Abstract
Antimicrobial peptides (AMPs) are short proteins with antimicrobial activity. A large portion of known AMPs originate from insects, and the number and diversity of these molecules in different species varies considerably. Insect AMPs represent a potential source of alternative antibiotics to address the limitation of current antibiotics, which has been caused by the emergence and spread of multidrug-resistant pathogens. To get more insight into AMPs, we investigated the diversity and evolution of insect AMPs by mapping their phylogenetic distribution, allowing us to predict the evolutionary origins of selected AMP families and to identify evolutionarily conserved and taxon-specific families. Furthermore, we highlight the use of the nematode Caenorhabditis elegans as a whole-animal model in high-throughput screening methods to identify AMPs with efficacy against human pathogens, including Acinetobacter baumanii and methicillin-resistant Staphylococcus aureus. We also discuss the potential medical applications of AMPs, including their use as alternatives for conventional antibiotics in ectopic therapies, their combined use with antibiotics to restore the susceptibility of multidrug-resistant pathogens, and their use as templates for the rational design of peptidomimetic drugs that overcome the disadvantages of therapeutic peptides.
The article is part of the themed issue ‘Evolutionary ecology of arthropod antimicrobial peptides’.
Keywords: antimicrobial peptides, anti-infective, drug development, evolution, immunity, insects
1. Introduction
Antimicrobial peptides (AMPs) are short immunity-related proteins that can act against bacteria, viruses, fungi or parasites. In insects, they are secreted from cells and tissues that contribute to host innate immunity such as the haemocytes or the fat body and they could be a valuable alternative to conventional antibiotics in the era of growing antimicrobial resistance [1–3]. AMPs participate in several defence-related processes, including the killing of pathogens, the ability to bind and neutralize endotoxins and to modulate the immune responses to infection [4]. The induction of AMPs in insects mediates a temporary humoral immune response characterized, for example, by enhanced AMP concentrations in the haemolymph, which is longer lasting than the initial cellular responses and which is believed to function as a back-up against persistent infections [5]. The functions of AMPs have been expanded beyond their role in defence against pathogens to include also the control of endosymbionts [6]. AMPs are ubiquitous among eukaryotes, but have been much more intensively studied in insects [7,8]. The non-ribosomally AMPs produced by bacteria and fungi are different from those contributing to the innate immunity of multicellular organisms which are ribosomally produced, and the term AMP usually refers specifically to these molecules [9]. A comprehensive list of known AMPs can be found in the Antimicrobial Peptide Database (APD) (http://aps.unmc.edu/AP).
In this work, we review the diversity of insect-derived AMPs, and evolution of insect AMPs by mapping their phylogenetic distribution. We highlight the use of the nematode Caenorhabditis elegans as a whole-animal model in high-throughput screening methods to identify AMPs with efficacy against human pathogens. We also discuss the potential medical applications of AMPs, emphasizing their roles as antimicrobials.
2. Insect antimicrobial peptides
(a). Classification of insect antimicrobial peptides
Insects produce a larger repertoire of AMPs than any other taxonomic group, and the number of individual AMPs produced by each species varies considerably. At one end of the scale, the invasive ladybird Harmonia axyridis is known to produce more than 50 AMPs [10]. At the other end, the pea aphid Acyrthosiphon pisum does not produce any known AMPs that act against bacteria [11]. The AMP repertoire is conceivably linked to the nature of the environmental threats faced by each species during evolution, i.e. those exposed to more pathogens, and more diverse pathogen species, would be expected to have evolved a broader repertoire of AMPs [12,13]. Insect AMPs display a remarkable evolutionary plasticity in terms of gain, loss and functional shifts of the coding genes. The last encompasses duplication and divergent evolution of AMPs, which can ultimately result in new functions of the resulting paralogues. This neo-functionalization enables adaptation to emerging pathogens, but also switches between immunity and non-immunity-related functions [14,15].
The increasing number of published insect genome and transcriptome datasets combined with the ability to probe haemolymph samples directly using proteomics techniques has resulted in the discovery of many new AMPs in the past few years [12]. Novel AMPs can be identified by homology to known peptides, but also by other features such as the presence of protease cleavage sites that release the mature peptide from propeptide precursors, and expression profiles that focus on immunocompetent cells and tissues such as haemocytes and the fat body [16,17].
Insect AMPs can be classified according to their structure or function. The three major structural classes are linear α-helical peptides without cysteine residues, peptides with a β-sheet globular structure stabilized by intramolecular disulfide bridges, and peptides that contain unusually high numbers of specific amino acid residues, such as proline or glycine [1,7,9]. Where secondary structures and disulfide bridges are present, these elements are often necessary for AMP activity [1,3,4,8]. The functional categorization of insect AMPs tends to be based on target pathogen range rather than any specific mechanism of action. Some have a broad range, whereas others show varying degrees of specificity towards Gram-positive or Gram-negative bacteria, fungi, parasites and even viruses [17,18].
The first insect AMP to be discovered was cecropin, so-called, because it is produced by larvae of the giant silk moth Hyalophora cecropia. This is the prototype α-helical linear AMP, and it is active against Gram-negative bacteria such as Escherichia coli [19,20]. Other cecropins have been identified more recently, as well as additional cecropin-like peptides such as sarcotoxins, hyphancin and enbocin, which can act against both Gram-positive and Gram-negative bacteria [21]. Most cecropins contain a tryptophan residue at or near the N-terminus, a long N-terminal amphiphilic α-helix, a shorter and more hydrophobic α-helix at the C-terminus and an amidated C-terminal residue.
Defensins represent the prototype for the second major structural class of insect AMPs [22]. They have a predominantly β-sheet globular structure, they are stabilized by intramolecular disulfide bridges [22], and they are widely distributed among different insect orders including ancient apterygote insects [23], hemimetabolous orders such as Hemiptera and Odonata [24], and holometabolous orders such as Coleoptera, Diptera, Hymenoptera and Lepidoptera [10,13,16,21,25]. The first insect peptides described as defensins were discovered in the flesh fly Phormia terranovae and were found to be active against Gram-positive bacteria [26], although similar peptides had already been identified in Sarcophaga peregrine [27]. Most insect defensins act against Gram-positive bacteria, although some also inhibit Gram-negative bacteria [10,21,25,26]. A small number of insect defensins act exclusively against filamentous fungi, e.g. gallerimycin from the greater wax moth Galleria mellonella [28].
A key example of the third structural group of AMPs is the proline-rich AMPs. As well as the characteristic multiple proline residues, these AMPs are 15–39 residues in length and feature two domains, one of which is highly conserved and confers general antimicrobial activity, whereas the other is more variable that mainly confers target specificity [29]. Proline-rich AMPs have been identified in the Diptera (drosocin and metchnikowin) [30], Hemiptera (pyrrhocoricin and metalnikowins) [31], Hymenoptera (apidaecins, abaecins and formaecins) [32,33] and Lepidoptera (lebocins) [34]. The proline-rich AMPs can be further divided into short-chain (20 residues or fewer) and long-chain (more than 20 residues) subfamilies, the former showing more potent activity against Gram-negative bacteria, whereas the latter are more active against Gram-positive bacteria and fungi [35,36]. Such differences in their specificity may be mediated by their distinct lipopolysaccharide binding activity or their ability to penetrate bacterial membranes, because at least some proline-rich AMPs are known to target intracellular molecules [31,34].
Another example of the third structural group is the glycine-rich AMPs. These peptides have been identified among the Coleoptera (coleoptericin, holotricin 2, holotricin 3, tenecin 3 and acaloleptin A), Diptera (diptericins, attacins and sarcotoxin II), Hemiptera (hemiptericin), Hymenoptera (hymenoptaecins) and Lepidoptera (attacins and gloverins) [7,10,13,17,37]. Most glycine-rich AMPs are highly specific for particular groups of Gram-negative bacteria, although honeybee hymenoptaecin also shows activity against the Gram-positive species Micrococcus lysodeikticus and Bacillus megaterium [38]. Our knowledge explaining the specificity of individual glycine-rich AMPs is fragmentary, but studies with attacins and gloverins from Lepidoptera implicate the presence of different targets. Attacin from Hylophora cecropia was found to inhibit outer membrane synthesis in E. coli [39,40], whereas gloverin from Manduca sexta binds to Gram-positive bacterial lipoteichoic acid, peptidoglycan and different moieties of lipopolysaccharide [41].
(b). Modes of action of insect antimicrobial peptides
Most insect AMPs have a net positive charge and contain up to 50% hydrophobic residues [1,7,9]. This leads to interaction of those AMPs with the negatively charged and lipophilic membranes of bacterial cells, reflecting the abundance of acidic phospholipids in the outer leaflet, in contrast to the membranes of eukaryotic cells which are dominated by zwitterionic and uncharged lipids [42]. AMPs are therefore electrostatically attracted to bacterial cell membranes, and once contact is established the hydrophobic residues promote integration, causing the outer leaflet of the membrane to expand and become thinner, ultimately creating pores or even causing lysis.
