL cases and can discriminate patients with different outcomes: (i) low risk patients have a normal karyotype or 13q deletion; (ii) intermediate risk patients have 11q deletion or trisomy 12; and (iii) high risk patients have 17p deletion or a complex karyotype [5]. Since many CLL cases show discordant prognostic factors, the identification of new parameters able to relate disease stage and clinical outcome is important for patient management. Recently, the presence of single polymorphisms, mutations, defects in processing, allele specific loss of transcription and epigenetic inactivation were shown to influence the expression of key microRNAs involved in CLL pathogenesis and affect the clinical course of disease [6–10]. Furthermore, recent studies suggest that the ability of malignant cells to (i) react to microenvironmental stimuli via B-cell receptor (BCR) signaling, (ii) interact with exosomes and accessory cells, and (iii) respond to the presence of viral microRNAs, can have a pivotal role in CLL onset/progression [11–13].
In this review, we discuss how microRNAs affect initiation and progression of CLLs and how these molecules can represent new diagnostic/prognostic markers and possible drugs for therapy.
Signatures of microRNAs in CLLs
MicroRNAs (miRNAs), a new class of endogenous small noncoding RNAs, have been associated with several types of cancer [14] and involved in various cellular processes, including DNA methylation, cellular growth, differentiation and apoptosis [15, 16]. Expression profiling revealed that microRNA signatures can distinguish normal B cells from malignant CLL cells and are associated with prognosis, progression, drug resistance, and BCR stimulation [11, 17, 18]. For instance, aggressive and indolent CLLs exhibit a different microRNAs profile [18] and a greater mortality rate is associated with high levels of miR-21 and miR-155 [19, 20], while miR-181b levels are decreased during CLL progression acting as a biomarker able to predict time to treatment [2]. Specific microRNA signatures can predict refractoriness to treatment, [17]. Indeed miR-148a, miR-222, and miR-21 exhibit a higher expression in fludarabine non-responder patients and lower levels of miR-34a were observed in patients resistant to therapy [21]. Finally, a signature of 39 differentially expressed miRNAs was described upon BCR activation [22]. In this study the expression of miR-155, which plays a role during T- and B-cell development [20, 23] was increased while the expression of miR-29c, miR-150, miR-181b, and miR-223 was reduced, as commonly observed in patients with shorter survival and/or time to treatment [22].
Role of specific microRNAs in CLL
MicroRNA-15a/16-1
The miR-15a/16-1 cluster was discovered in 2002 within the 13q14.3 deleted region in CLL [24] and several studies have demonstrated the central role of miR-15a/16-1 in CLL [25, 26]. In ~66% of CLL cases, miR-15a/16-1 expression is downregulated [24] and associates with the longest treatment-free period [27]. Loss of miR-15a/16-1 expression promotes mature B-cell expansion by deregulating the transition from G1 to S phase [28] and induces higher levels of the antiapoptotic proteins Bcl2 and Mcl1 [24, 29–32]. Indeed, an inverse correlation between miR-15a/16-1 and BCL2 expression levels was found in CLL, and miR-15a/16-1 downregulation in leukemic cell lines lead to an increased Bcl2 expression inhibiting apoptosis [29]. MCL1, an antiapoptotic BCL-2 family member associated with CLL cell survival and chemotherapy resistance, was also identified as a deregulated gene in CLL when comparing patients with high or low levels of miR-15a/16-1 [30].
The role of the second miR-15b/16-2 cluster located at chromosome 3q25 has been studied by Lovat et al [33]. MiR-15a is highly similar to miR-15b, and miR-16-1 is identical to miR-16-2; thus, both clusters could control a similar set of target genes and may have overlapping functions. To clarify the role of miR-15b/16-2 in vivo, knockout mouse was generated. MiR-15b/16-2 KO mice developed B-cell malignancy by age 15–18 mo with a penetrance of 60%. Mice showed enlarged spleens with abnormal B cell-derived white pulp enlargement. Flow cytometric analysis demonstrated an expanded CD19+ CD5+ population in the spleen of 40% knockout mice, a characteristic of the CLL phenotype in humans. This phenotype is comparable to that observed in the miR-15a/16-1 knockout mouse model [34] suggesting an important role of miR-15b/16-2 loss in CLL pathogenesis.
