Abstract
Transitional cell carcinoma (TCC), the most common cancer of the urinary bladder in dogs, is usually diagnosed at an advanced disease stage with limited response to chemotherapy. Commercial screening tests lack specificity and current diagnostic procedures are invasive. A proof of concept pilot project for analyzing the canine urinary proteome as a non-invasive diagnostic tool for TCC identification was conducted. Urine was collected from 12 dogs in three cohorts (healthy, urinary tract infection, TCC) and analyzed using liquid chromatography tandem mass spectrometry. The presence of four proteins (macrophage capping protein, peroxiredoxin 5, heterogeneous nuclear ribonucleoproteins A2/B, and apolipoprotein A1) was confirmed via immunoblot. Of the total 379 proteins identified, 96 were unique to the TCC group. A statistical model, designed to evaluate the accuracy of this multiplex biomarker approach for diagnosis of TCC, predicted the presence of disease with 90% accuracy.
Keywords: Canine, Biomarkers, Liquid chromatography, Tandem mass spectrometry, Transitional cell carcinoma
Introduction
Dogs are exposed to multiple stressors including pesticides, herbicides, chemotherapy, poor quality foods, and secondhand smoke; as in humans, these stressors can increase the risk of spontaneously developing transitional cell carcinoma (TCC).[1–3] Additionally, some breeds are genetically predisposed to TCC, with Shetland Sheepdogs, Collies, and West Highland White Terriers being among those with the highest prevalence.[2, 3] Canine TCC was found to resemble the same malignancy in humans when comparing histopathological characteristics, molecular features, and biological behavior. Clinical signs of TCC include pollakiuria, stanguria, hematuria, and tenesmus.[4] Dogs and humans are treated with similar chemotherapeutic protocols and, unfortunately, advanced disease stages in both species share limited response to medical therapy.[4–6] Because more than 90% of dogs exhibit progressive disease upon diagnosis, surgical treatment options are limited.[5–7] Chemotherapy and radiation treatments are frequently ineffective, with response rates of <35% and median survival time of <350 d.[8–10]
Tissue histology is the gold standard for TCC diagnosis, both in humans and dogs. However, obtaining samples involves general anesthesia and surgical biopsy, potentially causing tumor dissemination.[7, 11] Another option is the urine-based BARD bladder tumor antigen test [12]; however, it lacks specificity, often resulting in false positive results for patients with hematuria and proteinuria due to urinary tract infections (UTI). The use of single-biomarker diagnostics is problematic and there are presently no available assays for screening multiple biomarkers of symptomatic and non-symptomatic TCC.
Development of an improved screening test requires discovery of diagnostic and prognostic biomarkers specific to TCC. Recently, researchers have demonstrated the ability to detect soluble protein biomarkers secreted by cancer cells in vitro.[13–15] A proteomics-based approach enables the detection of proteins specific to early disease, which would assist patient staging and evaluation of disease progression.[13, 15, 16] In humans, high-throughput mass spectrometry (MS) of soluble protein found in urine is sensitive enough to differentiate healthy people from TCC patients; however, urine from UTI was not evaluated.[14, 17] We hypothesized that high-throughput MS can be used for identification of soluble proteins in TCC, UTI and healthy dogs, and that results from shotgun proteomic sequencing could be used to create a predictive statistical multiplex model for accurate diagnosis of canine TCC.
Our results demonstrated a reliable technique for the purification of soluble proteins from urine, protein identification using liquid chromatography tandem mass spectrometry (LC-MS/MS), and validation by antibody affinity. We predict that identifying canine TCC and UTI biomarkers will enable the development of tools for early detection and monitoring of disease progression, and may reveal novel therapeutic targets for both dogs and humans.
Materials and methods
Animals
The study included 12 dogs assigned to three equal-sized cohorts: healthy, UTI, and TCC. Recruitment was done with written consent from the dogs’ owners and in accordance with IACUC guidelines of Oregon State University (OSU).
Urine collection and fractionation
Urinary tracts of UTI and TCC dogs were prescreened with ultrasound scanning. Urine was collected in an aseptic manner (trans-abdominal cystocentesis for UTI and healthy dogs, urinary catheter for TCC dogs) and evaluated by urine analysis and bacterial culture and sensitivity test. Diagnosis of TCC was confirmed via cytology or histology.
After removing insoluble cellular debris by centrifugation, 3 mL urine was diluted with 12 mL deionized water and filtered through a 100 kDa molecular mass cutoff (MWCO) Macrosep column (Pall Corporation). The filtrate was centrifuged at 2000 g for 1 h at 4 °C, washed twice with 15 mL phosphate buffered saline and once with 15 mL distilled water, concentrated to 500 μL, dehydrated by vacuum centrifugation, and resolubilized in 300 μL of Laemelli buffer with 5% β-mercaptoethanol. In the same way, further size fractioning was done through 30 kDa and 3 kDa MWCO filters; filtrates were resuspended in 200 μL and 100 μL Laemelli buffer with 5% β-mercaptoethanol, respectively.
Protein separation and peptide preparation
Samples were separated by SDS-PAGE and proteins less than 50 kDa was excised and digested in-gel with trypsin and ProteaseMax surfactant (Promega), according to manufacturer’s protocols.