The ability of AMPs to increase membrane permeability has been confirmed in experiments involving dye-loaded bacteria exposed to Papilio xuthus cecropin A and papiliocin, resulting in extensive dye leakage [43]. Models that offer mechanistic explanations for the interaction between AMPs and bacterial membranes have been reviewed extensively elsewhere [4,9]. Although such models are attractive in their simplicity, they cannot yet explain how AMPs overcome barriers such as the peptidoglycan-rich bacterial cell wall [4,44]. The binding of AMPs to anionic lipopolysaccharides or teichoic acids appears essential for their activity, because charge neutralization makes bacteria more resistant to cationic AMPs [39–41,44]. Although most AMPs appear to function by increasing membrane porosity, the proline-rich AMPs (such as bumblebee abaecin) are instead thought to interact with intracellular targets representing the bacterial chaperone network such as DnaK or the protein synthesis apparatus [45]. For example, attacins have been shown to inhibit the synthesis of proteins which are components of the outer bacterial membrane [39,40]. Furthermore, some AMPs have been shown to inhibit cell wall synthesis by interfering with the corresponding enzymes or lipid phosphatidylethanolamines, or by delocalization of bacterial cell surface proteins [41]. Other insect AMPs such as the insect metalloprotease inhibitor (IMPI) neutralize specifically virulence-associated microbial metalloproteases [46].
An emerging aspect is that some co-occurring AMPs, which are, for example, simultaneously induced during immune responses, can enhance or enable the activity of others [45]. Besides such potentiation there are also examples for synergistic activity of AMPs. For example, a defensin (LSer-Def4) and a cecropin (LSer-Cec6) from the wound maggot Lucilia sericata display greater than additive antibacterial activity when tested in combination [21]. In agreement, synthesized bumblebee AMPs have recently been shown to display combinatorial activity against parasites such as the trypanosome Crithidia bombi [47].
3. Phylogeny of hexapod/insects and the evolution as well as distribution of antimicrobial peptides
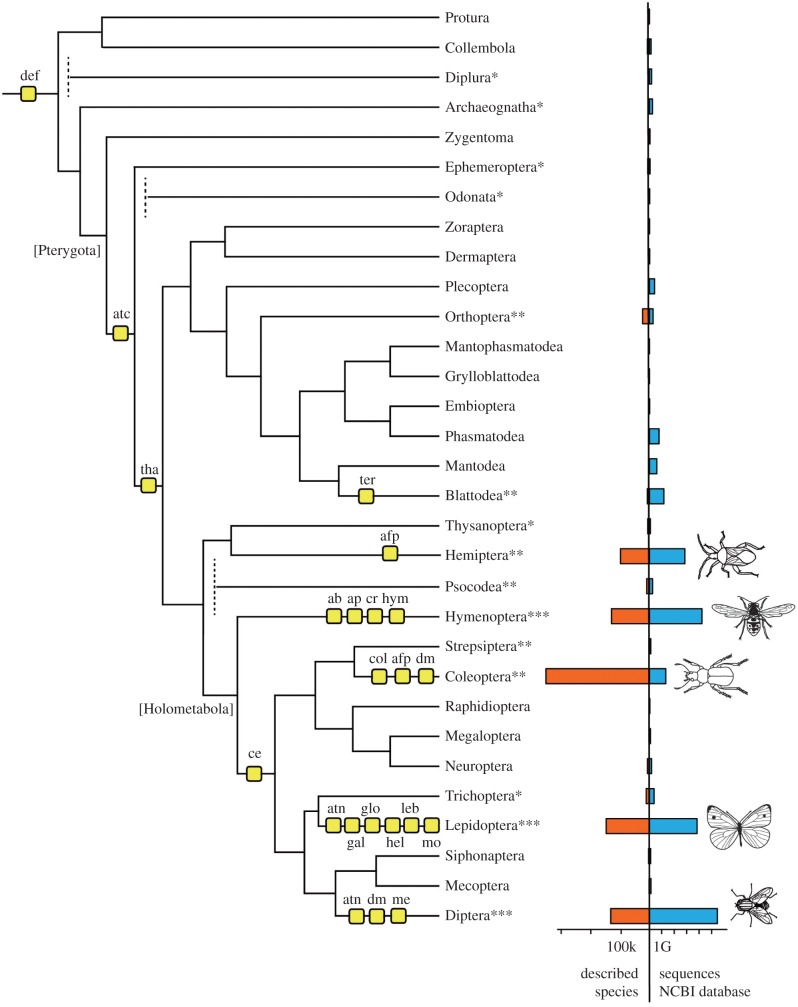
Study of evolution of novel genes participating in insect antimicrobial defence is a very interesting field. Hexapod AMPs may arise by gene duplication and subsequent diversification, by horizontal gene transfer or by de novo creation from non-coding sequences. A well-resolved, dated phylogeny of insects is available [48], providing an indispensable resource that can be used to analyse the evolutionary history of gene families including AMPs. However, the emergence of AMP genes mapped onto the insect phylogenetic tree (figure 1) shows [48] that most of the known AMPs are found in insect taxa with completed genome projects. This is not surprising because model insects with a completely sequenced genome are most intensively studied for a variety of biological questions, including AMPs and immunity. This suggests there is significant undiscovered AMP diversity in insects hidden among the less well-characterized insect taxa and the underrepresented families of the megadiverse orders. AMPs can arise de novo in restricted phylogenetic lineages over very short evolutionary timescales. Furthermore, tracing the evolution of AMPs in parallel with hexapod phylogeny involves two major obstacles. First, there is a substantial bias in favour of the insect taxa with abundant sequence data and against branches that are particularly underrepresented (figure 1). Most insect AMP genes have been discovered in the five megadiverse orders (Coleoptera, Diptera, Hemiptera, Hymenoptera and Lepidoptera), which encompass more than 90% of known insect species, 77% of the nucleotide sequences in the NCBI genomic and expressed sequence tag (EST) databases, and more than 95% of predicted protein sequences. Nevertheless, these megadiverse orders comprise only a part of hexapod diversity. They represent only the crown of the hexapod tree, spanning about 370 Myr of insect evolution, whereas the base of the tree is probably more than 100 Myr older and several early branching lineages remain to be studied in detail [48].
Figure 1.
Evolution of AMPs in insects. The topology largely follows a recent analysis of more than 150 species and concatenated alignments of 1478 genes derived from transcriptomic data [48]. Remaining controversies are depicted by dashed branches (Diplura, Odonata, Psocodea). Following modern taxonomy the Isoptera (termites) are included as a subclade of Blattodea, and Phthiraptera (true lice) are nested inside Psocodea. Asterisks mark taxa with genome projects (one asterisk: genome data available in database of i5k pilot project, but not yet officially published; two asterisks: one to three published genome projects; three asterisks: more than three published genome projects). The barplot shows the number of described species (orange bars to the left) and the number of publicly available nucleotide sequences (blue, to the right) as a measure for intensity of research (combined number of NCBI nucleotide and EST databases; evaluated on 14 October 2015). Hypothesized evolutionary origins of AMP families are mapped on the tree. Note that due to the incomplete representation of immune challenged transcriptomes or genomic data from insect orders, many AMPs may also have evolved on earlier branches of the tree. Abbreviations: ab, abaecin; afp, antifungal protein; ap; apidaecin; atc, attacin C-terminal domain; atn, attacin N-terminal domain; ce, cecropin; col, coleoptericin; cr, crustin; def, defensin; dm, drosomycin; gal, gallerimycin; glov, gloverin; hel, heliomicin; hym, hymenoptaecin; leb, lebocin; mor, moricin; ter, termicin; tha, thaumatin.
The second major obstacle is the recognition of homology among AMP genes. Their short length, substantial variation between species, as well as frequent gain and loss during evolution [12,14,15], hamper the identification of orthologues and often prevents the recognition and delimitation of AMP families. For example, the assignment of novel glycine-rich, proline-rich, defensin-like or cecropin-like peptides is often frustrated by the unconvincing homology revealed by sequence alignments or hidden Markov models (HMMs), as discussed in detail for the attacins in §3a. More groundwork is therefore required for the classification and delimitation of AMP families, and herein we therefore present only a brief overview of the evolutionary history of selected insect AMPs. We define AMP families by significant (best) hits with HMMER v. 3.1 [49] against domains listed in the PFAM protein family database (http://pfam.xfam.org). In those cases where PFAM cannot provide an HMM model, we define HMMs based on sequence alignments (e.g. lebocin and hymenoptaecin) or individual sequences (e.g. gallerimycin).
