Abstract
In this study, the secondary structure of the major ampullate silk from Peucetia viridans (Green Lynx) spiders is characterized by X-ray diffraction and solid-state NMR spectroscopy. From X-ray diffraction measurement, β-sheet nanocrystallites were observed and found to be highly oriented along the fiber axis, with an orientational order, fc ≈ 0.98. The size of the nanocrystallites was determined to be on average 2.5 nm × 3.3 nm × 3.8 nm. Besides a prominent nanocrystalline region, a partially oriented amorphous region was also observed with an fa ≈ 0.89. Two-dimensional 13C–13C through-space and through-bond solid-state NMR experiments were employed to elucidate structure details of P. viridans silk proteins. It reveals that β-sheet nanocrystallites constitutes 40.0 ± 1.2% of the protein and are dominated by alanine-rich repetitive motifs. Furthermore, based upon the NMR data, 18 ± 1% of alanine, 60 ± 2% glycine and 54 ± 2% serine are incorporated into helical conformations.
Keywords: Spider silk, Green Lynx, Peucetia viridans, Solid-state NMR, Wide-angle X-ray diffraction
1. Introduction
Spiders have evolved for millions of years and their ability to produce silk fibers is conserved for most of the species in the spider family [1]. Spiders can produce up to six different types of silk fibers and one type of glue and use them for a large variety of functions [2–4]. Compare to other natural or man-made fiber materials, the uniqueness of spider silk is the combination of the outstanding strength and extraordinary extensibility [2–6]. Along with its biocompatibility, spider silk is, without doubt, an attractive biomaterial and has been extensively studied by scientists from multiple disciplines [7–31].
Major ampullate spider silk, also commonly called dragline silk, has been frequently chosen as the model to investigate the molecular architecture because of its remarkable mechanical properties and its accessibility. For most Orbiculariae spiders, dragline silk typically contains two large proteins, named major ampullate spidroin 1 (MaSp1) and major ampullate spidroin 2 (MaSp2) [7,8]. The primary sequences for the two proteins have been partially or fully elucidated in previous studies [7,18,32–36]. The core regions, which accounts for more than 90% of the proteins, are highly repetitive with conserved amino acid motifs. X-ray diffraction studies revealed that dragline silk consists of nano-crystalline and amorphous regions [11–15,30,37,38]. The nano-crystallites are commonly observed by wide-angle X-ray diffraction to be aligned parallel to the fiber axis [11,12,15,30,34,37,38]. Furthermore, solid-state NMR results have shown that the nanocrystallites are β-sheet nano-structures primarily formed by poly(Ala) motifs [9,10,17,19–22,24,31,39,40]. And the amorphous regions, mainly comprised of disordered 31-helical and type II β-turn structures, are formed by Gly-Gly-X and Gly-Pro-Gly-X-X motifs, respectively [9,10,17,19–24,31,39,40]. Understanding protein structure provides critical insight into correlations between the molecular design of spider silk and the physical properties [23,41,42].
Green Lynx spiders belong to the RTA clade. Similar to orb-weaver spiders, the RTA clade is also one sub-family in the super family of Entelegynae [34,43–45]. The Green Lynx spiders can produce two types of silks similar to the major and minor ampullate silks of orb-weaver spiders. The mechanical properties of the major ampullate silk from Green Lynx spiders are comparable to that of the orb-weaver silks [46,47]. When wetted with water, the silk fibers contract up to 10% in length [46,48], which is also similar to the supercontraction phenomenon in the orb-weaver silks [5,49,50]. However, in contrast to the orb-weaver spiders, Green Lynx spiders do not spin webs but only lay silks on the ground as they walk [48]. In addition, cDNA analysis suggests that only one MaSp1-like protein exists in the Green Lynx major ampullate silk fibers [46,48]. Therefore, although the Green Lynx spider shares similarities with the orb-weaver spiders in their major ampullate silks, the molecular difference makes it interesting to compare the silk from Green Lynx spiders with the well studied orb-weaver dragline silks.
In this study, we combined wide-angle X-ray diffraction (WAXD) [30,31] and solid-state NMR techniques [24,31] to investigate protein secondary structure of the major ampullate (dragline) silk from the Green Lynx spiders. WAXD provides detailed information about the sizes of the nanocrystallites as well as the orientational orders of the crystalline and amorphous regions. Two-dimensional (2D) 13C–13C correlation experiments with dipolar-assisted rotational resonance (DARR) solid-state NMR detects inter-residue and intra-residue through space 13C–13C correlations. In conjunction with the DARR experiments, 2D 13C–13C double quantum/single quantum (DQ/SQ) refocused Incredible Natural Abundance DoublE QUAntum Transfer Experiment (INADEQUATE) provide additional through bond 13C–13C correlations. Conformations of amino acid residues in the major ampullate silk can be determined by the corresponding 13C chemical shifts extracted from the 2D experiments. Finally, the percentages for the amino acids in specific secondary structures are quantified and correlated with the amino acid motifs in the primary sequence of the silk protein.
2. Materials and methods
2.1. Sample preparation
Mature female Peucetia viridans (Green Lynx) spiders were fed with tap water and crickets once per week. Spiders were forcibly silked at a speed of 2 cm/s for 1 h every other day. The major ampullate silk (dragline silk) was separated from the minor ampullate silk (Fig. S1) with the aid of an optical microscope. To isotope enrich the dragline silk, the spiders were fed with ∼20 μl of U-[13C, 15N]-l-alanine or U-[13C, 15N]-l-proline solution (15% w/v) during silk collection [51]. A total of 10mg was collected for each isotope-enriched sample. In addition, a natural abundance (non-isotope-labeled) silk sample (8 mg) was collected. To prepare water-wetted silk samples, the dragline silk was soaked in 99.9% D2O for 12 h to allow water saturation in the silk fibers. Isotopes were purchased from Cambridge Isotopes Laboratories, Inc. and used as received.
2.2. Scanning electron microscopy
The dry dragline silk sample was taped on the sample holder and coated with Au/Pd in a Denton vacuum sputter coater desk II for 3 min under a pressure of 200 mTorr. The deposition rate was 5 nm/min with a current of 20 mA. The SEM image was taken using a FEI XL30 Environmental SEM-FEG. The SEM was operating at a vacuum pressure less than 9 × 10−5 mbar and a beam current of 7.00 kV.
2.3. Wide angle X-ray diffraction
X-ray experiments were carried out at Argonne National Laboratory Advanced Photon Source BioCars 14-ID-B.14-ID-B beam energy is 9KeV and the corresponding X-ray wavelength is 1.38 Å. The detector is Mar165 3072×3072 CCD from marXperts GmbH with a pixel pitch of 79 μm. Distance between detector and sample was fixed at 180 mm throughout the experiments. Silk sample was mounted to xyz-translation goniometer head with helium gas chamber placed between CCD detector and sample. The exposure time varies from 2 to 6 s depending on the size of silk bundle. For each sample we take 10 exposures. Background was taken by translating silk bundle away from X-ray beam. Multiple X-ray exposures were averaged and background subtracted using software Fit2D [52]. 1D WAXD profile was obtained by using azimuthal integration functionality in Fit2D. Automatic Gaussian peak searching was performed using lsqcurvefit optimization routine in MATLAB.