Mass spectrometry and protein annotation
Peptide sample analyses have been carried out using LTQ-FT mass spectrometer (Thermo Scientific) coupled to a nanoAcquity UPLC system (Waters) at the OSU Mass Spectrometry Facility. Dehydrated peptide samples were reconstituted in 20 μL of 3% acetonitrile (ACN) with 0.1% formic acid. Two microliters of the sample were injected on a trapping column (Cap Trap, Michrom) and separated using a C18 column (Agilent Zorbax 300SB-C18, 250 × 0.3 mm, 5 μm). Trapped peptides were washed with 3% ACN for 3 min at a flow rate of 5 μL/min and separated using a binary solvent gradient with 0.1% formic acid (A) and ACN (in 0.1% formic acid; B), with a flow rate of 4 μL/min. Solvent composition was increased from 3% B to 10% B in 3 min and to 30% B in 45 min. The ACN concentration was raised to 90% in 2 min followed by a 4 min hold and subsequent 6 min column re-equilibration at 3% ACN. LTQ-FT mass spectrometer was operated using data-dependent MS/MS acquisition mode in which MS precursor ion scan was performed in the ICR cell, from 350–2000 m/z with the resolving power set to 100,000 at m/z 400, and MS/MS scans were performed by the linear ion trap on the five most abundant doubly or triply charged precursor ions detected in the MS scan. All samples were run in triplicate.
Proteome Discoverer v1.3.0 was used to process raw data with Mascot v2.3 database searching algorithm against a canine protein database downloaded from a NCBI website (http://www.ncbi.nlm.nih.gov/) with automatic target decoy search (1% false discovery rate). The digestion enzyme was set to Trypsin/P with two missed cleavage sites and the precursor ion mass tolerance and fragment ion tolerance was set to 10 parts per million and 0.8 Da respectively. Carbamidomethyl (+57.02 Da) for cysteine, oxidation (+15.99 Da) of methionine and phosphorylation (+97.98 Da) of serine, threonine and tyrosine were used as dynamic modifications.
Immunoblot
Cellular extracts were probed with specific antibodies (Santa Cruz Biotechnology) against macrophage capping protein (sc-33084), peroxiredoxin 5 (PRX5) (sc-23977), heterogeneous nuclear ribonucleoproteins A2/B1 (sc-37405) and apolipoprotein (APO) A1 (sc-30089). Briefly, proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes with the iBlot platform (Life Technologies), blocked overnight with Odyssey blocking buffer (LI-COR), followed by binding of primary antibodies using dilutions recommended by the manufacturer. IRdye-conjugated secondary antibodies were used at a 1:10,000 dilution and membranes were scanned on an Odyssey platform (LI-COR).
PCR and sequencing
Bacterial infections were detected by amplification of the bacterial 16S ribosomal subunit using DNA from insoluble material isolated out of urine samples. Briefly, 250 μL urine was centrifuged at 13,000 g for 10 min. Cell pellets were resuspended in 100 μL deionized water followed by thermal cycling (96 °C 10s, 6 °C, 10 cycles) to lyse cells. Supernatants, containing DNA released from the cells, were clarified by centrifugation at 13,000 g for 10 min and used as template for amplification with primers Bac16SFor 5′GGCCCAGACTCCTACGGGAGGC3′ and Bac16SRev 5′GCGCTCGTTGCGGGACTTAACC3′ in a 50 μL reaction with HotStarTaq (Qiagen), following manufacturer’s protocol. Reactions were cycled (96 °C 30s, 56 °C 30s, 72 °C 60s) 35 times. A sample of cultured Escherichia coli was used as a positive control. Amplicons were separated on a 1% agarose gel stained with ethidium bromide, purified using PureLink gel extraction (Life Technologies), and analyzed by Sanger sequencing at the OSU Central Services Laboratory.
Statistical analysis
Using proteins identified from the LC-MS/MS analysis, we built a preliminary statistical model to test classifying TCC and non-TCC cases. Scaffold identified 379 proteins; the results of the 3 kD and 30 kD filtrations were treated as separate data points, giving 758 data points for each patient (Table 1). The model uses principal component analysis (PCA) to reduce data dimensionality and linear discriminant analysis (LDA) to classify cases into TCC or non-TCC.[18] The combined loadings from PCA and LDA were calculated to rank the importance of the proteins in discriminating between TCC and non-TCC (Table 1).
Table 1.
