Abstract
Many insects sustain long-term relationships with intracellular symbiotic bacteria that provide them with essential nutrients. Such endosymbiotic relationships likely emerged from ancestral infections of the host by free-living bacteria, the genomes of which experience drastic gene losses and rearrangements during the host–symbiont coevolution. While it is well documented that endosymbiont genome shrinkage results in the loss of bacterial virulence genes, whether and how the host immune system evolves towards the tolerance and control of bacterial partners remains elusive. Remarkably, many insects rely on a ‘compartmentalization strategy’ that consists in secluding endosymbionts within specialized host cells, the bacteriocytes, thus preventing direct symbiont contact with the host systemic immune system. In this review, we compile recent advances in the understanding of the bacteriocyte immune and cellular regulators involved in endosymbiont maintenance and control. We focus on the cereal weevils Sitophilus spp., in which bacteriocytes form bacteriome organs that strikingly evolve in structure and number according to insect development and physiological needs. We discuss how weevils track endosymbiont dynamics through at least two mechanisms: (i) a bacteriome local antimicrobial peptide synthesis that regulates endosymbiont cell cytokinesis and helps to maintain a homeostatic state within bacteriocytes and (ii) some cellular processes such as apoptosis and autophagy which adjust endosymbiont load to the host developmental requirements, hence ensuring a fine-tuned integration of symbiosis costs and benefits.
This article is part of the themed issue ‘Evolutionary ecology of arthropod antimicrobial peptides’.
Keywords: endosymbiosis, antimicrobial peptide, Coleoptericin, Sitophilus, bacteriocyte
1. Introduction
Among the striking attributes of insects is their ability to share long-term relationships with intracellular symbiotic bacteria (endosymbionts) [1]. Particularly frequent in species thriving on nutritionally unbalanced environments, such endosymbioses are believed to impact several aspects of host biology, including physiology, immunity and reproduction [2–8]. They ensure a nutritional complementation of the host diet by providing vitamins and essential amino acids [3,7,9,10], thus greatly improving host fitness [2,3,7,11,12]. Early on during insect embryogenesis, endosymbionts are housed within specific host cells, the bacteriocytes, which group together to form the bacteriome organ in some insect species. They also infect permanently the host germ cells, from which they are transmitted to progeny. While insect bacteriomes have been thoroughly investigated in terms of physiology and metabolism, little is known about the cellular functions and immune regulatory mechanisms operating within the bacteriocytes, which allow the tolerance of bacteria and the control of their density and location through the host life cycle.
Long-term host–symbiont relationships often lead to a complete interdependence of both partners over evolutionary time [13–16]. Once established within the host, endosymbionts generally experience severe genome size reduction due to relaxed evolutionary pressures on genes that are redundant with host functions [17–19] or become unnecessary for the new association [20]. Among the latter, virulence genes and genes involved in the biosynthesis of cell wall components, are prone to deletion [20–22]. As these elements constitute microbe-associated molecular patterns (MAMPs) that are central for bacterial perception by insect immune pattern recognition receptors (PRRs), the molecular cross-talk used by partners to ‘manage’ their coexistence may vary according to the age of the association, and hence to the level of bacterial genome reduction. In this evolutionary context, the study of recently established symbiotic associations involving endosymbionts presenting MAMPs on their cell wall may provide relevant insights into the mechanisms operating in insect endosymbiogenesis. Comparison of such recently established associations with ancient associations involving genome-reduced endosymbionts with degenerated MAMPs may help in understanding how the mechanisms of symbiont perception and control have evolved in relation to increased symbiont integration and dependence on the host.
Weevils from the Curculionoidea superfamily share a trophic endosymbiosis with γ-proteobacteria that have been integrating insect hosts at different evolutionary periods [23–25]. Candidatus Nardonella would be the ancestral symbiont (125 million years (Myr) of this superfamily and occurs in at least eight studied genera [24]. Candidatus Sodalis pierantonius symbiosis with the cereal weevils of the genus Sitophilus spp. is hypothesized to have been established recently (less than 1 Myr) [26], probably following Candidatus Nardonella's displacement [24,26,27]. Owing to its recent association as a symbiont, S. pierantonius is a valuable model for studying the early steps of insect endosymbiogenesis, in that its genome has not suffered any drastic size reduction [28–30]. In addition to the high prevalence of pseudogenes and transposable elements [29], which are thought to favour gene rearrangements and deletions, S. pierantonius genome encodes a functional type 3 secretion system (T3SS) [21], as well as the enzymes required to synthetize the cell wall peptidoglycan, a potent inducer of the insect immune response [30]. An injection of S. pierantonius bacteria into the weevil haemolymph results in the systemic induction of antimicrobial peptide (AMP) encoding gene expression [31], attesting that the host immune system has retained the ability to mount an immune defence against its bacterial partner, and that the seclusion of endosymbionts within the bacteriome protects them from the insect's humoral and cellular responses. Remarkably, this spatial symbiont compartmentalization seems to be a convergent evolutionary strategy for the maintenance and the regulation of mutualistic bacteria in both animals and plants. Examples include the trophosome of the giant tube worms Riftia pachyptila [32], root nodules in plants [33], the stratification and regionalization of the mammalian gut [34] and the development of the light organ in the squid Euprymna scolopes [35]. Symbiont compartmentalization could thus be considered as a ‘biological strategy’ that allows organisms to manage beneficial symbionts within a limited space, while host defences are maintained in the remaining host tissues where they prevent pathogenic infections.
While this compartmentalization protects the symbionts from direct exposure to the systemic immune response, it raises the question of the functional adaptation of the host immune system inside the bacteria-bearing compartment. Indeed, bacteriocytes must ensure: (i) the control of endosymbionts, i.e. ensuring they do not break free from these specialized housing cells; (ii) the maintenance of endosymbionts, i.e. their tolerance in high number; and (iii) the modulation of the endosymbiotic load according to the host developmental stages, which has been observed to different extents in many species. Here, we will review recent data on immune responses operating within the cereal weevil bacteriome organ, and their coordination with host cellular processes, including apoptosis and autophagy, ensuring symbiosis homeostasis.
2. The antimicrobial peptide Coleoptericin A keeps endosymbionts within the bacteriome organ
The weevil bacteriome was shown to mount a limited immune response, with few immune genes being expressed under physiological conditions [31,36]. Remarkably, this organ notably expresses only one AMP coding gene, coleoptericin A (colA) [31,37], while other AMP coding genes are slightly or not expressed, which presumably helps the endosymbionts to survive within this symbiotic tissue.
