Abstract
Pluripotent mouse embryonic stem cells (mESCs) show heterogeneous expression levels of transcription factors (TFs) involved in pluripotency regulation, among them Nanog and Rex1. The expression of both TFs can change dynamically between states of high and low activity, correlating with the cells' capacity for self-renewal. Stochastic fluctuations as well as sustained oscillations in gene expression are possible mechanisms to explain this behaviour, but the lack of suitable data hampered their clear distinction. Here, we present a systems biology approach in which novel experimental data on TF heterogeneity is complemented by an agent-based model of mESC self-renewal. Because the model accounts for intracellular interactions, cell divisions and heredity structures, it allows for evaluating the consistency of the proposed mechanisms with data on population growth and on TF dynamics after cell sorting. Our model-based analysis revealed that a bistable, noise-driven network model fulfils the minimal requirements to consistently explain Nanog and Rex1 expression dynamics in heterogeneous and sorted mESC populations. Moreover, we studied the impact of TF-related proliferation capacities on the frequency of state transitions and demonstrate that cellular genealogies can provide insights into the heredity structures of mESCs.
Keywords: Nanog heterogeneity, agent-based modelling, oscillations, flow cytometry, cellular genealogies
1. Introduction
Pluripotent mouse embryonic stem cells (mESCs) possess the remarkable capacity to self-renew indefinitely while retaining the potential to differentiate into all cell types of a mature organism. Thus, cultures of mESCs provide an unlimited source of unspecialized and specialized cells for basic research studies on the regulatory mechanisms of stem cell self-renewal and lineage specification.
A major stimulus for the differentiation of mESCs is autocrine Fgf4/Erk signalling (fibroblast growth factor 4/extracellular signal-regulated kinases) [1–3]. In conventional culture conditions containing the cytokine leukaemia inhibitory factor (LIF) and serum factors, Fgf4/Erk signalling is active and involved in the establishment of phenotypic and functionally different mESCs. In particular, mESCs cultured in LIF/serum display heterogeneous colony structures [4] as well as mosaic expression of several transcription factors (TFs), including Nanog, Rex1, Klf4 and Esrrb [5–9]. Because Nanog expression has a key role in the acquisition of ground state pluripotency in vivo and in vitro [10–12], its heterogeneity has been studied extensively. Nanog reporter cell lines show a bimodal fluorescence distribution with around 50–80% Nanog-high (NH) and 20–50% Nanog-low (NL) cells when analysed by flow cytometry [5,13–15]. In accordance with fluorescence measurements, quantitative measurements of Nanog mRNA molecules in single mESCs also show a broad and bimodal distribution of Nanog transcripts [15,16]. Most remarkable, mESCs possess the dynamic capacity to change between different Nanog expression states as demonstrated first by cell sorting experiments [5,13], and very recently also by live cell image analyses [15,17]. Although mESCs with low Nanog expression can sustain pluripotency and re-express Nanog, they possess a high propensity for differentiation [5,12,16,17]. In contrast, high Nanog expression shields mESCs from differentiation, leading to the concept of Nanog as gate-keeper in the control of mESC differentiation [11,18]. Whether Nanog is directly or indirectly involved in inhibition of differentiation cues is not yet clear. However, by means of a mathematical modelling approach, Muñoz-Descalzo et al. provided one potential explanation. In brief, they suggest that Nanog's gate-keeper function originates from its ability to buffer the differentiation-inducing activity of Oct4 through the formation of stable protein complexes [19]. Rex1, a downstream target of Nanog and a sensitive marker for undifferentiated mESCs, closely mimics Nanog expression dynamics [8,15,20,21], and thus serves as an experimentally accessible readout of the Nanog dynamics [22,23]. In particular, Rex1 reporter cell lines show a bimodal distribution of Rex1-high (RH) and Rex1-low (RL) cells, which are also able to convert into each other [8,24]. Several studies demonstrated that mESC subpopulations, which have been sorted either according to the expression of Nanog or Rex1, can reconstitute the initial bimodal distribution [5,8,9,13]. However, the temporal dynamics of this process has not been studied yet.
In recent years, a number of experimental strategies (reviewed in [25]) and computational models (reviewed in [26]) have been applied to reveal the molecular mechanisms and interactions underlying Nanog heterogeneity. Analysing the dynamic behaviour of small-scale TF network models, we previously identified two potential mechanisms that consistently account for bimodal Nanog distributions and reversible state transitions between different expression levels [22,27]. In the fluctuation model, mESCs can change their expression states stochastically owing to a bistable Nanog switch and transcriptional background noise [22,27]. Alternatively, in the oscillation model, a cyclic Nanog attractor is generated through an activator–repressor system [27]. This attractor causes periodic oscillations between high and low Nanog expression levels [27]. Although both network models establish a dynamic equilibrium of cells with high Nanog/Rex1 expression (NH/RH) and cells with low Nanog/Rex1 expression (NL/RL), the underlying mechanisms leading to this heterogeneous population phenotype differ substantially. We previously argued that the two models are distinguishable either on the basis of measurements of single-cell residence times in the two expression states, or based on the dynamics by which Nanog and Rex1 heterogeneity is re-established after cell sorting [27]. According to the latter option, we designed and performed a cell sorting experiment using a Rex1GFPd2 reporter mESC line maintained in LIF/serum conditions. This cell line has been used, because the reduced half-life of the destabilized GFPd2 (of 2 h) allows for monitoring changes in Rex1 expression on short time scales [24,28]. In particular, we purified RH and RL cells, cultured replicates of both subpopulations in identical conditions and monitored their cellular and molecular development over time by daily flow cytometry measurements. Thus, we obtained quantitative information on the expansion of the two populations, and on the dynamic re-establishment of Rex1 heterogeneity, which allows for evaluating the consistency of our model predictions.
In addition to intracellular mechanisms, cellular properties such as proliferation rates, survival and heredity structures might also impact the establishment and the maintenance of the population heterogeneity of self-renewing mESCs. However, existing intracellular network models represent mESCs solely in terms of their intracellular protein concentrations, neglecting any aspects of cell division, cell death or cellular relations. In order to evaluate the experimental cell sorting data also considering potential regulatory effects of cellular properties, a more comprehensive modelling framework is required.
Here, we present an agent-based model that integrates TF dynamics and cell signalling into autonomous, cellular agents with individual properties. Applying this model, relevant parameters such as cell cycle times and apoptosis rates can be derived directly from experimental results. In line with recent findings based on live cell image analyses [15,17], our model-based analysis provides strong evidence for stochastic fluctuations and bistability underlying Nanog heterogeneity. Moreover, our modelling results predict that state-specific proliferation capacities also impact on the expected frequency of state transitions and that this effect can be revealed by the analysis of pairs of daughter cells.