(a). Antimicrobial peptides that are widely distributed among insect taxa
Important examples of widely distributed AMPs derived from insects are defensin (will be discussed in §3c), cecropins and attacins. Cecropins (PFAM: PF00272) were first discovered in the Lepidoptera [50] and are found in the orders Coleoptera, Diptera and Lepidoptera, but not Hymenoptera. Given that Hymenoptera is a sister order for the other Holometabola, we predict that cecropins have probably evolved once (figure 1) and may be present in the Holometabola orders that remain to be characterized. Some AMPs found beyond the Holometabola have also been described as cecropins, but these share only superficial similarity with the cecropin domain defined by PFAM.
The attacins are represented by discrete N-terminal and C-terminal domains (PFAM: Attacin_N, PF03768; Attacin_C, PF03769). Full-length attacins containing both domains were first described in the Lepidoptera and other attacins were later found in certain brachyceran Diptera [51,52], but not in mosquitoes or chironomids. The alignment of dipteran and lepidopteran attacin N-terminal domains reveals no similarity, suggesting the HMM for Attacin_N is artificial. Furthermore, we have mined the transcriptomes of five Trichoptera species, four Mecoptera species and three Siphonaptera species without finding an attacin N-terminal domain (L. Podsiadlowski 2015, unpublished data). Given the distant phylogenetic relationship between Lepidoptera and Diptera–Brachycera, these N-terminal domains are likely to have evolved independently in the two taxa. By contrast, the C-terminal attacin domain is present in most of the Holometabola genomes studied thus far, and also in the orders Orthoptera (Locusta), Isoptera (Macrotermes) and Hemiptera (Rhodnius). In the latter case, the AMP was named prolixicin [53]. The diptericins [54] also show similarities with the Attacin_C domain, suggesting that an AMP resembling the attacin C-terminal domain was present at the base of the clade known as Neoptera (figure 1). We thus propose that a novel attacin consisting of N- and C-terminal domains evolved independently in the lineages Brachycera and Lepidoptera. This is in line with the hypothesis that the genes encoding three attacins and diptericin from Drosophila share a common ancestor [55].
(b). Antimicrobial peptides restricted to individual insect orders
Several AMP families have only been identified in a single insect order or even in a more restricted taxonomic group. For example, moricin (PFAM: PF06451) [56], glycine-rich gloverin (PFAM: PF10793), proline-rich lebocins [57] and the antifungal cysteine-rich peptides heliomicin (with similarity to PFAM toxin_3 domain, PF00537) [58] and gallerimycin [59] have only been found in the Lepidoptera.
Metchnikowin (PFAM: Antimicrobial10, PF08105) is a proline-rich AMP only found in the genus Drosophila [60], and it has been identified in all 12 Drosophila genomes sequenced thus far [61]. The coleoptericins (PF06286) are glycine- and proline-rich peptides which, as the name suggests, are only found in the Coleoptera. The first coleoptericin was discovered in larvae of the tenebrionid beetle Zophobas atratus [60], followed by similar peptides in other beetles such as Tribolium castaneum [62], the harlequin ladybird H. axyridis [10] and the longicorn beetle Acalolepta luxuriosa [37]. The latter has a remarkable coleoptericin gene comprising a multi-peptide precursor, which yields up to five mature peptides. Termicin (PFAM domain: Toxin_37, PF11415) is a knottin-type AMP discovered in termites [63], which is also found in their closest relatives, the cockroaches (e.g. Periplaneta americana EST library, NCBI FG130406, and Eupolyphaga sinensis cDNA, NCBI KR014250).
Several AMPs are also thought to be restricted to the Hymenoptera, including the proline-rich peptide abaecin (PFAM: Antimicrobial_5, PF08026) which is found in bees [64], ants [32,33,65], the genus Nasonia [66] and another pteromalid wasp [67]. Apidaecin (PF00807) is only found in bees (the genera Apis, Bombus, Megachila and Melipona) and may therefore represent a recent evolutionary adaptation [68]. Finally, hymenoptaecin [38] is a glycine-rich peptide, found only in bees, ants, and wasps from the genus Nasonia [66].
Crustins (containing a PFAM WAP-domain, PF00095) were discovered in crustaceans, but similar sequences have been identified in hymenopteran genomes [66]. They probably have a wider distribution among hexapods, e.g. we found short peptides with WAP domains in the genomes of the coleopteran T. castaneum (TC11324) and two termite species (Zootermopsis nevadensis ZNEV05303 [69] and Macrotermes natalensis MNAT10208).
(c). Antimicrobial peptides with a scattered distribution over unrelated taxa
The arthropod defensin family (PFAM: Defensin_2, PF01097) is the only hexapod AMP family that is broadly distributed beyond the insects, e.g. in ticks and scorpions, where modified defensins are a component of the toxin blend [70]. Other ‘defensins' are found in diverse invertebrates (e.g. molluscs and roundworms), as well as in vertebrate and plant species, but their sequence diversity and small size make it difficult to confirm a unique evolutionary origin. Arthropod defensins are found in diverse hexapod species, including the orders Zygentoma [23] and Odonata [24], clearly suggesting that an arthropod defensin was already present in the last common ancestor of all insects.
Drosomycin (PFAM: Gamma-thionin PF00304) was first identified in Drosophila melanogaster [71], but it is not present in all Drosophila species. Among the Diptera, this AMP is also found in Musca domestica but not in Glossina, Phlebotomus or any mosquito genome. Recently, a similar peptide was also discovered in two genera of beetles (Callosobruchus and Trox) [72], and there are also similarities with the cremycin family of nematode AMPs. The hypothesis that insect drosomycins and nematode cremycins may have been acquired from plants by horizontal gene transfer is supported by the patchy distribution pattern of homologues which are present in Ecdysozoa, absent in other animals, fungi and protozoa, but widespread in plants. The alternative explanation postulates the evolution of these antifungal peptides in the last common ancestor of all eukaryotes and independent loss in fungi and all Metazoa except Ecdysozoa [72].
Alo-3 (PFAM domain: PF11410) is a knottin-type antifungal peptide which was first identified in the beetle Acrocinus longimanus [73]. A related insect peptide was identified in the whitefly Bemisia tabaci (ABC40569-ABC40572). Both species are phytophagous. Because this domain is otherwise only found in plants and fungi, it may be another example of horizontal gene transfer from plants, and at least two independent events are likely to have occurred given the phylogenetic distance between Coleoptera and Hemiptera (figure 1).
Thaumatins are antifungal peptides (PFAM domain: PF00314) which have been identified in fungi, plants and animals, and in the last case they appear to be restricted to nematodes, ticks and insects. In a broad comparison of all eukaryotic thaumatins, all animal thaumatins form one clade, nested within plant thaumatins [74]. This suggests that animal thaumatins have a single origin, perhaps reflecting a horizontal gene transfer event early in the evolution of the Ecdysozoa (a clade including nematodes and arthropods). Thaumatins are found in many but not all insect orders. They are present in beetles [75], aphids [11] and termites [69], but among the Diptera, thaumatin has been identified in two chironomids (Polypedilum vanderplancki PVAN02763 and Polypedilum nubifer PNUB14148), but in none of the 32 complete genomes representing mosquitoes and flies. Thaumatin proteins are also scarce among the Lepidoptera and Hymenoptera, despite the availability of several genome datasets. Although the scattered distribution suggests independent acquisition events, the fact that all arthropod thaumatins are more closely related to each other than to fungal or plant thaumatins seems to rule out this hypothesis and favour multiple independent losses of thaumatin genes in several hexapod lineages. Despite the number of thaumatin encoding sequences in insects, only a thaumatin gene identified in T. castaneum has been heterologously expressed and confirmed to display antifungal activity [75].
4. Activity screening and medical applications of insect antimicrobial peptides
The number of infections caused by drug-resistant microbes is increasing, particularly those involving ‘ESKAPE’ bacteria (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumanii, Pseudomonas aeruginosa, Escherichia coli and Enterobacter species) that ‘escape’ the effects of current antimicrobial drugs, pose a substantial threat to public health, and contribute significantly to patient morbidity [76,77]. In addition to the ESKAPE bacteria, other pathogens including Clostridium difficile, Candida species [78] and multidrug-resistant/extensively drug-resistant mycobacteria provide further evidence that we are losing the war against emerging resistant microbes [76,77,79].
