Abstract
Cereal yellow dwarf virus (CYDV-RPV) causes a serious viral disease affecting small grain crops around the world. In the United States, it frequently is present in California where it causes significant yield losses, and when infections start early in development, plant death. CYDV is transmitted by aphids, and it has been a major impediment to developing malting barley in California. To identify chromosome locations associated with tolerance/resistance to CYDV, a segregating population of 184 recombinant inbred lines (RIL) from a cross of the California adapted malting barley line Butta 12 with the CYDV tolerant Madre Selva was used to construct a genetic map including 180 polymorphic markers mapping to 163 unique loci. Tolerance to CYDV was evaluated in replicated experiments where plants were challenged by aphid mediated inoculation with the isolate CYDV-RPV in a controlled environment. Quantitative trait loci (QTL) analysis revealed the presence of two major QTL for CYDV tolerance from Madre Selva on chromosomes 2H (Qcyd.MaBu-1) and 7H (Qcyd.MaBu-2), and 4 minor QTL from Butta 12 on chromosomes 3H, 4H, and 2H. This paper discusses the contribution of each QTL and their potential value to improve barley tolerance to CYDV.
INTRODUCTION
Barley and Cereal yellow dwarf viruses are ubiquitous and economically important viral pathogens of wheat, barley and oat around the world (Lister and Ranieri, 1995; Miller and Rasochova, 1997). Diseases caused by these viruses were first reported in California in 1951 following observations of yellow leaves and dwarfing of barley, wheat and oat (Oswald and Houston, 1951). Oswald and Houston named the new disease as “Yellow Dwarf” and identified the causal agent to be a virus transmitted by the prevalent aphid species present in California (Oswald and Houston, 1952). At least 25 aphid species have been reported as vectors of BYDV, with 10 species being the most common (Halbert and Voegtlin, 1995). Yellow Dwarf (YD) is caused by several distinct viruses (Rochow, 1969) found in two different genera of the family Luteoviridae. Barley yellow dwarf virus- PAV (BYDV-PAV) and Barley yellow dwarf virus- MAV (BYDV-MAV) belong to the genus Luteovirus; and Cereal yellow dwarf virus - RPV (CYDV-RPV), belongs to the genus Poleovirus. The optimum aphid vector for CYDV-RPV is Rhopalosiphum padi L. (Hemiptera: Aphididae), commonly known as bird cherry oat aphid (Rochow, 1961). The bird cherry oat aphid is also a frequent vector of BYDV (Halbert and Voegtlin, 1995). Suction traps at the University of California Davis field station (UCD, henceforth) revealed twelve different cereal aphid species, with the bird cherry oat aphid accounting for 43% of the captures (Pike et al. 1989). Griesbach et al. (1990) indicated that BYDV-PAV was the most common serotype in the Sacramento Valley of California, followed by CYDV-RPV.
Yellow Dwarf on barley results in several symptoms, including stunting of the plants due to reduced internode elongation, discoloration (yellowing), reduction in the numbers of tillers and kernels per spike, reduced kernel weight and root growth, delayed or reduced heading, and sterility (D'Arcy, 1995), resulting in significant reductions in grain yield and in severe cases, plant death. Some control of virus infection can be accomplished indirectly by controlling aphid populations with pesticides or altered planting dates, but plant resistance or tolerance is the most economically and environmentally sound approach for reducing economic losses caused by these viruses.
Whereas tolerance to BYDV-PAV has been well studied (Riedel et al., 2011) little is known about tolerance to CYDV. Sources of tolerance have been identified but no information on their inheritance is available (Capettini et al., 2002). At UCD vulnerability to CYDV has been a major impediment to the development of two-rowed malting barley varieties in California, since most of these varieties are susceptible to CYDV. The six-rowed feed barley program has a longer history of breeding at UCD and a lower incidence of CYDV.
The International Center for Agricultural Research in the Dry Areas (ICARDA), and the International Maize and Wheat Improvement Center (CIMMYT) barley breeding programs identified elite germplasm resistant to BYDV-PAV and MAV serotypes, and to CYDV-RPV-MEX (now RPS-MEX1 AF235168) (Capettini et al., 2002). The two-rowed tolerant cultivar “Madre Selva” selected at CIMMYT was added to the UCD crossing block and was used to generate segregating populations for breeding and mapping purposes.
