Abstract
SUMOylation plays critical roles during cell cycle progression. Many important cell cycle regulators, including many oncogenes and tumor suppressors, are functionally regulated via SUMOylation. The dynamic SUMOylation pattern observed throughout the cell cycle is ensured via distinct spatial and temporal regulation of the SUMO machinery. Additionally, SUMOylation cooperates with other post-translational modifications to mediate cell cycle progression. Deregulation of these SUMOylation and deSUMOylation enzymes causes severe defects in cell proliferation and genome stability. Different types of cancers were recently shown to be dependent on a functioning SUMOylation system, a finding that could potentially be exploited in anti-cancer therapies.
Keywords: SUMO, mitosis, SUMOylation, cancer, cell cycle
SUMO: a ubiquitin-like modifier that regulates nuclear processes
The complexity of eukaryotic proteomes is widely expanded by protein processing and a vast array of posttranslational modifications. The quick and reversible attachment of small modifiers is essential for all cellular processes and ensures dynamic and rapid responses to extracellular and intracellular stimuli. Apart from chemical modifications such as phosphorylation [1], glycosylation [2] and acetylation [3], small polypeptides can be attached to proteins, resulting in a change in the activity, localization, half-life or interactome of the target protein. Since the initial discovery of ubiquitin, the founding member of these small protein posttranslational modifications in 1975 [4], a large family of structurally related ubiquitin-like modifiers has been uncovered including SUMO, Nedd8, ISG15, FAT10, FUB1, UFM1, URM1, Atg12 and Atg8 [5–8]. The attachment of these small ubiquitin-like modifiers is catalysed by an enzymatic cascade consisting of an activating enzyme (E1), a conjugating enzyme (E2) and a ligase (E3), and can be reversed by specific proteases. In contrast to the amount of enzymes involved in regulating ubiquitylation, the number of identified enzymes regulating SUMOylation in human cells is limited (Table 1).
Table 1. Deregulation of SUMO machinery components in different cancer types.
| Proteins | Cancer type | Deregulation | References |
|---|---|---|---|
| SUMO proteins | |||
| SUMO1, SUMO2, SUMO3 |
liver | Overexpression of SUMO1 | [111] |
| colon | Overexpression of SUMO1 | [112] | |
| lip | Overexpression of SUMO1 | [113] | |
| gastric | Upregulation of SUMO1 pseudogene3 | [114] | |
| SUMO activating enzyme | |||
| SAE1/2 | gastric | Overexpression of SAE2 | [115] |
| lung | Overexpression of SAE2 | [116] | |
| breast | Low expression of SAE1 and SAE2 correlates with better survival | [69] | |
| SUMO conjugating enzyme | |||
| UBC9 | lung | Overexpression of UBC9 | [117] |
| primary colon and primary prostate | Overexpression of UBC9 | [64] | |
| metastatic breast, prostate and lung | Downregulation of UBC9 | [64] | |
| breast | High levels of UBC9 correlate with higher risk for cancer and poor response to chemotherapy | [118,119] | |
| multiple myeloma | Overexpression of UBC9 | [120] | |
| ovaries | Overexpression of UBC9 | [63] | |
| brain | Overexpression of UBC9 | [121] | |
| SUMO ligases | |||
| PIAS1, PIAS2, PIAS3, PIASγ (PIAS4) |
prostate | Overexpression of PIAS1 | [122,123] |
| Overexpression of PIAS3 | [124] | ||
| breast | Overexpression of PIAS1 | [125] | |
| Overexpression of PIAS3 | [124] | ||
| multiple myeloma | Overexpression of PIAS1 | [120] | |
| gastric | Downregulation of PIAS3 | [126] | |
| lung | Overexpression of PIAS3 | [124] | |
| brain | Overexpression of PIAS3 | [124] | |
| ovaries | Expression of PIASγ correlates with cancer aggressiveness | [127] | |
| pancreas | Overexpression of PIASγ | [128] | |
| RanBP2 | small cell lung cancer | Upregulation of RanBP2 compared to non-small cell lung cancer | [129] |
| leukemia | Expressed as RanBP2-ALK fusion protein | [130] | |
| SUMO proteases | |||
| SENP1, SENP1, SENP2, SENP3, SENP5, SENP6, SENP7 |
prostate | Overexpression of SENP1 | [131,132] |
| Overexpression of SENP3 | [101] | ||
| colon | Overexpression of SENP1 | [133] | |
| Overexpression of SENP3 | [134] | ||
| liver | Downregulation of SENP2 | [135] | |
| Overexpression of SENP6 | [136] | ||
| bladder | Downregulation of SENP2 | [137] | |
| gastric | Overexpression of SENP3 | [138] | |
| oral | Overexpression of SENP3 Overexpression of SENP5 |
[139–141] | |
| breast | Low expression of SENP5 contributes to better survival | [142] | |
| Downregulation of SENP6 | [143] | ||
| USPL1 | breast | Expression contributes to risk of Grade 3 breast tumors | [144] |
The SUMOylation pathway consists of the dimeric SUMO E1 SAE1/UBA2, the single E2 Ubc9 and E3 ligases including Protein Inhibitor of Activated STATs (PIAS)-family members, Ran Binding Protein 2 (RanBP2) and a few other E3 ligases. SUMOylation is reversible; SUMO proteases, originally named Sentrin-specific Proteases (SENPs) remove SUMOs from target proteins. Covalent SUMOylation of target proteins can enable their non-covalent interaction with partner proteins via SUMO Interaction Motifs (SIMs) in these proteins.
SUMO is predominantly found in the nucleus and plays a crucial role in many nuclear processes such as gene expression, genome stability [9], the DNA damage response [10], protein trafficking [11] and cell cycle control. It is therefore not surprising that SUMO signal transduction has been implicated in the development of several different cancer types, which could potentially be exploited in anti-cancer therapies. In this review we will focus on the role of SUMO in cell cycle regulation, specifically highlighting its physiological relevance and its deregulation in cancer. Recent progress includes the identification of many novel SUMO substrates with important roles in cell cycle progression using proteomic approaches, and the demonstration that different mouse cancer models are dependent on a functioning SUMOylation system. Switching of SUMOylation in these cancer models caused a proliferation block of the cancer cells, showing that SUMO conjugating enzymes are potential drug targets.
Twenty years of SUMO research in cell cycle control
SUMO was linked to cell cycle progression even before the identification of the small protein modifier itself. Twenty years ago, the yeast SUMO conjugating enzyme Ubc9 was first proposed to be a ubiquitin conjugating enzyme. Ubc9 was shown to be required for progression through mitosis by degrading M-phase cyclins [12]. Consequently, disrupting UBC9 in budding yeast resulted in large budded cells bearing only a single nucleus with a short spindle and replicated DNA, a hallmark of G2-M arrested cells. The yeast SUMO homologue, Suppressor of Mif Two 3 (Smt3), was also linked to cell cycle progression right from its identification. Smt3 was found as a functional suppressor of a mutation in the centromeric protein MIF2, a homologue of the mammalian Centromeric Protein-C (CENP-C). Similarly, disrupting SMT3 blocked yeast cells at the G2 to M transition [13]. Yeast cells lacking UBA2 or AOS1, both essential subunits of SUMO activating enzyme, or SMT3 itself, did not display sharp arrests in a particular phase of the cell cycle. Indeed null mutants derived from sporulation were found to undergo multiple cell divisions before they ceased growing [14,15]. These observations suggest that SUMOylation is not exclusively important during the G2/M transition in the yeast cell division cycle. Interestingly, early work of Erica Johnson revealed that the yeast septins are SUMOylated in a cell cycle dependent manner [16].
SUMOylation is also critical for cell cycle progression in mammals. Ubc9-deficient mouse embryos harboured severe mitotic defects, including larger metaphase plates due to hypocondensation, an increased number of anaphase bridges and an increased number of polyploid cells, leading to premature death at the early postimplantation stage [17]. Mammals harbour four different SUMO family members, SUMO1-4, only three of which appear to be conjugated, SUMO1-3. SUMO2 is the only essential SUMO family member. SUMO2-deficient mice died early during embryonic development (embryonic day 10.5), with severe developmental defects [18]. In contrast, SUMO3-/- mice were viable and did not show any overt defects [18]. Due to their high sequence similarity in their mature forms, it is unlikely that SUMO2 and SUMO3 have major different functional properties. However, the two proteins show a striking difference in their expression levels. The SUMO2 mRNA accounts for 80% of the entire SUMO mRNA pool in total mouse embryos at E7.5, whereas SUMO3 is only expressed at very low levels (2%) [18]. SUMO1-/- mice are also viable and fertile and lack any obvious phenotype, indicating that the SUMO2 pool is able to compensate for the absence of SUMO1 [19,20]. Mating experiments between the different knockout mice further indicated that the different SUMO family members functionally compensated for each other [18]. Thus, accurate mouse development appears to require the presence of a critical level of SUMO2.