Proteins that most discriminate TCC.
| GI accession | Protein | Filtration (kDa) | Model loading | Protein identification probability (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | UTI | TCC | |||||||||||||
| 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 | ||||
| 73971560 | Prostaglandin reductase 1 isoform 1 | 30 | 0.1142 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 |
| 356461044 | Peroxiredoxin-1 | 3 | 0.1142 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 |
| 73977992 | Fibrinogen gamma chain isoform 1 | 30 | 0.1142 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 |
| 17298186 | Heat shock protein 70 | 3 | 0.1141 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 97 | 100 | 100 | 100 |
| 157151714 | Collagen alpha-3(VI) chain precursor | 3 | 0.1141 | 100 | 100 | 100 | 100 | 100 | 100 | 99 | 100 | 0 | 0 | 0 | 0 |
| 345800929 | NAD(P)H dehydrogenase [quinone] 1 | 30 | 0.114 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 95 | 100 | 100 | 100 |
| 73974180 | 14-3-3 protein zeta/delta isoform 2 | 30 | 0.1123 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 95 | 95 | 100 | 100 |
| 114326321 | Phosphatidylethanolamine-binding protein 1 | 30 | 0.1095 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 100 | 100 |
| 73980918 | Macrophage-capping protein | 30 | 0.1095 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 100 | 100 |
| 223556019 | Carbonic anhydrase 2 | 3 | 0.1095 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 100 | 100 |
The full model was developed using all of the available data. The model was tested by using bootstrap resampling [18]. For each iteration, three TCC samples and six non-TCC (control or UTI) samples were used to train the model; one TCC and two non-TCC samples were used to test the model.
Results
The healthy control cohort consisted of four breeds (Argentinian, Newfoundland, Springer Spaniel and Bernese Mountain) ranging from ages 3 to 10 years. The UTI group consisted of two breeds (Anatolian Shepherd, Scottish Terrier) and two mixed-breed dogs ranging from ages 9 to 12 years. The TCC cohort consisted of three breeds (Scottish Terrier, American Eskimo, Welsh Corgi) and one mixed breed dog ranging from ages 10 to 13 years. One TCC patient had an apical mass and three TCC patients had bladder neck masses extending to the urethra, two of which showed ureter involvement. At the time of sample collection, two TCC dogs were undergoing chemotherapy treatment (Mitoxantrone 5 mg/M2 intravenous once every three weeks for five treatments and piroxicam 0.3 mg/kg oral daily). One dog was treated twice with piroxicam prior to urine collection and one dog had not yet received any treatment.
Urine culture and 16S PCR
No bacteria grew in urine cultures from healthy controls or the TCC group. From the UTI cohort, two samples were positive for E. coli, one sample for E. coli and Staphylococcus pseudintermedius, and one had evidence of bacteria on microscopic evaluation but did not yield a positive culture. The negative sample had a 16S PCR product identified as Methylobacterium spp. Although culture negative and asymptomatic, one sample obtained from the TCC group and one from the healthy control group were also positive for bacterial 16S DNA, suggesting that these patients may have a sub-clinical UTI or that samples had been contaminated during processing (Figure 1).
Figure 1.
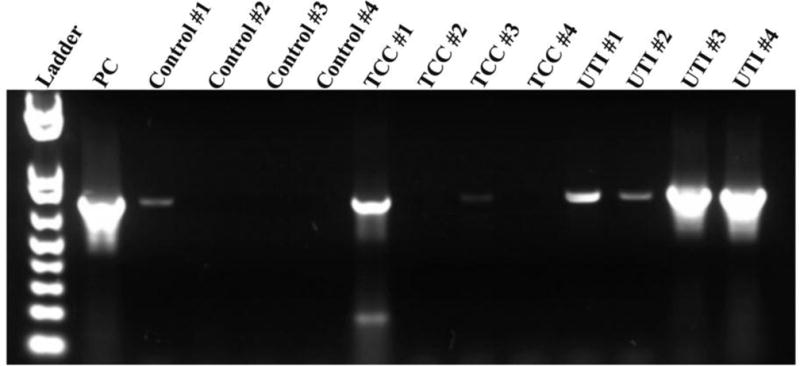
PCR of urine to detect bacterial presence. PCR was performed to confirm the absence of bacteria in urine from the TCC and control groups and to confirm the presence of bacteria in urine from the UTI group. The identity of the bacteria in the UTI group was found by sequencing. The results have demonstrated the presence of bacteria in two samples that were previously cultured negative (control 1 and TCC 1).
Identification of soluble proteins
Urine proteins were fractionated by ultra-filtration and visualized by SDS-PAGE. Following in-gel enzymatic digestion, resulting peptides were analyzed by LC-MS/MS. The majority of proteins identified were consistent between all samples within each group. Of the 379 proteins identified, 96 were detected in TCC, 39 in UTI, and 8 exclusively in the control group. A total of 131 proteins were shared by all groups (Figure 2, Table 1). Four proteins were selected based on LC-MS/MS and the availability of suitable antibodies: PRX5, macrophage capping protein, APO-A1, and heterogeneous nuclear ribonucleoproteins A2/B1 (Figure 3). Macrophage capping protein was confirmed in the 30 kDa filtrate of three TCC samples. PRX5 was present in all TCC samples. Immunoblot and LC-MS/MS results were largely correlative in the 3 kDa filtrate TCC samples; however, LC-MS/MS identified proteins in the 3 kDa fraction in one TCC sample that were not detected by immunoblot (PRX5 and APO-A1) (Figure 3). Heterogeneous nuclear ribonucleoproteins A2/B1 was detected in the 30 kDa filtrate of three TCC samples by immunoblot, in agreement with LC-MS/MS. The 3 kDa filtrate detected faint CapG and hnRNP in three TCC samples by immunoblot, but was only detected in two TCC samples by LC-MS/MS (Figure 3). APO-A1 in the 30 kDa filtrate had similar detection patterns in both fractions, excluding two TCC samples in which APO-A1 was identified only by immunoblot (Figure 3). One control 3 kDa filtrate revealed CapG by LC-MS/MS only (Figure 3).
