ABSTRACT
Fibroblast growth factor (FGF) signaling is important for skeletal development; however, cell-specific functions, redundancy and feedback mechanisms regulating bone growth are poorly understood. FGF receptors 1 and 2 (Fgfr1 and Fgfr2) are both expressed in the osteoprogenitor lineage. Double conditional knockout mice, in which both receptors were inactivated using an osteoprogenitor-specific Cre driver, appeared normal at birth; however, these mice showed severe postnatal growth defects that include an ∼50% reduction in body weight and bone mass, and impaired longitudinal bone growth. Histological analysis showed reduced cortical and trabecular bone, suggesting cell-autonomous functions of FGF signaling during postnatal bone formation. Surprisingly, the double conditional knockout mice also showed growth plate defects and an arrest in chondrocyte proliferation. We provide genetic evidence of a non-cell-autonomous feedback pathway regulating Fgf9, Fgf18 and Pthlh expression, which led to increased expression and signaling of Fgfr3 in growth plate chondrocytes and suppression of chondrocyte proliferation. These observations show that FGF signaling in the osteoprogenitor lineage is obligately coupled to chondrocyte proliferation and the regulation of longitudinal bone growth.
KEY WORDS: FGF signaling, PTHLH, IHH, Skeletal development, Endochondral bone formation, Osteoblast, Chondrocyte, Mouse
Summary: Conditional ablation of Fgfr1 and Fgfr2 in the osteoblast lineage in mouse reveals a feed-forward loop controlling FGFR3 signaling in growth plate chondrocytes.
INTRODUCTION
Human genetic disease and conditional gene inactivation experiments in mice have demonstrated essential roles for FGFR1 and FGFR2 in development of the appendicular and axial skeleton (Ornitz and Marie, 2002, 2015; Su et al., 2014). Although both receptors are expressed in the osteoprogenitor lineage, redundant functions of these FGFRs and mechanisms that couple FGFR signaling in the osteoprogenitor lineage to chondrogenesis and longitudinal bone growth are not known.
In mice, Fgfr1 has been targeted with a range of Cre drivers including brachyury (T), Ap2 (Tfap2a), Prx1 (Prrx1), Col2a1, Col1, osteocalcin (OC; Bglap) and Dmp1 (Jacob et al., 2006; Karolak et al., 2015; Li et al., 2005; Verheyden et al., 2005; Xiao et al., 2014; Yu and Ornitz, 2008; Zhang et al., 2014). With the exception of Col1-Cre, OC-Cre and Dmp1-Cre, which target relatively late stages of development, inactivation of Fgfr1 was in multiple cell lineages that include condensing mesenchyme, chondrocytes and osteoprogenitors. Observed phenotypes for Prx1-Cre and T-Cre include impaired limb bud development, increased cell death and reduced size of mesenchymal condensations (Li et al., 2005; Verheyden et al., 2005; Yu and Ornitz, 2008). Col2a1-Cre targets chondrocytes and osteoblasts, and inactivation of Fgfr1 resulted in an expanded hypertrophic chondrocyte zone (Jacob et al., 2006; Karolak et al., 2015); however, whether this was a cell-autonomous function of FGFR1 in hypertrophic chondrocytes or a non-cell-autonomous effect of inactivation of Fgfr1 in the osteoblast lineage could not be determined from these experiments. Use of Col1-Cre or OC-Cre to target Fgfr1 in mature osteoblasts resulted in increased bone mass and osteoblast number and no reported effect on bone length (Jacob et al., 2006; Zhang et al., 2014). Use of Dmp1-Cre to target Fgfr1 in osteocytes resulted in decreased osteocyte-specific gene expression but no overt skeletal phenotype (Xiao et al., 2014).
Mice in which the Fgfr2c splice variant has been inactivated (Fgfr2c−/−) were viable but showed reduced postnatal growth (Eswarakumar et al., 2002). Fgfr2 has also been conditionally targeted with a Dermo1 (Twist2) Cre driver or has been suppressed using RNA interference in limb bud mesenchyme. Inactivation of Fgfr2 with Dermo1-Cre, which effectively targets the chondrocyte and osteoblast lineage, also showed that Fgfr2 is necessary for postnatal bone growth (Yu et al., 2003). Suppression of Fgfr2 expression in limb bud mesenchyme in the Ap2-Cre lineage showed that FGFR2 is important for digit and tarsal bone development and ossification (Coumoul et al., 2005). None of the Fgfr2 gene inactivation studies provided a mechanism to explain the decreased bone growth.
Fgfr1 and Fgfr2 have considerable overlap in their expression patterns in developing limb bud and bone (Orr-Urtreger et al., 1991; Peters et al., 1992; Yu et al., 2003). Inactivation of Fgfr1 and Fgfr2 in limb mesenchyme with Prx1-Cre resulted in severe skeletal hypoplasia (Yu and Ornitz, 2008). Analysis of phenotypes in distal limb bud mesenchyme identified a role for FGFR signaling in regulating cell survival but not proliferation (Yu and Ornitz, 2008). The severity of the phenotype in the limb bud precluded analysis of embryonic or postnatal skeletal development.
Fgfr3 is expressed in proliferating and prehypertrophic chondrocytes and functions to inhibit postnatal chondrogenesis (Chen et al., 2001; Havens et al., 2008; Naski et al., 1998; Ornitz and Marie, 2015; Su et al., 2014). Loss of function of FGFR3, either globally or specifically in chondrocytes, leads to skeletal overgrowth in mice, sheep and humans (Beever et al., 2006; Colvin et al., 1996; Deng et al., 1996; Makrythanasis et al., 2014; Ornitz and Marie, 2015; Toydemir et al., 2006; Zhou et al., 2015). The inhibitory activity of FGFR3 on growth plate chondrocytes explains the pathogenic consequences of gain-of-function mutations in FGFR3 in suppressing pre-pubertal skeletal growth in achondroplasia and related chondrodysplastic disorders (Laederich and Horton, 2012; Naski et al., 1998, 1996). The signaling mechanisms by which FGFR3 suppresses chondrogenesis involve activation of STAT1, ERK1/2 (MAPK3/1) and p38 (MAPK14), increased expression of Snail1 (Snai1), decreased expression of AKT, and activation of protein phosphatase 2a (PP2a), which dephosphorylates (activates) the retinoblastoma family members p107 (RBL1) and p130 (RBL2). Activation of p107 (and p130) and increased expression of the cell cycle inhibitor p21Waf1/Cip1 (CDKN1A) function to directly suppress chondrocyte proliferation (Aikawa et al., 2001; Cobrinik et al., 1996; Dailey et al., 2003; de Frutos et al., 2007; Kolupaeva et al., 2013, 2008; Kurimchak et al., 2013; Laplantine et al., 2002; Legeai-Mallet et al., 2004; Priore et al., 2006; Raucci et al., 2004; Su et al., 1997). Although much is known about signals downstream of FGFR3 in chondrocytes, the mechanisms that regulate FGFR3 expression and activation and that coordinate osteogenesis and chondrogenesis are poorly understood.
Here we investigate cell-autonomous FGFR1 and FGFR2 signaling in the osteoprogenitor lineage. We show that inactivation of FGFR1 and FGFR2 with Osx-Cre (Rodda and McMahon, 2006) (Osx is also known as Sp7) results in decreased bone mass. Unexpectedly, we found that loss of FGFR1/2 in the osteoprogenitor lineage has a profound effect on chondrogenesis and postnatal longitudinal bone growth. The mechanism by which osteoprogenitor FGFR1/2 signaling regulates chondrogenesis involves activation of FGFR3 expression and signaling in chondrocytes through reduction in the expression of Pthlh and increased expression of Fgf9 and Fgf18, which encode ligands that normally regulate endochondral bone growth.
RESULTS
Postnatal growth defects in mice lacking Fgfr1 and Fgfr2 in the osteoprogenitor lineage
Fgfr1 and Fgfr2 are expressed in the perichondrium and periosteum during skeletal development (Yu et al., 2003). FGFR1 and FGFR2 have similar in vitro signaling potency and ligand response profiles to FGF9 and FGF18 (Zhang et al., 2006), ligands that have key roles in regulating skeletal development (Hung et al., 2016, 2007; Liu et al., 2007, 2002; Ohbayashi et al., 2002). In several tissues, including the limb bud, palate, lung, kidney, liver, cerebellum, epidermis and inner ear, Fgfr1 and Fgfr2 show significant functional redundancy (Böhm et al., 2010; Huh et al., 2015; Meyer et al., 2012; Ornitz and Itoh, 2015; Poladia et al., 2006; Sims-Lucas et al., 2011; Smith et al., 2012; White et al., 2006; Yang et al., 2010; Yu et al., 2015; Yu and Ornitz, 2008). To study the roles of FGFR signaling in the osteoprogenitor lineage, the Osx-GFP::Cre (Osx-Cre) allele was crossed to floxed alleles of Fgfr1 and Fgfr2 (Rodda and McMahon, 2006; Trokovic et al., 2003; Yu et al., 2003). Osx-Cre efficiently targets the osteoprogenitor lineage (trabecular bone and cortical bone), bone marrow stroma, a small percentage of chondrocytes, and some other non-skeletal cell types (Chen et al., 2014a; Rodda and McMahon, 2006).
Osx-Cre;Fgfr1f/f;Fgfr2f/f double conditional knockout (abbreviated here as Osx-Cre;DCKO), Fgfr1f/f;Fgfr2f/f double floxed control (abbreviated here as DFF), and Osx-Cre control mice appeared normal at birth. Body weight was not significantly different between Osx-Cre;DCKO, DFF and Osx-Cre control mice before postnatal day (P) 4 (Fig. 1A, Fig. S1A). Inactivation of Fgfr1 and Fgfr2 in the Osx-Cre lineage was confirmed by qRT-PCR evaluation of mRNA isolated from cortical bone from P21 DFF and Osx-Cre;DCKO mice (Fig. S2). Histological evaluation of embryonic day (E) 18.5 Osx-Cre;DCKO proximal tibia showed an increase in height of the hypertrophic chondrocyte zone and narrowing of the growth plate and diaphysis, but no other changes in cortical, trabecular or growth plate histology (Fig. 1B). Furthermore, bone architecture of Osx-Cre;DCKO mice, as determined by Alizarin Red and Alcian Blue staining of P0 skeletons, also showed slightly narrowed long bones, but normal mineralized regions and cartilaginous growth plates (Fig. 1C).
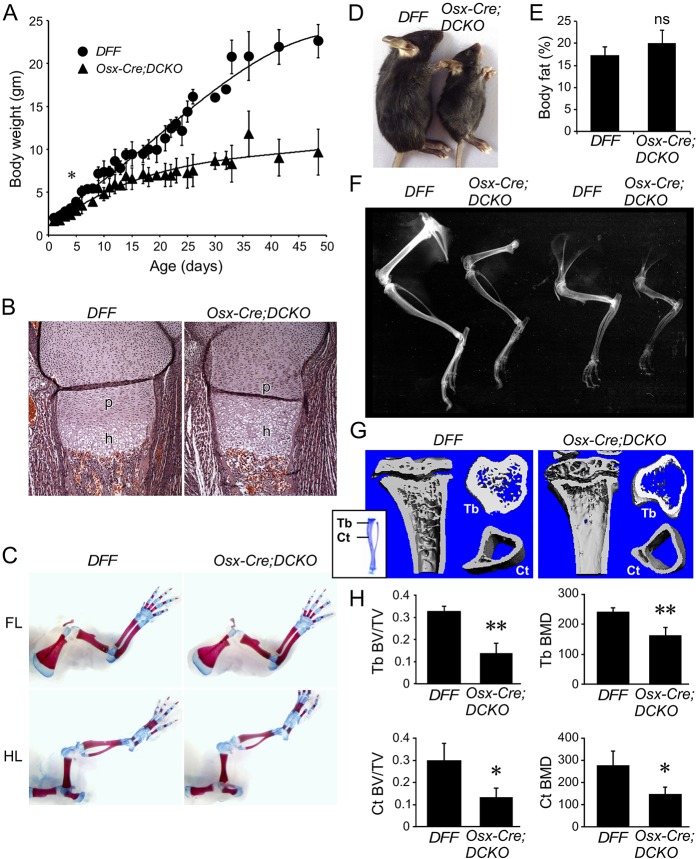
Fig. 1.
