Abstract
West Nile virus (WNV) is an emerging pathogen that has decimated bird populations and caused severe outbreaks of viral encephalitis in humans. Currently, little is known about the within-host viral kinetics of WNV during infection. We developed mathematical models to describe viral replication, spread and host immune response in wild-type and immunocompromised mice. Our approach fits a target cell-limited model to viremia data from immunocompromised knockout mice and an adaptive immune response model to data from wild-type mice. Using this approach, we first estimate parameters governing viral production and viral spread in the host using simple models without immune responses. We then use these parameters in a more complex immune response model to characterize the dynamics of the humoral immune response. Despite substantial uncertainty in input parameters, our analysis generates relatively precise estimates of important viral characteristics that are composed of nonlinear combinations of model parameters: we estimate the mean within-host basic reproductive number, R0, to be 2.3 (95% of values in the range 1.7–2.9); the mean infectious virion burst size to be 2.9 plaque-forming units (95% of values in the range 1.7–4.7); and the average number of cells infected per infectious virion to be between 0.3 and 0.99. Our analysis gives mechanistic insights into the dynamics of WNV infection and produces estimates of viral characteristics that are difficult to measure experimentally. These models are a first step towards a quantitative understanding of the timing and effectiveness of the humoral immune response in reducing host viremia and consequently the epidemic spread of WNV.
Keywords: West Nile virus infection, within-host viral dynamics, ordinary differential equation models, parameter estimation, biologically relevant parameters, basic reproductive number
1. Introduction
West Nile virus (WNV) is a flavivirus that is a significant cause of viral encephalitis in humans [1–3]. It is not only maintained in an enzootic cycle between mosquitoes and birds [4], but can also infect and cause disease in other vertebrates including mice. After its discovery in the USA in 1999, WNV spread rapidly across North America, and also has been reported in Central and South America [5–7]. Between 1999 and 2010, there were a total of 1.8 million human illnesses, 1308 of which resulted in death [8]. Although vaccines are available for animal use, no vaccines or specific therapies for WNV are currently approved for humans [9].
WNV is an enveloped virus with a single-stranded, positive sense, 11-kb RNA genome [9] that was first isolated in 1937 [10]. While much is known about the biology of WNV including the immune response it elicits and its epidemiology (reviewed in [9]), information regarding the detailed kinetics of within-host WNV infection is limited. Following mosquito-borne transmission, WNV initially infects keratinocytes and epidermal Langerhans cells [11]. Infected Langerhans cells then migrate to the draining lymph node where macrophages are infected [9]. From the draining lymph node, WNV spreads to the spleen, kidney and spinal cord and ultimately breaches the blood–brain barrier to infect neurons [9].
WNV infection in mice is characterized by an initial exponential growth of viral titre that peaks 3–4 days post-infection (DPI), followed by an exponential decline that leads to undetectable levels of virus by 6–8 DPI. Peak viral replication occurs before the virus has spread to the spleen and kidneys (analysis of data in mice from [12]). The initial targets of WNV replication are Langerhans cells, macrophages and keratinocytes [11].
Both T and B cell responses are important in controlling WNV infection [9,12,13]. During a primary response, viral-specific-induced immunoglobulin M (IgM) is detected just before the virus titre peak, approximately 2–4 DPI, and immunoglobulin G (IgG) is detected at approximately 8 DPI [12] well after viremia has been reduced to almost undetectable levels in serum.
The effect of induced IgM is to lower peak viremia and reduce the time to peak viremia [12]. In mice that lack B cells, WNV is 100% lethal at doses of 100 plaque-forming units (PFU) and higher and 50% lethal following infection with 1 PFU [13]. Thus, innate responses are unable to protect mice in the absence of induced IgM, which is critical for reduction of peak viremia and host survival. In wild-type mice, cytotoxic T cell responses do not appear until 6 DPI, well after the peak of virus replication [14]. Our goal is to quantitatively describe the interplay between viral dynamics and the B cell response in the early phase of WNV infection.
Mathematical models can provide important insights into viral dynamics and immune response. For example, human immunodeficiency virus (HIV) infection was modelled to analyse the kinetics of viral load decline in patients receiving antiretroviral therapy. These analyses led to the first in vivo estimates of the rate of HIV replication, the number of virus particles produced and cleared daily, and the average lifespan of productively infected cells [15–18]. These models further elucidated the cause of the rapid appearance of drug resistance to single HIV drugs and helped lead-in the era of combination antiretroviral therapy [19,20]. Viral dynamic models also have been used to analyse the effects of interferon and ribavirin treatment as well as direct-acting antiviral agents on hepatitis C virus (HCV) dynamics [21–26], and the effects of therapy on hepatitis B virus (HBV) kinetics [27–29]. HIV, HCV and HBV infections produce prolonged chronic diseases, and viral load changes can be studied over periods of days, months and years. In contrast, WNV produces an acute infection of short duration. Previous work has used modelling to understand the kinetics of acute infection in influenza [30–33]. However, to date, there are no within-host models of WNV infection.
Our primary goal is to characterize the dynamics of WNV replication and the adaptive humoral response in murine hosts. Based on data obtained during experimental infection of mice, we develop kinetic models of primary WNV infection. We estimate the basic reproductive number (R0, the number of cells infected by each infected cell early in infection when target cells are not limiting), and other aspects of viral replication. We characterize not just the most likely value of R0 and other relevant parameters, but also the range of possible values given measured empirical data and the dynamics assumed in our models. We also characterize the timing and effectiveness of IgM at clearing the virus.
We characterize viral replication and immune response based on predictions from a set of differential equation models. We estimate model parameters based on published data from laboratory experiments of WNV infection. We compare model predictions with measured viremia in wild-type mice and immunocompromised IgM knockout mice to identify what parameters governing viral kinetics and immune responses could have generated the observed viremia values. The problem is computationally challenging, because there is a paucity of quantitative experimental data that we can use to estimate parameter values, and because there are many parameters to be estimated. Further, there are trade-offs between model parameters that complicate parameter estimation. Thus, our approach is to first identify the plausible parameter space and then to sample that space to more precisely estimate the range of parameter values consistent with experimental data.
We first use empirical data to estimate a parameter characterizing the rate of clearance of WNV by innate immune responses in the first 90 min of infection. Next, we analyse data from a knockout experiment in which there was no humoral (secreted IgM) response in order to characterize WNV infection dynamics in the absence of the secreted IgM response. Then we incorporate these parameters into a model to characterize the kinetics of the humoral response from experimental data in wild-type mice that have an intact IgM response. Using an approach similar to that of Handel et al. [34] for influenza, we estimate some parameters in isolation, and then incorporate those parameters into a more complex model that includes an adaptive immune response. Thus, we avoid the problem of needing to estimate more parameters in our final model than can be identified from the data.
2. Material and methods
2.1. Study data
We use data from three WNV experimental infection studies [12,13,35]. In order to estimate the infectious virus decay rate, we use data from a viral decay study in which wild-type (C57BL/6) mice were intravenously inoculated with 105 PFU of WNV 3356 NY2000 [35]. Serum was collected 5, 15, 45 and 90 min post-infection. Serum was tested for virus by plaque assay on Vero cells (African green monkey kidney epithelial cells).
The second set of studies (wild-type and knockout mouse studies) were based on experimental infections of wild-type and IgM knockout mice. In these studies, wild-type and knockout (sIgM–/– C57BL/6 J) mice were subcutaneously inoculated with 100 PFU of a WNV strain closely related to the one used in the viral decay study (WNV 3000.0259 NY2000) [11]. Serum was collected every other day until 10 DPI and titrated for virus by plaque assay on BHK-21 cells (baby hamster kidney fibroblast cells).