MicroRNA-29 and microRNA-181
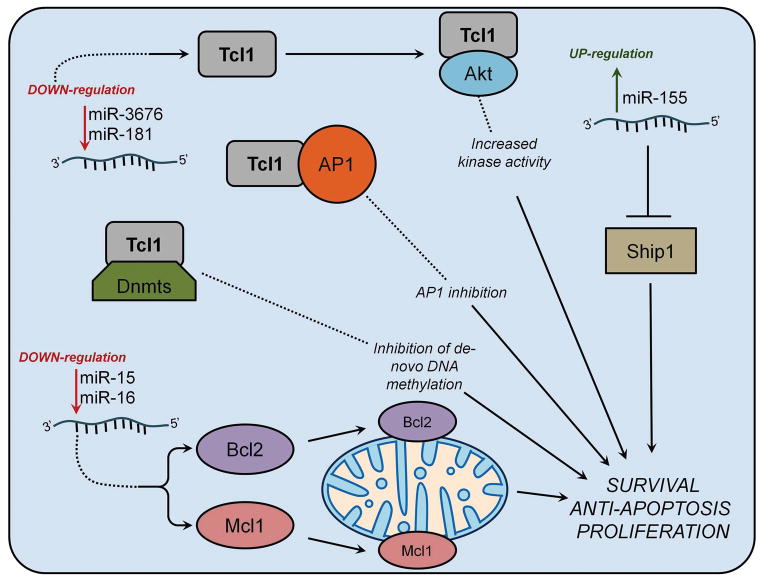
In both indolent and aggressive CLLs, miR-29 is overexpressed when compared to normal B cells, suggesting a possible role as an oncogene in CLL. On the other hand, miR-29 expression is down-regulated in aggressive versus indolent CLLs [35, 36]. To clarify the role of miR-29 in CLL, we designed a transgenic mouse model overexpressing miR-29 in mouse B-cells. These mice developed a disease resembling the indolent form of human CLL. We observed an increase in CD5+ CD19+ IgM+ B-cell populations (a hallmark of CLL) and the percentage of leukemic cells increased with age, indicating a gradual progression of indolent CLL [36]. In aggressive CLL, miR-29 down-regulation appears to be involved in Tcl1 over-expression, along with miR-181 [35]. Activation of the TCL1 oncogene is a central initiating event in the pathogenesis of aggressive CLL, and high Tcl1 expression correlates with aggressive phenotype [37]. TCL1 functions as a coactivator of the cell survival kinase AKT and inhibits de novo DNA methylthansferases Dnmt3A and Dnmt3B leading to a decrease methylation of DNA in CLL with higher Tcl1 expression [38]. TCL1 is a predicted target of miR-29 and expression levels of TCL1 and miR-29 are inversely correlated in CLL. MiR-181b is also down-regulated in aggressive CLLs and predicted to target TCL1. Pekarsky et al demonstrated that co-expression of TCL1 with miR-29 and miR-181 significantly decreased Tcl1 expression [35]. Thus, the role of miR-29 in CLLs can be explained according to its effect on Tcl1. MiR-29 up-regulation in indolent CLLs has no effect on TCL1 expression since TCL1 is not expressed in indolent CLLs and miR-29 is not sufficient to cause aggressive CLL [36], In contrast, up-regulation of Tcl1 is required for the initiation of the aggressive form of CLL and down-regulation of miR-29 in aggressive CLL (compared to the indolent form) contributes to up-regulation of Tcl1 [39]. In both indolent and aggressive CLLs, miR-181 is downregulated compared to normal B-cells and shows higher expression levels in indolent vs aggressive cases [35, 40] Furthermore, miR-181b expression diminishes during CLL progression when evaluated in sequential samples from the same patients, suggesting that this microRNA could be used as markers to track disease progression [2].
MicroRNA-34a and microRNA-34b/c
In CLL, 11q deleted region includes the miR-34b/c cluster [41], deletion of 17p contains the TP53 tumor suppressor [42], and 13q deletion causes miR15a/16-1 downregulation [30]. Thus, we investigated weather, miR-34b/c cluster, tumor protein p53, and miR-15a/16-1 cluster, share a molecular pathway that could clarify the prognostic implications of 11q, 17p, and 13q deletions in CLL [41]. The regions upstream miR-15a/16-1, miR-34b/c, and miR-34a contain several TP53 binding sites. Thus, TP53 can induce the expression of these microRNAs. On the other hand, miR-15a/16-1 target TP53 and BCL2, while miR-34 members target ZAP70 [41]. Loss of miR-15a/16-1 in 13q deleted patients, leads to higher levels of both Bcl2 and p53 [29]. In this scenario, high levels of Bcl2 cause a decrease in the number of apoptotic cells; however high levels of p53 keep the tumor burden relatively low, explaining the indolent course of 13q deleted CLL patients, and boost the transactivation of miR-34b/c leading to lower levels of ZAP70 [4]. In CLL patients with 11q deletion, since miR-15a/16-1 are not deleted, TP53 is not upregulated thus offering a lower control of apoptosis. Furthermore, TP53 transactivation of miR-34b/c is ineffective, since this microRNA is deleted [41] leading to a higher expression of ZAP70 [4]. Lastly, deletion of 17p highly correlates with unfavorable outcomes and response to treatment is often poor. Indeed, the majority of chemotherapy-resistant patients show 17p deletions and TP53 mutations. However, the lack of p53 expression alone cannot explain almost half of the refractory cases. In order to clarify this observation Zenz et al. studied miR-34a expression in chemotherapy-resistant CLL with and without 17p deletion or TP53 mutation and observed a low expression of miR-34a in all cases [21, 43]. Thus, miR-34a is associated with chemotherapy-refractory regardless of 17p deletion/TP53 mutation.