Coleoptericins have been found only in coleopteran insects so far; in cereal weevils, two peptides, ColA and ColB, were identified from transcriptomic data [31,36]. Coleoptericins display a bacteriostatic activity on a broad spectrum of bacteria, including both Gram-positive and Gram-negative bacteria [38–40]. In vitro incubation of Escherichia coli with weevil ColA impairs bacterial cell division and leads to cell gigantism [39,40]. It is noteworthy that endosymbiotic bacteria have been observed as long and filamentous cells in cereal weevils and other coleopteran symbiotic species [27,41]. Bacterial cell elongation was also described in plant systems, raising the question of whether plants and animals use a similar strategy to interact with their symbionts. In 2010, Van de Velde et al. demonstrated that the factors inhibiting Rhizobium division in legumes are the nodule-specific cysteine-rich peptides (NCRs) [42]. NCRs govern rhizobia differentiation, which results in the gigantism of the bacterial cells and their inability to multiply in vitro. Size elongation has been interpreted as a terminal maturation stage of the bacterium, which results in repeated chromosome DNA replication without cell cytokinesis [43]. To determine whether ColA AMP is responsible for endosymbiont gigantism in Sitophilus weevils, and whether this process is also irreversible, RNA interference (RNAi) has been used to suppress colA expression in vivo [40]. Unlike what has been observed with the NCR-induced gigantism of Rhizobium in plants, which is irreversible, alteration of colA gene expression induced a significant size reduction of S. pierantonius. More importantly, colA extinction resulted in a loss of spatial control of endosymbionts, which gained the ability to exit the bacteriocytes and invade surrounding tissues [40]. These results, along with the relatively high amount of ColA peptide observed at the border of symbiotic tissues, led to the conclusion that ColA acts as a ‘border patrol agent’ that prevents endosymbionts from leaving the bacteriome, ensuring bacteriocyte homeostasis.
Moreover, ColA specifically targets weevil endosymbiont cytokinesis and does not inhibit DNA replication, thus leading to the production of giant polyploid bacterial cells. Both S. pierantonius and Nardonella, the ancestral endosymbiont of weevils, exhibit high polyploidy that correlates with bacterial size [40]. For example, Nardonella cells from the palm weevil Rhynchophorus ferrugineus can reach 200 µm and contain 120 chromosomes [40]. This endosymbiont polyploidy may be beneficial for the host, as it would ensure a high level of bacterial protein synthesis and metabolic capacity. It is likely one of the adaptive features generated by host–symbiont coevolution: ColA spatially restricts endosymbionts to bacteriocytes and inhibits their cytokinesis without impairing their metabolic activity and their ability to supply the host with nutritional components, highlighting the concept of ‘endosymbiont domestication’ [40,44].
To get insights into how the Sitophilus colA gene may have evolved towards these symbiotic functions, the weevil colB paralogue was analysed. Although ColB presents strong sequence similarity with ColA, both peptides have distinct functions. ColB has a low basal expression in bacteriocytes [31], and it does not generate any bacterial gigantism phenotype when incubated in vitro with E. coli. Furthermore, far-Western experiments showed interactions of ColA with the E. coli membrane proteins OmpA and OmpC, RpL2 and EF-Ts elongation factors, as well as with the chaperonin GroEL [40]. groEL deficient E. coli mutants display a filamentous bacterial phenotype resembling that of S. pierantonius, hinting that ColA–GroEL interaction may mediate the observed bacterial cytokinesis inhibition in E. coli and S. pierantonius. Interestingly, ColB was shown to interact with OmpC and elongation factors but not with the chaperonin GroEL [40], suggesting that the affinity of ColA for this protein may have been acquired during divergent evolution of these coleoptericins. Furthermore, ColA does not interact with HSP60, the eukaryotic homologue protein of GroEL, indicating that ColA strictly targets prokaryotic proteins [40].
Another interesting question with regard to colA relates to how the expression of this gene is regulated in the bacteriome. In Drosophila melanogaster, AMP synthesis relies on the activation of the Toll and Imd signalling pathways, the two main controls in the immune-related activation of NF-κB transcription factors [45–47]. NF-κB-independent expression has also been described for the local expression of some AMPs in specific regions of Drosophila epithelia [47,48]. On an evolutionary perspective, it would be interesting to figure out whether colA expression involves a NF-κB-dependent or -independent mechanism under physiological conditions. Moreover, we have recently noticed that colA transcript levels are highly correlated with S. pierantonius population dynamics in adult weevils (see §4). This suggests that colA gene regulation is adjusted to endosymbiont load, which could involve either the direct sensing of bacteria, e.g. through the recognition of bacterial MAMPs, or the indirect perception of their activity, e.g. through the detection of bacterial metabolites.
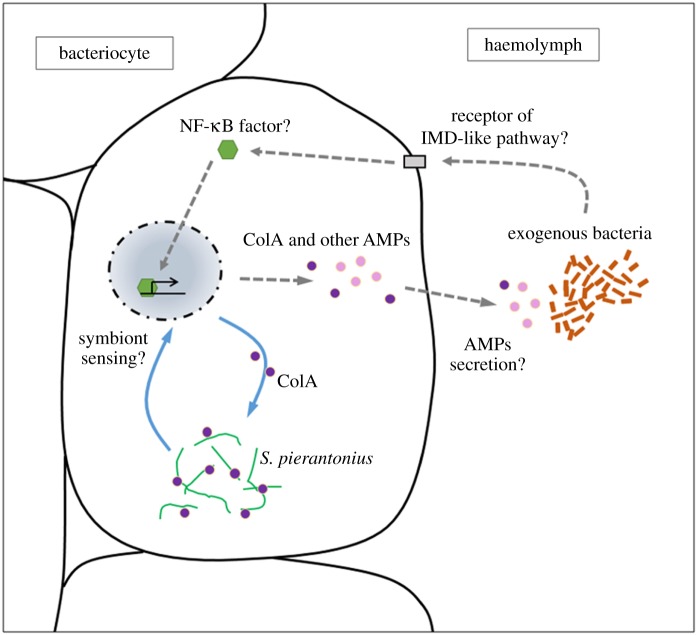
Lastly, the expression of colA in the bacteriome is also modulated by systemic challenges of insects by exogenous bacteria [49]. An injection of Gram-negative or Gram-positive bacteria into the weevil's haemolymph triggers an immune response of the bacteriome, attested by the induction of several AMP coding genes, including colB and colA [49]. We have qualified this response as ‘external’, since it is triggered by exogenous bacteria, in contrast to the ‘internal’ immune response that is directed towards endosymbionts (figure 1). Although it remains to be confirmed that induced AMPs are secreted outside the bacteriome to cope with systemic infections, these findings show that both ColA and ColB AMPs have conserved a host defence function upon infection, in addition to their divergent functions under physiological conditions in the bacteriome organ. Remarkably, colA induction in the bacteriome by systemic infection does not interfere with the endosymbiont load, implying another level of regulation that remains to be explored.
