Abstract
Microtubules are required throughout plant development for a wide variety of processes, and different strategies have evolved to visualize and analyze them. This chapter provides specific methods that can be used to analyze microtubule organization and dynamic properties in plant systems and summarizes the advantages and limitations for each technique. We outline basic methods for preparing samples for immunofluorescence labelling, including an enzyme-based permeabilization method, and a freeze-shattering method, which generates microfractures in the cell wall to provide antibodies access to cells in cuticle-laden aerial organs such as leaves. We discuss current options for live cell imaging of MTs with fluorescently tagged proteins (FPs), and provide chemical fixation, high pressure freezing/freeze substitution, and post-fixation staining protocols for preserving MTs for transmission electron microscopy and tomography.
Keywords: microtubules, EB1, GFP, kinesin, MAP4, MBD, MOR1, ARK1, electron tomography, live cell imaging, correlative light and electron microscopy, immunofluorescence
1. Introduction
Microtubules (MTs) are a unifying feature of eukaryotic cells. Studying them in plants is not only important for understanding mechanisms of plant growth and development but also of broader interest for understanding the mechanisms that generate MT spatial organization, including the role of accessory proteins in MT dynamics. The current literature describes the role of MTs in spindle formation and division plane organization [1–3], transition from division to expansion [4,5], cell wall formation and morphogenesis in diffusely expanding organs [6–13], tip growing root hairs and pollen tubes [14–19], movement of stomata [20], endomembranes [21–23] or nucleus [24–27], chloroplast organization and positioning [28–30] or in association with amyloplasts [31]. This research requires a variety of experimental strategies, ranging from basic description of orientation patterns, MT dynamics and associations with other cellular structures, plant hormones, mechanical stress to pharmacological and genetic perturbations. With cytoskeleton research increasing in relevance to different plant research fields, it is especially important for researchers to be provided with a variety of standard techniques and tools for imaging MTs in plant cells. In this chapter, we outline such preparation methods for light and electron microscopy. Methods for imaging root MT and actin filament arrays are provided in chapter XX by E. Blancaflor and colleagues “Fluorescence Imaging of the Cytoskeleton in Plant Roots“.
Immunofluorescence Options
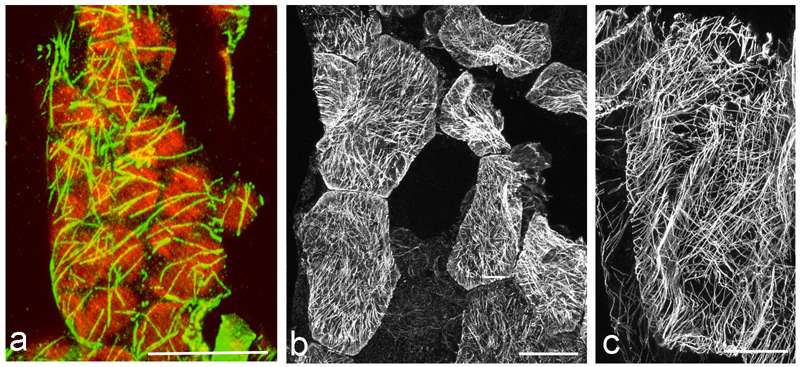
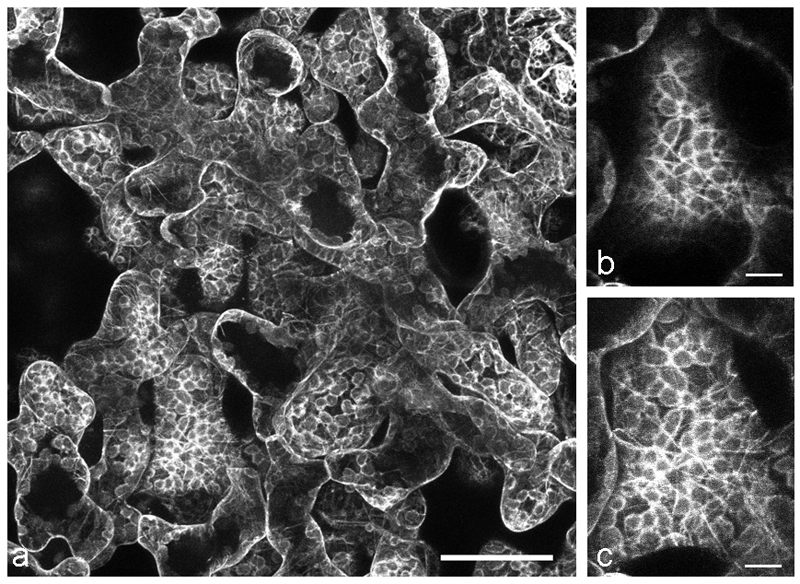
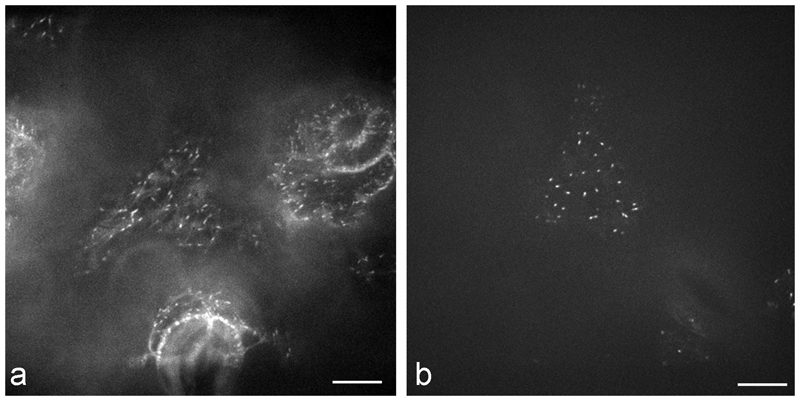
Immunofluorescence microscopy remains a useful approach for MT imaging at the level of resolution of visible light [32]. Despite the convenience of live cell imaging with transgene-introduced fluorescent reporter proteins (eg. [33,34]), immunofluorescence remains essential under many circumstances. It can be utilized immediately on any interesting plant material because it does not require the laborious and time-consuming cloning, transformation and selection procedures required for introducing transgenic reporter proteins or. This is especially relevant for non-model systems for which genetic transformation is not yet feasible or when wild material is collected in its natural habitat. We demonstrate this first point with images of MTs in leaf cells of the high alpine plant Oxyria digyna (Fig. 1 a-c) for which the “freeze shattering” method [35] was adapted to examine MTs in relation to stress-induced chloroplast protuberances [29]. Previous work had demonstrated that MTs were a critical factor in the establishment of chloroplast stromules in the model system Arabidopsis thaliana [36]. Immunofluorescence enabled MTs in Oxyria leaves to be documented [29] without the need to develop protocols for transformation of this non-model system.
Fig. 1.
Immunolocalization of MTs in the alpine vascular plant Oxyria digyna: a merge of MT staining (green) and chloroplast autofluorescence showing the close vicinity of MTs with chloroplasts. b MTs in epidermal cells, c MTs in parenchyma cells; note the very dense network in z-stack projections. The image is composed of approximately 100 images, comprising a depth of about 20 µm; bars 20 µm. Figs. b-c reprinted from [29] with permission of Thieme Verlag KG.
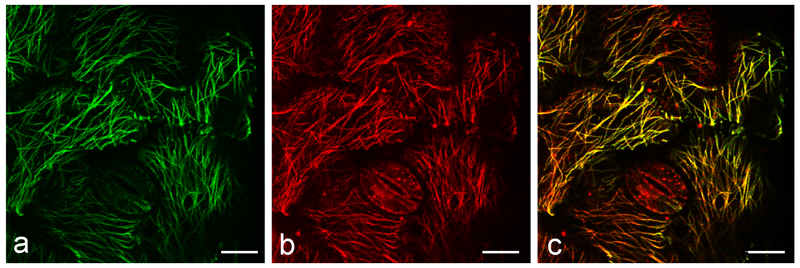
Immunofluorescence microscopy is also a useful adjunct technology for corroborating evidence from live cell strategies, where, as discussed below, artifact needs to be ruled out (Fig. 2). It is the best option for detecting low abundance proteins, which require high-intensity light and long dwell times or when large series of optical sections need to be collected because these procedures are toxic to living cells. Although immuno- procedures kill the samples, after fixation and appropriate fade protection, immunolabelled material can be scanned repeatedly and stored for months with little loss of image quality. Immunofluorescence is also an excellent method for describing the distribution of MT-associated proteins (MAPs) along MTs using double labelling strategies (Fig. 3). Finally, at present, immunofluorescence is the only practical method to use when applying one of the exciting new super-resolution technologies, which approach transmission electron microscopy in terms of resolution but generally require extensive dwell times.
Fig. 2.
Comparison of wild-type, wt (a, b) and mor1-1 (c, d) MT bundles via immunolocalization (a,c) and GFP-MBD live cell imaging (b, d) in Arabidopsis thaliana at 31°C. Plants were grown at 21º for 5 days and then transferred to 31ºC. a wt immunolabeled (fixed in a fixative that was preheated to 31ºC and labeled with anti-tubulin) and b wt GFP-MBD expressing root. Images were taken using a temperature-controlled stage that kept the specimens at 31ºC, 2-3.5 h after temperature shifts; c immunolabeled mor 1-1, d mor1-1 at 31°C showing apparent MT bundles in the GFP-MBD fusion protein expressing line. GFP-MBD labeling identifies bright, bundle-like structures at 31°C. Although some evidence for this is observed in wild-type cells, MT remodeling is much more extensive in mor1-1. These thickened MT structures are not observed with immunolabeling, suggesting this putative bundling of MTs is dependent on the GFP-MBD fusion protein. Bars, 10 μm.
Fig. 3.
Immuno-labeling of tubulin (a) and MOR1 (b) in wild-type background at 31ºC.
For immuno-labeling, 11 day-old wild-type plants grown at 21ºC were cultured at 31ºC for one day and fixed in fixatives that were preheated to 31ºC and then double immuno-labeled. a tubulin, b MOR1, c merge of a and b. The abaxial sides of first leaves are shown in all images. Bars, 10 µm.