The objective of this study was to map QTL for tolerance/resistance to CYDV-RPV in the recombinant inbreed lines (RIL) derived from the cross Madre Selva x Butta 12 and to evaluate the potential values of these QTL to improve tolerance/resistance to CYDV-RPV.
MATERIALS AND METHODS
Plant Material
A mapping population was created by crossing Butta 12 to Madre Selva. Both parental genotypes are two-rowed spring barley (Hordeum vulgare L.). Butta 12 is an UCD malting line originated from the cross of BU27 (an Oregon State University line) by a UCD selection from the F2 population of Triumph/Tyra//Arupo ‘S’*2/Abyssinian, provided by ICARDA/CIMMYT, Mexico. Madre Selva was selected from a list of lines reported as resistant to CYDV-RPV-MEX (Capettini et al., 2002) and originated from the cross Roland-BAR/EH11//Esc.11.72.83.3E.7E.5E.1E/3/Arupo*3/Abyssinian-BAR/4/Aleli. An Ethiopian landrace is probably the source of resistance in Madre Selva. Starting in the F2 generation, the population was derived by single seed descent (SSD) to near-homozygosis (F5) resulting in 184 recombinant-inbreed lines (RIL).
Linkage Map Construction
To construct a linkage map and perform a QTL analysis of tolerance to CYDV, the 184 RIL were genotyped using an Illumina VeraCode custom assay with 384 single nucleotide polymorphisms (SNP) from the Illumina GoldenGate BOPA1 and BOPA2 assays (Close et al., 2009) selected for distribution throughout the barley genome. To increase marker density in the QTL regions, polymorphic SNP were identified by genotyping the parents of the population with the Illumina 9K barley iSelect assay (Comadran et al., 2012). The population was then genotyped using KASP assays (LGC Genomics, Middlesex, UK) developed from source sequences of the selected SNP. After merging completely linked markers, 170 unique loci were assigned to seven linkage groups based on previous consensus maps (Close et al., 2009; Muñoz-Amatriain et al., 2011). Utilizing the Onemap package in the statistical software R (Margarido et al., 2007), a genetic map was constructed with genetic distances calculated with the Kosambi function. Markers were initially grouped using a logarithm of odds (LOD) score of 3, but groups known to map to the same chromosome were then linked at lower LOD scores (these regions are indicated as gaps in the map).
Virus Inoculation and Disease Assessment
The Cereal yellow dwarf virus-RPV (CYDV-RPV) and Rhopalosiphum padi L. aphids were collected from symptomatic Avena fatua L. plants at UCD. Reverse transcription polymerase chain reaction (RT-PCR), using oligonucleotide primers S2a-F 5’-TCACCTTCGGGCCGTCTCTATCAG-3’ and Yan-R 5’-TGTTGAGGAGTCTACCTATTTG-3’ (Robertson and French, 2007), and nucleotide sequence analysis were used to confirm that source plants contained only CYDV-RPV. The virus was maintained using Avena sativa L. cv. “California Red” plants in a greenhouse at ambient light and temperature maintained between 20°C – 25°C. The aphid vector, R. padi, used for inoculation was also maintained on A. sativa cv. “California Red” plants in a growth room at 16 hour light and 22°C.
CYDV-RPV inoculations of seedlings from the RIL population were done using viruliferous aphids (R. padi). Aphids were allowed a 48 hour access acquisition period on infected tissue and a 96 hour inoculation access period (IAP) to the experimental seedlings. Seven to ten days old RIL seedlings were separately caged with approximately 50 aphids per plant for the duration of the IAP. The plants were then sprayed with Safari (Valent U.S.A. Corp.) to kill the aphids, and moved to a greenhouse under ambient light at 20°C – 25°C. After symptoms developed (3 – 4 weeks post inoculation) selected plants were then tested again by RT-PCR with both S2a-F and Yan-R for CYDV-RPV (Robertson and French, 2007) coupled with primers CYDV-forward 5’-TCTTACACATAAACCCAACAN-3’ and CYDV-reverse 5’-CATTCTGGAATGCCGGATCAN-3’ for CYDV-RPS (Pallett et al., 2010) paired with additional sequence analysis to confirm that inoculated plants contained only CYDV-RPV.