Postnatal growth defects in mice lacking Fgfr1 and Fgfr2 in the osteoprogenitor lineage. (A) Growth curve of control (DFF) and Osx-Cre;DCKO mice showing reduced growth of Osx-Cre;DCKO mice after P4. Data are pooled from 35 DFF mice and 26 Osx-Cre;DCKO mice. Not all mice were weighed at every time point. All data points have error estimation but some are not visible because they are smaller than the data point symbol. (B) Proximal tibia histology (H&E staining) at E18.5 showing a normal proliferating chondrocyte zone (p) and an expanded hypertrophic chondrocyte zone (h) in Osx-Cre;DCKO mice. (C) Alizarin Red and Alcian Blue staining of whole skeleton at P0 showing similar skeletal architecture of control and Osx-Cre;DCKO mice. FL, forelimb; HL, hindlimb. (D) Osx-Cre;DCKO mice are smaller than control (DFF) mice at P21. (E) Whole-body DEXA analysis of DFF and Osx-Cre;DCKO mice (age 24-26 days, n=4) showing normal body fat content. ns, not significant. (F) Radiographic images of hindlimb (left) and forelimb (right) of 3-month-old mice showing reduced bone density of Osx-Cre;DCKO compared with DFF control mice. (G) Micro-CT analysis at P21 showing reduced trabecular (Tb) and cortical (Ct) bone in Osx-Cre;DCKO mice. (H) Quantification of micro-CT data showing reduced ratio of cortical and trabecular bone volume to total bone volume (BV/TV) and bone mineral density (BMD) in Osx-Cre;DCKO mice (n=3). Error bars, s.d.; *P<0.05, **P<0.01.
Osx-Cre;DCKO mice failed to gain normal body weight compared with DFF or Osx-Cre control mice. This growth defect became statistically significant (P<0.05) after P4 (Fig. 1A). By 3 weeks of age, Osx-Cre;DCKO mice were approximately half normal size but otherwise healthy (Fig. 1A,D). Because Osx-Cre is active in some non-skeletal lineages, including stromal cells, adipocytes, perivascular cells in the bone marrow, olfactory glomerular cells, and a subset of gastric and intestinal epithelial cells (Chen et al., 2014a), we questioned whether inactivation of Fgfr1 and Fgfr2 with Osx-Cre could influence growth by affecting the nutritional or hormonal status of the mice. Analysis of bone density and total body fat content, using dual-energy X-ray absorptiometry (DEXA), showed a 29±2% (n=4, P<0.01) decrease in bone mineral content in Osx-Cre;DCKO compared with DFF mice, but no significant change in body fat content (Fig. 1E). Additionally, litters were placed on a high-fat, high-calorie diet at birth until 5 weeks of age. On this diet, Osx-Cre;DCKO and DFF mice both showed an elevated (19±1%, n=4, P<0.05) body fat content but Osx-Cre;DCKO mice still showed a decrease in bone mineral content (30±1%, n=4, P<0.01). We conclude that the growth defect in Osx-Cre;DCKO mice is most likely a consequence of impaired FGF signaling in Osx-Cre-targeted cell lineages within skeletal tissue and not a consequence of extrinsic hormonal or nutritional changes.
The Osx-Cre allele, by itself, has been reported to have variable effects on skeletal growth that could depend on the genetic background (Huang and Olsen, 2015; Wang et al., 2015). To evaluate a potential contribution of the Osx-Cre allele in the mixed C57BL/6J;129X1 background used in these studies, wild-type hybrid mice were compared with littermate Osx-Cre mice by following growth and by endpoint skeletal micro-CT and histological analysis. Growth curves for wild-type and Osx-Cre mice revealed a slight delay in Osx-Cre mice at P30 that normalized after P36 (Fig. S1A). Micro-CT analysis of cortical and trabecular bone showed no significant difference in the bone volume to total volume (BV/TV) ratio or in bone mineral density (BMD) between P21 wild-type and Osx-Cre mice (Fig. S1B,C). Growth plate histology and Fgfr3 expression at P21 were also similar between wild-type and Osx-Cre mice (Fig. S1D,E). These studies show that the Osx-Cre allele has a minimal effect on bone growth in the genetic background used in these studies.
Decreased bone formation in Osx-Cre;DCKO mice
Radiographic analysis of intact skeletons of 3-month-old mice revealed that Osx-Cre;DCKO mice had shorter bones and reduced bone density compared with control mice (Fig. 1F). The overall shape of the bones was normal. Micro-CT analysis of intact long bones (femur, tibia) revealed that the Osx-Cre;DCKO mice had reduced trabecular and cortical bone (Fig. 1G). This was reflected in a significantly reduced trabecular and cortical BV/TV ratio and BMD (Fig. 1H). Consistent with the micro-CT analysis, von Kossa-stained histological sections of P21 tibia revealed a reduced area of mineralized cortical bone, trabecular bone (primary spongiosa), and secondary ossification centers in Osx-Cre;DCKO mice (Fig. 2A). Although Osx-Cre;DCKO mice clearly have less mineralized trabecular and cortical bone and thus decreased numbers of osteoblasts, histological analysis of the trabecular region revealed normal osteoblast density and a similar intensity of type I collagen (Col1) expression in osteoblasts (Fig. 2B,C). Consistent with this, histomorphometric analysis revealed a normal number of osteoblasts (N.Ob) and osteoblast surface area (Ob.S) when normalized to bone surface area (Fig. 2D).
Fig. 2.
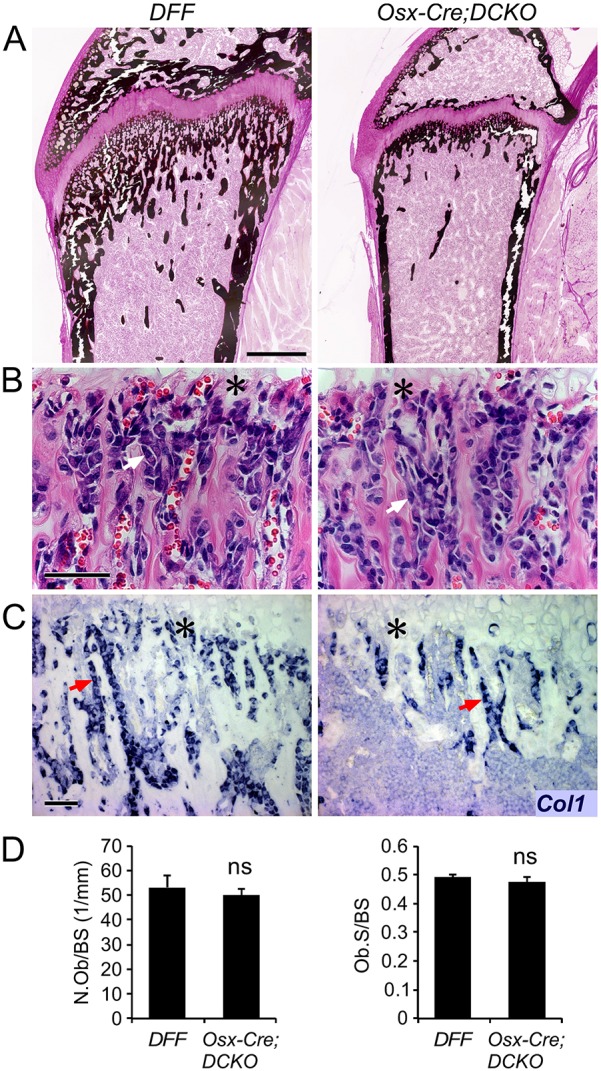
Decreased cortical and trabecular bone formation in Osx-Cre;DCKO mice. (A) Histology of the proximal tibia at P21, showing reduced mineralized bone (von Kossa stain) in Osx-Cre;DCKO mice. (B) Histology (H&E staining) showing normal osteoblast morphology in the trabecular region adjacent to the chondro-osseous junction (asterisk) in Osx-Cre;DCKO mice. (C) Type I collagen (Col1) expression detected by in situ hybridization in DFF and Osx-Cre;DCKO mice. (D) Histomorphometry of DFF and Osx-Cre;DCKO mice (n=3) showing normal osteoblast number per bone surface (BS) area and normal osteoblast surface per bone surface. Arrows (B,C) indicate osteoblasts. ns, non significant. Scale bars: A, 500 µm; B,C, 50 µm.
Decreased growth plate size in Osx-Cre;DCKO mice
Growth plate histology of P21 Osx-Cre;DCKO mice compared with DFF controls showed a significant decrease in the overall length of the growth plate and the length of the proliferating (columnar) chondrocyte zone (24% and 36%, respectively; P<0.02) (Fig. 3A,B). At this stage of postnatal development, the hypertrophic chondrocyte zone, which was expanded at E18.5, was not significantly different from that of controls. Normalization of the length of the hypertrophic zone could be due to compensatory changes in the number of available input cells (assessed by chondrocyte proliferation) and changes in the distal loss of hypertrophic chondrocytes through apoptosis, degradation of the extracellular matrix, or differentiation into trabecular osteoblasts.
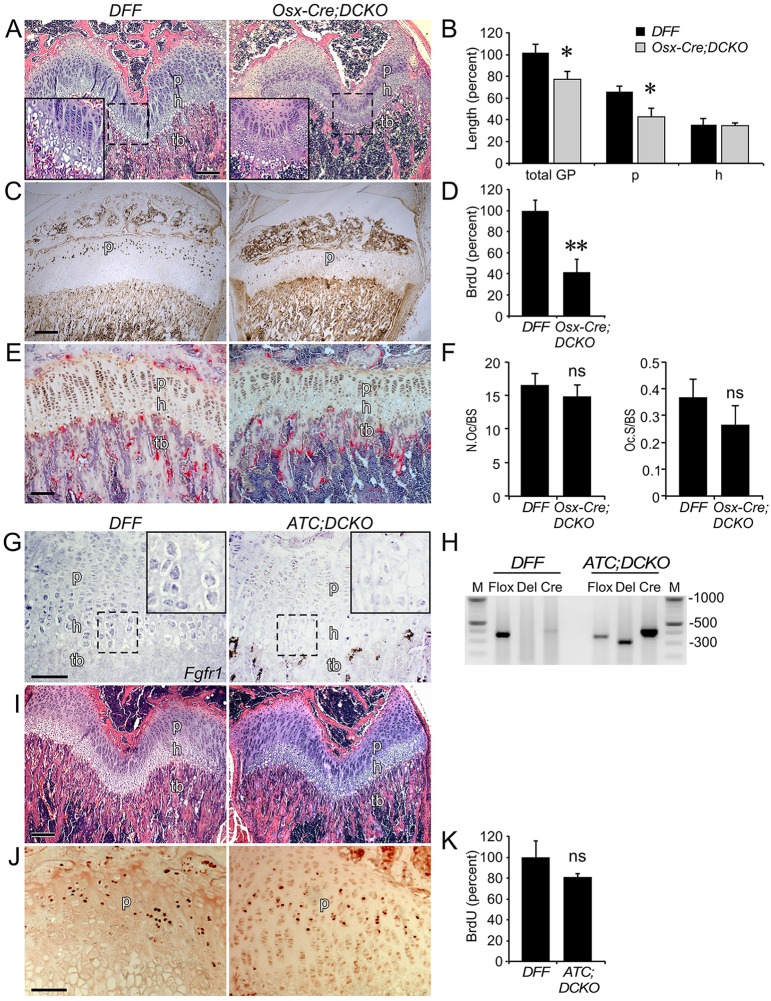
Fig. 3.