2.4. Solid-state NMR
Solid-state NMR experiments were conducted on a Varian VNMRS 400 MHz wide-bore spectrometer with a 1.6 mm triple resonance magic angle spinning (MAS) probe configured in 1H/13C/15N mode. All 1D and 2D solid-state NMR experiments were collected under a MAS frequency of 35 kHz. 13C direct detect (DD) MAS NMR experiments were performed on natural abundance, U-[13C, 15N]-l-alanine and U-[13C, 15N]-l-proline labeled dragline silk samples with 135 kHz 1H two-pulse phased-modulated (TPPM) decoupling [53] during acquisition. The spectra were collected with 25 kHz sweep width, 10s recycle delay for dry samples and 1 s for water-wetted samples. A fully relaxed 13CDD MAS NMR spectrum was collected on the U-[13C, 15N]-l-alanine labeled water-wetted dragline silk sample with 50 s recycle delay and 1024 transients. 1H–13C cross-polarization (CP) MAS NMR experiments were conducted on all dry dragline silk samples with 25 kHz sweep width, 3 s recycle delay and 135 kHz 1H TPPM decoupling during acquisition. 1H–13C CP condition was matched with a ramped 1H rf field from ∼141 to ∼149 kHz and 13C rf field of 100 kHz. The contact time during CP was 1 ms. The same CP condition was utilized for all other CP-based 2D experiments in this study.
2D DARR experiments were collected on the [13C, 15N]-enriched dragline silk with a sweep width of 25 kHz in both dimensions. 512 and 256 data points were acquired for direct and indirect dimensions, respectively. The 1H–13CCP condition was the same as described above. Continuous wave irradiation was applied on the 1H channel with a rf matching the 35 kHz rotary frequency. Spectra were collected with a DARR mixing time of 150 ms and 1 s to gain through space intra- and/or inter-residue correlations. Spectra were acquired with 32 transients, 3 s recycle delay and 135 kHz 1H TPPM decoupling field strength.
Two versions of 2D refocused DQ/SQINADEQUATE NMR experiment were performed on [13C, 15N]-enriched dragline silk samples. A CP-based version [39,54] was applied on the dry silk samples, while another 13C direct polarized version [21] was used on the water-wetted silk samples. The spectra were collected with 25 kHz sweep width and 1024 complex points in the direct dimension and 35 kHz sweep width in the indirect dimension. The number of complex points collected in the indirect dimension ranged from 128 (for dry silk sample) to 256 (for wetted silk sample) to ensure that fully decayed signals can be recorded. 1 s and 3 s recycle delays were utilized for the water-wetted silk sample to superimpose different components in the wetted silk and 3 s recycle delay was used for the dry silk sample. Chemical shifts and linewidths of the carbonyl peaks for Ala and Gly were extracted from the refocused INADEQUATE spectra of water-wetted U-[13C, 15N]-l-alanine labeled dragline silk. With the chemical shifts and linewidths for each component, the carbonyl region of the fully relaxed 13C DD MAS spectrum was fit to extract peak heights to determine the percentages of Ala and Gly in β-sheet structure. The aliphatic region was fit using the chemical shifts for each peak obtained from INADEQUATE spectra and the Ser Cβ peak was fit into two components, one is for β-sheet structure and the other is for helical structure. Spectrum deconvolution was performed with MestReNova (version 8).
2.5. Structural composition prediction and quantification
Percentages of Ala, Gly and Ser in β-sheet conformation were predicted based on the partial amino acid sequence of MaSp1 protein for Green Lynx dragline silk (GenBank: GU306168). Amino acids were predicted to adopt β-sheet conformation if they were in the motifs matching the following rules: (i) any poly(Ala) regions with four or more Ala were counted as β-sheet; (ii) any poly(Gly-X) (X = Ala or Ser) regions were counted as β-sheet; (iii) any Gly, Ser preceding or terminating the poly(Ala) or poly(Gly-X) regions were considered as β-sheet; (iv) in the nonrepresentative domain, any region that consists of four or more amino acids that include only Ala, Gly and Ser was counted as β-sheet. This quantification method has been previously published using Nephila clavipes dragline spider silk [24]. In this work, we apply the same method to the partial sequence of Green Lynx dragline silk.
3. Results
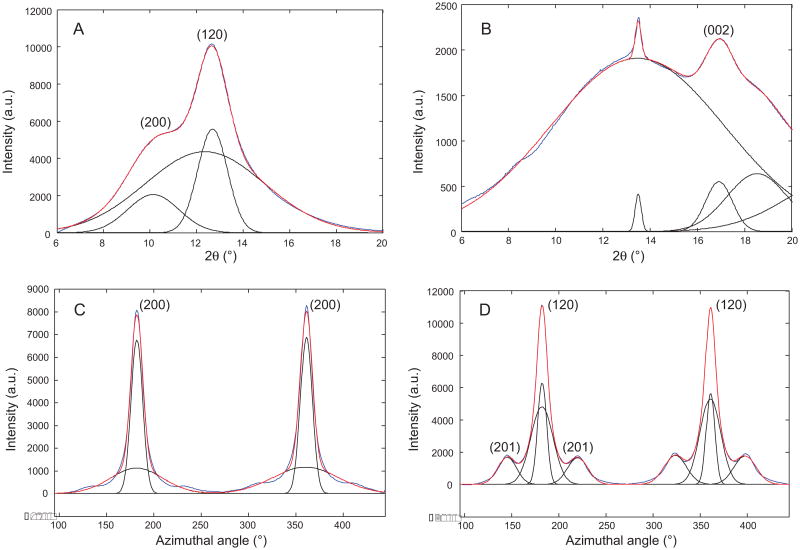
Mature female Green Lynx spider (Fig. 1A) was forcibly silked and bundles of dragline silk (Fig. 1B) were collected for solid-state NMR or wide angle X-ray diffraction. The forcibly silked Green Lynx dragline silk fibers are uniformly shaped with a diameter of ∼1 μm as measured by SEM (Fig. 1C). WAXD experiments were performed on the dragline silk sample to characterize the secondary structure. 2D WAXD pattern of a vertically aligned P. viridans (Green Lynx) major ampullate spider silk fiber bundle is shown in Fig. 2. The beamstop shadow and CCD detector lines (yellow) have been masked for integration. In addition, the diffraction intensity has been color-coded by Fit2D to assist visualizing the pattern. The observed diffraction pattern is comparable to previously published major ampullate spider silks [30,31]. The equatorial reflections are commonly assigned to β-sheet nanocrystalline structures with the reflections 30° on either side of the axis arising from the electron density of the surrounding amorphous regions [55]. Fig. 3A shows the azimuthal integration of an equatorial wedge 15° on either side of the horizontal axis deconvoluted to demonstrate the Gaussian fitting procedure results used to optimize the reflection positions. These two strongest reflections are assigned to the (200) and (120) reflections as indicated in the figure. The meridian wedge without the beamstop shadow was integrated to obtain parameters for the final dimension of the unit cell arising from the (002) reflection (Fig. 3B). The additional narrow peak occurring around 13.5 degrees in 2θ space is assigned to the superlattice reflection and is not usually associated with individual β-sheet nanocrystals. The fiber bundle was well aligned and had sufficient fiber proximity that many higher order reflections were observed in this 2D pattern. The 12 most intense unique and well-differentiated reflections were iteratively used to calculate an orthorhombic unit cell of dimensions 6.6 Å by 9.7 Å by 10.9 Å. The radial broadening of the crystalline fractions in 2θ space was used to approximate the nanocrystallite dimensions through Scherrer's formula, L = 0.9 × λ/(FWHM × cosθ). These data illustrate that the nano-crystal dimensions were 2.5 nm by 3.3 nm by 3.8 nm as calculated from the (200), (120), and (002) reflections. The integration of the equatorial (200) and (120) reflection comprise the majority of the component relating to the β-sheet nanocrystallites and as a percentage of the total diffraction intensity, we can approximate the crystalline component of the silk at 34%. Radial integration of a narrow ring around the (200) and (120) reflections can be used to approximate the orientation factor of the β-sheet nanocrystallites within the spider silk fiber using Herman's orientation function: fc = (3(cos2φ) – 1)/2, where φ is the FWHM of the resulting Gaussian-fit (Fig. 3C). The obtained orientation factor of 0.98 indicates there is nearly perfect alignment of these nanocrystallites within the fiber along the fiber axis. The small component of disorder likely arises from imperfect alignment of the fibers within the bundle. The amorphous component can also be aligned when adjacent disordered strands are parallel to each other. The orientation factor can also be applied to the amorphous component to assess the extent of ordering among the fraction of the polypeptide backbone not arising from the β-sheet nanocrystallites. The (201) reflection demonstrated in Fig. 3D is associated with the amorphous component of the spider silk fibers and equates to an amorphous orientation factor of 0.89.
