Abstract
Insects show long-lasting antimicrobial immune responses that follow the initial fast-acting cellular processes. These immune responses are discussed to provide a form of phrophylaxis and/or to serve as a safety measure against persisting infections. The duration and components of such long-lasting responses have rarely been studied in detail, a necessary prerequisite to understand their adaptive value. Here, we present a 21 day proteomic time course of the mealworm beetle Tenebrio molitor immune-challenged with heat-killed Staphylococcus aureus. The most upregulated peptides are antimicrobial peptides (AMPs), many of which are still highly abundant 21 days after infection. The identified AMPs included toll and imd-mediated AMPs, a significant number of which have no known function against S. aureus or other Gram-positive bacteria. The proteome reflects the selective arena for bacterial infections. The results also corroborate the notion of synergistic interactions in vivo that are difficult to model in vitro.
This article is part of the themed issue ‘Evolutionary ecology of arthropod antimicrobial peptides’.
Keywords: Tenebrio molitor, antimicrobial peptides, long-lasting immunity, proteomics
1. Introduction
Persistent infections are not only of medical importance, but are also very common throughout the animal kingdom. Studying organisms that use antimicrobial peptides (AMPs) as important components of their defence cascade can provide interesting insights into understanding persistent infections. Many pathogens establish persistent infections. Examples from insects include gregarines [1], Plasmodium [2] and a variety of intracellular bacteria such as Wolbachia [3]. While intracellular pathogens are in immune-privileged sites, even pathogens exposed to the immune system sometimes go unrecognized. Examples include the bacterium Spiroplasma, which can be prevalent in the haemolymph of Drosophila, but is cleared if the immune system is upregulated by another challenge [4]. Microsporidia can form persistent infections in the haemocoel, sometimes without apparent fitness costs to the host [5].
Persistent associations between hosts and microbes are often not pathogenic; mutualists in insects, for example, establish persistent colonization of host organs and provide benefits to the hosts [6]. Such persistent associations by mutualists are maintained by AMPs [7].
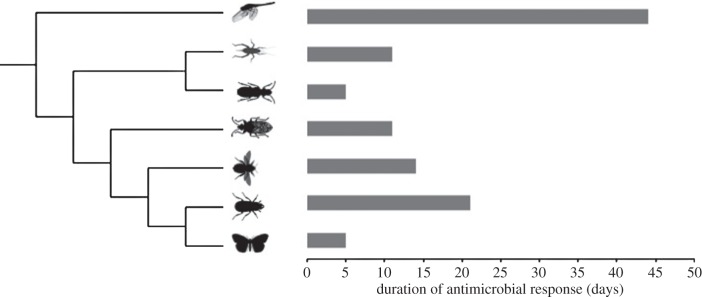
Infections, persistent or not, can result in long-lasting upregulation of immune defences, as exemplified by a study in the mealworm beetle, Tenebrio molitor. Upon infection, antimicrobial activity in the haemolymph is elevated for at least three weeks [8], which constitutes a third of adult lifespan before the onset of senescence under laboratory conditions [9]. Upregulation of antimicrobial defences for more than five days has only been described for a very limited number of insect species from several orders (figure 1). Most of these studies were not designed to estimate the duration of upregulated immune function and hence immune responses reported there lasted at least as long, but almost certainly longer, than reported. For most insect species that are commonly used to study immunity or infection, data on the duration of antimicrobials are not available. Given that the available data are spread across different taxa, we propose that the phenomenon of long-lasting antimicrobial responses is rather common.
Figure 1.
Long-lasting immune responses in insects. These are reported data that depict the minimum duration, as all but one of the studies have not been designed to investigate the duration of antimicrobial immune responses (references for data are in [10]). Insect silhouettes from phylopic.org.
Given that maintaining an immune response is costly [11], what are the adaptive benefits of such sustained immune responses? Using T. molitor, Moret & Siva-Jothy [12] found that beetles immune-challenged with LPS (bacterial cell wall components) better survived fungal infections that were applied up to seven days after the immune challenge. They proposed some form of ‘adaptive prophylaxis’. This notion hinges on the assumption that a first infection is an honest signal for a secondary infection: an infection mirrors the risk of future infections in the individual's environment. An alternative explanation is to assume that wounding, which happens not only during artificial infections but, for example, also during parasitoid attack, may result in opportunistic bacterial infections.