Figure 1.
Internal and external immune responses of S. oryzae bacteriocytes. The internal immune response (solid arrows) consists of the intracellular activity of ColA on endosymbionts. Endosymbiont density is sensed by a still unknown mechanism, and ColA is produced accordingly, which ensures bacterial intracellular seclusion. The external immune response (dotted arrows) consists of the induction of AMP coding gene expression, including colA expression, after recognition of exogenous bacteria. This response is likely to involve an IMD-like pathway. In case of an infection, both responses coexist and do not seem to interfere with each other.
Taken together, these findings provide an insight into how some innate immune system components, originally involved in fighting pathogens, can evolve adaptively and participate in isolating cooperative bacteria from the systemic immune responses, hence allowing the maintenance of mutualistic associations.
3. Endosymbiont tolerance would require antimicrobial peptide expression to stay low
Apart from colA, the bacteriome immune response is marked by a low expression of the other AMP genes studied so far, despite the massive presence of bacteria inside bacteriocytes [31,37]. This restrained local immune response in bacteriocytes, or ‘tolerance’ towards the bacterial partner, is likely to be essential for endosymbiont maintenance. Such a modulated immune response was also noticed in carpenter ants Camponotus floridanus, the bacteriocyte-bearing midgut of which displays a low expression of genes coding for hymenoptaecin and defensin-1 AMPs and for lysozymes [50]. In this insect model, genes coding for the PRR peptidoglycan recognition protein 2 (PGRP-2) and Gram-negative binding protein (GNBP), i.e. proteins involved in the recognition of bacteria upstream of the immune response, are less expressed in the midgut when compared with other tissues [50]. Furthermore, genes coding for the enzymatic PGRPs, PGRP-LB and PGRP-SC2, display a high expression level in the midguts of late pupae when endosymbiont population expands. PGRP-LB and -SC2 may degrade endosymbiont immunogenic peptidoglycan, highlighting a possible mechanism of tolerance that would rely on a low detection of endosymbionts by the immune system [50]. In cereal weevils, the findings that an immune response is activated in the bacteriome following larval challenge with exogenous free-living bacteria [49] indicate that immune pathways are functional in this organ, supporting the idea of an active mechanism of AMP gene repression under physiological conditions, a repression that would be cancelled in case of infections, allowing the defence of the bacteriome and its associated endosymbionts.
Genetic negative regulations of local immune responses in tissues in contact with beneficial bacterial communities, such as the gut epithelium, are well described in insects. As mentioned above, elicitation of immune pathways can be reduced by secreted PGRPs with amidase activity, which degrade immunogenic peptidoglycans [51–53]. Signal transduction can also be decreased by intracellular regulators such as Pirk, which binds to the IMD pathway membrane receptor and causes its internalization [54,55], or by the products of genes such as caudal, which represses the NF-κB-dependent AMP gene expression in the gut, therefore allowing the tolerance of gut microbiota in Drosophila [56]. However, no orthologues of known negative immune regulators have been shown to be highly expressed in the S. oryzae bacteriome. As an example, pirk and caudal basal expression levels are, respectively, two and three times lower in the bacteriome than in other weevil tissues ([49] and unpublished RNAseq data).
An alternative hypothesis to explain AMP gene repression in the bacteriome organ would involve epigenetic mechanisms. Recently, Goto et al. [57] have provided evidence of an epigenetic control on insect immunity by the DNA methyltransferase 1 associated protein 1 (DMAP1). This protein has been identified in Drosophila as an immune regulator that interferes downstream of NF-κB factors, probably acting on chromatin remodelling and increasing the intensity of immune response upon infection [57]. DNA methylation has been one of the first broadly described mechanisms of epigenetic regulation of gene expression in several models [58–60], including the coleopteran Tribolium castaneum [61], but the degree of methylation of insect genomes appears low compared with that observed in mammals, suggesting a lesser impact of DNA methylation on insects. On the other hand, histone methylation has been described in Drosophila and in Planococcus mealybugs as a significant regulatory mechanism for gene expression [62–64]. Such a highly conserved mechanism could be at work in the S. oryzae bacteriome, and would allow a rapid switch on or off in gene expression depending on environmental and physiological conditions, i.e. AMP gene repression under physiological conditions and induction upon infection.
4. Cellular processes take the lead over antimicrobial peptides during endosymbiont recycling
In addition to endosymbiont protection against humoral and cellular immune responses, the compartmentalization of endosymbionts is also reminiscent of an organization in units of production of metabolic compounds essential for the host. We recently have demonstrated that symbiont compartmentalization in the adult stage allows the insect to modulate the symbiont load according to its physiological needs, and to promptly eliminate these ‘factory units’ when they are no longer beneficial [65].
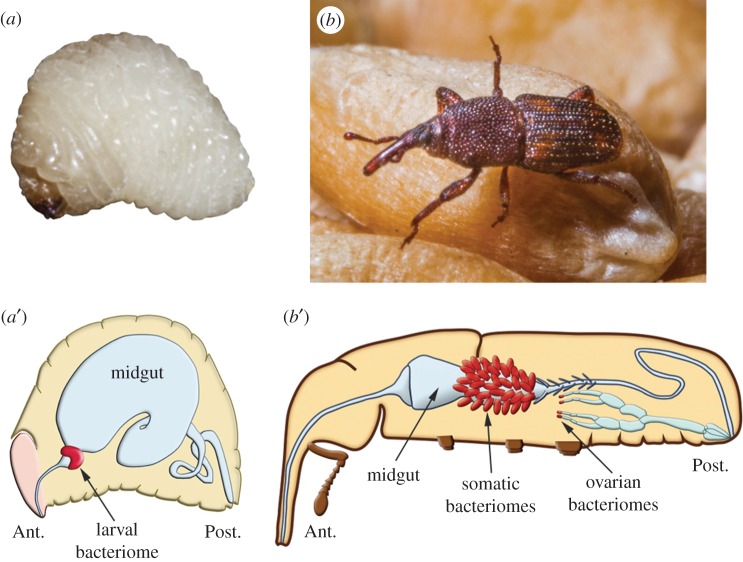
At metamorphosis, the weevil larval bacteriome dissociates and multiple small bacteriomes are formed at the apex of the adult mesenteric caeca [66] (figure 2). During the first week following the adult moulting, these mesenteric bacteriomes grow drastically in size and the endosymbiont population quickly expands (more than 10-fold during the first 5 days). Strikingly, bacteriomes then regress rapidly during the second week until the symbiotic bacteria are completely eliminated by day 15 (figure 3). It is noteworthy that the population of endosymbionts associated with the ovaries is not dislodged, indicating that different regulatory mechanisms are operating according to host tissues. Modulations of the endosymbiotic load during progression from one insect life stage to another have been reported in several insect models. In the pea aphid Acyrthosiphon pisum and in the ant C. floridanus, the endosymbiont population also varies during host development, although symbiont dynamics are weaker than in S. oryzae [68–70]. In mealybugs of the genus Planococcus, co-primary symbionts (i.e. two coexisting species of obligate endosymbionts) each undergo their own dynamics, indicating that each symbiont population may be controlled by separate mechanisms [71].