The wild-type and knockout mouse studies measured viremia in terms of both viral RNA and PFU, but the latter had fewer measurements. Because the viral decay study measured infectious virus (PFU), we converted all RNA measurements in the wild-type and knockout mouse study to PFU. We calculated the ratio of RNA copies per ml of blood for knockout mice to the viral titre in PFU per ml of blood for knockout mice at day 2 and 4 post-infection from the experimental data. We found that approximately 500 RNA copies corresponded to an infectious unit (PFU). This accounts for the fact that not all viral particles produce infections.
Finally, in order to determine the effect of the antibody response, we used an antibody titre study in which wild-type (C57BL/6J) mice were subcutaneously inoculated with 100 PFU of WNV 3000.0259 NY2000 [13]. Serum was collected every other day until 12 DPI and neutralizing antibody titres were determined by a standard plaque reduction neutralization assay [36]. The amount of antibody (measured in units of plaque reduction neutralization titre for 50% inhibition of plaques, PRNT50) was determined and reported in the study.
2.2. Fitting models to data
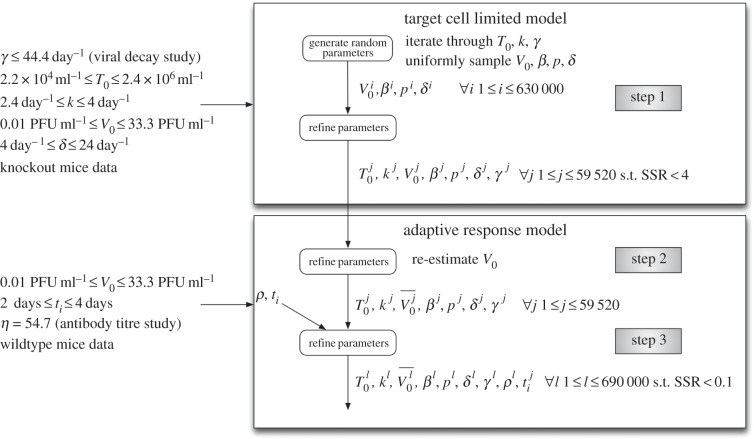
The ordinary differential equations describing our viral kinetic models (equations (2.1)–(2.4) and (2.6)–(2.9)) were solved numerically in Matlab [37]. A fourth-order Runge–Kutta method of integration was used with a step size of 0.0004 h. The curve-fitting method uses nonlinear least-squares regression that minimizes the sum of the squared residuals (SSR) between the experimentally measured virus titres and model-predicted values (measured as log10 PFU ml−1 of serum). We weighted all the data points equally in our fitting procedure, because no uncertainty was provided for the experimental viral titres. We sample parameter values uniformly at random within the biologically plausible parameter space to determine which combinations of parameters could feasibly have generated the observed viremia curves. Our aim is to determine the plausible range of model parameters that produce viremia curves consistent with observed data, rather than the single set of parameters that give the best fit. The computational approach is outlined in figure 1, and a detailed description of our parameter sampling approach is described in the electronic supplementary material.
Figure 1.
Outline of the computational approach. The model parameters are T0 (initial target cell density), k (rate of transition from latently infected to productively infected cells), γ (WNV clearance rate), V0 (inoculated virus density), β (rate constant of infection), δ (death rate of productively infected cells), p (infectious virus production rate), ρ (efficacy of antibody neutralization) and ti (time of initiation of the IgM response).
2.3. A target cell-limited model
The goal of the target cell-limited model is to estimate parameters governing WNV dynamics in IgM knockout mice incapable of mounting an IgM antibody response. We assume that the infection is target cell-limited in these mice as the viral titre decline from the peak occurs well before the IgG response is detected at day 8. Models of target cell-limited acute infection have been developed for both HIV [38] and influenza A virus infection [30]. Here, we use a target cell-limited model with an eclipse phase, given by the following differential equations:
| 2.1 |
| 2.2 |
| 2.3 |
| 2.4 |
Target cells (T) become infected by virus (V) at rate βTV, where β is the rate constant characterizing infection. The initial viral titre and the initial density of target cells are denoted V0 and T0, respectively. The initial density of infected cells (I1 and I2) is assumed to be zero. The separation of infected cells into two classes, I1 cells that are infected but not yet producing virus and I2 cells that produce virus, was proposed earlier for influenza infection [30] and for HIV [16]. This separation increases the realism of the model [39–41], because delays in the production of virus after the time of initial infection are part of the viral life cycle (the eclipse phase).
The parameter 1/k is the average transition time from I1 to I2. Productively infected cells (I2) release virus at an average rate p per cell and die at rate δ per cell, where 1/δ is the average lifespan of a productively infected cell. Free infectious virus is cleared at rate γ per infectious unit per day, for example by phagocytosis or loss of infectivity. Virus also disappears from blood by entering cells during the infection process at rate βTV. The effects of innate immune responses and T cell responses are not explicitly described in this model, but are implicitly included in the clearance rate of virus (γ) and the death rate of infected cells (δ).
We use this model to identify values of V0, β, p and δ that are consistent with observed viral kinetics from IgM knockout mice.
2.4. Adaptive immune response model
In the antibody titre study [13], wild-type mice were subcutaneously infected with WNV and titres of neutralizing antibody were measured during a primary response (figure 2). The data are well described by the following empirical piecewise linear function:
| 2.5 |
Figure 2.
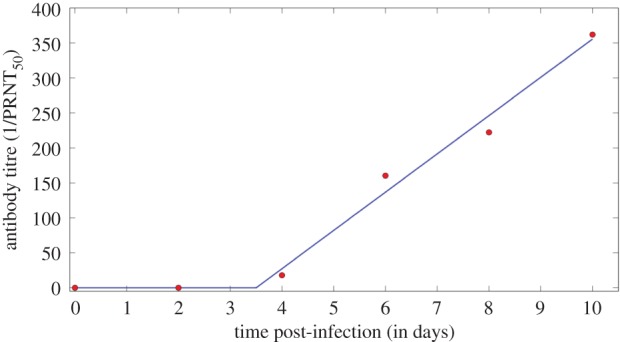
Fit of the function A(t) given by equation (2.5) to the neutralizing antibody data (circles) from the antibody titre study [13]. (Online version in colour.)
The level of neutralizing antibody at time t, A(t), measured by the plaque reduction neutralization test (PRNT) is 0 (undetectable) before time ti and afterwards increases linearly at rate η. This equation was fitted to the antibody titre data (figure 2) and η and ti were estimated (table 1). We note the caveat that our model assumes IgM is absent before time ti, but IgM could exist at low levels undetectable by the assay and models that account for this would be the subject of future investigation. In order to have a simple model and because the data also exhibit piecewise linear effects, we have not explicitly modelled the interaction between the immune system and virus in the form of the equation for the antibody response. The immune response (represented by IgM) and its interaction with the virus is modelled explicitly by an additional clearance term in the equation for virus (equation (2.9)).
Table 1.
Input parameters to model.
| parameters | description | estimated ranges | source |
|---|---|---|---|
| γ | WNV clearance rate | ≤44.4 day−1 (95% of values in interval 29.5–54) also varied with values 10 day−1, 20 day−1 and 44.4 day−1 | fit to viral decay study [35] |
| t1/2 (=ln2/γ) | half-life of free virus | ≥22.5 min (95% of values in interval 18.5–33.8) | calculated |
| η | rate of IgM production | 54.7 day−1 | fit to antibody titre study [13] |
| T0 | initial target cell density | 2.2 × 104–2.4 × 107 ml−1 | estimated from [42] |
| k | rate of transition from I1 to I2 | 2.4–4 day−1 | estimated from [43,44] |
We include the antibody response (equation (2.5)) in the target cell-limited model given by equations (2.1)–(2.4). We assume that neutralizing antibody, A, binds virus, V and neutralizes it with rate constant ρ. The model for infectious virus is altered to become equation (2.9):
| 2.6 |
| 2.7 |
| 2.8 |
| 2.9 |
We use this model to estimate ρ, the rate at which IgM neutralizes virus.