This has led us to propose, using the beetle T. molitor as a model, that the function of long-lasting antimicrobial immunity and hence a main role of AMPs is mopping up and policing persistent infections, a proposal already mentioned by Dunn [13]. Haine et al. [14] reported that the vast majority of a high-dose Staphylococcus aureus infection is cleared within minutes and the inducible AMP-related response is only measurable after several hours but subsequently maintained for several days. Johnston et al. [15] using RNA-seq supported this view.
This view also suggests a two-stage immune response, where cellular and constitutive components act as the fast-reacting means, and the inducible AMP-based immune response deals with the surviving pathogens (a view recently also considered for vertebrate innate immunity [16]). Recent work in Drosophila on growth-blocking peptides (GBP) supports this notion [17]. GBP mediates cellular immune responses and simultaneously suppresses AMP expression via Pvf2 or Pvr. Noh et al. [18] studied the upregulation of apolipohorin at 72 h in T. molitor and found also patterns consistent with a fast-acting cellular response separated from the antimicrobial response. Work in other insects has also shown that apolipophorin contributes to the induction of AMP and suppression of nodule formation.
AMPs were first identified in T. molitor over 20 years ago. The known repertoire of AMPs varies in the specificity of their antimicrobial activity, but collectively exhibits activity against Gram-negative and -positive bacteria, as well as fungi. Tenecin-1, an AMP which is active specifically against Gram-positive bacteria, was first identified by purifying the peptide from a haemolymph fraction showing antimicrobial activity [19]. Tenecin-2 was identified simultaneously [20] and is active against Gram-negative bacteria. Tenecin-3 is antifungal [21], while Tenecin-4 is anti-Gram negative [22]. These inducible humoral defences complement the activity of constitutive immune effectors such as haemocytes and oxidative enzymes [23], including components of the phenoloxidase system, which have been characterized biochemically more completely in T. molitor than in other insects [24]. The discovery of long-lasting immune responses prompted more systematic study of T. molitor's immune system, complementing what had been discovered by biochemical studies. By sequencing mRNAs that were over-expressed in Tenebrio larvae infected with S. aureus, Dobson et al. [25] identified an upregulation of number of novel immune-induced transcripts, including two AMPs (a coleoptericin and an attacin), a range of redox enzymes, and some genes that may suppress bacterial growth metabolically, e.g. iron-chelating ferritins and transferrins. The coupled upregulation of AMPs that kill both Gram-positive and -negative bacteria, in response to infection with only a Gram-positive bacterium, indicates that expression of the Imd and Toll pathway may be molecularly coupled.
Long-lasting immunity in insects at the transcriptomic level has hardly been studied. Also, transcriptomics of immune function and proteomics have surprisingly rarely been studied in the same system [26]. While the transcriptomic approach certainly contributes to our understanding of the host side of an infection, a proteomic approach, by determining the amounts and concentrations of antimicrobials, allows better to capture the selective arena in which pathogenic bacteria are situated. Here, we provide a three-week proteomic time course of immunoproteins in T. molitor infected with S. aureus from whole beetle homogenates and relate these findings to a transcriptomic study over seven days [15].
2. Material and methods
(a). Insect culturing
Final-instar T. molitor larvae purchased from a commercial supplier (Futtertier-shop, Germany) were reared in open boxes containing wheat bran (Wilhelm Ströh jun. GMBH & Co. KG, Lübeck, Germany) supplemented with fresh apple at 30°C under dark conditions. When the animals reached the pupal stage, females were selected and placed individually into compartments of lidded grid boxes. Emergence of adults was checked bi-daily. After emergence, each beetle was provided with wheat bran, 5 × 5 mm2 of filter paper and a 2 × 2 mm slice of apple. Animals aged 7–14 days post-eclosion were used for the immune challenge experimental procedures.
(b). Bacterial cultures
Heat-killed S. aureus strain SH1000 was used as inoculum for bacterial injections. Bacterial culture of S. aureus was grown overnight at 37°C and 220 r.p.m. in LB medium, then centrifuged at 10 000 g for 10 min at 4°C, washed, and resuspended in the same volume with sterile PBS, heat-killed at 95°C for 5 min, dispensed into 1-ml aliquots, and stored at −80°C until further use.