Live Cell Imaging with Microtubule Reporter Proteins
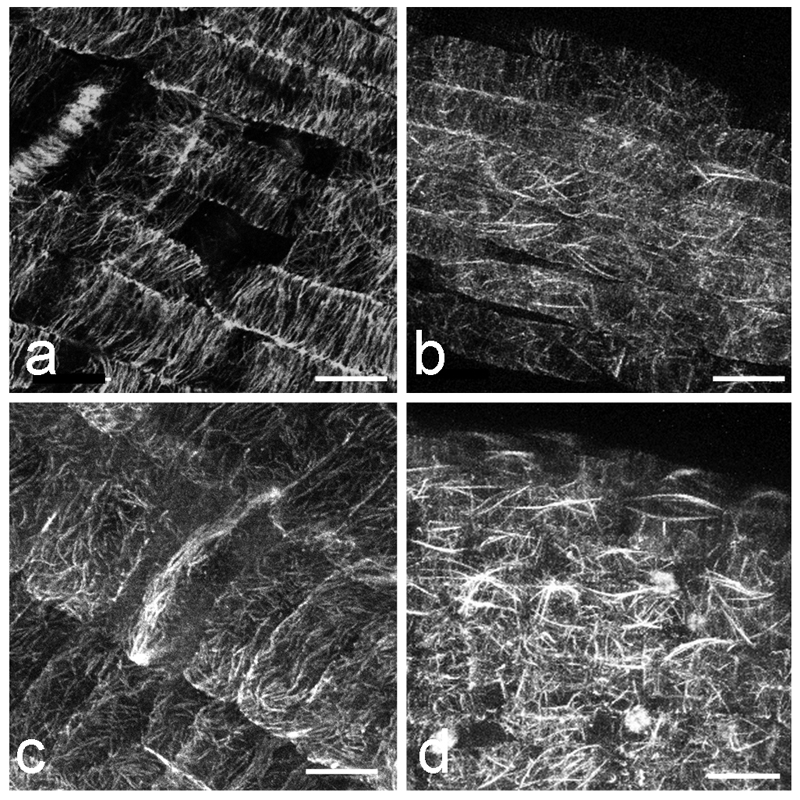
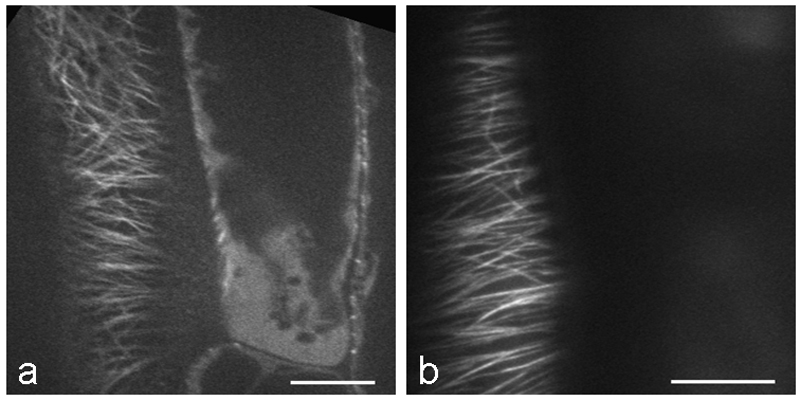
Ever since the successful exploitation of green fluorescent protein (GFP) in the early 1990s, a vast body of literature has been generated on the use of this and other intrinsically fluorescent proteins as reporters of structural proteins [33]. Many constructs, cell and plant lines are now available for observing MTs in living plant cells including the MT-binding domain (MBD) of mammalian MAP4, various tubulins (TUB), EB1, and other MAPs (Table 1). There are clear advantages to being able to follow MTs in living cells. Time-lapse imaging enables the growth and shrinkage of MTs to be followed in near real-time [37–39]. Due to the difficulties in obtaining clear MT images from deep tissue, most MT dynamics data come from observing cortical MTs in epidermal cells. Spinning disk confocal microscopy is especially suitable for study of MT dynamics because of the quick image acquisition possible compared to conventional laser scanning confocal microscopy. Another option would be to use total internal reflection fluorescence (TIRF) microscopy or more precisely near-TIRF (also referred to as variable angle epifluorescence) microscopy. TIRF and near-TIRF allow fluorescence to be captured close to the cover slip, such as in the cell cortex of epidermal cells. By adjusting the incident illumination angle, the excitation depth can be changed, eliminating or decreasing the cytoplasmic background fluorescence caused by free tubulin (Fig. 4). However, due to the limitation in depth of excitation, endoplasmic MTs or MTs in deep tissues cannot be acquired with TIRF and near-TIRF microscopy. Multiphoton fluorescence microscopy is ideal for such cases, and can also be used to acquire images from deep tissue such as spongy mesophyll cells in the leaf (Fig. 5). Conventional laser scanning confocal microscopy also remains a powerful tool for obtaining optical sections in thick samples and for following processes such as mitosis in the root [40,41] as well as quantitative analysis for co-localization of MT-associated proteins with MTs [42]. There are, however, several limitations to live cell imaging of microtubules and great care needs to be exercised to avoid acquisition of erroneous information. Some of these problems are outlined in detail by Shaw, 2006 [43]. In the Notes section, we outline several important issues to consider when live cell imaging MTs, such as phototoxicity, choice of promoter, fusion protein reporter construction and culture temperature.
Table 1.
Transgenic reporter lines for observing microtubules in live cells of Arabidopsis thaliana
| Tubulin marker line | promoter | Note | Reference | ||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| CFP-TUA1 | 35S | [81] | |||||
| YFP-TUA5 | 35S | [6] | |||||
| mCherry-TUA5 | 35S | [89] | |||||
| GFP-TUA6 | 35S | Right handed helical growth, no expression in roots | [65] | ||||
| GFP-TUB6 | 35S | No expression in roots | [58] | ||||
| mRFP-TUB6 | 35S | [7] | |||||
| mRFP-TUB6 | UBQ1 | [60] | |||||
| Microtubule binding protein marker line | promoter | Note | Reference | ||||
|
|
|
||||||
| GFP-MBD (from mammalian MAP4) | 35S | MT bundling | [68,80] | ||||
| mRFP-MBD | 35S | [90] | |||||
| mCherry-MAP4-MBD | 35S | [89] | |||||
| GFP-EB1 | 35S | Overexpressing EB1 extensively bind MTs including minus end of MTs and endomembrane. | [78,79] | ||||
| EB1b-GFP | EB1 | MT plus end marker | [70] | ||||
| ARK1-GFP | |||||||
| ARK1 ΔARM-GFP | ARK1 | Expressing all cell types and label MT plus end | [91] | ||||
Fig. 4.
Images of RFP-TUB6 labelled MTs in Arabidopsis hypocotyl cells acquired using spinning disk confocal microscopy (a) and near-TIRF microscopy (b). a Single frame image was acquired with a spinning disk confocal microscope (Perkin Elmer UltraView Vox) with a 63x NA 1.3 glycerol immersion lens. In the image, MTs are in focus in the left cell, and free tubulin in the cytoplasm is shown in the focal plane in the right cell. Occasionally we observe free tubulin as background signal. b This single frame image was acquired with a near-TIRF microscope (Zeiss Laser TIRFIII) with a 63x NA 1.46 oil immersion lens. MTs are in focus in only one cell. Due to the thin excitation depth, MTs in neighboring cells are not captured. Bars, 10 µm.
Figure 5.
MTs observed in Arabidopsis inner leaf tissues using two-photon imaging a Spongy mesophyll cells expressing GFP-TUB6. Leaf was mounted in perfluoroperhydrophenanthrene (PP11) and the image was acquired with a two-photon fluorescence microscope (Olympus FV1000 MPE) with a 25x NA 1.05 water immersion lens. GFP-TUB6 is localized around chloroplasts and MTs are observed in some mesophyll cells. b MTs in spongy mesophyll cell from a, focal plane in the cell cortex, c same cell as in b, maximum Z-projection of 5 slices (5 µm total thickness), encompassing the outer periclinal cortex of a spongy mesophyll cell . Bars, 50 µm (a), 10 µm (b-c).
Super resolution microscopy
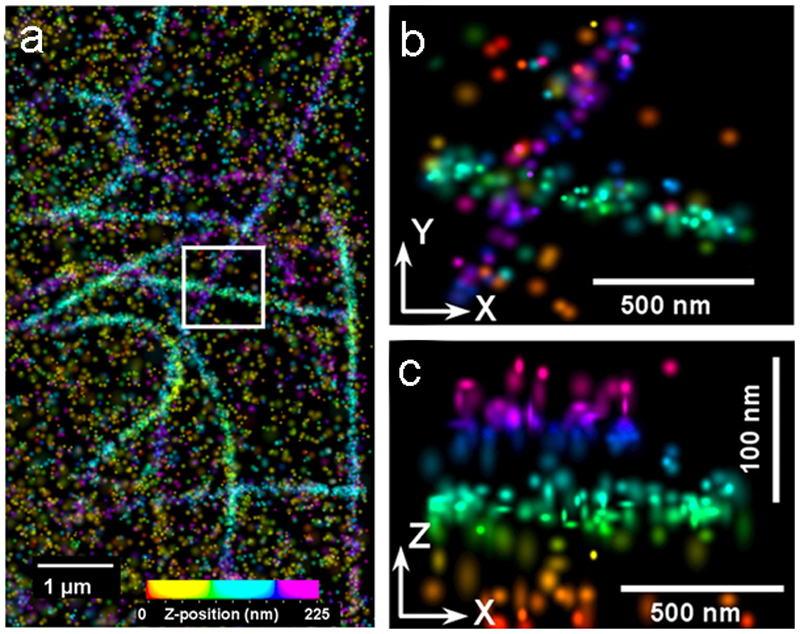
Standard diffraction-limited optical microscopy techniques cannot resolve cellular details that are closer together than ~200 nm in the focal plane or ~450 nm along the optical axis. This is far from resolving MTs, which have a 25-nm diameter. Scanning tunnelling, atomic force, or electron microscopes provide much higher resolutions but are restricted to cell surfaces or fixed specimens. In contrast, super-resolution light microscopy techniques increase resolution to below the diffraction limit with the possibility to explore a whole cell in 3D [44]. Compared to traditional confocal microscopy, a dramatically increased lateral resolution is achieved by Stimulated Emission Depletion (STED) and Ground State Depletion (GSD) microscopy. These techniques are particularly suitable to visualize MTs after standard immunofluorescence preparation [45–47]. Stochastic optical reconstruction microscopy (STORM) or photoactivation localization microscopy (PALM), combined with single-particle tracking, allow mapping transport trajectories of, for example, lysosomes [48] or other cargo on individual MTs by using photoactivatable fluorescent proteins. Interferometric photoactivated localization microscopy (iPALM) has even been applied successfully to measure the 25-nm MT diameter by visualizing a fluorescent tagged α-tubulin (Fig. 6 a-c; [49]). Until now, these methods have mainly been restricted to mammalian cells, as super resolution microscopes are very sensitive to auto-fluorescent molecules such as chlorophylls and accessory pigments. Their excitation with class 4 lasers at 592 or 660 nm (eg. as applied by STED microscopy) causes excessive heating and structural damage. However, in the future, these limitations could be bypassed by irradiating samples at a higher wavelength (775 nm) to avoid excessive autofluorescence. In plants, structured illumination microscopy (SIM) of GFP-fused MT-associated proteins was used to visualize MT dynamics in hypocotyl epidermal cells of Arabidopsis thaliana at the subdiffraction level [50]. Although still mainly practical for fixed material, super resolution techniques are of great value for MT imaging, as they bridge the gap between cellular ultrastructure and superstructure.
Fig. 6.
Superresolution iPALM image of fluorescent labelled microtubules (m-KikGR fused to α-tubulin), rendered with z axis color-coding. (a) Large area overview. (b) X-Y projection and corresponding (c) Z–Y projection of the area bound by the white box in (a). From [49], National Academy of Sciences, with permission.