For the model with a humoral response, we estimated the parameters V0, ρ and ti. We re-estimated V0 in this model, because a different experiment was analysed. We also tested a variant of the model in which neutralizing antibody reduces the infectivity of WNV as 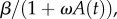 where ω is the efficacy of antibody-mediated reduction in infectivity (in this case assuming ρ = 0). Antibody bound to virus in the plaque assay could also reduce viremia measured in PFU in a manner dependent on the concentration of antibody. Hence, we also rescaled viremia measured in PFU as
where ω is the efficacy of antibody-mediated reduction in infectivity (in this case assuming ρ = 0). Antibody bound to virus in the plaque assay could also reduce viremia measured in PFU in a manner dependent on the concentration of antibody. Hence, we also rescaled viremia measured in PFU as 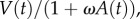 where V(t) represents infectious virions given by equation (2.9). These models produce fits to data that are indistinguishable from the simpler model; therefore, we analyse the simpler model (equations (2.6)–(2.9)). Lastly, because more than one infectious virion may correspond to a PFU, we multiplied the βTV term in equation (2.9) by v, where v = 1 implies one infectious virion is a PFU and v < 1 allows for the possibility that more than one infectious virion constitutes a PFU [34]. To determine if this is an important effect, we scanned through a set of fixed values for 1/v = 1, 2, 3, 5, 10, 100, 1000. We found significantly fewer parameter sets consistent with the data when v < 1, and with v < 0.1 there were no such solutions found. In addition, the quality of the fits did not improve, measured by the minimum SSR, except when 1/v = 2, in which case there was a 1% improvement. Given this minor effect at the cost of an additional parameter, we assumed v = 1 in the remainder of the work.
where V(t) represents infectious virions given by equation (2.9). These models produce fits to data that are indistinguishable from the simpler model; therefore, we analyse the simpler model (equations (2.6)–(2.9)). Lastly, because more than one infectious virion may correspond to a PFU, we multiplied the βTV term in equation (2.9) by v, where v = 1 implies one infectious virion is a PFU and v < 1 allows for the possibility that more than one infectious virion constitutes a PFU [34]. To determine if this is an important effect, we scanned through a set of fixed values for 1/v = 1, 2, 3, 5, 10, 100, 1000. We found significantly fewer parameter sets consistent with the data when v < 1, and with v < 0.1 there were no such solutions found. In addition, the quality of the fits did not improve, measured by the minimum SSR, except when 1/v = 2, in which case there was a 1% improvement. Given this minor effect at the cost of an additional parameter, we assumed v = 1 in the remainder of the work.
2.5. Constraints on model parameters
In order to use the models to identify feasible parameter values, we simulate the model dynamics with different parameters and compare the model-predicted viremia to experimentally observed viremia. These comparisons are computationally expensive, so we reduced the parameter space that needs to be explored based on experimental data. In this section, we summarize how we infer plausible ranges of parameter values from experimental studies.
1/k: the duration of the eclipse phase for neuronal cells infected in vitro with WNV is 6–8 h [43]. Experimental data in vivo suggest that WNV completes one round of replication in dendritic cells within 12 h in mice [44]. Hence, the duration of the eclipse phase in vivo is less than 12 h in mice. We constrained the eclipse phase to be between 6 and 10 h.
1/δ: we allow for a lengthy viral budding period in the model by letting 1/δ, the productively infected cell lifespan, vary from 1 h to 3 days based on the lifespan of productively infected cells in other lytic viral infections, such as HIV [45] and influenza [30].
T0: estimating initial target cell density (T0) requires first identifying which cells are likely to be infected in the first days after inoculation. Subcutaneously inoculated WNV initially infects Langerhans cells [46] and keratinocytes [47] in skin. Langerhans cells then migrate to the draining lymph node and lead to infection of cells that have not been definitely identified but are thought to be macrophages and follicular dendritic cells [46]. A study silencing early viral replication in dendritic cells and macrophages showed suppression of WNV replication [48] (M. Swamy 2014, personal communication) suggesting that these two populations are likely important target cells for WNV.
We estimate T0 by estimating the density of dendritic cells, Langerhans cells and keratinocytes, and then include additional variation reflecting uncertainty in these estimates and the density of other potentially infectable cell types. The density of Langerhans cells in human skin is approximately 1.4 × 103 per mm2 [49]. The density of keratinocytes in human skin is higher: 7 × 104 per mm2, given 2 × 103 melanocytes per mm2 of skin [49] and a 1 : 36 ratio of melanocytes to keratinocytes [49]. WNV is initially localized within a 1 cm2 region of tissue surrounding the site of mosquito inoculation [35]. Thus, we estimate the number of infectable Langerhans cells as 1.4 × 105 and keratinocytes as 7 × 106 in skin.
Because WNV is measured as the number of PFU per millilitre of blood serum we reparametrize our model, so that target cells (T) and infected cells (I1 and I2) correspond to the number of cells required to produce virus in 1 ml, as has previously been done in models of HCV infection [50]. In order to convert these infectable numbers of cells into a density of target cells, we divide by the mouse blood volume (3 ml, based on blood volume of 95 ml kg−1 of body mass [51] and 30 g mass of C57BL/6 J mice [52]), giving a density of the order of 106 ml−1. We note that this is likely a slight underestimate of density, because the serum volume is less than the total blood volume. Given the uncertainty in these estimates, we vary T0 by approximately an order of magnitude above and two orders of magnitude below the estimated density (2.3 × 104–2.3 × 107 ml−1).
V0: bounds on the initial density of inoculated virus (V0) were set in the following manner. In the wild-type and knockout mice studies, 100 PFU of WNV were inoculated into the mouse footpad. Assuming complete absorption into blood (3 ml in mice) gives an upper bound for V0 of approximately 33.3 PFU ml−1. For the lower bound, we assume 1% absorption into whole tissue (30 ml−1 in mice) and obtain approximately 0.01 PFU ml−1. To allow for other possible uncertainties, we allow V0 to vary between 10−4 and 3.3 × 103 PFU ml−1.
ti: the time of initiation of the IgM response (ti) was constrained using data from the antibody titre study [13]. The study measured antibody titres on alternate DPI and the first measurable antibody titre occurred at 4 DPI [13]. Hence, we constrained ti to be between 2 and 4 days.
γ: We estimate γ, the rate of viral decay in equation (2.4), using a simple model and data from the viral decay study [35]. Because virus requires some time to infect cells, produce progeny and lead to the death of infected cells, immediately after intravenous inoculation virus titres decline. We describe virus that is not yet being produced but is still being cleared by setting p = 0 in equation (2.4), giving
| 2.10 |
where 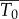 is the density of infectible target cells in blood at the time of infection and
is the density of infectible target cells in blood at the time of infection and 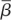 is the infection rate constant of WNV for such target cells. Thus, immediately after infection, we expect an exponential decline in viral titre with rate
is the infection rate constant of WNV for such target cells. Thus, immediately after infection, we expect an exponential decline in viral titre with rate 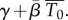 Viral titres in serum within the first 90 min following intravenous inoculation of mice with 105 PFU of WNV were reported in fig. 7 of the viral decay study [35]. Using these data, we estimate
Viral titres in serum within the first 90 min following intravenous inoculation of mice with 105 PFU of WNV were reported in fig. 7 of the viral decay study [35]. Using these data, we estimate 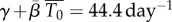 (figure 3).
(figure 3).