MicroRNA-155
MiR-155 is increasingly overexpressed as normal B-cells progress toward a monoclonal B-cell lymphocytosis and to CLL, thus it represents a biomarker for the risk of progression [20, 44]. Furthermore, relative expression levels of miR-155 in plasma collected from patients before therapy were significantly lower in patients who achieved complete remission after treatment than in those who had poorer treatment responses. Hence miR-155 expression can identify CLL patients who may not respond well to therapy [44]. A transgenic mouse expressing mmu-miR-155 in B-cells was generated to study its role in B-cell development and lymphomagenesis [45]. Eμ-mmu-miR-155 mice showed an initial preleukemic pre-B-cell proliferation followed by a frank B-cell malignancy, indicating that miR-155 is able to induce polyclonal expansion and suggesting that miR-155 is directly implicated in the initiation/progression of B-cell lymphomas [45].
MiR-155 also interferes with the BCR induced signaling pathways by modulating the expression of SHIP1 in CLL. SHIP1 encodes for Src homology-2 domain containing inositol 5-phosphatase 1, a phosphatase that may suppress BCR signaling. MiR-155 targets SHIP1 and, by reducing its expression, enhances the sensitivity of B-cells to BCR stimulation [20]. Furthermore T-cells or accessory cells of the lymphoid tissue can augment miR-155 expression by stimulating the BCR response via CD154/CD40 or BAFF/APRIL interaction. Indeed, a reduced expression of SHIP1 and enhanced responsiveness to BCR ligation was observed in vitro in CLL cells stimulated with CD154 or BAFF/APRIL. Similar effects were also observed in normal B cells, suggesting that miR-155 could play a physiological role in the regulation of B-cell response to BCR ligation [20].
MicroRNA-17/92 cluster
MiR-17/92 is a polycistronic microRNA cluster able to inhibit the expression of the tumor suppressor PTEN and the proapoptotic protein Bim [46]. This cluster is overexpressed in many lymphoid malignancies including CLL. To study the role of this microRNA cluster in lymphomagenesis, a mouse model expressing high levels of miR-17/92 in lymphocytes was generated [46]. These mice died prematurely as a consequence of a developed autoimmunity followed by a lymphoproliferative disorder characterized by lymphocytes with a high proliferation rate and decreased apoptosis. Later, a transgenic mouse overexpressing miR-17/92 specifically in B-cells was generated [47] to investigate the effect of miR-17/92 in B cells malignancies. Eighty percent of mir-17/92 transgenics developed a B-cell malignancy characterized by expansion of CD19+ B cells. Forty-four microRNAs and 680 genes were differentially expressed in malignant B-cells compared to controls. Eleven downregulated miRs were correlated with 66 upregulated target mRNAs and 8 upregulated miRs were correlated with 101 downregulated target mRNAs [47]. A very recent study showed that in aggressive UM-IgVH CLL, BCR response is achieved through upregulation of miR-17/92. Accordingly to this study, the induction of miR-17/92 in UM-IgVH CLLs is driven by BCR activation [48].
Role of SNPs and mutations in the expression of microRNAs
MicroRNA expression can be modulated by single nucleotide polymorphisms (SNPs). For instance, the regulation of miR-34a expression is subjected to the presence of a single nucleotide polymorphism in the intronic region of the promoter of ubiquitin ligase MDM2 (SNP 309) [6]. TP53 transactivates miR-34a enhancing apoptosis and cycle arrest [49–51] and miR-34a has been implicated in the CLL response to DNA damage through a p53-mediated induction [42, 43, 52]. In patients with intact p53, the presence of SNP 309 in MDM2 promoter induces down-regulation of miR-34a [6]. Indeed, SNP 309 leads to increased expression of MDM2, which binds p53 inhibiting its transactivation effects on miR-34a [6]. The presence of SNPs in pre-miRNAs and/or miRNA processing genes also contributes to predisposition for CLL [53]. Indeed, 57 out of 91 SNPs genotyped in 107 CLL patients and 350 cancer-free controls were located in miRNA processing genes (62.6%) and 34 in pre-miRNAs (37.4%) sequences. Nine of these SNPs were significantly associated with CLL risk; seven of them were located in six miRNA processing genes and two of them in pre-miRNAs. Thus, SNPs located in genes involved in miRNAs biogenesis pathway or in pre-miRNAs contribute to CLL [53].
We recently showed that that miR-3676, a potent inhibitor of TCL1 expression, is mutated in CLL [7]. DNA sequencing of 545 CLL samples and 146 samples from healthy individuals revealed that six CLL samples (~1%) contained five different mutations within the miR-3676 gene. Interestingly, two of these mutations significantly inhibited expression of miR-3676, confirming that these are loss-of-function mutations [7]. Since mature miR-3676 starts exactly at the end tRNA-Thr and ends at a transcription termination stop for RNA polymerase III, it cannot be excluded the possibility that miR-3676 represents a member of a new class of small RNAs, tRNA-derived small RNAs (tsRNAs), generated during tRNA processing [54, 55].