Fig. 1.
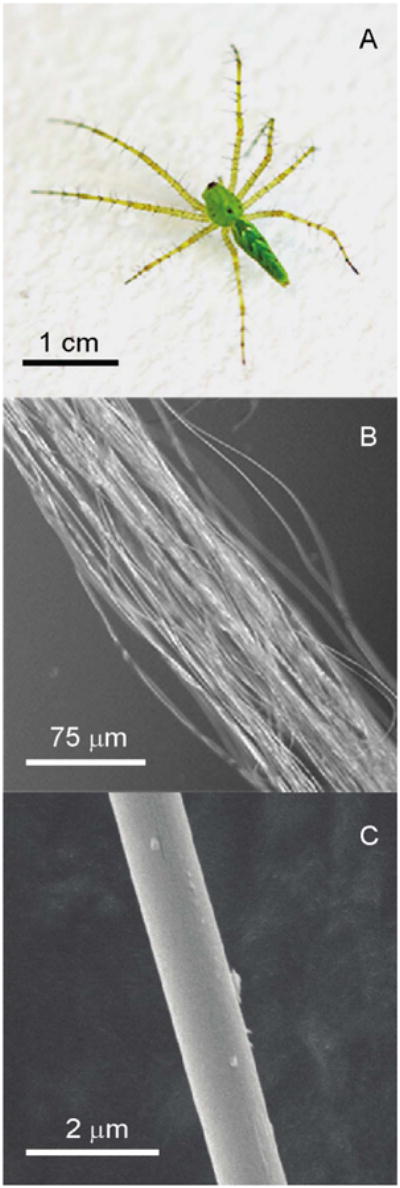
(A) Picture of a mature female Peucetia viridans spider; (B) optical microscopy image of Peucetia viridans spider dragline silk bundle; (C) SEM image of a single dragline silk fiber.
Fig. 2.
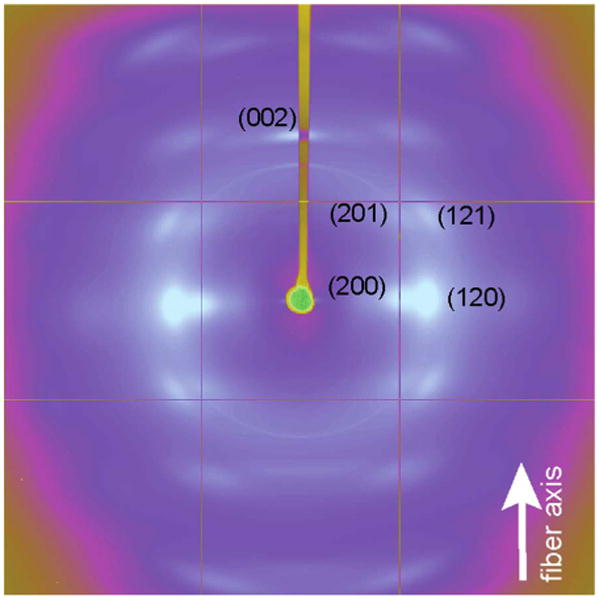
2D WAXD pattern of Peucetia viridans spider major ampullate silk.
Fig. 3.
1D WAXD radial intensity profiles for Green Lynx major ampullate silk fibers. The (120/200) equatorial peaks (A) and the (002) meridian peak (B) are deconvoluted into broad amorphous and narrow crystalline components (black line). The (200) and (120) peak (C and D) are also radially deconvoluted (black line). Blue line is the experimental data while the red line is the total fit.
Previous reports on the partial sequence of the MaSp1-like protein do not provide accurate amino acid composition of Green Lynx dragline silk [46]. Here, amino acid analysis was applied on natural abundance silk sample utilizing a 1H solution-state NMR technique as previously described (Fig. S2) [56]. The molar percentages of 15 amino acids in the silk proteins was measured and shown in Table S1. As a member from the RTA clade, Green Lynx spiders were predicted to use only the MaSp1-like protein in their dragline silk [48]. However, the existence of Pro with 3.0% molar percentage was observed in the hydrolyzed silk solution. Pro was by far exclusively found in MaSp2-like protein in spider dragline silk [34]. Therefore, the determined Pro content suggests that Green Lynx dragline silk consists of both MaSp1 and MaSp2 proteins.
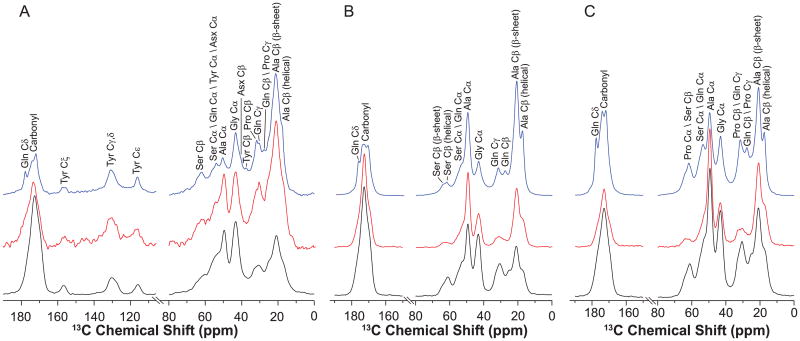
1H–13C CP MAS NMRspectrum of natural abundance Green Lynx dragline silk is displayed in Fig. 4A (black). Signals were observed for amino acid with composition higher than 3%, including Ala, Gly, Gln, Asx (aspartic acid and aspartate) and Ser. Also, Tyr (2.7%) side chain can be unambiguously identified because of the distinct chemical shifts from 100 to 160 ppm. As shown in Figs. 3C and 4B, the level of 13C enrichments for Ala and Pro were significantly elevated, when the spider silk was isotope-enriched by feeding the spiders with uniformly 13C labeled Ala or Pro solutions. In addition to Ala and Pro, 13C was also enriched in Gly, Gln and Ser [56].
Fig. 4.