The Centers for Disease Control and Prevention (CDC) report that two million people per year in the USA acquire serious infections due to bacteria that are resistant to one or more current antibiotics (www.cdc.gov/). This results in 23 000 deaths as a direct result of these infections and many more due to complications, costing up to US$20 billion in excess direct healthcare costs and annual productivity losses exceeding US$35 billion. Similarly, the European Centre for Disease Prevention and Control (ECDC) claim that antibiotic resistance costs the European Union approximately €1.5 billion per year (ecdc.europa.eu/). Agencies such as the US President's Council of Advisors on Science and Technology (PCAST), have highlighted the need to focus on antibiotic drug discovery to address the issue of drug-resistant pathogens [80]. Accordingly, the UK has recently launched the Five Year Antimicrobial Resistance Strategy 2013–2018 (https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/244058/20130902_UK_5_year_AMR_strategy.pdf), a collaboration involving multiple agencies including government departments and academic centres. Importantly, one of the successful approaches to identifying potential antimicrobials is the high-throughput screening method using the nematode C. elegans as alternative host. This method was further applied to identify AMPs as alternative antimicrobials.
(a). Caenorhabditis elegans as a high-throughput screening model to identify antimicrobial peptides with activity against human pathogens
The identification of compounds with in vivo activity against human pathogens and their development as new drugs is challenging, but high-throughput screening provides a more efficient way to identify such compounds quickly. High-throughput screening was first applied to drug development in the 1980s and the same concept can be used today to screen libraries of AMPs [81,82].
The nematode C. elegans is a model organism with a completely sequenced genome [83,84]. It is a small, free-living, transparent worm, which is approximately 1 mm in length and comprises about 1000 cells. It has a lifespan of two to three weeks, and many thousands of individual worms can be propagated on plates containing nematode growth medium agar spiked with lawns of non-pathogenic bacteria [85]. Caenorhabditis elegans is widely used for the in vivo investigation of host–pathogen interactions, including the study of microbial pathogenesis and innate immune responses [86–88]. Notably, several evolutionarily conserved innate immune response mechanisms have been described in C. elegans, including p38 mitogen-activated protein kinase (MAPK) pathways [89,90]. Moreover, various antimicrobial agents have been shown to prolong the survival of infected worms, confirming that this model organism is suitable for antimicrobial discovery [87,91,92]. Furthermore, the C. elegans–microbe liquid assay allows the identification of agents that directly kill microbes or that possess immunomodulatory and anti-virulence effects. This host–pathogen system has been further developed to enable high-throughput experiments in an automated robotic system which is used to dispense medium, worms and compounds onto the assay plates, and which monitors nematode survival manually or by image-based automated screening [93,94].
The suitability of C. elegans for the high-throughput screening of antimicrobial compounds was first demonstrated using Enterococcus faecalis as a model pathogen [93]. More recently, high-throughput screens using C. elegans have identified antimicrobial agents that work against challenging pathogens such as methicillin-resistant Staphylococcus aureus (MRSA), Pseudomonas aeruginosa and Candida albicans [94,95]. Strikingly, using this assay in a screening of 68 synthetic insect-derived AMPs, we determined that defensin 1 from the red flour beetle T. castaneum displays synergistic activity with the antibiotics Telavancin and Daptomycin against multidrug-resistant S. aureus [96]. These results open a new avenue for the application of insect-derived AMPs. In combination with antibiotics they could be used to restore the susceptibility of multidrug-resistant pathogens.
We have established a C. elegans–Acinetobacter baumannii assay to conduct a pilot screen on a library of 68 insect-derived AMPs. This screen identified 15 cecropins and cecropin-like AMPs that prolonged the survival of C. elegans infected with A. baumannii [97]. One of the identified AMPs (BR003-cecropin A), isolated from the mosquito Aedes aegypti was found to be effective against multiple species of Gram-negative bacteria and to act with a low minimum inhibitory concentration against different A. baumannii strains [97].
(b). Potential medical applications of antimicrobial peptides
Previous reports have highlighted several potential medical roles for AMPs [3,8,17,98,99]. As stated above, they have diverse mechanisms of action including the inhibition of gene expression or protein synthesis (e.g. targeting ribosomal proteins, RNA polymerase, or directly binding to DNA), the inhibition of cell wall synthesis (e.g. by targeting the corresponding enzymes or lipid phosphatidylethanolamines), or the delocalization of bacterial cell surface proteins [98,99]. Some AMPs have demonstrated immune-stimulatory effects by inducing cell migration, cell proliferation or the release of cytokines and chemokines [100]. Other insect-derived AMPs such as Harmoniasin from the harlequin ladybird H. axyridis may be useful leads for the development of anti-cancer drugs [101].
Indeed, there is an increasing number of insect-derived AMPs shown to inhibit human pathogens. Examples of susceptible bacteria include multidrug-resistant A. baumannii, Bacillus coagulans, Citrobacter freundii, Enterobacter aerogenes, E. coli, Francisella tularensis, K. pneumonia, Legionella dumoffei, Listeria monocytogenes, Proteus vulgaris, S. aureus and Streptococcus sanguinis [17,21,53,96,97,102,103]. Examples of fungi which are susceptible to insect AMPs include Aspergillus fumigatus, Alternaria spp., Botrytis cinerea, C. albicans, Cryptococcus neoformans, Fusarium spp., Neurospora crassa, Pichia pastoris, Trichoderma viridae [17,28,37,104]. Of note is that various insect AMPs have been shown to inhibit viruses including a mellitin derivative (hecate) from the honeybee (Apis mellifera) which is active against Herpes simplex virus 1 (HSV-1) [100], and two alloferons from the blowfly Calliphora vicina which are active against Human influenza viruses A and B [105].
There is a debate about the ability of bacteria to evolve resistance against AMPs. On the one hand, many researchers argue that AMPs such as defensins retained their antibacterial activity over millions of years and only modest resistance development has been observed under in vitro selection pressure [106]. Further, exposure to AMPs elicits neither stress responses nor increased mutation rates in treated bacteria [107]. On the other hand, there are several mechanisms described which can mediate bacterial resistance against AMPs [108].
Bacteria-derived gramicidins represent the first peptide antibiotics to become commercially available [109] and numerous reports have highlighted that insect-derived AMPs are attractive candidates to be developed as alternatives to conventional antibiotics [3,8,17,98,99,106]. Despite worldwide efforts, there are no insect-derived AMPs on the market yet. Their development as antibiotics requires solutions to some obstacles which will be briefly addressed.
The testing of insect-derived AMPs displaying potent in vivo and in vitro activity in preclinical and clinical studies requires amounts which are difficult to produce economically [110]. However, the costs for the synthesis of short peptides (up to approx. 80 residues) have decreased markedly in recent years [111], enabling at least the production of insect-derived AMPs which are too small to be immunogenic or allergenic. For the larger insect AMPs, particularly those with complex three-dimensional structures that are stabilized by intramolecular disulfide bonds, we need cost-efficient heterologous expression systems, and insect cell lines in particular have proved to be promising tools for the production of functional insect-derived recombinant peptides [112]. The process development of cost-efficient insect cell-based protein production systems has become a major challenge in insect biotechnology [113] and the recently achieved solutions are groundbreaking, but still exceed the manufacturing costs of conventional drugs [114].
The bioavailability of drugs depends on their stability. The susceptibility of insect AMPs to host proteases differs markedly. Basically, linear peptides are generally more proteolytically degradable than those AMPs with an intramolecular structure stabilized by disulfide bonds such as that known from defensins [106]. The antibacterial activity of the latter hampers their heterologous production in bacteria which also usually lack the ability to synthesize properly folded functional peptides. These limitations can be overcome by the use of advanced insect cell-based expression systems [112–114]. The design of functional analogues or peptidomimetics which are more resistant to hydrolysis by host proteases, has emerged as another strategy to develop novel antibiotics using insect-derived AMPs as leads [115,116]. Further, only limited amounts of particular AMPs are required for systemic application if they are used to restore the susceptibility of pathogens to conventional antibiotics [117]. Consequently, the combinatorial use of insect-derived AMPs together with antibiotics has become another avenue of research aiming to implement these natural products in therapeutic approaches.
The above-mentioned obstacles for systemic application of insect-derived AMPs have favoured their development for ectopic applications which do require less demanding preclinical and clinical research [118]. A prime example is the development of AMPs from medicinal maggots of L. sericata which are formulated in hydrogels to test the efficacy of synthetic counterparts in wound dressings and as cosmetic ingredients to deter dermatological pathogens [21,104,119]. Other insect-derived AMPs such as the IMPI from G. mellonella [120] are currently also being developed for the therapy of chronic wounds [121]. Further promising medicinal applications of insect-derived AMPs are currently being explored in therapies to cure eye, lung and urogenital infections [117,122,123]. For example, it has been demonstrated that a defensin from G. mellonella or AMPs of medicinal maggots are active against causative agents of lung infection [21,117]. Recombinant analogues of insect-derived AMPs can be delivered to the lung bound on inhalable microparticles and the simultaneous application of AMPs displaying synergistic activity is expected to reduce the amounts required for therapeutic approaches.