Role of defects in microRNA processing and allele specific transcription of microRNAs
Primary transcripts (pri-miRNAs) of miR-15a/-16/-15b are elevated and the processing intermediates (precursor miRNAs) are reduced in cells from CLL patients compared with non-malignant B-cells, indicating a block of miRNA maturation at the DROSHA processing step [8]. A defect in the post-transcriptional processing of tumor suppressor pri-miRNA transcripts is an interesting novel miRNA-related tumor escape mechanism and a possible underlying cause for miRNAs downregulation whenever levels of mature miRNAs are reduced.
Recently, Veronese et al. identified a novel allele-specific mechanism that involve RNA polymerase III (RNAPIII) for miR-15a/16-1 transcription [9]. This mechanism is independent of the DLEU2 host gene, which is typically transcribed monoallellically by RNA Polymerase II (RNAPII). Usually, one allele of miR-15a/16-1 is transcribed by RNAPII along with DLEU2 host gene, the other one by RNAPIII. In a subset of 13q14 deleted CLL patients characterized by high expression of ZAP70, miR-15a/16-1 is transcribed exclusivelly by RNAPIII. Thus, a double allele-specific transcriptional regulation of the miR-15a/16-1 locus represents a mechanisms that may distinguish at onset aggressive from indolent forms of CLL and provides a basis for the clinical heterogeneity of the CLL patients carrying 13q14 deletions [9]. Indeed, in a CLL case of monozygotic twins that differed in ZAP70 status and clinical features, transcription of primiR-15a/16-1 was driven by RPIII in the aggressive ZAP70-positive patient and by RPII in the indolent ZAP70-negative case [9].
Epigenetic regulation of microRNAs
Deregulation of epigenetic processes can modify microRNA expression, and microRNAs can be involved in epigenetic regulation of genes [56]. The histone deacetylases (HDACs) promote chromatin compaction, epigenetic gene silencing [57] and affect microRNAs expression. Indeed, miR-15a/16-1 is silenced by epigenetic mechanisms in 30%-35% of CLL samples and in samples with monoallelic 13q14 deletion HDACs repressed miR-15a/16-1 expression of the residual allele. Moreover, HDAC1-3 are over-expressed in CLL but not in normal lymphocytes [56]. The two copies of the 13q14.3 critical region replicate asynchronously, suggesting differential chromatin packaging. Monoallelic expression originates from either the maternal or paternal copy, which excludes an imprinting mechanism, and that one CpG island of the region is methylated. DNA demethylation of this CpG island and global histone hyperacetylation induced biallelic expression, whereas replication timing was not affected. Thus, differential replication timing represents an early epigenetic mark that distinguishes the two copies of 13q14.3, resulting in differential chromatin packaging and monoallelic expression [10].
NF-kB signaling is enhanced in CLL as a consequence of epigenetic inactivation of NF-kB targeting microRNAs. Baer et al. found that miR-708 strongly represses NF-kB pathway and identified an enhancer region downstream of the miR-708 promoter that displays a distinct DNA methylation status in CLL. High enhancer methylation significantly correlates with lower miR-708 expression and is predominantly found in patients with poor prognosis and shorter time to treatment [58]. MiR-9-3 was also revealed as a hypermethylated tumor suppressor miRNA in CLL, able to down-regulate the NF-κB1pathway. MiR-9-3 methylation is tumor-specific in CLL cell lines, and tumor suppressor activity of miR-9-3 was demonstrated in CLL [59]. Indeed, restoration of miR-9-3 in I83-E95 cells (cell line harboring complete methylation of miR-9-3) led to reduced cellular proliferation and enhanced apoptosis together with downregulation of NF-kB. Moreover, miR-9-3 methylation was associated with advanced disease stage. Therefore, miR-708 and miR-9-3 methylation may account for constitutive upregulation of NF-kB signaling pathway in CLL [58, 59]. Lastly, the miR-34b/c cluster located within the commonly deleted region at 11q is also epigenetically regulated [60]. The expression and methylation status of these miRs was investigated in CLLs with or without 11q-deletion. The MiR-34b/c promoter was found aberrantly hypermethylated in 48% of CLL cases and miR-34b/c expression was inversely correlated to DNA methylation. Furthermore, increased miR-34b/c methylation inversely correlated with the presence of 11q-deletion, indicating that methylation and del(11q) independently silence these miRs [60].
Microenvironment, exosomes and viral microRNAs
MicroRNAs can interact with BCR signaling and microenvironment. BCR stimulation can alter the expression of certain microRNAs, contributing to B cell proliferation and/or apoptosis, and certain microRNAs can influence BCR signaling and immunoglobulin production [20, 23]. A signature of 39 differentially expressed miRNAs was found upon BCR stimulation [11] and BCR activation lead to reduced levels of miR-29c, miR-181b, or miR-223, frequently observed in patients with shorter survival and/or time to treatment [22, 61]. MiR-155 is instead upregulated in response to BCR ligation and plays a role in T- and B-cell development [20, 23].