Transmission electron microscopy
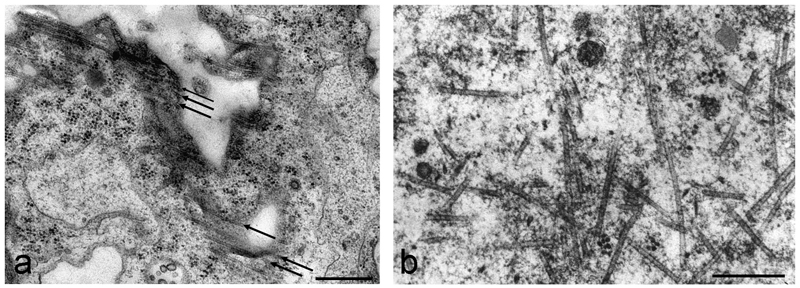
Despite the emergence of super resolution imaging, transmission electron microscopy (TEM) remains a valuable tool when high resolution analysis of individual MTs is required, especially when other cellular structures need to also be visualized. Since the 1960s, when MTs were first observed in plant electron micrographs [51], there has been debate over the best fixation protocol. Chemical fixation does a reasonable job, though after glutaraldehyde-osmium fixation (Fig. 7 a) or potassium hexacyanoferrate fixation, MTs appear slightly wrinkled (though well-preserved). Presently, high-pressure freezing followed by freeze substitution is considered the best method for preserving plant cellular ultrastructure (eg. [52–55]) but involves specialized apparatuses and cannot be performed in many research facilities. We therefore also describe plunge freezing as an alternative, which can preserve cortical regions of cells up to thicknesses of ~ 20 µm very well. This method is acceptable for examining the ultrastructure of cortical MTs in single cells (Fig. 7 b).
Fig. 7.
TEM visualization of MTs in different algal cells: a chemical fixation protocol in Klebsormidium crenulatum visualizing cortical MTs in an adult cell. The arrows indicate the parallel orientation of the MTs. b plunge-frozen and freeze-substituted MTs in Micrasterias muricata. MTs in association with a migrating nucleus show various orientations in a MT organizing centre. Note the clear and sharp outer surfaces of the MTs. Bars, 0.2 µm
Despite the static picture of MT organization provided, TEM remains essential when resolution beyond the limit of conventional fluorescence microscopy is required, such as when examining the higher order structure of MT bundles, the presence of crosslinking proteins and the interaction of MTs with endomembrane components and other structures. Using gold-coupled antibodies, it is also possible to analyze how proteins colocalize [56], identify motor proteins of the kinesin family [24] and investigate how crosslinking MAP65s interact with MTs [57]. It is important to note, however, that immunolocalization by TEM requires specialized preparation to preserve antigenicity of the protein epitopes. Best results are obtained after high pressure freeze fixation, freeze substitution and embedding in LR white resin [24]. Within the context of TEM, we have provided a short section on electron tomography for plant MT imaging. Electron tomography is a three-dimensional TEM imaging technique in which the internal ultrastructure of a sample is captured including, for example, MT orientation, bundling, and association with MAPs, within a thin resin-embedded or frozen section. Electron tomography has the potential to provide a complete picture of MT organization within the cortical cytoplasm.
Correlative microscopy
Finally, a chapter on imaging techniques would not be complete without mention of correlative microscopy. Correlative microscopy combines, in the broadest sense, two or more different microscopy techniques in order to obtain complementary information about a sample. In practice, it often involves the use of light and electron microscopy (and is therefore termed ‘correlative light and electron microscopy’ or CLEM). Given proper preparation, a sample can be imaged with fluorescence microscopy to identify a location of interest (for example, a fluorescently-tagged MT-associated protein) and the same sample subsequently imaged with TEM or tomography to obtain high resolution 2D or 3D information at the location of interest. CLEM has tremendous potential to combine information gained by specific tagging, localization and dynamics studies with 3D cellular ultrastructure. The reader is referred to Barton et al., 2009 (45) for more information.
2. Materials
2.1. Plant material
Arabidopsis thaliana seeds, constructs: seed stocks, including transgenic and mutant lines, as well as some DNA stocks, can generally be obtained from the Arabidopsis Biological Resource Centre. The Arabidopsis Information Resource (TAIR) website provides details and ordering information (http://www.arabidopsis.org/abrc/index.jsp).
Plant leaf material (this can vary according to the researchers’demands). Immunofluorescence and TEM protocols generally work with every type of plant leaf.
Plant cell cultures (eg. suspension cell cultures, unicellular green algae).
Specific transgenic reporter lines for observing MTs in live cells of Arabidopsis thaliana are listed in Table 1 (see Note 1):
2.2. Reporter Lines (see Note 1 and Table 1)
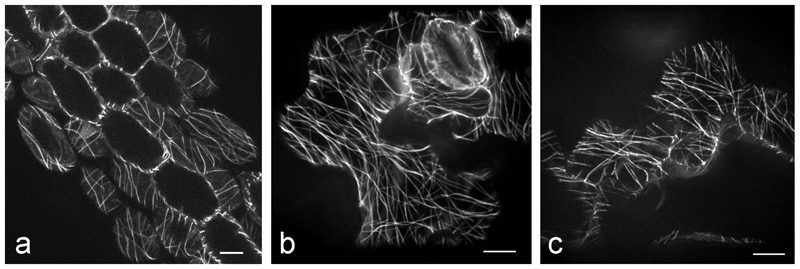
35Spro-GFP-TUB6: This construct from the Hashimoto lab (NAIST, Japan; contact Dr. T. Hashimoto: hasimoto@bs.naist.jp) fuses GFP to the N-terminus of the β tubulin 6 (At5g12550) isoform of Arabidopsis thaliana [58] (Fig. 8 b, c) (see Note 2).
35Spro:GFP-MBD: This construct was developed in Dr. Richard Cyr’s lab (Pennsylvania State University; rjc8@psu.edu) and is a heterologous MT reporter that works in plants, constructed from the MT binding domain (amino acid residues 935-1084) of the mammalian MAP4 [59] (Figs. 2 b, d, 7 a) (see Note 3).
EB1bpro:EB1b-GFP: This reporter generates comet-like fluorescent patterns at the growing plus ends of MTs and can be used to measure MT growth and to analyze MT polarity within an array (Fig. 9) (see Note 4).
UBQ1pro:mRFP-TUB6: Engineered for and first described in Ambrose et al. 2011 [60] by the Wasteneys Lab (Department of Botany, University of British Columbia; contact geoffrey.wasteneys@ubc.ca), Also available through the Arabidopsis Biological Resource Centre. This reporter is useful when the localization of a GFP- or YFP-tagged protein needs to be assessed in relation to MTs. It has several advantages over previous constructs (See Note 5).
ARK1pro:ARK1-GFP and ARK1pro:ARK1ΔARM-GFP: This construct is from the Wasteneys lab [91] . ARMADILLO-REPEAT KINESIN 1 (ARK1) is a MT plus-end tracking protein. These reporter lines show asymmetric plus-end labeling of MTs in many cell types in seedlings.
Fig. 8.
Comparison of different GFP-transgenic lines in wild-type background of Arabidopsis thaliana. a GFP-MBD hypocotyl of a 10 day-old plant grown at 21°C, b 11 day-old GFP-TUA, c 12 day-old GFP-TUB expressing plants grown at 21ºC were imaged within a few hours after the temperature was increased to 31ºC. Bars, 10 µm
Fig. 9.
Images of 35Spro:GFP-EB1 (a) and EB1pro:EB1-GFP (b) expressing Arabidopsis thaliana cells at 21ºC. a 35Spro:GFP-EB1, GFP-EB1 forms a comet-like shape and weakly associates with the side walls of MTs, and the background fluorescence is high. b EB1pro:EB1-GFP, plus-end labeling by EB1-GFP is more concentrated showing dot-like forms and the background signal is very low. Abaxial surface of 1st leaves from 12 day-old plants were used. Plants were grown at 21ºC and imaged with a Quorum Wave FX Spinning Disc Confocal System (Quorum, Guelph, Ontario, Canada) with a 63x NA 1.3 glycerol-immersion lens. A Bionomic Controller BC-110 stage together with a Heat Exchanger HEC-400 kept the specimens at 21ºC. Bars, 10 µm
2.3. Technical Equipment
Temperature-controlled stage: Bionomic Controller BC-110 together with a Heat Exchanger HEC-400, a Bionomic Controller BC-100 (20-20 Technology Inc., Wilmington, NC, USA).
Objective lens heater (BIOPTECHS, Butler, PS, USA).
Culture chambers for live cell imaging (obtained from Electron Microscopy Sciences, Hatfield, USA).
Thermocouple device (FLUKE52; John Fluke MFG. Co., Inc., Everett, Washington, USA).
High pressure freezer (formerly Balzers HPM 010, later taken over by BAL-TEC; then produced by ABRA Fluid AG, Widnau, Switzerland; currently available as “High Pressure Freezing Machine HPM 010” by Boeckeler Instruments Inc., Tucson/AZ, USA).
Freeze substitution device (LEICA EM AFS, Leica Microsysteme GmbH, Vienna, Austria).
Plunge-freezing device (LEICA EM GP)
2.4. Chemicals and Reagents
2.4.1. Chemicals and Reagents for Fluorescence Microscopy
All buffers are stored at 4°C but should be at room temperature when in use, as MTs depolymerize at cold temperatures.
2x PME buffer: 50 mM PIPES, 5 mM EGTA, 1 mM manganese sulfate, pH 7.2
Fixative: 0.5% glutaraldehyde, 1.5% formaldehyde in 1 x PME buffer, pH 7.2. Prepare before use. Use of 25% glutaraldehyde in ampules (EM grade) is preferred. Due to its tendency to polymerize easily, make aliquots of 25% glutaraldehyde and store at −20 °C. Freeze-thawing of glutaraldehyde will induce polymer formation, which affects the fixation efficiency. Use of 16% formaldehyde in ampules (EM grade) is preferred. Store at 4°C or room temperature.
1 x PMET buffer: 0.05% Triton X-100 in 1 x PME buffer, pH 7.2
1 x PBS buffer: 137 mM sodium chloride, 2.7 mM potassium chloride, 4.3 mM sodium hydrogen phosphate, 1.4 mM potassium dihydrogen phosphate, pH 7.5
Cell wall digestion enzyme solution: 0.05% pectolyase (Pectolyase Y-23, MP Biomedicals, USA), 0.4 M mannitol, 1% BSA in 1 × PBS. Solution can be stored at −20 °C
Permeabilization buffer: PBS, 1% Triton X-100 in 1 x PBS, pH 7.5
Sodium borohydride solution: 1 mg/mL sodium borohydride in 1 x PBS (needs to be fresh, prepare before use)
Incubation buffer: 50 mM glycine in 1 x PBS
Blocking buffer: 1% BSA, 50 mM glycine in 1 x PBS
Primary antibody solution: primary antibody in blocking buffer. For example, mouse anti-α-tubulin, clone B 512 (Sigma) works well at a 1:1000 dilution.
Secondary antibody solution: secondary antibody in blocking buffer. For mouse anti-α-tubulin antibody B 512, Alexa 488-conjugated goat anti-mouse IgG (GE Healthcare) could be used at a 1:100 dilution.
Citifluor AF1 antifading agent (Citifluor Ltd. UK).
2.4.2. Chemicals and Reagents for Live Cell Imaging
Agar (plant cell culture tested), 1.5%
-
Hoagland's growth media or MS media
Hoaglands’ growth media: 2 mM KNO3, 5 mM Ca(NO3)2, 2 mM MgSO4, 1 mM KH2PO4, 90 μM EDTA, 46 μM H3BO4, 9.2 μM MnCl2, 0.77 μM ZnSO4, 0.32 μM CuSO4, 0.11 μM MoO3, autoclave the media
MS media: Dissolve 4.31g of Murashige and Skoog (MS) basal salt mixture, 0.5 g of 2-(N- Morpholino) ethanesulfonic acid (MES) in 1L water, Adjust pH to 5.7 using KOH, autoclave the media.