Disease readings were taken 3-4 weeks after inoculation, or when symptoms developed (Fig.1), using a 0 to 5 scale (0= no symptoms, 5= yellowing over the whole plant). The yellowing started on the tip of the leaves and spread to the base. Experiments were repeated in two seasons: the first experiment was in July 2012 (Year 1) and the second in April 2013 (Year 2).
Figure 1. Variation in resistance to CYDV.
RIL from the Butta 12/Madre Selva population, three weeks post-inoculation with CYDV-RPV-viruliferous aphids. Left: tolerant (disease score = 1). Right: susceptible (disease score = 3).
QTL Analyses
QTL for tolerance to CYDV were detected using a multiple interval mapping approach in the program MultiQTL (http://www.multiqtl.com). This method allows for increased precision and power by using multiple marker intervals as covariates in the model to account for more of the phenotypic variability (Kao et al., 1999). The analysis was performed separately on the average disease score measurements for the two experiments.
Markers at the peaks of each of the QTL were used as factors in a factorial ANOVA (SAS Institute, 2011). The model included the main effects of the QTL and their first order interactions. A Levene test for homogeneity of variance and a Shapiro-Wilks test for normality were performed to test the assumptions of the ANOVA.
RESULTS AND DISCUSSION
Linkage Map
The linkage map consists of 180 markers mapping to 170 unique loci and spans a distance of 1268.8 cM, with an average marker spacing of 7.9 cM (Fig.2). Locus order and distance are in agreement with previously published maps (Close et al., 2009; Muñoz-Amatriain et al., 2011). The marker spacing on each chromosome correlates highly with the 2011 consensus map (Muñoz-Amatriain et al.), with R2 values ranging from 0.83 on chromosome 2H to 0.97 on chromosome 3H and 7H.
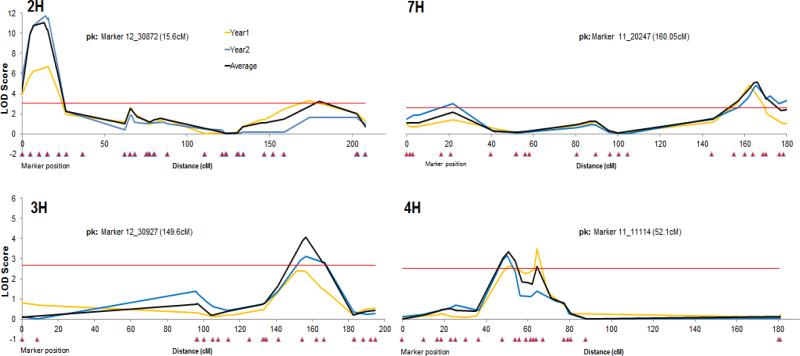
Figure 2. Linkage map and QTL for CYDV resistance.
SNP map of Butta 12/Madre Selva RIL. The QTL labeled with solid black are contributed by Madre Selva. The ovals in the ideograms represent the approximate centromere location. Markers grouped at LOD scores <3 but known to be located in the same arm are separated by a gap flanked by two diagonal lines.
There are three gaps in the map that separate groups of markers previously known to be part of the same chromosome at LOD threshold of three (chromosomes 2HL, 3HS, and 4HL, Fig.2). The gaps on chromosomes 3HS and 4HL span large regions with many of the markers from the 9K iSelect SNP assay being non-polymorphic between the parents, Butta 12 and Madre Selva. The two gaps include 61 and 43 non-polymorphic markers respectively, which represent a distance of 41.4 and 35.9 cM according to the consensus map. The probability of finding no polymorphism in such a large number of adjacent markers by chance is 8e−14 and 5.9e−10 respectively, suggesting that these two gaps are due to identity by descent between the parents. The gap on chromosome 2HL spans a region including several markers from the 9K iSelect SNP assay that are polymorphic between Butta 12 and Madre Selva (Muñoz-Amatriain et al., 2011) indicating that the gap in this chromosome segment is not identical by descent. Our limited attempts to develop KASP assays for SNP in this region failed, and since no QTL was detected with the markers flanking this region, we focused our marker development efforts in the QTL regions.
QTL Analysis
In both replications there was a wide range of disease phenotypes among the RIL, some with very severe symptoms and others with a robust tolerance that showed little to no symptoms. The distribution of CYDV disease scores did not deviate from normality according to the Shapiro-Wilks test, suggesting that multiple loci with additive effects contribute to this trait. Results of QTL analysis, using a multiple interval mapping approach indicated that tolerance to CYDV was determined by multiple loci. In addition, the analyses showed that both parents contribute alleles for increased tolerance (Fig. 3, Table 1), which explains the transgressive segregation observed in lines carrying alleles from both parents.