BCR activation in CLL also induces the secretion of exosomes, whereas inhibition of BCR reduces CLL exosome release. Many reports showed that miRNAs are released by donor cells through circulating exosomes and that plasma of CLL patients presents a different amount of extracellular miRNAs when compared to healthy controls [62, 63]. Expression of miR-150 and miR-155 in exosomes was elevated in CLL-derived exosomes vs normal B cells and further increased in response to BCR activation, indicating that the BCR signaling is involved in CLL exosome secretion [63]. Furthermore, CLL-derived exosomes are incorporated by endothelial and mesenchymal stem cells contributing to a tumor-supportive microenvironment [12]. Since therapy often reduced the pool of malignant cells but does not affect plasma features, it is likely that exosomes continue to induce abnormal gene profiles. Therefore, miRNA profiling in plasma could reflect another mechanism of malignant B-cell proliferation [12]. Additionally, in CLL derived exosome, packaging of miR-202-3p results in enhanced expression a Hedgehog signaling intermediate in the parental CLL cells thus altering the transcriptome and behavior of recipient cells [64].
Lastly, several studies investigated the role of Epstein-Barr Virus (EBV) in CLL [65]. EBV is a human herpes virus associated with a subclinical latent infection of B cells in healthy individuals and a variety of B-cell lymphomas [66]. EBV infection may influence the expression of cellular miRNAs and in lymphoblastoid B-cells is able to induce miR-155 expression [67]. Furthermore, EBV encodes for two viral miRNAs: BHRF1 and BART [68]. BHRF1-1 expression levels in plasma from CLL patients are higher than in plasma of healthy donors, associated with shorter survival and correlated with tumor burden, markers, and outcome of patients with relapsed CLL, suggesting a role of this viral miRNA in CLL onset/progression. Moreover, in primary malignant B-cells, exogenous expression of BHRF1-1 lead to reduced levels of TP53, indicating that EBV viral miRNAs can target cellular genes. Lastly, the expression levels of EBV miRNAs in plasma vs B-cell from CLL patients showed a discrepancy. The exosome phenomenon is likely to explain this finding. Indeed, malignant B-cells are actively secreting exosomes containing viral miRNAs and EBV infects not only B lymphocytes, but also epithelial cells, smooth muscle cells, T- and NK cells which can release BHRF1-1 into the plasma via exosomes. Thus, latent EBV-infected cells communicate with the surrounding cells by releasing viral miRNAs embedded in exosomes [69].
Therapeutic implications
The contribution of microRNAs to CLL onset/progression suggests the possibility of developing therapeutic approaches based on miRNAs. The Eμ-TCL1 transgenic mouse model mimicking the aggressive form of human CLL was used to test miR-181b as a therapeutic agent. In vitro enforced expression of miR-181b induced apoptosis in human B-cell lines and in mouse Eμ-TCL1 leukemic splenocytes. MiR-181b affected the expression of Tcl1, Bcl2 and Mcl1, Akt and phospho-Erk1/2 indicating that miR-181b exerts a broad range of actions that affect proliferation, survival and apoptosis, and can potentially be used to reduce expansion of CLL cells. Interestingly, an anti-TCL1 siRNA was not as effective as miR-181b. Thus, the anti-leukemic effect of miR-181b can be effective also for not TCL1-driven CLLs. Furthermore, in vivo studies demonstrated that miR-181b reduces leukemic cell expansion and increases mice survival [70]. Lastly, a synergic effect of miR-181b with fludarabine was observed in human primary CLL cells [71], providing an additional evidence for a potential role of miR-181b as a therapeutic agent in CLL.
Finally, Dereani et al designed specific oligonucleotides to target endogenous miR-17 (antagomiR17). In vitro administration of antagomiR17, reduced miR-17 expression and the proliferation of CLL-like MEC-1 cells. When injected in vitro in tumors genetated by the MEC-1 cells in SCID mice, antagomiR17 reduced tumor growth and increased mice survival, providing the rationale for the use of antagomiR17 as a novel potential therapeutic tool in CLL [72].
Concluding remarks
The ability of microRNAs to modulate gene expression is essential to provide fine control of several cell processes and deregulation of microRNAs can be involved in CLL development/progression (Fig. 1). Deregulation of microRNAs can originate from chromosomal alteration, epigenetic modulation, allele selection, aberrant precursor processing, or interaction with other gene. MiR-15a/16-1 deletion, an initializing step in CLL, leads to an increase of Bcl2 expression. MiR-34 family members are involved in a fine-regulated feedback circuitry with p53 and miR-15a/16-1, suggesting bidirectional interplay between microRNAs and genes. Downregulation of miR-29 and miR-181b in aggressive CLL contributes to overexpression of Tcl1. Furthermore, study of miR-181b and miR-17/92 may provide useful insights into drug design, delivery, resistance mechanisms, and microenvironmental responses [70–72]. Given these results, we can conclude that microRNAs have a deep impact on CLL development/progression (Table 1).
Figure 1. Role of microRNAs in CLL pathogenesis.
miR-15/16 deletion results in Bcl2 and Mcl1 overexpression leading to B-cell transformation. Down-regulation of miR-181 and miR-3676 causes Tcl1 overexpression, increased Akt activity and inhibition of de novo DNA methylation. miR-155 up-regulation causes B-cell malignancies through the targeting of SHIP1.