2. Material and methods
2.1. Cell culture and cell counts
In Rex1GFPd2 mESCs (described in [28]), a destabilized GFP protein is expressed from the Rex1 locus. We used this cell line, because the construct ensures a comparable half-life of 2 h between the GFP and Rex1 protein, which is essential to quantitatively monitor the dynamic behaviour of mESCs also over short time scales. Rex1GFPd2 cells were cultured without feeders on plastic coated with 0.1% gelatin in LIF/serum conditions (DMEM (glutamax high-glucose, Gibco) media supplemented with 10% FBS (Gibco), 0.055 mM β-mercaptoethanol (Gibco), 1 × MEM non-essential amino acids (Invitrogen), 5000 µ ml−1 penicillin–streptomycin (Invitrogen) and 16 ng ml−1 LIF (generated by the MPI-CBG, Dresden)). Cells were seeded at a density of 1.4 × 104 cells per cm2 and split after 3 days in culture. To determine the number of living and dead cells, cultured cells were treated with trypsin, stained with trypan blue and counted using a Countess automated cell counter at different time points.
2.2. Flow cytometry measurements
For the cell sorting experiment, Rex1GFPd2 mESCs were sorted into RH and RL cells using a BD FACSAria III sorter. Subsequently, six replicates of each subpopulation were cultured under LIF/serum conditions. Flow cytometry analyses of the sorted populations were performed every day over a period of 11 days using a BD FACSCalibur cytometer. Data were analysed using BD CellQuest Pro Software.
2.3. The intracellular network models
The transcriptional regulation of the pluripotency factors Oct4, Sox2, Nanog and Rex1 has been modelled by two different interaction networks. The main difference between the two models is how autocrine Fgf4/Erk signalling integrates into the core pluripotency network. The topology of the fluctuation model is outlined in figure 1a. This model has been demonstrated to consistently describe culture-dependent Nanog expression patterns observed in mESCs populations [22]. The topology of the oscillation model is shown in figure 1d. We previously showed that an activator–repressor system between Nanog and a hypothetical factor X can establish sustained oscillations [27]. Here, we assume that Fgf4/Erk is part of the regulatory cycle (i.e. it replaces X), because Fgf4 has been shown to be secreted by cells with high Nanog expression [29], and Erk has been demonstrated to repress Nanog transcription [12,30].
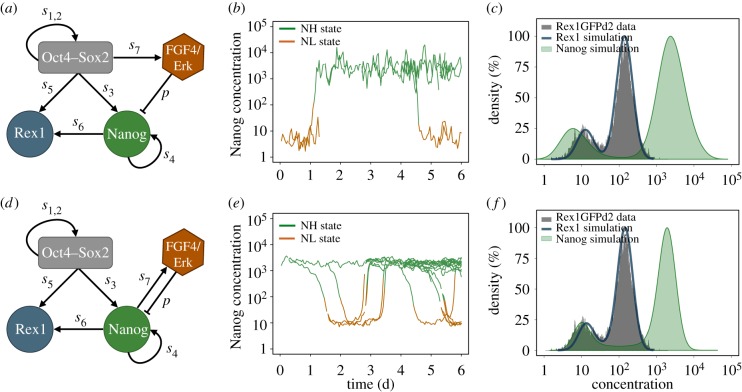
Figure 1.
Comparison of the agent-based fluctuation and oscillation model. (a) Network scheme of the fluctuation model. The negative regulation of Nanog by Fgf4/Erk captures mESCs in a bistable setting. The repression rate is termed p; transcription rates are denoted by si. (b) Stochastic fluctuations between the NH and the NL state of one simulated cell and its progeny. (c) Bimodal Nanog and Rex1 distributions of a simulated cell population adapted to flow cytometry data of Rex1GFPd2 mESCs (grey histogram). (d) Network scheme of the oscillation model. The negative feedback loop between Nanog and Fgf4/Erk generates sustained oscillations. (e) Oscillations between NH and NL levels of one simulated cell and its progeny. (f) Bimodal Nanog and Rex1 distributions of a simulated cell population adapted to flow cytometry data of Rex1GFPd2 mESCs (grey histogram). (Online version in colour.)
In both models, regulatory interactions between Oct4–Sox2 heterodimers, Nanog and Rex1 proteins, and Fgf4/Erk signalling are modelled by a set of coupled stochastic differential equations. The equations describing the temporal changes of Oct4–Sox2, Nanog and Rex1 concentrations (denoted as [OS], [N] and [R]) are equal for the fluctuation and the oscillation model and are given by
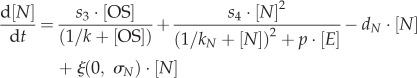 |
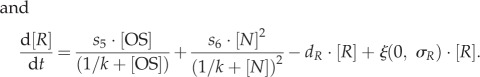 |
The regulation of Fgf4/Erk signalling is specific to the network models. In the fluctuation model, the Fgf4/Erk cascade (denoted as [E]) is assumed to be activated by Oct4–Sox2 heterodimers (as described in [31–33]):
In the oscillation model, Fgf4/Erk is proposed to be activated by Nanog homodimers (as suggested in [29])
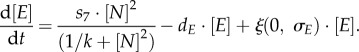 |
Transcription rates are referred to as si (with 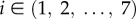 ). The regulation of the Oct4–Sox2 heterodimer is described by a combined transcription rate termed s1,2, which is composed of two transcription rates s1 and s2 for Oct4 and Sox2, respectively, and a formation rate for the heterodimer (cf. [27]). Binding rates are denoted as k and the repression rate of Nanog as p. All proteins and protein complexes are assumed to degrade by first-order kinetics with protein-specific degradation rates dj (with
). The regulation of the Oct4–Sox2 heterodimer is described by a combined transcription rate termed s1,2, which is composed of two transcription rates s1 and s2 for Oct4 and Sox2, respectively, and a formation rate for the heterodimer (cf. [27]). Binding rates are denoted as k and the repression rate of Nanog as p. All proteins and protein complexes are assumed to degrade by first-order kinetics with protein-specific degradation rates dj (with 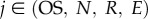 ). Furthermore, we assume that the expression dynamics of all factors are affected by a background noise termed ξ. The stochastic noise is implemented as a zero-mean Gaussian process, which is multiplied by the respective protein concentration. Modelled in this way, the noise increases linearly with gene expression to account for the lower variability of protein levels within the NL/RL state compared with the NH/RH state (cf. width of the two peaks of the fluorescence distributions in figure 1c). The parameter σj (with
). Furthermore, we assume that the expression dynamics of all factors are affected by a background noise termed ξ. The stochastic noise is implemented as a zero-mean Gaussian process, which is multiplied by the respective protein concentration. Modelled in this way, the noise increases linearly with gene expression to account for the lower variability of protein levels within the NL/RL state compared with the NH/RH state (cf. width of the two peaks of the fluorescence distributions in figure 1c). The parameter σj (with 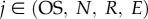 ) defines the protein-specific noise amplitude. One time unit t corresponds to 1 h in an experimental setting. Further details on the network models can be found in [22,27]. Parameters of both intracellular network models have been adjusted previously to fit bimodal Nanog and Rex1 distributions as obtained from flow cytometry analysis of mESCs cultured in LIF/serum conditions. Further details and parameter values are given in electronic supplementary material, table S1.