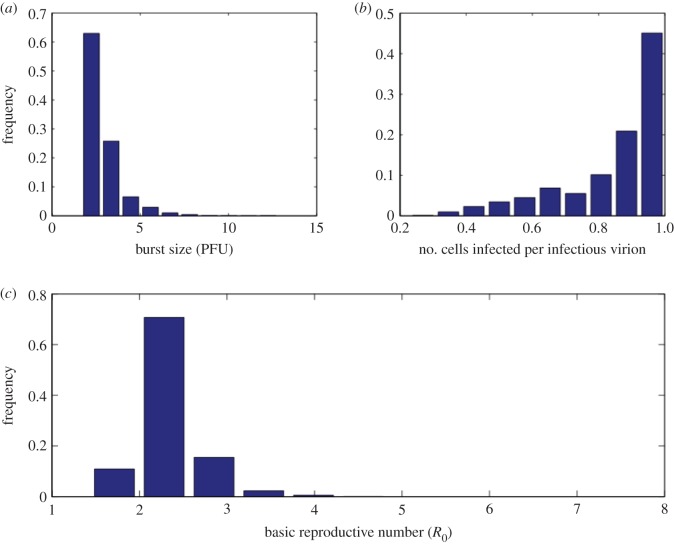
Figure 7.
Histograms of biologically relevant quantities estimated by our approach. (a) Infectious virion burst size (in PFU). (b) Average number of cells infected per infectious virion. (c) Basic reproductive number (R0). (Online version in colour.)
Figure 3.
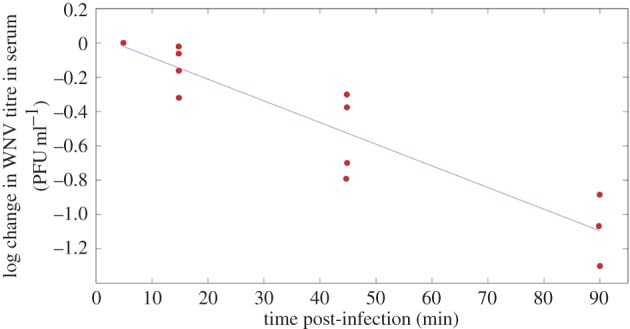
Equation (2.10) fitted to data from [35] (viral decay study) where viral titres in serum were measured within the first 90 min following intravenous inoculation of mice with WNV. Circles, experimental data points from [35]. Solid line, best fit to log-transformed data using linear least-squares regression. r2 = 0.85 and p-value = 2 × 10−5. (Online version in colour.)
Because the target cells in the first minutes after infection are different from those that migrate to lymph nodes, we cannot reliably estimate 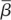 or
or 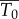 in the footpad inoculation study. Thus, we constrain γ to be less than 44.4 per day (the half-life of free infectious virus (t1/2 = ln 2/γ) is ≥22 min).
in the footpad inoculation study. Thus, we constrain γ to be less than 44.4 per day (the half-life of free infectious virus (t1/2 = ln 2/γ) is ≥22 min).
Because the viral decay study used intravenous inoculation and measured viral titre in blood until 90 min post-infection, the infectible cell types are different from a study that uses footpad inoculation as the route of infection. It is also difficult to estimate what proportion of virus gets into organs and lymph nodes and the proportion of all infectible cells that are reached in the first 90 min after inoculation. Hence, the target cell density for intravenous inoculation (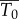 ) in the viral decay study is probably different from the target cell density for footpad inoculation in the wild-type and knockout mice (T0). Because the rate constant of infection depends on the infectible cell types, the corresponding rate constants of infection (β and
) in the viral decay study is probably different from the target cell density for footpad inoculation in the wild-type and knockout mice (T0). Because the rate constant of infection depends on the infectible cell types, the corresponding rate constants of infection (β and 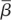 ) are also probably different in the two settings. Because we are unaware of any data that would allow us to independently estimate
) are also probably different in the two settings. Because we are unaware of any data that would allow us to independently estimate 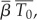 we fixed γ at various values between 44.4 and 10 day−1 and examined the sensitivity of our estimates of other model parameters to this choice.
we fixed γ at various values between 44.4 and 10 day−1 and examined the sensitivity of our estimates of other model parameters to this choice.
η: in order to estimate the rate of induced IgM production (η), we fitted equation (2.5) to data from the antibody titre study [13] and found η = 54.7 day−1.
ρ: in order to estimate the efficacy of induced IgM-mediated virus neutralization, we set a minimum of 1 and explored up to 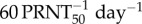 (we also explored values up to 100 with no change in results).
(we also explored values up to 100 with no change in results).
β: in order to estimate the rate constant of infection, we set a minimum of 10−5 and explored up to 10−2 ml−1 day−1.
The variation in model-predicted viremia from the points from step 1 of the computational approach (knockout mice: SSR less than 4 and wild-type mice: SSR less than 0.1) is shown in figure 4 (dotted lines). A visual representation of the model-predicted virus trajectories for an SSR of 4 and 0.1 is also shown in figure 4 (solid lines).
Figure 4.
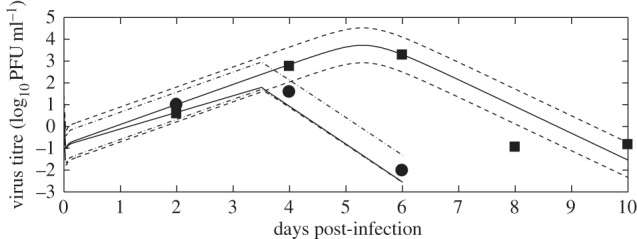
The numerical solutions of the models (virus titre) and variation in model fits. The variation (dotted lines) is from the points from step 1 of the computational approach (knockout mice: SSR less than 4 and wild-type mice: SSR less than 0.1). A visual representation of the model-predicted virus trajectory with SSR of 4 and 0.1 is also shown (solid lines).
3. Results
3.1. Estimating parameters from a target cell-limited model
We set up a framework to sample parameter values within the biologically plausible parameter space to determine which combinations of parameters could feasibly have generated the observed viremia curves.
Our goal is to determine the plausible range of biological parameters rather than the single set of parameters that give the best fit. Hence, we sample the parameter space uniformly at random from the intervals listed in table 2. Each set of sampled parameters was given as input to a subroutine that we developed to identify sets of model parameters that produce viremia curves consistent with observed data. The final output of this computational procedure is an ensemble of model parameters. The computational approach is outlined in figure 1 and explained in greater detail in electronic supplementary material.
Table 2.
Imposed biological constraints and estimated output ranges, mean and standard deviation of model parameters. V0, inoculated virus density (PFU ml−1); β, rate constant of infection (ml day−1); δ, death rate of productively infected cells (day−1); p, infectious virus production rate (day−1); ρ, efficacy of antibody neutralization (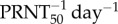 ); ti, time of initiation of the IgM response (days post-infection); R0, basic reproductive number (equation (3.1)); p/δ, infectious burst size (average number of infectious virus particles produced over the lifetime of an infected cell, PFU).