A conditional knockout of Ubc9 in chicken DT40 cells did not arrest cells in a specific cell cycle stage, but led to an increased number of apoptotic cells with multiple or fragmented nuclei [21]. Similarly, knockdowns of Ubc9 or the SUMO E1 subunit SAE2 in human cells severely reduced the cell proliferation rate but did not arrest cells in a specific cell cycle stage [22,23]. This could be explained by SUMO having essential functions during all cell cycle phases in mammalian cells, not just during the G2/M transition. This multitude of SUMO signals that are essential for efficient cell cycle progression will be detailed in this Review.
Redistribution of the SUMO machinery during mitosis
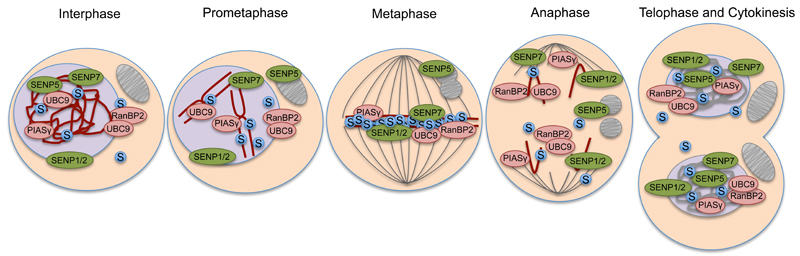
SUMO localizes at centromeres, kinetochores and mitotic and meiotic chromosomes in different organisms including humans and frogs [24–26]. In Caenorhabditis elegans, SUMO accumulates at the metaphase plate but its presence decreases during anaphase [27], which is regulated via the interplay between the SUMO ligase GEX3 interacting protein 17 (GEI-17) and the SUMO protease Ubiquitin-Like Protease 4 (ULP-4). These observations demonstrate that SUMO target proteins that are essential for cell cycle progression need to be SUMOylated in an accurate temporal and spatial manner. This is predominantly achieved via changes in the localization and activity of SUMO ligases and proteases. Consistently, several SUMO ligases and proteases are redistributed during mitosis (Figure 1).
Figure 1. Redistribution of the Small Ubiquitin-like Modifier (SUMO) machinery during mitosis.
During interphase, SUMO2/3 (S in blue circles) and UBC9 are mostly present inside the nucleus (purple) and also exhibit specific functions in the cytoplasm. During early mitosis, the chromosomes (dark red) condense and align at the equator of the cell. Similarly, SUMO2/3 accumulates at the metaphase plate and disappears again during anaphase after the sister chromatids have separated. The RanGap-SUMO1/UBC9/RanBP2 complex and the SUMO proteases SENP1 and SENP2 are mostly located at the nuclear envelope and the nuclear pores during interphase and redistribute to the centromeres and kinetochores during early mitosis. SUMO1 remains associated with the SUMO E3 ligase RanBP2 during mitosis and is therefore also present at mitotic chromosomes. Similarly, the SUMO ligase PIASγ and the SUMO protease SENP7 are known to accumulate at centromeric and pericentric regions during metaphase. The SUMO protease SENP5, by contrast, translocates from the nucleoli to the mitochondria (grey) at the early onset of mitosis prior to nuclear breakdown. The SUMO E2 UBC9 and SUMO E3 ligases are shown in pink and SUMO proteases (SENPs) are shown in green.
The SUMO E3 ligase PIASγ is specifically targeted to mitotic chromatin [28]. Furthermore, the stable complex of RanGAP1-SUMO1, the SUMO E3 ligase RanBP2 and the SUMO E2 Ubc9, present at the nuclear envelope during interphase, translocates to the kinetochores and the spindle apparatus during mitosis [29,30]. Downregulation of RanBP2 in mice leads to severe defects in chromosomal segregation resulting in increased aneuploidy, underlining its essential role during mitotic progression [31].
Similar to RanBP2, the SUMO proteases SENP1 and SENP2 are mostly localized at the nuclear envelope and nuclear pore complexes during interphase [32]. During mitosis, green fluorescent protein (GFP)-fusions of both SENP proteins were found to accumulate at outer kinetochores [33]. Endogenous SENP1 was also found at centromeres and spindle microtubules. Interestingly, overexpression of SENP2 led to cell cycle arrest at the pro-metaphase stage, caused by defects in targeting the microtubule motor protein CENP-E to kinetochores [34]. Several studies showed that the N-terminal domains of SENP proteases determine their subcellular localization and interaction partners [35–37]. Indeed, a comparison between different chimeric fusion proteins of SENP1 and SENP2 revealed that the N-terminus of SENP2 is essential for its unique features. This domain is required to bind the nuclear pore subcomplex Nup107-160 and karyopherin α, which move to the outer kinetochores during prometaphase [33]. Whereas only SENP2 overexpression blocks cells in metaphase, SENP1 but not SENP2 depletion caused slower progression through mitosis due to delayed chromosome separation, suggesting an essential role for SENP1 during the spindle assembly checkpoint and the disintegration of cohesins [33]. SENP7 directly interacts with and stabilizes the heterochromatin protein 1 (HP1) at pericentric regions in mouse cells, a step needed for the formation of centromeric organization and accurate chromosomal segregation [38,39]. Accordingly, SENP7 is able to deSUMOylate HP1 in vivo and depletion of SENP7 leads to prolonged time spent in mitosis.
Interestingly, SENP5 plays a cytoplasmic role during mitosis. SENP5 translocates to mitochondria at the onset of mitosis, where its function is needed to promote mitochondrial fragmentation [40]. Additionally, SENP5 plays a role in cytokinesis [41].
These examples highlight the functional redistribution of the SUMO machinery during specific cell cycle stages to enable cell cycle progression. It would be of great interest to further investigate the dynamic signals required for accurate localization of the SUMO machinery during cell cycle progression.
SUMOylation and Cyclin-dependent kinases in concert
The activity and localization of the SUMO machinery can be influenced by interaction partners and by crosstalk with other posttranslational modifications. Interestingly, SENP3 is heavily phosphorylated during mitosis, suggesting its regulation via mitotic kinases [42]. Additionally, kinases that are active at defined moments during cell cycle progression can also influence SUMOylation by cooperating with Ubc9. A subset of SUMO targets is regulated via an internal Phosphorylation Dependent SUMOylation Motif (PDSM) ΨKxExxSP, where Ψ is a large hydrophobic residue [43]. Phosphorylation of these sites enhances the binding of Ubc9 to stimulate SUMOylation of these targets [44]. This PDSM motif partly overlaps with the motif [S/T]Px[K/R], the preferential phosphorylation site for cyclin-dependent kinases (CDKs), which are key players in cell cycle progression [45]. Combined, this suggests that CDKs might regulate SUMO target proteins via the consensus motif ΨKxExxSPx[K/R].
The degradation of the human Flap endonuclease 1 (FEN1) serves as example for the strictly defined hierarchy of SUMOylation and other PTM events needed at a specific point in time during cell cycle progression. FEN1 is involved in the processing of Okazaki fragments during DNA replication in S phase, and is phosphorylated by cyclin A-dependent kinases, which are only active at this specific stage of the cell cycle [46]. Although lacking a regular PDSM motif, the three-dimensional structure of the protein reveals that a phosphorylated serine residue lies adjacent to the SUMO acceptor site [47]. Phosphorylation is a prerequisite for SUMOylation, which in turn promotes polyubiquitylation as a third posttranslational modification and eventually leads to degradation of the protein at the end of S phase, when it is no longer needed. It would be interesting to overlap the targets of cyclin-dependent kinases with known SUMO substrates, specifically as recent advances in proteomic approaches allow us to identify both phosphorylation and SUMOylation sites [48–52]. This might lend insight into the substrates that are co-regulated by SUMOylation and CDKs.
The mutual interaction between CDKs and SUMOylation furthermore includes the regulation of the activity of CDKs by SUMOylation. SUMOylation of CDK6 during mitosis has been described to block ubiquitylation and subsequent degradation of the enzyme, ensuring its presence during G1/S transition [53]. Additional CDKs and cyclins have been identified as SUMO targets in recent proteomic screens and targeted approaches, including CDK1, CDK9, CDK11 and Cyclin-E, further highlighting crosstalk between SUMOylation and phosphorylation during cell cycle regulation [23,48,54] (Figure 2).