Figure 2.
Sitophilus spp. endosymbiont localization. (a) Fourth instar larva and (a’) cartoon larva illustration. The shape and localization of the bacteriome of Sitophilus vary with insect development. In the larva, which develops inside a cereal grain, a bilobular bacteriome is attached to the junction between the foregut and the midgut. (b) Adult and (b’) cartoon adult illustration. The endosymbionts are housed in multiple bacteriomes located at the end of midgut mesenteric caeca, and in the female reproductive tract. Adapted from Vigneron et al. [65].
Figure 3.
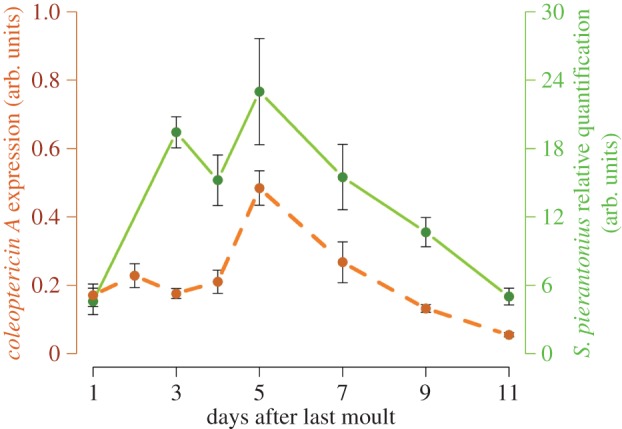
colA gene expression in S. oryzae young adult is correlated wtih endosymbiont dynamics. colA expression was monitored by real-time quantitative PCR and normalized with the expression of housekeeping genes rpl29 and gapdh as described by Masson et al. [67]. Sodalis pierantonius density was assessed by quantitative PCR as described by Vigneron et al. [65]. Both dynamics are correlated (Spearman's rank correlation ρ = 0.7857; pv = 0.0480), suggesting that colA function towards endosymbionts is maintained during host adult life.
In cereal weevils, we have shown that the symbiotic burst in the first days of adulthood matches host need for the amino acids tyrosine and phenylalanine which are produced by the endosymbiont and transformed by the insect into 3,4-dihydroxyphenylalanine (DOPA), which is required for strengthening and stabilizing the newly synthesized cuticle [65]. Once the formation and stabilization of the new adult cuticle has been finished, endosymbionts are rapidly eliminated. The importance of endosymbiosis in the process of cuticle synthesis can be appreciated through the analysis of aposymbiotic weevils (i.e. insects that have been experimentally deprived of their endosymbionts), whose adults carry a much thinner cuticle than symbiotic weevils [65]. The ability of weevils to maintain a stable symbiotic state in the larval stage, and to manipulate the endosymbiont population and adjust its load to the insect's physiological needs in the adult stage suggests complex control mechanisms that probably involve the bacteriome local immune response. The rapid proliferation of symbionts in the period after metamorphosis, followed by their rapid and total elimination, raises different questions depending on the phase being considered: which mechanisms allow endosymbiont maintenance during the growing phase? Is immunity involved in symbiont clearance? Are any other mechanisms entailed?
Recent data have shown that colA expression in the gut of symbiotic insects is correlated with symbiont density (figure 3, [67]) and that bacteria remain intracellular during the whole elimination process, avoiding thereby tissue inflammation and systemic immune activation [67]. These findings suggest that, similar to the larval stage, ColA continues to target and regulate endosymbiont cell division as long as bacteria are present in adults. Remarkably, the other AMP coding genes, including colB, are not transcriptionally activated during bacterial dynamics [67]. This, in addition to the high expression of the negative immune regulator pirk gene during the whole dynamics, suggests an active clamping of the immune pathways that may be relevant not only for endosymbiont tolerance, but also for permitting their rapid growth at the initial phase of the adult stage [67]. Taking into consideration the downregulation of AMP encoding genes during the elimination phase, AMPs are unlikely to be involved in this endosymbiont dynamics process. Symbiont elimination was instead shown to involve two cellular processes: these are apoptosis, or programmed cell death, and autophagy, which is a conserved cellular mechanism allowing eukaryotic cells to recycle cell components and organelles and to preserve cellular homeostasis [72,73]. These cell processes may allow the host to minimize the cost inherent to symbiont growth in the initial adult phase. The recycling of endosymbiont cell components, which is highlighted by the accumulation of massive lamellar bodies in the cells [65], would enable the host to recover a part of the energy invested in endosymbiont growth, control and maintenance.
Autophagy is classically activated in response to organelle damage and nutritional deficiency. This process is also involved in many host–bacterial interactions, notably in the clearance of the intracellular pathogen Listeria monocytogenes in Drosophila [74], and in the regulation of Wolbachia populations in nematodes, crustaceans and insects [75,76]. It also plays a critical role in the elimination of dinoflagellate symbionts in the sea anemone Aiptasia pallida [77,78]. In this cnidarian model, endosymbiont autophagy is triggered during the bleaching phenomenon as a response to thermal stress, the precise function of which remains poorly understood. The common thread in these examples is that autophagy is activated upon stress generated either by bacteria or by the environment. In cereal weevils, symbiont recycling appears to be a programmed process that is set up during insect development. Nevertheless, and similar to the aforementioned models, the rapid bacterial multiplication in emerging weevil adults could be considered as a bacterial or metabolic stress that could trigger the transcription of autophagy effector genes. In line with this hypothesis, we have shown recently that genes from the autophagy-related gene (ATG) family [79] are induced during the symbiotic burst, several days before the effective autophagic digestion of endosymbionts [67]. This also raises the question of which signals link endosymbiont perception by the host to the effector processes. More accurately, two signals could be expected: one triggering ATG expression around the third day of adult life, and the other triggering the activation of recycling through autophagy. The latter has been proposed as DOPA by Vigneron et al. [65]. DOPA, which is produced from tyrosine and phenylalanine, is critical for the sclerotization and melanization of the cuticle [80]. Once cuticle formation has been completed, around the sixth day of adult weevil life, DOPA has been shown to accumulate in insect tissues and may act as a signal to trigger symbiont recycling. Further research may unravel the nature of the signals activating ATG transcription, as well as the pathways involved in DOPA-signal integration.