Agarose (low gelling temperature) 2%
Perfluoroperhydrophenanthrene (56919 FLUKA, Sigma) as a mounting media for leaves (see Note 6)
2.4.3. Chemicals and Reagents for Transmission Electron Microscopy
Cacodylic buffer: 50 mM cacodylic acid, sodium salt, pH 7.2
Cacodylate-glutaraldehyde: 50 mM cacodylate buffer, 1% glutaraldehyde (EM-grade), pH 6.8
Cacodylate-glutaraldehyde-calcium chloride: 50 mM cacodylate buffer, 1% glutaraldehyde, 5 mM calcium chloride, pH 7.2
Cacodylate-osmium: 50 mM cacodylate buffer, 1% osmium tetroxide
Cacodylate-osmium-ferricyanide: 50 mM cacodylate buffer, 1% osmium tetroxide, 0.8% potassium hexacyanoferrate III (ferricyanide), pH 7.2
2% aqueous uranyl acetate dihydrate.
Ethanol solutions: 15%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 95%, 100%
1,2- propylene oxide reagent
Propylene oxide-ethanol mixture: 1,2 propylene oxide and ethanol (1:1 v/v)
A suitable embedding resin: Embed 812, Araldite 502, Spurr’s resin, Agar Low Viscosity Resin (LV) (Agar Scientific Ltd. Essex CM24 8DA, England) (See Note 7)
Formvar 1595 E
Chloroform
Reynold's lead citrate
2.4.4. Chemicals and Reagents for High Pressure Freezing and Freeze Substitution
Lecithin: L-a-phosphatidyl-choline; 100 mg/mL dissolved in chloroform
Sucrose: 100 mM sucrose
Acetone-osmium-tannic acid: 2% osmium tetroxide and 0.1% tannic acid in acetone
Acetone-osmium-uranyl acetate: 2% osmium tetroxide and 0.05% uranyl acetate in acetone
2.4.5. Chemicals and Reagents for Immunogold labelling TEM
TBS: 50 mM Tris buffered saline, Trizma pre-set crystals; pH 7.5
TBS-blocking solution: TBS, 1% BSA (fraction V), 1% acetylated BSA (BSAac), 0.2% Tween-20
TBS-incubation solution: TBS, 1% BSA, 0.1% BSAac, 0.01% Tween-20
TBS-BSA: TBS, 1% BSA
Primary Antisera/Antibodies (eg. monoclonal antibody directed against bovine brain kinesin, K-1005; anti-α tubulin, cloneB-5-1-2, T 6074, Sigma)
10 nm - collodial gold conjugated corresponding secondary antibody (eg. goat anti-rabbit IgG, G-7402; goat anti-mouse IgM, G-5652, Sigma)
Formvar-coated nickel grids
3. Methods
3.1. Microtubule Staining of Fixed Material
3.1.1 Standard MT labelling method using wall-degrading enzymes; suitable for plant organs lacking thick waxy cuticles such as roots.
Ensure that samples are always in solution. Dried areas increase the chance of getting non-specific binding of antibodies. This whole mount protocol is modified from [61]. The root-tip squash method is available in [40].
Fix samples for 40 min in 0.5% glutaraldehyde, 1.5% formaldehyde fixative solution. Make sure that the sample is immersed in the fixative.
Wash 3 times for 10 min in PMET buffer.
Incubate the sample with cell wall digestion enzyme solution for 20 min.
Rinse 3 times for 10 min in PMET on the shaker.
Incubate the sample with permeabilization buffer for 3 hrs at room temperature on the shaker.
Rinse 3 times for 10 min in PBS, and make sodium borohydride solution.
Incubate the sample in sodium borohydride solution for 20 min. This additional step helps to reduce aldehyde-derived autofluorescence by reducing free aldehydes groups.
Discard sodium borohydride solution and add blocking buffer. Incubate for 30 min.
Place the primary antibody solution in a microscope well slide, which allows use of less antibody solution (50 – 100 μl). Place the sample in the antibody solution and incubate in a humid box (place wet kimwipes in the box) at 4°C overnight.
Transfer samples to a small Petri dish using a brush and rinse 5 times for 10 min each in incubation buffer on the shaker.
Incubate in blocking buffer for 30 min. Another option for blocking buffer would be to use the non-immune serum from the animal species are used to raise the secondary antibody. For example, 5% normal goat serum (Pierce Biotechnology) in incubation buffer.
Place secondary antibody solution in the well slide and samples. Incubate in the humid box wrapped with aluminum foil for 3 hrs at 37°C.
Transfer samples to small Petri dish using a brush and rinse 3 times for 10 min in 1x PBS on the shaker. Cover the Petri dish to exclude light.
Mount sample in CitiFluor AF1 anti-fading mounting reagent.
Seal the cover slip with non-fluorescent nail polish.
The slides can be stored at −20°C for long time storage.
3.1.2 MT staining using the freeze shattering method [35] with useful modifications according to [29]; suitable for aerial organs.
Fix samples for 40 min in 0.5% glutaraldehyde, 1.5% formaldehyde fixative solution. Large organs should be cut into smaller pieces to facilitate infiltration of fixative.
Wash in 1 x PMET buffer.
Blot samples to remove excess buffer, place between two glass slides and dip into liquid nitrogen. Remove and quickly apply pressure by compressing the slides with a pair of pliers before the tissues thaw (see Note 8).
Transfer sample to permeabilization buffer containing PBS for 1 h.
Transfer samples to PBS (pH 7.4) for 10 min, followed by 20 min incubation in PBS containing 1 mg/mL sodium borohydride.
Incubate in primary antibody overnight at 4°C.
Wash extensively in buffer to remove any unbound antibody (e.g. 3 times 10 min in PBS).
Incubate in secondary antibody (Alexa488-conjugated goat anti-mouse IgG, 1:200) for 1h at 37°C.
Mount samples in Citifluor AF1 antifade agent.
Examine with a Confocal Laser Scanning Microscope (CLSM), using a high Numerical Aperture (NA) objective lens (eg. Zeiss Axiovert, 63x magnification, 1.4 NA).
Generate excitation with an Argon laser at 488 nm.
Collect long pass (LP) 560 nm filtered emission and band pass (BP) 505-530 nm filtered emission simultaneously in two channels; if possible collect a DIC image with another channel.
Capture Z-stacks according to requirements.
Generate projections eg. using ImageJ software (freeware; http://www.imageJ).
3.2. Fluorescent Proteins to Visualize the MT Cytoskeleton
3.2.1 This section describes a standard method of preparing aerial organs such as leaves or cotyledons of Arabidopsis thaliana for live imaging on an inverted microscope. It can be used for any organs including roots, stems and floral organs and can of course be modified for any plant expressing a fluorescent reporter.
Plant surface sterilized seeds on Petri plates containing nutrient medium in 1.5% agar. Wrap plates with porous surgical tape.
Store plates at 4°C for 3-5 days to synchronize germination, then transfer to a growth cabinet, keeping the plants growing under constant light (80 µmol m−2 second−1) or in an 16 h light, 8 h dark cycle at 21°C for 11-12 days.
Excise whole leaves or pieces of them and place them on the coverslip that forms the bottom of microscope culture dishes (see Note 9). We obtain dishes suitable for this purpose from Electron Microscopy Sciences, Hatfield, USA. If you are using an upright microscope, the leaves can be placed directly onto a glass slide and covered with cover slips held in place at corners with small amounts of vacuum grease.
Mount leaf cuttings in water or perfluoroperhydrophenanthrene (see Note 6), taking care to ensure that the surface of interest is facing the coverslip. A small square of 2% agarose can be placed on top of the leaf to stabilize it from drifting during imaging.
Allow samples to equilibrate under standard conditions for at least 1 hour prior to observation.
Place culture chambers on microscope stage and ensure that images are recorded according to the spectral properties of the fluorescent protein. Images can be taken every 8 seconds over 3 to 5 minutes for GFP-TUB and every 5 or 8 seconds over 40 to 60 seconds for EB1b-GFP (see Notes 10 and 11).
To keep the temperature stable around the mounted samples, use a temperature-controlled stage: Bionomic controller BC-110 equipped with Heat exchanger HEC-400 (20/20 Technology Inc.) (see Note 12). An objective lens heater (Bioptechs) is sometime also required, especially when using immersion lenses. Monitor the temperature of the sample immediately after imaging by measuring the temperature of glycerol on the coverslip using a thermocouple device.
Avoid recording images over periods longer than 4 hours.
3.3. TEM Preparations to Visualize MTs in Plant Cells
3.3.1. Glutaraldehyde-osmium fixation protocol for ultrastructure studies (with modifications after [29,59])
Cut small blocks of tissue or cells and place in glass vials. Fix tissue or cells in 10-50 mM cacodylate buffer (see Note 13),) containing 1% glutaraldehyde for 30 - 90 min (see Note 14 and Fig. 7 a, Klebsormidium crenulatum).
Remove fixative and wash cells 3 times 5 min in cacodylate buffer.
Postfix cells in cacodylate-osmium for 2 h at room temperature or 12 h at 4°C.
Wash cells 3 times 5 min with distilled water.
Dehydrate the cells in increasing concentrations of cold ethanol, 15 min per concentration: 15%, 30%, 40%, 50%, 70%, 80%, 90%, 95%, 100%. Keep in 100% for 30 min.
Transfer the cells to a 1:1 mixture of propylene oxide:ethanol and allow to equilibrate for 10 min.
Transfer to propylene oxide for 10 min.
Transfer the cells to a 1:1 mixture of propylene oxide:embedding resin.
Rotate the cells in this mixture for 24 h in order to allow the propylene oxide to evaporate and for sufficient penetration of the cells with the resin.
Transfer the cells to freshly prepared resin in a polymerization chamber or aluminum dish.
Incubate in a desiccator for 4 days.
Polymerize the resin for 16 h at 60°C.
Select samples and section using an ultramicrotome. Section thickness should generally be in the range of 50 – 200 nm. The ideal section thickness for transmission electron microscopy with accelerating voltages between 50 kV and 120 kV is about 30–100 nm. Thicker sections (up to 200 nm) can be visualized using a 200 kV accelerating voltage and may be beneficial when considering tomography, in which case a larger reconstructed volume can provide more information about MT configuration within the sample.
Collect the sections on formvar-coated copper grids (see Notes 15 and 16).
Counter stain the sections by floating them, section-side down, on small 10 µl droplets of 2% aqueous uranyl acetate for 30 min (see Note 17).
Wash for 2 min with distilled water and allow to dry fully.
Next, counterstain on droplets of with Reynold´s lead citrate (see Note 18).
Wash for 2 min with distilled water and allow to dry fully.
Investigate and image with a transmission electron microscope. Some considerations for electron tomography are included below.
3.3.2. Considerations for electron tomography
When preparing grids for tilt series acquisition, one must take the following points into consideration:
Copper grids, which are non-ferromagnetic, should be used (and not nickel grids, see Note 15). Nickel grids distort the magnetic field of the objective lens and reduce the lens’ ability to reproduce the object plane.