13C DD MAS spectra of dry (red) and wet (blue) major ampullate silk and 13C CP MAS spectra (black) of dry major ampullate silk from (A) natural; (B) U-[13C, 15N]-l-Ala labeled and (C) U-[13C, 15N]-l-Pro labeled Peucetia viridans spiders. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
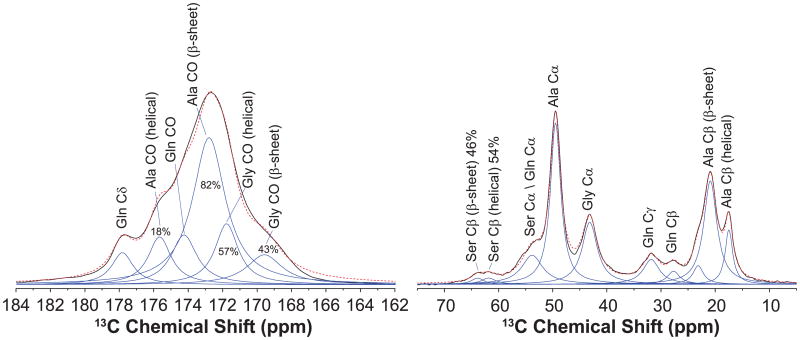
Ala Cβ and Ser Cβ peaks exhibit considerable asymmetry, indicating that Ala and Ser are incorporated into two or more distinct secondary structures. The multiple components can be further resolved in spectra collected on water-wetted silk samples. Interacting with water leads to the plasticization of the non-β-sheet regions and the structure reconfiguration in silk fibers. As a result, the anisotropic morphology of the nanocrystallites is changed, primarily due to the amorphous regions becoming less restricted and dynamic [17,57–59]. Therefore, the plasticizing effect of water on spider dragline silk can be utilized to increase the resolution of its NMR spectra [24,31,40]. In Fig. 4, 13C DD MAS NMR spectra of three silk samples were collected in dry (red) and water-wetted (blue) conditions. Spectral resolution in the carbonyl region is significantly improved for the wet silk compared with the dry samples.
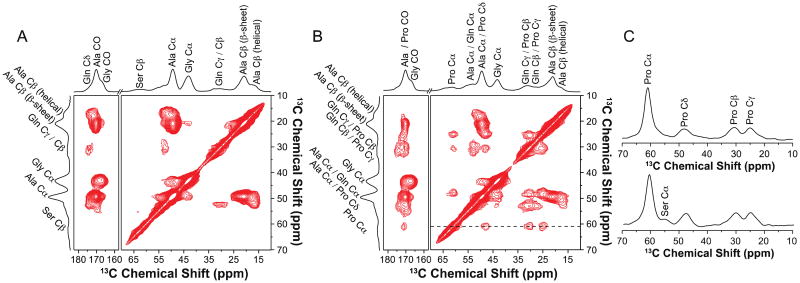
2D 13C–13C correlation spectra of the U-[13C, 15N]-l-Ala and U-[13C, 15N]-l-Pro labeled Green Lynx dragline silk are shown in Figs.4B and 5A. The spectrum collected with a DARR mixing time of 150 ms provides two sets of intra-residue correlation between Ala Cα, Cβ and CO for both silk samples. The Ala Cα, Cβ and CO with chemical shifts of 49.2, 20.9 and 172.6 ppm show an intense correlation. The other correlation with less intensity was observed with Ala Cα at 50.3 ppm, Ala Cβ at 17.2 ppm and Ala CO at 175.5 ppm. Based on their chemical shifts, the two correlations were assigned to Ala residues adopting β-sheet and helical secondary structures, respectively. In Fig. 5B, the 13C–13C correlations within the Pro spin system was identified. The extracted chemical shifts are 60.7, 30.7, 25.0, 47.9 and 173.0 ppm for Pro Cα, Cβ, Cγ, Cδ and CO, respectively. In addition, a weak inter-residue correlation between Ala Cα and Gly Cα was observed as displayed in Fig. 5A. However, as shown in Fig. 5C, no inter-residue correlation between Pro Cα and Gly Cα can be observed in the slices extracted at Pro Cα (60.7 ppm) even when 1 s of DARR mixing time was applied (Fig. 5C, bottom).
Fig. 5.
2D 13C–13C correlation NMR spectrum of (A) U-[13C, 15N]-l-Ala labeled and (B) U-[13C, 15N]-l-Pro labeled Peucetia viridans spider dry major ampullate silk with DARR mixing time of 150 ms. (C) Slices at Pro Cα were extracted from 2D 13C–13C DARR experiments of U-[13C, 15N]-l-Pro labeled dry major ampullate silk with 150 ms (top) and 1 s (bottom) mixing times.
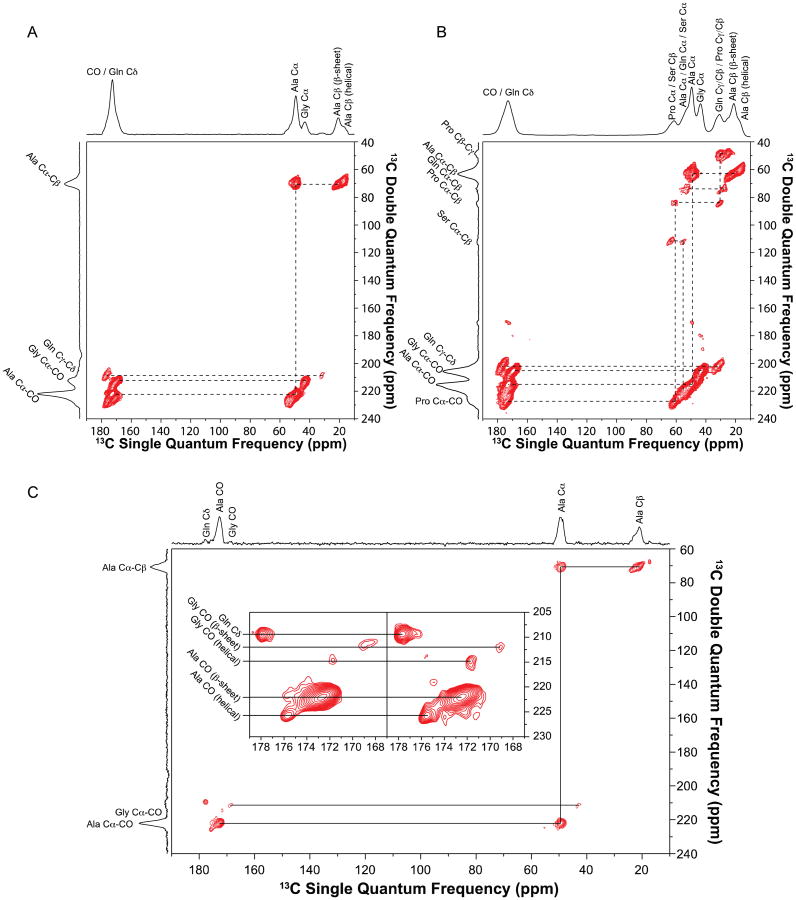
Additional 2D through-bond DQ/SQ refocused INADEQUATE NMR spectra were collected for 13C-enriched Green Lynx dragline silk. Cross-polarization was utilized to detect the 13C spins in the dry silk samples (Fig. 6A and B). Double quantum correlations of Ala Cα-Cβ, Ala Cα-CO, Gly Cα-CO and Gln Cγ-Cδ were presented in Fig. 6A. Fig. 6B displays the correlations between Pro Cα-Cβ, Pro Cβ-Cγ, Pro Cα-CO as well as the correlation between Gln Cα-Cβ. These spectra further confirm the chemical shift assignments determined from the through-space experiments as discussed above. In addition, another two sets of correlations between Ser Cα-Cβ were observed, indicating two secondary structure environments for Ser. To better resolve the overlapped carbonyl region, refocused INADEQUATE NMR experiments were applied on the wet U-[13C, 15N]-l-Ala labeled silk. As 1H–13C CP efficiency is reduced by the increasing mobility of silk proteins in the wet sample, direct-polarization 13C experiment was used instead. Ala and Gly each show two distinct carbonyl environments in the inset of Fig. 6C. The rigid β-sheet structures have a longer 13C T1 relaxation time compared with the relative mobile helical structures, hence the intensity of the β-sheet peaks will be enhanced by using longer recycle delay (3s compared to 1s) [24]. Based upon this phenomenon, the Ala CO at 172.6 ppm and Gly CO at 169.6 ppm were identified as β-sheet conformation while Ala CO at 175.5 ppm and Gly CO at 171.8 ppm were determined to be in helical secondary structure.