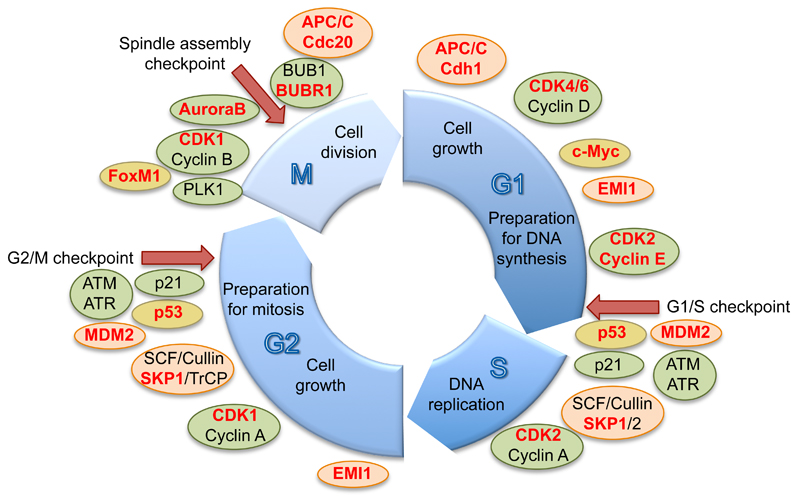
Figure 2. SUMOylation of important cell cycle regulators.
The cell cycle exhibits four different cell cycle phases. In G1 phase, the cell prepares for DNA replication, which takes place during S phase. In the subsequent G2 phase, the cell undergoes further preparations to finally be able to enter mitosis, in which the chromosomes segregate and the cell divides. Several checkpoints throughout the cell cycle ensure the integrity of the genome and the proper division of the cell. A multitude of transcription factors (yellow) and enzymes regulating phosphorylation (green) and ubiquitylation (red) events are important guards of these checkpoints or influence other essential steps for cell cycle progression at specific time points. This figure summarizes some of the most important cell cycle regulators, many of which have been described to be SUMOylated and are highlighted in red font.
These proteomic approaches have also greatly helped in identifying other SUMO target proteins essential for cell cycle progression and have uncovered global SUMO dynamics throughout the cell cycle [23,55]. It would be interesting to identify groups of specific substrates regulated by the different dynamic SUMO machinery components. The identification of target subsets for each SUMO E3 enzyme and each SUMO protease could provide more insight into the specific roles played by these enzymes in cell cycle progression. In addition, further functional analysis of specific SUMO targets is of high importance, as the exact effect of SUMOylation on specific target proteins with essential roles in cell cycle regulation has only been described in a few cases. The effect of SUMOylation on the activity of Topoisomerase IIα, for example, has been studied in relatively more detail (Box 1).
Text Box 1. SUMOylation regulates Topoisomerase IIα in mitosis.
A key SUMO target protein in cell cycle progression is Topoisomerase IIα (TOPO IIα), which is needed for decatenation of sister centromeres to prevent the formation of anaphase bridges during chromosomal segregation. TOPO IIα is SUMOylated both in S. cerevisiae and in higher eukaryotes [31,106,107]. SUMOylation of TOPO IIα takes place preferentially during mitosis and inhibits the decatenation activity of the enzyme in vitro [107]. The SUMO ligase RanBP2 modifies TOPO IIα in mammalian cells and is essential for proper TOPO IIα localization at the inner centromere [31]. By contrast PIASγ, but not RanBP2, is needed for SUMOylation of TOPO IIα in Xenopus egg extracts. Recruitment of PIASγ to centromeric regions during metaphase is executed by the Rod/Zw10/Zwilch (RZZ) complex [108] (Figure 3). At the onset of anaphase, as soon as the sister chromosomes are accurately aligned, the RZZ is released, which leads to translocation of PIASγ away from the kinetochores. Due to the absence of PIASγ and the presence of multiple SENPs, TOPO IIα is no longer modified via SUMO during anaphase, resulting in increased decatenation activity and proper chromosome segregation.
Interestingly, the Polo-like kinase 1- interacting checkpoint helicase (PICH) interacts with SUMOylated TOPO IIα at mitotic chromosomes of Xenopus egg extracts [109]. In addition, PICH also binds to SUMOylated PARP-1 at mitotic chromosomes and SUMOylation of PICH itself inhibits its loading onto DNA. These observations strongly suggest that PICH localization and activity at mitotic chromosomes are tightly regulated by SUMOylation. SUMOylation of PARP-1, however, did not alter the activity or localization of the enzyme. Still, inhibition of the SUMO pathway strongly increases PARylation at mitotic chromosomes, demonstrating that these two posttranslational modifications harbour interconnected roles during chromosomal segregation [110].
Protein group modification at mitotic chromosomes
In many cases, several subunits of the same regulatory complexes are targeted via SUMOylation. This is consistent with proteomic studies, showing that SUMO frequently modifies entire functional groups of proteins [56]. Protein group SUMOylation, potentially enhanced by the formation of longer SUMO2/3 chains on the target proteins, can trigger the formation of complexes at centromeric regions and might therefore be essential for chromosome alignment and segregation. Interestingly, both SENP6, a SUMO protease with a preference for poly-SUMO chains, and Ring Finger Protein 4 (RNF4), known to ubiquitylate proteins bearing poly-SUMO chains, are essential regulators of mitotic progression [57]. In human HeLa cells, depletion of SENP6 leads to defects in spindle assembly and chromosome condensation during mitosis by decreasing the stability of essential complexes, particularly the CENP-H/I/K complex, at the kinetochores [57]. Interestingly, this phenotype can partially be rescued by depletion of RNF4, suggesting antagonistic functions of these two proteins at the inner kinetochores during mitosis (Figure 3).
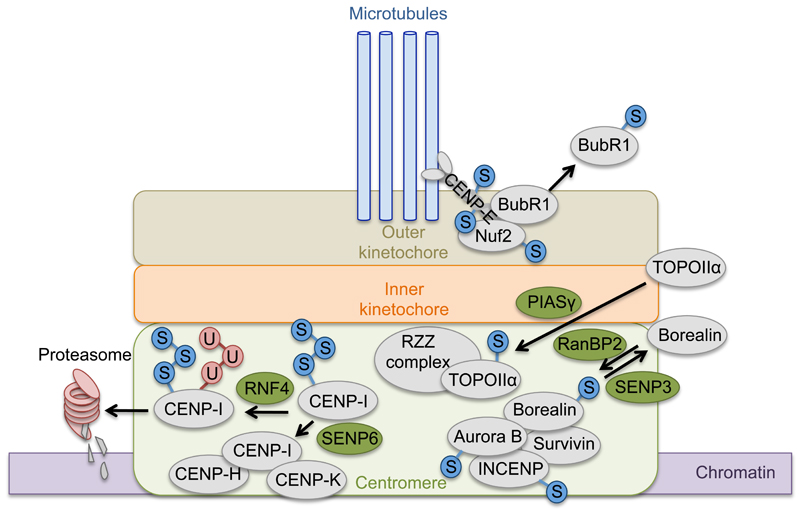
Figure 3. SUMO target proteins at centromeres and kinetochores.
Several SUMOylation events have been identified to be essential for accurate chromosomal alignment and segregation. This figure depicts the localization of important SUMO targets and several interaction partners (light grey circles) at the centromeric region and the kinetochores during mitosis, and highlights several enzymes responsible for these modification events (green circles). SUMO (S) is shown in blue circles, ubiquitin (U) is depicted in red circles. Abbreviations used: Budding Uninhibited by Benzimidazoles-Related (BUBR1), Centromere Protein (CENP), Inner Centromere Protein (INCENP), Kinetochore protein (Nuf2), Protein Inhibitor of Activated STATs (PIAS), Ran Binding Protein 2 (RanBP2), Ring Finger Protein 4 (RNF4), Rod-Zwilch-Zw10 complex (RZZ complex), Sentrin-specific Protease (SENP), Topoisomerase II α (TOPOIIα).
The formation of complexes including multiple SUMO substrates is additionally aided by the presence of SUMO Interaction Motifs (SIMs) within complex subunits. For example the microtubule motor protein CENP-E, which is essential during chromosome alignment, is a SUMO target protein and contains a SUMO interaction motif that is required for targeting the protein to the outer kinetochore [34]. Interestingly, two proteins interacting with CENP-E, kinetochore protein Nuf2 and Budding Uninhibited by Benzimidazoles-Related 1 (BubR1), have also been identified as SUMO2/3 target proteins, indicating that these modifications might be involved in targeting CENP-E to the kinetochore. BubR1 is an essential part of the spindle assembly checkpoint (SAC) and is localized at unattached kinetochores during pro-metaphase. In metaphase, however, it needs to be released to enable cells to enter anaphase and SUMOylation has been described to promote this process. An ectopically expressed SUMO deficient mutant of BubR1 is inhibited in its removal from the kinetochores during metaphase, leading to mitotic delay and increased chromosomal missegregation [58] (Figure 3).