(c). Immune challenge experiments
Adult female beetles aged 7–14 days were split into three treatment groups—non-injected control group, procedural control (PBS-injected) group and immune-challenged (injected with heat-killed S. aureus) group—and were followed for 21 days. Five microlitres of inoculum or sterile PBS were injected using sterile glass capillary needles into each beetle's haemocoel into the space between the second and third abdominal sternites that were first swabbed with 70% ethanol. Injected beetles and healthy controls were maintained individually as described above until collection. Ten individuals per time point per treatment were collected at 30 min, 7 days and 21 days post-inoculation, flash-frozen in liquid nitrogen and stored at −80°C until further use.
(d). Protein extraction
Frozen beetles (two individuals per biological replicate) were pulverized in liquid nitrogen. Approximately 20 µg of ground tissue were transferred into 1.5 ml tubes containing 200 µl of denaturation buffer (6 M urea/2 M thiourea in 10 mM HEPES, pH 8.0) supplemented with 20 mM DTT, incubated for 5 min and centrifuged for 10 min at 4°C and 20 000 g. Ten microlitres of the supernatant were used for sample preparation for mass spectrometry.
(e). Preparation of protein samples for mass spectrometry
Four biological replicates per treatment per group were used for mass spectrometry. In total, 20 µl of denaturation buffer were added to 10 µl of protein samples and used for in-solution protein digestion as described previously [27]. Briefly, proteins, solubilized in denaturation buffer, were reduced with 10 mM DTT for 30 min, followed by alkylation with 55 mM iodoacetamide for 20 min, and overnight digestion with 1 µg lysyl endopeptidase (LysC) (catalogue number 125-05061, Wako, Japan), resuspended in 50 mM ammonium bicarbonate (ABC). After pre-digestion with LysC, protein samples were diluted fourfold with 50 mM ABC and subjected to overnight trypsin digestion using 1 µg of sequencing-grade modified trypsin (catalogue number V5111, Promega, Madison, USA), diluted in 50 mM ABC buffer. All in-solution protein digestion steps were performed at room temperature and after addition of iodoacetamide the samples were protected from light. After trypsin digestion overnight at room temperature, the reaction was stopped by adding an equal volume of Buffer A* (5% acetonitrile, 3% trifluoroacetic acid). Samples were then desalted using Empore C18 SD 4 mm/1 ml solid phase extraction (SPE) cartridges (catalogue number 66871-U, SIGMA-Aldrich, Taufkirchen, Germany).
(f). Mass spectrometry and statistical analyses
Peptides were separated by reverse-phase chromatography using a Dionex Ultimate 3000 nanoLC (Thermo Fisher Scientific, Bremen, Germany) with a 5–60% acetonitrile gradient (90 min) and 0.1% formic acid at a flow rate of 350 nl min−1 on in-house manufactured 25 cm fritless silica microcolumns with an inner diameter of 100 µm packed with ReproSil-Pur C18-AQ 3 µm resin (Dr Maisch GmbH, Ammerbuch-Entringen, Germany). Eluting peptides were ionized online by electrospray ionization and transferred into an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). The LTQ Orbitrap was operated in the positive mode to simultaneously measure full scan MS spectra (from m/z 300–1700) in the Orbitrap analyser at resolution R = 60 000 following isolation and fragmentation of the 20 most intense ions in the LTQ part by collision-induced dissociation (CID).
MS and MS/MS data from each LC/MS run were analysed using MaxQuant software (v. 1.5.3.30). Identification of proteins was performed using the MaxQuant-implemented Andromeda peptide search engine and the in-house prepared protein database of T. molitor, which was based on the reverse-translated RNA-seq data for predicted proteins [15]. Label-free quantification of proteins was performed using the label-free quantification algorithm MaxLFQ of the MaxQuant software package [10]. Statistical analysis was performed with Perseus software (v. 1.5.2.4), using the Kolmogorov–Smirnov test, and all values with p ≤ 0.05 were considered significant.