Decreased growth plate size and decreased chondrocyte proliferation in Osx-Cre;DCKO mice. (A) Histology (H&E staining) of the distal femur showing smaller growth plate and reduced trabecular bone thickness in P21 Osx-Cre;DCKO compared with DFF mice. Inset, 2× magnification of boxed region. (B) Growth plate measurements showing reduced total growth plate and proliferative zone length, and normal hypertrophic zone length, in P21 Osx-Cre;DCKO compared with DFF mice. Data are normalized to the total growth plate height of DFF mice (n=3). (C) BrdU immunohistochemistry showing reduced chondrocyte proliferation in Osx-Cre;DCKO compared with DFF mice. (D) Quantification of BrdU-labeled cells in the proliferating chondrocyte zone of DFF and Osx-Cre;DCKO growth plates (n=6), expressed as percent of control. (E) TRAP staining (red) of DFF and Osx-Cre;DCKO proximal tibia. (F) Histomorphometric analysis showing no difference in osteoclast number per bone surface, and normal osteoclast surface per bone surface, of DFF and Osx-Cre;DCKO mice. (G) Expression of Fgfr1, assessed by in situ hybridization, in the proximal tibia showing reduced expression in hypertrophic chondrocytes in ATC;DCKO compared with DFF mice. Inset, 2× magnification of boxed region. (H) Confirmation of deletion of Fgfr1 in the growth plate of ATC;DCKO mice. Flox, unrecombined Fgfr1 flox allele; Del, Fgfr1 deleted allele; Cre, Cre recombinase allele; M, markers (bp). (I) Histology (H&E staining) of distal femur showing similar growth plate size in DFF and ATC;DCKO mice. (J) BrdU immunohistochemistry showing no difference in chondrocyte proliferation in DFF and ATC;DCKO growth plate. (K) Quantification of BrdU-labeled cells in the proliferating chondrocyte zone of DFF and ATC;DCKO growth plates (n=3), expressed as percent of control. p, proliferating chondrocytes; h, hypertrophic chondrocytes; tb, trabecular bone; GP, growth plate. Error bars, s.d.; *P<0.02, **P<0.001. ns, not significant. Scale bars: A,C,I, 200 µm; E,G,J, 100 µm.
Chondrocyte proliferation in P21 mice was evaluated by BrdU labeling. Osx-Cre;DCKO mice showed a 58% reduction in chondrocyte proliferation (Fig. 3C,D). Cell death, as evaluated by activated caspase 3 immunostaining, was decreased in distal hypertrophic chondrocytes in Osx-Cre;DCKO mice (Fig. S3A,B), and matrix degradation potential, as evaluated by measuring osteoclast number (N.Oc) and osteoclast surface (Oc.S) per bone surface area, did not significantly differ between DFF and Osx-Cre;DCKO mice (Fig. 3E,F). Collectively, these data suggest that normalization of the hypertrophic chondrocyte zone in P21 Osx-Cre;DCKO mice results from decreased chondrocyte proliferation that is partially compensated for by decreased cell death in distal hypertrophic chondrocytes.
Decreased chondrocyte proliferation is due to effects of non-cell-autonomous loss of Fgfr1 and Fgfr2
Given that Osx-Cre targets a small percentage of prehypertrophic and hypertrophic chondrocytes (Chen et al., 2014a) and that Fgfr1 is expressed in hypertrophic chondrocytes, it was necessary to determine whether inactivation of Fgfr1 (and Fgfr2) in growth plate chondrocytes could contribute to the observed decrease in chondrocyte proliferation. The aggrecan enhancer-driven, tetracycline-inducible Cre (ATC) transgene allele, which efficiently targets proliferating and hypertrophic chondrocytes during embryonic development (Dy et al., 2012), was used to inactivate floxed alleles of Fgfr1 and Fgfr2. Female mice carrying ATC;Fgfr1f/f;Fgfr2f/f (ATC;DCKO) embryos were placed on doxycycline throughout gestation and pups were maintained on doxycycline until P21. In situ hybridization shows Fgfr1 expression in hypertrophic chondrocytes in DFF control mice and decreased expression in ATC;DCKO mice (Fig. 3G). PCR analysis of isolated growth plates from P21 mice demonstrated inactivation of Fgfr1 (Fig. 3H). However, at P21, DFF control mice and ATC;DCKO mice were of similar weight and showed no difference in growth plate histology (Fig. 3I) or chondrocyte proliferation (Fig. 3J,K). We conclude from these data that FGFR1 (and FGFR2, which is not expressed in chondrocytes) does not have a major cell-autonomous impact on embryonic or postnatal chondrogenesis.
Increased expression of Fgf9 and Fgf18 in Osx-Cre;DCKO mice
We hypothesized that inactivation of Fgfr1 and Fgfr2 in the Osx-Cre lineage could lead to a compensatory upregulation of Fgf9 or Fgf18, which encode ligands that are each necessary for normal embryonic skeletal development (Hung et al., 2007; Liu et al., 2007, 2002; Ohbayashi et al., 2002) and together display marked redundancy in skeletal development (Hung et al., 2016). Because FGF9 and FGF18 are also thought to function as ligands that signal to FGFR3 during postnatal bone growth to negatively regulate chondrocyte proliferation, compensatory upregulation of Fgf9 or Fgf18 expression due to loss of FGFR1/2 signaling in the osteoprogenitor lineage could aberrantly activate FGFR3 in the growth plate and suppress chondrocyte proliferation. To test this hypothesis, we performed in situ hybridization analysis of paraffin-fixed intact bone tissues and qRT-PCR on distal bone tissue from DFF and Osx-Cre;DCKO mice. In situ analysis revealed that Fgf9 expression was induced in perichondrial tissue, adjacent connective tissue, reserve, proliferating and prehypertrophic chondrocytes of Osx-Cre;DCKO mice (Fig. 4A). Consistent with the in situ expression data, qRT-PCR analysis of distal bone tissue showed a ∼3.5-fold increase in Fgf9 expression in tissue from Osx-Cre;DCKO compared with DFF mice (Fig. 4B). Analysis of Fgf18 by in situ hybridization showed increased expression in reserve, proliferating and prehypertrophic chondrocytes in Osx-Cre;DCKO compared with DFF mice (Fig. 4C). Consistent with these data, qRT-PCR showed a ∼1.5-fold increase in Fgf18 expression in Osx-Cre;DCKO compared with DFF distal bone tissue (Fig. 4D).
Fig. 4.
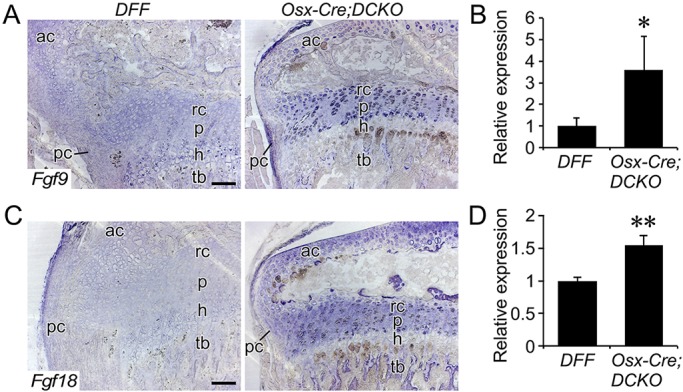
Increased expression of Fgf9 and Fgf18 in Osx-Cre;DCKO mice. (A) Expression of Fgf9, assessed by in situ hybridization, in proximal tibia of P21 mice showing increased expression in the perichondrium, reserve, proliferating and prehypertrophic chondrocytes of Osx-Cre;DCKO mice. (B) qRT-PCR analysis of Fgf9 expression in DFF and Osx-Cre;DCKO proximal tibia metaphysis (n=3). (C) Expression of Fgf18, assessed by situ hybridization, in P21 proximal tibia showing increased expression in articular cartilage, proliferating and prehypertrophic chondrocytes in the growth plate and in trabecular bone of Osx-Cre;DCKO mice. (D) qRT-PCR analysis of Fgf18 expression in DFF and Osx-Cre;DCKO proximal tibia metaphysis (n=3). rc, reserve chondrocytes; p, proliferating chondrocytes; h, hypertrophic chondrocytes; ac, articular chondrocytes; tb, trabecular bone; pc, perichondrium. Error bars, s.d.; *P<0.05, **P<0.002. Scale bars: 100 µm
Increased Fgfr3 expression and signaling in Osx-Cre;DCKO growth plate
In situ hybridization revealed a striking increase in Fgfr3 expression in Osx-Cre;DCKO compared with DFF mice in both proliferating and prehypertrophic chondrocytes (Fig. 5A). This increase was confirmed by qRT-PCR analysis of distal bone tissue from P21 distal femur and proximal tibia (Fig. 5B). The Snail1 transcription factor is induced by FGFR3 and is required for the activation of both the STAT1 and MAPK branches of the FGFR3 signaling pathway (de Frutos et al., 2007). Consistent with increased FGFR3 expression and signaling, Snail1 expression was strongly increased in Osx-Cre;DCKO compared with DFF mice (Fig. 5C). Immunostaining for the chondrocyte-specific transcription factor SOX9 showed mildly elevated levels of expression in Osx-Cre;DCKO compared with DFF mice (Fig. 5D).
Fig. 5.
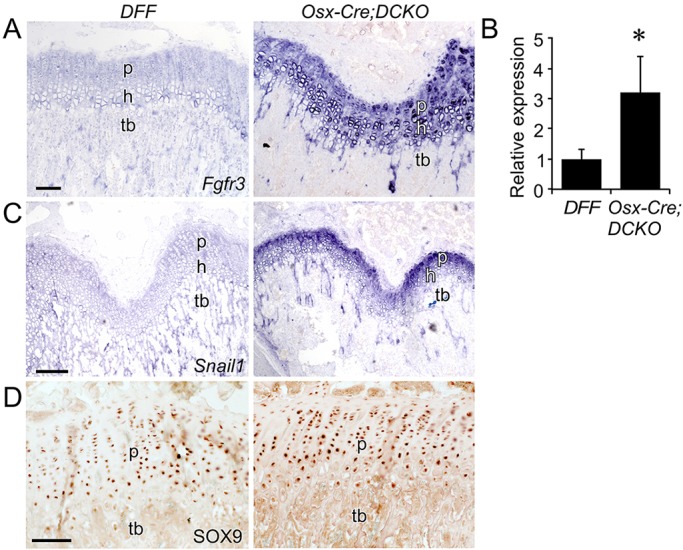
Increased Fgfr3 expression and signaling in Osx-Cre;DCKO mice. (A,C) Expression, assessed by in situ hybridization, of Fgfr3 (A) and Snail1 (C) in DFF and Osx-Cre;DCKO distal femur. (B) qRT-PCR analysis of Fgfr3 expression in DFF and Osx-Cre;DCKO proximal tibia and distal femur metaphysis (n=3). (D) Immunohistochemistry of DFF and Osx-Cre;DCKO proximal tibia showing mildly increased SOX9 in Osx-Cre;DCKO compared with DFF mice. p, proliferating chondrocytes; h, hypertrophic chondrocytes; tb, trabecular bone. Error bars, s.d.; *P<0.05. Scale bars: 100 µm.