) defines the protein-specific noise amplitude. One time unit t corresponds to 1 h in an experimental setting. Further details on the network models can be found in [22,27]. Parameters of both intracellular network models have been adjusted previously to fit bimodal Nanog and Rex1 distributions as obtained from flow cytometry analysis of mESCs cultured in LIF/serum conditions. Further details and parameter values are given in electronic supplementary material, table S1.
2.4. The agent-based model framework
In the agent-based model, each living cell is characterized by a set of attributes including a lifetime, a cell fate and an intrinsic cell state that is defined based on the expression of a particular set of genes. In this study, the intrinsic state of a mESC depends on the expression of Nanog and Rex1 determined either by the fluctuation or by the oscillation model described above. Cell fate decisions and protein concentrations are evaluated in discrete time steps mimicking 1 h in an experimental setting.
To simulate a cell's life cycle, we assumed that each cell can die with a rate d at any time. In order to estimate the death rate of mESCs cultured in LIF/serum conditions in an unperturbed situation, we counted dead cells at various time points after seeding. Dead cells are degraded constantly, i.e. they are only visible (countable) for a certain time period. We found that the proportion of dead cells in culture is about 10% (±1.6%). Assuming that dead cells are degraded within 8 h (a rough estimated based on protein half-lives), a death rate d of 0.014 (cells per hour) in the agent-based model is required to account for a constant proportion of about 10% visible dead cells. The death rate has been kept constant for all model scenarios and conditions.
Cell division has been modelled in two different ways (cf. electronic supplementary material, figure S1). In the first scenario, the cell's lifetime persists until a predefined cell cycle time has been reached. Then, the cell divides into two daughter cells, which inherit the protein concentrations from the mother cell and get individual cell cycle times. Because the cell sorting experiment indicated differences in the proliferation capacity of RH and RL cells (cf. Results), in this scenario, cell cycle times of daughter cells are assumed to depend on the Rex1 concentration of the mother cell just before division. Technically, individual cell cycle times were randomly chosen either from a normal distribution with mean cctRH, if the mother cell is RH, or from a normal distribution with mean cctRL, if the mother cell is RL (cf. electronic supplementary material, figure S1a). Furthermore, we assumed that a cell retains the chosen cell cycle time until it dies or divides, even if the cell changes its expression state in between. In contrast, in the second division scenario, cells divide with a certain probability. To account for differences in the cellular turnover of RH and RL cells, in this scenario, the probability of cell division depends on its current Rex1 concentration and increases with the cell's lifetime (cf. electronic supplementary material, figure S1b). Thus, the probability for a cell to divide dynamically changes according to its actual expression state. Also in this scenario, protein concentrations are inherited equally to the two daughter cells.
Owing to the assumption that Rex1 is a direct target gene of Nanog, Rex1 and Nanog expression strongly correlates such that almost 100% of simulated mESCs possess either high levels of both proteins or low levels of both proteins (i.e. mESCs are either NH/RH or NL/RL, cf. electronic supplementary material, figure S3 [15]). Thus, mechanisms that are proposed to depend on the expression state of a cell can be linked to Nanog or Rex1 expression without changing the model results.
2.5. Parameter estimations based on cell counts
To estimate Rex1-related mean cell cycle times (i.e. the model parameters cctRH and cctRL) of mESCs cultured in LIF/serum conditions simultaneous, we performed parameter screenings for the fluctuation and the oscillation model. Therefore, we simulated mESC growth for a broad range of cctRH and cctRL with the objective to minimize the residual sum of squares (RSS) between measured and simulated cell counts. Because differential cell cycle times impact the proportion of RH and RL cells (cf. Results), we adapted the intensity of Nanog repression by Fgf4/Erk (i.e. the model parameter p) prior to the parameter screenings to achieve a final proportion of 75% RH and 25% RL cells similar to the experimental results. Apart from the repression intensity p, all parameters of the intracellular network models remained unchanged. Details on parameter screenings are given in the electronic supplementary material.
2.6. Statistical analysis of cellular genealogies
To investigate whether different assumptions on intracellular mechanisms (e.g. fluctuations versus oscillations) or on cellular properties (e.g. Rex1-related proliferation rates and their origin) result in detectable correlation structures between maternally related cells, simulated cellular genealogies (as shown in figure 3a) have been evaluated statistically. Therefore, the approach described by [34] has been adapted to measure similarities in the cell cycle times between related cells, and correlations in the number of state transitions from the NH into the NL state or vice versa. Briefly, observed differences between related cells in simulated genealogies have been averaged and compared with null distributions that comprise expected differences under the assumption that there are no correlations between related cells. Null distributions are obtained by a randomization procedure, which is explained in detail in the electronic supplementary material. An empirical p-value has been calculated as the fraction of expected values from the null distribution that are less or equal to the observed value. A significant result, defined by a p-value less than 0.05, indicates that related cells are more similar in their cell cycle behaviour or in their expression changes than expected assuming a random cell cycle distribution or uncorrelated transitions.
2.7. Software
The agent-based model has been implemented in Java programing language. Simulation and experimental results have been evaluated using R [35] and Mathematica (Wolfram Research Inc., Champaign, IL).