5. Conclusion
An ever increasing number of AMPs are being found in insects. The corresponding genes display a remarkable evolutionary plasticity in terms of gain, loss and neo-functionalization. Mapping the presence of AMPs on the phylogenetic tree of insects reveals the existence of widespread and taxon-specific AMP families. Recent in vitro and in vivo screening using surrogate model hosts such as C. elegans has shown that insect-derived AMPs display promising activity against human pathogens that could make them suitable as alternatives to conventional antibiotics. However, their development must address the limitations associated with the application of peptide-based drugs.
Acknowledgements
We thank Richard M. Twyman for editing the manuscript.
Authors' contributions
All authors reviewed and approved the final manuscript. A.V. researched and wrote the section covering the diversity and mode of action of AMPs; L.P. researched and wrote the section covering the evolution and phylogenetic distribution of insect AMPs; E.M., M.M. and A.V. researched and wrote the section covering the medical applications of insect AMPs.
Competing interests
We declare we have no competing interests.
Funding
A.V. acknowledges generous funding by the Hessen State Ministry of Higher Education, Research and the Arts (HMWK) via the ‘LOEWE Center for Insect Biotechnology and Bioresources'. L.P. acknowledges support from Leibnitz Graduate School ‘Genomic Biodiversity Research (GBR).
References
- 1.Bulet P, Stocklin R, Menin L. 2004. Anti-microbial peptides: from invertebrates to vertebrates. Immunol. Rev. 198, 169–184. ( 10.1111/j.0105-2896.2004.0124.x) [DOI] [PubMed] [Google Scholar]
- 2.Vale N, Aguiar L, Gomes P. 2014. Antimicrobial peptides: a new class of antimalarial drugs? Front. Pharmacol. 5, 275 ( 10.3389/fphar.2014.00275) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hancock RE, Sahl HG. 2006. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24, 1551–1557. ( 10.1038/nbt1267) [DOI] [PubMed] [Google Scholar]
- 4.Brogden KA. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3, 238–250. ( 10.1038/nrmicro1098) [DOI] [PubMed] [Google Scholar]
- 5.Makarova O, Rodriguez-Rojas A, Eravci M, Weise C, Dobson A, Johnston P, Rolff J. 2016. Antimicrobial defence and persistent infection in insects revisited. Phil. Trans. R. Soc. B 371, 20150296 ( 10.1098/rstb.2015.0296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masson F, Zaidman-Rémy A, Heddi A. 2016. Antimicrobial peptides and cell processes tracking endosymbiont dynamics. Phil. Trans. R. Soc. B 371, 20150298 ( 10.1098/rstb.2015.0298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bulet P, Stocklin R. 2005. Insect antimicrobial peptides: structures, properties and gene regulation. Protein Pept. Lett. 12, 3–11. ( 10.2174/0929866053406011) [DOI] [PubMed] [Google Scholar]
- 8.Yi HY, Chowdhury M, Huang YD, Yu XQ. 2014. Insect antimicrobial peptides and their applications. Appl. Microbiol. Biotechnol. 98, 5807–5822. ( 10.1007/s00253-014-5792-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiesner J, Vilcinskas A. 2010. Antimicrobial peptides: the ancient arm of the human immune system. Virulence 1, 440–464. ( 10.4161/viru.1.5.12983) [DOI] [PubMed] [Google Scholar]
- 10.Vilcinskas A, Mukherjee K, Vogel H. 2013. Expansion of the antimicrobial peptide repertoire in the invasive ladybird Harmonia axyridis. Proc. R. Soc. B 280, 20122113 ( 10.1098/rspb.2012.2113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerardo NM, et al. 2010. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 11, R21 ( 10.1186/gb-2010-11-2-r21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vilcinskas A. 2013. Evolutionary plasticity of insect immunity. J. Insect Physiol. 59, 123–129. ( 10.1016/j.jinsphys.2012.08.018) [DOI] [PubMed] [Google Scholar]
- 13.Altincicek B, Vilcinskas A. 2007. Analysis of the immune-related transcriptome from microbial stress resistant, rat-tailed maggots of the drone fly Eristalis tenax. BMC Genomics 8, 326 ( 10.1186/1471-2164-8-326) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sackton TB, Lazzaro BP, Schlenke TA, Evans JD, Hultmark D, Clark AG. 2007. Dynamic evolution of the innate immune system in Drosophila. Nat. Genet. 39, 1461–1468. ( 10.1038/ng.2007.60) [DOI] [PubMed] [Google Scholar]
- 15.Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC. 2007. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science 316, 1738–1743. ( 10.1126/science.1139862) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogel H, Altincicek B, Glöckner G, Vilcinskas A. 2011. A comprehensive transcriptome and immune-gene repertoire of the Lepidopteran model mini-host Galleria mellonella. BMC Genomics 12, 308 ( 10.1186/1471-2164-12-308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vilcinskas A. 2011. Anti-infective therapeutics from the Lepidopteran model host Galleria mellonella. Curr. Pharm. Des. 17, 1240–1245. ( 10.2174/138161211795703799) [DOI] [PubMed] [Google Scholar]
- 18.Pretzel J, Mohring F, Rahlfs S, Becker K. 2013. Antiparasitic peptides. Adv. Biochem. Eng. Biotechnol. 135, 157–192. ( 10.1007/10_2013_191) [DOI] [PubMed] [Google Scholar]
- 19.Steiner H, Hultmark D, Engstrom A, Bennich H, Boman HG. 1981. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 292, 246–248. ( 10.1038/292246a0) [DOI] [PubMed] [Google Scholar]
- 20.Hultmark D, Steiner H, Rasmuson T, Boman HG. 1980. Insect immunity. Purification and properties of three inducible bactericidal proteins from hemolymph of immunized pupae of Hyalophora cecropia. Eur. J. Biochem. 106, 7–16. ( 10.1111/j.1432-1033.1980.tb05991.x) [DOI] [PubMed] [Google Scholar]
- 21.Pöppel AK, Vogel H, Wiesner J, Vilcinskas A. 2015. Antimicrobial peptides expressed in medicinal maggots of the blow fly Lucilia sericata show combinatorial activity against bacteria. Antimicrob. Agents Chemother. 59, 2508–2514. ( 10.1128/AAC.05180-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao BC, Lin HC, Yang D, Ye X, Li ZG. 2015. Disulfide bridges in defensins. Curr. Top. Med. Chem. 16, 206–219. ( 10.2174/1568026615666150701115911) [DOI] [PubMed] [Google Scholar]
- 23.Altincicek B, Vilcinskas A. 2007. Identification of immune-related genes from an apterygote insect, the firebrat Thermobia domestica. Insect Biochem. Mol. Biol. 37, 726–731. ( 10.1016/j.ibmb.2007.03.012) [DOI] [PubMed] [Google Scholar]
- 24.Johnston P, Rolff J. 2013. Immune- and wound-dependent differential gene expression in an ancient insect. Dev. Comp. Immunol. 40, 320–324. ( 10.1016/j.dci.2013.01.012) [DOI] [PubMed] [Google Scholar]
- 25.Tonk M, Knorr E, Cabezas-Cruz A, Valdes J, Kollewe C, Vilcinskas A. 2015. Tribolium castaneum defensins are primarily active against Gram-positive bacteria. J. Invertebr. Pathol. 132, 208–215. ( 10.1016/j.jip.2015.10.009) [DOI] [PubMed] [Google Scholar]
- 26.Lambert J, et al. 1989. Insect immunity: isolation from immune blood of the dipteran Phormia terranovae of two insect antibacterial peptides with sequence homology to rabbit lung macrophage bactericidal peptides. Proc. Natl Acad. Sci. USA 86, 262–266. ( 10.1073/pnas.86.1.262) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuyama K, Natori S. 1988. Purification of three antibacterial proteins from the culture medium of NIH-Sape-4, an embryonic cell line of Sarcophaga peregrina. J. Biol. Chem. 263, 17 112–17 116. [PubMed] [Google Scholar]
- 28.Langen G, Imani J, Altincicek B, Kieseritzky G, Kogel KH, Vilcinskas A. 2006. Transgenic expression of gallerimycin, a novel antifungal insect defensin from the greater wax moth Galleria mellonella, confers resistance to pathogenic fungi in tobacco. Biol. Chem. 387, 549–557. ( 10.1515/BC.2006.071) [DOI] [PubMed] [Google Scholar]