Activation of FGF9 in the perichondrium suppresses chondrocyte proliferation
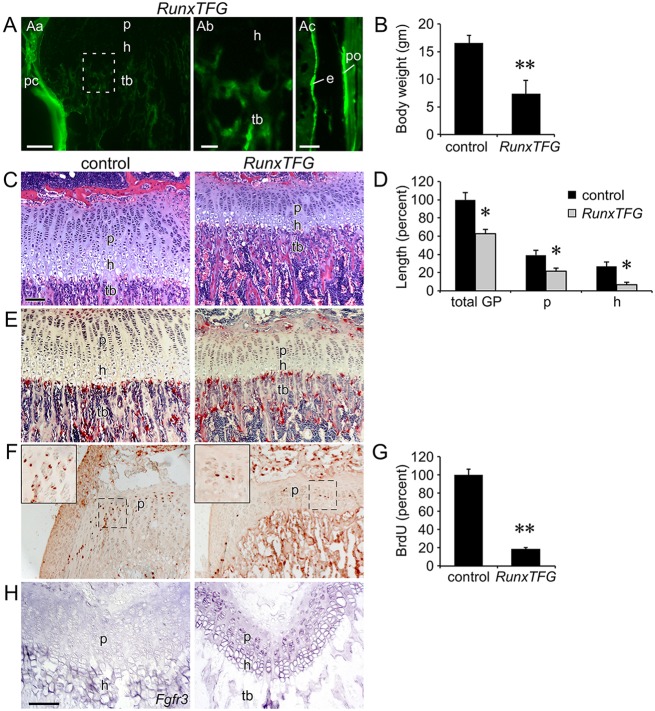
The ability of FGF9 to signal from perichondrial tissue to growth plate chondrocytes has been inferred from phenotypes seen in Fgf9−/− embryos (Hung et al., 2007). Additionally, transgenic mice that overexpressed FGF9 in chondrocytes (Col2a1-Fgf9) showed short limbs and a smaller growth plate and died by 5 weeks of age (Garofalo et al., 1999). However, whether FGF9 has the capacity to signal from periosteal and trabecular osteoblasts to growth plate chondrocytes during prepubertal growth was not known. To conditionally overexpress Fgf9 in periosteal and trabecular osteoblasts, Runx2-rtTA (Chen et al., 2014b) and TRE-Fgf9-ires-eGFP (White et al., 2006) transgenic mice were mated to generate biallelic Runx2-rtTA;TRE-Fgf9-ires-eGFP (RunxTFG) mice. In the presence of doxycycline, GFP fluorescence was observed in the perichondrium, periosteum and trabecular bone of RunxTFG mice, but not in proliferating or hypertrophic chondrocytes (Fig. 6A). Compared with control (single-transgenic mouse), RunxTFG transgenic mice showed a significantly (P<0.01) reduced body weight at P21 (Fig. 6B). Growth plate histology revealed that, compared with the control, RunxTFG transgenic mice had significantly (P<0.01) smaller proliferating and hypertrophic chondrocyte zones (Fig. 6C,D). The height of the trabecular zone in RunxTFG transgenic mice was reduced but otherwise histologically normal, and osteoclast numbers and morphology appeared normal (Fig. 6C,E). Most notably, chondrocyte proliferation was significantly (P<0.001) reduced in RunxTFG transgenic mice compared with the control (Fig. 6F,G). Finally, in situ hybridization revealed that activation of Fgf9 in the perichondrium/periosteum and trabecular bone induced the expression of Fgfr3 in proliferating chondrocytes (Fig. 6H).
Fig. 6.
Activation of Fgf9 in the perichondrium suppresses chondrocyte proliferation. (A) Fluorescence imaging of induced GFP expression in trabecular bone (tb), cortical bone (e, endosteum; po, periosteum) and perichondrium (pc) of RunxTFG mice. GFP was not observed in hypertrophic chondrocytes (h). The boxed region in Aa is magnified 2x in Ab; Ac shows GFP expression in endosteal and periosteal cortical bone. (B) Decreased body weight of P21 RunxTFG mice (n=3) compared with Runx2-rtTA single-transgenic control (n=4). (C) Histology (H&E staining) of the proximal tibia showing a smaller growth plate in P21 RunxTFG compared with Runx2-rtTA single-transgenic control. (D) Growth plate measurements showing reduced total growth plate, proliferative zone and hypertrophic zone size in P21 RunxTFG mice. (E) TRAP staining of P21 control and RunxTFG mice showing normal osteoclast number. (F) BrdU immunohistochemistry showing reduced chondrocyte proliferation in RunxTFG compared with control P21 mice. (G) Quantification of BrdU-labeled cells in the proliferating chondrocyte zone of P21 control and RunxTFG growth plates (n=3). (H) Expression of Fgfr3, assessed by in situ hybridization, in P21 control and RunxTFG distal femur. rc, reserve chondrocytes; p, proliferating chondrocytes; h, hypertrophic chondrocytes; tb, trabecular bone. Error bars, s.d.; *P<0.01, **P<0.001. Scale bars: Aa,Ac,C-H, 100 µm; Ab, 20 µm.
PTHLH links Osx-Cre lineage FGFR1/2 signaling to Fgfr3 expression and chondrocyte proliferation in the postnatal growth plate
Indian hedgehog (IHH) and parathyroid hormone-like peptide (PTHLH) are crucial regulators of endochondral bone growth (Kozhemyakina et al., 2015; Long and Ornitz, 2013). IHH stimulates chondrocyte proliferation and Pthlh expression, while PTHLH suppresses chondrocyte maturation and Ihh expression. Because we have observed apparent non-cell-autonomous effects of loss of Osx-Cre lineage FGFR1 and FGFR2 on chondrocyte growth, it was important to examine the potential activity of other signaling pathways that regulate growth plate function. Compared with controls, Ihh was decreased in the P21 growth plate of Osx-Cre;DCKO mice (Fig. 7A,B). Interestingly, we found that Pthlh expression was also reduced in reserve chondrocytes in Osx-Cre;DCKO mice (Fig. 7C). qRT-PCR analysis of distal bone tissue showed an overall reduction in Pthlh mRNA (Fig. 7D). Consistent with FGFR3 signaling suppressing Ihh-Pthlh expression (Chen et al., 2001; Li et al., 2010; Minina et al., 2002; Naski et al., 1998), in mice induced to overexpress Fgf9, expression of Pthlh was reduced in reserve zone chondrocytes (Fig. 7E).
Fig. 7.
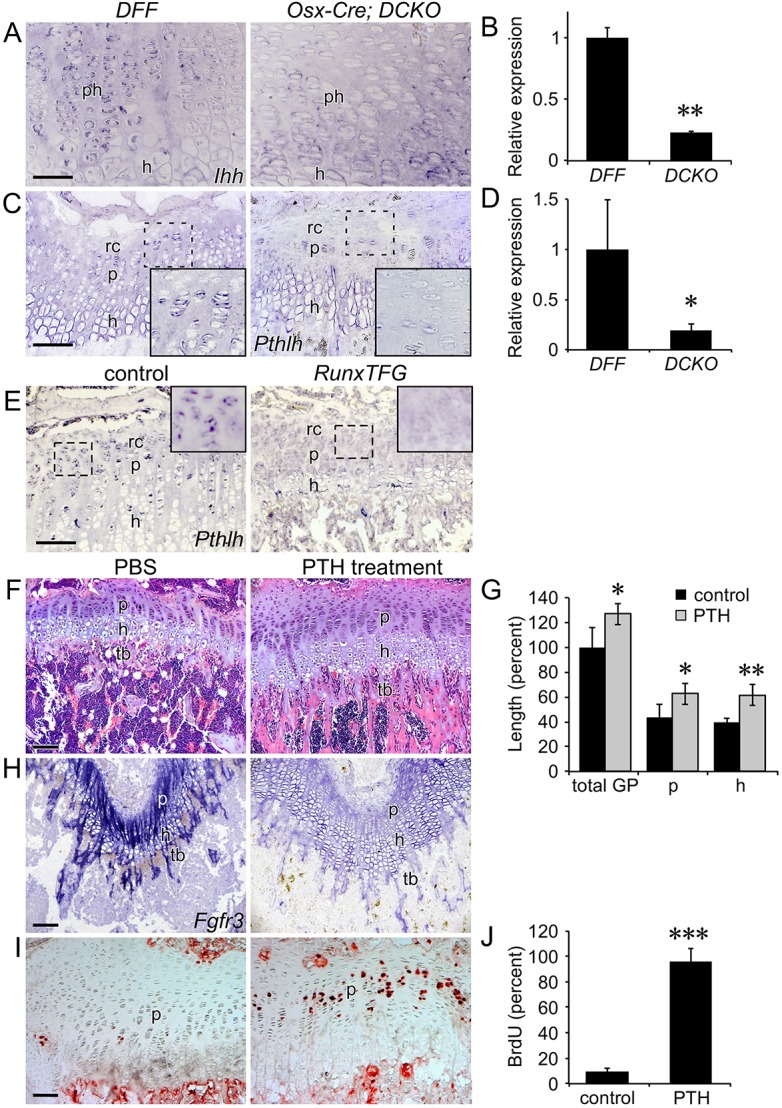
Rescue of the Osx-Cre;DCKO growth plate phenotype by administration of PTH(1-34). (A) Expression of Ihh, assessed by in situ hybridization, in P21 distal femur showing decreased expression in the growth plate of Osx-Cre;DCKO mice. (B) qRT-PCR analysis of Ihh expression in DFF and Osx-Cre;DCKO proximal tibia metaphysis (n=3). (C) Expression of Pthlh, assessed by in situ hybridization, in P21 distal femur showing decreased expression in the peripheral growth plate in Osx-Cre;DCKO mice. Inset, 2× magnification. (D) qRT-PCR analysis of Pthlh expression in DFF and Osx-Cre;DCKO proximal tibia metaphysis (n=3). (E) Expression of Pthlh in P21 control and RunxTFG proximal tibia. (F) Histology (H&E staining) of the proximal tibia showing a larger growth plate and increased trabecular bone in P21 PTH-treated compared with PBS-treated (control) Osx-Cre;DCKO mice. (G) Growth plate measurements showing increased total growth plate, proliferative and hypertrophic zone size in PTH-treated (n=3) compared with PBS-treated (n=4) mice. (H) Expression of Fgfr3, assessed by in situ hybridization, in the distal femur of P21 PTH-treated compared with PBS-treated Osx-Cre;DCKO mice. (I) BrdU immunohistochemistry showing increased chondrocyte proliferation in P21 PTH-treated compared with PBS-treated Osx-Cre;DCKO mice. (J) Quantification of BrdU-labeled cells in the proliferating chondrocyte zone of PTH-treated compared with PBS-treated Osx-Cre;DCKO mice (n=3). rc, reserve chondrocytes; p, proliferating chondrocytes; ph, prehypertrophic chondrocytes; h, hypertrophic chondrocytes; tb, trabecular bone. Error bars, s.d.; *P<0.05, **P<0.005, ***P<0.001. Scale bars: A,C, 50 µm; E,F,H,I, 100 µm.