Figure 3. QTL for CYDV resistance consistent in the two experiments.
The upper panel includes QTL on chromosomes 2H and 7H contributed by Madre Selva. The lower panel includes the QTL on chromosomes 3H and 4H contributed by Butta 12.
Table 1.
Factorial ANOVA- including the markers at the peaks of the 6 QTL identified in this study. All interactions were non-significant and are not included in the table.
| Year 1 | Year 2 | Average | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Major QTL | Peak Marker | PEV1 | P | P | P | LOD Score | |||
| Q2Ha | 12_30872 | 20.0 | <0.0001 | *** | <0.0001 | **** | <0.0001 | **** | 11.07 |
| Q7H | 11_20247 | 9.6 | 0.0008 | ** | 0.0001 | ** | <0.0001 | **** | 5.43 |
| Q3H | 12_30927 | 5.9 | 0.0063 | ** | 0.0022 | ** | 0.0006 | *** | 4.19 |
| Q4H | 11_11114 | 6.7 | 0.0037 | ** | 0.0027 | ** | 0.0009 | *** | 3.34 |
| Minor QTL | |||||||||
| Q2Hb | 12_30259 | 3.0 | 0.0422 | * | 0.1775 | NS | 0.054 | NS | 2.54 |
| Q2Hc | 12_31506 | 3.5 | 0.0125 | * | 0.0606 | NS | 0.01 | * | 3.12 |
| R2 Major | 0.30 | 0.33 | 0.37 | ||||||
| R2 All | 0.35 | 0.37 | 0.42 | ||||||
PEV= percent of explained variation
The QTL with the largest effect on CYDV tolerance, designated Qcyd.MaBu-1, was mapped on chromosome 2HS and showed a peak at marker 12_30872. This region will be designated 2Ha hereafter, to differentiate it from two other minor QTL mapped on the same chromosome (described below). This resistant allele was contributed by Madre Selva and explains 20 % of the phenotypic variation in the full ANOVA model. The second major QTL, designated Qcyd.MaBu-2, was mapped on chromosome 7HL, with a peak at marker 11_20247. The resistant allele also derives from Madre Selva and explains 9.6 % of the variation.
The resistant alleles for additional smaller QTL mapped on chromosomes 3H, 4H, and two regions of 2H (2Hb and 2Hc) (Fig. 3, Table 1) were contributed by Butta 12. The QTL on chromosome 3HL, peak marker 12_30927, explained 5.9 % of the variation and was highly significant in both years. The QTL on chromosome 4H, peak marker 11_11114, was also significant (P ≤ 0.05) during the two seasons and explained 6.7 % of the variation. The minor QTL on chromosome 2H, labeled as 2Hb and 2Hc, were less consistent, being significant in one experiment and barely or non-significant in the other. These two loci explain 3 and 3.5 % of the variation, respectively. The analysis of variance (ANOVA) including the markers at the peaks of the 6 QTL showed no significant interactions between all possible pairs of these six QTL. Based on these results we concluded that the combined effects of these QTL are mainly additive.