Fig. 6.
2D 13C–13C through-bond double quantum/single quantum (DQ/SQ) refocused INADEQUATE NMR spectrum of (A) U-[13C, 15N]-l-Ala labeled and (B) U-[13C, 15N]-l-Pro labeled Peucetia viridans spider dry major ampullate silk. (C) 2D INADEQUATE spectrum of U-[13C, 15N]-l-Ala labeled Peucetia viridans spider wet major ampullate silk. Inset shows the CO regions of the 2D INADEQUATE spectra collected with 1s (right) and 3 s (left) recycle delays.
With the chemical shift and the full-width at half-maximum (FWHM) extracted from Fig. 6C, the percentage of the secondary structures can be quantified by fitting the carbonyl region of a fully relaxed 13C DD NMR spectrum. As shown in the left panel of Fig. 7, the carbonyl region was fitted into several components according to the corresponding chemical shift and FWHM. 82 ± 1% of Ala and 40 ± 2% of Gly are determined to present in β-sheet regions. The Ala and Gly in helical conformation are found to be 18 ± 1% and 60 ± 2%, respectively. The fit of Ala Cβ peak in the right panel gives a percentage of 20 ± 3% helical and 80 ± 3% β-sheet, which is consistent with the results obtained from fitting the carbonyl region. In addition, the asymmetric Ser Cβ peak was fit into two components. The secondary structure assignment for Ser Cβ was conducted by comparing the relative intensity of the two components in the 13C DD NMR spectra of the wet and dry silk collected with 1 s recycle delay (data no shown) [24,31]. The Ser Cβ at 61.8 ppm shows enhanced signal in the wet silk, indicating belongs to the mobile helical component. Thus, 54 ± 2% of Ser are in helical structure and 46 ± 2% are in β-sheet structure.
Fig. 7.
The carbonyl and aliphatic regions from the fully relaxed 13C direct detect (DD) MAS NMR spectrum ofthe U-[13C, 15N]-l-alanine labeled Peucetia viridans spider wet major ampullate silk. The spectrum was deconvoluted to extract the percentages of Ala, Gly and Ser residues that are incorporated into β-sheet or helical conformations.
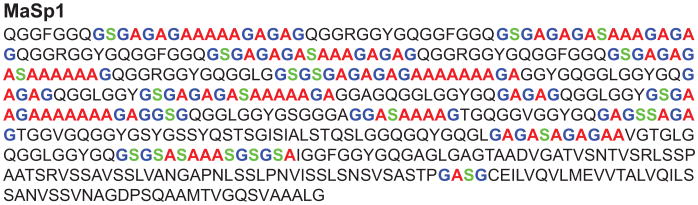
Partial amino acid sequence of the Green Lynx MaSp1 silk protein is displayed in Fig. 8. It was used to estimate the percentage of the secondary structures based on the amino acid motif model described in the method section. All the color-coded amino acids were counted as adopting β-sheet conformation while the rest are considered as being in helical conformation. Since only MaSp1 silk protein has been identified in the Green Lynx dragline silk [46], there is no information available for the MaSp1:MaSp2 ratio at this moment. Due to the similarity of the amino acid compositions in the two spider silk proteins, the partial sequence of MaSp1 protein is a reasonable representative of the entire Green Lynx spider dragline silk protein. As shown in Table 1, based on this model in Fig. 8, 78% of Ala, 38% of Gly and 42% of Ser were predicted to present in β-sheet regions.
Fig. 8.
Partial primary amino acid sequence for the Peucetia viridans spider MaSp1 protein. The Ala, Gly and Ser residues predicted by the primary sequence to adopt β-sheet conformation were colored in red, blue and green, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Percent of β-sheet and helical conformations in the Peucetia viridans spider major ampullate silk predicted by the primary amino acid sequence (AA%) and determined from deconvolution ofthe fully relaxed 13C DD-MAS NMR spectrum (NMR%).
| Residue | β-Sheet | Helical | ||
|---|---|---|---|---|
|
|
|
|||
| AA (%) | NMR (%) | AA (%) | NMR (%) | |
| Ala | 78 | 82 ± 1 | 22 | 18 ± 1 |
| Gly | 38 | 40 ± 3 | 62 | 60 ± 3 |
| Ser | 42 | 46 ± 2 | 58 | 54 ± 2 |
4. Discussion
Compare to the dragline silk from Nephila or Black Widow spiders, Green Lynx spiders produce much thinner fibers. But the dragline silk from Green Lynx spiders are still comparable to other dragline silks in terms of mechanical properties [47,48]. This can be mainly attributed to the similar construction and topology of the β-sheet crystals observed from the WAXD data, and the persistent presence of mass fractal hierarchical structures of the nano-crystallites indicated by the small-angle X-ray scattering (SAXS) structure factors. The SAXS structure factor shows perfect power law relation in the q range of 0.16–0.6 nm−1 (Fig. S4), indicating that the nano-crystals in Green Lynx is mass fractal with hierarchical spacial arrangement, forming a mechanically robust self-similar beta-sheets network [60–62]. According to the WAXD data, the crystalline region and amorphous region are both highly oriented based on their orientational orders. However, the size of the nanocrystalline components is relatively smaller, especially in the direction along the fiber axis [38]. The result implies that the length of the backbones in the β-sheet structures is relatively shorter, which in turn leads to the lower strength of the silk fiber [63]. This can be explained by the fewer numbers of Ala in the poly(Ala) motif in the partial sequence of the MaSp1 protein of Green Lynx dragline silk.
Solid-state NMR data provides detailed information about the secondary structures for the building blocks in the dragline silk protein. Ala, Gly and Ser in Green Lynx dragline silk are shown to adopt both β-sheet and helical structures. Most Ala are in the β-sheet conformation, which can be ascribed to the poly(Ala) and poly(Gly-Ala) motifs. As comparison, the majority of Gly are in the Gly-Gly-X (X can be Gln, Tyr, Arg, Phe, Leu, Val and Ala) motifs, which contribute to ≥60% of Gly in helical regions. As fewer Ser are adjacent to the motifs of poly(Ala) and poly(Gly-Ala), the percentage of Ser in β-sheet is smaller than that in helical conformation. The percentage of crystallinity for the Green Lynx dragline silk is 40.0 ± 1.2% estimated from the percentage of Ala, Gly and Ser that are in β-sheet conformation. The crystallinity from WAXD is smaller than that calculated from the NMR data [24,30]. This discrepancy likely results from the difference of the two methods in counting the percentage of crystals. The percentage from WAXD data is calculated based on the volume of the crystalline region while the one from NMR data is determined based on the molar percent of the involved amino acids. Since Ala, Gly and Ser are the amino acids that have smaller molar volumes among the 15 amino acids in the spider silk, the crystallinity derived from the molar percent of the three amino acids should be equivalent to a smaller value if it is converted to the by-volume percentage. Comparing with the previous results for Black Widow and N. clavipes dragline silks [24,31]. There is no direct correlation that can be found between the percent of crystallinity and a certain parameter in mechanical properties.