Beyond the signalling mechanisms, these findings show that an adapted cross-talk between metabolic and cellular functions has been selected through host–symbiont coevolution. This cross-talk participates in optimizing the cost of symbiosis and in speeding up completion of a protective exoskeleton during the critical phase when insects emerge from the cereal grains and face new environmental challenges.
5. Conclusion
The serial integration of intracellular mutualistic bacteria from free-living potentially pathogenic bacteria remains a puzzling phenomenon. As early as 1933, the insect pathologist André Paillot reported that ‘Symbiosis can originally be considered as a pathogenic bacterial infection. The long adaptation of these bacteria to the same organism progressively decreased their virulence until they became harmless to the host’ [81]. This visionary theory, which was postulated when symbiosis was viewed as a biological curiosity, is now being supported by accumulating findings on endosymbiont comparative genomics. Among these findings there are: the identification of secretion systems and virulence-encoding genes in recently established mutualistic bacteria, the deletions of genes encoding MAMPs in long-established endosymbionts, and the complete metabolic dependence between associated partners. However, what still remains to be determined is whether and how the host genome evolves in parallel with bacterial genome shrinkage during host–symbiont coevolution, and how the host immune system is involved in the tolerance and control of the bacterial partners in number and localization. Recent research on S. oryzae and C. floridanus has revealed that the host immune system is a central player in endosymbiont control. In S. oryzae in particular, studies on ColA have highlighted the constrained adaptive evolution of host AMP structure and activity, which participate in endosymbiont seclusion within the bacteriocytes. In spite of the functionality of immune pathways in the bacteriome, endosymbiont massive presence does not trigger the activation of immune effectors in this tissue, indicating that not only the sequence of ColA may have been shaped through host–symbiont coevolution, but also the regulatory mechanisms of the other AMPs. Furthermore, the contrast between colA and other AMP gene expression profiles in the bacteriome reinforces the assumption that AMP expression is a critical player in endosymbiont tolerance and maintenance. Nevertheless, findings on young adult weevils attest that AMP-based immunity is not the only player in endosymbiosis control and homeostasis. Cellular processes such as autophagy and apoptosis are indeed major effectors in the regulation of endosymbiont dynamics, especially adjusting the endosymbiont population to the host's physiological needs, which in fine improves the cost/benefit ratio of endosymbiosis. This illustrates the concept of symbiotic trade-off: the insect offers ‘board and lodging’ to the endosymbionts as long as it relies on their metabolic supply, but when the cost of symbiont maintenance overcomes the benefit provided, the insect cellular machinery specifically recycles endosymbionts from the gut bacteriocytes, while preserving ovary-associated endosymbionts involved in the reproductive physiology of the host and symbiont transmission.
In conclusion, recent data provide evidence that at least three mechanisms have been selected along with host–symbiont coevolution to ensure a fine-tuned regulation of endosymbiosis homeostasis and dynamics: (i) the compartmentalization of symbionts, which isolates them from the host systemic immune response, (ii) the use of AMPs as ‘symbiont shepherds’ and (iii) the activation of two cell-autonomous processes, autophagy and apoptosis, which allow rapid and tissue-specific adjustment of the symbiont load at a given stage of physiological maturation. Deciphering how these mechanisms are interconnected and integrated with the host metabolism may open up a new avenue for the understanding of holobiont function and evolution.
Acknowledgements
We sincerely thank our collaborators Carole Vincent-Monégat, Agnès Vallier, Séverine Balmand, Nicolas Parisot, Aurélien Vigneron and Justin Maire, who participated in putting data on S. oryzae together and who allowed the emergence of new ideas through their constructive discussion. We acknowledge Jens Rolff, Paul Schmid-Hempel and the editorial team of Philosophical Transactions of Royal Society B for their kind invitation to participate in this volume. We also deeply thank Stuart Reynolds for his constructive critical reading of the manuscript.
Competing interests
We have no competing interests.
Funding
This work was supported by the French ANR-10 BSV7-170101-545 03 (ImmunSymbArt) and ANR-13-BSV7-0016-01 (IMetSym). F.M. was also funded by a doctoral grant from the French Ministry of Higher Education and Research.