Many of the subunits of the chromosomal passenger complex (CPC) have been shown to be SUMOylated. The CPC promotes the selective disassembly of microtubules from misattached chromosomes and is therefore pivotal for accurate chromosome alignment during early mitosis. It consists of the Aurora B kinase, Inner centromeric protein (INCENP), Survivin and Borealin, of which Aurora B, Borealin and the yeast Survivin homolog Bir1 have been shown to be SUMOylated [42,59,60]. A SUMOylation-deficient mutant of Aurora B was unable to rescue the severe defects in chromosome alignment after depletion of endogenous Aurora B and failed to re-localize from the outer chromosomal arms to the inner centromeres at the transition from pro-metaphase to metaphase [61]. Apart from affecting the localization of the enzyme, SUMOylation has also been identified to promote auto-phosphorylation of Aurora B, which is essential for its activation during early mitosis [59]. Another CPC subunit, Borealin, is highly SUMOylated by RanBP2 during metaphase, and its SUMOylation levels drop at the transition to anaphase due to the activity of SENP3 [42]. However, the functional relevance of Borealin SUMOylation is currently unknown.
Apart from stabilizing complexes, polySUMOylation of proteins can also destabilize complexes, as demonstrated for the cohesin complex in yeast [62]. Functional inactivation of the sister chromatid cohesin protein Pds5 promotes SUMOylation of the cohesin subunit Mcd1, which leads to its ubiquitylation by the SUMO targeted ubiquitin ligase Slx5-Slx8 and subsequent degradation. These results demonstrate that polySUMOylation destabilizes the cohesion complex and that Psd5 activity antagonizes this process. Thus, it is important to analyse the effect of SUMOylation for each complex individually, as SUMOylation can have greatly varying effects.
SUMOylation in tumorigenesis
Deregulation of the SUMO machinery in cancer cells
Related to the essential role of SUMOylation in maintaining chromosome integrity and regulating cell proliferation, evidence is accumulating for a key role of SUMOylation in cancer. Many components of the SUMO machinery are highly expressed in cancer tissues, suggesting that activated SUMOylation is linked to tumor growth (Table 1). Overexpression of the SUMO conjugating enzyme Ubc9 occurs in many types of cancer, including ovarian [63], colon and prostate cancer [64], and promotes cell invasion and metastasis [65]. Interestingly, overexpression of the SUMO proteases SENP1 and SENP5 occurs in malignant cancers, suggesting that SUMOylation needs to be tightly regulated to prevent malignant progression and cell proliferation [66,67].
Interestingly, knockdown of the SUMO E1 subunit SAE2 strongly impaired colon tumor growth in mice, showing the functional relevance of SUMOylation for tumor growth [68]. The SUMO E1 enzyme is also essential for Myc-driven tumors in mice [69]. Consistently, low levels of the SUMO activating enzyme in patients suffering from Myc-dependent breast cancer correlated with longer metastasis-free survival [69]. These findings underline the potential of targeting the SUMO machinery for cancer therapy.
SUMOylation-mediated regulation of oncogenes and tumor suppressors
The transcription factor c-Myc is a prime example of an oncogene involved in cell proliferation and apoptosis. Amplification of c-Myc is one of the most frequent events in a wide variety of different cancer types [70]. Recently, c-Myc was found as a SUMO target specifically after heat shock and inhibition of the proteasome, suggesting that SUMO-modified c-Myc is rapidly degraded by the proteasome [48,71,72] (Figure 4). Alternatively, c-Myc is specifically SUMOylated in response to protein-stress as a signalling mechanism. Knocking down the SUMO-activating enzyme SAE2 blocked cancer progression in a mouse model of c-Myc driven breast cancer [69]. However, it is currently unclear, how SUMOylation exactly promotes c-Myc-driven tumorigenesis. Knocking down SUMO ligase PIAS1 reduced the SUMOylation levels of c-Myc and led to increased expression of a c-Myc driven reporter gene [73], however inhibiting SAE2 repressed genes normally induced by c-Myc [69]. Despite these mechanistic questions, the finding that c-Myc driven tumors are dependent on a functioning SUMOylation system open up exciting new therapeutic opportunities to block this key oncogene [74].
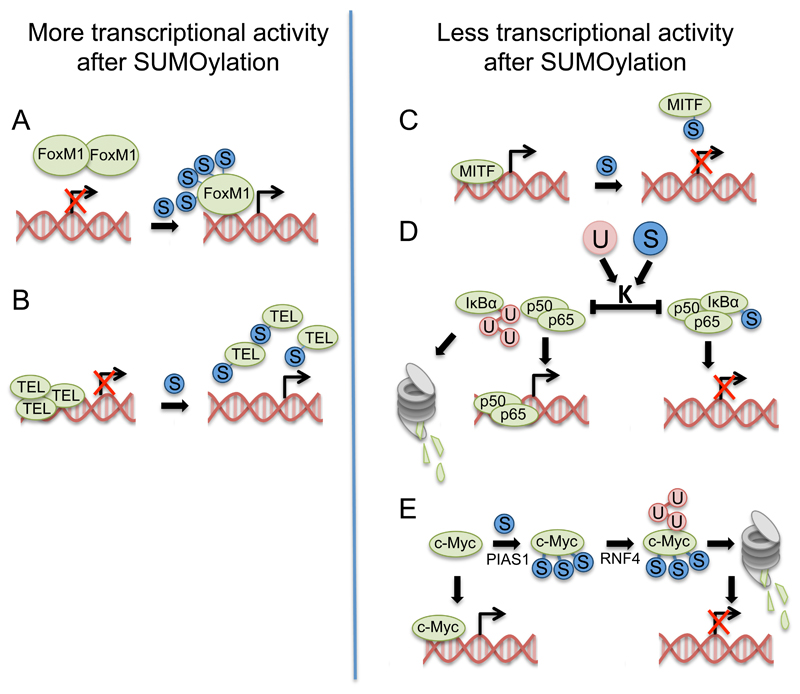
Figure 4. SUMOylation modulates the activity of transcription factors.
The majority of SUMO targets identified so far are involved in transcriptional regulation and chromatin remodelling. The effect of SUMOylation on the target protein can differ greatly and therefore has to be examined for each SUMO target and each SUMO site individually. This figure shows some of the mechanisms identified for transcription factors involved in cancer development. A) SUMO (S in blue circles) is known to affect protein-protein interactions and can therefore either obstruct binding to transcriptional inhibitors or promote the formation of inhibitory protein complexes. SUMOylation of FoxM1, for example, has been described to inhibit the formation of protein dimers and thereby induce transcriptional activity [23]. B) Similar to FoxM1, SUMOylation of the transcriptional repressor TEL/ETV6 blocks the formation of the multimer, which is needed to repress transcriptional activity at promoter regions. Therefore SUMOylation of TEL also promotes gene expression [85]. C) A SUMO-deficient mutant of MITF shows enhanced binding to DNA and increased expression of the target gene HIF1α. Therefore it has been suggested that SUMOylation of MITF blocks DNA binding and leads to decreased transcriptional activity [79] D) SUMOylation can also influence the stability of a target protein. SUMO and ubiquitin (U in red circles) can compete for the same lysine (K) residues as described for IκBα, a repressor of the multimeric transcription factor NFκB. While ubiquitylation of IκBα leads to proteasomal degradation and the release of the two subunits p65 and p50 and therefore enhances transcriptional activity, SUMOylation of IκBα stabilizes the protein and blocks gene expression [105]. E) However, the presence of multiple SUMO moieties can promote ubiquitylation via SUMO targeted ubiquitin ligases (Stubl) as described as a potential mechanism for c-Myc. SUMOylation via PIAS1 promotes ubiquitylation via the Stubl RNF4 and subsequent proteasomal degradation, thereby reducing transcriptional activity [73].
Another key proliferation driver, the Forkhead transcription factor 1 (FoxM1), is also regulated by SUMOylation [23] (Figure 4). FoxM1 is overexpressed in many types of solid tumors such as breast, colon, lung, prostate and liver cancer [75]. Similar to c-Myc, it is still not completely understood how SUMOylation affects FoxM1 activity. Contradicting results have been published for the different FoxM1 SUMOylation mutants used in these studies [23,76]. Specifically, modification of FoxM1 with SUMO1 was described to inhibit its transcriptional activity and resulted in increased APC/CDH1-dependent ubiquitylation and subsequent degradation of FoxM1 and therefore reduced mitotic progression [76]. By contrast, SUMOylation of FoxM1 with SUMO2 was found to increase its transcriptional activity to promote cell proliferation [23]. These differences could be explained by the use of a FoxM1-Ubc9 fusion construct in one study [76], potentially resulting in SUMOylation of different acceptor lysines compared to non-fused FoxM1[23].
Interestingly, phosphorylation of FoxM1 by Polo-like kinase 1 appeared to alter its SUMOylation levels, highlighting an additional FoxM1 regulatory step [77]. SUMOylated FoxM1 has been detected in human gastric cancer cells [78], but whether SUMOylation levels of FoxM1 are generally altered in tumor tissues and whether this affects tumor cell growth remains to be elucidated.
In addition to c-Myc and FoxM1, another key SUMOylated oncogene is the melanoma-lineage-specific microphthalmia-associated transcription factor (MITF). A MITF Mi-E318K germline mutation increased predisposition to sporadic melanoma and renal cell carcinoma. This mutation disrupts a SUMO consensus site, and lack of SUMOylation increased the transcriptional activity of MITF, increasing the levels of other tumor promoting factors such as hypoxia-inducible factor 1 α HIF1α [79,80]. Thus, SUMOylation of MITF prevents tumor initiation and progression (Figure 4).