); ti, time of initiation of the IgM response (days post-infection); R0, basic reproductive number (equation (3.1)); p/δ, infectious burst size (average number of infectious virus particles produced over the lifetime of an infected cell, PFU).
| statistic | V0 | β | δ | p | ρ | ti | R0 | p/δ |
|---|---|---|---|---|---|---|---|---|
| imposed constraint | [10−4, 3.3 × 103] | [10−5, 10−2] | [0.3, 24] | [0.3, 120] | [1, 60] | [2, 4] | [1, 30] | [1, 30] |
| estimated range | [0.01, 3.3 × 103] | [2 × 10−4, 9 × 10−3] | [1.7, 24] | [5.2, 118.2] | [2.1, 60] | [3.1, 3.9] | [1.4, 7.1] | [1.7, 12.8] |
| mean ± s.d. | 5 ± 4 | 1.3 × 10−3 ± 1.8 | 11.8 ± 1.7 | 32.8 ± 1.6 | 28.2 ± 18.3 | 3.6 ± 0.1 | 2.3 ± 0.3 | 2.9 ± 1 |
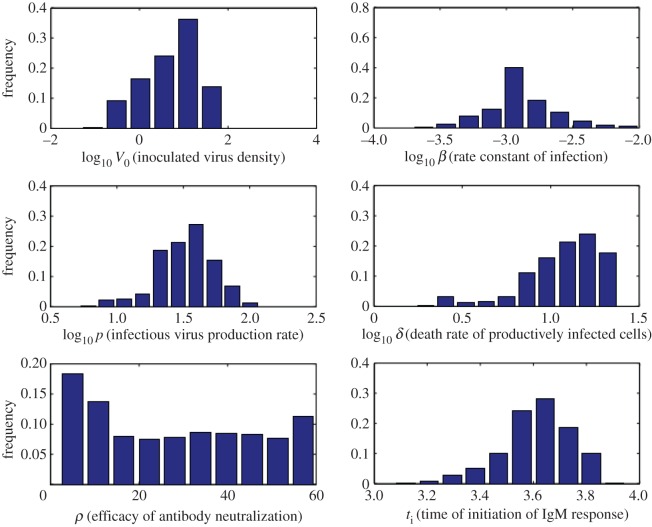
The numerical solution of the target cell-limited model (equations (2.1)–(2.4)) is plotted in figure 5 (black solid line) together with the data from the knockout mice experiment using one representative parameter set from our estimation algorithm. The corresponding target cell depletion is shown by the black dashed line. As one might expect, for an infection with 100 PFU, which is 100% lethal in these mice, target cell depletion is profound. However, in wild-type mice that mount a humoral immune response, the peak titre is significantly lower (red solid line) and the target cell depletion is minimal (red dashed line). The mean, standard deviation and ranges of model parameters determined from the estimation procedure are reported in tables 1 and 2, and the histograms of the estimated model parameters are shown in figure 6.
Figure 5.
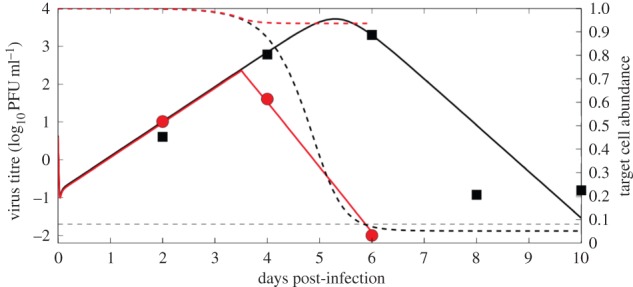
The numerical solutions of the models (virus titre) together with data from the wild-type and knockout study [12]. The solid black line is the numerical solution of the virus titre from the target cell-limited model (equations (2.1)–(2.4)) using one representative parameter set from our estimation algorithm and the black squares are the data from knockout mice. The black dashed line is the predicted corresponding target cell dynamics (fraction of target cells remaining). The red solid line is the numerical solution of the virus titre from the model including a humoral response (equations (2.6)–(2.9)) and the red circles are the data for wild-type mice. The red dashed line is the corresponding predicted target cell dynamics. The limit of detection (LOD) of the assay is indicated by the horizontal dashed line. The day 6 data point for wild-type mice was below the LOD and set to half the LOD [53]. The parameters used to generate the plot are: T0 = 2.3 × 105 ml−1, V0 = 4.3487 PFU ml−1, β = 4 × 10−4 ml day−1, δ = 23.0341 day−1, p = 57.88 PFU day−1, γ = 44.43 day−1, V0 for WT = 4 PFU ml−1,  , ti = 3.5 days.
, ti = 3.5 days.
Figure 6.
Histograms of estimated model parameters (V0, β, p, δ, ρ, ti) from the target cell-limited model and model with humoral response. (Online version in colour.)
From the feasible parameter values in table 2, we calculated the possible values of the basic reproductive number, R0, the average number of second-generation infections produced by a single infected cell in a population of susceptible cells. If R0 is greater than 1, then an infection can be established, whereas an infection rapidly dies out if R0 is less than 1. For the target cell-limited model (equations (2.1)–(2.4)), R0 is given by
| 3.1 |
The average number of infectious virions produced over the lifetime of a productively infected cell is p/δ, and these infectious virions have an average lifetime given by 1/(γ + βT0). The number of cells that these infectious virions can infect over their lifespan is derived by multiplying the previous quantities with the rate constant of infectivity (β) and the initial density of target cells (T0).
Despite considerable uncertainty in imposed bounds on model parameters (e.g. the initial target cell density, T0, is allowed to vary by three orders of magnitude and γ is allowed to vary from 10 to 44.43 day−1), we are able to estimate that R0 has a mean of 2.3 (95% of values in interval 1.7–3; table 2 and figure 7). The infectious virion burst size (p/δ) or the average number of infectious virions released over the lifespan of a productively infected cell was estimated to have a mean of 2.9 PFU (95% of values in interval 1.7–4.7 PFU; figure 7). Because we estimated the ratio of WNV RNA copies to PFU to be approximately 500, this implies that on average an infected cell produces approximately 850–2350 WNV RNA copies over its productively infected lifetime. We also estimated the average number of cells infected by an infectious virion (βT0/(γ + βT0)) to be between 0.3 and 0.99 with higher values being observed more often in our simulations (figure 7).
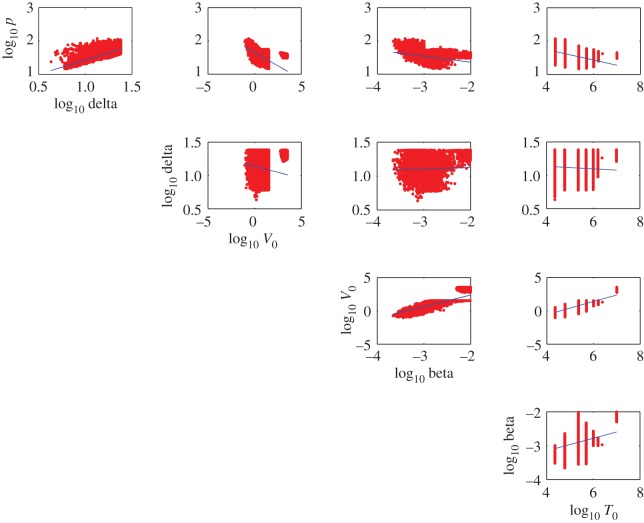
Our computational procedure reveals correlations between parameters (figure 8) which indicate trade-offs between model parameters. For example, the rate constant for infection, β, has a statistically significant relationship with the initial density of inoculated virus, V0 (figure 8, p-value < 0.001, r2 = 0.12). Such trade-offs preclude estimation of unique model parameter values and imply that there is a family of solutions. The estimated ranges of model parameters in conjunction with the correlation plots between model parameters define the family of solutions that are in agreement with data.
Figure 8.
Correlation between target cell-limited model parameters (T0, V0, β, p, δ).
3.2. Estimating parameters from a model incorporating an adaptive immune response
Because the antibody response takes a few days to develop, we assumed the basic viral dynamic parameters were the same early in infection in wild-type and knockout mice.
However, the model with an adaptive antibody response has two additional parameters that affect viremia after antibody response: ρ (efficacy of adaptive IgM neutralization, 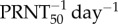 ) and ti (the time of initiation of the adaptive IgM response, DPI). We sample the parameters ρ and ti uniformly at random from the intervals listed in table 2. Each set of sampled parameters was given as input to a computational procedure outlined in figure 1 and explained in electronic supplementary material. The output of this computational procedure is an ensemble of model parameters. The initial density of inoculated virus, V0, was also re-estimated as this was an independent experiment.