References
- 1.Buchner P. 1965. Endosymbiosis of animals with plant microorganisms. New York, NY: Interscience. [Google Scholar]
- 2.Heddi A, Lefebvre F, Nardon P. 1993. Effect of endocytobiotic bacteria on mitochondrial enzymatic activities in the weevil Sitophilus oryzae (Coleoptera: Curculionidae). Insect Biochem. Mol. Biol. 23, 403–411. ( 10.1016/0965-1748(93)90024-m) [DOI] [Google Scholar]
- 3.Heddi A, Grenier AM, Khatchadourian C, Charles H, Nardon P. 1999. Four intracellular genomes direct weevil biology: nuclear, mitochondrial, principal endosymbiont, and Wolbachia. Proc. Natl Acad. Sci. USA 96, 6814–6819. ( 10.1073/pnas.96.12.6814) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Douglas P. 1998. Host benefit and the evolution of specialization in symbiosis. Heredity (Edinb). 81, 599–603. ( 10.1046/j.1365-2540.1998.00455.x) [DOI] [Google Scholar]
- 5.Pais R, Lohs C, Wu Y, Wang J, Aksoy S. 2008. The obligate mutualist Wigglesworthia glossinidia influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl. Environ. Microbiol. 74, 5965–5974. ( 10.1128/AEM.00741-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiss BL, Wang J, Aksoy S. 2011. Tsetse immune system maturation requires the presence of obligate symbionts in larvae. PLoS Biol. 9, e1000619 ( 10.1371/journal.pbio.1000619) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michalkova V, Benoit JB, Weiss BL, Attardo GM, Aksoy S. 2014. Vitamin B6 generated by obligate symbionts is critical for maintaining proline homeostasis and fecundity in tsetse flies. Appl. Environ. Microbiol. 80, 5844–5853. ( 10.1128/AEM.01150-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss B, Maltz M, Aksoy S. 2012. Obligate symbionts activate immune system development in the tsetse fly. J. Immunol. 188, 3395–3403. ( 10.4049/jimmunol.1103691.Obligate) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wicker C. 1983. Differential vitamin and choline requirements of symbiotic and aposymbiotic Sitophilus oryzae (Coleoptera: Curculionidae). Comp. Biochem. Physiol. A 76, 177–182. ( 10.1016/0300-9629(83)90311-0) [DOI] [Google Scholar]
- 10.Akman Gunduz E, Douglas AE. 2009. Symbiotic bacteria enable insect to use a nutritionally inadequate diet. Proc. R. Soc. B 276, 987–991. ( 10.1098/rspb.2008.1476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nardon P, Grenier AM. 1991. Serial endosymbiosis theory and weevil evolution: the role of symbiosis. In Symbiosis as a source of evolutionary innovation: speciation and morphogenesis (eds Margulis L, Fester R), pp. 153–168. Cambridge, MA: MIT Press. [PubMed] [Google Scholar]
- 12.Douglas AE, Prosser WA. 1992. Synthesis of the essential amino acid tryptophan in the pea aphid (Acyrthosiphon pisum) symbiosis. J. Insect Physiol. 38, 565–568. ( 10.1016/0022-1910(92)90107-O) [DOI] [Google Scholar]
- 13.Degnan PH, Lazarus AB, Brock CD, Wernegreen JJ. 2004. Host–symbiont stability and fast evolutionary rates in an ant-bacterium association: cospeciation of Camponotus species and their endosymbionts, Candidatus blochmannia. Syst. Biol. 53, 95–110. ( 10.1080/10635150490264842) [DOI] [PubMed] [Google Scholar]
- 14.Baumann L, Baumann P. 2005. Cospeciation between the primary endosymbionts of mealybugs and their hosts. Curr. Microbiol. 50, 84–87. ( 10.1007/s00284-004-4437-x) [DOI] [PubMed] [Google Scholar]
- 15.Mazzon L, Martinez-Sañudo I, Simonato M, Squartini A, Savio C, Girolami V. 2010. Phylogenetic relationships between flies of the Tephritinae subfamily (Diptera, Tephritidae) and their symbiotic bacteria. Mol. Phylogenet. Evol. 56, 312–326. ( 10.1016/j.ympev.2010.02.016) [DOI] [PubMed] [Google Scholar]
- 16.Hosokawa T, Nikoh N, Koga R, Sato M, Tanahashi M, Meng XY, Fukatsu T. 2012. Reductive genome evolution, host–symbiont co-speciation and uterine transmission of endosymbiotic bacteria in bat flies. ISME J. 6, 577–587. ( 10.1038/ismej.2011.125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407, 81–86. ( 10.1038/35024074) [DOI] [PubMed] [Google Scholar]
- 18.Latorre A, Gil R, Silva FJ, Moya A. 2005. Chromosomal stasis versus plasmid plasticity in aphid endosymbiont Buchnera aphidicola. Heredity (Edinb). 95, 339–347. ( 10.1038/sj.hdy.6800716) [DOI] [PubMed] [Google Scholar]
- 19.Russell CW, Bouvaine S, Newell PD, Douglas AE. 2013. Shared metabolic pathways in a coevolved insect-bacterial symbiosis. Appl. Environ. Microbiol. 79, 6117–6123. ( 10.1128/AEM.01543-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomez-Valero L. 2004. The evolutionary fate of nonfunctional DNA in the bacterial endosymbiont Buchnera aphidicola. Mol. Biol. Evol. 21, 2172–2181. ( 10.1093/molbev/msh232) [DOI] [PubMed] [Google Scholar]
- 21.Dale C, Plague GR, Wang B, Ochman H, Moran NA. 2002. Type III secretion systems and the evolution of mutualistic endosymbiosis. Proc. Natl Acad. Sci. USA 99, 12 397–12 402. ( 10.1073/pnas.182213299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dale C, Jones T, Pontes M. 2005. Degenerative evolution and functional diversification of type-III secretion systems in the insect endosymbiont Sodalis glossinidius. Mol. Biol. Evol. 22, 758–766. ( 10.1093/molbev/msi061) [DOI] [PubMed] [Google Scholar]
- 23.Heddi A, Charles H, Khatchadourian C, Bonnot G, Nardon P. 1998. Molecular characterization of the principal symbiotic bacteria of the weevil Sitophilus oryzae: a peculiar G+C content of an endocytobiotic DNA. J. Mol. Evol. 47, 52–61. ( 10.1007/PL00006362) [DOI] [PubMed] [Google Scholar]
- 24.Lefèvre C, Charles H, Vallier A, Delobel B, Farrell B, Heddi A. 2004. Endosymbiont phylogenesis in the Dryophthoridae weevils: evidence for bacterial replacement. Mol. Biol. Evol. 21, 965–973. ( 10.1093/molbev/msh063) [DOI] [PubMed] [Google Scholar]
- 25.Toju H, Hosokawa T, Koga R, Nikoh N, Meng XY, Kimura N, Fukatsu T. 2010. ‘Candidatus Curculioniphilus buchneri’, a novel clade of bacterial endocellular symbionts from weevils of the genus Curculio. Appl. Environ. Microbiol. 76, 275–282. ( 10.1128/AEM.02154-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clayton AL, Oakeson KF, Gutin M, Pontes A, Dunn DM, von Niederhausern AC, Weiss RB, Fisher M, Dale C. 2012. A novel human-infection-derived bacterium provides insights into the evolutionary origins of mutualistic insect–bacterial symbioses. PLoS Genet. 8, e1002990 ( 10.1371/journal.pgen.1002990) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conord C, Despres L, Vallier A, Balmand S, Miquel C, Zundel S, Lemperiere G, Heddi A. 2008. Long-term evolutionary stability of bacterial endosymbiosis in Curculionoidea: additional evidence of symbiont replacement in the Dryophthoridae family. Mol. Biol. Evol. 25, 859–868. ( 10.1093/molbev/msn027) [DOI] [PubMed] [Google Scholar]