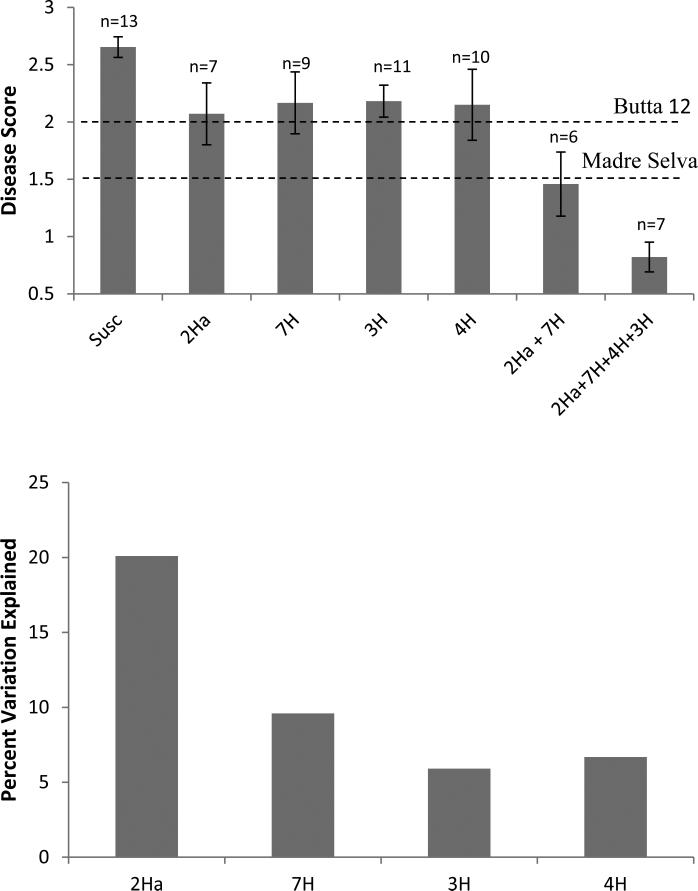
Figure 4 shows the relative tolerance to CYDV in the lines carrying only the QTL on 2H (Qcyd.MaBu-1), 7H (Qcyd.MaBu-2), 3H, and 4H; as well as the effect of the combinations of Qcyd.MaBu-1 and Qcyd.MaBu-2, and all four QTL loci. Comparison of least-square means for single QTL indicate a reduction in disease scores of 33 % on 2H (Qcyd.MaBu-1); and of 26 % on 7H (Qcyd.MaBu-2). The combination of these two major QTL reduces disease score further by 48 %. Finally, the combination of the four QTL reduces disease score 69 % relative to the wild type susceptible plants but only 30 % relative to the lines with the two major QTL.
Figure 4. Effect of individual QTL.
A) Mean disease severity in plants carrying no QTL (Susc.), single QTL (2Ha= Qcyd.MaBu-1, 7H= Qcyd.MaBu-2, 3H and 4H) individually, and combinations of the two or four major QTL. Bars indicate Standard Errors of the means and “n” indicates the number of lines in the segregating population carrying that particular allele combination. Madre Selva and Butta 12 (parental lines) average disease score marked with dash lines. B) Percent of variation explained by each QTL from a factorial ANOVA including the four major QTL (PROC VARCOMP, SAS 9.3)
The QTL for CYDV tolerance discovered in this study are distinct from the named genes for resistance to BYDV described before. Several genes have been reported as contributing tolerance/resistance to BYDV: ryd1, a recessive gene identified in the spring barley cultivar ‘Rojo’ (Suneson, 1955); Ryd2 and Ryd3, identified in Ethiopian landraces (Schaller et al., 1964; Niks et al., 2004). Ryd2 is the most widely used gene for BYDV tolerance in commercial breeding of barley (Burnett et al., 1995; Ovesna et al., 2002; Šíp et al., 2004). It has been located on chromosome 3HL (Collins et al., 1996) and it is associated with reduction in virus concentration for BYDV-PAV and MAV, but not for CYDV-RPV (Baltenberger et al., 1987; Banks et al., 1992; Ranieri et al., 1993; Capettini et al., 2002). While there are no common markers between these studies and our study, our 3HL QTL is approximately 60 cM from the centromere and therefore is unlikely to be identical to Ryd2, which is located 0.5 cM from the centromere (Collins et al., 1996). Ryd3 has been located on chromosome 6H (Niks et al., 2004). Riedel et al. (2011) reported that pyramiding these two genes in doubled haploid lines resulted in a significant reduction of virus titer, or quantitative resistance to BYDV-PAV. Ryd4 Hb, on chromosome 3HL, has been reported as contributing complete resistance to BYDV. Ryd4 Hb has been transferred to common barley from the wild relative Hordeum bulbosum L. (Johnston et al., 2009; Scholz et al., 2009; von Bothmer et al., 1995) but has not been used in barley breeding programs due to linkage drag.
In addition to the four named BYDV resistance genes there are several reports for BYDV resistance QTL (Toojinda et al., 2000; Scheurer et al., 2001; Kraakman et al., 2006). A doubled haploid population from the cross Shyri x Galena was used to map QTL for BYDV-PAV and BYDV-MAV tolerance on chromosomes 7H, 4H, and 1H (Toojinda et al., 2000). Similarly, Scheurer et al. (2001), working with a cross between the BYDV tolerant cultivar Post to Vixen (Ryd2) and Post to Nixe, found a QTL for BYDV resistance at the telomeric region of chromosome 2HL and another one in the Ryd2 region. Additional QTL were found on chromosomes 7H and 4H that together explained about 40% of the phenotypic variation. Unfortunately, the absence of common markers between the different BYDV mapping studies and our CYDV SNP map limits the ability to compare the QTL regions located in the same chromosomes.