According to our amino acid analysis result (Table S1), there is 3.0 ± 0.2% of proline within the Green Lynx dragline silk proteins. When the silk proteins were 13C enriched by feeding the spiders with 13C labeled Pro solution, the distinct 13C correlation pattern for Pro was observed in the 2D 13C–13C DARR spectrum. This result confirms the presence of Pro in the Green Lynx dragline silk. Additionally, the presence of inter-residue Pro Cα–Ser Cα correlation and the absence of Pro Cα–Gly Cα correlation indicate that the common GPGXX motif is not in the Green Lynx silk protein (Fig. 5C) [23]. However, the 13C chemical shifts indicate that Pro is in the type II β-turn structure, which is the common secondary structure adopted by the GPGXX motif found in most MaSp2 proteins for dragline spider silk [64,65]. The relationship between proline content and mechanical properties has been well demonstrated [49,66]. Based on the Pro content, the stress-strain curves for the supercontracted dragline silk from different spider species have been plotted by Liu et al. [49]. The stress–strain curve for the dragline silk of Peucetia rubrolineata spiders (a close sibling to the Green Lynx spiders) reported by Pérez-Rigueiro et al. [46] can be well fit into the stress–strain curve pattern [49] based on 3% of Pro content in the silk. Therefore, Pro is still a key amino acid in determining the physical properties of spider silks. The lack of Gly-Pro through space correlations is a motivating factor for further experiments to determine the Pro containing motif in Green Lynx dragline spider silk.
Green Lynx spiders along with other RTA clade spiders are believed to have only one type of spidroin protein (MaSp1) in their dragline silk [43,46–48]. However, the discovery of Pro in Green Lynx dragline silk cannot be explained by the small amount of Pro in the terminal region of the MaSp1 sequence (Fig. 8). Therefore, the result strongly indicates that there is a second spidroin (MaSp2) in the dragline silk, which has been overlooked. Using the percentages of Pro within the MaSp2 proteins of several Orbiculariae spiders, the MaSp2 content in Green Lynx dragline silk is estimated to be 20+%. Assuming Green Lynx dragline silk is similar to other dragline silks and contains two proteins that are similar to MaSp1 and 2 in composition, molecular weight and composition, then we can guess that the MaSp1:MaSp2 ratio in Green Lynx dragline silk would be very close to that of the N. clavipes and Black Widow dragline silks [18,67]. The two spidroins, besides Pro content, show high similarity in amino acid composition, amino acid repetitive motifs and molecular architecture [33,43]. The slight difference in the mechanical properties of the two spidroins [26] ensures the spiders can tailor the physical properties of their silk fibers to adapt to the changes of environment by varying the ratio of the two spidroins. Since Green Lynx spiders are a more ancient species than the Orbic-ulariaes, it seems that the spiders realized the benefits of using two spidroins in their silk much earlier than we thought. The picture of how the two spidroins correlate and interact with each other still remains unclear but very intriguing.
5. Conclusions
The secondary structure of the Green Lynx spider dragline silk protein was characterized by WAXD and solid-state NMR spectroscopy. β-Sheet nanocrystallites were observed to be embedded in the amorphous region by WAXD and the size of the crystals is determined to be 2.5, 3.3 and 3.8 nm. Both crystalline and amorphous regions were found to be highly oriented and aligned parallel to the dragline silk fiber. The solid-state NMR data indicated that Ala, Gly and Ser are incorporated into both β-sheet and helical structures. The percentages of each amino acid in β-sheet conformation were calculated by fitting the carbonyl region of the fully relaxed 13C DD NMR spectrum. 78% of Ala, 38% of Gly and 42% of Ser were determined to be in β-sheet secondary structure. This agrees with the theoretical prediction based on the primary amino acid sequence of the silk protein. Therefore, a quantitative correlation between the secondary structures and primary amino acid sequence was established. Furthermore, 3.0% of Pro was detected in the Green Lynx dragline silk and chemical shift analysis indicated that Pro is in β-turn conformation. The large amount of Pro in the dragline silk implies the possible existence of a second silk protein, MaSp2. Further, detailed structural information at the molecular level was revealed for the dragline silk protein of a spider in the RTA clade. Green Lynx dragline silk presents a molecular architecture very similar with that from Orbiculariaes spiders.
Supplementary Material
Footnotes
Appendix A. Supplementary data: Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.ijbiomac.2015.07.048
References
- 1.Shear WA, Palmer JM, Coddington JA, Bonamo PM. A devonian spinneret – early evidence of spiders and silk use. Science. 1989;246:479–481. doi: 10.1126/science.246.4929.479. [DOI] [PubMed] [Google Scholar]
- 2.Gosline JM, Demont ME, Denny MW. The structure and properties of spider silk. Endeavour. 1986;10:37–43. [Google Scholar]
- 3.Altman GH, Diaz F, Jakuba C, Calabro T, Horan RL, Chen JS, Lu H, Richmond J, Kaplan DL. Silk-based biomaterials. Biomaterials. 2003;24:401–416. doi: 10.1016/s0142-9612(02)00353-8. [DOI] [PubMed] [Google Scholar]
- 4.Lewis RV. Spider silk: ancient ideas for new biomaterials. Chem Rev. 2006;106:3762–3774. doi: 10.1021/cr010194g. [DOI] [PubMed] [Google Scholar]
- 5.Gosline JM, Denny MW, Demont ME. Spider silk as rubber. Nature. 1984;309:551–552. [Google Scholar]
- 6.Vollrath F, Porter D. Spider silk as archetypal protein elastomer. Soft Matter. 2006;2:377–385. doi: 10.1039/b600098n. [DOI] [PubMed] [Google Scholar]
- 7.Xu M, Lewis RV. Structure of a protein superfiber – spider dragline silk. Proc Natl Acad Sci U S A. 1990;87:7120–7124. doi: 10.1073/pnas.87.18.7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinman MB, Lewis RV. Isolation of a clone encoding a 2nd dragline silk fibroin – Nephila clavipes dragline silk is a 2-protein fiber. J Biol Chem. 1992;267:19320–19324. [PubMed] [Google Scholar]
- 9.Simmons A, Ray E, Jelinski LW. Solid-state 13C NMR of Nephila clavipes dragline silk establishes structure and identity of crystalline regions. Macromolecules. 1994;27:5235–5237. [Google Scholar]
- 10.Kummerlen J, van Beek JD, Vollrath F, Meier BH. Local structure in spider dragline silk investigated by two-dimensional spin-diffusion nuclear magnetic resonance. Macromolecules. 1996;29:2920–2928. [Google Scholar]