- 28.Charles H, Heddi A, Guillaud J, Nardon C, Nardon P. 1997. A molecular aspect of symbiotic interactions between the weevil Sitophilus oryzae and its endosymbiotic bacteria: over-expression of a chaperonin. Biochem. Biophys. Res. Commun. 239, 769–774. ( 10.1006/bbrc.1997.7552) [DOI] [PubMed] [Google Scholar]
- 29.Gil R, et al. 2008. Massive presence of insertion sequences in the genome of SOPE, the primary endosymbiont of the rice weevil Sitophilus oryzae. Int. Microbiol. 11, 41–48. [PubMed] [Google Scholar]
- 30.Oakeson KF, et al. 2014. Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol. Evol. 6, 76–93. ( 10.1093/gbe/evt210) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anselme C, Perez-Brocal V, Vallier A, Vincent-Monegat C, Charif D, Latorre A, Moya A, Heddi A. 2008. Identification of the weevil immune genes and their expression in the bacteriome tissue. BMC Biol. 6, 43 ( 10.1186/1741-7007-6-43) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bright M, Klose J, Nussbaumer AD. 2013. Giant tubeworms. Curr. Biol. 23, R224–R225. ( 10.1016/j.cub.2013.01.039) [DOI] [PubMed] [Google Scholar]
- 33.Tóth K, Stacey G. 2015. Does plant immunity play a critical role during initiation of the legume-rhizobium symbiosis? Front. Plant Sci. 06, 1–7. ( 10.3389/fpls.2015.00401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hooper LV, Littman DR, Macpherson AJ. 2012. Interactions between the microbiota and the immune system. Science 336, 1268–1273. ( 10.1126/science.1223490) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McFall-Ngai M. 2014. Divining the essence of symbiosis: insights from the squid-vibrio model. PLoS Biol. 12, e1001783 ( 10.1371/journal.pbio.1001783) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heddi A, Vallier A, Anselme C, Xin H, Rahbe Y, Wackers F. 2005. Molecular and cellular profiles of insect bacteriocytes: mutualism and harm at the initial evolutionary step of symbiogenesis. Cell Microbiol. 7, 293–305. ( 10.1111/j.1462-5822.2004.00461.x) [DOI] [PubMed] [Google Scholar]
- 37.Vigneron A, Charif D, Vincent-Monegat C, Vallier A, Gavory F, Wincker P, Heddi A. 2012. Host gene response to endosymbiont and pathogen in the cereal weevil Sitophilus oryzae. BMC Microbiol. 12(Suppl 1), S14 ( 10.1186/1471-2180-12-S1-S14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bulet P, Cociancich S, Dimarcq JL, Lambert J, Reichhart JM, Hoffmann D, Hetru C, Hoffmann JA. 1991. Insect immunity: isolation from a coleopteran insect of a novel inducible antibacterial peptide and of new members of the insect defensin family. J. Biol. Chem. 266, 24 520–24 525. ( 10.1016/j.chom.2009.07.008) [DOI] [PubMed] [Google Scholar]
- 39.Sagisaka A, Miyanoshita A, Ishibashi J, Yamakawa M. 2001. Purification, characterization and gene expression of a glycine and proline-rich antibacterial protein family from larvae of a beetle, Allomyrina dichotoma. Insect Mol. Biol. 10, 293–302. ( 10.1046/j.0962-1075.2001.00261.x) [DOI] [PubMed] [Google Scholar]
- 40.Login FH, Balmand S, Vallier A, Vincent-Monégat C, Vigneron A, Weiss-Gayet M, Rochat D, Heddi A. 2011. Antimicrobial peptides keep insect endosymbionts under control. Science 334, 362–365. ( 10.1126/science.1209728) [DOI] [PubMed] [Google Scholar]
- 41.Nardon P, Nardon C. 1998. Endocytobiote control by the host in the weevil Sitophilus oryzae, Coleoptera, Curculionidae. Symbiosis 25, 237–250. [Google Scholar]
- 42.Van de Velde W, et al. 2010. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327, 1122–1126. ( 10.1126/science.1184057) [DOI] [PubMed] [Google Scholar]
- 43.Mergaert P, et al. 2006. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc. Natl Acad. Sci. USA 103, 5230–5235. ( 10.1073/pnas.0600912103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Login FH, Heddi A. 2013. Insect immune system maintains long-term resident bacteria through a local response. J. Insect Physiol. 59, 232–239. ( 10.1016/j.jinsphys.2012.06.015) [DOI] [PubMed] [Google Scholar]
- 45.Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart JM, Hoffmann JA. 1995. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc. Natl Acad. Sci. USA 92, 9465–9469. ( 10.1073/pnas.92.21.9465) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. 1996. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86, 973–983. ( 10.1016/S0092-8674(00)80172-5) [DOI] [PubMed] [Google Scholar]
- 47.Lemaitre B, Hoffmann J. 2007. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743. ( 10.1146/annurev.immunol.25.022106.141615) [DOI] [PubMed] [Google Scholar]
- 48.Tzou P, Ohresser S, Ferrandon D, Capovilla M, Reichhart JM, Lemaitre B, Hoffmann JA, Imler JL. 2000. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity 13, 737–748. ( 10.1016/S1074-7613(00)00072-8) [DOI] [PubMed] [Google Scholar]
- 49.Masson F, Vallier A, Vigneron A, Balmand S, Vincent-Monégat C, Zaidman-Rémy A, Heddi A. 2015. Systemic infection generates a local-like immune response of the bacteriome organ in insect symbiosis. J. Innate Immun. 7, 290–301. ( 10.1159/000368928) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ratzka C, Gross R, Feldhaar H. 2013. Gene expression analysis of the endosymbiont-bearing midgut tissue during ontogeny of the carpenter ant Camponotus floridanus. J. Insect Physiol. 59, 611–623. ( 10.1016/j.jinsphys.2013.03.011) [DOI] [PubMed] [Google Scholar]
- 51.Zaidman-Rémy A, et al. 2006. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity 24, 463–473. ( 10.1016/j.immuni.2006.02.012) [DOI] [PubMed] [Google Scholar]
- 52.Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, Royet J. 2006. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2, e14 ( 10.1371/journal.ppat.0020014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paredes JC, Welchman DP, Poidevin M, Lemaitre B. 2011. Negative regulation by amidase PGRPs shapes the Drosophila antibacterial response and protects the fly from innocuous infection. Immunity 35, 770–779. ( 10.1016/j.immuni.2011.09.018) [DOI] [PubMed] [Google Scholar]