SUMOylation furthermore plays a critical role in Acute Promyelocytic Leukemia (APL). APL is caused by the promyelocytic leukemia protein-retinoic acid acceptor α PML-RARα fusion oncoprotein [81]. Mouse models expressing this fusion gene display an APL-like phenotype. Interestingly, a single point mutation in PML-RARα, disrupting the K160 SUMOylation site in the PML moiety, is sufficient to block the APL-like phenotype [82].
Proteomic studies have revealed that a large fraction of the SUMOylated proteome is involved in transcriptional regulation and chromatin remodelling. Many transcription factors, such as p53 [83], c-Jun [84], translocation Ets leukemia (TEL) [85] and cAMP response element-binding protein CREB [86], are known oncogenes and tumor suppressors, deregulated in a multitude of human cancers. Whereas the effect of SUMOylation on the activity of these regulators is partially identified, it is unknown whether SUMOylation of these target proteins is actually deregulated in human cancers. Therefore, it would be of great interest to efficiently purify SUMO targets from these tissues, which is currently a major challenge in the SUMO field. A start to develop relevant methodology has been made [49,87].
Targeting the SUMO system
As described above, many components of the SUMO machinery are overexpressed in cancer tissues and knockdown of the SUMO pathway blocks cell proliferation and induces apoptosis. It is therefore of great interest to develop compounds that specifically block the activity of the SUMOylation machinery. Drugs targeting the SUMO activating enzyme are currently under investigation. Chemical inhibitors, such as ginkgolic acid and anacardic acid, bind to the SUMO activating enzyme and block the E1-SUMO intermediate [88]. Treatment of NOTCH1-activated breast epithelial cells with ginkgolic acid, for example, reduces cell proliferation and induces apoptosis, suggesting a potential effect of this SUMOylation inhibitor on NOTCH1-driven breast cancer [89]. Reactive oxygen species (ROS) can inhibit both the SUMO activating and SUMO conjugating enzymes in a different manner, via the formation of reversible disulfide bridges between the catalytic cysteine residues [90]. It has been reported that several chemotherapeutic drugs used to treat acute myeloid leukemias induce the formation of ROS, thereby inhibiting the SUMO pathway and subsequently reducing tumor growth [91]. Additionally, small SUMO-mimicking peptides can act as SUMOylation inhibitors [92]. Another interesting compound in this context is arsenic trioxide. This ancient Chinese drug was identified to induce SUMOylation and subsequent degradation of PML-RARα, a fusion protein responsible for acute promyelocytic leukemia [93–96].
Interestingly, elevated levels of SENP expression have also been linked to cancer development, underlining that disruption of the equilibrium between SUMOylation and deSUMOylation leads to abnormal cell proliferation. Several SENPs are known to regulate important oncogenes and tumor supressors. SENP1, for example, is involved in the deSUMOylation and stabilization of HIF1α [97], a transcription factor known to be upregulated in many human cancers and highly important for tumor survival [98]. The specific inhibition of SENP1 via potent drugs would therefore be of great interest for the development of new cancer therapies [99,100].
Despite the potential of targeting the SUMO machinery for cancer treatment, it remains highly challenging to develop specific small molecule inhibitors to selectively inhibit only the enzyme of interest. Furthermore, both the SUMO activating enzyme and SUMO conjugating enzyme regulate overall SUMOylation levels and therefore also cellular processes independent from cancer development [101]. It has to be taken into account that targeting the entire SUMO conjugation pathway might have severe side effects. Some of the recent discoveries on the regulation and specificity of Ubc9 after acetylation, however, might raise new possibilities in developing drugs to target only distinct Ubc9 functions [102]. These findings highlight the need to further understand the regulatory pathways and exact mechanisms of the SUMO machinery and the signals that trigger SUMOylation of specific SUMO targets involved in cancer progression.
Concluding remarks and future perspectives
Precisely timed post-translational modifications are essential to ensure accurate progression through the cell cycle. We now know extensive sets of ubiquitylation and phosphorylation events that are important for the transition from one cell cycle phase to the next. By contrast, we are limited in our understanding of SUMOylation events during cell cycle progression. Interestingly, inhibition of the SUMO pathway leads to cell cycle arrest in yeast and to decreased cell cycle progression and severe chromosomal defects in mouse and human cells, demonstrating key contributions of this posttranslational modification to cell cycle progression. Recent proteomic analyses have uncovered hundreds of target proteins differentially SUMOylated throughout the cell cycle. It is believed that these dynamic SUMO signals are ensured via differences in activity and localization of the SUMO machinery at different cell cycle phases. As many of the identified SUMO target proteins are known oncogenes and tumor suppressors, deregulation of these pathways via overexpression of the SUMO system is known to contribute to increased cell proliferation and cell invasion and reduced apoptosis in tumors. These findings will trigger further investigations into the regulatory mechanisms underlying the dynamic distribution and activity of the SUMO machinery. Identification of the effects of SUMOylation on important cell cycle regulators will provide additional functional insights into the role of SUMOylation in cancer.
Evidence exists that some tumors are dependent on a functional SUMO pathway. Interfering with SUMO signal transduction has been shown to block the growth of these tumors. Whether SUMOylation is more broadly required for tumor growth remains to be established. A more global investigation of the expression levels of different members of the SUMO machinery and overall SUMOylation levels in a broad panel of tumors needs to be carried out. Additionally, it will be interesting to study the SUMOylation levels of specific target proteins like TOPO IIα in these tumors. This knowledge can be used to select cancer types that could potentially be treated by blocking the SUMOylation machinery. An additional challenge will be to selectively interfere with SUMOylation in cancer cells, to avoid toxicity in healthy tissues. Rapidly growing cells in the gut, the bone marrow and elsewhere might similarly be dependent on a functioning SUMOylation system [103]. Nevertheless, it will be interesting to further investigate whether interfering with SUMOylation could be a valid anti-cancer strategy (Outstanding Questions box). Similar approaches to interfere with Neddylation, another ubiquitin-like modification, are showing promising results [5,104].
Outstanding Questions Box.
What is the functional relevance of SUMOylation for all novel SUMO substrates which have been recently identified? A wealth of novel SUMO target proteins has been identified in a site-specific manner using proteomics approaches. These targets include important cell cycle regulators.
What is the identity of the SUMO target protein subset for each SUMOylation and deSUMOylation enzyme? The SUMOylation and deSUMOylation machinery consist of a relatively small number of enzymes. Given the complexity of the SUMO proteome, each SUMO E3 and SUMO protease must be responsible for regulating a relatively large number of SUMO target proteins.
How do different post-translational modifications cooperate in a cell-wide manner to drive cell cycle progression? Similar to SUMOylation, ubiquitylation, phosphorylation and other post-translational modifications play key roles during cell cycle progression.
What are the mechanisms behind the redistribution of the SUMO machinery during cell cycle progression?
Different types of cancer are dependent on a functioning SUMOylation system. Is it possible to develop SUMOylation inhibitors and deSUMOylation inhibitors to treat cancer? Will healthy cells be able to tolerate these inhibitors?
Trends.
-
-
Knocking down SUMO conjugating enzymes in eukaryotic cells causes an overall delay in cell cycle progression.
-
-
SUMO co-modifies groups of proteins throughout all phases of the cell cycle to regulate cell cycle progression.
-
-
Cyclin-dependent kinases and SUMOylation can act in concert to regulate cell cycle progression.
-
-
The dependence of cancer cells on a functioning SUMOylation system can be exploited as anti-cancer therapy. Inhibitors of SUMO conjugating enzymes could be used for this purpose.
Acknowledgements
The laboratory of ACOV is supported by the European Research Council (ERC) and the Netherlands Organisation for Scientific Research (NWO).