- 11.Bram A, Branden CI, Craig C, Snigireva I, Riekel C. X-ray diffraction from single fibres of spider silk. J Appl Crystallogr. 1997;30:390–392. [Google Scholar]
- 12.Grubb DT, Jelinski LW. Fiber morphology of spider silk: the effects of tensile deformation. Macromolecules. 1997;30:2860–2867. [Google Scholar]
- 13.Riekel C, Vollrath F. Spider silk fibre extrusion: combined wide- and small-angle X-ray microdiffraction experiments. Int J Biol Macromol. 2001;29:203–210. doi: 10.1016/s0141-8130(01)00166-0. [DOI] [PubMed] [Google Scholar]
- 14.Yang Z, Grubb DT, Jelinski LW. Small-angle X-ray scattering of spider dragline silk. Macromolecules. 1997;30:8254–8261. [Google Scholar]
- 15.Riekel C, Branden C, Craig C, Ferrero C, Heidelbach F, Muller M. Aspects of X-ray diffraction on single spider fibers. Int J Biol Macromol. 1999;24:179–186. doi: 10.1016/s0141-8130(98)00084-1. [DOI] [PubMed] [Google Scholar]
- 16.van Beek JD, Hess S, Vollrath F, Meier BH. The molecular structure of spider dragline silk: folding and orientation of the protein backbone. Proc Natl Acad Sci U S A. 2002;99:10266–10271. doi: 10.1073/pnas.152162299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holland GP, Lewis RV, Yarger JL. WISE NMR characterization of nanoscale heterogeneity and mobility in supercontracted Nephila clavipes spider dragline silk. J Am Chem Soc. 2004;126:5867–5872. doi: 10.1021/ja031930w. [DOI] [PubMed] [Google Scholar]
- 18.Ayoub NA, Garb JE, Tinghitella RM, Collin MA, Hayashi CY. Blueprint for a high-performance biomaterial: full-length spider dragline silk genes. PLoS ONE. 2007;2:e514. doi: 10.1371/journal.pone.0000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcotte I, van Beek JD, Meier BH. Molecular disorder and structure of spider dragline silk investigated by two-dimensional solid-state NMR spectroscopy. Macromolecules. 2007;40:1995–2001. [Google Scholar]
- 20.Holland GP, Creager MS, Jenkins JE, Lewis RV, Yarger JL. Determining secondary structure in spider dragline silk by carbon–carbon correlation solid-state NMR spectroscopy. J Am Chem Soc. 2008;130:9871–9877. doi: 10.1021/ja8021208. [DOI] [PubMed] [Google Scholar]
- 21.Holland GP, Jenkins JE, Creager MS, Lewis RV, Yarger JL. Quantifying the fraction of glycine and alanine in beta-sheet and helical conformations in spider dragline silk using solid-state NMR. Chem Commun. 2008:5568–5570. doi: 10.1039/b812928b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holland GP, Jenkins JE, Creager MS, Lewis RV, Yarger JL. Solid-state NMR investigation of major and minor ampullate spider silk in the native and hydrated states. Biomacromolecules. 2008;9:651–657. doi: 10.1021/bm700950u. [DOI] [PubMed] [Google Scholar]
- 23.Jenkins JE, Creager MS, Butler EB, Lewis RV, Yarger JL, Holland GP. Solid-state NMR evidence for elastin-like beta-turn structure in spider dragline silk. Chem Commun. 2010;46:6714–6716. doi: 10.1039/c0cc00829j. [DOI] [PubMed] [Google Scholar]
- 24.Jenkins JE, Creager MS, Lewis RV, Holland GP, Yarger JL. Quantitative correlation between the protein primary sequences and secondary structures in spider dragline silks. Biomacromolecules. 2010;11:192–200. doi: 10.1021/bm9010672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazaris A, Arcidiacono S, Huang Y, Zhou JF, Duguay F, Chretien N, Welsh EA, Soares JW, Karatzas CN. Spider silk fibers spun from soluble recombinant silk produced in mammalian cells. Science. 2002;295:472–476. doi: 10.1126/science.1065780. [DOI] [PubMed] [Google Scholar]
- 26.Brooks AE, Nelson SR, Jones JA, Koenig C, Hinman M, Stricker S, Lewis RV. Distinct contributions of model MaSp1 and MaSp2 like peptides to the mechanical properties of synthetic major ampullate silk fibers as revealed in silico. Nanotechnol Sci Appl. 2008;1:9–16. doi: 10.2147/nsa.s3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang YS, Hu JH, Miao YG, Zhao AC, Zhao TF, Wu DY, Liang LF, Miikura AM, Shiomi K, Kajiura Z, Nakagaki M. Expression of EGFP-spider dragline silk fusion protein in BmN cells and larvae of silkworm showed the solubility is primary limit for dragline proteins yield. Mol Biol Rep. 2008;35:329–335. doi: 10.1007/s11033-007-9090-6. [DOI] [PubMed] [Google Scholar]
- 28.Askarieh G, Hedhammar M, Nordling K, Saenz A, Casals C, Rising A, Johansson J, Knight SD. Self-assembly of spider silk proteins is controlled by a pH-sensitive relay. Nature. 2010;465:236–238. doi: 10.1038/nature08962. [DOI] [PubMed] [Google Scholar]
- 29.Hagn F, Eisoldt L, Hardy JG, Vendrely C, Coles M, Scheibel T, Kessler H. A conserved spider silk domain acts as a molecular switch that controls fibre assembly. Nature. 2010;465:239–242. doi: 10.1038/nature08936. [DOI] [PubMed] [Google Scholar]
- 30.Sampath S, Isdebski T, Jenkins JE, Ayon JV, Henning RW, Orgel JPRO, Antipoa O, Yarger JL. X-ray diffraction study of nanocrystalline and amorphous structure within major and minor ampullate dragline spider silks. Soft Matter. 2012;8:6713–6722. doi: 10.1039/C2SM25373A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenkins JE, Sampath S, Butler E, Kim J, Henning RW, Holland GP, Yarger JL. Characterizing the secondary protein structure of black widow dragline silk using solid-state NMR and X-ray diffraction. Biomacromolecules. 2013;14:3472–3483. doi: 10.1021/bm400791u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi CY, Shipley NH, Lewis RV. Hypotheses that correlate the sequence, structure, and mechanical properties of spider silk proteins. Int J Biol Macromol. 1999;24:271–275. doi: 10.1016/s0141-8130(98)00089-0. [DOI] [PubMed] [Google Scholar]
- 33.Hayashi CY, Lewis RV. Molecular architecture and evolution of a modular spider silk protein gene. Science. 2000;287:1477–1479. doi: 10.1126/science.287.5457.1477. [DOI] [PubMed] [Google Scholar]
- 34.Gatesy J, Hayashi C, Motriuk D, Woods J, Lewis R. Extreme diversity, conservation, and convergence of spider silk fibroin sequences. Science. 2001;291:2603–2605. doi: 10.1126/science.1057561. [DOI] [PubMed] [Google Scholar]
- 35.Pouchkina-Stantcheva NN, McQueen-Mason SJ. Molecular studies of a novel dragline silk from a nursery web spider, Euprosthenops sp. (Pisauridae) Comp Biochem Phys B. 2004;138:371–376. doi: 10.1016/j.cbpc.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 36.Motriuk-Smith D, Smith A, Hayashi CY, Lewis RV. Analysis of the conserved N-terminal domains in major ampullate spider silk proteins. Biomacromolecules. 2005;6:3152–3159. doi: 10.1021/bm050472b. [DOI] [PubMed] [Google Scholar]
- 37.Riekel C, Muller M, Vollrath F. In situ X-ray diffraction during forced silking of spider silk. Macromolecules. 1999;32:4464–4466. [Google Scholar]