3. Results
3.1. Two mechanistic explanations for Nanog heterogeneity in self-renewing mouse embryonic stem cells
The intracellular, transcriptional regulation of the pluripotency factors, Oct4, Sox2, Nanog and Rex1, has been modelled by two different interaction networks. The fluctuation model integrates autocrine Fgf4/Erk signalling into the core pluripotency network as illustrated in figure 1a. We have demonstrated that the negative regulation of Nanog by Fgf4/Erk can lead to the coexistence of two stable expression states (bistability), whereas a sufficiently strong transcriptional noise is capable to induce changes between them [22]. Thus, the activity of Fgf4/Erk signalling renders mESCs susceptible to state transitions, but it does not trigger them. Integrated in an agent-based model framework, it is now possible to analyse stochastic fluctuations between high and low Nanog expression levels in related cells. In particular, simulated single cell time courses can reveal dynamics of protein expression as illustrated for one selected mother cell and its progeny in figure 1b, whereas snapshots at different time points provide distributions of the expression levels that can compared with flow cytometry data. As shown in figure 1c, the parameters of the fluctuation model can be adapted to resemble flow cytometry data on fluorescently labelled populations of mESCs cultured in LIF/serum conditions (cf. electronic supplementary material and [22]). However, noise-driven fluctuations are not the only possible explanation for reversible state transitions leading to bimodal distributions of Nanog and Rex1 expression. In the oscillation model, Nanog and Fgf4/Erk are assumed to be part of an activator–repressor system as illustrated in figure 1d. We previously demonstrated that such a network structure can, in general, account for the existence of a cyclic attractor such that Nanog expression levels change periodically [27]. To incorporate latest findings on Fgf4/Erk signalling, we amended this former network model assuming that high levels of Nanog activate Fgf4/Erk signalling [29], which, in turn, inhibits Nanog transcription [12,30]. Consequently, in the oscillation model, Nanog is downregulated in mESC that express high levels of Nanog. Lower levels of Nanog, however, lead to a decline in Fgf4/Erk signalling, its repressive activity diminishes, mESCs can regain Nanog and the regulatory cycle starts again as shown in figure 1e. Thus, in contrast to the fluctuation model, Fgf4/Erk signalling is actively involved in the dynamics of state transitions. Similar to the fluctuation model, Rex1 mimics the oscillatory behaviour of Nanog and the model features characteristic properties of mESCs cultured in LIF/serum conditions (figure 1f).
In the following, we evaluate the consistency of the two agent-based models with the new experimental data on the reestablishment of Rex1 heterogeneity in sorted subpopulations.
3.2. Model assessment regarding transcriptional dynamics
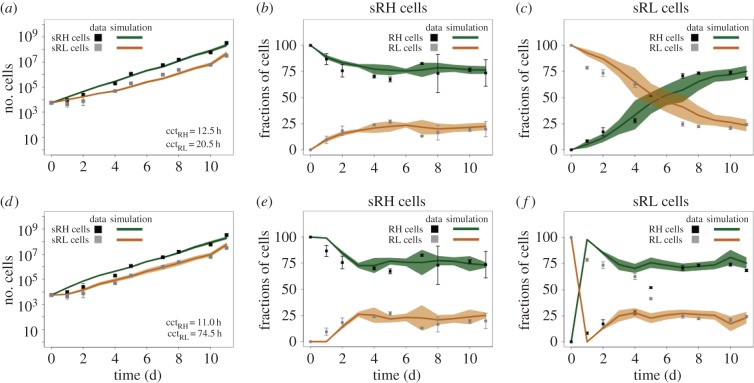
Rex1GFPd2 mESCs cultured in LIF/serum conditions establish a stable bimodal distribution of 70–80% RH and 20–30% RL cells (figure 1c and [21,24,36]). Separating RH and RL cells by flow cytometry and replating the purified subpopulations in identical conditions, the number of mESCs increases exponentially in initially sorted RH (sRH) and sorted RL (sRL) populations as shown in figure 2a. However, the increase is apparently lower in sRL compared with sRH populations. As the number of dying cells fluctuates around 10% likewise in both populations (cf. electronic supplementary material, figure S2), the difference in expansion has to result from differences in the proliferative capacity of the cells. To evaluate this hypothesis, we applied the agent-based model to estimate the mean cell cycle time of the mESCs in the RH state (cctRH) and of mESCs in the RL state (cctRL) based on these cell counts. Therefore, we simulated a comparable cell sorting experiment for a wide range of parameter combinations using both the fluctuation model and the oscillation model. Figure 2 summarizes the experimental and the model-based results for the fluctuation model (upper row, figure 2a–c) and for the oscillation model (bottom row, figure 2d–f).
Figure 2.
Experimental and model results on the expansion and on expression profiles of sorted Rex1GFPd2 mESCs. (a,d) Cell counts of living cells in initially sorted RH (sRH, black squares) and sorted RL (sRL, grey squares) populations together with simulation results (lines) for the best-fit parameters using the fluctuation model (a) and the oscillation model (d). (b,e) Percentages of measured RH (black squares) and RL (grey squares) cells in sRH populations together with simulation results of the fluctuation model (b) and the oscillation model (e). (c,f) Percentages of measured RH (black squares) and RL (grey squares) cells in sRL populations and simulation results of the fluctuation model (c) and the oscillation model (f). The mean cell counts and percentages of cultured mESCs are depicted as squares ± standard deviation (whisker). The mean number and proportions of the simulated replicates are depicted as lines ± standard deviation (bands). (Online version in colour.)
Estimating the two mean cell cycle times cctRH and cctRL using the fluctuation model (cf. Materials and methods), we obtained mean cell cycle times of 12.5 and 20.5 h for sRH and sRL, respectively (figure 2a). In the course of the experiment, the sorted populations revert into the original population mixture of RH and RL cells. The corresponding fractions of RH cells and RL cells in the sorted populations are shown in figure 2b,c. Starting with 100% RH cells (sRH populations), the initial distribution of about 70% RH and 30% RL cells is restored by day 3–4 after seeding both in culture and in the model (figure 2b). Starting with 100% RL cells (sRL populations), the initial equilibrium is also re-established, but considerably slower, because 70% RH cells have to be ‘produced’ either through state transitions of RL cells or through the proliferation of RH cells (figure 2c). Around day 5–6, the proportion of both subpopulations is about 50%. 70% RH cells have been reached by day 11 both in vitro and in silico. Without any additional adjustments, the simulation results of the fluctuation model are highly consistent with the experimental data.
Applying the oscillation model to estimate the two mean cell cycle times cctRH and cctRL, we obtained a much larger difference between the two subpopulations. In particular, we estimated mean cell cycle times of 11.0 and 74.5 h for sRH and sRL, respectively (figure 2d). However, comparing the corresponding predicted cell fractions with the experimental measurements (figure 2e,f), it turns out that the oscillation model fails in reproducing the dynamic re-establishment of sRL populations. The reason for this is the underlying cyclic Nanog attractor. In such a dynamic system, sorting cells according to their expression levels corresponds to a selection of only those cells that are at the same ‘position’ of the regulatory cycle (i.e. that have a clear tendency for the direction of expression changes). More precisely, the oscillation model predicts almost 100% of the sorted RL cells to express low levels of Nanog (cf. electronic supplementary material, figure S3a). Thus, the concentration of Fgf4/Erk in all these cells declines and Nanog transcription can be reinforced. Culturing RL cells in identical conditions, Nanog expression is upregulated almost simultaneously in all cells leading to a conversion in the proportions of RL/NL and RH/NH cells as illustrated in figure 2f. This synchronization is lost over time because of the transcriptional background noise.