) and ti (the time of initiation of the adaptive IgM response, DPI). We sample the parameters ρ and ti uniformly at random from the intervals listed in table 2. Each set of sampled parameters was given as input to a computational procedure outlined in figure 1 and explained in electronic supplementary material. The output of this computational procedure is an ensemble of model parameters. The initial density of inoculated virus, V0, was also re-estimated as this was an independent experiment.
The numerical solution of the adaptive antibody response model (equations (2.6)–(2.9)) is plotted in figure 5 (red solid line) together with the predicted target cell depletion (red dashed line) using one representative parameter set from our estimation algorithm. The variation in the model-generated numerical solutions is shown in figure 4. The feasible range of ti is from 3.1 to 3.9 DPI (table 2). The efficacy of IgM-mediated virus neutralization (ρ) has trade-offs with ti. We found that any value of ρ above 2.1 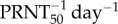 and within the feasible range could have generated the observed viremia curves in wild-type mice (table 2).
and within the feasible range could have generated the observed viremia curves in wild-type mice (table 2).
4. Discussion
WNV is a flavivirus that can cause viral encephalitis in humans. Other related flaviviruses such as dengue virus, Zika virus and HCV are also human pathogens of public health relevance. While the transmission of WNV between hosts has been modelled [54–57], little attention has been paid to modelling the within-host dynamics of WNV. Here we have modified a within-host model first developed for influenza A infection [30] in order to describe WNV infection in mice. Owing to limited data, we developed a method to determine characteristics of within-host WNV infection even though the existing experimental data do not allow us to precisely estimate model parameters.
A humoral immune response contributes to the successful clearing of WNV infection [12]. However, a mathematical model incorporating a humoral response has more parameters than we can estimate reliably from the available data. A basic target cell-limited model has fewer parameters, but fails to capture the effects of a humoral response in limiting the infection. In order to reliably estimate parameters, we first analysed data from the early stages of infection in immunocompromised IgM knockout mice, which allowed us to estimate key parameters in a target cell-limited model related to pathogen replication. Then, we applied a more sophisticated model incorporating a humoral immune response to analyse data from wild-type mice. By combining data from immunocompromised and wild-type mice with a set of mathematical models, we are able to estimate important features of within-host WNV infection, such as the basic reproductive number and the infectious virion burst size.
We used data from laboratory experiments to constrain model parameters to be within biologically realistic ranges. Then, we designed a computational procedure to sample within those ranges to find which combinations of parameter values could have feasibly generated the observed viremia curves in wild-type and immunocompromised mice. The parameter estimates required solving differential equations millions of times. Such computationally hard problems have been solved using other approaches for other infectious diseases [58–60]. Work has also been done in looking at the ensemble of solutions that describe experimental data in intracellular regulatory networks [61] with the result that some parameters are intrinsically unidentifiable compared with others that can be estimated with greater precision [62].
Through our computational procedure, we were able to reduce the uncertainty of some model parameters. We estimated the range of the productively infected cell lifespan (1/δ) to be between 1 and 14 h, and the time of initiation of the IgM response to be between 3.1 and 3.9 days. In order to validate our prediction of 1/δ, we compared it with independent estimates using additional data from in vitro experiments (described in detail in electronic supplementary material). Briefly, analysis of in vitro infection data in dendritic cells and keratinocytes produces estimates that are consistent with our estimates of productively infected cell lifespans: approximately 4–8 h in dendritic cells in vitro (electronic supplementary material, figure S1) and 4–6 h in keratinocytes in vitro (electronic supplementary material, figure S2). We note the caveat that the in vitro estimates may depend on factors such as experimental conditions and the cells and media used, but nonetheless, the in vitro results give us some confidence in our predictions.
Most significantly, we found that important features of viral spread could be estimated with relatively tight bounds. We were able to make these predictions because the computational procedure reveals correlations between estimated model parameters (figure 8). Such correlations indicate trade-offs among parameters that make it difficult to estimate individual parameters precisely, but certain ratios of parameters (which give meaningful characterizations of the virus infection) are more constrained. From the biological constraints on individual parameters, we initially estimated that the basic reproductive number, R0, could vary from 1 to 30, but fitting the model to data constrained R0 to be between 1.7 and 3 (95% CI). Similarly, we initially assumed that the infectious virion burst size could vary from 1 to 30 PFU, but with fitting found the variation is 1.7–4.7 PFU (95% CI; figure 7).
Our approach could be applicable to other emerging diseases that have similar uncertainty in model parameters, pathogens where similar wild-type and knockout experimental data are available or pathogens for which there is scant experimental data. Similar analyses can also be done for other disease models of host dynamics and natural infections by progressively building more complex models on simpler ones. Given the potentially enormous complexity of disease dynamics, parameter estimation is difficult. Parameters estimated from simpler models provide the scaffolding for more complex models that may be able to capture more biological realism. Even when there are correlations among model parameters, this process can reveal constraints on biologically relevant parameters such as the basic reproductive number and infectious virion burst size.
Bayesian methods could be also be valuable: such methods can elegantly incorporate information such as phylogeny or life history as biologically motivated priors [63]. Alternatively, parameter estimates from a previous set of experimental data can be used as priors for a Bayesian model, which would be useful in incorporating both uncertainty in parameters and data. With a suitable choice of priors, Bayesian methods may also help in parameter identifiability as has been shown previously in models of HIV [64].
The humoral immune response has a critical role in conferring protection in WNV-infected hosts [12]. Our model predicted that the effect of the humoral immune response is to protect target cells from infection and death, as only 10% of target cells remain in knockout mice versus 90% in wild-type mice (figure 5). Indeed, there is 100% mortality in knockout mice highlighting the importance of the humoral immune response [12]. The humoral response also reduces the peak viral titre in blood as well as the time taken to reach peak viremia (figure 5). Viral titre above a certain level is an important epidemiological determinant for the spread of WNV [65], which is mosquito-borne. Above a threshold of viremia of 105 PFU ml−1 in blood, a host is capable of infecting an uninfected mosquito, which in turn can infect other uninfected hosts and maintain WNV spread [65]. Hence, hosts that can sustain viremia above this threshold and for a longer duration are pathogen reservoirs. As evidenced in figure 5, in the presence of an induced IgM response, viral titres above a threshold and the time to reach that peak viral titres above a certain level are both reduced compared with the kinetics seen in the IgM-deficient mice. The models presented here are an important step towards a quantitative understanding of the role of the humoral immune response in reducing host infectivity and the epidemic spread of WNV.
Supplementary Material
Acknowledgements
We thank Dr Michael Diamond for sharing his experimental data with us and Dr Mah Lee Ng, Dr Kristen Bernard and Dr Manjunath Swamy for helpful discussions.
Authors' contributions
S.B. performed the experiments. S.B., J.G., R.R., M.M. and A.S.P. analysed the data. S.B., R.R., A.S.P. and M.M. wrote the manuscript. All authors approved the final manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by National Institutes of Health grants (R01-AI104373, R01-AI028433, RR018754 and R01-OD011095), a National Science Foundation grant (NSF EF 1038682), NIH contract HHSN272201000055C and a James S. McDonnell Foundation Complex Systems Scholar Award. Portions of this work were done under the auspices of the US Department of Energy under contract DE-AC52-06NA25396.