Analysis of Fgfr3 promoter function in vitro shows that Fgfr3 expression could be directly regulated (suppressed) by PTHLH activation of protein kinase A (PKA) (McEwen et al., 1999). To test whether parathyroid hormone (PTH) signaling could suppress Fgfr3 expression in vivo in Osx-Cre;DCKO mice that highly overexpress Fgfr3, Osx-Cre;DCKO mice were injected intermittently (daily) with PTH [PTH(1-34) peptide] from P15 to P21, a treatment regimen known to stimulate the anabolic effects of PTH signaling on bone (Esen et al., 2015; Xie et al., 2012). Compared with control Osx-Cre;DCKO mice that were only injected with PBS, PTH-injected Osx-Cre;DCKO mice showed an increase in the size of the growth plate, increased thickness of trabecular bone, decreased expression of Fgfr3, and increased chondrocyte proliferation (Fig. 7F-J).
DISCUSSION
The growth plate is a transient component of developing endochondral bone that mediates longitudinal bone growth from late stages of embryonic development through puberty (Hunziker and Schenk, 1989; Noonan et al., 1998). FGFR3 is a well-established negative regulator of postnatal bone growth, functioning in the growth plate in proliferating and prehypertrophic chondrocytes. Activating mutations in FGFR3 are responsible for achondroplasia, the most common form of dwarfism in humans (Horton et al., 2007; Ornitz and Marie, 2015). As signaling pathways that function downstream of FGFR3 are well established (Ornitz and Itoh, 2015), the identification of non-cell-autonomous mechanisms that regulate FGFR3 expression and signaling and postnatal growth plate function are essential for further elucidating the complex regulatory networks that control endochondral bone formation.
Inactivation of Fgfr1 and Fgfr2 in the Osx-Cre lineage disrupted a non-cell-autonomous feedback loop, resulting in activation of FGFR3 signaling in growth plate chondrocytes and suppression of chondrocyte proliferation and longitudinal bone growth (Fig. 8). The precise cell type(s) that maintains this feedback loop is not known; however, it is likely to be an immature osteoprogenitor, as similar phenotypes are not observed when Fgfr1 and Fgfr2 are inactivated in mature osteoblasts with the OC-Cre allele (our unpublished data). A likely early event eliciting this phenotype is increased expression of Fgf9 in osteoprogenitor cells in the perichondrium, resulting in increased signaling through FGFR3 in adjacent chondrocytes. Activation of FGFR3 inhibits Ihh expression and signaling in prehypertrophic chondrocytes (Naski et al., 1998), a factor that is required to maintain Pthlh expression in reserve and articular chondrocytes (Hilton et al., 2005; Koziel et al., 2005; St-Jacques et al., 1999; Vortkamp et al., 1996). Propagating events include increased Fgfr3 expression and signaling in the growth plate, which may further suppress Ihh and Pthlh and increase Fgf9 and Fgf18 expression. This non-cell-autonomous signaling pathway thus coordinates osteoprogenitor development and longitudinal bone growth.
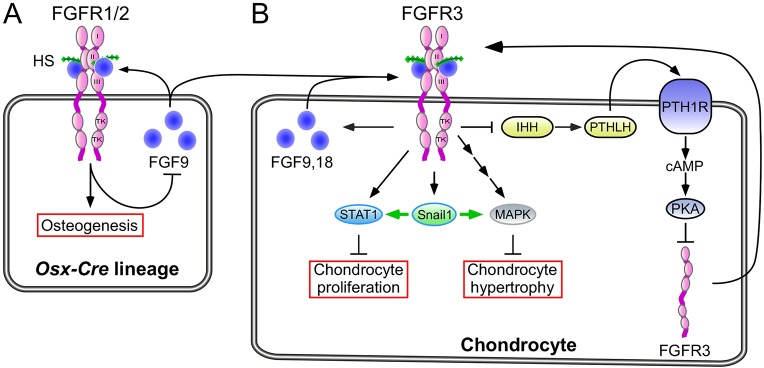
Fig. 8.
Model of FGF-regulated interactions between osteoprogenitor lineages and growth plate chondrocytes in postnatal endochondral bone growth. (A) FGFR1 and FGFR2 in the osteoprogenitor lineage are regulated by FGF9 expressed in osteoprogenitors and adjacent connective tissue and periosteum. (B) Inactivation of FGFR1 and FGFR2 results in compensatory increased expression of Fgf9, which aberrantly activates FGFR3 and downstream Snail1 to suppress chondrocyte proliferation and hypertrophy. Increased FGFR3 signaling also promotes Fgf9 and Fgf18 expression in chondrocytes and suppresses expression of Ihh and Pthlh. PTHLH functions to suppress Fgfr3 expression, and reduced Pthlh contributes to increased Fgfr3 expression. The aberrant activation of FGFR3 (expression and signaling in chondrocytes) might initiate a feed-forward signaling loop in chondrocytes that functions to terminate chondrogenesis. HS, heparan sulfate; TK, tyrosine kinase domain; I,II,III, immunoglobulin-like domain.
FGFR1/2 function in the osteoprogenitor lineage
Although FGFR1 and FGFR2 signaling have robust functions in limb bud mesenchyme, the effect of disrupting their function in the osteoprogenitor lineage during embryonic development is surprisingly mild. Osx-Cre;DCKO mice were born alive and showed no patterning defects in the appendicular skeleton. However, Osx-Cre;DCKO mice exhibited a calvarial ossification defect at birth (data not shown) and a postnatal reduction in cortical bone growth, which indicates that osteoprogenitor lineage FGFR signaling is required for osteoblast growth and maturation that is independent of chondrogenesis. The precise role of FGFR signaling in osteoblasts will require further investigation.
FGFR signaling in osteoprogenitor cells indirectly affects growth plate activity
The most striking feature of Osx-Cre;DCKO mice is the profound reduction in chondrocyte proliferation and longitudinal bone growth. We posited that this phenotype resulted from non-cell-autonomous changes in chondrocytes that are secondary to loss of FGFR1 and FGFR2 signaling in osteoprogenitor cells. Because Osx-Cre targets a small percentage of chondrocytes (Chen et al., 2014a), the possibility remained that the observed phenotype could result from inactivation of Fgfr1 and Fgfr2 in chondrocytes. However, this is unlikely because Fgfr1 expression is restricted to hypertrophic chondrocytes and Fgfr2 is not expressed in proliferating or hypertrophic chondrocytes. Nevertheless, to rule out cell-autonomous effects of FGFR1 and FGFR2 in chondrocytes, these genes were inactivated specifically in chondrocytes using the ATC allele. The normal development of ATC;DCKO mice demonstrated that inactivation of Fgfr1 and Fgfr2 in proliferating and hypertrophic chondrocytes does not significantly affect chondrogenesis or prepubertal longitudinal bone growth.
A second feature of the Osx-Cre;DCKO phenotype is the prominent increase in Fgfr3 expression in proliferating and hypertrophic chondrocytes. In vitro analysis of the Fgfr3 promoter identified a regulatory sequence that results in decreased promoter activity in response to cAMP (McEwen et al., 1999). These in vitro data suggested that the observed decrease in Pthlh expression could contribute to increased Fgfr3 expression. In support of this model, intermittent injection of Osx-Cre;DCKO mice with PTH(1-34) peptide suppressed Fgfr3 expression in chondrocytes and increased chondrocyte proliferation (Fig. 7).
A third feature of the Osx-Cre;DCKO phenotype is reduced bone volume and density. This could result from cell-autonomous effects of FGFR signaling in osteoblasts, or be due to the reduced levels of Pthlh expression. Haploinsufficiency of Pthlh results in osteopenia in mice (Miao et al., 2005), with similar morphologies to Osx-Cre;DCKO mice.
Regulation of embryonic versus postnatal growth plate
The experiments presented here focus on the postnatal growth plate of 21-day-old Osx-Cre;DCKO mice. Although the Osx-Cre allele used to target Fgfr1 and Fgfr2 is active as early as E12.5 (Ono et al., 2014; Rodda and McMahon, 2006), the embryonic phenotype appears to be limited to expansion of the hypertrophic chondrocyte zone, similar to the phenotype observed when the Col2-Cre allele was used to inactivate Fgfr1 (Jacob et al., 2006). Thus, FGFR1/2 signaling either does not have a major role in the osteoprogenitor lineage prior to the establishment of a secondary ossification center and formation of a mature growth plate, or the non-cell-autonomous mechanism that we identified is not activated during embryonic development. Most studies investigating skeletal development focus on the embryonic growth plate. However, the postnatal growth plate is the developmental structure that accounts for the majority of organismal skeletal growth and, yet, gene expression patterns and the molecular and cellular mechanisms that regulate the postnatal growth plate are poorly defined.
In the embryonic growth plate, IHH is involved in a feedback loop that regulates Pthlh expression in the distal periarticular perichondrium (Kronenberg, 2003). However, in postnatal bone there is a reorganization of the growth plate, Pthlh expression shifts to reserve zone chondrocytes, and IHH signaling (GLI1) and Pth1r expression remain prominent in reserve/proliferating and prehypertrophic chondrocytes, respectively (Chau et al., 2011; Chen et al., 2008; Koziel et al., 2004). Thus, in the postnatal growth plate, PTHLH- and IHH-responsive cells overlap with Fgfr3 expression patterns.
Loss of FGFR1/2 signaling in perichondrial and osteoprogenitor cells might disrupt growth plate homeostasis by initially triggering increased expression of FGF9 and FGF18. We posit that these events lead to increased FGFR3 expression and signaling (modeled by forced expression of Fgf9 in perichondrial cells and osteoblasts). Secondarily, increased FGFR3 signaling could suppress Ihh expression and signaling and lead to the suppression of Pthlh in chondrocytes, in turn leading to an aberrant feed-forward signal that further increases the expression of Fgfr3 (Fig. 8). The ability to block this feed-forward loop by administration of PTH(1-34) supports a model in which PTHLH regulates communication between osteoprogenitors, chondroprogenitors and growth plate chondrocytes in a mature postnatal growth plate.
Termination of skeletal growth
Osx-Cre;DCKO mice show increased expression of Fgf9 and Fgf18 in reserve, proliferating and prehypertrophic chondrocytes and in cells at the periphery of the growth plate that may include chondroprogenitors in the groove of Ranvier. This might represent an amplification of a normal feed-forward induction of Fgf9 and Fgf18 that could function to permanently suppress growth plate chondrocyte proliferation at puberty and suppress articular chondrocyte proliferation and differentiation in adults. This model is consistent with the continued expression of endogenous Fgf18 in the postnatal growth plate and perichondrium and in adult articular chondrocytes (Ellsworth et al., 2002; Lazarus et al., 2007; Mori et al., 2014).