These results argue against an oscillatory system as a potential explanation for reversible Nanog and Rex1 expression changes, and clearly favour the noise-driven, bistable system underlying Nanog heterogeneity. Moreover, the analysis revealed that mESCs with high Nanog/Rex1 expression need to have a higher proliferation capacity compared with cells with low Nanog/Rex1 levels. Consequently, we next studied how differential cycle times can impact the dynamic equilibrium of unbiased and primed mESCs using the agent-based fluctuation model.
3.3. The effect of state-specific proliferation capacities on population heterogeneity
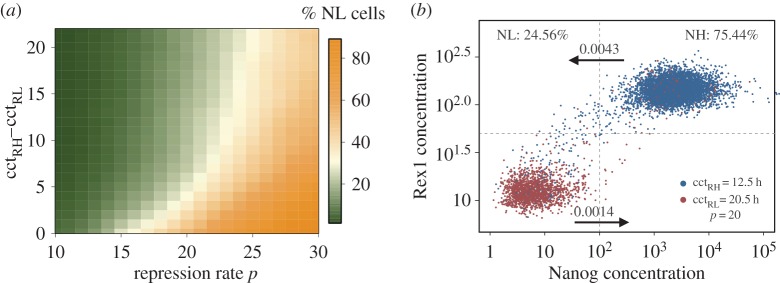
Differential cell cycle times shift the proportion of cells in favour of the subpopulation with the lower cycle time, in this case towards undifferentiated RH/NH cells (table 1). For example, assuming identical mean cell cycle times for both subpopulations (e.g. cctRH = cctRL = 12.5 h), the fraction of NH cells is about 75%. In contrast, differential cell cycle times, as estimated from the experimental data (i.e. cctRH = 12.5 h and cctRL = 20.5 h), increase the fraction of NH cells to 90% under identical conditions. However, the strength of Nanog repression (modelled by repression rate p) has been shown to modulate the frequency of state transitions between the high and the low expression pattern [22] and can therefore antagonize the effect of differential cell proliferation. As illustrated by the heat map in figure 3a, an increase in Nanog repression enhances the proportion of NL cells (horizontal green to orange colour shift) independent of the particular cell cycle times or cell cycle time difference (i.e. cctRH−cctRL). However, the larger the difference between the two mean cell cycle times, the less impact has Fgf4/Erk signalling on the composition of mESCs populations (vertical orange to green shift in figure 3a). Notably, in mESCs populations with differential cell cycle times, the frequency of transitions from the NH to the NL state is, in fact, increased compared with populations with homogeneous proliferation capacities. However, elevated transitions are compensated through the higher proliferation of cells with high Nanog and Rex1 expression (table 1).
Table 1.
The dependency of NH cells and state transition (per cell cycle) on cell cycle times and on Nanog repression by Fgf4/Erk signalling.
| identical mean cell cycle times |
differential mean cycle times |
|||||
|---|---|---|---|---|---|---|
| cctRH = cctRL = 12.5 h |
cctRH = 12.5 h and cctRL = 20.5 h |
|||||
| fractions | state transitions |
fractions | state transitions |
|||
| % of NH | NH to NL | NL to NH | % of NH | NH to NL | NL to NH | |
| repression | ||||||
| p = 10 | 97.77 | 0.0005 | 0.0003 | 98.72 | 0.0005 | 0.0003 |
| p = 15 | 74.03 | 0.0019 | 0.0010 | 90.41 | 0.0018 | 0.0009 |
| p = 20 | 34.41 | 0.0033 | 0.0020 | 75.44 | 0.0043 | 0.0014 |
| p = 25 | 23.97 | 0.0041 | 0.0023 | 53.97 | 0.0067 | 0.0019 |
Figure 3.
The effect of cell cycle time differences on population heterogeneity. (a) Heat map of the proportion of NL cells depending on the difference between the two mean cell cycle times with cctRH = 12.5, and on the repression rate p. (b) Simulation result using the estimated, differential cell cycle time and a repression rate of p = 20. (Online version in colour.)
Summarizing, a higher Nanog repression is required to establish a dynamically stabilized mESC population with around 75% NH/RH and 25% NL/RL cells considering differential cell cycle times of 12.5 and 20.5 h for cells with high and low expression levels, respectively (figure 3b). Because it is largely unknown how mESCs acquire such TF-related cell cycle times, in the last section, we speculate about potential mechanisms and evaluate their implications on correlation structures between related mESCs.
3.4. Analysis of cellular genealogies reveals correlation structures between single mouse embryonic stem cells
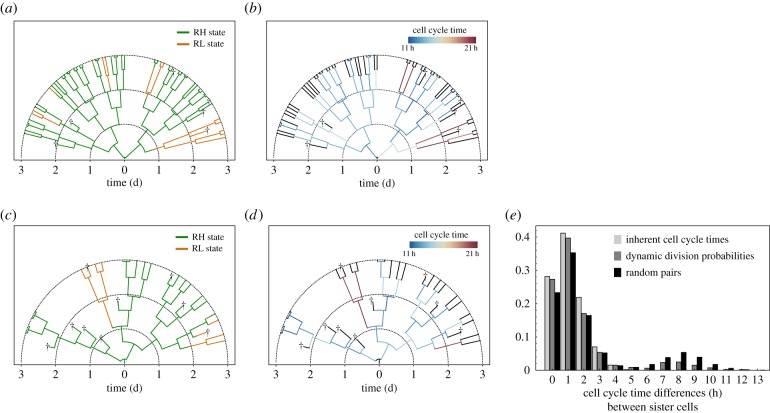
Cellular genealogies as shown in figure 4a,b are suitable instruments to visualize the divisional history of a single cell together with individual properties such as cell cycle times or gene expression levels. Experimentally reconstructed cellular genealogies from live cell image data (like in [17]) successfully demonstrated this approach. However, based on these data, it is not yet possible to directly recover the underlying gene regulatory network and the mechanisms behind the emergence of clonal relationships. Here, we demonstrate that the agent-based model can be used to evaluate the outcome of different hypothesis on underlying mechanisms by providing in silico genealogies. In particular, we show how different model scenarios on the establishment of differential cell cycle times can be distinguished based on a sophisticated statistical analysis of cellular genealogies (Materials and methods; [34,37]).
Figure 4.
Comparison of two cell division scenarios using cellular genealogies. (a,b) Simulated cellular genealogy of one mESCs clone assuming maternally inherent cell cycle times. Cells are depicted as branches starting with the founding cell at the root. Dead cells are marked by crosses. Time is depicted radially. The colour code in (a) depicts Rex1 expression, whereas in (b), individual cell cycle times are illustrated. Black branches indicate that cell cycle times are not measurable owing to cell death, or incomplete cell tracks. (c,d) Simulated cellular genealogy of one mESCs clone assuming dynamic division probabilities. (e) A comparison of observed differences in the cell cycle times of simulated sister cells using the two model scenarios (grey bars) with those of random pairs (black bars). Large cell cycle time differences more than 5 h are extremely rare among sister cells with inherent cell cycle times (light grey), but can be observed between sister cells with dynamic probabilities (dark grey). (Online version in colour.)