Figure 2.
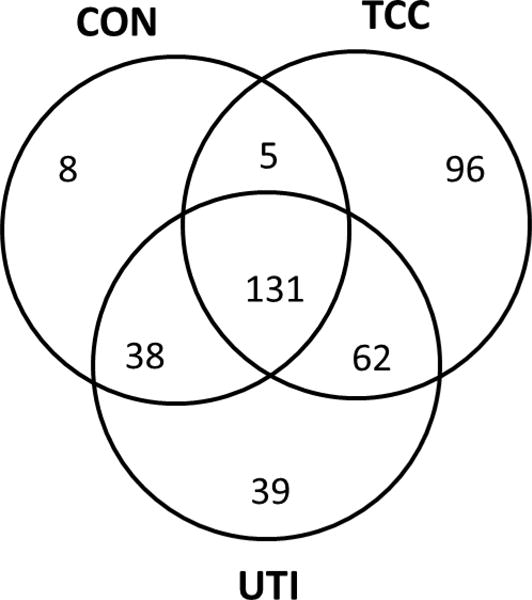
Venn diagram showing the distribution of identified proteins between the groups. A total of 379 proteins were identified. The TCC group had the highest amount of proteins that were exclusive to this group (96), followed by the UTI group (39), and the control group (CON) (8).
Figure 3.
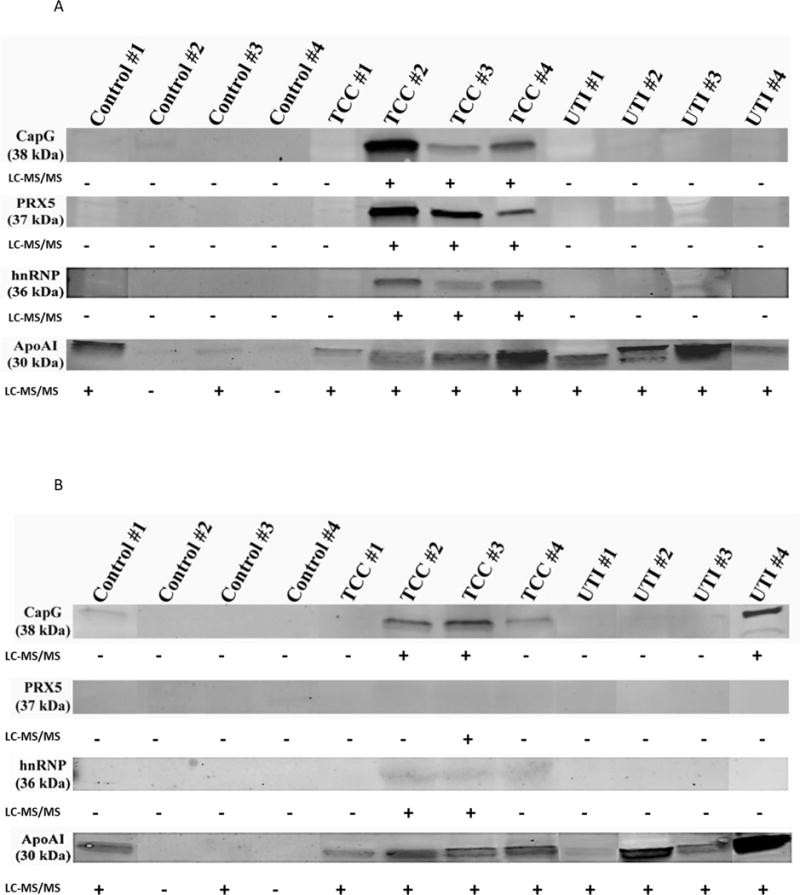
A and B: Immunoblots of selected proteins at 30 KDa (1A) and 3 KDa (1B) filtrates in all examined samples. The +/− signs are annotated to reflect the LC-MS/MS results.
Statistical analysis
PCA followed by LDA produced a robust model to classify the data into TCC and non-TCC cases (Figure 4). Re-substitution of the data into the model correctly classified all but a single case (TCC 1). Testing of the model using bootstrap resampling showed high accuracy (mean 90.6%, 95% CI [89.5%, 91.6%]).
Figure 4.