MATERIALS AND METHODS
Mice
Mice were housed in a pathogen-free facility and handled in accordance with standard use protocols, animal welfare regulations, and the NIH Guide for the Care and Use of Laboratory Animals. All protocols were approved by the Washington University Animal Studies Committee. Osx-GFP::Cre (Osx-Cre) (Rodda and McMahon, 2006), Fgfr1f/f (Trokovic et al., 2003), Fgfr2f/f (Yu et al., 2003), aggrecan enhancer-driven, tetracycline-inducible Cre (ATC) (Dy et al., 2012), Runx2-rtTA (Chen et al., 2014b) and TRE-Fgf9-ires-eGFP (White et al., 2006) have been described previously.
Homozygous floxed alleles of Fgfr1 and Fgfr2 were maintained as double floxed mice (DFF) and outbred to hybrid C57BL/6J;129X1 mice every second generation and then backcrossed to homozygosity. Double conditional knockout breeding males (Osx-Cre;Fgfr1f/f;Fgfr2f/f) were generated by crossing Osx-Cre mice with DFF mice, backcrossing to DFF and suppressing the Cre activity of Osx-Cre with doxycycline. To inactivate Fgfr1/2 in the osteoprogenitor lineage, DFF female mice were crossed with Osx-Cre;DCKO breeder male mice resulting in a 50% yield of Osx-Cre;DCKO mice and DFF controls. Osx-Cre control mice were generated by crossing Osx-Cre mice to wild-type hybrid mice. A similar breeding strategy was used to generate ATC;DCKO mice. To express Fgf9 in the osteoblast lineage, Runx2-rtTA mice (Chen et al., 2014b) were crossed to TRE-Fgf9-ires-eGFP (White et al., 2006) to generate RunxTFG double-transgenic mice. Females were induced with doxycycline chow (Bio-Serv, S3888; 200 mg/kg green pellets) from E0 to P21. High-fat, high-calorie diet included breeder chow (PicoLab, Mouse Diet 20) supplemented with Nutri-Cal (Patterson Veterinary Supply) from birth to 5 weeks of age.
Body weights were measured for multiple litters two to three times per week until animals were sacrificed for analysis. Growth curves represent cumulative pooled data from multiple litters and overlapping time points covering the entire timecourse.
Histology, immunohistochemistry and immunofluorescence
For histological analysis of long bones, intact femur and tibia were isolated, fixed in 4% PFA/PBS overnight at 4°C or fixed in 10% buffered formalin overnight at room temperature. Bones were rinsed in water several times and decalcified in 14% EDTA/PBS for 2 weeks. Paraffin-embedded tissue sections (5 µm) were stained with Hematoxylin and Eosin (H&E), tartrate-resistant acid phosphatase (TRAP), von Kossa or Alizarin Red.
For immunohistochemistry, paraffin sections or cryosections were rehydrated and treated with 0.3% hydrogen peroxide in methanol for 15 min to suppress endogenous peroxidase activity. Antigen retrieval was achieved by microwaving the sections in 10 mM citrate buffer (pH 6.0) for 10 min followed by gradual cooling to room temperature. Sections were incubated overnight at 4°C with the following primary antibodies: anti-SOX9 (Millipore, AB5535, rabbit polyclonal; 1:100), anti-active caspase 3 (BD Pharmingen, 559565; 1:100). Secondary antibody was Alexa Fluor 488 donkey anti-rabbit (Life Technologies, A-21206; 1:1000). Colorimetric detection used the ABC Kit (Invitrogen, 95-9943). Immunofluorescence imaging was performed on a Zeiss Apotome fluorescence microscope. Data are representative of at least three independent experiments.
For in situ hybridization analysis, tissues were fixed and decalcified at 4°C. For frozen sections, the tissues were fixed as described above and decalcified for 3 days, transferred to 30% sucrose (Sigma, S0389) for 24 h, embedded in OCT compound (Tissue-Tek), sectioned at 5 µm and stored at −20°C until analysis. Non-radioactive in situ hybridization was performed as previously described (Naski et al., 1998). In situ probes: Fgf9 (Colvin et al., 1999), Fgf18 (Liu et al., 2002), Fgfr3 (Peters et al., 1993), Snail1 (Vega et al., 2004), Pthlh (Lee et al., 1996; Long et al., 2001), Ihh (Bitgood and McMahon, 1995) and Col1 (Rossert et al., 1995). Data are representative of at least three independent experiments. Where necessary, image adjustments (to brightness/contrast) were made equally to allow clearer visualization of cellular expression in both control and knockout images.
Cell proliferation was determined by injecting BrdU (5-bromo-2′-deoxyuridine; Sigma, 9285) at 0.1 mg/g body weight 2 h before tissues were harvested. Anti-BrdU mouse monoclonal (BD Biosciences, 347580) was used at 1:200. BrdU labeling was normalized to the total number of cells in the proliferating zone or to the area of the proliferating zone. Data were then normalized to that of DFF control mice. At least three mice and two or three sections per mouse were analyzed for each genotype.
Histomorphometry
H&E- and TRAP-stained sections were used for quantification of osteoblast and osteoclast number and surface, using BioQuant OSTEO 2010 software. Measurements of growth plate length in H&E-stained sections were made using Canvas X software (ACD Systems). All lengths were normalized to the total length of the DFF control growth plate. Statistical analysis (Student's t-test) was based on measurements of tissue samples from at least three control and three experimental mice.
Micro-CT and DEXA analysis
For micro-CT, intact long bones were isolated and fixed in 70% ethanol overnight at 4°C and then stored at −20°C until analysis. Bones were embedded in 1.5% agarose and scanned (µCT40, SCANCO Medical). Micro-CT analysis of trabecular and cortical bone was performed as follows. For trabecular bone, 100 to 150 sections were selected below the growth plate for reconstruction and quantification. For cortical bone quantification, 50 to 100 sections were selected from the mid-diaphysis of the femur or tibia. Quantification was performed using SCANCO Medical micro-CT systems software. DEXA (GE/Lunar PIXImus) was used for measurements of whole-body bone density and body fat content. Data are representative of at least three mice per genotype.
Real-time quantitative PCR (RT-qPCR)
Distal bone, containing the growth plate, perichondrium and trabecular bone, was dissected. Immediately after isolation, the tissues were individually frozen in liquid nitrogen and stored at −80°C until analysis. Frozen tissues were pulverized in a dry ice-cooled stainless steel flask with a ball bearing in a Micro Dismembrator (Sartorius) at 2000 rpm for 20 s. RNA was stabilized with TRIzol (Ambion) and total RNA isolation was prepared according to the manufacturer's instructions. cDNA was synthesized using the iScript Select cDNA Synthesis Kit (#170-8841, Bio-Rad). mRNA expression was measured using TaqMan Fast Advanced Master Mix (4444557, Life Technologies) and TaqMan assay probes for Ihh, Pthlh, Fgf9, Fgf18 and Fgfr3. Hprt was used as a normalization control.
PTH treatment
For in vivo treatment of mice with PTH, 15-day-old Osx-Cre;DCKO mice were injected intraperitoneally once per day (morning) with synthetic PTH-related peptide (1-34) (H-6630, Bachem) at 80 µg/kg body weight or with PBS (control). Mice were injected for 5 days and then sacrificed at P21.
Statistics
Data are reported as mean±s.d. Data were analyzed using a two-tailed Student's t-test and P<0.05 considered statistically significant. Numbers of mice per group per experiment are stated in the figure legends.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization and methodology, K.K., K.Y., F.L. and D.M.O.; investigation, K.K., K.Y., J.L., J.C., C.S. and D.M.O.; writing of original draft, K.K. and D.M.O.; manuscript review and editing, K.K., K.Y., F.L. and D.M.O.; funding acquisition, resources and supervision, F.L. and D.M.O.
Funding
This work was supported by National Institutes of Health (NIH) grants [HD049808 to D.M.O., AR055923 to F.L., DE025077 to K.Y.]; the Washington University Musculoskeletal Research Center [NIH P30 AR057235]; and by internal funds from the Craniofacial Center of Seattle Children's Hospital and Center for Developmental Biology and Regenerative Medicine of Seattle Children's Research Institute (K.Y.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.131722/-/DC1
References
- Aikawa T., Segre G. V. and Lee K. (2001). Fibroblast growth factor inhibits chondrocytic growth through induction of p21 and subsequent inactivation of cyclin E-Cdk2. J. Biol. Chem. 276, 29347-29352. 10.1074/jbc.M101859200 [DOI] [PubMed] [Google Scholar]
- Beever J. E., Smit M. A., Meyers S. N., Hadfield T. S., Bottema C., Albretsen J. and Cockett N. E. (2006). A single-base change in the tyrosine kinase II domain of ovine FGFR3 causes hereditary chondrodysplasia in sheep. Anim. Genet. 37, 66-71. 10.1111/j.1365-2052.2005.01398.x [DOI] [PubMed] [Google Scholar]
- Bitgood M. J. and McMahon A. P. (1995). Hedgehog and Bmp genes are coexpressed at many diverse sites of cell–cell interaction in the mouse embryo. Dev. Biol. 172, 126-138. 10.1006/dbio.1995.0010 [DOI] [PubMed] [Google Scholar]
- Böhm F., Speicher T., Hellerbrand C., Dickson C., Partanen J. M., Ornitz D. M. and Werner S. (2010). FGF receptors 1 and 2 control chemically induced injury and compound detoxification in regenerating livers of mice. Gastroenterology 139, 1385-1396.e8. 10.1053/j.gastro.2010.06.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau M., Forcinito P., Andrade A. C., Hegde A., Ahn S., Lui J. C., Baron J. and Nilsson O. (2011). Organization of the Indian hedgehog - parathyroid hormone-related protein system in the postnatal growth plate. J. Mol. Endocrinol. 47, 99-107. 10.1530/JME-10-0177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Li C., Qiao W., Xu X. and Deng C. (2001). A Ser(365)-->Cys mutation of fibroblast growth factor receptor 3 in mouse downregulates Ihh/PTHrP signals and causes severe achondroplasia. Hum. Mol. Genet. 10, 457-465. 10.1093/hmg/10.5.457 [DOI] [PubMed] [Google Scholar]
- Chen X., Macica C. M., Nasiri A. and Broadus A. E. (2008). Regulation of articular chondrocyte proliferation and differentiation by indian hedgehog and parathyroid hormone-related protein in mice. Arthritis Rheum. 58, 3788-3797. 10.1002/art.23985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Shi Y., Regan J., Karuppaiah K., Ornitz D. M. and Long F. (2014a). Osx-Cre targets multiple cell types besides osteoblast lineage in postnatal mice. PLoS ONE 9, e85161 10.1371/journal.pone.0085161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Tu X., Esen E., Joeng K. S., Lin C., Arbeit J. M., Rüegg M. A., Hall M. N., Ma L. and Long F. (2014b). WNT7B promotes bone formation in part through mTORC1. PLoS Genet. 10, e1004145 10.1371/journal.pgen.1004145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobrinik D., Lee M. H., Hannon G., Mulligan G., Bronson R. T., Dyson N., Harlow E., Beach D., Weinberg R. A. and Jacks T. (1996). Shared role of the pRB-related p130 and p107 proteins in limb development. Genes Dev. 10, 1633-1644. 10.1101/gad.10.13.1633 [DOI] [PubMed] [Google Scholar]