In fact, we considered two different scenarios on how Rex1-related cell cycle times can be established and propagated in simulated mESC populations. In the first scenario, we supposed that cell cycle times of daughter cells are determined by the Rex1 concentration of the mother cell and, thus, remain predefined over a cell's lifetime (cf. Material and methods). The internal clock of a cell increases until cell cycle is completed or until the cell dies. One simulated genealogy assuming maternally inherited cell cycle times is shown in figure 4a. The colour code indicates the expression state of a cell. Only a few mESCs switch from the RH (green) into the RL (orange) state, or vice versa, within 3 days of in silico culture. The individual cell cycle times of the cells are illustrated in figure 4b demonstrating that changes in the proliferation capacity owing to state transitions consolidate only after cell division.
Alternatively to a fixed, imprinted cell cycle time, mESCs may rather divide with a certain probability independent of their history. In the second scenario, we therefore assumed that the division probability of a cell depends on the lifetime and the current Rex1 expression level of the cell itself, whereby RH cells have an overall higher division probability than RL cells (cf. Material and methods). Thus, the proliferation capacity of a cell can change dynamically according to with the Rex1/Nanog expression. A simulated genealogy of this alternative scenario is depicted in figure 4c. A few mESCs change their expression state from RH to RL, which leads to a simultaneous decrease of their division probabilities. However, the intrinsic probability of a cell to divide cannot be measured experimentally and differences in the cell cycle times can be observed only after cell division as shown in figure 4d, very similar to the first model scenario (cf. figure 4). Thus, a direct comparison of measured cell cycle distributions (e.g. from single-cell tracking data) will not allow to distinguish between the two model scenarios.
However, the assumption of inherent cell cycle times implies that related cells are rather similar in their cell cycle behaviour, a phenomenon that has already been observed in haematopoietic stem cell populations [38]. Analysing the differences in cell cycle times between sister cells using a number of simulated genealogies from this model scenario (cf. Material and methods), only small differences are observed as shown by the light grey bars in figure 4e. A second peak at larger differences is impossible owing to the heredity structure. In contrast, cell cycle times of sister cells with individual division probabilities can differ more clearly, because mESCs that change the expression state adapt their proliferation rate even before the cell cycle is completed (dark grey bars in figure 4e). Thus, cell cycle times of sister cells can diverge from each other more clearly. A similar bimodal distribution of cell cycle time differences is expected, if cycle times are assigned randomly as shown by the black bars in figure 4e (cf. randomization procedure in the electronic supplementary material).
In conclusion, a comparison of cell cycle time differences between sister cells based on cellular genealogies can indicate whether proliferation capacities are more likely maternally defined and inherited or regulated in a cell-autonomous fashion. However, agent-based models are required for the development of testable predictions (e.g. reference genealogies) to which experimental results can be compared.
4. Discussion
We established an agent-based modelling framework to discriminate between noise-driven fluctuations and deterministic oscillations as potential reasons for the observed Nanog heterogeneity in mESC cultures. Comparing model predictions on the expansion of mESCs and on the establishment of molecular heterogeneity with experimental data, we found stochastic transitions between two stable Nanog expression states to be consistent with distributions of Nanog and Rex1 expression in both heterogeneous and sorted mESC populations. An oscillatory system, in contrast, fails to describe the dynamic reestablishment of Rex1 heterogeneity in presorted cell populations, owing to a synchronization effect induced by the sorting procedure. Thus, we conclude that pluripotent mESCs can express high and low levels of Nanog because of an intrinsic bistable regulation pattern of Nanog, which is retained in LIF/serum conditions through the activity of Fgf4/Erk signalling. The ability of mESCs to reversibly change between the different expression patterns is facilitated by random fluctuations in gene expression (i.e. noise-driven).
This conclusion is supported by recent experimental findings showing the stochastic nature of Nanog state transitions by means of quantitative single-cell expression analysis [15–17]. In particular, Abranches et al. and Singer et al. compared Nanog expression dynamics of single mESCs in different culture conditions revealing that Nanog fluctuations are an inherent property of pluripotent mESCs [14–16]. Additionally, Singer et al. [15] demonstrated that mESCs cultured in LIF/serum conditions establish two distinct Nanog expression states, in which they remain for multiple cell cycles. During these intrastate periods, expression levels have been shown to vary on shorter time scales owing to transcriptional bursting [15,16]. Functional state transitions from the NH to the NL state have been found to occur more often than vice versa [15], similar to the predictions of our fluctuation model (cf. table 1). Although Filipczyk et al. [17] did not observe a clear bimodal Nanog fluorescence distribution, they also reported the existence of relatively stable states of high and low Nanog expression that are influenced by biological noise. Analysing expression levels of a huge number of related mESCs, Filipczyk et al. [17] confirmed that Nanog state transitions are rare and uncorrelated events following no deterministic pattern (cf. our statistical analysis of simulated genealogies, electronic supplementary material, figure S5). Moreover, in agreement with the cell sorting data presented here, the establishment of the observed Nanog distribution required many generations, and no evidence for oscillations could be documented [17].
An alternative noise-driven system that can account for reversible transitions in Nanog expression has been suggested by Kalmar et al. [13]. In contrast to the fluctuation model proposed by us, the authors suggested the existence of only one stable expression state at high Nanog levels (monostability). Excursions from this state towards a region of low Nanog expression are likewise induced by a background noise [13]. However, as no second stable state exists, cells rapidly return to their origin leading to very short residence times of mESCs in the ‘NL state’. This prediction contradicts experimental findings on rather long residence times in both Nanog expression states [15,17]. In contrast, residence times for both Nanog expression states are predicted to be substantially longer in a bistable system, exceeding typical cell cycle times of self-renewing mESCs [22,27].
A hallmark of self-renewing mECSs is their fast cell cycle progression (about 10–16 h) owing to a very short G1 phase [39]. In particular, mESCs omit the early G1 phase, which is known to be the crucial period for cell fate decision processes as differentiation trigger, such as Erk signalling, become active [39,40]. Upon differentiation, however, the duration of G1 increases substantially resulting in longer cell cycle times, as shown for mouse and human ESCs [40,41], and neural stem cells (NSCs) [40]. In this regard, it seems highly relevant that our model analysis reveals differential mean cell cycle times of 12.5 h for NH/RH and 20.5 h for NL/RL cells. Although the molecular basis for this increase is not yet clear, our model analysis proposes a (direct) link between Nanog expression and cell cycle regulation as a consistent explanation of the experimental data. We hypothesize that Nanog downregulation prolongs the cell cycle duration of mESCs such that cells in the NL/RL state are exposed to differentiation cues longer compared with NH/RH cells. This could be another explanation for the observed tendency for NL/RL cells to differentiate (gate-keeper function). Moreover, parameter screens indicated that the model results are more robust against variations in the cell cycle time of RL cells compared with RH cells. While the quality of the model adaptation (measured by a least-square error approach) is rather insensitive against variations of the mean cycle times of RL cells in the range between 16.5 and 24.5 h, the mean doubling time of RH cells can only vary in the order of about 0.5 h to retain the overall quality of fit (cf. electronic supplementary material, table S2). These findings suggest that cells in the NL/RL state display a higher diversity in their proliferation capacities compared with mESCs in the NH/RH state.