CONCLUSIONS
To our knowledge, there are no reported QTL for tolerance to CYDV. Therefore, the identification of the chromosomal locations of two major loci for CYDV tolerance, Qcyd.MaBu-1 and Qcyd.MaBu-2, as well as other minor QTL derived from Butta 12 provide the first molecular tools to pyramid QTL for CYDV tolerance in barley. The pyramiding of the two major QTL seems sufficient to reduce the disease severity by 48%, a substantial reduction that will translate to improved grain yield and malting-quality barley production. We have initiated the construction of high-density genetic maps of the main QTL for CYDV tolerance (Qcyd.MaBu-1 and Qcyd.MaBu-2) with the long term objective of cloning these genes. A better understanding of the genes involved in the resistance to CYDV may help breeders develop new strategies to control these economically damaging viruses. In the future, it will be interesting to test if the QTL for tolerance to CYDV identified in this study, are also able to confer tolerance to some BYDV serotypes.
Abbreviations
- ANOVA
analysis of variance
- BYDV
barley yellow dwarf virus
- CYDV
cereal yellow dwarf virus
- IAP
inoculation access period
- LOD
logarithm of odds
- PEV
percent of explained variation
- QTL
quantitative trait locus or quantitative trait loci depending on context
- RIL
recombinant inbreed lines
- RT-PCR
reverse transcription polymerase chain reaction
- SSD
single seed descent
- SNP
single nucleotide polymorphism
Contributor Information
Isabel A. del Blanco, Dept. of Plant Sciences, University of California, Davis, CA 95616.
Joshua Hegarty, Dept. of Plant Sciences, University of California, Davis, CA 95616..
L. Gallagher, Dept. of Plant Sciences, University of California, Davis, CA 95616.
B. W. Falk, Dept of Plant Pathology, University of California, Davis, CA 95616.
G. Brown-Guedira, USDA-ARS, NCSU, Raleigh, NC, 27695-7620
E. Pellerin, Dept of Plant Pathology, University of California, Davis, CA 95616.
J. Dubcovsky, Dept. of Plant Sciences, University of California, Davis, CA 95616.; Howard Hughes Medical Institute, Chevy Chase, MD.
REFERENCES
- Baltenberger DE, Ohm HW, Foster JE. Reactions of oat, barley, and wheat to infection with barley yellow dwarf virus isolates. Crop Sci. 1987;27:195–198. [Google Scholar]
- Banks PM, Waterhouse PM, Larkin PJ. Pathogenicity of three RPV isolates of Barley Yellow Dwarf Virus on barley, wheat and wheat alien addition lines. Ann appl Biol. 1992;121:305–314. [Google Scholar]
- Burnett PA, Comeau A, Qualset CO. Host plant tolerance or resistance for control of barley yellow dwarf. In: D'Arcy CJ, Burnett PA, editors. Barley yellow dwarf: 40 Years of Progress. APS Press; St. Paul: 1995. pp. 321–343. [Google Scholar]
- Capettini F, Henry M, Vivar H. Breeding BYDV resistant barley with an agronomically improved background. In: Henry M, McNab M, editors. Barley Yellow Dwarf Disease: Recent advances and future strategies. CIMMYT; Mexico: 2002. pp. 117–118. [Google Scholar]
- Close TJ, Bhat PR, Lonardi S, Wu YH, Rostoks N, Ramsay L, Druka A, Stein N, Svensson JT, Wanamaker S, Bozdag S, Roose ML, Moscou MJ, Chao SAM, Varshney RK, Szucs P, Sato K, Hayes PM, Matthews DE, Kleinhofs A, Muehlbauer GJ, DeYoung J, Marshall DF, Madishetty K, Fenton RD, Condamine P, Graner A, Waugh R. Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics. 2009;10:582. doi: 10.1186/1471-2164-10-582. doi 10.1186/1471-2164-10-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins NC, Paltridge NG, Ford CM, Symons RH. The Yd2 gene for Barley yellow dwarf virus resistance maps close to the centromere on the long arm of barley chromosome 3. Theor Appl Genet. 1996;92:858–864. doi: 10.1007/BF00221898. [DOI] [PubMed] [Google Scholar]