3. Results
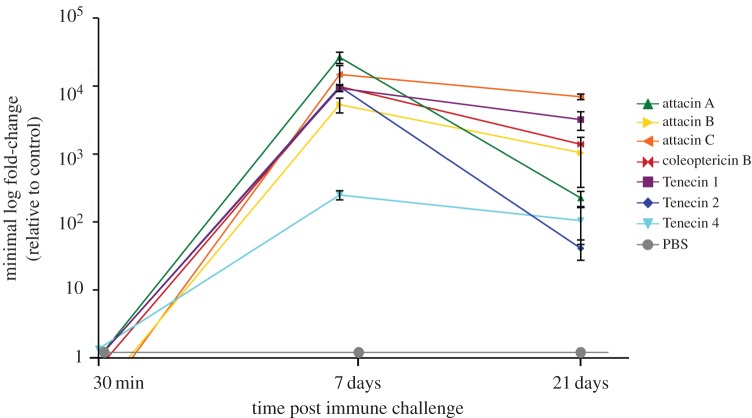
The proteome analysis identified 1669 proteins in total, of which 306 proteins were differentially expressed when comparing samples from infected beetles and procedural controls with no treatment controls. Of those differentially expressed proteins, 184 were upregulated and 117 were downregulated in infected beetles, whereas only 3 and 2 were up- and downregulated, respectively, in the procedural controls. Thirty minutes after infection only two proteins were up- or downregulated in infected beetles. At day 7 post-inoculation, 87 proteins were upregulated and 80 proteins were downregulated, while 97 and 35 proteins were up- and downregulated at 21 dpi, respectively. However, in terms of AMP expression, the strongest effects were visible on day 7 (figure 2) in infected beetles, while no expression of AMPs was detected in sham-injected individuals.
Figure 2.
Peptide abundance of AMPs in T. molitor over 21 days after immune challenge with heat-killed S. aureus. Depicted are the known AMPs Tenecin 1–4 and putative AMPs.
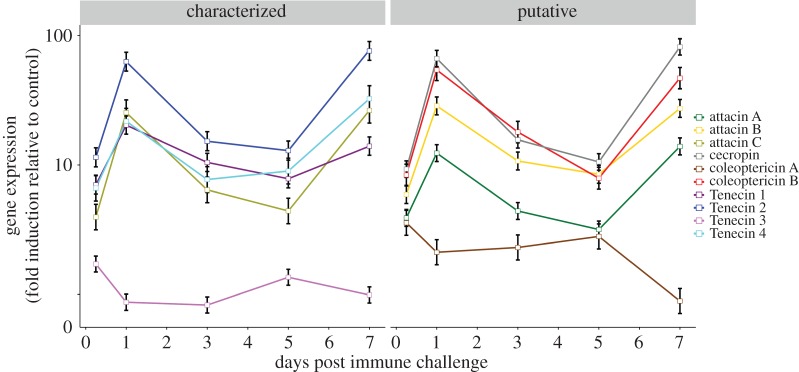
Here, we focus on the expression of AMPs (figure 2; electronic supplementary material, tables S1–S3). All three previously described inducible Tenecins (1,2,4) were highly expressed at day 7. In addition, our study revealed four other AMPs, mostly attacins. These AMPs overlapped with the transcriptomic data on day 7 (figure 3), though it has to be noted that the proteomic data were based on whole-body lysate. The expression of AMPs dropped on day 21 but remained significantly upregulated, whereas phenoloxidases were no longer differentially expressed (see electronic supplementary material, figure S1).
Figure 3.
Gene expression of AMPs in T. molitor over a 7 day time course of infection with S. aureus. Depicted are the known AMPs Tenecin 1–4 in for which functional data are available (see text), and putative AMPs and an attacin described [25]. Adapted from Johnston et al. [15].
A gene ontology analysis for the proteins with increased and reduced abundance yielded four main groupings (figure 4). The two categories capturing the most abundant peptides were defence against bacteria and regulation of haemocyte differentiation. ‘Fatty acid metabolic process' and ‘cellular amino acid metabolism’ accounted for the majority of proteins with decreased abundance (for a more detailed description, see electronic supplementary material, table S4).
Figure 4.