Generally, slot grids are preferred. Plant tissue sections are often larger than the mesh size on a mesh grid; using a mesh grid may result in some of the sample being obstructed by the grid bar. When using a slot grid, tilt series acquisition can be performed along the axis of the slot.
Thicker (200 nm) sections are advised in order to maximize the reconstructed volume in the final tomogram.
Electron dose must be taken into consideration during tilt series acquisition, especially when performing CEMOVIS. For vitreous samples, a “low-dose” imaging protocol is necessary to avoid excessive electron irradiation of the sample.
For further information, including operating protocols, the reader is referred to [62].
3.3.3. High pressure freeze fixation protocols to preserve microtubules
The protocol described here uses a commercially available hyperbaric freezing device. The methods basically follow the methods of ([52]Fig. 10), with the addition of notes from own experience and information gained from several other works as stated in Notes 19 to 26.
Dip specimen cups in lecithin (1-a-phosphatidyl-cholin; 100 mg/mL dissolved in chloroform) and allow to dry until specimen holders are covered with lecithin.
Prepare samples in their regular culture media; for certain plant tissues it may be useful to transfer samples to 100 mM sucrose (see Note 19).
Transfer the samples into gold or aluminum specimen cups.
Ensure that no air bubbles are present in the preparation as they will damage the samples with the high pressures involved in this process.
Close two cups face to face or, depending on the sample, orient the face toward the bottom.
Mount the cups in the specimen holder and transfer to the High Pressure Freezing Machine HPM 010 (see Note 20).
Freeze samples in liquid nitrogen according to the manufacturer’s instructions.
Remove samples immediately after the freezing procedure and transfer them to liquid nitrogen, where they can be stored until further processing, either by freeze substitution followed by sectioning (as outlined below), or by cryo-sectioning (CEMOVIS) (see Note 21).
Freeze substitute samples in 2% OsO4 in acetone or 0.1% tannic acid in acetone at −80°C for 24 h and a mixture of 2% OsO4 and 0.05% uranyl acetate in acetone at −80°C for 28 h (see Note 22).
Raise the temperature to −30°C in the same medium over the course of 10 h. Bring to room temperature and rinse in acetone.
Embed and further process the samples as described in Subheading 3.3.1., step 7
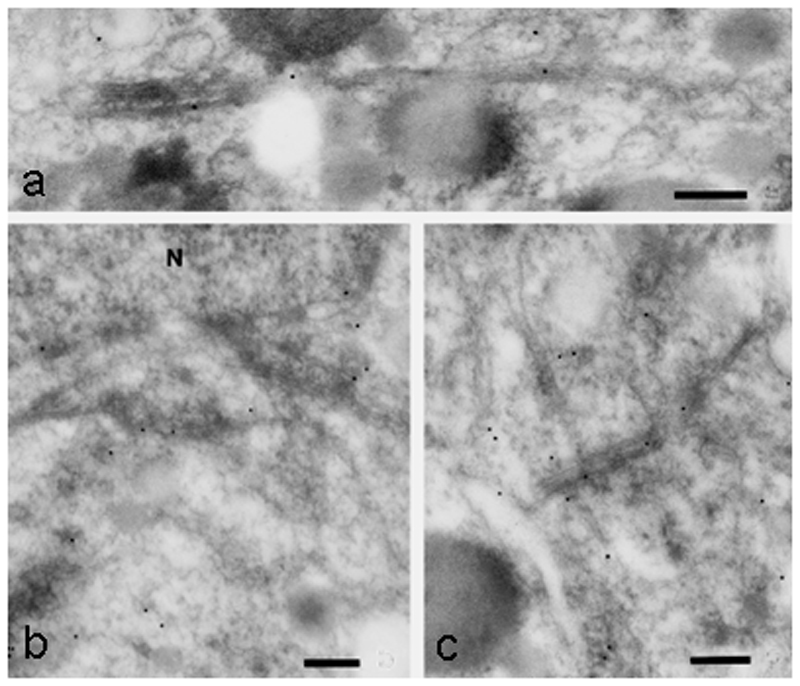
Fig. 10.
High pressure-frozen MTs in the vicinity of a migrating nucleus in the green alga Micrasteris denticulata immuno-gold (10 nm) stained with an antibody to brain kinesin. Bars, 0.2 µm. Reprinted from [24], Elsevier, with permission.
3.3.4. Plunge Freezing of thin samples
As an alternative to high-pressure freezing, we describe plunge freezing [63]. Protocol is based on [53,64].
Prepare the cryo-fixation device: switch on the plunge-freezing device (Leica EM GP) and fill the water container for the humidifier with 60 mL distilled water. Mark a cryo-storage box for identification of the sample and position this box in the plunge-freezing device transfer container. Fill the liquid nitrogen dewar with (1.5 L) liquid nitrogen. When the ethane container reaches liquid nitrogen temperature, fill the (black) secondary cryogen container with ethane or ethane/propane (Tivol, Briegel, & Jensen, 2008) using the cryogen liquefier.
Set the vitrification parameters: using the control panel, set the environmental chamber temperature to 22 °C and the humidity (Hr) to 95%. Set the blotting time to zero seconds (blotting is used to remove excess liquid from an EM grid before plunging a liquid sample and is not necessary for solid plant samples). Set the liquid ethane temperature to − 181 °C when liquid ethane is used, or to − 191 °C when a liquid ethane/propane mixture is used.
Sample vitrification: mount a thin sample onto the plunge device forceps (see Note 23). Using the plunge freezer, rapidly plunge the sample into the liquid ethane, or liquid propane/ethane mixture.
After the freezing step, transfer the sample into the cryo-storage box prepared. Plunge more samples, if desired. When finished, store the cryo-storage box in liquid nitrogen before further manipulation (either freeze substitution followed by sectioning or CEMOVIS).
3.3.5. Immunolabelling TEM Protocol
The procedures described are modified from [56] and can be adjusted for basically any new MT-associated protein or for other cytoskeletal components (Fig. 9).
Fix samples. Follow the Subheading 3.3.1. Step 1 to 5, except for osmium fixation (Step 3 and 4). Osmium fixation would mask the epitopes. After dehydration (Step 5 in Subheading 3.3.1.), make LR white resin: acetone mix to start resin infiltration. The resin concentration should gradually increase such as 1 drop of resin in 20 ml acetone, 10 %, 30 %, 50 %, 70 %, 90 %, 95 %, 100 % resin. Exchange 100 % resin for three times. Polymerize resin under UV light.
Thin section should be collected on formvar-coated nickel grids.
Pre-incubate sections in TBS-blocking solution for 1 hour (see Note 24). This can be done by floating the grids on drops of the solution.
Incubate sections in primary antibodies (e.g. monoclonal antibody directed against bovine brain kinesin; Sigma K-1005) diluted 1:50, 1:100 in TBS, 1% BSA, 0.1% BSAac, 0.01% Tween-20 (TBST) for 20–24 h at 4°C. Antibody concentrations for immunoTEM generally need to be higher than those used for immunofluorescence.
Wash in droplets of 50 mM TBS, 1 % BSA, 4 times, 15 minutes.
Transfer into gold conjugated secondary antibody in TBS-incubation solution in a dilution of 1:200 or 1:400 and incubate for 1.5 or 2 h at room temperature.
For control purposes omit the first antibody on some sections or incubate in antigen-saturated primary antibody.
After the incubation with secondary antibody, rinse the sections extensively with TBST to remove any unbound antibody (see Note 25).
Continue as in Subheading 3.3.1., step 15 (see Note 26) for uranyl acetate and lead citrate staining.
4. Notes
Fusion protein reporter construction: Not surprisingly, fusing a fluorescent reporter to a protein can alter its function. The GFP-TUA6 construct (GFP is fused to the N-terminus of alpha tubulin 6) was the first tubulin-based fluorescent reporter to work in plant cells [65]. It generates bright labeling of MTs in aerial tissue (it is expressed but apparently does not incorporate into MTs in root tissues) but causes right-handed organ twisting, a phenomenon attributed to inhibition of the tagging of alpha tubulin at its N-terminus, which may interfere with its GTP hydrolysis-promoting activity [66]. After testing several different fluorescent reporters for imaging MTs, we recommend the constructs listed in the text. These constructs are available upon request from the laboratory of origin or from the Arabidopsis stock centres. Most constructs are also available with other fluorescent tags such as yellow fluorescent protein (YFP) and red fluorescent protein (RFP).
The expression of 35Spro:GFP-TUB6 in stably transformed lines of the Columbia ecotype is regulated by the cauliflower mosaic virus 35S promoter [58]. In comparison to some other MT reporters, this one generates relatively low fluorescence but there are no detectable developmental or morphological phenotypes [66,67]. This line cannot be used reliably to examine MTs in root cells though some labelling of MTs can be observed in dividing cells.
The 35Spro:GFP-MBD construct is good for examining MTs in root tissues, where it has been used to document MTs in division [41,40] and elongation zones [68] as well as during root hair development [15,69]. Nevertheless, caution needs to be exercised with this reporter. Plants stably expressing GFP-MBD under the CMV35S promoter frequently have severe developmental defects and low seed yield, and MT bundle formation has been observed under certain conditions (for more details see Notes on choice of promoter, Note 10). We suggest confining the use of GFP-MBD to roots, and then to select specimens that have low level of expression. This can be achieved by selecting heterozygotes from an F2 segregating population [15].
There are many different fluorescent EB1 reporters available but the EB1bpro:EB1-GFP construct from the Cyr lab (Pennsylvania State University, USA) uses the promoter element from the Arabidopsis thaliana EB1b isoform to drive moderate expression levels [70], which confines comets to the growing plus ends. Even so, we have found that MT growth rates are slightly higher when measured with this reporter compared to GFP-TUB [39], a finding that underscores the importance of carefully regulating the expression of some reporters (for further details see Notes on choice of promoter, Note 10). EB1 reporters seem to work equally well with the fluorescent tag at either the C- or N-terminus.
Using the UBQ1 promoter to drive mRFP-TUB expression prevents gene silencing, which is common with 35S promoters. This reporter also works well in root tissues.
Perfluoroperhydrophenanthrene (PP11) is a non-toxic solution, which infiltrates the air space between mesophyll cells of the leaf, enabling a homogenous refractive index through the tissue. The image quality and resolution of the leaf inner tissue is greatly improved with little effect on physiology [71–73].
For ultrasctructural studies, Spurr’s resin (TedPella Inc, USA) [74] may be the best choice among other epoxide resins when different degrees of hardness have to be achieved according to the needs for different plant tissues. Its low viscocity property facilitates to infiltrate into the plant tissue quickly. The kit “Agar Low Viscosity Resin (LV)” from Agar Scientific is another alternative. For immunogold labeling, use LR white resin (London Resin Company, medium grade). Acrylic resins including LR white resin polymerize under UV light and that prevent epitopes from heat damage that usually associated with the polymerization of epoxide resin. Their hydrophilic properties and rough surface facilitate the epitope recognition by antibodies.
Some delicate samples can be effectively permeabilized by repeated freeze-thaw cycles and do not require compression in the frozen state.