References
- 1.Johnson LN. The regulation of protein phosphorylation. Biochem Soc Trans. 2009;37:627–641. doi: 10.1042/BST0370627. [DOI] [PubMed] [Google Scholar]
- 2.Moremen KW, et al. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol. 2012;13:448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choudhary C, et al. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 4.Goldstein G, et al. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc Natl Acad Sci U S A. 1975;72:11–15. doi: 10.1073/pnas.72.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soucy TA, et al. The NEDD8 Conjugation Pathway and Its Relevance in Cancer Biology and Therapy. Genes Cancer. 2010;1:708–716. doi: 10.1177/1947601910382898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao C, et al. Interferon-induced ISG15 pathway: an ongoing virus-host battle. Trends Microbiol. 2013;21:181–186. doi: 10.1016/j.tim.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hipp MS, et al. FAT10, a ubiquitin-independent signal for proteasomal degradation. Mol Cell Biol. 2005;25:3483–3491. doi: 10.1128/MCB.25.9.3483-3491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basler M, et al. The ubiquitin-like modifier FAT10 in antigen processing and antimicrobial defense. Mol Immunol. 2015 doi: 10.1016/j.molimm.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Muller S, et al. SUMO: a regulator of gene expression and genome integrity. Oncogene. 2004;23:1998–2008. doi: 10.1038/sj.onc.1207415. [DOI] [PubMed] [Google Scholar]
- 10.Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 11.Melchior F, et al. SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem Sci. 2003;28:612–618. doi: 10.1016/j.tibs.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Seufert W, et al. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- 13.Dieckhoff P, et al. Smt3/SUMO and Ubc9 are required for efficient APC/C-mediated proteolysis in budding yeast. Mol Microbiol. 2004;51:1375–1387. doi: 10.1046/j.1365-2958.2003.03910.x. [DOI] [PubMed] [Google Scholar]
- 14.Johnson ES, et al. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dohmen RJ, et al. An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J Biol Chem. 1995;270:18099–18109. doi: 10.1074/jbc.270.30.18099. [DOI] [PubMed] [Google Scholar]
- 16.Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147:981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nacerddine K, et al. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9:769–779. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, et al. SUMO2 is essential while SUMO3 is dispensable for mouse embryonic development. EMBO Rep. 2014;15:878–885. doi: 10.15252/embr.201438534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evdokimov E, et al. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J Cell Sci. 2008;121:4106–4113. doi: 10.1242/jcs.038570. [DOI] [PubMed] [Google Scholar]
- 20.Zhang FP, et al. Sumo-1 function is dispensable in normal mouse development. Mol Cell Biol. 2008;28:5381–5390. doi: 10.1128/MCB.00651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi T, et al. Ubc9 is essential for viability of higher eukaryotic cells. Exp Cell Res. 2002;280:212–221. doi: 10.1006/excr.2002.5634. [DOI] [PubMed] [Google Scholar]
- 22.Neyret-Kahn H, et al. Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome Res. 2013;23:1563–1579. doi: 10.1101/gr.154872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schimmel J, et al. Uncovering SUMOylation Dynamics during Cell-Cycle Progression Reveals FoxM1 as a Key Mitotic SUMO Target Protein. Mol Cell. 2014;53:1053–1066. doi: 10.1016/j.molcel.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Ayaydin F, Dasso M. Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol Biol Cell. 2004;15:5208–5218. doi: 10.1091/mbc.E04-07-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown PW, et al. Small ubiquitin-related modifier (SUMO)-1, SUMO-2/3 and SUMOylation are involved with centromeric heterochromatin of chromosomes 9 and 1 and proteins of the synaptonemal complex during meiosis in men. Hum Reprod. 2008;23:2850–2857. doi: 10.1093/humrep/den300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vigodner M. Sumoylation precedes accumulation of phosphorylated H2AX on sex chromosomes during their meiotic inactivation. Chromosome Res. 2009;17:37–45. doi: 10.1007/s10577-008-9006-x. [DOI] [PubMed] [Google Scholar]
- 27.Pelisch F, et al. Dynamic SUMO modification regulates mitotic chromosome assembly and cell cycle progression in Caenorhabditis elegans. Nat Commun. 2014;5:5485. doi: 10.1038/ncomms6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Azuma Y, et al. PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. EMBO J. 2005;24:2172–2182. doi: 10.1038/sj.emboj.7600700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joseph J, et al. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J Cell Biol. 2002;156:595–602. doi: 10.1083/jcb.200110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joseph J, et al. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr Biol. 2004;14:611–617. doi: 10.1016/j.cub.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 31.Dawlaty MM, et al. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133:103–115. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hang J, Dasso M. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J Biol Chem. 2002;277:19961–19966. doi: 10.1074/jbc.M201799200. [DOI] [PubMed] [Google Scholar]
- 33.Cubenas-Potts C, et al. SENP1 and SENP2 Affect Spatial and Temporal Control of Sumoylation in Mitosis. Mol Biol Cell. 2013;24:3483–3495. doi: 10.1091/mbc.E13-05-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang XD, et al. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol Cell. 2008;29:729–741. doi: 10.1016/j.molcel.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolli N, et al. Distribution and paralogue specificity of mammalian deSUMOylating enzymes. Biochem J. 2010;430:335–344. doi: 10.1042/BJ20100504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garvin AJ, et al. The deSUMOylase SENP7 promotes chromatin relaxation for homologous recombination DNA repair. EMBO Rep. 2013;14:975–983. doi: 10.1038/embor.2013.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goeres J, et al. The SUMO-specific isopeptidase SENP2 associates dynamically with nuclear pore complexes through interactions with karyopherins and the Nup107-160 nucleoporin subcomplex. Mol Biol Cell. 2011;22:4868–4882. doi: 10.1091/mbc.E10-12-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maison C, et al. The SUMO protease SENP7 is a critical component to ensure HP1 enrichment at pericentric heterochromatin. Nat Struct Mol Biol. 2012;19:458–460. doi: 10.1038/nsmb.2244. [DOI] [PubMed] [Google Scholar]
- 39.Romeo K, et al. The SENP7 SUMO-Protease Presents a Module of Two HP1 Interaction Motifs that Locks HP1 Protein at Pericentric Heterochromatin. Cell Rep. 2015;10:771–782. doi: 10.1016/j.celrep.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 40.Zunino R, et al. Translocation of SenP5 from the nucleoli to the mitochondria modulates DRP1-dependent fission during mitosis. J Biol Chem. 2009;284:17783–17795. doi: 10.1074/jbc.M901902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Bacco A, et al. The SUMO-specific protease SENP5 is required for cell division. Mol Cell Biol. 2006;26:4489–4498. doi: 10.1128/MCB.02301-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein UR, et al. RanBP2 and SENP3 function in a mitotic SUMO2/3 conjugation-deconjugation cycle on Borealin. Mol Biol Cell. 2009;20:410–418. doi: 10.1091/mbc.E08-05-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hietakangas V, et al. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci U S A. 2006;103:45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mohideen F, et al. A molecular basis for phosphorylation-dependent SUMO conjugation by the E2 UBC9. Nat Struct Mol Biol. 2009;16:945–952. doi: 10.1038/nsmb.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asghar U, et al. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov. 2015;14:130–146. doi: 10.1038/nrd4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henneke G, et al. Phosphorylation of human Fen1 by cyclin-dependent kinase modulates its role in replication fork regulation. Oncogene. 2003;22:4301–4313. doi: 10.1038/sj.onc.1206606. [DOI] [PubMed] [Google Scholar]
- 47.Guo Z, et al. Sequential posttranslational modifications program FEN1 degradation during cell-cycle progression. Mol Cell. 2012;47:444–456. doi: 10.1016/j.molcel.2012.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hendriks IA, et al. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat Struct Mol Biol. 2014;21:927–936. doi: 10.1038/nsmb.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hendriks IA, et al. System-wide identification of wild-type SUMO-2 conjugation sites. Nat Commun. 2015;6:7289. doi: 10.1038/ncomms8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hendriks IA, et al. SUMO-2 Orchestrates Chromatin Modifiers in Response to DNA Damage. Cell Rep. 2015;10:1778–1791. doi: 10.1016/j.celrep.2015.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olsen JV, Mann M. Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol Cell Proteomics. 2013;12:3444–3452. doi: 10.1074/mcp.O113.034181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tammsalu T, et al. Proteome-Wide Identification of SUMO2 Modification Sites. Sci Signal. 2014;7:rs2. doi: 10.1126/scisignal.2005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bellail AC, et al. SUMO1 modification stabilizes CDK6 protein and drives the cell cycle and glioblastoma progression. Nat Commun. 2014;5:4234. doi: 10.1038/ncomms5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bonne-Andrea C, et al. SUMO2/3 modification of cyclin E contributes to the control of replication origin firing. Nat Commun. 2013;4:1850. doi: 10.1038/ncomms2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cubenas-Potts C, et al. Identification of SUMO-2/3-modified proteins associated with mitotic chromosomes. Proteomics. 2015;15:763–772. doi: 10.1002/pmic.201400400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jentsch S, Psakhye I. Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu Rev Genet. 2013;47:167–186. doi: 10.1146/annurev-genet-111212-133453. [DOI] [PubMed] [Google Scholar]