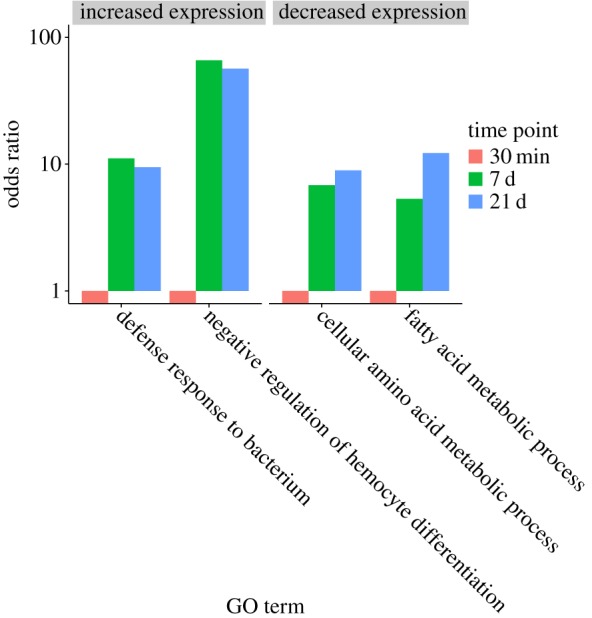
GO terms for proteins with significant changes in abundance over the time course. (Online version in colour.)
4. Discussion
The main inducible and characterized AMPs of Tenebrio, as well a significant number of putative AMPs, were highly abundant for up to three weeks. This corroborates the results of Haine et al. [8], who found, using zone of clearance assays, that the haemolymph after infection shows long-lasting antimicrobial activity mostly resulting from a cocktail of AMPs. Such long-lasting upregulation results in an antimicrobial environment for persistent infections. A very small proportion (more than 1% of the original inoculum) of S. aureus persists for at least 21 days in T. molitor, both in phagocytes and in the haemocoel, [28]. We have shown before at the transcript level [15] that AMPs are expressed for at least 7 days, and studies in other insects (figure 1) based on mostly functional clearance assays found long-lasting immunity. Here, we present the first comprehensive study on AMP abundance over a three-week time course.
The AMPs identified here show a strong overlap with the AMPs identified in the transcriptomic study by Johnston et al. [15]. Though both studies were carried out in Tenebrio, the proteomics used whole-body homogeneates, while the transcriptomic study was based on fat bodies, the organ that synthesizes AMPs. The number of AMPs discovered by both, proteomics and RNA-seq, is comparable to the number of AMPs in many other insect species [29]. Recently, a new group of very short AMPs was described in Drosophila dubbed Bomanins [30]. We could not find any evidence for the existence of this type of peptides either in our transcriptomic or in our proteomic data.
That several AMPs are simultaneously expressed upon infection has been shown in earlier studies. Sun et al. [31], for example, reported co-expression of attacins in a moth. If AMPs occur in cocktails, this could be explained either by a lack of specific pathogen recognition or alternatively by synergistic interactions between AMPs. Strain-specific responses have been described in bumblebees [32], yet the degree and mechanism of specificity is unknown. Work by Westerhoff et al. [33] found synergistic interactions between AMPs on frog skin; recent work in insects has demonstrated synergisms between AMPs against bacteria as well as trypanosomes [34,35]. These studies, in the light of our results showing long-lasting high abundance of AMPs in Tenebrio, are consistent with the idea that the composition of the antimicrobial cocktails reflects synergistic interactions. Using a suite of commercially available AMPs, including cecropin and mellitin as insect AMPs, it was found that AMPs are broadly synergistic [36]. Synergism was even more pronounced in mixtures of three AMPs in comparisons with combinations of two AMPs. In this issue, Baeder et al. [37] expand on these findings and develop a theoretical approach to capture the nature of interactions between AMPs.
AMP genes are exceptional immune genes, as they do not display any signatures of fast evolution that is typical for immune genes [38]. Lazzaro & Unckless by reanalysing data from several Drosophila species found that AMPs are under stabilizing selection. The nature of the selective forces is currently unclear. Synergisms between AMPs might well contribute to such stabilizing selection. An intriguing aspect of our proteomics data and the one-week transcriptomic time course [15] is that some of the AMPs that are upregulated have no known activity against the infective agent in our experiments. This is of interest because the toll-regulated coleoptericin Tenecin-2 seems to interact synergistically in vivo with Tenecin 1 (C. Zanchi, P. Johnston, J. Rolff, unpublished data, 2016).