Excising organs from plants will generate a wound response that may alter MT behaviour at a site distal to the wound. Other physically-induced responses include touch responses, gravitropisms and photoinduction. Forcing cells close to the coverslip for optimal viewing may generate a touch response that manifests itself in aberrant MT behaviour. Some plant organs, such as hypocotyls and roots, are strongly gravity- or light-responsive and when placed on a conventional horizontal microscope stage will undergo differential flank growth in an attempt to resume vertical growth. Horizontally placed roots, for example, will be stimulated to grow more rapidly on the upper flank and growth will be inhibited on the lower flank. With an inverted stage microscope, this will mean that the growth of cells closest the lens will be reduced or prematurely arrested. Upon placement in the horizontal position, cell division in Arabidopsis roots has been shown to be temporarily suspended [40]. Hypocotyls are exquisitely light responsive and rapid switches in MT orientation, likely involving altered dynamics, have been noted when etiolated hypocotyls are exposed to blue light or treated with hormones [75,76].
Phototoxicity: The risk of phototoxicity from the high intensity light used to excite fluorophores is the first consideration when live cell imaging [77]. Scan times need to be limited to avoid bleaching the fluorescence or impairing the function of the tagged protein. This makes it far more challenging to collect the same level of 3-D information that is possible on fixed, immunolabelled material. Choosing fluorescent proteins that can be excited by low energy, longer wavelength light (such as RFP) is one strategy but special attention to autofluorescence is then required. Probably the most important way to limit phototoxicity is to choose the most light-sensitive camera possible, and to minimize the intensity of the excitation light source. Spinning disc scan heads can also reduce dwell times, reducing both phototoxicity and photobleaching.
Choice of promoter: To overcome the need for high intensity excitation, transgenic lines are typically chosen that have high expression levels, usually achieved by using the 35S cauliflower mosaic virus (CMV) or Ubiquitin promoter elements, which drive constitutive and generally high expression in most tissues of higher plants. Depending on where the transgene inserts, and whether multiple insertions take place, the level of expression of MT reporter proteins can vary considerably. The 35S CMV promoter has been used effectively for most MT reporters, but in some cases the high level of expression generates artefacts by changing the dynamics of MTs or promoting bundle formation. For example, 35S-driven EB1-GFP can decorate both the plus and minus ends of growing MTs and is even found along the MT lattice and endomembranes [78,79], whereas it is distributed to the growing plus end when an endogenous promoter is used to drive its expression [70]. The GFP-MBD reporter fusion was constructed from the MT binding domain (amino acid residues 935-1084) of the mammalian MAP4 [80]. Under control of the 35S promoter, GFP-MBD expression decorates MTs in plant cells very effectively (Fig. 1 d) but can cause significant developmental defects, suggesting that this heterologous protein (there is no MAP4 homologue known in plant cells) interferes with normal function of MTs. In different genetic backgrounds, the 35Spro:GFP-MBD has been shown to cause unusual bundle formation that is not observed in the same genotypes with other reporters or by immunofluorescence [81,82,40]. Interestingly, in a recent study in which the IRX7 promoter element was used to control GFP-MBD expression, expression was restricted to vascular and epidermal cells, and no developmental defects were reported [83].
Culture temperature: It has been reported that temperature shifts significantly alter MT dynamics in Arabidopsis epidermal cells [39]. If the intention is to measure MT dynamics (growth and shrinkage velocity, time spent in growth, shrinkage and pause) then a temperature-controlled stage is necessary. The large proportion of metal components in microscopes makes it difficult to manipulate temperatures. Lenses are good temperature sinks and special lens heaters may be required if the desired culture temperature is above ambient.
The buffer concentration has to be selected carefully according to the physiological demand of the cells (10-50 mM is suitable for most plant cell types). When the buffer concentration is too high, osmotic phenomena may occur that lead to a detachment of the plasma membrane from the cell wall.
Cells can be alternatively fixed in 50 mM cacodylate buffer, 1% glutaraldehyde, washed, postfixed for 2 h in a mixture of buffered 1% osmium tetroxide, 0.8 % potassium hexacyanoferrate and then post-stained in aqueous 2% uranyl acetate according to [84].
Copper grids rapidly conduct heat away from the support film and help prevent thermal expansion and hence movement of the specimen under thermal radiation. During immunolabelling, however, Cu2+ ions may have an inhibitory effect on antibody functioning, so nickel grids are used in immuno- applications. Ni grids, however, are ferromagnetic, and distort the magnetic field of the objective lens, which is especially of concern during tilt series acquisition for tomography, when image alignment is important.
Grids can be either coated with formvar in the lab, or purchased pre-coated. Briefly, to prepare formvar coated grids in the lab, dip a clean coverslip in 0.3% formvar dissolved in chloroform, remove gently and allow to dry. Next, cut the edges on top of the cover slip with a sharp pin or razor blade and allow the thin film to float on a water surface (MilliQ water in a glass dish) by gently dipping the coverslip in the water at an angle. Place grids on this film and finally collect from the water surface with ParafilmTM.
Prior to counterstaining, spin down (> 5 minutes) the uranyl acetate and lead citrate solutions to remove any precipitates. During incubation in uranyl acetate solution it is necessary to keep the sections in darkness by covering the dish with aluminum foil. Light causes precipitation of the compound.
During incubation in lead citrate it is necessary to place some NaOH pellets in the staining dish (near the sections) to prevent precipitation of the lead citrate on the sections due to exposure to CO2. The following procedure has been found useful for the preparation of Reynolds lead citrate: allow distilled water to boil for 10 min to remove air and carbon dioxide, let cool and use only this water for the subsequent steps. Dissolve 1.33 g lead nitrate and 1.76 g sodium citrate in 30 mL of the boiled distilled water, shake vigorously for 1 min (the salts will not dissolve right away, but will remain in a white solution) and then let stand for 30 min. Next, add 8 ml of 1 M sodium hydroxide until the solution becomes clear. Wait. If necessary, continue adding drops of sodium hydroxide until the solution clears. Finally, fill to 50 mL with boiled distilled water.
Samples may be adjusted to increasing sucrose concentrations by cultivating them in medium containing gradually elevating sucrose [54].
Alternatively samples can be frozen in a Leica EMPACT high pressure freezer (Leica Microsysteme GmbH, Vienna, Austria).
CEMOVIS, or cryo-electron microscopy of vitreous sections, enables direct visualization of high pressure or plunge frozen samples in their native state, avoiding the possible artifacts induced by dehydration and staining during freeze substitution [85]. It requires a cryo-microtome device with which vitreous sections can be cut from the frozen block of tissue. These can then be directly imaged at cryogenic conditions with the TEM.
Some researchers recommend adding a certain proportion of water to the samples to improve the quality of the final preservation (eg. [86,87]). Added water (5% (v/v)) saturates the polar solvent during warming, which is beneficial for sample quality because it maintains structural water layers around cellular macromolecules (and stabilizes the hydrophobic effect). It is always necessary to add water (5% (v/v)) to high-pressure freezing and freeze substitution protocols for STED and PALM/STORM in order to maintain fluorescence of fluorescent tagged proteins [88]. Moreover, it may be useful to prolong the substitution process for several days or up to a week according to the size of the plant samples (A. Staehelin, personal communication).
This method is only suitable when the samples are extremely thin, and it is expected that only the first ~ 20 µm are preserved well, whereas with high pressure freezing up to ~ 200 µm thick samples are preserved well.
Depending on the desired blocking, alternatively the following “blocking buffers” can be used: (a) TBS, 1% BSA, 0.1% Tween-20 for 20 min followed by incubation in TBS, 1% acetylated BSA; or (b) TBS containing 1% BSA, 20% bovine fetal serum followed by incubation in TBS, 1% BSA, 1% BSAac, 0.5% Tween-20; or (c) TBS, 5% non-fat milk, 0.1% Tween-20 followed by TBS, 1% BSA, 0.2% gelatine; or (d) 10 mm phosphate-buffered saline (PBS), pH=7.4, 50 mm glycine (Merck), followed by PBS containing 1% BSA, 2% gelatine (Merck).
To wash the grids, follow this procedure: rinse the grids containing the sections with a mild spray of TBS from a Pasteur pipette, and then transfer onto small droplets of TBS for 2 min. Next, rinse a second time with TBS and complete the washing step with a mild spray of double-distilled water from a plastic spray bottle.
Counterstaining may either be omitted or the duration reduced according to the desired density of staining.
Acknowledgements
This chapter has been supported by Austrian Science Fund (FWF) project P24242-B16 to AH and funding from the Natural Sciences and Engineering Research Council and the Canadian Institutes of Health Research to GOW. The Thieme Verlag KG is acknowledged for their kind permission to reproduce Fig. 1 b-c, Fig. 6 a-c is reproduced with permission of the National Academy of Sciences of the United States of America, and Dr. Jennifer Lippincott-Schwartz, National Institute of Child Health and Human Development, Bethesda, MD. Elsevier Inc. for reproduction of Fig. 10.