- 29.Li W, Tailhades J, O'Brien-Simpson NM, Separovic F, Otvos L Jr, Hossain MA, Wade JD. 2014. Proline-rich antimicrobial peptides: potential therapeutics against antibiotic-resistant bacteria. Amino Acids 46, 2287–2294. ( 10.1007/s00726-014-1820-1) [DOI] [PubMed] [Google Scholar]
- 30.Levashina EA, Ohresser S, Bulet P, Reichhart JM, Hetru C, Hoffmann JA. 1995. Metchnikowin, a novel immune-inducible proline-rich peptide from Drosophila with antibacterial and antifungal properties. Eur. J. Biochem. 233, 694–700. ( 10.1111/j.1432-1033.1995.694_2.x) [DOI] [PubMed] [Google Scholar]
- 31.Taniguchi M, et al. In press. Pyrrhocoricin, a proline-rich antimicrobial peptide derived from insect, inhibits the translation process in the cell-free Escherichia coli protein synthesis system. J. Biosci. Bioeng. ( 10.1016/j.jbiosc.2015.09.002) [DOI] [PubMed] [Google Scholar]
- 32.Ratzka C, Förster F, Liang C, Kupper M, Dandekar T, Feldhaar H, Gross R. 2012. Molecular characterization of antimicrobial peptide genes of the carpenter ant Camponotus floridanus. PLoS ONE 7, e32559 ( 10.1371/journal.pone.0043036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta SK, Kupper M, Ratzka C, Feldhaar H, Vilcinskas A, Gross R, Dandekar T, Förster F. 2015. Scrutinizing the immune defence inventory of Camponotus floridanus applying total transcriptome sequencing. BMC Genomics 16, 540 ( 10.1186/s12864-015-1748-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rao XJ, Xu XX, Yu XQ. 2012. Functional analysis of two lebocin-related proteins from Manduca sexta. Insect Biochem. Mol. Biol. 42, 231–239. ( 10.1016/j.ibmb.2011.12.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahnamaeian M, Langen G, Imani J, Khalifa W, Altincicek B, von Wettstein D, Kogel K-H, Vilcinskas A. 2009. Insect peptide metchnikowin confers on barley a selective capacity for resistance to fungal ascomycetes pathogens. J. Exp. Bot. 60, 4105–4114. ( 10.1093/jxb/erp240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahnamaeian M, Vilcinskas A. 2012. Defense gene expression is potentiated in transgenic barley expressing antifungal peptide Metchnikowin throughout powdery mildew challenge. J. Plant Res. 125, 115–124. ( 10.1007/s10265-011-0420-3) [DOI] [PubMed] [Google Scholar]
- 37.Imamura M, Wada S, Ueda K, Saito A, Koizumi N, Iwahana H, Sato R. 2009. Multipeptide precursor structure of acaloleptin A isoforms, antibacterial peptides from the Udo longicorn beetle, Acalolepta luxuriosa. Dev. Comp. Immunol. 33, 1120–1127. ( 10.1016/j.dci.2009.06.004) [DOI] [PubMed] [Google Scholar]
- 38.Casteels P, Ampe C, Jacobs F, Tempst P. 1993. Functional and chemical characterization of Hymenoptaecin, an antibacterial polypeptide that is infection-inducible in the honeybee (Apis mellifera). J. Biol. Chem. 268, 7044–7054. [PubMed] [Google Scholar]
- 39.Carlsson A, Engstrom P, Palva ET, Bennich H. 1991. Attacin, an antibacterial protein from Hyalophora cecropia, inhibits synthesis of outer membrane proteins in Escherichia coli by interfering with omp gene transcription. Infect Immun. 59, 3040–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carlsson A, Nystrom T, de Cock H, Bennich H. 1998. Attacin—an insect immune protein—binds LPS and triggers the specific inhibition of bacterial outer-membrane protein synthesis. Microbiology 144, 2179–2188. ( 10.1099/00221287-144-8-2179) [DOI] [PubMed] [Google Scholar]
- 41.Xu XX, Zhong X, Yi HY, Yu XQ. 2012. Manduca sexta gloverin binds microbial components and is active against bacteria and fungi. Dev. Comp. Immunol. 38, 255–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown KL, Hancock RE. 2006. Cationic host defense (antimicrobial) peptides. Curr. Opin. Immunol. 18, 24–30. ( 10.1016/j.coi.2005.11.004) [DOI] [PubMed] [Google Scholar]
- 43.Kim JK, et al. 2011. Structure and function of papiliocin with antimicrobial and anti-inflammatory activities isolated from the swallowtail butterfly, Papilio xuthus. J. Biol. Chem. 286, 41 296–41 311. ( 10.1074/jbc.M111.269225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilmes M, Cammue B, Sahl HG, Thevissen K. 2011. Antibiotic activities of host defense peptides: more than lipid bilayer perturbation. Nat. Prod. Rep. 8, 1350–1358. ( 10.1039/c1np00022e) [DOI] [PubMed] [Google Scholar]
- 45.Rahnamaeian M, et al. 2015. Insect antimicrobial peptides show potentiating functional interactions against Gram-negative bacteria. Proc. R. Soc. B 282, 20150293 ( 10.1098/rspb.2015.0293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clermont A, Wedde M, Seitz V, Podsiadlowski L, Lenze D, Hummel M, Vilcinskas A. 2004. Cloning and expression of an inhibitor of microbial metalloproteinases from insects contributing to innate immunity. Biochem. J. 382, 315–322. ( 10.1042/BJ20031923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marxer M, Vollenweider V, Schmid-Hempel P. 2016. Insect antimicrobial peptides act synergistically to inhibit a trypanosome parasite. Phil. Trans. R. Soc. B 371, 20150302 ( 10.1098/rstb.2015.0302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Misof B, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science 346, 763–767. ( 10.1126/science.1257570) [DOI] [PubMed] [Google Scholar]
- 49.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195 ( 10.1371/journal.pcbi.1002195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hultmark D, Engstrom A, Bennich H, Kapur R, Boman HG. 1982. Insect immunity: isolation and structure of cecropin D and four minor antibacterial components from Cecropia pupae. Eur. J. Biochem. 127, 207–217. ( 10.1111/j.1432-1033.1982.tb06857.x) [DOI] [PubMed] [Google Scholar]
- 51.Hao Z, Kasumba I, Lehane MJ, Gibson WC, Kwon J, Aksoy S. 2001. Tsetse immune responses and trypanosome transmission: implications for the development of tsetse-based strategies to reduce trypanosomiasis. Proc. Natl Acad. Sci. USA 98, 12 648–12 653. ( 10.1073/pnas.221363798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asling B, Dushay MS, Hultmark D. 1995. Identification of early genes in the Drosophila immune response by PCR-based differential display: the Attacin A gene and the evolution of attacin-like proteins. Insect Biochem. Mol. Biol. 25, 511–518. ( 10.1016/0965-1748(94)00091-C) [DOI] [PubMed] [Google Scholar]
- 53.Ursic-Bedoya R, Buchhop J, Joy JB, Durvasula R, Lowenberger C. 2011. Prolixicin: a novel antimicrobial peptide isolated from Rhodnius prolixus with differential activity against bacteria and Trypanosoma cruzi. Insect Mol. Biol. 20, 775–786. ( 10.1111/j.1365-2583.2011.01107.x) [DOI] [PubMed] [Google Scholar]
- 54.Dimarcq JL, et al. 1988. Insect immunity. Purification and characterization of a family of novel inducible antibacterial proteins from immunized larvae of the dipteran Phormia terranovae and complete amino-acid sequence of the predominant member, diptericin A. Eur. J. Biochem. 171, 17–22. ( 10.1111/j.1432-1033.1988.tb13752.x) [DOI] [PubMed] [Google Scholar]
- 55.Hedengren M, Borge K, Hultmark D. 2000. Expression and evolution of the Drosophila Attacin/Diptericin gene family. Dev. Comp. Immunol. 279, 574–581. ( 10.1006/bbrc.2000.3988) [DOI] [PubMed] [Google Scholar]
- 56.Hara S, Yamakawa M. 1995. Moricin, a novel type of antibacterial peptide isolated from the silkworm, Bombyx mori. J. Biol. Chem. 270, 29 923–29 927. ( 10.1074/jbc.270.13.7142) [DOI] [PubMed] [Google Scholar]
- 57.Hara S, Yamakawa M. 1995. A novel antibacterial peptide family isolated from the silkworm, Bombyx mori. Biochem. J. 310, 651–656. ( 10.1042/bj3100651) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lamberty M, Ades S, Uttenweiler-Joseph S, Brookhart G, Bushey D, Hoffmann JA, Bulet P. 1999. Insect immunity. Isolation from the lepidopteran Heliothis virescens of a novel insect defensin with potent antifungal activity. J. Biol. Chem. 274, 9320–9326. ( 10.1074/jbc.274.14.9320) [DOI] [PubMed] [Google Scholar]