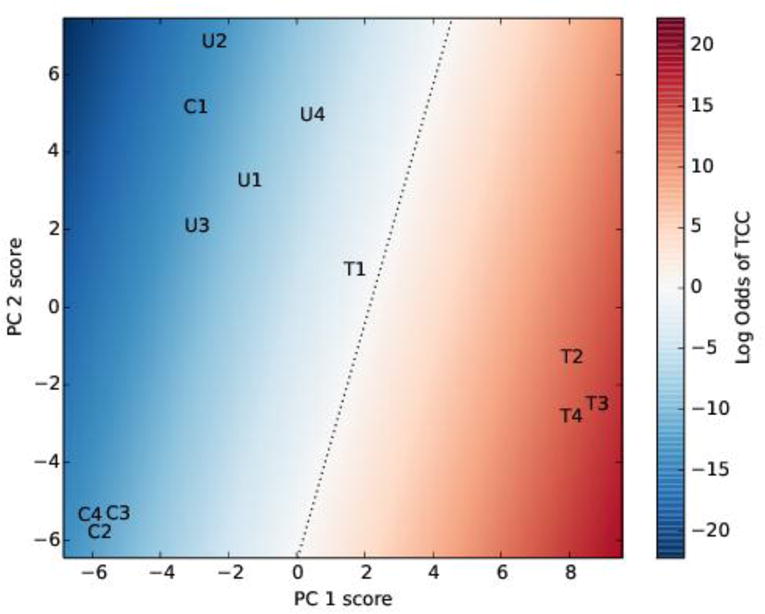
Principal component analysis and linear discriminant analysis of the initial data. The data were transformed using principal component analysis, keeping two components, followed by linear discriminant analysis. The letters C, U and T represent the control, UTI and TCC samples, respectively. The background color indicates the model odds of TCC; the dotted black line is where the odds of TCC is 1 (i.e. the log odds of TCC is 0). PC is principal component.
The model accuracy can be improved further using receiver operating characteristic (ROC) analysis.[19] Using the line where model predicts even odds of TCC and non-TCC (Figure 4) as the classification threshold results in incorrect classification of one case. Refining the model by decreasing the threshold odds from 1 to between 0.2265 and 0.0016 (moving the dotted line towards the upper left in Figure 4) leads to the correct classification of all cases. Because perfect classification of the data is possible by adjusting the threshold, the area under the ROC curve (AUC) is 1. Bootstrap testing showed very high AUC values were robust: the AUC was 1 in 995 of the 1000 bootstrap samples, 0.5 in 4 samples, and 0 in 1 sample. Therefore, the model is likely to be able to correctly classify cases with high accuracy.
The identified proteins were ranked according to their contribution to the statistical classification model (Table 1). The most influential were the seven proteins identified with high likelihood in all TCC samples and in none of the non-TCC samples, or vice versa. An additional 30 proteins were identified with high likelihood in three of the TCC samples and not identified in TCC 1 or in any non-TCC samples. These proteins are likely responsible for the large difference between model results for TCC 1 and for the other TCC samples.
Discussion
Early diagnosis of TCC is essential for effective treatment and delay of disease progression. Characterizing proteins in urine from humans with TCC has identified novel disease biomarkers and highlighted the potential of a multiplex analysis to enhance the sensitivity and specificity of screening assays.[20] While cancer research has traditionally focused on cellular biomarkers, recent efforts have broadened to include soluble biomarkers in blood, sputum and urine.[16, 20, 21] Even though there are several available TCC diagnostic tests in human medicine, in veterinary medicine only the BARD test was assessed, exhibiting high sensitivity with lower specificity.[12] While we did not perform the BARD test on our samples, the complement factor H (CFH) related protein was identified with 100% probability. In our study, CFH related protein, the target of the BARD test, was detected in two TCC samples and one control sample, supporting the previous reports suggesting lower credibility of CFH related protein as an independent marker for TCC.
Potential biomarkers abundant in the urine of TCC patients include a collection of secreted waste material, metabolic byproducts, cancer-cell secretomes, cell lysates, and other material from the interaction between the tumor and its environment. High-throughput LC-MS/MS is effective for identifying a broad array of biomarkers and potential therapeutic targets, many of which have not yet been investigated or identified.[16, 17] Our preliminary study establishes a canine urine protein signature that can define canine TCC, resulting in the identification of proteins that may discriminate between healthy patients and those with TCC or UTIs.
A total of 379 proteins were identified across all cohorts, with 96 being unique to TCC (Figure 2). Previous studies have found a total of 295–387 unique proteins in urine collected from people with noninvasive bladder cancer.[22, 23] However 40% of those were identical to proteins from urine of healthy people identified by another study.[24] The smaller number of unique proteins identified in this study can be explained by our additional control group (dogs with UTI) and comparison of our data to a limited annotated canine library. Because TCC and UTI are both accompanied by severe inflammation, their urine protein profiles share similarities. To our knowledge, studies analyzing human urine did not include UTI controls. This point can help explain the limited number of TCC-specific proteins identified in our study.
Four proteins were selected as potential TCC biomarkers based on their significance in cancer development, identification by LC-MS/MS specifically in TCC samples, and the availability of commercial antibodies that are compatible with canine proteins. Macrophage capping protein regulates cell motility and its up-regulation has been correlated with tumor invasion. LC-MS/MS detected this protein in three of the four TCC samples, while none was found in the healthy control or UTI samples. Future studies should address macrophage capping protein as a potential diagnostic biomarker and investigate its role in canine TCC. PRXs reduce oxides such as hydrogen peroxide; PRX1, PRX5 and PRX6 are associated with several malignancies including cancers of the breast, bladder and colon. High PRX1 and PRX6 expression levels correlate with development and recurrence of TCC; high PRX5 expression level is linked to mammary cancer.[25] We identified PRX1, PRX5 and PRX6 with LC-MS/MS analysis and further confirmed the presence of PRX5 via immunoblot. Heterogeneous nuclear ribonucleoprotein A2/B1 is an RNA binding protein and has an essential role in post-transcriptional regulation of mRNA in that changes in its expression have been linked to lung and colon cancers.[26] Both immunoblot and LC-MS/MS detected ribonucleoprotein A2/B1; although it has not yet been linked to TCC, it may serve as a novel biomarker in dogs.