References
- 1.Ceausu E, et al. 1997. Clinical manifestations in the West Nile virus outbreak. Rom. J. Virol. 48, 3–11. [PubMed] [Google Scholar]
- 2.Petersen LR, Marfin AA. 2002. West Nile virus: a primer for the clinician. Ann. Intern. Med. 137, 173–179. ( 10.7326/0003-4819-137-3-200208060-00009) [DOI] [PubMed] [Google Scholar]
- 3.Sejvar JJ et al. 2003. Neurologic manifestations and outcome of West Nile virus infection. J. Am. Med. Assoc. 290, 511–515. ( 10.1001/jama.290.4.511) [DOI] [PubMed] [Google Scholar]
- 4.Hayes EB, Komar N, Nasci RS, Montgomery SP, O'Leary DR, Campbell GL. 2005. Epidemiology and transmission dynamics of West Nile virus disease. Emerg. Infect. Dis. 11, 1167–1173. ( 10.3201/eid1108.050289a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deardorff E, et al. 2006. Introductions of West Nile virus strains to Mexico. Emerg. Infect. Dis. 12, 314–318. ( 10.3201/eid1202.050871) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komar N, Clark GG. 2006. West Nile virus activity in Latin America and the Caribbean. Rev. Panam. Salud Publica 19, 112–117. ( 10.1590/S1020-49892006000200006) [DOI] [PubMed] [Google Scholar]
- 7.Lanciotti RS, et al. 1999. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 286, 2333–2337. ( 10.1126/science.286.5448.2333) [DOI] [PubMed] [Google Scholar]
- 8.Kilpatrick AM. 2011. Globalization, land use, and the invasion of West Nile virus. Science 334, 323–327. ( 10.1126/science.1201010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samuel MA, Diamond MS. 2006. Pathogenesis of West Nile virus infection: a balance between virulence, innate and adaptive immunity, and viral evasion. J. Virol. 80, 9349–9360. ( 10.1128/JVI.01122-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smithburn KC, Taylor RM, Rizk F, Kader A. 1954. Immunity to certain arthropod-borne viruses among indigenous residents of Egypt. Am. J. Trop. Med. Hyg. 3, 9–18. [DOI] [PubMed] [Google Scholar]
- 11.Suthar MS, Diamond MS, Gale M Jr. 2013. West Nile virus infection and immunity. Nat. Rev. Microbiol. 11, 115–128. ( 10.1038/nrmicro2950) [DOI] [PubMed] [Google Scholar]
- 12.Diamond MS, Sitati EM, Friend LD, Higgs S, Shrestha B, Engle M. 2003. A critical role for induced IgM in the protection against West Nile virus infection. J. Exp. Med. 198, 1853–1862. ( 10.1084/jem.20031223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diamond MS, Shrestha B, Marri A, Mahan D, Engle M.. 2003. B cells and antibody play critical roles in the immediate defense of disseminated infection by West Nile encephalitis virus. J. Virol. 77, 2578–2586. ( 10.1128/JVI.77.4.2578). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shrestha B, Diamond M.. 2004. Role of CD8+ T cells in control of West Nile virus infection. J. Virol. 78, 8312–8321. ( 10.1128/JVI.78.15.8312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373, 123–126. ( 10.1038/373123a0) [DOI] [PubMed] [Google Scholar]
- 16.Perelson AS, Kirschner DE, De Boer R. 1993. Dynamics of HIV infection of CD4+ T cells. Math. Biosci. 114, 81–125. ( 10.1016/0025-5564(93)90043-A) [DOI] [PubMed] [Google Scholar]
- 17.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271, 1582–1586. ( 10.1126/science.271.5255.1582) [DOI] [PubMed] [Google Scholar]
- 18.Perelson AS, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho DD. 1997. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387, 188–191. ( 10.1038/387188a0) [DOI] [PubMed] [Google Scholar]
- 19.Perelson AS. 2002. Modelling viral and immune system dynamics. Nat. Rev. Immunol. 2, 28–36. ( 10.1038/nri700) [DOI] [PubMed] [Google Scholar]
- 20.Perelson AS, Ribeiro RM. 2013. Modeling the within-host dynamics of HIV infection. BMC Biol. 11, 96 ( 10.1186/1741-7007-11-96) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dixit N, Layden-Almer J, Layden T. 2004. Modelling how ribavirin improves interferon response rates in hepatitis C virus infection. Nature 432, 922–924. ( 10.1038/nature03153) [DOI] [PubMed] [Google Scholar]
- 22.Neumann AU, Lam NP, Dahari H, Davidian M, Wiley TE, Mika BP, Perelson AS, Layden TJ. 2000. Differences in viral dynamics between genotypes 1 and 2 of hepatitis C virus. J. Infect. Dis. 182, 28–35. ( 10.1086/315661) [DOI] [PubMed] [Google Scholar]
- 23.Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, Perelson AS. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282, 103–107. ( 10.1126/science.282.5386.103) [DOI] [PubMed] [Google Scholar]
- 24.Rong L, Dahari H, Ribeiro RM, Perelson AS. 2010. Rapid emergence of protease inhibitor resistance in hepatitis C virus. Sci. Transl. Med. 2, 30ra32. ( 10.1126/scitranslmed.3000544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rong L, Guedj J, Dahari H, Coffield DJ Jr, Levi M, Smith P, Perelson AS. 2013. Analysis of hepatitis C virus decline during treatment with the protease inhibitor Danoprevir using a multiscale model. PLoS Comput. Biol. 9, e1002959 ( 10.1371/journal.pcbi.1002959) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guedj J, Dahari H, Rong L, Sansone ND, Nettles RE, Cotler SJ, Layden TJ, Uprichard SL, Perelson AS. 2013. Modeling shows that the NS5A inhibitor daclatasvir has two modes of action and yields a shorter estimate of the hepatitis C virus half-life. Proc. Natl Acad. Sci. USA 110, 3991–3996. ( 10.1073/pnas.1203110110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsiang M, Rooney JF, Toole JJ, Gibbs CS. 1999. Biphasic clearance kinetics of hepatitis B virus from patients during adefovir dipivoxil therapy. Hepatology 29, 1863–1869. ( 10.1002/hep.510290626) [DOI] [PubMed] [Google Scholar]
- 28.Nowak MA, Bonhoeffer S, Hill AM, Boehme R, Thomas HC, McDade H. 1996. Viral dynamics in hepatitis B virus infection. Proc. Natl Acad. Sci. USA 93, 4398–4402. ( 10.1073/pnas.93.9.4398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewin SR, Ribeiro RM, Walters T, Lau GK, Bowden S, Locarnini S, Perelson AS. 2001. Analysis of hepatitis B viral load decline under potent therapy: complex decay profiles observed. Hepatology 34, 1012–1020. ( 10.1053/jhep.2001.28509) [DOI] [PubMed] [Google Scholar]
- 30.Baccam P, Beauchemin C, Macken C, Hayden F, Perelson A.. 2006. Kinetics of influenza A virus infection in humans. J. Virol. 80, 7590–7599. ( 10.1128/JVI.01623-05) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith AM, Perelson AS. 2011. Influenza A virus infection kinetics: quantitative data and models. Wiley Interdiscip. Rev. Syst. Biol. Med. 3, 429–445. ( 10.1002/wsbm.129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beauchemin CAA, Handel A. 2011. A review of mathematical models of influenza A infections within a host or cell culture: lessons learned and challenges ahead. BMC Public Health 11, S7 ( 10.1186/1471-2458-11-S1-S7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pawelek KA, Huynh GT, Quinlivan M, Cullinane A, Rong L, Perelson AS. 2012. Modeling within-host dynamics of influenza virus infection including immune responses. PLoS Comput. Biol. 8, e1002588 ( 10.1371/journal.pcbi.1002588) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Handel A, Longini IM, Antia R. 2010. Towards a quantitative understanding of the within-host dynamics of influenza A infections. J. R. Soc. Interface 7, 35–47. ( 10.1098/rsif.2009.0067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Styer L, Kent K, Albright R, Bennett C, Kramer L, Bernard K. 2007. Mosquitoes inoculate high doses of West Nile virus as they probe and feed on live hosts. PLoS Pathog. 3, e132 ( 10.1371/journal.ppat.0030132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halstead S, Venkateshan C, Gentry M, Larsen L. 1984. Heterogeneity of infection enhancement of dengue 2 strains by monoclonal antibodies. J. Immunol. 132, 1529–1532. [PubMed] [Google Scholar]
- 37.The MathWorks Inc. 2011. MATLAB version 7.13.0.564.