The gene ontology analysis revealed that the majority of peptides with reduced abundance are categorized either as ‘metabolic’ or ‘catabolic’. This finding is highly consistent with the notion of a metabolic cost of mounting an immune response [11]. Again, the data are very consistent with the findings of our previous transcriptomic study.
Persistent infections are causing major problems in the treatment of infectious diseases [39,40]. While more and more mechanisms enabling bacterial cells to persist in antimicrobial environments have been elucidated [41], much less is known about effective treatments of persistent infections. One important avenue of current research aims to use drugs that target the bacterial membranes [39]. This is based on the notion that it is harder to evolve resistance against drugs targeting the membranes, and also because some recent studies yielded promising results in treating biofilms with membrane-targeting drugs such as lipoglycopeptides and AMPs. Studying the interactions of AMPs in natural systems such as the insect immune system has the potential to inform our understanding and application of AMPs.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
For mass spectrometry (M.E. and C.W.), we would like to acknowledge the assistance of the Core Facility BioSupraMol supported by the Deutsche Forschungsgemeinschaft (DFG).
Data accessibility
The proteomics data are available on request.
Authors' contributions
O.M., A.R.R. and J.R. conceived the study. O.M., A.R.R., M.E., C.W. and J.R. designed the experiment. O.M., A.R.R., M.E. and C.W. carried out the experiment. M.E., O.M., A.R.R. and P.R.J. analysed the data. All authors contributed to the writing.
Competing interests
We have no competing interests.
Funding
J.R., O.M. and P.R.J. were funded by ERCgrant 260986 EVORESIN. A.R.R. and J.R. were funded by SFB 973 (DFG), M.E. was funded by SFB 958 (DFG).
References
- 1.Siva-Jothy MT, Plaistow SJ. 1999. A fitness cost of eugregarine parasitism in a damselfly. Ecol. Entomol. 25, 465–470. ( 10.1046/j.1365-2311.1999.00222.x) [DOI] [Google Scholar]
- 2.Michel K, Kafatos FC. 2005. Mosquito immunity against Plasmodium. Insect Biochem. Mol. Biol. 35, 677–689. ( 10.1016/j.ibmb.2005.02.009) [DOI] [PubMed] [Google Scholar]
- 3.Zug R, Hammerstein P. 2014. Bad guys turned nice? A critical assessment of Wolbachia mutualisms in arthropod hosts. Biol. Rev. 90, 89–111. ( 10.1111/brv.12098) [DOI] [PubMed] [Google Scholar]
- 4.Hurst GDD, Anbutsu H, Kutsukake M, Fukatsu T. 2003. Hidden from the host: Spiroplasma bacteria infecting Drosophila do not cause an immune response, but are suppressed by ectopic immune activation. Insect Mol. Biol. 12, 93–97. ( 10.1046/j.1365-2583.2003.00380.x) [DOI] [PubMed] [Google Scholar]
- 5.Vilcinskas A, Stoecker K, Schmidtberg H, Röhrich CR, Vogel H. 2013. Invasive harlequin ladybird carries biological weapons against native competitors. Science 340, 862–863. ( 10.1126/science.1234032) [DOI] [PubMed] [Google Scholar]
- 6.Masson F, Zaidman-Rémy A, Heddi A. 2016. Antimicrobial peptides and cell processes tracking endosymbiont dynamics. Phil. Trans. R. Soc. B 371, 20150298 ( 10.1098/rstb.2015.0298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Login FH, Balmand S, Vallier A, Vincent-Monegat C, Vigneron A, Weiss-Gayet M, Rochat D, Heddi A. 2011. Antimicrobial peptides keep insect endosymbionts under control. Science 334, 362–365. ( 10.1126/science.1209728) [DOI] [PubMed] [Google Scholar]
- 8.Haine ER, Pollitt LC, Moret Y, Siva-Jothy MT, Rolff J. 2008. Temporal patterns in immune responses to a range of microbial insults (Tenebrio molitor). J. Insect Physiol. 54, 1090–1097. ( 10.1016/j.jinsphys.2008.04.013) [DOI] [PubMed] [Google Scholar]