In the agent-based model, we were faced with the question if and how the Rex1-related proliferation rates are passed on after cell division. Because experimental references on that issue are missing, we applied the agent-based model to contrast two opposing scenarios. In the first model scenario, mESCs possess an innate cell cycle time, which is determined by the expression state of the mother cell at the time of division. In the second model, mESCs are assumed to divide with a certain probability according to their current expression state. By means of cellular genealogies as shown in figure 4, we were able to analyse correlation structures for these different assumptions, which can eventually be compared with experimental data and thus provide new insights into heredity structures among self-renewing mESCs. However, neither completely inherited nor a completely dynamic cell cycle regulation might be the most realistic scenario. Heredity structures are supposedly more complex and subject to external perturbations such that an imprinted cellular programme may be overridden, e.g. through individual cell–cell interactions or other spatio-temporal effects. Related to the latter reasoning, we recently demonstrated that agent-based models can be extended by a spatial dimension based on live cell imaging data [23]. A comparison of in vitro with in silico colonies with respect to spatial fluorescence patterns revealed that TF-related cell cycle times and cell–cell adhesions favour the clustering of mESCs with high Rex1 expression in the interior of colony structures [23]. Whether differential cell properties result from intrinsic cell states or whether individual properties promote a certain state themselves (e.g. owing to the establishment of cell–cell interactions) has yet to be resolved.
In conclusion, experimental and computational findings on pluripotent mESCs reveal that Fgf4/Erk signalling generates a multistable regime in which cells with different intracellular and cellular properties can coexist. In the presence of the cytokine LIF, a dynamic equilibrium of cells with high and low Nanog expression is maintained. However, we argue that upon LIF removal, this equilibrium is disturbed such that all mESCs gradually switch into the NL state. In the NL state, cell cycle times become prolonged and cell–cell interactions are relaxed, potentially facilitating lineage commitment into one of the three germ layers. How pluripotency regulation and lineage-specific differentiation are mechanistically linked is largely unknown and needs to be investigated more extensive. Quantitative multiscale models can support this process by providing hypotheses and evaluating their consistency with experimental data.
Supplementary Material
Acknowledgement
We thank Tüzer Kalkan for providing Rex1GFPd2 embryonic stem cells.
Authors' contributions
M.H. and I.G. designed the models and conceived the experiments; M.H. implemented the models; M.H. and M.W. performed the experiments; M.H. and T.Z. analysed the data; F.B. and M.W. contributed reagents/materials/analysis tools; M.H. and I.G. wrote the paper; T.Z., I.R., M.W. and F.B. contributed manuscript input; I.G. and I.R. supervised the research.
Competing interests
We declare we have no competing interests.
Funding
This research was supported by the DFG priority programme SPP1356 Pluripotency and Cellular Reprogramming (RO3500/2-1). M.W. was supported by an EMBO LTF and co-funded by the European Commission FP7 (Marie Curie Actions, EMBOCOFUND2010, GA-2010-267146).
References
- 1.Burdon T, Stracey C, Chambers I, Nichols J, Smith A. 1999. Suppression of SHP-2 and Erk signalling promotes self-renewal of mouse embryonic stem cells. Dev. Biol. 210, 30–43. ( 10.1006/dbio.1999.9265) [DOI] [PubMed] [Google Scholar]
- 2.Kunath T, Saba-El-Leil MK, Almousailleakh M, Wray J, Meloche S, Smith A. 2007. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development 134, 2895–2902. ( 10.1242/dev.02880) [DOI] [PubMed] [Google Scholar]
- 3.Stavridis MP, Lunn JS, Collins BJ, Storey KG. 2007. A discrete period of FGF-induced Erk1/2 signalling is required for vertebrate neural specification. Development 134, 2889–2894. ( 10.1242/dev.02858) [DOI] [PubMed] [Google Scholar]
- 4.Scherf N, Herberg M, Thierbach K, Zerjatke T, Kalkan T, Humphreys P, Smith A, Glauche I, Roeder I. 2012. Imaging, quantification and visualization of spatio-temporal patterning in mESC colonies under different culture conditions. Bioinformatics 28, i556–i561. ( 10.1093/bioinformatics/bts404) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers I, et al. 2007. Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230–1234. ( 10.1038/nature06403) [DOI] [PubMed] [Google Scholar]
- 6.Singh AM, Hamazaki T, Hankowski KE, Terada N. 2007. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells 25, 2534–2542. ( 10.1634/stemcells.2007-0126) [DOI] [PubMed] [Google Scholar]
- 7.Hayashi K, Lopes SM, Tang F, Surani MA. 2008. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell 3, 391–401. ( 10.1016/j.stem.2008.07.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. 2008. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development 135, 909–918. ( 10.1242/dev.017400) [DOI] [PubMed] [Google Scholar]
- 9.Canham MA, Sharov AA, Ko MS, Brickman JM. 2010. Functional heterogeneity of embryonic stem cells revealed through translational amplification of an early endodermal transcript. PLoS Biol. 8, e1000379 ( 10.1371/journal.pbio.1000379) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642. ( 10.1016/S0092-8674(03)00393-3) [DOI] [PubMed] [Google Scholar]
- 11.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. 2003. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, 643–655. ( 10.1016/S0092-8674(03)00392-1) [DOI] [PubMed] [Google Scholar]