Table 1.
MicroRNAs in CLL can function as oncogenes or tumor suppressors.
| miRNA | Function | References |
|---|---|---|
| miR-155 | Oncogenic function in CLL Activates of BCR response by targeting SHIP1 |
20, 44, 45 |
| miR-17/92 | Oncogenic function in CLL Targets tumor suppressor genes PTEN and BIM Trigged by BCR response in aggressive CLL |
46, 47, 48 |
| BHRF1-1 | Viral microRNA with oncogenic function Targets still under investigation |
68, 69 |
| miR-29 | Oncogenic function in indolent CLL (upregulated vs normal B cells) Tumor suppressor function in aggressive CLLs (down regulated vs indolent CLL) Targets TCL1 oncogene in aggressive CLL |
35, 36 |
| miR-15a/16-1 | Tumor suppressor function in CLL Targets antiapoptotic genes BCL2 and MCL1 |
24, 29–32, 41 |
| miR-15b/16-2 | Tumor suppressor function in CLL Targets still under investigation (likely same targets as miR-15a/16-1) |
33 |
| miR-181b | Tumor suppressor function in CLL Targets TCL1 oncogene Marker for CLL progression |
2, 25, 40 |
| miR-34a | Tumor suppressor function in CLL Low levels associate with chemotherapy-refractory Transactivated by p53 |
6, 21, 43 |
| miR-34b/c | Tumor suppressor function in CLL Targets ZAP70 (marker for CLL progression) Transactivated by p53 |
41, 60 |
| miR-3676 | Tumor suppressor function in CLL Targets TCL1 oncogene |
7 |
| miR-708 | Tumor suppressor in CLL Targets IKKβ (activator of NF-kB) Epigenetically inactivated in aggressive CLL patients |
58 |
| miR-9-3 | Tumor suppressor in CLL Targets NF-kB Epigenetically inactivated in aggressive CLL patients |
59 |
As discussed above, numerous studies were carried out to uncover molecular mechanisms related to microRNA expression, regulation and targeting. However the ways to practically utilize this knowledge is still under development. It remains to be seen if microRNAs from CLL cells or exosomes can be used in clinical practices to track CLL. The same applies to the usage of microRNAs as drugs, as high costs and a lack of reliable delivery systems represent significant obstacles in the development of microRNA based therapies.
Trends.
Many microRNAs show disregulated expression in CLL.
MicroRNAs function as oncogenes or tumor suppressors in CLL.
MicroRNAs can serve as markers for CLL classification and progression.
Outstanding questions.
Can microRNA signatures predict time to treatment and clinical outcomes of CLL?
Are alterations in microRNA expression initiating events in CLL pathogenesis, or they occur later during the progression of the disease?
Are other classes of small non-coding RNAs (like tsRNAs or piRNAs) also involved in CLL pathogenesis?
References
- 1.Bullrich F, Croce C. Molecular Biology of Chronic Lymphocytic Leukemia. In: Cheson Bruce., editor. Chronic Lymphocytic Leukemias. 2. Marcel Dekker, Inc; New York: 2001. pp. 9–32. Revised and Expanded. [Google Scholar]
- 2.Visone R, et al. miR-181b is a biomarker of disease progression in chronic lymphocytic leukemia. Blood. 2011;118:3072–3079. doi: 10.1182/blood-2011-01-333484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orchard JA, et al. ZAP-70 expression and prognosis in chronic lymphocytic leukaemia. Lancet. 2004;363:105–111. doi: 10.1016/S0140-6736(03)15260-9. [DOI] [PubMed] [Google Scholar]
- 4.Rassenti LZ, et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N Engl J Med. 2004;351:893–901. doi: 10.1056/NEJMoa040857. [DOI] [PubMed] [Google Scholar]
- 5.Moreno C, Montserrat E. Genetic lesions in chronic lymphocytic leukemia: what’s ready for prime time use? Haematologica. 2010;95:12–15. doi: 10.3324/haematol.2009.016873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asslaber D, et al. microRNA-34a expression correlates with MDM2 SNP309 polymorphism and treatment-free survival in chronic lymphocytic leukemia. Blood. 2010;115:4191–4197. doi: 10.1182/blood-2009-07-234823. [DOI] [PubMed] [Google Scholar]
- 7.Balatti V, et al. TCL1 targeting miR-3676 is codeleted with tumor protein p53 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:2169–2174. doi: 10.1073/pnas.1500010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allegra D, et al. Defective DROSHA processing contributes to downregulation of MiR-15/-16 in chronic lymphocytic leukemia. Leukemia. 2014;28:98–107. doi: 10.1038/leu.2013.246. [DOI] [PubMed] [Google Scholar]
- 9.Veronese A, et al. Allele-specific loss and transcription of the miR-15a/16-1 cluster in chronic lymphocytic leukemia. Leukemia. 2015;29:86–95. doi: 10.1038/leu.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mertens D, et al. Allelic silencing at the tumor-suppressor locus 13q14.3 suggests an epigenetic tumor-suppressor mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7741–7746. doi: 10.1073/pnas.0600494103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kluiver JL, Chen CZ. MicroRNAs regulate B-cell receptor signaling-induced apoptosis. Genes Immun. 2012;13:239–244. doi: 10.1038/gene.2012.1. [DOI] [PubMed] [Google Scholar]