- Colvin J. S., Bohne B. A., Harding G. W., McEwen D. G. and Ornitz D. M. (1996). Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat. Genet. 12, 390-397. 10.1038/ng0496-390 [DOI] [PubMed] [Google Scholar]
- Colvin J. S., Feldman B., Nadeau J. H., Goldfarb M. and Ornitz D. M. (1999). Genomic organization and embryonic expression of the mouse fibroblast growth factor 9 gene. Dev. Dyn. 216, 72-88. [DOI] [PubMed] [Google Scholar]
- Coumoul X., Shukla V., Li C., Wang R.-H. and Deng C.-X. (2005). Conditional knockdown of Fgfr2 in mice using Cre-LoxP induced RNA interference. Nucleic Acids Res. 33, e102 10.1093/nar/gni100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey L., Laplantine E., Priore R. and Basilico C. (2003). A network of transcriptional and signaling events is activated by FGF to induce chondrocyte growth arrest and differentiation. J. Cell Biol. 161, 1053-1066. 10.1083/jcb.200302075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Frutos C. A., Vega S., Manzanares M., Flores J. M., Huertas H., Martínez-Frías M. L. and Nieto M. A. (2007). Snail1 is a transcriptional effector of FGFR3 signaling during chondrogenesis and achondroplasias. Dev. Cell 13, 872-883. 10.1016/j.devcel.2007.09.016 [DOI] [PubMed] [Google Scholar]
- Deng C., Wynshaw-Boris A., Zhou F., Kuo A. and Leder P. (1996). Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell 84, 911-921. 10.1016/S0092-8674(00)81069-7 [DOI] [PubMed] [Google Scholar]
- Dy P., Wang W., Bhattaram P., Wang Q., Wang L., Ballock R. T. and Lefebvre V. (2012). Sox9 directs hypertrophic maturation and blocks osteoblast differentiation of growth plate chondrocytes. Dev. Cell 22, 597-609. 10.1016/j.devcel.2011.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellsworth J. L., Berry J., Bukowski T., Claus J., Feldhaus A., Holderman S., Holdren M. S., Lum K. D., Moore E. E., Raymond F. et al. (2002). Fibroblast growth factor-18 is a trophic factor for mature chondrocytes and their progenitors. Osteoarthritis Cartilage 10, 308-320. 10.1053/joca.2002.0514 [DOI] [PubMed] [Google Scholar]
- Esen E., Lee S. Y., Wice B. M. and Long F. (2015). PTH promotes bone anabolism by stimulating aerobic glycolysis via IGF signaling. J. Bone Miner. Res. 30, 2137 10.1002/jbmr.2714 [DOI] [PubMed] [Google Scholar]
- Eswarakumar V. P., Monsonego-Ornan E., Pines M., Antonopoulou I., Morriss-Kay G. M. and Lonai P. (2002). The IIIc alternative of Fgfr2 is a positive regulator of bone formation. Development 129, 3783-3793. [DOI] [PubMed] [Google Scholar]
- Garofalo S., Kliger-Spatz M., Cooke J. L., Wolstin O., Lunstrum G. P., Moshkovitz S. M., Horton W. A. and Yayon A. (1999). Skeletal dysplasia and defective chondrocyte differentiation by targeted overexpression of fibroblast growth factor 9 in transgenic mice. J. Bone Miner. Res. 14, 1909-1915. 10.1359/jbmr.1999.14.11.1909 [DOI] [PubMed] [Google Scholar]
- Havens B. A., Velonis D., Kronenberg M. S., Lichtler A. C., Oliver B. and Mina M. (2008). Roles of FGFR3 during morphogenesis of Meckel's cartilage and mandibular bones. Dev. Biol. 316, 336-349. 10.1016/j.ydbio.2008.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton M. J., Tu X., Cook J., Hu H. and Long F. (2005). Ihh controls cartilage development by antagonizing Gli3, but requires additional effectors to regulate osteoblast and vascular development. Development 132, 4339-4351. 10.1242/dev.02025 [DOI] [PubMed] [Google Scholar]
- Horton W. A., Hall J. G. and Hecht J. T. (2007). Achondroplasia. Lancet 370, 162-172. 10.1016/S0140-6736(07)61090-3 [DOI] [PubMed] [Google Scholar]
- Huang W. and Olsen B. R. (2015). Skeletal defects in Osterix-Cre transgenic mice. Transgenic Res. 24, 167-172. 10.1007/s11248-014-9828-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh S.-H., Warchol M. E. and Ornitz D. M. (2015). Cochlear progenitor number is controlled through mesenchymal FGF receptor signaling. eLife 4, e05921 10.7554/eLife.05921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung I. H., Yu K., Lavine K. J. and Ornitz D. M. (2007). FGF9 regulates early hypertrophic chondrocyte differentiation and skeletal vascularization in the developing stylopod. Dev. Biol. 307, 300-313. 10.1016/j.ydbio.2007.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung I. H., Schoenwolf G. C., Lewandoski M. and Ornitz D. M. (2016). A combined series of Fgf9 and Fgf18 mutant alleles identifies unique and redundant roles in skeletal development. Dev. Biol. 411, 72-84. 10.1016/j.ydbio.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunziker E. B. and Schenk R. K. (1989). Physiological mechanisms adopted by chondrocytes in regulating longitudinal bone growth in rats. J. Physiol. 414, 55-71. 10.1113/jphysiol.1989.sp017676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob A. L., Smith C., Partanen J. and Ornitz D. M. (2006). Fibroblast growth factor receptor 1 signaling in the osteo-chondrogenic cell lineage regulates sequential steps of osteoblast maturation. Dev. Biol. 296, 315-328. 10.1016/j.ydbio.2006.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolak M. R., Yang X. and Elefteriou F. (2015). FGFR1 signaling in hypertrophic chondrocytes is attenuated by the Ras-GAP neurofibromin during endochondral bone formation. Hum. Mol. Genet. 24, 2552-2564. 10.1093/hmg/ddv019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva V., Laplantine E. and Basilico C. (2008). PP2A-mediated dephosphorylation of p107 plays a critical role in chondrocyte cell cycle arrest by FGF. PLoS ONE 3, e3447 10.1371/journal.pone.0003447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva V., Daempfling L. and Basilico C. (2013). The B55alpha regulatory subunit of protein phosphatase 2A mediates fibroblast growth factor-induced p107 dephosphorylation and growth arrest in chondrocytes. Mol. Cell. Biol. 33, 2865-2878. 10.1128/MCB.01730-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozhemyakina E., Lassar A. B. and Zelzer E. (2015). A pathway to bone: signaling molecules and transcription factors involved in chondrocyte development and maturation. Development 142, 817-831. 10.1242/dev.105536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziel L., Kunath M., Kelly O. G. and Vortkamp A. (2004). Ext1-dependent heparan sulfate regulates the range of Ihh signaling during endochondral ossification. Dev. Cell 6, 801-813. 10.1016/j.devcel.2004.05.009 [DOI] [PubMed] [Google Scholar]
- Koziel L., Wuelling M., Schneider S. and Vortkamp A. (2005). Gli3 acts as a repressor downstream of Ihh in regulating two distinct steps of chondrocyte differentiation. Development 132, 5249-5260. 10.1242/dev.02097 [DOI] [PubMed] [Google Scholar]
- Kronenberg H. M. (2003). Developmental regulation of the growth plate. Nature 423, 332-336. 10.1038/nature01657 [DOI] [PubMed] [Google Scholar]
- Kurimchak A., Haines D. S., Garriga J., Wu S., De Luca F., Sweredoski M. J., Deshaies R. J., Hess S. and Graña X. (2013). Activation of p107 by fibroblast growth factor, which is essential for chondrocyte cell cycle exit, is mediated by the protein phosphatase 2A/B55alpha holoenzyme. Mol. Cell. Biol. 33, 3330-3342. 10.1128/MCB.00082-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laederich M. B. and Horton W. A. (2012). FGFR3 targeting strategies for achondroplasia. Expert Rev. Mol. Med. 14, e11 10.1017/erm.2012.4 [DOI] [PubMed] [Google Scholar]
- Laplantine E., Rossi F., Sahni M., Basilico C. and Cobrinik D. (2002). FGF signaling targets the pRb-related p107 and p130 proteins to induce chondrocyte growth arrest. J. Cell Biol. 158, 741-750. 10.1083/jcb.200205025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus J. E., Hegde A., Andrade A. C., Nilsson O. and Baron J. (2007). Fibroblast growth factor expression in the postnatal growth plate. Bone 40, 577-586. 10.1016/j.bone.2006.10.013 [DOI] [PubMed] [Google Scholar]
- Lee K., Lanske B., Karaplis A. C., Deeds J. D., Kohno H., Nissenson R. A., Kronenberg H. M. and Segre G. V. (1996). Parathyroid hormone-related peptide delays terminal differentiation of chondrocytes during endochondral bone development. Endocrinology 137, 5109-5118. [DOI] [PubMed] [Google Scholar]
- Legeai-Mallet L., Benoist-Lasselin C., Munnich A. and Bonaventure J. (2004). Overexpression of FGFR3, Stat1, Stat5 and p21Cip1 correlates with phenotypic severity and defective chondrocyte differentiation in FGFR3-related chondrodysplasias. Bone 34, 26-36. 10.1016/j.bone.2003.09.002 [DOI] [PubMed] [Google Scholar]
- Li C., Xu X., Nelson D. K., Williams T., Kuehn M. R. and Deng C.-X. (2005). FGFR1 function at the earliest stages of mouse limb development plays an indispensable role in subsequent autopod morphogenesis. Development 132, 4755-4764. 10.1242/dev.02065 [DOI] [PubMed] [Google Scholar]
- Li M., Seki Y., Freitas P. H. L., Nagata M., Kojima T., Sultana S., Ubaidus S., Maeda T., Shimomura J., Henderson J. E. et al. (2010). FGFR3 down-regulates PTH/PTHrP receptor gene expression by mediating JAK/STAT signaling in chondrocytic cell line. J. Electron Microsc. 59, 227-236. 10.1093/jmicro/dfq002 [DOI] [PubMed] [Google Scholar]
- Liu Z., Xu J., Colvin J. S. and Ornitz D. M. (2002). Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes Dev. 16, 859-869. 10.1101/gad.965602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Lavine K. J., Hung I. H. and Ornitz D. M. (2007). FGF18 is required for early chondrocyte proliferation, hypertrophy and vascular invasion of the growth plate. Dev. Biol. 302, 80-91. 10.1016/j.ydbio.2006.08.071 [DOI] [PubMed] [Google Scholar]
- Long F. and Ornitz D. M. (2013). Development of the endochondral skeleton. Cold Spring Harb. Perspect. Biol. 5, a008334 10.1101/cshperspect.a008334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F., Zhang X. M., Karp S., Yang Y. and McMahon A. P. (2001). Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development 128, 5099-5108. [DOI] [PubMed] [Google Scholar]
- Makrythanasis P., Temtamy S., Aglan M. S., Otaify G. A., Hamamy H. and Antonarakis S. E. (2014). A novel homozygous mutation in FGFR3 causes tall stature, severe lateral tibial deviation, scoliosis, hearing impairment, camptodactyly, and arachnodactyly. Hum. Mutat. 35, 959-963. 10.1002/humu.22597 [DOI] [PubMed] [Google Scholar]
- McEwen D. G., Green R. P., Naski M. C., Towler D. A. and Ornitz D. M. (1999). Fibroblast growth factor receptor 3 gene transcription is suppressed by cyclic adenosine 3′,5′-monophosphate: Identification of a chondrocytic regulatory element. J. Biol. Chem. 274, 30934-30942. 10.1074/jbc.274.43.30934 [DOI] [PubMed] [Google Scholar]