- 59.Schuhmann B, Seitz V, Vilcinskas A, Podsiadlowski L. 2003. Cloning and expression of gallerimycin, an antifungal peptide expressed in immune response of greater wax moth larvae, Galleria mellonella. Arch. Insect. Biochem. Physiol. 53, 125–133. ( 10.1002/arch.10091) [DOI] [PubMed] [Google Scholar]
- 60.Bulet P, et al. 1991. Insect immunity. Isolation from a coleopteran insect of a novel inducible antibacterial peptide and of new members of the insect defensin family. J. Biol. Chem. 266, 24 520–24525. [PubMed] [Google Scholar]
- 61.Hahn MW, Han MV, Han SG. 2007. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 3, e197 ( 10.1371/journal.pgen.0030197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zou Z, Evans JD, Lu Z, Zhao P, Williams M, Sumathipala N, Hetru C, Hultmark D, Jiang H. 2007. Comparative genomic analysis of the Tribolium immune system. Genome Biol. 8, R177 ( 10.1186/gb-2007-8-8-r177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lamberty M, Zachary D, Lanot R, Bordereau C, Robert A, Hoffmann JA, Bulet P. 2001. Insect immunity. Constitutive expression of a cysteine-rich antifungal and a linear antibacterial peptide in a termite insect. J. Biol. Chem. 276, 4085–4092. ( 10.1074/jbc.M002998200) [DOI] [PubMed] [Google Scholar]
- 64.Casteels P, Ampe C, Riviere L, Van Damme J, Elicone C, Fleming M, Jacobs F, Tempst P. 1990. Isolation and characterization of abaecin, a major antibacterial response peptide in the honeybee (Apis mellifera). Eur. J. Biochem. 187, 381–386. ( 10.1111/j.1432-1033.1990.tb15315.x) [DOI] [PubMed] [Google Scholar]
- 65.Zhang Z, Zhu S. 2012. Comparative genomics analysis of five families of antimicrobial peptide-like genes in seven ant species. Dev. Comp. Immunol. 38, 262–274. ( 10.1016/j.dci.2012.05.003) [DOI] [PubMed] [Google Scholar]
- 66.Tian C, Gao B, Fang Q, Ye G, Zhu S. 2010. Antimicrobial peptide-like genes in Nasonia vitripennis: a genomic perspective. BMC Genomics 11, 187 ( 10.1186/1471-2164-11-187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shen X, Ye G, Cheng X, Yu C, Altosaar I, Hu C. 2010. Characterization of an abaecin-like antimicrobial peptide identified from a Pteromalus puparum cDNA clone. J. Invertebr. Pathol. 105, 24–29. ( 10.1016/j.jip.2010.05.006) [DOI] [PubMed] [Google Scholar]
- 68.Casteels P, Ampe C, Jacobs F, Vaeck M, Tempst P. 1989. Apidaecins: antibacterial peptides from honeybees. EMBO J. 8, 2387–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Terrapon N, et al. 2015. Molecular traces of alternative social organization in a termite genome. Nat. Comm. 5, 3636. [DOI] [PubMed] [Google Scholar]
- 70.Tonk M, Cabezas-Cruz A, Valdés J, Rego ROM, Grubhoffer L, Estrada-Peña A, Vilcinskas A, Kotsyfakis M, Rahnamaeian M. 2015. Ixodes ricinus defensins attack distantly-related pathogens. Dev. Comp. Immunol. 53, 358–365. ( 10.1016/j.dci.2015.08.001) [DOI] [PubMed] [Google Scholar]
- 71.Fehlbaum P, et al. 1994. Insect immunity. Septic injury of Drosophila induces the synthesis of a potent antifungal peptide with sequence homology to plant antifungal peptides. J. Biol. Chem. 269, 33 159–33 163. [PubMed] [Google Scholar]
- 72.Zhu S, Gao B. 2014. Nematode-derived drosomycin-type antifungal peptides provide evidence for plant-to-ecdysozoan horizontal transfer of a disease resistance gene. Nat. Commun. 5, 3154. [DOI] [PubMed] [Google Scholar]
- 73.Barbault F, Landon C, Guenneugues M, Meyer JP, Schott V, Dimarcq JL, Vovelle F. 2003. Solution structure of Alo-3: a new knottin-type antifungal peptide from the insect Acrocinus longimanus. Biochemistry 42, 14 434–14 442. ( 10.1021/bi035400o) [DOI] [PubMed] [Google Scholar]
- 74.Petre B, Major I, Rouhier N, Duplessis S. 2011. Genome-wide analysis of eukaryote thaumatin-like proteins (TLPs) with an emphasis on poplar. BMC Plant Biol. 11, 33 ( 10.1186/1471-2229-11-33) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Altincicek B, Knorr E, Vilcinskas A. 2008. Beetle immunity: identification of immune-inducible genes from the model insect Tribolium castaneum. Dev. Comp. Immunol. 32, 585–595. ( 10.1016/j.dci.2007.09.005) [DOI] [PubMed] [Google Scholar]
- 76.Laxminarayan R, et al. 2013. Antibiotic resistance—the need for global solutions. Lancet Infect Dis. 13, 1057–1098. ( 10.1016/S1473-3099(13)70318-9) [DOI] [PubMed] [Google Scholar]
- 77.Ziakas PD, Zacharioudakis IM, Zervou FN, Mylonakis E. 2015. Methicillin-resistant Staphylococcus aureus prevention strategies in the ICU: a clinical decision analysis. Crit. Care Med. 43, 382–393. ( 10.1097/CCM.0000000000000711) [DOI] [PubMed] [Google Scholar]
- 78.Kontoyiannis DP, et al. 2010. Prospective surveillance for invasive fungal infections in hematopoietic stem cell transplant recipients, 2001–2006: overview of the Transplant-Associated Infection Surveillance Network (TRANSNET) Database. Clin. Infect Dis. 50, 1091–1100. ( 10.1086/651263) [DOI] [PubMed] [Google Scholar]
- 79.Rice LB. 2008. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect Dis. 197, 1079–1081. ( 10.1086/533452) [DOI] [PubMed] [Google Scholar]
- 80.Roberts RR, et al. 2009. Hospital and societal costs of antimicrobial-resistant infections in a Chicago teaching hospital: implications for antibiotic stewardship. Clin. Infect Dis. 49, 1175–1184. ( 10.1086/605630) [DOI] [PubMed] [Google Scholar]
- 81.Moy TI, Conery AL, Larkins-Ford J, Wu G, Mazitschek R, Casadei G, Lewis K, Carpenter AE, Ausubel FM. 2009. High-throughput screen for novel antimicrobials using a whole animal infection model. ACS Chem. Biol. 4, 527–533. ( 10.1021/cb900084v) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Segalat L. 2006. Drug discovery: here comes the worm. ACS Chem. Biol. 1, 277–278. ( 10.1021/cb600221m) [DOI] [PubMed] [Google Scholar]
- 83.Chen N, Lawson D, Bradnam K, Harris TW, Stein LD. 2004. WormBase as an integrated platform for the C. elegans ORFeome. Genome Res. 14(10B), 2155–2161. ( 10.1101/gr.2521304) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Harris TW, et al. 2010. WormBase: a comprehensive resource for nematode research. Nucleic Acids Res. 38(Database issue), D463–D467. ( 10.1093/nar/gkp952) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.O'Rourke EJ, Conery AL, Moy TI. 2009. Whole-animal high-throughput screens: the C. elegans model. Methods Mol. Biol. 486, 57–75. ( 10.1007/978-1-60327-545-3_5) [DOI] [PubMed] [Google Scholar]
- 86.Irazoqui JE, Urbach JM, Ausubel FM. 2010. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat. Rev. Immunol. 10, 47–58. ( 10.1038/nri2689) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Muhammed M, Coleman JJ, Mylonakis E. 2012. Caenorhabditis elegans: a nematode infection model for pathogenic fungi. Methods Mol. Biol. 845, 447–454. ( 10.1007/978-1-61779-539-8_31) [DOI] [PubMed] [Google Scholar]
- 88.Dierking K, Yang W, Schulenburg H. 2016. Antimicrobial effectors in the nematode Caenorhabditis elegans: an outgroup to the Arthropoda. Phil. Trans. R. Soc. B 371, 20150299 ( 10.1098/rstb.2015.0299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schulenburg H, Kurz CL, Ewbank JJ. 2004. Evolution of the innate immune system: the worm perspective. Immunol. Rev. 198, 36–58. ( 10.1111/j.0105-2896.2004.0125.x) [DOI] [PubMed] [Google Scholar]
- 90.Muhammed M, Fuchs BB, Wu MP, Breger J, Coleman JJ, Mylonakis E. 2012. The role of mycelium production and a MAPK-mediated immune response in the C. elegans-Fusarium model system. Med. Mycol. 50, 488–496. ( 10.3109/13693786.2011.648217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tampakakis E, Okoli I, Mylonakis E. 2008. A C. elegans-based, whole animal, in vivo screen for the identification of antifungal compounds. Nat. Protoc. 3, 1925–1931. ( 10.1038/nprot.2008.193) [DOI] [PubMed] [Google Scholar]