All UTI samples exhibited microscopic presence of bacteria. Three samples cultured positive for E. coli, and the fourth was positive for Methylobacterium spp by PCR. This mismatch can be explained by the fact that UTI caused by Methylobacterium spp. has been shown to be under-diagnosed with standard culture techniques.[27] PCR also identified bacteria in one healthy sample and one TCC sample. Interestingly, both of these samples were shifted towards the UTI group when applied to the multiplex model.
One of the challenges in processing urine samples for proteomics was removing abundant proteins and other components, such as albunin and urea, prior to LC-MS/MS. These routinely clog the 3 kDa membrane, potentially masking less abundant proteins, increasing sample-sample variability, and resulting in suboptimal results. Additionally, the composition of urine from different patients is highly variable. By serially fractioning the urine with 100 kDa, 30 kDa and 3 kDa filters, running the 30 kDa and 3 kDa fractions on SDS-PAGE, and in-gel digestion of bands less than 50 kDa, we were able to minimize the presence of albumin, urea and other salts and increase replicate consistency. We observed some inconsistencies between LC-MS/MS and immunoblot, primarily in the 3 kDa filtrate. While higher molecular mass proteins leaked through the 3 kDa filter and detected by immunoblot, it is possible that the concentrations of these proteins were too low to be detected by LC-MS/MS. Furthermore, differences in identified proteins could result from modification or degradation of antibody binding epitopes. Looking forward, an enzyme-linked immunosorbent assay approach is likely to be more sensitive and quantitative than immunoblot.
Statistical analysis demonstrates the importance of a multiplex approach for detection of proteins uniquely present in TCC and defining a clear separation between the TCC cohort and other two groups. Three healthy controls had similar protein compositions; however the fourth control was more similar to the UTI cohort (C1 in Figure 4). Likewise, one TCC sample (TCC 1) was more similar to the UTI cohort. These outliers did not exhibit positive bacterial cultures, however PCR revealed the presence of bacteria. Sub-acute infections in those patients may explain the differences. Taken together, statistical analysis of the LC-MS/MS results enabled the development of a robust and accurate model to categorize samples as TCC or non-TCC, and identified proteins that are most useful for classifying samples. Our small sample size demands caution, but the robustness and accuracy of our model suggests that statistical classification is possible, particularly with more data to train models. Subsequent studies that leverage larger sample sizes should facilitate a more precise definition of highly-predictive biomarkers.
Conclusion
We have shown that the combination of ultra-filtration and LC-MS/MS is a reliable and useful approach for characterizing the proteome of canine urine. Our pilot study identified potential biomarkers that were unique to each of our three cohorts: healthy, TCC and UTI. We validated selected proteins by immunoblot and created a statistical model using a biomarker multiplex that can distinguish cohorts.
While multiplex protein analysis via LC-MS/MS predicted disease with 90% confidence in this pilot study, further analysis of a larger cohort will highlight the significance of the identified proteins and potentially yield a focused perspective on important biomarkers relevant to the diagnosis of TCC in dogs. Additionally, developing a more direct assay for the detection of specific proteins, such as an enzyme-linked immunosorbent assay approach, will make the proposed multiplex screen for canine TCC more feasible and bring it closer to clinical utility. Finally, our novel approach to biomarker discovery can be applied to studies in humans to determine whether the same biomarkers can predict TCC in humans and dogs, and if not, which biomarkers are unique to urine from human TCC patients.
Figure 5.