- 54.Kleino A, Myllymaki H, Kallio J, Vanha-aho LM, Oksanen K, Ulvila J, Hultmark D, Valanne S, Ramet M. 2008. Pirk is a negative regulator of the Drosophila Imd pathway. J. Immunol. 180, 5413–5422. ( 10.4049/jimmunol.180.8.5413) [DOI] [PubMed] [Google Scholar]
- 55.Lhocine N, et al. 2008. PIMS modulates immune tolerance by negatively regulating Drosophila innate immune signaling. Cell Host Microbe 4, 147–158. ( 10.1016/j.chom.2008.07.004) [DOI] [PubMed] [Google Scholar]
- 56.Ryu JH, et al. 2008. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319, 777–782. ( 10.1126/science.1149357) [DOI] [PubMed] [Google Scholar]
- 57.Goto A, Fukuyama H, Imler J-L, Hoffmann JA. 2014. The chromatin regulator DMAP1 modulates activity of the nuclear factor κB (NF-κB) transcription factor relish in the Drosophila innate immune response. J. Biol. Chem. 289, 20 470–20 476. ( 10.1074/jbc.C114.553719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bird AP, Taggart MH. 1980. Variable patterns of total DNA and rDNA methylation in animals. Nucleic Acids Res. 8, 1485–1497. ( 10.1093/nar/8.7.1485) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li E, Beard C, Jaenisch R. 1993. Role for DNA methylation in genomic imprinting. Nature 366, 362–365. ( 10.1038/366362a0) [DOI] [PubMed] [Google Scholar]
- 60.Robinson KL, Tohidi-Esfahani D, Lo N, Simpson SJ, Sword GA. 2011. Evidence for widespread genomic methylation in the migratory locust, Locusta migratoria (Orthoptera: Acrididae). PLoS ONE 6, e28167 ( 10.1371/journal.pone.0028167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feliciello I, Parazajder J, Akrap I. 2013. First evidence of DNA methylation in insect Tribolium castaneum: environmental regulation of DNA methylation within heterochromatin. Epigenetics 8, 534–541. ( 10.4161/epi.24507) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kouzarides T. 2007. Chromatin modifications and their function. Cell 128, 693–705. ( 10.1016/j.cell.2007.02.005) [DOI] [PubMed] [Google Scholar]
- 63.Eissenberg JC, Reuter G. 2009. Cellular mechanism for targeting heterochromatin formation in Drosophila. Int. Rev. Cell Mol. Biol. 273, 1–47. ( 10.1016/S1937-6448(08)01801-7) [DOI] [PubMed] [Google Scholar]
- 64.Bongiorni S, Pugnali M, Volpi S, Bizzaro D, Singh PB, Prantera G. 2009. Epigenetic marks for chromosome imprinting during spermatogenesis in coccids. Chromosoma 118, 501–512. ( 10.1007/s00412-009-0214-8) [DOI] [PubMed] [Google Scholar]
- 65.Vigneron A, et al. 2014. Insects recycle endosymbionts when the benefit is over. Curr. Biol. 24, 2267–2273. ( 10.1016/j.cub.2014.07.065) [DOI] [PubMed] [Google Scholar]
- 66.Nardon P, Wicker C. 1981. La symbiose chez le genre Sitophilus (Coléoptère, Curculionidae). Principaux aspects morphologiques, physiologiques et génétiques. Ann. Biol. 20, 327–373. [Google Scholar]
- 67.Masson F, et al. 2015. Weevil endosymbiont dynamics is associated with a clamping of immunity. BMC Genomics 16, 819 ( 10.1186/s12864-015-2048-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koga R, Tsuchida T, Fukatsu T. 2003. Changing partners in an obligate symbiosis: a facultative endosymbiont can compensate for loss of the essential endosymbiont Buchnera in an aphid. Proc. R. Soc. B 270, 2543–2550. ( 10.1098/rspb.2003.2537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sakurai M, Koga R, Tsuchida T, Meng XY, Fukatsu T. 2005. Rickettsia symbiont in the pea aphid Acyrthosiphon pisum: novel cellular tropism, effect on host fitness, and interaction with the essential symbiont Buchnera. Appl. Environ. Microbiol. 71, 4069–4075. ( 10.1128/AEM.71.7.4069-4075.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stoll S, Feldhaar H, Fraunholz MJ, Gross R. 2010. Bacteriocyte dynamics during development of a holometabolous insect, the carpenter ant Camponotus floridanus. BMC Microbiol. 10, 308 ( 10.1186/1471-2180-10-308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kono M, Koga R, Shimada M, Fukatsu T. 2008. Infection dynamics of coexisting beta- and gammaproteobacteria in the nested endosymbiotic system of mealybugs. Appl. Environ. Microbiol. 74, 4175–4184. ( 10.1128/AEM.00250-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Levine B, Mizushima N, Virgin HW. 2011. Autophagy in immunity and inflammation. Nature 469, 323–335. ( 10.1038/nature09782) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yorimitsu T, Klionsky DJ. 2005. Autophagy: molecular machinery for self-eating. Cell Death Differ. 12(Suppl 2), 1542–1552. ( 10.1038/sj.cdd.4401765) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yano T, et al. 2008. Autophagic control of Listeria through intracellular innate immune recognition in Drosophila. Nat. Immunol. 9, 908–916. ( 10.1038/ni.1634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Le Clec'h W, Braquart-Varnier C, Raimond M, Ferdy J-B, Bouchon D, Sicard M. 2012. High virulence of Wolbachia after host switching: when autophagy hurts. PLoS Pathog. 8, e1002844 ( 10.1371/journal.ppat.1002844) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Voronin D, Cook DAN, Steven A, Taylor MJ. 2012. Autophagy regulates Wolbachia populations across diverse symbiotic associations. Proc. Natl Acad. Sci. USA 109, E1638–E1646. ( 10.1073/pnas.1203519109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dunn SR, Schnitzler CE, Weis VM. 2007. Apoptosis and autophagy as mechanisms of dinoflagellate symbiont release during cnidarian bleaching: every which way you lose. Proc. R. Soc. B 274, 3079–3085. ( 10.1098/rspb.2007.0711) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dunn SR, Weis VM. 2009. Apoptosis as a post-phagocytic winnowing mechanism in a coral-dinoflagellate mutualism. Environ. Microbiol. 11, 268–276. ( 10.1111/j.1462-2920.2008.01774.x) [DOI] [PubMed] [Google Scholar]
- 79.Klionsky DJ, et al. 2003. A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5, 539–545. ( 10.1016/S1534-5807(03)00296-X) [DOI] [PubMed] [Google Scholar]
- 80.Andersen SO. 2010. Insect cuticular sclerotization: a review. Insect Biochem. Mol. Biol. 40, 166–178. ( 10.1016/j.ibmb.2009.10.007) [DOI] [PubMed] [Google Scholar]
- 81.Paillot A. 1933. L’infection chez les insectes: immunité et symbiose. Paris, France: Patissier. [Google Scholar]