- 12.Paggetti J, et al. Exosomes released by chronic lymphocytic leukemia cells induce the transition of stromal cells into cancer-associated fibroblasts. Blood. 2015;126:1106–1117. doi: 10.1182/blood-2014-12-618025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolcetti R, Carbone A. Epstein-Barr virus infection and chronic lymphocytic leukemia: a possible progression factor? Infect Agent Cancer. 2010;5:22. doi: 10.1186/1750-9378-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Fabbri M, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, Chen Y. New insight into the role of miRNAs in leukemia. Sci China C Life Sci. 2009;52:224–231. doi: 10.1007/s11427-009-0036-1. [DOI] [PubMed] [Google Scholar]
- 17.Ferracin M, et al. MicroRNAs involvement in fludarabine refractory chronic lymphocytic leukemia. Mol Cancer. 2010;9:123. doi: 10.1186/1476-4598-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calin GA, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossi S, et al. microRNA fingerprinting of CLL patients with chromosome 17p deletion identify a miR-21 score that stratifies early survival. Blood. 2010;116:945–952. doi: 10.1182/blood-2010-01-263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui B, et al. MicroRNA-155 influences B-cell receptor signaling and associates with aggressive disease in chronic lymphocytic leukemia. Blood. 2014;124:546–554. doi: 10.1182/blood-2014-03-559690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zenz T, et al. Detailed analysis of p53 pathway defects in fludarabine-refractory chronic lymphocytic leukemia (CLL): dissecting the contribution of 17p deletion, TP53 mutation, p53-p21 dysfunction, and miR34a in a prospective clinical trial. Blood. 2009;114:2589–2597. doi: 10.1182/blood-2009-05-224071. [DOI] [PubMed] [Google Scholar]
- 22.Li S, et al. MicroRNA expression profiling identifies activated B cell status in chronic lymphocytic leukemia cells. PLoS One. 2011;6:e16956. doi: 10.1371/journal.pone.0016956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vigorito E, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calin GA, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raveche ES, et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109:5079–5086. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Migliazza A, et al. Nucleotide sequence, transcription map, and mutation analysis of the 13q14 chromosomal region deleted in B-cell chronic lymphocytic leukemia. Blood. 2001;97:2098–2104. doi: 10.1182/blood.v97.7.2098. [DOI] [PubMed] [Google Scholar]
- 27.Dohner H, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez-Beato M, et al. Cell cycle deregulation in B-cell lymphomas. Blood. 2003;101:1220–1235. doi: 10.1182/blood-2002-07-2009. [DOI] [PubMed] [Google Scholar]
- 29.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calin GA, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci U S A. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pekarsky Y, Croce CM. Role of miR-15/16 in CLL. Cell Death Differ. 2015;22:6–11. doi: 10.1038/cdd.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nature reviews. Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 33.Lovat F, et al. miR-15b/16-2 deletion promotes B-cell malignancies. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:11636–11641. doi: 10.1073/pnas.1514954112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein U, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 35.Pekarsky Y, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 36.Santanam U, et al. Chronic lymphocytic leukemia modeled in mouse by targeted miR-29 expression. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12210–12215. doi: 10.1073/pnas.1007186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herling M, et al. TCL1 shows a regulated expression pattern in chronic lymphocytic leukemia that correlates with molecular subtypes and proliferative state. Leukemia. 2006;20:280–285. doi: 10.1038/sj.leu.2404017. [DOI] [PubMed] [Google Scholar]
- 38.Palamarchuk A, et al. Tcl1 protein functions as an inhibitor of de novo DNA methylation in B-cell chronic lymphocytic leukemia (CLL) Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2555–2560. doi: 10.1073/pnas.1200003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pekarsky Y, Croce CM. Is miR-29 an oncogene or tumor suppressor in CLL? Oncotarget. 2010;1:224–227. doi: 10.18632/oncotarget.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calin GA, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 41.Fabbri M, et al. Association of a microRNA/TP53 feedback circuitry with pathogenesis and outcome of B-cell chronic lymphocytic leukemia. JAMA : the journal of the American Medical Association. 2011;305:59–67. doi: 10.1001/jama.2010.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dijkstra MK, et al. 17p13/TP53 deletion in B-CLL patients is associated with microRNA-34a downregulation. Leukemia. 2009;23:625–627. doi: 10.1038/leu.2008.264. [DOI] [PubMed] [Google Scholar]
- 43.Zenz T, et al. miR-34a as part of the resistance network in chronic lymphocytic leukemia. Blood. 2009;113:3801–3808. doi: 10.1182/blood-2008-08-172254. [DOI] [PubMed] [Google Scholar]