- 9.Pursall ER, Rolff J. 2011. Immune responses accelerate ageing: proof-of-principle in an insect model. PLoS ONE 6, e19972 ( 10.1371/journal.pone.0019972) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. 2014. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526. ( 10.1074/mcp.M113.031591) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bajgar A, Kucerova K, Jonatova L, Tomcala A, Schneedorferova I, Okrouhlik J, Dolezal T. 2015. Extracellular adenosine mediates a systemic metabolic switch during immune response. PLoS Biol. 13, e1002135 ( 10.1371/journal.pbio.1002135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moret Y, Siva-Jothy MT. 2003. Adaptive innate immunity? Responsive-mode prophylaxis in the mealworm beetle, Tenebrio molitor. Proc. R. Soc. Lond. B 270, 2475–2480. ( 10.1098/rspb.2003.2511) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn E. 1986. Biochemical aspects of insect immunology. Ann. Rev. Entomol. 31, 321–339. [Google Scholar]
- 14.Haine ER, Moret Y, Siva-Jothy MT, Rolff J. 2008. Antimicrobial defense and persistent infection in insects. Science 322, 1257–1259. ( 10.1126/science.1165265) [DOI] [PubMed] [Google Scholar]
- 15.Johnston PR, Makarova O, Rolff J. 2013. Inducible defenses stay up late: temporal patterns of immune gene expression in Tenebrio molitor. G3 (Bethesda) 4, 947–955. ( 10.1534/g3.113.008516) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mills CD, Ley K, Buchmann K, Canton J. 2015. Sequential immune responses: the weapons of immunity. J. Innate Immun. 7, 443–449. ( 10.1159/000380910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsuzuki S, et al. 2014. Switching between humoral and cellular immune responses in Drosophila is guided by the cytokine GBP. Nat. Commun. 5, 4628 ( 10.1038/ncomms5628) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noh JY, et al. 2014. Genomic organization, sequence characterization and expression analysis of Tenebrio molitor apolipophorin-III in response to an intracellular pathogen, Listeria monocytogenes. Gene 534, 204–217. ( 10.1016/j.gene.2013.10.058) [DOI] [PubMed] [Google Scholar]
- 19.Moon HJ, Al E. 1994. Purification and molecular cloning of cDNA for an inducible antibacterial protein from larvae of the coleopteran, Tenebrio molitor. J. Biochem. 116, 53–58. [DOI] [PubMed] [Google Scholar]
- 20.Roh K-B, et al. 2009. Proteolytic cascade for the activation of the insect toll pathway induced by the fungal cell wall component. J. Biol. Chem. 284, 19474–19 481. ( 10.1074/jbc.M109.007419) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim DH. 1998. Bacterial expression of Tenecin 3, an insect antifungal protein isolated from Tenebrio molitor, and its efficient purification. Mol. Celss 8, 786–789. [PubMed] [Google Scholar]
- 22.Chae J-H, Kurokawa K, So Y-I, Hwang HO, Kim M-S, Park J-W, Jo Y-H, Lee YS, Lee BL. 2011. Purification and characterization of tenecin 4, a new anti-Gram-negative bacterial peptide, from the beetle Tenebrio molitor. Dev. Comp. Immunol. 36, 540–546. ( 10.1016/j.dci.2011.09.010) [DOI] [PubMed] [Google Scholar]
- 23.Siva-Jothy MT, Moret Y, Rolff J. 2005. Insect immunity: an evolutionary ecology perspective. Adv. Insect Phys. 32, 1–48. ( 10.1016/S0065-2806(05)32001-7) [DOI] [Google Scholar]
- 24.Kan H, et al. 2008. Molecular control of phenoloxidase-induced melanin synthesis in an insect. J. Biol. Chem. 283, 25 316–25 323. ( 10.1074/jbc.M804364200) [DOI] [PubMed] [Google Scholar]
- 25.Dobson AJ, Johnston PR, Vilcinskas A, Rolff J. 2012. Identification of immunological expressed sequence tags in the mealworm beetle Tenebrio molitor. J. Insect Physiol. 58, 1556–1561. ( 10.1016/j.jinsphys.2012.09.009) [DOI] [PubMed] [Google Scholar]
- 26.Yang W, Dierking K, Esser D, Tholey A, Leippe M, Rosenstiel P, Schulenburg H. 2015. Overlapping and unique signatures in the proteomic and transcriptomic responses of the nematode Caenorhabditis elegans toward pathogenic Bacillus thuringiensis. Dev. Comp. Immunol. 51, 1–9. ( 10.1016/j.dci.2015.02.010) [DOI] [PubMed] [Google Scholar]