- 57.Mukhopadhyay D, et al. The SUMO protease SENP6 is essential for inner kinetochore assembly. J Cell Biol. 2010;188:681–692. doi: 10.1083/jcb.200909008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang F, et al. BubR1 is modified by sumoylation during mitotic progression. J Biol Chem. 2012;287:4875–4882. doi: 10.1074/jbc.M111.318261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ban R, et al. Mitotic kinase Aurora-B is regulated by SUMO-2/3 conjugation/deconjugation during mitosis. Genes Cells. 2011;16:652–669. doi: 10.1111/j.1365-2443.2011.01521.x. [DOI] [PubMed] [Google Scholar]
- 60.Montpetit B, et al. Sumoylation of the budding yeast kinetochore protein Ndc10 is required for Ndc10 spindle localization and regulation of anaphase spindle elongation. J Cell Biol. 2006;174:653–663. doi: 10.1083/jcb.200605019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fernandez-Miranda G, et al. SUMOylation modulates the function of Aurora-B kinase. J Cell Sci. 2010;123:2823–2833. doi: 10.1242/jcs.065565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.D'Ambrosio LM, Lavoie BD. Pds5 prevents the PolySUMO-dependent separation of sister chromatids. Curr Biol. 2014;24:361–371. doi: 10.1016/j.cub.2013.12.038. [DOI] [PubMed] [Google Scholar]
- 63.Mo YY, et al. A role for Ubc9 in tumorigenesis. Oncogene. 2005;24:2677–2683. doi: 10.1038/sj.onc.1208210. [DOI] [PubMed] [Google Scholar]
- 64.Moschos SJ, et al. Expression analysis of Ubc9, the single small ubiquitin-like modifier (SUMO) E2 conjugating enzyme, in normal and malignant tissues. Hum Pathol. 2010;41:1286–1298. doi: 10.1016/j.humpath.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 65.Zhu S, et al. Ubc9 promotes breast cell invasion and metastasis in a sumoylation-independent manner. Oncogene. 2010;29:1763–1772. doi: 10.1038/onc.2009.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang C, et al. Tumor-suppressive microRNA-145 induces growth arrest by targeting SENP1 in human prostate cancer cells. Cancer Sci. 2015;106:375–382. doi: 10.1111/cas.12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang K, Zhang XC. Inhibition of SENP5 suppresses cell growth and promotes apoptosis in osteosarcoma cells. Exp Ther Med. 2014;7:1691–1695. doi: 10.3892/etm.2014.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He X, et al. Characterization of the loss of SUMO pathway function on cancer cells and tumor proliferation. PLoS One. 2015;10:e0123882. doi: 10.1371/journal.pone.0123882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kessler JD, et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335:348–353. doi: 10.1126/science.1212728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sabo A, et al. SUMOylation of Myc-family proteins. PLoS One. 2014;9:e91072. doi: 10.1371/journal.pone.0091072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kalkat M, et al. Identification of c-MYC SUMOylation by mass spectrometry. PLoS One. 2014;9:e115337. doi: 10.1371/journal.pone.0115337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gonzalez-Prieto R, et al. c-Myc is targeted to the proteasome for degradation in a SUMOylation-dependent manner, regulated by PIAS1, SENP7 and RNF4. Cell Cycle. 2015;14:1859–1872. doi: 10.1080/15384101.2015.1040965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Evan G. Cancer. Taking a back door to target Myc. Science. 2012;335:293–294. doi: 10.1126/science.1217819. [DOI] [PubMed] [Google Scholar]
- 75.Wierstra I. FOXM1 (Forkhead box M1) in tumorigenesis: overexpression in human cancer, implication in tumorigenesis, oncogenic functions, tumor-suppressive properties, and target of anticancer therapy. Adv Cancer Res. 2013;119:191–419. doi: 10.1016/B978-0-12-407190-2.00016-2. [DOI] [PubMed] [Google Scholar]
- 76.Myatt SS, et al. SUMOylation inhibits FOXM1 activity and delays mitotic transition. Oncogene. 2013;33:4316–4329. doi: 10.1038/onc.2013.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang J, et al. Polo-like Kinase 1-mediated Phosphorylation of Forkhead Box Protein M1b Antagonizes Its SUMOylation and Facilitates Its Mitotic Function. J Biol Chem. 2015;290:3708–3719. doi: 10.1074/jbc.M114.634386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shao DF, et al. High-level SAE2 promotes malignant phenotype and predicts outcome in gastric cancer. Am J Cancer Res. 2015;5:140–154. [PMC free article] [PubMed] [Google Scholar]
- 79.Bertolotto C, et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature. 2011;480:94–98. doi: 10.1038/nature10539. [DOI] [PubMed] [Google Scholar]
- 80.Yokoyama S, et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature. 2011;480:99–103. doi: 10.1038/nature10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.De TH, et al. The cell biology of disease: Acute promyelocytic leukemia, arsenic, and PML bodies. J Cell Biol. 2012;198:11–21. doi: 10.1083/jcb.201112044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu J, et al. A sumoylation site in PML/RARA is essential for leukemic transformation. Cancer Cell. 2005;7:143–153. doi: 10.1016/j.ccr.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 83.Gostissa M, et al. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Muller S, et al. c-Jun and p53 activity is modulated by SUMO-1 modification. J Biol Chem. 2000;275:13321–13329. doi: 10.1074/jbc.275.18.13321. [DOI] [PubMed] [Google Scholar]
- 85.Roukens MG, et al. Identification of a new site of sumoylation on Tel (ETV6) uncovers a PIAS-dependent mode of regulating Tel function. Mol Cell Biol. 2008;28:2342–2357. doi: 10.1128/MCB.01159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Comerford KM, et al. Small ubiquitin-related modifier-1 modification mediates resolution of CREB-dependent responses to hypoxia. Proc Natl Acad Sci U S A. 2003;100:986–991. doi: 10.1073/pnas.0337412100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Becker J, et al. Detecting endogenous SUMO targets in mammalian cells and tissues. Nat Struct Mol Biol. 2013;20:525–531. doi: 10.1038/nsmb.2526. [DOI] [PubMed] [Google Scholar]
- 88.Fukuda I, et al. Ginkgolic acid inhibits protein SUMOylation by blocking formation of the E1-SUMO intermediate. Chem Biol. 2009;16:133–140. doi: 10.1016/j.chembiol.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 89.Licciardello MP, et al. NOTCH1 activation in breast cancer confers sensitivity to inhibition of SUMOylation. Oncogene. 2014 doi: 10.1038/onc.2014.319. [DOI] [PubMed] [Google Scholar]
- 90.Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell. 2006;21:349–357. doi: 10.1016/j.molcel.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 91.Bossis G, et al. The ROS/SUMO axis contributes to the response of acute myeloid leukemia cells to chemotherapeutic drugs. Cell Rep. 2014;7:1815–1823. doi: 10.1016/j.celrep.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 92.Zhao B, et al. SUMO-mimicking peptides inhibiting protein SUMOylation. Chembiochem. 2014;15:2662–2666. doi: 10.1002/cbic.201402472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lallemand-Breitenbach V, et al. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- 94.Tatham MH, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 95.Zhang XW, et al. Arsenic trioxide controls the fate of the PML-RARalpha oncoprotein by directly binding PML. Science. 2010;328:240–243. doi: 10.1126/science.1183424. [DOI] [PubMed] [Google Scholar]
- 96.Weisshaar SR, et al. Arsenic trioxide stimulates SUMO-2/3 modification leading to RNF4-dependent proteolytic targeting of PML. FEBS Lett. 2008;582:3174–3178. doi: 10.1016/j.febslet.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 97.Cheng J, et al. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell. 2007;131:584–595. doi: 10.1016/j.cell.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Keith B, et al. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2012;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu J, et al. SENP1 inhibition induces apoptosis and growth arrest of multiple myeloma cells through modulation of NF-kappaB signaling. Biochem Biophys Res Commun. 2015;460:409–415. doi: 10.1016/j.bbrc.2015.03.047. [DOI] [PubMed] [Google Scholar]