- 12.Silva J, et al. 2009. Nanog is the gateway to the pluripotent ground state. Cell 138, 722–737. ( 10.1016/j.cell.2009.07.039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalmar T, Lim C, Hayward P, Muñoz-Descalzo S, Nichols J, Garcia-Ojalvo J, Martinez Arias A. 2009. Regulated fluctuations in Nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 7, e1000149 ( 10.1371/journal.pbio.1000149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abranches E, Bekman E, Henrique D. 2013. Generation and characterization of a novel mouse embryonic stem cell line with a dynamic reporter of Nanog expression. PLoS ONE 8, e59928 ( 10.1371/journal.pone.0059928) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singer ZS, Yong J, Tischler J, Hackett JA, Altinok A, Surani MA, Cai L, Elowitz MB. 2014. Dynamic heterogeneity and DNA methylation in embryonic stem cells. Mol. Cell 55, 319–331. ( 10.1016/j.molcel.2014.06.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abranches E, Guedes AM, Moravec M, Maamar H, Svoboda P, Raj A, Henrique D. 2014. Stochastic Nanog fluctuations allow mouse embryonic stem cells to explore pluripotency. Development 141, 2770–2779. ( 10.1242/dev.108910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filipczyk A, et al. 2015. Network plasticity of pluripotency transcription factors in embryonic stem cells. Nat. Cell Biol. 17, 1235–1246. ( 10.1038/ncb3237) [DOI] [PubMed] [Google Scholar]
- 18.Silva J, Smith A. 2008. Capturing pluripotency. Cell 132, 532–536. ( 10.1016/j.cell.2008.02.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muñoz-Descalzo S, Rue P, Faunes F, Hayward P, Jakt LM, Balayo T, Garcia-Ojalvo J, Martinez Arias A. 2013. A competitive protein interaction network buffers OCT4-mediated differentiation to promote pluripotency in embryonic stem cells. Mol. Syst. Biol 9, 694 ( 10.1038/msb.2013.49) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi W, Wang H, Pan G, Geng Y, Guo Y, Pei D. 2006. Regulation of the pluripotency marker Rex-1 by Nanog and SOX2. J. Biol. Chem., 281 23 319–23 325. ( 10.1074/jbc.M601811200) [DOI] [PubMed] [Google Scholar]
- 21.Marks H, et al. 2012. The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149, 590–604. ( 10.1016/j.cell.2012.03.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herberg M, Kalkan T, Glauche I, Smith A, Roeder I. 2014. A modelbased analysis of culture-dependent phenotypes of mESCs. PLoS ONE 9, e92496 ( 10.1371/journal.pone.0092496) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herberg M, Zerjatke T, de Back W, Glauche I, Roeder I. 2015. Image-based quantification and mathematical modeling of spatial heterogeneity in ESC colonies. Cytometry Part A 87, 481–490. ( 10.1002/cyto.a.22598) [DOI] [PubMed] [Google Scholar]
- 24.Kalkan T, Smith A. 2014. Mapping the route from naive pluripotency to lineage specification. Phil. Trans. R. Soc. B 369, 20130540 ( 10.1098/rstb.2013.0540) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torres-Padilla ME, Chambers I. 2014. Transcription factor heterogeneity in pluripotent stem cells: a stochastic advantage. Development 141, 2173–2181. ( 10.1242/dev.102624) [DOI] [PubMed] [Google Scholar]
- 26.Herberg M, Roeder I. 2015. Computational modelling of embryonic stem cell fate control. Development 142 ( 10.1242/dev.116343) [DOI] [PubMed] [Google Scholar]
- 27.Glauche I, Herberg M, Roeder I. 2010. Nanog variability and pluripotency regulation of embryonic stem cells–insights from a mathematical model analysis. PLoS ONE 5, e11238 ( 10.1371/journal.pone.0011238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wray J, Kalkan T, Gomez-Lopez S, Eckardt D, Cook A, Kemler R, Smith A. 2011. Inhibition of glycogen synthase kinase-3 alleviates TCF3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat. Cell Biol. 13, 838–845. ( 10.1038/ncb2267) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frankenberg S, Gerbe F, Bessonnard S, Belville C, Pouchin P, Bardot O, Chazaud C. 2011. Primitive endoderm differentiates via a three-step mechanism involving Nanog and RTK signaling. Dev. Cell 21, 1005–1013. ( 10.1016/j.devcel.2011.10.019) [DOI] [PubMed] [Google Scholar]
- 30.Hamazaki T, Kehoe SM, Nakano T, Terada N. 2006. The Grb2/Mek pathway represses Nanog in murine embryonic stem cells. Mol. Cell Biol. 26, 7539–7549. ( 10.1128/MCB.00508-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ambrosetti DC, Basilico C, Dailey L. 1997. Synergistic activation of the fibroblast growth factor 4 enhancer by SOX2 and OCT-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol. Cell. Biol. 17, 6321–6329. ( 10.1128/MCB.17.11.6321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor OCT4. Cell 95, 379–391. ( 10.1016/S0092-8674(00)81769-9) [DOI] [PubMed] [Google Scholar]
- 33.Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P. 2005. Transcriptional regulation of Nanog by OCT4 and SOX2. J. Biol. Chem., 280 24 731–24 737. ( 10.1074/jbc.M502573200) [DOI] [PubMed] [Google Scholar]
- 34.Bach E, Zerjatke T, Herklotz M, Scherf N, Niederwieser D, Roeder I, Pompe T, Cross M, Glauche I. 2014. Elucidating functional heterogeneity in haematopoietic progenitor cells: a combined experimental and modelling approach. Exp. Hematol. 42, 826–837. ( 10.1016/j.exphem.2014.05.011) [DOI] [PubMed] [Google Scholar]
- 35.R Core Team. 2015. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. See https://www.R-project.org.
- 36.Wray J, Kalkan T, Smith AG. 2010. The ground state of pluripotency. Biochem. Soc. Trans. 38, 1027–1032. ( 10.1042/BST0381027) [DOI] [PubMed] [Google Scholar]
- 37.Glauche I, Lorenz R, Hasenclever D, Roeder I. 2009. A novel view on stem cell development: analysing the shape of cellular genealogies. Cell Prolif. 42, 248–263. ( 10.1111/j.1365-2184.2009.00586.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scherf N, Franke K, Glauche I, Kurth I, Bornhauser M, Werner C, Pompe T, Roeder I. 2012. On the symmetry of siblings: automated single-cell tracking to quantify the behavior of hematopoietic stem cells in a biomimetic setup. Exp. Hematol. 40, 119–130. ( 10.1016/j.exphem.2011.10.009) [DOI] [PubMed] [Google Scholar]
- 39.White J, Dalton S. 2005. Cell cycle control of embryonic stem cells. Stem Cell Rev. 1, 131–138. ( 10.1385/SCR:1:2:131) [DOI] [PubMed] [Google Scholar]
- 40.Roccio M, Schmitter D, Knobloch M, Okawa Y, Sage D, Lutolf MP. 2013. Predicting stem cell fate changes by differential cell cycle progression patterns. Development 140, 459–470. ( 10.1242/dev.086215) [DOI] [PubMed] [Google Scholar]
- 41.Filipczyk AA, Laslett AL, Mummery C, Pera MF. 2007. Differentiation is coupled to changes in the cell cycle regulatory apparatus of human embryonic stem cells. Stem Cell Res. 1, 45–60. ( 10.1016/j.scr.2007.09.002) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.