References
- 1.Rasmussen CG, Wright AJ, Muller S. The role of the cytoskeleton and associated proteins in determination of the plant cell division plane. Plant J. 2013;75(2):258–269. doi: 10.1111/tpj.12177. [DOI] [PubMed] [Google Scholar]
- 2.Louveaux M, Hamant O. The mechanics behind cell division. Current Opinion in Plant Biology. 2013;16(6):774–779. doi: 10.1016/j.pbi.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 3.Masoud K, Herzog E, Chaboute ME, Schmit AC. Microtubule nucleation and establishment of the mitotic spindle in vascular plant cells. Plant J. 2013;75(2):245–257. doi: 10.1111/tpj.12179. [DOI] [PubMed] [Google Scholar]
- 4.Ruan Y, Wasteneys GO. CLASP: a microtubule-based integrator of the hormone-mediated transitions from cell division to elongation. Curr Opin Plant Biol. 2014;22:149–158. doi: 10.1016/j.pbi.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Ambrose C, Wasteneys GO. Cell edges accumulate gamma tubulin complex components and nucleate microtubules following cytokinesis in Arabidopsis thaliana. PLoS One. 2011;6(11):e27423. doi: 10.1371/journal.pone.0027423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paredez AR, Somerville CR, Ehrhardt DW. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 2006;312(5779):1491–1495. doi: 10.1126/science.1126551. [DOI] [PubMed] [Google Scholar]
- 7.Fujita M, Himmelspach R, Hocart CH, Williamson RE, Mansfield SD, Wasteneys GO. Cortical microtubules optimize cell-wall crystallinity to drive unidirectional growth in Arabidopsis. Plant J. 2011;66(6):915–928. doi: 10.1111/j.1365-313X.2011.04552.x. [DOI] [PubMed] [Google Scholar]
- 8.Lei L, Li SD, Bashline L, Gu Y. Dissecting the molecular mechanism underlying the intimate relationship between cellulose microfibrils and cortical microtubules. Front Plant Sci. 2014;5:8. doi: 10.3389/fpls.2014.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujita M, Lechner B, Barton DA, Overall RL, Wasteneys GO. The missing link: do cortical microtubules define plasma membrane nanodomains that modulate cellulose biosynthesis? Protoplasma. 2012;249:S59–S67. doi: 10.1007/s00709-011-0332-z. [DOI] [PubMed] [Google Scholar]
- 10.Oda Y, Fukuda H. The dynamic interplay of plasma membrane domains and cortical microtubules in secondary cell wall patterning. Front Plant Sci. 2013;4:6. doi: 10.3389/fpls.2013.00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sampathkumar A, Yan A, Krupinski P, Meyerowitz EM. Physical Forces Regulate Plant Development and Morphogenesis. Curr Biol. 2014;24(10):R475–R483. doi: 10.1016/j.cub.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivakov A, Persson S. Plant cell shape: modulators and measurements. Front Plant Sci. 2013;4(13) doi: 10.3389/fpls.2013.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang CH, Halsey LE, Szymanski DB. The development and geometry of shape change in Arabidopsis thaliana cotyledon pavement cells. BMC Plant Biol. 2011;11:13. doi: 10.1186/1471-2229-11-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bibikova TN, Blancaflor EB, Gilroy S. Microtubules regulate tip growth and orientation in root hairs of Arabidopsis thaliana. Plant J. 1999;17(6):657–665. doi: 10.1046/j.1365-313X.1999.00415.x. [DOI] [PubMed] [Google Scholar]
- 15.Sakai T, van der Honing H, Nishioka M, Uehara Y, Takahashi M, Fujisawa N, Saji K, Seki M, Shinozaki K, Jones MA, Smirnoff N, et al. Armadillo repeat-containing kinesins and a NIMA-related kinase are required for epidermal-cell morphogenesis in Arabidopsis. Plant J. 2008;53(1):157–171. doi: 10.1111/j.1365-313X.2007.03327.x. [DOI] [PubMed] [Google Scholar]
- 16.Sieberer B, Timmers A. Microtubules in plant root hairs and their role in cell polarity and tip growth in root hairs. In: Emons A, Ketelaar T, editors. Plant Cell Monograph. Vol. 12. Springer; Berlin: 2009. pp. 233–248. [Google Scholar]
- 17.Ambrose C, Wasteneys GO. Microtubule Initiation from the Nuclear Surface Controls Cortical Microtubule Growth Polarity and Orientation in Arabidopsis thaliana. Plant Cell Physiol. 2014;55(9):1636–1645. doi: 10.1093/pcp/pcu094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rounds CM, Bezanilla M. Growth Mechanisms in Tip-Growing Plant Cells. Annu Rev Plant Biol. 2013;64:243–265. doi: 10.1146/annurev-arplant-050312-120150. [DOI] [PubMed] [Google Scholar]
- 19.Chebli Y, Kroeger J, Geitmann A. Transport Logistics in Pollen Tubes. Mol Plant. 2013;6(4):1037–1052. doi: 10.1093/mp/sst073. [DOI] [PubMed] [Google Scholar]
- 20.Yu R, Huang RF, Wang XC, Yuan M. Microtubule dynamics are involved in stomatal movement of Vicia faba L. Protoplasma. 216(1–2):113–118. doi: 10.1007/Bf02680138. [DOI] [PubMed] [Google Scholar]
- 21.Brandizzi F, Wasteneys GO. Cytoskeleton-dependent endomembrane organization in plant cells: an emerging role for microtubules. Plant J. 2013;75(2):339–349. doi: 10.1111/tpj.12227. [DOI] [PubMed] [Google Scholar]
- 22.Foissner I, Menzel D, Wasteneys GO. Microtubule-dependent motility and orientation of the cortical endoplasmic reticulum in elongating characean internodal cells. Cell Motil Cytoskel. 2009;66(3):142–155. doi: 10.1002/Cm.20337. [DOI] [PubMed] [Google Scholar]
- 23.Hamada T, Ueda H, Kawase T, Hara-Nishimura I. Microtubules contribute to tubule elongation and anchoring of endoplasmic reticulum, resulting in high network complexity in Arabidopsis. Plant physiology. 2014;166(4):1869–1876. doi: 10.1104/pp.114.252320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holzinger A, Lütz-Meindl U. Kinesin-like proteins are involved in postmitotic nuclear migration of the unicellular green alga Micrasterias denticulata. Cell Biology International. 2002;26(8):689–697. doi: 10.1006/cbir.2002.0920. [DOI] [PubMed] [Google Scholar]
- 25.Holzinger A, Lütz-Meindl U. Evidence for kinesin- and dynein-like protein function in circular nuclear migration in the green alga Pleurenterium tumidum: Digital time lapse analysis of inhibitor effects. Journal of Phycology. 2003;39(1):106–114. doi: 10.1046/j.1529-8817.2003.02074.x. [DOI] [Google Scholar]
- 26.Miki T, Nishina M, Goshima G. RNAi screening identifies the armadillo repeat-containing kinesins responsible for microtubule-dependent nuclear positioning in Physcomitrella patens. Plant Cell Physiol. 2015 doi: 10.1093/pcp/pcv002. [DOI] [PubMed] [Google Scholar]
- 27.Tatout C, Evans DE, Vanrobays E, Probst AV, Graumann K. The plant LINC complex at the nuclear envelope. Chromosome Research. 2014;22(2):241–252. doi: 10.1007/s10577-014-9419-7. [DOI] [PubMed] [Google Scholar]
- 28.Kandasamy MK, Meagher RB. Actin-organelle interaction: Association with chloroplast in Arabidopsis leaf mesophyll cells. Cell Motil Cytoskel. 1999;44(2):110–118. doi: 10.1002/(Sici)1097-0169(199910)44:2<110::Aid-Cm3>3.0.Co;2-O. [DOI] [PubMed] [Google Scholar]
- 29.Holzinger A, Wasteneys GO, Lütz C. Investigating cytoskeletal function in chloroplast protrusion formation in the arctic-alpine plant Oxyria digyna . Plant Biology. 2007;9(3):400–410. doi: 10.1055/s-2006-924727. [DOI] [PubMed] [Google Scholar]
- 30.Holzinger A, Kwok EY, Hanson MR. Effects of arc3, arc5 and arc6 mutations on plastid morphology and stromule formation in green and nongreen tissues of Arabidopsis thaliana. Photochem Photobiol. 2008;84(6):1324–1335. doi: 10.1111/j.1751-1097.2008.00437.x. [DOI] [PubMed] [Google Scholar]
- 31.Zaffryar S, Zimerman B, Abu-Abied M, Belausov E, Lurya G, Vainstein A, Kamenetsky R, Sadot E. Development-specific association of amyloplasts with microtubules in scale cells of Narcissus tazetta. Protoplasma. 2007;230(3–4):153–163. doi: 10.1007/s00709-006-0238-3. [DOI] [PubMed] [Google Scholar]
- 32.Gilroy S. Fluorescence microscopy of living plant cells. Annual Review of Plant Physiology and Plant Molecular Biology. 1997;48:165–190. doi: 10.1146/annurev.arplant.48.1.165. [DOI] [PubMed] [Google Scholar]
- 33.Mathur J. The illuminated plant cell. Trends in Plant Science. 2007;12(11):506–513. doi: 10.1016/j.tplants.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 34.Dixit R, Cyr R, Gilroy S. Using intrinsically fluorescent proteins for plant cell imaging. Plant J. 2006;45(4):599–615. doi: 10.1111/j.1365-313X.2006.02658.x. [DOI] [PubMed] [Google Scholar]
- 35.Wasteneys GO, WillingaleTheune J, Menzel D. Freeze shattering: a simple and effective method for permeabilizing higher plant cell walls. Journal of Microscopy-Oxford. 1997;188:51–61. doi: 10.1046/j.1365-2818.1977.2390796.x. [DOI] [PubMed] [Google Scholar]
- 36.Kwok EY, Hanson MR. Microfilaments and microtubules control the morphology and movement of non-green plastids and stromules in Nicotiana tabacum. Plant J. 2003;35(1):16–26. doi: 10.1046/j.1365-313X.2003.01777.x. [DOI] [PubMed] [Google Scholar]
- 37.Shaw SL, Kamyar R, Ehrhardt DW. Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science. 2003;300(5626):1715–1718. doi: 10.1126/science.1083529. [DOI] [PubMed] [Google Scholar]
- 38.Chan J, Calder G, Fox S, Lloyd C. Cortical microtubule arrays undergo rotary movements in Arabidopsis hypocotyl epidermal cells. Nature Cell Biology. 2007;9(2):171–U157. doi: 10.1038/Ncb1533. [DOI] [PubMed] [Google Scholar]
- 39.Kawamura E, Wasteneys GO. MOR1, the Arabidopsis thaliana homologue of Xenopus MAP215, promotes rapid growth and shrinkage, and suppresses the pausing of microtubules in vivo. Journal of Cell Science. 2008;121(24):4114–4123. doi: 10.1242/Jcs.039065. [DOI] [PubMed] [Google Scholar]
- 40.Kawamura E, Himmelspach R, Rashbrooke MC, Whittington AT, Gale KR, Collings DA, Wasteneys GO. MICROTUBULE ORGANIZATION 1 regulates structure and function of microtubule arrays during mitosis and cytokinesis in the Arabidopsis root. Plant physiology. 2006;140(1):102–114. doi: 10.1104/pp.105.069989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pastuglia M, Azimzadeh J, Goussot M, Camilleri C, Belcram K, Evrard JL, Schmit AC, Guerche P, Bouchez D. gamma-tubulin is essential for microtubule organization and development in Arabidopsis. Plant Cell. 2006;18(6):1412–1425. doi: 10.1105/039644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang HY, Smertenko AP, Igarashi H, Dixon DP, Hussey PJ. Dynamic interaction of NtMAP65-1a with microtubules in vivo. Journal of Cell Science. 2005;118(14):3195–3201. doi: 10.1242/Jcs.02433. [DOI] [PubMed] [Google Scholar]
- 43.Shaw SL. Imaging the live plant cell. Plant J. 2006;45(4):573–598. doi: 10.1111/j.1365-313X.2006.02653.x. [DOI] [PubMed] [Google Scholar]
- 44.Hell SW. Toward fluorescence nanoscopy. Nat Biotechnol. 2003;21(11):1347–1355. doi: 10.1038/nbt895. [DOI] [PubMed] [Google Scholar]
- 45.Dyba M, Jakobs S, Hell SW. Immunofluorescence stimulated emission depletion microscopy. Nat Biotechnol. 2003;21(11):1303–1304. doi: 10.1038/nbt897. [DOI] [PubMed] [Google Scholar]
- 46.Hell SW. Far-field optical nanoscopy. Science. 2007;316(5828):1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 47.Folling J, Bossi M, Bock H, Medda R, Wurm CA, Hein B, Jakobs S, Eggeling C, Hell SW. Fluorescence nanoscopy by ground-state depletion and single-molecule return. Nat Methods. 2008;5(11):943–945. doi: 10.1038/nmeth.1257. [DOI] [PubMed] [Google Scholar]