- 44.Ferrajoli A, et al. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood. 2013;122:1891–1899. doi: 10.1182/blood-2013-01-478222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Costinean S, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sandhu SK, et al. B-cell malignancies in microRNA Emu-miR-17~92 transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:18208–18213. doi: 10.1073/pnas.1315365110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bomben R, et al. The miR-17 approximately 92 family regulates the response to Toll-like receptor 9 triggering of CLL cells with unmutated IGHV genes. Leukemia. 2012;26:1584–1593. doi: 10.1038/leu.2012.44. [DOI] [PubMed] [Google Scholar]
- 49.Bommer GT, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 50.Chang TC, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Molecular cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tarasov V, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 52.Mraz M, et al. miR-34a, miR-29c and miR-17-5p are downregulated in CLL patients with TP53 abnormalities. Leukemia. 2009;23:1159–1163. doi: 10.1038/leu.2008.377. [DOI] [PubMed] [Google Scholar]
- 53.Martin-Guerrero I, et al. Genetic variants in miRNA processing genes and pre-miRNAs are associated with the risk of chronic lymphocytic leukemia. PLoS One. 2015;10:e0118905. doi: 10.1371/journal.pone.0118905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haussecker D, et al. Human tRNA-derived small RNAs in the global regulation of RNA silencing. Rna. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee YS, et al. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes & development. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sampath D, et al. Histone deacetylases mediate the silencing of miR-15a, miR-16, and miR-29b in chronic lymphocytic leukemia. Blood. 2012;119:1162–1172. doi: 10.1182/blood-2011-05-351510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nature genetics. 1999;23:474–478. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- 58.Baer C, et al. Epigenetic silencing of miR-708 enhances NF-kappaB signaling in chronic lymphocytic leukemia. International journal of cancer. Journal international du cancer. 2015;137:1352–1361. doi: 10.1002/ijc.29491. [DOI] [PubMed] [Google Scholar]
- 59.Wang LQ, et al. Epigenetic inactivation of miR-9 family microRNAs in chronic lymphocytic leukemia--implications on constitutive activation of NFkappaB pathway. Mol Cancer. 2013;12:173. doi: 10.1186/1476-4598-12-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deneberg S, et al. microRNA-34b/c on chromosome 11q23 is aberrantly methylated in chronic lymphocytic leukemia. Epigenetics. 2014;9:910–917. doi: 10.4161/epi.28603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mraz M, Kipps TJ. MicroRNAs and B cell receptor signaling in chronic lymphocytic leukemia. Leukemia & lymphoma. 2013;54:1836–1839. doi: 10.3109/10428194.2013.796055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cortez MA, et al. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yeh YY, et al. Characterization of CLL exosomes reveals a distinct microRNA signature and enhanced secretion by activation of BCR signaling. Blood. 2015;125:3297–3305. doi: 10.1182/blood-2014-12-618470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Farahani M, et al. CLL Exosomes Modulate the Transcriptome and Behaviour of Recipient Stromal Cells and Are Selectively Enriched in miR-202-3p. PLoS One. 2015;10:e0141429. doi: 10.1371/journal.pone.0141429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsimberidou AM, et al. Epstein-Barr virus in patients with chronic lymphocytic leukemia: a pilot study. Leukemia & lymphoma. 2006;47:827–836. doi: 10.1080/10428190500398856. [DOI] [PubMed] [Google Scholar]
- 66.Maeda E, et al. Spectrum of Epstein-Barr virus-related diseases: a pictorial review. Jpn J Radiol. 2009;27:4–19. doi: 10.1007/s11604-008-0291-2. [DOI] [PubMed] [Google Scholar]
- 67.Linnstaedt SD, et al. Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein-Barr virus. J Virol. 2010;84:11670–11678. doi: 10.1128/JVI.01248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim do N, Lee SK. Biogenesis of Epstein-Barr virus microRNAs. Mol Cell Biochem. 2012;365:203–210. doi: 10.1007/s11010-012-1261-7. [DOI] [PubMed] [Google Scholar]
- 69.Ferrajoli A, et al. Epstein-Barr Virus MicroRNAs are Expressed in Patients with Chronic Lymphocytic Leukemia and Correlate with Overall Survival. EBioMedicine. 2015;2:572–582. doi: 10.1016/j.ebiom.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bresin A, et al. miR-181b as a therapeutic agent for chronic lymphocytic leukemia in the Emicro-TCL1 mouse model. Oncotarget. 2015;6:19807–19818. doi: 10.18632/oncotarget.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu DX, et al. miR-181a/b significantly enhances drug sensitivity in chronic lymphocytic leukemia cells via targeting multiple anti-apoptosis genes. Carcinogenesis. 2012;33:1294–1301. doi: 10.1093/carcin/bgs179. [DOI] [PubMed] [Google Scholar]
- 72.Dereani S, et al. Potential therapeutic role of antagomiR17 for the treatment of chronic lymphocytic leukemia. J Hematol Oncol. 2014;7:79. doi: 10.1186/s13045-014-0079-z. [DOI] [PMC free article] [PubMed] [Google Scholar]