- 27.Sury MD, McShane E, Hernandez-Miranda LR, Birchmeier C, Selbach M. 2015. Quantitative proteomics reveals dynamic interaction of c-Jun N-terminal Kinase (JNK) with RNA transport granule proteins splicing factor proline- and glutamine-rich (Sfpq) and non-POU domain-containing octamer-binding protein (nono) during neuronal differ. Mol. Cell. Proteomics 14, 50–65. ( 10.1074/mcp.M114.039370) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGonigle JE, Purves J, Rolff J. 2016. Intracellular survival of Staphylococcus aureus during persistent infection in the insect Tenebrio molitor. Dev. Comp. Immunol. 59, 34–38. ( 10.1016/j.dci.2016.01.002) [DOI] [PubMed] [Google Scholar]
- 29.Viljakainen L. 2015. Evolutionary genetics of insect innate immunity. Brief. Funct. Genomics 14, 407–412. ( 10.1093/bfgp/elv002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clemmons AW, Lindsay SA, Wasserman SA. 2015. An effector peptide family required for Drosophila toll-mediated immunity. PLoS Pathog. 11, e1004876 ( 10.1371/journal.ppat.1004876) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun SC, Lindström I, Lee JY, Faye I. 1991. Structure and expression of the attacin genes in Hyalophora cecropia. Eur. J. Biochem. 196, 247–254. ( 10.1111/j.1432-1033.1991.tb15811.x) [DOI] [PubMed] [Google Scholar]
- 32.Barribeau SM, Sadd BM, du Plessis L, Schmid-Hempel P. 2014. Gene expression differences underlying genotype-by-genotype specificity in a host-parasite system. Proc. Natl Acad. Sci. USA 111, 3496–3501. ( 10.1073/pnas.1318628111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westerhoff HV, Zasloff M, Rosner JL, Hendler RW, De Waal A, Vaz Gomes A, Jongsma PM, Riethorst A, Juretić D. 1995. Functional synergism of the magainins PGLa and magainin-2 in Escherichia coli, tumor cells and liposomes. Eur. J. Biochem. 228, 257–264. ( 10.1111/j.1432-1033.1995.0257n.x) [DOI] [PubMed] [Google Scholar]
- 34.Rahnamaeian M, et al. 2015. Insect antimicrobial peptides show potentiating functional interactions against Gram-negative bacteria. Proc. R. Soc. B 282, 20150293 ( 10.1098/rspb.2015.0293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marxer M, Vollenweider V, Schmid-Hempel P. 2016. Insect antimicrobial peptides act synergistically to inhibit a trypanosome parasite. Phil. Trans. R. Soc. B 371, 20150302 ( 10.1098/rstb.2015.0302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu G, Baeder DY, Regoes RR, Rolff J. 2016. The more the better? Combination effects of antimicrobial peptides. Antimicrob. Agents Chemother. 60, 1–9. ( 10.1128/AAC.01428-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baeder DY, Yu G, Hozé N, Rolff J, Regoes RR. 2016. Antimicrobial combinations: Bliss independence and Loewe additivity derived from mechanistic multi-hit models. Phil. Trans. R. Soc. B 371, 20150294 ( 10.1098/rstb.2015.0294) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Unckless RL, Lazzaro BP. 2016. The potential for adaptive maintenance of diversity in insect antimicrobial peptides. Phil. Trans. R. Soc. B 371, 20150291 ( 10.1098/rstb.2015.0291) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hurdle JG, O'Neill AJ, Chopra I, Lee RE. 2011. Targeting bacterial membrane function: an underexploited mechanism for treating persistent infections. Nat. Rev. Microbiol. 9, 62–75. ( 10.1038/nrmicro2474) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levin BR, Rozen DE. 2006. Non-inherited antibiotic resistance. Nat. Rev. Microbiol. 4, 556–562. ( 10.1038/nrmicro1445) [DOI] [PubMed] [Google Scholar]
- 41.Lewis K. 2010. Persister cells. Annu. Rev. Microbiol. 64, 357–372. ( 10.1146/annurev.micro.112408.134306) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The proteomics data are available on request.