- 92.Pukkila-Worley R, Holson E, Wagner F, Mylonakis E. 2009. Antifungal drug discovery through the study of invertebrate model hosts. Curr. Med. Chem. 16, 1588–1595. ( 10.2174/092986709788186237) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moy TI, Ball AR, Anklesaria Z, Casadei G, Lewis K, Ausubel FM. 2006. Identification of novel antimicrobials using a live-animal infection model. Proc. Natl Acad. Sci. USA 103, 10 414–10 419. ( 10.1073/pnas.0604055103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Breger J, Fuchs BB, Aperis G, Moy TI, Ausubel FM, Mylonakis E. 2007. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog. 3, e18 ( 10.1371/journal.ppat.0030018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Desalermos A, Muhammed M, Glavis-Bloom J, Mylonakis E. 2011. Using C. elegans for antimicrobial drug discovery. Expert Opin. Drug Discov. 6, 645–652. ( 10.1517/17460441.2011.573781) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rajamuthiah R, Jayamani E, Conery AL, Fuchs BB, Kim W, Johnston T, Vilcinskas A, Ausubel FM, Mylonakis E. 2015. A defensin from the model beetle Tribolium castaneum acts synergistically with Telavancin and Daptomycin against multidrug resistant Staphylococcus aureus. PLoS ONE 10, e0128576 ( 10.1371/journal.pone.0128576) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jayamani E, Rajamuthiah R, Larkins-Ford J, Fuchs BB, Conery AL, Vilcinskas A, Ausubel FM, Mylonakis E. 2015. Insect-derived cecropins display activity against Acinetobacter baumannii in a whole-animal high-throughput Caenorhabditis elegans model. Antimicrob. Agents Chemother. 59, 1728–1737. ( 10.1128/AAC.04198-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baltzer SA, Brown MH. 2011. Antimicrobial peptides: promising alternatives to conventional antibiotics. J. Mol. Microbiol. Biotechnol. 20, 228–235. ( 10.1159/000331009) [DOI] [PubMed] [Google Scholar]
- 99.Narayana JL, Chen JY. 2015. Antimicrobial peptides: possible anti-infective agents. Peptides 72, 88–94. ( 10.1016/j.peptides.2015.05.012) [DOI] [PubMed] [Google Scholar]
- 100.Kruse T, Kristensen HH. 2008. Using antimicrobial host defense peptides as anti-infective and immunomodulatory agents. Expert Rev. Anti-Infect Ther. 6, 887–895. ( 10.1586/14787210.6.6.887) [DOI] [PubMed] [Google Scholar]
- 101.Kim IW, Lee JH, Kwon YN, Yun EY, Nam SH, Ahn MY, Kang DC, Hwang JS. 2013. Anticancer activity of a synthetic peptide derived from harmoniasin, an antibacterial peptide from the ladybug Harmonia axyridis. Int. J. Oncol. 43, 622–628. [DOI] [PubMed] [Google Scholar]
- 102.Mukherjee K, et al. 2011. Anti-Listeria activities of Galleria mellonella hemolymph proteins. Appl. Environ. Microbiol. 77, 4237–4240. ( 10.1128/AEM.02435-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vonkavaara M, Pavel ST, Holzl K, Nordfelth R, Sjostedt A, Stoven S. 2013. Francisella is sensitive to insect antimicrobial peptides. J. Innate Immun. 5, 50–59. ( 10.1159/000342468) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pöppel AK, Koch A, Kogel KH, Vogel H, Kollewe C, Wiesner J, Vilcinskas A. 2014. Lucimycin, an antifungal peptide from the therapeutic maggot of the common green bottle fly Lucilia sericata. Biol. Chem. 395, 649–656. ( 10.1515/hsz-2013-0263) [DOI] [PubMed] [Google Scholar]
- 105.Chernysh S, Kim SI, Bekker G, Pleskach VA, Filatova NA, Anikin VB, Platonov VG, Bulet P. 2002. Antiviral and antitumor peptides from insects. Proc. Natl Acad. Sci. USA 99, 12 628–12 632. ( 10.1073/pnas.192301899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wilmes M, Sahl HG. 2014. Defensin-based anti-infective strategies. Int. J. Med. Microbiol. 304, 93–99. ( 10.1016/j.ijmm.2013.08.007) [DOI] [PubMed] [Google Scholar]
- 107.Rodriguez-Rojas A, Makarova O, Rolff J. 2014. Antimicrobials, stress and mutagenesis. Plos Path. 10, e1004445 ( 10.1371/journal.ppat.1004445) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Joo H-S, Fu C-L, Otto M. 2016. Bacterial strategies of resistance to antimicrobial peptides. Phil. Trans. R. Soc. B 371, 20150292 ( 10.1098/rstb.2015.0292) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Prenner EJ, Lewis RN, McElhaney RN. 1999. The interaction of the antimicrobial peptide gramicidin S with lipid bilayer model and biological membranes. Biochim. Biophys. Acta 1462, 201–221. ( 10.1016/S0005-2736(99)00207-2) [DOI] [PubMed] [Google Scholar]
- 110.Hilpert K, Volkmer-Engert R, Walter T, Hancock RE. 2005. High-throughput generation of small antibacterial peptides with improved activity. Nat. Biotech. 23, 1008–1012. ( 10.1038/nbt1113) [DOI] [PubMed] [Google Scholar]
- 111.Vlieghe P, Losowski V, Martinez J, Khrestchatisky M. 2010. Synthetic therapeutic peptides: science and market. Drug Disc. Today 15, 40–56. ( 10.1016/j.drudis.2009.10.009) [DOI] [PubMed] [Google Scholar]
- 112.Kollewe C, Vilcinskas A. 2013. Production of recombinant proteins in insect cells. Am. J. Biochem. Biotechnol. 9, 255–271. ( 10.3844/ajbbsp.2013.255.271) [DOI] [Google Scholar]
- 113.Druzinec D, Salzig D, Brix A, Kraume M, Vilcinskas A, Kollewe C, Czermak P. 2013. Optimization of insect cell based protein production processes—online monitoring, expression systems, scale up. Adv. Biochem. Eng. Biotechnol. 136, 65–100. ( 10.1007/10_2013_205) [DOI] [PubMed] [Google Scholar]
- 114.Müller H, Salzig D, Czermak P. 2015. Considerations for the process development of insect-derived antimicrobial peptide production. Biotechnol. Prog. 3, 1–11. ( 10.1002/btpr.2002) [DOI] [PubMed] [Google Scholar]
- 115.Berthold N, Czihal P, Fritsche S, Sauer U, Schiffer G, Knappe D, Alber G, Hoffmann R. 2013. Novel apidaecin 1b analogs with superior serum stabilities for treatment of infections by Gram-negative pathogens. Antimicrob. Agents Chemother. 57, 402–409. ( 10.1128/AAC.01923-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Baghian A, Jaynes J, Enright F, Kousoulas KG. 1997. An amphipathic alpha-helical synthetic peptide analogue of melittin inhibits herpes simplex virus-1 (HSV-1)-induced cell fusion and virus spread. Peptides 18, 177–183. ( 10.1016/S0196-9781(96)00290-2) [DOI] [PubMed] [Google Scholar]
- 117.Palusińska-Szysz M, Zdybicka-Barabas A, Pawlikowska-Pawlęga B, Mak P, Cytryńska M. 2012. Anti-Legionella dumoffii activity of Galleria mellonella defensin and apolipophorin III. Int. J. Mol. Sci. 13, 17 048–17 064. ( 10.3390/ijms131217048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Steinstraesser L, et al. 2008. Host defense peptides in wound healing. Mol. Med. 14, 528–537. ( 10.2119/2008-00002.Steinstraesser) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rahnamaeian M, Vilcinskas A. 2015. Short antimicrobial peptides as cosmetic ingredients to deter dermatological pathogens. App. Microbiol. Biotechnol. 99, 8847–8855. ( 10.1007/s00253-015-6926-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wedde M, Weise C, Nuck R, Altincicek B, Vilcinskas A. 2007. The insect metalloproteinase inhibitor gene of the lepidopteran Galleria mellonella encodes two distinct inhibitors. Biol. Chem. 388, 119–127. ( 10.1515/BC.2007.013) [DOI] [PubMed] [Google Scholar]
- 121.Eisenhardt M, et al. 2015. Development of an insect metalloproteinase inhibitor drug carrier system for application in chronic wound infections. J. Pharm. Pharmacol. 67, 1481–1491. ( 10.1111/jphp.12452) [DOI] [PubMed] [Google Scholar]
- 122.Lo JCY, Lange D. 2015. Current and potential applications of host-defense peptides and proteins in urology. Biomed. Res. Int. 2015, 189016 ( 10.1155/2015/189016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brandt CR. 2014. Peptide therapeutics for treating ocular surface infections. J. Ocular Pharmacol. Therpeutics 30, 691–699. ( 10.1089/jop.2014.0089) [DOI] [PMC free article] [PubMed] [Google Scholar]