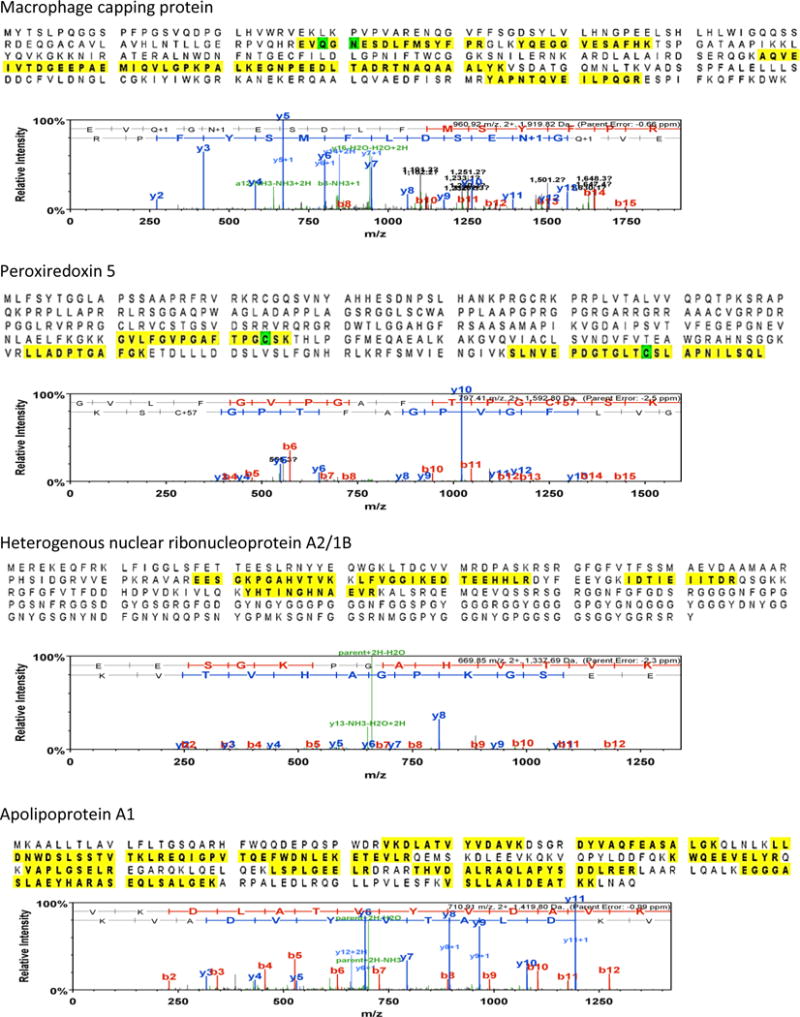
Protein sequence of the four selected proteins used for the validation of our results. The highlighted areas demonstrate the peptides that were detected and identified by LC-MS/MS. Although the peptide threshold was set at 2, it is evident that LC-MS/MS identified many more peptides (highlighted) in these proteins, increasing the confidence of the protein identity.
Acknowledgments
The OSU mass spectrometry facility and core lab is supported in part by a grant from the National Institute of Environmental Health Sciences (P30 ES0000210).
Footnotes
Abbreviations: TCC, UTI, LC-MS/MS, MWCO, PRX, PCA, LDA, ROC, AUC
Conflict of interest statement
None of the authors has any financial or personal relationships that could inappropriately influence or bias the content of this manuscript.
References
- 1.Macy DW, Withrow SJ, Hoopes J. Transitional cell-carcinoma of the bladder associated with cyclophosphamide administration. Journal of the American Animal Hospital Association. 1983;19:965–969. [Google Scholar]
- 2.Glickman LT, Raghavan M, Knapp DW, Bonney PL, Dawson MH. Herbicide exposure and the risk of transitional cell carcinoma of the urinary bladder in Scottish Terriers. Journal of the American Veterinary Medical Association. 2004;224:1290–1297. doi: 10.2460/javma.2004.224.1290. [DOI] [PubMed] [Google Scholar]
- 3.SF Glickman LT, McKee LJ, Reif JS, Goldschmidt MH. Epidemiologic study of insecticide exposures, obesity, and risk of bladder cancer in household dogs. Journal of Toxicology and Environmental Health. 1989;28:407–414. doi: 10.1080/15287398909531360. [DOI] [PubMed] [Google Scholar]
- 4.Valli VE, Norris A, Jacobs RM, Laing E, Withrow S, Macy D, Tomlinson J, Mccaw D, Ogilvie GK, Pidgeon G, Henderson RA. Pathology of canine bladder and urethral cancer and correlation with tumor progression and survival. Journal of Comparative Pathology. 1995;113:113–130. doi: 10.1016/s0021-9975(05)80027-1. [DOI] [PubMed] [Google Scholar]
- 5.Knapp DW, Glickman NW, DeNicola DB, Bonney PL, Lin TL, Glickman LT. Naturally-occurring canine transitional cell carcinoma of the urinary bladder A relevant model of human invasive bladder cancer. Urologic Oncology: Seminars and Original Investigations. 2000;5:47–59. doi: 10.1016/s1078-1439(99)00006-x. [DOI] [PubMed] [Google Scholar]
- 6.Mutsaers AJ, Widmer WR, Knapp DW. Canine transitional cell carcinoma. Journal of Veterinary Internal Medicine. 2003;17:136–144. doi: 10.1892/0891-6640(2003)017<0136:ctcc>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 7.Stone EA, George TF, Gilson SD, Page RL. Partial cystectomy for urinary bladder neoplasia: surgical technique and outcome in 11 dogs. Journal of Small Animal Practice. 1996;37:480–485. doi: 10.1111/j.1748-5827.1996.tb01745.x. [DOI] [PubMed] [Google Scholar]
- 8.Henry CJ, McCaw DL, Turnquist SE, Tyler JW, Bravo L, Sheafor S, Straw RC, Dernell WS, Madewell BR, Jorgensen L, Scott MA, Higginbotham ML, Chun R. Clinical evaluation of mitoxantrone and piroxicam in a canine model of human invasive urinary bladder carcinoma. Clinical Cancer Research. 2003;9:906–911. [PubMed] [Google Scholar]
- 9.Boria PA, Glickman NW, Schmidt BR, Widmer WR, Mutsaers AJ, Adams LG, Snyder PW, DiBernardi L, De Gortari AE, Bonney PL, Knapp DW. Carboplatin and piroxicam therapy in 31 dogs with transitional cell carcinoma of the urinary bladder. Veterinary and Comparative Oncology. 2005;3:73–80. doi: 10.1111/j.1476-5810.2005.00070.x. [DOI] [PubMed] [Google Scholar]
- 10.Poirier VJ, Forrest LJ, Adams WM, Vail DM. Piroxicam, mitoxantrone, and coarse fraction radiotherapy for the treatment of transitional cell carcinoma of the bladder in 10 dogs: A pilot study. Journal of the American Animal Hospital Association. 2004;40:131–136. doi: 10.5326/0400131. [DOI] [PubMed] [Google Scholar]
- 11.Anderson WI, Dunham BM, King JM, Scott DW. Presumptive subcutaneous surgical transplantation of a urinary-bladder transitional cell-carcinoma in a dog. Cornell Veterinarian. 1989;79:263–266. [PubMed] [Google Scholar]