- Meyer M., Muller A.-K., Yang J., Moik D., Ponzio G., Ornitz D. M., Grose R. and Werner S. (2012). FGF receptors 1 and 2 are key regulators of keratinocyte migration in vitro and in wounded skin. J. Cell Sci. 125, 5690-5701. 10.1242/jcs.108167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao D., He B., Jiang Y., Kobayashi T., Sorocéanu M. A., Zhao J., Su H., Tong X., Amizuka N., Gupta A. et al. (2005). Osteoblast-derived PTHrP is a potent endogenous bone anabolic agent that modifies the therapeutic efficacy of administered PTH 1-34. J. Clin. Invest. 115, 2402-2411. 10.1172/JCI24918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minina E., Kreschel C., Naski M. C., Ornitz D. M. and Vortkamp A. (2002). Interaction of FGF, Ihh/Pthlh, and BMP signaling integrates chondrocyte proliferation and hypertrophic differentiation. Dev. Cell 3, 439-449. 10.1016/S1534-5807(02)00261-7 [DOI] [PubMed] [Google Scholar]
- Mori Y., Saito T., Chang S. H., Kobayashi H., Ladel C. H., Guehring H., Chung U.-I. and Kawaguchi H. (2014). Identification of fibroblast growth factor-18 as a molecule to protect adult articular cartilage by gene expression profiling. J. Biol. Chem. 289, 10192-10200. 10.1074/jbc.M113.524090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naski M. C., Wang Q., Xu J. and Ornitz D. M. (1996). Graded activation of fibroblast growth factor receptor 3 by mutations causing achondroplasia and thanatophoric dysplasia. Nat. Genet. 13, 233-237. 10.1038/ng0696-233 [DOI] [PubMed] [Google Scholar]
- Naski M. C., Colvin J. S., Coffin J. D. and Ornitz D. M. (1998). Repression of hedgehog signaling and BMP4 expression in growth plate cartilage by fibroblast growth factor receptor 3. Development 125, 4977-4988. [DOI] [PubMed] [Google Scholar]
- Noonan K. J., Hunziker E. B., Nessler J. and Buckwalter J. A. (1998). Changes in cell, matrix compartment, and fibrillar collagen volumes between growth-plate zones. J. Orthop. Res. 16, 500-508. 10.1002/jor.1100160416 [DOI] [PubMed] [Google Scholar]
- Ohbayashi N., Shibayama M., Kurotaki Y., Imanishi M., Fujimori T., Itoh N. and Takada S. (2002). FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. 16, 870-879. 10.1101/gad.965702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono N., Ono W., Nagasawa T. and Kronenberg H. M. (2014). A subset of chondrogenic cells provides early mesenchymal progenitors in growing bones. Nat. Cell Biol. 16, 1157-1167. 10.1038/ncb3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz D. M. and Itoh N. (2015). The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 4, 215-266. 10.1002/wdev.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz D. M. and Marie P. J. (2002). FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev. 16, 1446-1465. 10.1101/gad.990702 [DOI] [PubMed] [Google Scholar]
- Ornitz D. M. and Marie P. J. (2015). Fibroblast growth factor signaling in skeletal development and disease. Genes Dev. 29, 1463-1486. 10.1101/gad.266551.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Urtreger A., Givol D., Yayon A., Yarden Y. and Lonai P. (1991). Developmental expression of two murine fibroblast growth factor receptors, flg and bek. Development 113, 1419-1434. [DOI] [PubMed] [Google Scholar]
- Peters K. G., Werner S., Chen G. and Williams L. T. (1992). Two FGF receptor genes are differentially expressed in epithelial and mesenchymal tissues during limb formation and organogenesis in the mouse. Development 114, 233-243. [DOI] [PubMed] [Google Scholar]
- Peters K., Ornitz D., Werner S. and Williams L. (1993). Unique expression pattern of the FGF receptor 3 gene during mouse organogenesis. Dev. Biol. 155, 423-430. 10.1006/dbio.1993.1040 [DOI] [PubMed] [Google Scholar]
- Poladia D. P., Kish K., Kutay B., Hains D., Kegg H., Zhao H. and Bates C. M. (2006). Role of fibroblast growth factor receptors 1 and 2 in the metanephric mesenchyme. Dev. Biol. 291, 325-339. 10.1016/j.ydbio.2005.12.034 [DOI] [PubMed] [Google Scholar]
- Priore R., Dailey L. and Basilico C. (2006). Downregulation of Akt activity contributes to the growth arrest induced by FGF in chondrocytes. J. Cell Physiol. 207, 800-808. 10.1002/jcp.20620 [DOI] [PubMed] [Google Scholar]
- Raucci A., Laplantine E., Mansukhani A. and Basilico C. (2004). Activation of the ERK1/2 and p38 mitogen-activated protein kinase pathways mediates fibroblast growth factor-induced growth arrest of chondrocytes. J. Biol. Chem. 279, 1747-1756. 10.1074/jbc.M310384200 [DOI] [PubMed] [Google Scholar]
- Rodda S. J. and McMahon A. P. (2006). Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 133, 3231-3244. 10.1242/dev.02480 [DOI] [PubMed] [Google Scholar]
- Rossert J., Eberspaecher H. and de Crombrugghe B. (1995). Separate cis-acting DNA elements of the mouse pro-alpha 1(I) collagen promoter direct expression of reporter genes to different type I collagen-producing cells in transgenic mice. J. Cell Biol. 129, 1421-1432. 10.1083/jcb.129.5.1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims-Lucas S., Cusack B., Baust J., Eswarakumar V. P., Masatoshi H., Takeuchi A. and Bates C. M. (2011). Fgfr1 and the IIIc isoform of Fgfr2 play critical roles in the metanephric mesenchyme mediating early inductive events in kidney development. Dev. Dyn. 240, 240-249. 10.1002/dvdy.22501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. M., Williamson T. L., Schwartz M. L. and Vaccarino F. M. (2012). Impaired motor coordination and disrupted cerebellar architecture in Fgfr1 and Fgfr2 double knockout mice. Brain Res. 1460, 12-24. 10.1016/j.brainres.2012.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Jacques B., Hammerschmidt M. and McMahon A. P. (1999). Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 13, 2072-2086. 10.1101/gad.13.16.2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W.-C. S., Kitagawa M., Xue N., Xie B., Garofalo S., Cho J., Deng C., Horton W. A. and Fu X.-Y. (1997). Activation of Stat1 by mutant fibroblast growth-factor receptor in thanatophoric dysplasia type II dwarfism. Nature 386, 288-292. 10.1038/386288a0 [DOI] [PubMed] [Google Scholar]
- Su N., Jin M. and Chen L. (2014). Role of FGF/FGFR signaling in skeletal development and homeostasis: learning from mouse models. Bone Res. 2, 14003 10.1038/boneres.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toydemir R. M., Brassington A. E., Bayrak-Toydemir P., Krakowiak P. A., Jorde L. B., Whitby F. G., Longo N., Viskochil D. H., Carey J. C. and Bamshad M. J. (2006). A novel mutation in FGFR3 causes camptodactyly, tall stature, and hearing loss (CATSHL) syndrome. Am. J. Hum. Genet. 79, 935-941. 10.1086/508433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trokovic R., Trokovic N., Hernesniemi S., Pirvola U., Vogt Weisenhorn D. M., Rossant J., McMahon A. P., Wurst W. and Partanen J. (2003). FGFR1 is independently required in both developing mid- and hindbrain for sustained response to isthmic signals. EMBO J. 22, 1811-1823. 10.1093/emboj/cdg169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega S., Morales A. V., Ocaña O. H., Valdés F., Fabregat I. and Nieto M. A. (2004). Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 18, 1131-1143. 10.1101/gad.294104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyden J. M., Lewandoski M., Deng C., Harfe B. D. and Sun X. (2005). Conditional inactivation of Fgfr1 in mouse defines its role in limb bud establishment, outgrowth and digit patterning. Development 132, 4235-4245. 10.1242/dev.02001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vortkamp A., Lee K., Lanske B., Segre G. V., Kronenberg H. M. and Tabin C. J. (1996). Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science 273, 613-622. 10.1126/science.273.5275.613 [DOI] [PubMed] [Google Scholar]
- Wang L., Mishina Y. and Liu F. (2015). Osterix-Cre transgene causes craniofacial bone development defect. Calcif. Tissue Int. 96, 129-137. 10.1007/s00223-014-9945-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White A. C., Xu J., Yin Y., Smith C., Schmid G. and Ornitz D. M. (2006). FGF9 and SHH signaling coordinate lung growth and development through regulation of distinct mesenchymal domains. Development 133, 1507-1517. 10.1242/dev.02313 [DOI] [PubMed] [Google Scholar]
- Xiao Z., Huang J., Cao L., Liang Y., Han X. and Quarles L. D. (2014). Osteocyte-specific deletion of Fgfr1 suppresses FGF23. PLoS ONE 9, e104154 10.1371/journal.pone.0104154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Su N., Jin M., Qi H., Yang J., Li C., Du X., Luo F., Chen B., Shen Y. et al. (2012). Intermittent PTH (1-34) injection rescues the retarded skeletal development and postnatal lethality of mice mimicking human achondroplasia and thanatophoric dysplasia. Hum. Mol. Genet. 21, 3941-3955. 10.1093/hmg/dds181 [DOI] [PubMed] [Google Scholar]
- Yang J., Meyer M., Müller A.-K., Böhm F., Grose R., Dauwalder T., Verrey F., Kopf M., Partanen J., Bloch W. et al. (2010). Fibroblast growth factor receptors 1 and 2 in keratinocytes control the epidermal barrier and cutaneous homeostasis. J. Cell Biol. 188, 935-952. 10.1083/jcb.200910126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K. and Ornitz D. M. (2008). FGF signaling regulates mesenchymal differentiation and skeletal patterning along the limb bud proximodistal axis. Development 135, 483-491. 10.1242/dev.013268 [DOI] [PubMed] [Google Scholar]
- Yu K., Xu J., Liu Z., Sosic D., Shao J., Olson E. N., Towler D. A. and Ornitz D. M. (2003). Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development 130, 3063-3074. 10.1242/dev.00491 [DOI] [PubMed] [Google Scholar]
- Yu K., Karuppaiah K. and Ornitz D. M. (2015). Mesenchymal fibroblast growth factor receptor signaling regulates palatal shelf elevation during secondary palate formation. Dev. Dyn. 244, 1427-1438. 10.1002/dvdy.24319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ibrahimi O. A., Olsen S. K., Umemori H., Mohammadi M. and Ornitz D. M. (2006). Receptor specificity of the fibroblast growth factor family: the complete mammalian FGF family. J. Biol. Chem. 281, 15694-15700. 10.1074/jbc.M601252200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Su N., Luo F., Wen X., Tang Y., Yang J., Chen S., Jiang W., Du X. and Chen L. (2014). Deletion of Fgfr1 in osteoblasts enhances mobilization of EPCs into peripheral blood in a mouse endotoxemia model. Int. J. Biol. Sci. 10, 1064-1071. 10.7150/ijbs.8415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Xie Y., Tang J., Huang J., Huang Q., Xu W., Wang Z., Luo F., Wang Q., Chen H. et al. (2015). FGFR3 deficiency causes multiple chondroma-like lesions by upregulating hedgehog signaling. PLoS Genet. 11, e1005214 10.1371/journal.pgen.1005214 [DOI] [PMC free article] [PubMed] [Google Scholar]