- 48.Balint S, Verdeny Vilanova I, Sandoval Alvarez A, Lakadamyali M. Correlative live-cell and superresolution microscopy reveals cargo transport dynamics at microtubule intersections. Proc Natl Acad Sci U S A. 2013;110(9):3375–3380. doi: 10.1073/pnas.1219206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shtengel G, Galbraith JA, Galbraith CG, Lippincott-Schwartz J, Gillette JM, Manley S, Sougrat R, Waterman CM, Kanchanawong P, Davidson MW, Fetter RD, et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc Natl Acad Sci U S A. 2009;106(9):3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Komis G, Mistrik M, Samajova O, Doskocilova A, Ovecka M, Illes P, Bartek J, Samaj J. Dynamics and organization of cortical microtubules as revealed by superresolution structured illumination microscopy. Plant physiology. 2014;165(1):129–148. doi: 10.1104/pp.114.238477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ledbetter MC, Porter KR. A Microtubule in Plant Cell Fine Structure. Journal of Cell Biology. 1963;19(1):239–&. doi: 10.1083/Jcb.19.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meindl U, Lancelle S, Hepler PK. Vesicle production and fusion during lobe formation in Micrasterias Visualized by high-pressure freeze fixation. Protoplasma. 1992;170(3–4):104–114. doi: 10.1007/Bf01378786. [DOI] [Google Scholar]
- 53.Holzinger A. Aspects of cell development in Micrasterias muricata (Desmidiaceae) revealed by cryofixation and freeze substitution. Nova Hedwigia. 2000;70(3–4):275–287. [Google Scholar]
- 54.Segui-Simarro JM, Austin JR, White EA, Staehelin LA. Electron tomographic analysis of somatic cell plate formation in meristematic cells of Arabidopsis preserved by high-pressure freezing. Plant Cell. 2004;16(4):836–856. doi: 10.1105/Tpc.017749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eder M, Lutz-Meindl U. Pectin-like carbohydrates in the green alga Micrasterias characterized by cytochemical analysis and energy filtering TEM. Journal of Microscopy. 2008;231(2):201–214. doi: 10.1111/j.1365-2818.2008.02036.x. [DOI] [PubMed] [Google Scholar]
- 56.Holzinger A, Valenta R, Lutz-Meindl U. Profilin is localized in the nucleus-associated microtubule and actin system and is evenly distributed in the cytoplasm of the green alga Micrasterias denticulata. Protoplasma. 2000;212(3–4):197–205. doi: 10.1007/Bf01282920. [DOI] [Google Scholar]
- 57.Gaillard J, Neumann E, Van Damme D, Stoppin-Mellet V, Ebel C, Barbier E, Geelen D, Vantard M. Two Microtubule-associated Proteins of Arabidopsis MAP65s Promote Antiparallel Microtubule Bundling. Molecular Biology of the Cell. 2008;19(10):4534–4544. doi: 10.1091/mbc.E08-04-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakamura M, Naoi K, Shoji T, Hashimoto T. Low concentrations of propyzamide and oryzalin alter microtubule dynamics in Arabidopsis epidermal cells. Plant Cell Physiol. 2004;45(9):1330–1334. doi: 10.1093/Pcp/Pch300. [DOI] [PubMed] [Google Scholar]
- 59.Holzinger A, Karsten U, Lutz C, Wiencke C. Ultrastructure and photosynthesis in the supralittoral green macroalga Prasiola crispa from Spitsbergen (Norway) under UV exposure. Phycologia. 2006;45(2):168–177. doi: 10.2216/05-20.1. [DOI] [Google Scholar]
- 60.Ambrose C, Allard JF, Cytrynbaum EN, Wasteneys GO. A CLASP-modulated cell edge barrier mechanism drives cell-wide cortical microtubule organization in Arabidopsis. Nat Commun. 2011;2:430. doi: 10.1038/ncomms1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sugimoto K, Williamson RE, Wasteneys GO. New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant physiology. 2000;124(4):1493–1506. doi: 10.1104/pp.124.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frank J, SpringerLink ebooks - Biomedical and Life Sciences (2006) Electron tomography methods for three-dimensional visualization of structures in the cell. 2006 [Google Scholar]
- 63.Nitta K, Kaneko Y. Simple plunge freezing applied to plant tissues for capturing the ultrastructure close to the living state. Journal of Electron Microscopy. 2004;53(6):677–680. doi: 10.1093/jmicro/dfh092. [DOI] [PubMed] [Google Scholar]
- 64.Koning RI, Celler K, Willemse J, Bos E, van Wezel GP, Koster AJ. Correlative cryo-fluorescence light microscopy and cryo-electron tomography of Streptomyces. Methods Cell Biol. 2014;124:217–239. doi: 10.1016/B978-0-12-801075-4.00010-0. [DOI] [PubMed] [Google Scholar]
- 65.Ueda K, Matsuyama T, Hashimoto T. Visualization of microtubules in living cells of transgenic Arabidopsis thaliana. Protoplasma. 1999;206(1–3):201–206. doi: 10.1007/Bf01279267. [DOI] [Google Scholar]
- 66.Abe T, Hashimoto T. Altered microtubule dynamics by expression of modified alpha-tubulin protein causes right-handed helical growth in transgenic Arabidopsis plants. Plant J. 2005;43(2):191–204. doi: 10.1111/j.1365-313X.2005.02442.x. [DOI] [PubMed] [Google Scholar]
- 67.Ambrose JC, Wasteneys GO. CLASP modulates microtubule-cortex interaction during self-organization of acentrosomal microtubules. Molecular Biology of the Cell. 2008;19(11):4730–4737. doi: 10.1091/mbc.E08-06-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Granger CL, Cyr RJ. Spatiotemporal relationships between growth and microtubule orientation as revealed in living root cells of Arabidopsis thaliana transformed with green-fluorescent-protein gene construct GFP-MBD. Protoplasma. 2001;216(3–4):201–214. doi: 10.1007/BF02673872. [DOI] [PubMed] [Google Scholar]
- 69.Van Bruaene N, Joss G, Van Oostveldt P. Reorganization and in vivo dynamics of microtubules during Arabidopsis root hair development. Plant physiology. 2004;136(4):3905–3919. doi: 10.1104/pp.103.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dixit R, Chang E, Cyr R. Establishment of polarity during organization of the acentrosomal plant cortical microtubule array. Molecular Biology of the Cell. 2006;17(3):1298–1305. doi: 10.1091/mbc.E05-09-0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Littlejohn GR, Gouveia JD, Edner C, Smirnoff N, Love J. Perfluorodecalin enhances in vivo confocal microscopy resolution of Arabidopsis thaliana mesophyll. New Phytol. 2010;186(4):1018–1025. doi: 10.1111/j.1469-8137.2010.03244.x. [DOI] [PubMed] [Google Scholar]
- 72.Littlejohn GR, Love J. A simple method for imaging Arabidopsis leaves using perfluorodecalin as an infiltrative imaging medium. J Vis Exp. 2012;(59) doi: 10.3791/3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Littlejohn GR, Mansfield JC, Christmas JT, Witterick E, Fricker MD, Grant MR, Smirnoff N, Everson RM, Moger J, Love J. An update: improvements in imaging perfluorocarbon-mounted plant leaves with implications for studies of plant pathology, physiology, development and cell biology. Front Plant Sci. 2014;5:140. doi: 10.3389/fpls.2014.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spurr AR. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969;26(1):31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- 75.Sambade A, Pratap A, Buschmann H, Morris RJ, Lloyd C. The influence of light on microtubule dynamics and alignment in the Arabidopsis hypocotyl. Plant Cell. 2012;24(1):192–201. doi: 10.1105/tpc.111.093849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vineyard L, Elliott A, Dhingra S, Lucas JR, Shaw SL. Progressive transverse microtubule array organization in hormone-induced Arabidopsis hypocotyl cells. Plant Cell. 2013;25(2):662–676. doi: 10.1105/tpc.112.107326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dixit R, Cyr R. Cell damage and reactive oxygen species production induced by fluorescence microscopy: effect on mitosis and guidelines for non-invasive fluorescence microscopy. Plant J. 2003;36(2):280–290. doi: 10.1046/j.1365-313X.2003.01868.x. [DOI] [PubMed] [Google Scholar]
- 78.Chan J, Calder GM, Doonan JH, Lloyd CW. EB1 reveals mobile microtubule nucleation sites in Arabidopsis. Nature Cell Biology. 2003;5(11):967–971. doi: 10.1038/Ncb1057. [DOI] [PubMed] [Google Scholar]
- 79.Mathur J, Mathur N, Kernebeck B, Srinivas BP, Hulskamp M. A novel localization pattern for an EB1-like protein links microtubule dynamics to endomembrane organization. Curr Biol. 2003;13(22):1991–1997. doi: 10.1016/j.cub.2003.10.033. [DOI] [PubMed] [Google Scholar]
- 80.Marc J, Granger CL, Brincat J, Fisher DD, Kao TH, McCubbin AG, Cyr RJ. A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell. 1998;10(11):1927–1939. doi: 10.1105/tpc.10.11.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.DeBolt S, Gutierrez R, Ehrhardt DW, Melo CV, Ross L, Cutler SR, Somerville C, Bonetta D. Morlin, an inhibitor of cortical microtubule dynamics and cellulose synthase movement. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(14):5854–5859. doi: 10.1073/pnas.0700789104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stoppin-Mellet V, Gaillard J, Vantard M. Katanin's severing activity favors bundling of cortical microtubules in plants. Plant J. 2006;46(6):1009–1017. doi: 10.1111/j.1365-313X.2006.02761.x. [DOI] [PubMed] [Google Scholar]
- 83.Wightman R, Turner SR. Severing at sites of microtubule crossover contributes to microtubule alignment in cortical arrays. Plant J. 2007;52(4):742–751. doi: 10.1111/j.1365-313X.2007.03271.x. [DOI] [PubMed] [Google Scholar]
- 84.Holzinger A, Meindl U. Jasplakinolide, a novel actin targeting peptide, inhibits cell growth and induces actin filament polymerization in the green alga Micrasterias. Cell Motil Cytoskel. 1997;38(4):365–372. doi: 10.1002/(Sici)1097-0169(1997)38:4<365::Aid-Cm6>3.0.Co;2-2. [DOI] [PubMed] [Google Scholar]
- 85.Al-Amoudi A, Chang JJ, Leforestier A, McDowall A, Salamin LM, Norlen LP, Richter K, Blanc NS, Studer D, Dubochet J. Cryo-electron microscopy of vitreous sections. EMBO J. 2004;23(18):3583–3588. doi: 10.1038/sj.emboj.7600366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Buser C, Walther P. Freeze-substitution: the addition of water to polar solvents enhances the retention of structure and acts at temperatures around -60 degrees C. J Microsc. 2008;230(Pt 2):268–277. doi: 10.1111/j.1365-2818.2008.01984.x. [DOI] [PubMed] [Google Scholar]
- 87.Walther P, Ziegler A. Freeze substitution of high-pressure frozen samples: the visibility of biological membranes is improved when the substitution medium contains water. Journal of Microscopy. 2002;208:3–10. doi: 10.1046/j.1365-2818.2002.01064.x. [DOI] [PubMed] [Google Scholar]
- 88.Watanabe S, Punge A, Hollopeter G, Willig KI, Hobson RJ, Davis MW, Hell SW, Jorgensen EM. Protein localization in electron micrographs using fluorescence nanoscopy. Nat Methods. 2011;8(1):80–84. doi: 10.1038/nmeth.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gutierrez R, Lindeboom JJ, Paredez AR, Emons AM, Ehrhardt DW. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol. 2009;11(7):797–806. doi: 10.1038/ncb1886. [DOI] [PubMed] [Google Scholar]
- 90.Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof YD, Schumacher K, Gonneau M, Hofte H, Vernhettes S. Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell. 2009;21(4):1141–1154. doi: 10.1105/tpc.108.065334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Eng RC, Wasteneys GO. The microtubule plus-end tracking protein ARMADILLO-REPEAT KINESIN1 promotes microtubule catastrophe in Arabidopsis. Plant Cell. 2014;26(8):3372–3386. doi: 10.1105/tpc.114.126789. [DOI] [PMC free article] [PubMed] [Google Scholar]