- Comadran J, Kilian B, Russell J, Ramsay L, Stein N, Ganal M, Shaw P, Bayer M, Thomas W, Marshall D, Hedley P, Tondelli A, Pecchioni N, Francia E, Korzun V, Walther A, Waugh R. Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat. Genet. 2012;44:1388–92. doi: 10.1038/ng.2447. [DOI] [PubMed] [Google Scholar]
- D'Arcy CJ. Symptomatology and host range of barley yellow dwarf. In: D'Arcy CJ, Burnett PA, editors. Barley yellow dwarf: Forty years of progress. APS Press; St. Paul: 1995. pp. 9–28. [Google Scholar]
- Griesbach J, Falk B, Valverde R. Incidence of BYDV in California cereals. Plant Dis. 1990;74:111–114. [Google Scholar]
- Halbert S, Voegtlin D. Biology and taxonomy of vectors of barley yellow dwarf virus. In: D'Arcy CJ, Burnett PA, editors. Barley yellow dwarf: 40 Years of Progress. APS Press; St. Paul: 1995. pp. 217–258. [Google Scholar]
- Johnston PA, Timmerman-Vaughan GM, Farnden KJF, Pickering R. Marker development and characterization of Hordeum bulbosum introgression lines: A resource for barley improvement. Theor Appl Genet. 2009;118:1429–1437. doi: 10.1007/s00122-009-0992-7. [DOI] [PubMed] [Google Scholar]
- Kao CH, Zeng ZB, Teasdale RD. Multiple interval mapping for quantitative trait loci. Genetics. 1999;152:1203–1216. doi: 10.1093/genetics/152.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraakman ATW, Martinez F, Mussiraliev B, van Eeuwijk FA, Niks RE. Linkage disequilibrium mapping of morphological, resistance, and other agronomically relevant traits in modern spring barley cultivars. Mol Breeding. 2006;17:41–58. [Google Scholar]
- Lister RM, Ranieri R. Distribution and economic importance of Barley yellow dwarf. In: D'Arcy CJ, Burnett PA, editors. Barley yellow dwarf: 40 Years of Progress. APS Press; St. Paul: 1995. pp. 59–53. [Google Scholar]
- Margarido GRA, Souza AP, Garcia AAF. OneMap: software for genetic mapping in outcrossing species. Hereditas. 2007;144:78–79. doi: 10.1111/j.2007.0018-0661.02000.x. doi: 10.1111/j.2007.0018-0661.02000.x. [DOI] [PubMed] [Google Scholar]
- Miller WA, Rasochova L. Barley yellow dwarf viruses. Annu Rev Phytopathol. 1997;35:167–190. doi: 10.1146/annurev.phyto.35.1.167. [DOI] [PubMed] [Google Scholar]
- Muñoz-Amatriain M, Moscou MJ, Bhat PR, Svensson JT, Bartoš J, Suchánková P, Šimková H, Endo TR, Fenton RD, Lonardi S, Castillo AM, Chao SM, Cistué L, Cuesta-Marcos A, Forrest KL, Hayden MJ, Hayes PM, Horsley RD, Makoto K, Moody D, Sato K, Vallés MP, Wulff BBH, Muehlbauer GJ, Doležel J, Close TJ. An Improved Consensus Linkage Map of Barley Based on Flow-Sorted Chromosomes and Single Nucleotide Polymorphism Markers. Plant Genome. 2011;4:238–249. doi: 10.3835/plantgenome2011.08.0023. [Google Scholar]
- Niks RE, Habekuss A, Bekele B, Ordon F. A novel major gene on chromosome 6H for resistance of barley against the Barley yellow dwarf virus. Theor Appl Genet. 2004;109:1536–1543. doi: 10.1007/s00122-004-1777-7. [DOI] [PubMed] [Google Scholar]
- Oswald JW, Houston BR. A new virus disease of cereals, transmissible by aphids. Plant Dis Rep. 1951;35:471–475. [Google Scholar]
- Oswald JW, Houston BR. Barley yellow dwarf, a virus disease of barley, wheat and oats readily transmitted by four species of aphids. Phytopathology. 1952;42:15. [Google Scholar]