- 38.Stafford M, Cao Y, Ho D, Corey L. 2000. Modeling plasma virus concentration and CD4+ T cell kinetics during primary HIV infection. J. Theor. Biol. 203, 285–301. ( 10.1006/jtbi.2000.1076) [DOI] [PubMed] [Google Scholar]
- 39.Herz A, Bonhoeffer S, Anderson RM, May RM, Nowak MA. 1996. Viral dynamics in vivo: limitations on estimates of intracellular delay and virus decay. Proc. Natl Acad. Sci. USA 93, 7247–7251. ( 10.1073/pnas.93.14.7247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mittler JE, Sulzer B, Neumann AU, Perelson AS. 1998. Influence of delayed viral production on viral dynamics in HIV-1 infected patients. Math. Biosci. 152, 143–163. ( 10.1016/S0025-5564(98)10027-5) [DOI] [PubMed] [Google Scholar]
- 41.Holder BP, Beauchemin CA. 2011. Exploring the effect of biological delays in kinetic models of influenza within a host or cell culture. BMC Public Health 11, S10 ( 10.1186/1471-2458-11-S1-S10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marti P, Ruiz SR, Marti G, Anjuere F, Vargas HH, Lopez-bravo M, Ardavi C.. 2002. Dramatic increase in lymph node dendritic cell number during infection by the mouse mammary tumor virus occurs by a CD62 L-dependent blood-borne DC recruitment. Blood 99, 1282–1288. ( 10.1182/blood.V99.4.1282) [DOI] [PubMed] [Google Scholar]
- 43.Cheeran MC-J, Hu S, Sheng WS, Rashid A, Peterson PK, Lokensgard JR. 2005. Differential responses of human brain cells to West Nile virus infection. J. Neurovirol. 11, 512–524. ( 10.1080/13550280500384982) [DOI] [PubMed] [Google Scholar]
- 44.Purtha W, Chachu K, Virgin H, Diamond MS. 2008. Early B-cell activation after West Nile virus infection requires alpha/beta interferon but not antigen receptor signaling. J. Virol. 82, 10 964–10 974. ( 10.1128/JVI.01646-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Markowitz M, Louie M, Hurley A, Sun E, Di Mascio M, Perelson AS, Ho DD. 2003. A novel antiviral intervention results in more accurate assessment of human immunodeficiency virus type 1 replication dynamics and T-cell decay in vivo. J. Virol. 77, 5037–5038. ( 10.1128/JVI.77.8.5037-5038.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diamond MS, Shrestha B, Mehlhop E. 2003. Innate and adaptive immune responses determine protection against disseminated infection by West Nile encephalitis virus. Viral Immunol. 16, 259–278. ( 10.1089/088282403322396082) [DOI] [PubMed] [Google Scholar]
- 47.Lim P-Y, Behr MJ, Chadwick CM, Shi P-Y, Bernard KA. 2011. Keratinocytes are cell targets of West Nile virus in vivo. J. Virol. 85, 5197–5201. ( 10.1128/JVI.02692-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye C, Abraham S, Wu H, Shankar P, Manjunath N. 2011. Silencing early viral replication in macrophages and dendritic cells effectively suppresses avivirus encephalitis. PLoS ONE 6, e17889 ( 10.1371/journal.pone.0017889) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoath SB, Leahy DG. 2003. The organization of human epidermis: functional epidermal units and phi proportionality. J. Invest. Dermatol. 121, 1440–1446. ( 10.1046/j.1523-1747.2003.12606.x) [DOI] [PubMed] [Google Scholar]
- 50.Dahari H, Layden-Almer JE, Kallwitz E, Ribeiro RM, Cotler SJ, Layden TJ, Perelson AS. 2009. A mathematical model of hepatitis C virus dynamics in patients with high baseline viral loads or advanced liver disease. Gastroenterology 136, 1402–1409. ( 10.1053/j.gastro.2008.12.060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riches AC, Sharp JG, Thomas DB, Smith SV. 1973. Blood volume determination in the mouse. J. Physiol. 228, 279–284. ( 10.1113/jphysiol.1973.sp010086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson DE, Ruff S. 1999. The Smithsonian book of North American mammals. Washington, DC: Smithsonian Institution Press. [Google Scholar]
- 53.Lynn HS. 2001. Maximum likelihood inference for left-censored HIV RNA data. Stat. Med. 20, 33–45. () [DOI] [PubMed] [Google Scholar]
- 54.Jiang J, Qiu Z, Wu J, Zhu H. 2009. Threshold conditions for West Nile virus outbreaks. Bull. Math. Biol. 71, 627–647. ( 10.1007/s11538-008-9374-6) [DOI] [PubMed] [Google Scholar]
- 55.Lewis M, Rencławowicz J, van den Driessche P. 2006. Traveling waves and spread rates for a West Nile virus model. Bull. Math. Biol. 68, 3–23. ( 10.1007/s11538-005-9018-z) [DOI] [PubMed] [Google Scholar]
- 56.Simpson JE, Hurtado PJ, Medlock J, Molaei G, Andreadis TG, Galvani AP, Diuk-Wasser MA. 2012. Vector host-feeding preferences drive transmission of multi-host pathogens: West Nile virus as a model system. Proc. R. Soc. B 279, 925–933. ( 10.1098/rspb.2011.1282) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pawelek KA, Niehaus P, Salmeron C, Hager EJ, Hunt GJ. 2014. Modeling dynamics of Culex pipiens complex populations and assessing abatement strategies for West Nile virus. PloS ONE 9, e108452 ( 10.1371/journal.pone.0108452) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Smith AM, Adler FR, McAuley JL, Gutenkunst RN, Ribeiro RM, McCullers JA, Perelson AS. 2011. Effect of 1918 PB1-F2 expression on influenza A virus infection kinetics. PLoS Comput. Biol. 7, e1001081 ( 10.1371/journal.pcbi.1001081) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hancioglu B, Swigon D, Clermont G. 2007. A dynamical model of human immune response to influenza A virus infection. J. Theor. Biol. 246, 70–86. ( 10.1016/j.jtbi.2006.12.015) [DOI] [PubMed] [Google Scholar]
- 60.Dahari H, Ribeiro RM, Rice CM, Perelson AS. 2007. Mathematical modeling of subgenomic hepatitis C virus replication in Huh-7 cells. J. Virol. 81, 750–760. ( 10.1128/JVI.01304-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brown KS, Sethna JP. 2003. Statistical mechanical approaches to models with many poorly known parameters. Phys. Rev. E 68, 021904 ( 10.1103/PhysRevE.68.021904) [DOI] [PubMed] [Google Scholar]
- 62.Gutenkunst RN, Waterfall JJ, Casey FP, Brown KS, Myers CR, Sethna JP. 2007. Universally sloppy parameter sensitivities in systems biology models. PLoS Comput. Biol. 3, 1871–1878. ( 10.1371/journal.pcbi.0030189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banerjee S. 2013. Scaling in the immune system. PhD thesis, University of New Mexico, USA.
- 64.Han C, Chaloner K, Perelson AS. 2002. Bayesian analysis of a population HIV dynamic model. In Case studies in Bayesian statistics. Lecture Notes in Statistics, vol. 167, pp. 223–238. New York, NY: Springer. [Google Scholar]
- 65.Komar N, Langevin S, Hinten S, Nemeth N, Edwards E, Hettler D, Davis B, Bowen R, Bunning M. 2003. Experimental infection of North American birds with the New York 1999 strain of West Nile virus. Emerg. Infect. Dis. 9, 311–322. ( 10.3201/eid0903.020628) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.