- 100.Kumar A, Zhang KY. Advances in the development of SUMO specific protease (SENP) inhibitors. Comput Struct Biotechnol J. 2015;13:204–211. doi: 10.1016/j.csbj.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bawa-Khalfe T, Yeh ET. SUMO Losing Balance: SUMO Proteases Disrupt SUMO Homeostasis to Facilitate Cancer Development and Progression. Genes Cancer. 2010;1:748–752. doi: 10.1177/1947601910382555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hsieh YL, et al. Ubc9 acetylation modulates distinct SUMO target modification and hypoxia response. EMBO J. 2013;32:791–804. doi: 10.1038/emboj.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Demarque MD, et al. Sumoylation by Ubc9 regulates the stem cell compartment and structure and function of the intestinal epithelium in mice. Gastroenterology. 2011;140:286–296. doi: 10.1053/j.gastro.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 104.Soucy TA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 105.Desterro JM, et al. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 106.Bachant J, et al. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol Cell. 2002;9:1169–1182. doi: 10.1016/s1097-2765(02)00543-9. [DOI] [PubMed] [Google Scholar]
- 107.Ryu H, et al. PIASy-dependent SUMOylation regulates DNA topoisomerase IIalpha activity. J Cell Biol. 2010;191:783–794. doi: 10.1083/jcb.201004033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ryu H, Azuma Y. Rod/Zw10 complex is required for PIASy-dependent centromeric SUMOylation. J Biol Chem. 2010;285:32576–32585. doi: 10.1074/jbc.M110.153817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sridharan V, et al. SUMOylation Regulates Polo-like Kinase 1-interacting Checkpoint Helicase (PICH) during Mitosis. J Biol Chem. 2015;290:3269–3276. doi: 10.1074/jbc.C114.601906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ryu H, et al. PIASy mediates SUMO-2/3 conjugation of poly(ADP-ribose) polymerase 1 (PARP1) on mitotic chromosomes. J Biol Chem. 2010;285:14415–14423. doi: 10.1074/jbc.M109.074583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guo WH, et al. Overexpression of SUMO-1 in hepatocellular carcinoma: a latent target for diagnosis and therapy of hepatoma. J Cancer Res Clin Oncol. 2011;137:533–541. doi: 10.1007/s00432-010-0920-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang H, et al. Over-Expression of Small Ubiquitin-Related Modifier-1 and Sumoylated p53 in Colon Cancer. Cell Biochem Biophys. 2013;67:1081–1087. doi: 10.1007/s12013-013-9612-x. [DOI] [PubMed] [Google Scholar]
- 113.Oliveira Alves MG, et al. Study of MDM2 and SUMO-1 expression in actinic cheilitis and lip cancer. Arch Dermatol Res. 2014;306:837–841. doi: 10.1007/s00403-014-1500-8. [DOI] [PubMed] [Google Scholar]
- 114.Mei D, et al. Up-regulation of SUMO1 pseudogene 3 (SUMO1P3) in gastric cancer and its clinical association. Med Oncol. 2013;30:709. doi: 10.1007/s12032-013-0709-2. [DOI] [PubMed] [Google Scholar]
- 115.Shao DF, et al. High-level SAE2 promotes malignant phenotype and predicts outcome in gastric cancer. Am J Cancer Res. 2015;5:589–602. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 116.Liu X, et al. Knockdown of SUMO-activating enzyme subunit 2 (SAE2) suppresses cancer malignancy and enhances chemotherapy sensitivity in small cell lung cancer. J Hematol Oncol. 2015;8:67. doi: 10.1186/s13045-015-0164-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li H, et al. Ubc9 promotes invasion and metastasis of lung cancer cells. Oncol Rep. 2013;29:1588–1594. doi: 10.3892/or.2013.2268. [DOI] [PubMed] [Google Scholar]
- 118.Wozniak K, et al. Polymorphism of UBC9 Gene Encoding the SUMO-E2-Conjugating Enzyme and Breast Cancer Risk. Pathol Oncol Res. 2013;20:67–72. doi: 10.1007/s12253-013-9659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen SF, et al. Ubc9 expression predicts chemoresistance in breast cancer. Chin J Cancer. 2011;30:638–644. doi: 10.5732/cjc.011.10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Driscoll JJ, et al. The sumoylation pathway is dysregulated in multiple myeloma and is associated with adverse patient outcome. Blood. 2010;115:2827–2834. doi: 10.1182/blood-2009-03-211045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yang W, et al. Small ubiquitin-like modifier 1-3 conjugation [corrected] is activated in human astrocytic brain tumors and is required for glioblastoma cell survival. Cancer Sci. 2013;104:70–77. doi: 10.1111/cas.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hoefer J, et al. PIAS1 is increased in human prostate cancer and enhances proliferation through inhibition of p21. Am J Pathol. 2012;180:2097–2107. doi: 10.1016/j.ajpath.2012.01.026. [DOI] [PubMed] [Google Scholar]
- 123.Toropainen S, et al. SUMO ligase PIAS1 functions as a target gene selective androgen receptor coregulator on prostate cancer cell chromatin. Nucleic Acids Res. 2015;43:848–861. doi: 10.1093/nar/gku1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang L, Banerjee S. Differential PIAS3 expression in human malignancy. Oncol Rep. 2004;11:1319–1324. [PubMed] [Google Scholar]
- 125.Liu B, et al. PIAS1 regulates breast tumorigenesis through selective epigenetic gene silencing. PLoS One. 2014;9:e89464. doi: 10.1371/journal.pone.0089464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li J, et al. PIAS3, an inhibitor of STAT3, has intensively negative association with the survival of gastric cancer. Int J Clin Exp Med. 2015;8:682–689. [PMC free article] [PubMed] [Google Scholar]
- 127.Sun L, et al. PIASy mediates hypoxia-induced SIRT1 transcriptional repression and epithelial-to-mesenchymal transition in ovarian cancer cells. J Cell Sci. 2013;126:3939–3947. doi: 10.1242/jcs.127381. [DOI] [PubMed] [Google Scholar]
- 128.Chien W, et al. PIAS4 is an activator of hypoxia signalling via VHL suppression during growth of pancreatic cancer cells. Br J Cancer. 2013;109:1795–1804. doi: 10.1038/bjc.2013.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Horio Y, et al. Relationship of mRNA expressions of RanBP2 and topoisomerase II isoforms to cytotoxicity of amrubicin in human lung cancer cell lines. Cancer Chemother Pharmacol. 2010;66:237–243. doi: 10.1007/s00280-009-1151-1. [DOI] [PubMed] [Google Scholar]
- 130.Lim JH, et al. RANBP2-ALK fusion combined with monosomy 7 in acute myelomonocytic leukemia. Cancer Genet. 2014;207:40–45. doi: 10.1016/j.cancergen.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 131.Bawa-Khalfe T, et al. SENP1 induces prostatic intraepithelial neoplasia through multiple mechanisms. J Biol Chem. 2010;285:25859–25866. doi: 10.1074/jbc.M110.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang Q, et al. SUMO-specific protease 1 promotes prostate cancer progression and metastasis. Oncogene. 2013;32:2493–2498. doi: 10.1038/onc.2012.250. [DOI] [PubMed] [Google Scholar]
- 133.Xu Y, et al. SUMO-specific protease 1 regulates the in vitro and in vivo growth of colon cancer cells with the upregulated expression of CDK inhibitors. Cancer Lett. 2011;309:78–84. doi: 10.1016/j.canlet.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 134.Han Y, et al. SENP3-mediated de-conjugation of SUMO2/3 from promyelocytic leukemia is correlated with accelerated cell proliferation under mild oxidative stress. J Biol Chem. 2010;285:12906–12915. doi: 10.1074/jbc.M109.071431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shen HJ, et al. SENP2 regulates hepatocellular carcinoma cell growth by modulating the stability of beta-catenin. Asian Pac J Cancer Prev. 2012;13:3583–3587. doi: 10.7314/apjcp.2012.13.8.3583. [DOI] [PubMed] [Google Scholar]
- 136.Qian J, et al. Inhibition of SENP6-induced radiosensitization of human hepatocellular carcinoma cells by blocking radiation-induced NF-kappaB activation. Cancer Biother Radiopharm. 2013;28:196–200. doi: 10.1089/cbr.2012.1288. [DOI] [PubMed] [Google Scholar]
- 137.Tan MY, et al. SUMO-Specific Protease 2 Suppresses Cell Migration and Invasion through Inhibiting the Expression of MMP13 in Bladder Cancer Cells. Cell Physiol Biochem. 2013;32:542–548. doi: 10.1159/000354458. [DOI] [PubMed] [Google Scholar]
- 138.Ren YH, et al. De-SUMOylation of FOXC2 by SENP3 promotes the epithelial-mesenchymal transition in gastric cancer cells. Oncotarget. 2014;5:7093–7104. doi: 10.18632/oncotarget.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sun Z, et al. Overexpression of SENP3 in oral squamous cell carcinoma and its association with differentiation. Oncol Rep. 2013;29:1701–1706. doi: 10.3892/or.2013.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cheng Y, et al. Sentrin/small ubiquitin-like modifier-specific protease 5 protects oral cancer cells from oxidative stress-induced apoptosis. Mol Med Rep. 2015;12:2009–2014. doi: 10.3892/mmr.2015.3662. [DOI] [PubMed] [Google Scholar]
- 141.Ding X, et al. Overexpression of SENP5 in oral squamous cell carcinoma and its association with differentiation. Oncol Rep. 2008;20:1041–1045. [PubMed] [Google Scholar]
- 142.Cashman R, et al. SENP5 mediates breast cancer invasion via a TGFbetaRI SUMOylation cascade. Oncotarget. 2014;5:1071–1082. doi: 10.18632/oncotarget.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mooney SM, et al. Sumoylation of p68 and p72 RNA helicases affects protein stability and transactivation potential. Biochemistry. 2010;49:1–10. doi: 10.1021/bi901263m. [DOI] [PubMed] [Google Scholar]
- 144.Bermejo JL, et al. Exploring the association between genetic variation in the SUMO isopeptidase gene USPL1 and breast cancer through integration of data from the population-based GENICA study and external genetic databases. Int J Cancer. 2013;133:362–372. doi: 10.1002/ijc.28040. [DOI] [PubMed] [Google Scholar]