- Ovesna J, Šíp V, Chrpova J, Vacke J, Skorpǐk M, Polakova K, Kucera L. Exploitation of detected BYDV resistance genes in barley breeding. In: Henry M, McNab M, editors. Barley Yellow Dwarf Disease: Recent advances and future strategies. CIMMYT; Mexico: 2002. pp. 78–81. [Google Scholar]
- Pallett DW, Ho T, Cooper I, Wang H. Detection of Cereal yellow dwarf virus using small interfering RNAs and enhanced infection rate with Cocksfoot streak virus in wild cocksfoot grass (Dactylis glomerata). J Virol Methods. 2010;168:223–227. doi: 10.1016/j.jviromet.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Pike KS, Allison D, Boydston L, Qualset CO, Vogt HE, Summers CG. Suction trap reveals 60 wheat aphid species, including Russian wheat aphid. Calif. Agric. 1989;43:22–24. [Google Scholar]
- Ranieri R, Lister RM, Burnett PA. Relationships between Barley yellow dwarf virus titer and symptom expression in barley. Crop Sci. 1993;33:968–973. [Google Scholar]
- Riedel C, Habekuss A, Schliephake E, Niks R, Broer I, Ordon F. Pyramiding of Ryd2 and Ryd3 conferring tolerance to a German isolate of Barley yellow dwarf virus-PAV (BYDV-PAV-ASL-1) leads to quantitative resistance against this isolate. Theor Appl Genet. 2011;123:69–76. doi: 10.1007/s00122-011-1567-y. [DOI] [PubMed] [Google Scholar]
- Robertson NL, French RC. Genetic analysis of a novel Alaska barley yellow dwarf virus in the family Luteoviridae. Arch Virol. 2007;152:369–82. doi: 10.1007/s00705-006-0846-4. [DOI] [PubMed] [Google Scholar]
- Rochow WF. Vector specificity of three strains of barley yellow dwarf virus. Phytopathology. 1961;51:578–579. [PubMed] [Google Scholar]
- Rochow WF. Biological properties of four isolates of barley yellow dwarf virus. Phytopathology. 1969;59:1580–1589. [PubMed] [Google Scholar]
- SAS Institute . SAS system for Windows release 9.3. Cary, NC.: 2011. [Google Scholar]
- Schaller CW, Qualset CO, Rutger JN. Inheritance and linkage of the Yd2 gene conditioning resistance to the Barley yellow dwarf disease in barley. Crop Sci. 1964;4:544–548. [Google Scholar]
- Scheurer KS, Friedt W, Huth W, Waugh R, Ordon F. QTL analysis of tolerance to a German strain of BYDV-PAV in barley (Hordeum vulgare L.). Theor Appl Genet. 2001;103:1074–1083. [Google Scholar]
- Scholz M, Ruge-Wehling B, Habekuss A, Schrader O, Pendinen G, Fischer K, Wehling P. Ryd4Hb: A novel resistance gene introgressed from Hordeum bulbosum into barley and conferring complete and dominant resistance to the Barley yellow dwarf virus. Theor Appl Genet. 2009;119:837–849. doi: 10.1007/s00122-009-1093-3. [DOI] [PubMed] [Google Scholar]
- Šíp V, Chrpová J, Vacke J, Ovesná J. Possibility of exploiting the Yd2 resistance to BYDV in spring barley breeding. Plant Breeding. 2004;123:24–29. [Google Scholar]
- Suneson CA. Breeding for resistance to yellow dwarf virus in barley. Agron J. 1955;47:283. [Google Scholar]
- Toojinda T, Broers LH, Chen XM, Hayes PM, Kleinhofs A, Korte J, Kudrna D, Leung H, Line RF, Powell W, Ramsay L, Vivar H, Waugh R. Mapping quantitative and qualitative disease resistance genes in a doubled haploid population of barley (Hordeum vulgare L.). Theor Appl Genet. 2000;101:580–589. [Google Scholar]
- von Bothmer R, Jacobsen N, Baden C, Jørgensen RB, Linde-Laursen I. Systematic and Ecogeographic Studies on Crop Genepools 7. International Plant Genetic Resources Institute; Rome, Italy: 1995. An ecogeographical study of the genus Hordeum. [Google Scholar]