- 38.Du N, Liu XY, Narayanan J, Li LA, Lim MLM, Li DQ. Design of superior spider silk: from nanostructure to mechanical properties. Biophys J. 2006;91:4528–4535. doi: 10.1529/biophysj.106.089144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lesage A, Auger C, Caldarelli S, Emsley L. Determination of through-bond carbon–carbon connectivities in solid-state NMR using the INADEQUATE experiment. J Am Chem Soc. 1997;119:7867–7868. [Google Scholar]
- 40.Creager MS, Jenkins JE, Thagard-Yeaman LA, Brooks AE, Jones JA, Lewis RV, Holland GP, Yarger JL. Solid-state NMR comparison of various spiders' dragline silk fiber. Biomacromolecules. 2010;11:2039–2043. doi: 10.1021/bm100399x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee SM, Pippel E, Gosele U, Dresbach C, Qin Y, Chandran CV, Brauniger T, Hause G, Knez M. Greatly increased toughness of infiltrated spider silk. Science. 2009;324:488–492. doi: 10.1126/science.1168162. [DOI] [PubMed] [Google Scholar]
- 42.Keten S, Xu ZP, Ihle B, Buehler MJ. Nanoconfinement controls stiffness, strength and mechanical toughness of beta-sheet crystals in silk. Nat Mater. 2010;9:359–367. doi: 10.1038/nmat2704. [DOI] [PubMed] [Google Scholar]
- 43.Starrett J, Garb JE, Kuelbs A, Azubuike UO, Hayashi CY. Early events in the evolution of spider silk genes. PLoS ONE. 2012;7:e38084. doi: 10.1371/journal.pone.0038084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bond JE, Garrison NL, Hamilton CA, Godwin RL, Hedin M, Agnarsson I. Phylogenomics resolves a spider backbone phylogeny and rejects a prevailing paradigm for orb web evolution. Curr Biol. 2014;24:1765–1771. doi: 10.1016/j.cub.2014.06.034. [DOI] [PubMed] [Google Scholar]
- 45.Fernández R, Hormiga G, Giribet G. Phylogenomic analysis of spiders reveals nonmonophyly of orb weavers. Curr Biol. 2014;24:1772–1777. doi: 10.1016/j.cub.2014.06.035. [DOI] [PubMed] [Google Scholar]
- 46.Perez-Rigueiro J, Plaza GR, Torres FG, Hijar A, Hayashi C, Perea GB, Elices M, Guinea GV. Supercontraction of dragline silk spun by lynx spiders (Oxyopidae) Int J Biol Macromol. 2010;46:555–557. doi: 10.1016/j.ijbiomac.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 47.Boutry C, Rezac M, Blackledge TA. Plasticity in major ampullate silk production in relation to spider phylogeny and ecology. PLoS ONE. 2011;6:e22467. doi: 10.1371/journal.pone.0022467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boutry C, Blackledge TA. Evolution of supercontraction in spider silk: structure–function relationship from tarantulas to orb-weavers. J Exp Biol. 2010;213:3505–3514. doi: 10.1242/jeb.046110. [DOI] [PubMed] [Google Scholar]
- 49.Liu Y, Sponner A, Porter D, Vollrath F. Proline and processing of spider silks. Biomacromolecules. 2008;9:116–121. doi: 10.1021/bm700877g. [DOI] [PubMed] [Google Scholar]
- 50.Liu Y, Shao ZZ, Vollrath F. Elasticity of spider silks. Biomacromolecules. 2008;9:1782–1786. doi: 10.1021/bm7014174. [DOI] [PubMed] [Google Scholar]
- 51.Shi XY, Yarger JL, Holland GP. Probing site-specific 13C/15N-isotope enrichment of spider silk with liquid-state NMR spectroscopy. Anal Bioanal Chem. 2013;405:3997–4008. doi: 10.1007/s00216-013-6802-8. [DOI] [PubMed] [Google Scholar]
- 52.Hammersley AP, Svensson SO, Hanfland M, Fitch AN, Hausermann D. Two-dimensional detector software: from real detector to idealised image or two-theta scan. High Press Res. 1996;14:235–248. [Google Scholar]
- 53.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decoupling in rotating solids. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
- 54.Cadars S, Sein J, Duma L, Lesage A, Pham TN, Baltisberger JH, Brown SP, Emsley L. The refocused INADEQUATE MAS NMR experiment in multiple spin-systems: interpreting observed correlation peaks and optimizing lineshapes. J Magn Reson. 2007;188:24–34. doi: 10.1016/j.jmr.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 55.Warwicker JO. Comparative studies of fibroins. 2. Crystal structures of various fibroins. J Mol Biol. 1960;2:350. doi: 10.1016/s0022-2836(60)80046-0. [DOI] [PubMed] [Google Scholar]
- 56.Shi XY, Holland GP, Yarger JL. Amino acid analysis of spider dragline silk using 1H NMR. Anal Biochem. 2013;440:150–157. doi: 10.1016/j.ab.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 57.Blackledge TA, Boutry C, Wong SC, Baji A, Dhinojwala A, Sahni V, Agnarsson I. How super is supercontraction? Persistent versus cyclic responses to humidity in spider dragline silk. J Exp Biol. 2009;212:1980–1988. doi: 10.1242/jeb.028944. [DOI] [PubMed] [Google Scholar]
- 58.Eles PT, Michal CA. Strain dependent local phase transitions observed during controlled supercontraction reveal mechanisms in spider silk. Macromolecules. 2004;37:1342–1345. [Google Scholar]
- 59.Termonia Y. Molecular modeling of spider silk elasticity. Macromolecules. 1994;27:7378–7381. [Google Scholar]
- 60.Teixeira J. Small-angle scattering by fractal systems. J Appl Crystallogr. 1988;21:781–785. [Google Scholar]
- 61.Schaefer DW. Polymers, fractals, and ceramic materials. Science. 1989;243:1023–1027. doi: 10.1126/science.243.4894.1023. [DOI] [PubMed] [Google Scholar]
- 62.Mou Q, Benmore CJ, Weber WS, Yarger JL. Insights into the hierarchical structure of spider dragline silk fibers: evidence for fractal clustering of β-sheet nano-crystallites. 2015 arxiv:1507.04321. [Google Scholar]
- 63.Cetinkaya M, Xiao SB, Grater F. Effects of crystalline subunit size on silk fiber mechanics. Soft Matter. 2011;7:8142–8148. [Google Scholar]
- 64.Yao XL, Hong M. Structure distribution in an elastin-mimetic peptide (VPGVG) 3 investigated by solid-state NMR. J Am Chem Soc. 2004;126:4199–4210. doi: 10.1021/ja036686n. [DOI] [PubMed] [Google Scholar]
- 65.Ohgo K, Ashida J, Kumashiro KK, Asakura T. Structural determination of an elastin-mimetic model peptide, (Val-Pro-Gly-Val-Gly)(6), studied by 13C CP/MAS NMR chemical shifts, two-dimensional off magic angle spinning spin-diffusion NMR, rotational echo double resonance, and statistical distribution of torsion angles from protein data bank. Macromolecules. 2005;38:6038–6047. [Google Scholar]
- 66.Savage KN, Gosline JM. The role of proline in the elastic mechanism of hydrated spidersilks. J Exp Biol. 2008;211:1948–1957. doi: 10.1242/jeb.014225. [DOI] [PubMed] [Google Scholar]
- 67.Brooks AE, Steinkraus HB, Nelson SR, Lewis RV. An investigation of the divergence of major ampullate silk fibers from Nephila clavipes and Argiope aurantia. Biomacromolecules. 2005;6:3095–3099. doi: 10.1021/bm050421e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.