- 12.Henry CJ, Tyler JW, McEntee MC, Stokol T, Rogers KS, Chun R, Garrett LD, McCaw DL, Higginbotham ML, Flessland KA, Stokes PK. Evaluation of a bladder tumor antigen test as a screening test for transitional cell carcinoma of the lower urinary tract in dogs. American Journal of Veterinary Research. 2003;64:1017–1020. doi: 10.2460/ajvr.2003.64.1017. [DOI] [PubMed] [Google Scholar]
- 13.Vlahou A, Schellhamrner PF, Mendrinos S, Patel K, Kondylis FI, Gong L, Nasim S, Wright GL. Development of a novel proteomic approach for the detection of transitional cell carcinoma of the bladder in urine. American Journal of Pathology. 2001;158:1491–1502. doi: 10.1016/S0002-9440(10)64100-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y-F, Wu D-L, Guan M, Liu W-W, Wu Z, Chen Y-M, Zhang W-Z, Lu Y. Tree analysis of mass spectral urine profiles discriminates transitional cell carcinoma of the bladder from noncancer patient. Clinical Biochemistry. 2004;37:772–779. doi: 10.1016/j.clinbiochem.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 15.Lin CY, Tsui KH, Yu CC, Yeh CW, Chang PL, Yung BYM. Searching cell-secreted proteomes for potential urinary bladder tumor markers. Proteomics. 2006;6:4381–4389. doi: 10.1002/pmic.200600066. [DOI] [PubMed] [Google Scholar]
- 16.Schwamborn K, Krieg RC, Grosse J, Reulen N, Weiskirchen R, Knuechel R, Jakse G, Henkel C. Serum proteomic profiling in patients with bladder cancer. European Urology. 2009;56:989–997. doi: 10.1016/j.eururo.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 17.Vlahou A, Schellhammer PF, Mendrinos S, Patel K, Kondylis FI, Gong L, Nasim S, Wright GL., Jr Development of a novel proteomic approach for the detection of transitional cell carcinoma of the bladder in urine. The American journal of pathology. 2001;158:1491–1502. doi: 10.1016/S0002-9440(10)64100-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richard O Duda PEH, Stork David G. Pattern Classification. 2nd. Wiley; New York: 2001. [Google Scholar]
- 19.Fielding AH. Cluster and classification techniques for the biosciences. Cambridge; New York: 2007. [Google Scholar]
- 20.Issaq HJ, Nativ O, Waybright T, Luke B, Veenstra TD, Issaq EJ, Kravstov A, Mullerad M. Detection of bladder cancer in human urine by metabolomic profiling using high performance liquid chromatography/mass spectrometry. The Journal of Urology. 2008;179:2422–2426. doi: 10.1016/j.juro.2008.01.084. [DOI] [PubMed] [Google Scholar]
- 21.Wu J-Y, Yi C, Chung H-R, Wang D-J, Chang W-C, Lee S-Y, Lin C-T, Yang Y-C, Yang W-CV. Potential biomarkers in saliva for oral squamous cell carcinoma. Oral Oncology. 2010;46:226–231. doi: 10.1016/j.oraloncology.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 22.Linden M, Lind SB, Mayrhofer C, Segersten U, Wester K, Lyutvinskiy Y, Zubarev R, Malmstrom PU, Pettersson U. Proteomic analysis of urinary biomarker candidates for nonmuscle invasive bladder cancer. Proteomics. 2012;12:135–144. doi: 10.1002/pmic.201000810. [DOI] [PubMed] [Google Scholar]
- 23.Niu HT, Dong Z, Jiang G, Xu T, Liu YQ, Cao YW, Zhao J, Wang XS. Proteomics research on muscle-invasive bladder transitional cell carcinoma. Cancer Cell Int. 2011;11:17. doi: 10.1186/1475-2867-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7:R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura K, Ojima H, Kubota D, Sakumoto M, Nakamura Y, Tomonaga T, Kosuge T, Kondo T. Proteomic identification of the macrophage-capping protein as a protein contributing to the malignant features of hepatocellular carcinoma. Journal of Proteomics. 2012:362–373. doi: 10.1016/j.jprot.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 27.Lee C-H, Tang Y-F, Liu J-W. Underdiagnosis of urinary tract infection caused by Methylobacterium species with current standard processing of urine culture and its clinical implications. Journal of Medical Microbiology. 2004;53:755–759. doi: 10.1099/jmm.0.05435-0. [DOI] [PubMed] [Google Scholar]


