ABSTRACT
The orchestrated division of cardiomyocytes assembles heart chambers of distinct morphology. To understand the structural divergence of the cardiac chambers, we determined the contributions of individual embryonic cardiomyocytes to the atrium in zebrafish by multicolor fate-mapping and we compare our analysis to the established proliferation dynamics of ventricular cardiomyocytes. We find that most atrial cardiomyocytes become rod-shaped in the second week of life, generating a single-muscle-cell-thick myocardial wall with a striking webbed morphology. Inner pectinate myofibers form mainly by direct branching, unlike delamination events that create ventricular trabeculae. Thus, muscle clones assembling the atrial chamber can extend from wall to lumen. As zebrafish mature, atrial wall cardiomyocytes proliferate laterally to generate cohesive patches of diverse shapes and sizes, frequently with dominant clones that comprise 20-30% of the wall area. A subpopulation of cardiomyocytes that transiently express atrial myosin heavy chain (amhc) contributes substantially to specific regions of the ventricle, suggesting an unappreciated level of plasticity during chamber formation. Our findings reveal proliferation dynamics and fate decisions of cardiomyocytes that produce the distinct architecture of the atrium.
KEY WORDS: Atrium, Brainbow, Cardiomyocyte, Clonal analysis, Heart development, Zebrafish
Summary: Lineage tracing of cardiomyocytes in the zebrafish heart reveal their proliferation dynamics and fate decisions, and distinguish atrial and ventricular morphogenesis at the cellular level.
INTRODUCTION
The heart achieves its form through the specification, proliferation and organization of cardiac muscle cells and their supporting tissues. Congenital defects in these events are relatively common and resultant structural abnormalities can be pathologic (van der Linde et al., 2011; van der Bom et al., 2011; Bruneau, 2008). Thus, delineating the cellular and molecular steps required for normal cardiac chamber development has been a crucial research objective.
From its origin as a simple, contractile tube, the heart undergoes significant changes in size and shape as it adapts to the hemodynamic demands of a growing animal (Christoffels et al., 2000). Previous studies have identified cardiac genetic programs as well as external cues from the environment that organize early heart morphogenesis (Rana et al., 2013; Bakkers, 2011). To track the fates of cardiac precursors as their progeny contribute to the primitive chambers, several studies have used injectable dyes or viral and genetic labeling systems (Keegan et al., 2004; Stainier et al., 1993; Abu-Issa and Kirby, 2008; Mikawa et al., 1992; Meilhac et al., 2004). Although these clonal analysis studies have provided valuable retrospective snapshots of the proliferative abilities of single cells, they are limited in their ability to shed light on proliferation dynamics that generate final cardiac structure.
Recent advances in genetically encoded fluorescent reporters and imaging techniques have improved the ability to simultaneously visualize and quantify the proliferative behaviors of many individual cells and their progeny within developing hearts (Devine et al., 2014; Lescroart et al., 2014). In particular, Brainbow multicolor genetic labeling technology (Livet et al., 2007) has yielded new insights into the cellular mechanisms by which cardiomyocyte proliferation drives maturation of the zebrafish cardiac ventricle. To initiate growth of the inner cardiac trabeculae, larval cardiomyocytes delaminate from the ventricular wall, seed typically at a non-adjacent site on the wall and proliferate (Liu et al., 2010; Gupta and Poss, 2012). As larvae mature into juveniles, the ventricular wall grows as a single-cardiomyocyte-thick patchwork of muscle clones, built by continued lateral expansion of several dozen embryonic cardiomyocytes. To create the final, cortical layer of adult muscle, juvenile trabecular myocytes derived from a rare group of ∼8 clonally dominant embryonic cardiomyocytes breach the wall and expand on the chamber surface (Gupta and Poss, 2012).
Whereas multicolor fate-mapping technology has deepened our understanding of ventricular morphogenesis, how the atrial chamber is built has not been explored at a comparable level. Thus, we have an incomplete view of how differential regulation of cardiomyocyte division might account for distinct chamber morphologies. Here, we used multicolor fate-mapping to visualize and quantify the contributions to zebrafish heart morphogenesis by embryonic atrial cardiomyocytes. We describe dynamic morphologic changes in atrial structure during development and we identify the cellular mechanism by which atrial pectinate muscle arises. We also determine the number, size and distribution of muscle clones in the chamber wall – analysis that reveals heterogeneous expansion and emergence of dominant cardiac muscle clones. Finally, we demonstrate a developmental window in which a subpopulation of cardiomyocytes that transiently express the archetypical atrial marker atrial myosin heavy chain [amhc; also known as myosin heavy chain 6 (myh6)] contributes cardiac muscle to the apex of the growing ventricle. Our findings distinguish atrial and ventricular morphogenesis at new cellular resolution and provide a platform for molecular dissection of cardiac chamber specification.
RESULTS
Multicolor labeling of atrial cardiomyocytes
Zebrafish amhc encodes a major component of atrial myofibrils that is detected in the atrium as early as 18 hours post fertilization (hpf) (Berdougo et al., 2003; Stainier et al., 1996). amhc has been used extensively as an indicator of atrial identity (Chen and Fishman, 1996; Targoff et al., 2013). Recently, Zhang and colleagues generated a transgenic line employing a tamoxifen-inducible Cre recombinase driven by amhc regulatory sequences as a means to genetically tag and trace atrial cardiomyocytes and their progeny (Zhang et al., 2013).
To simultaneously fate-map the contributions of most or all amhc-expressing cardiomyocytes during chamber morphogenesis, we crossed transgenic amhc:CreER zebrafish with animals transgenic for the Cre-inducible multicolor reporter strain Tg(β-act2:Brainbow1.0L)pd49 (priZm) (Fig. 1A) (Gupta and Poss, 2012). Embryos containing both transgenes were treated at 3 days post fertilization (dpf) with 4-hydroxytamoxifen (4-HT) to achieve mosaic recombination of priZm cassettes in amhc+ cardiomyocytes, before assessment by confocal imaging at different ages. No color recombination was observed in vehicle-treated amhc:CreER; priZm embryos or those lacking the amhc:CreER transgene (Fig. S1A-C) and 4-HT-activated atrial recombination was comparable when amhc:CreER was paired with the single-color β-actin2:RSG reporter transgene (Fig. S1D). We noted similar degrees of color recombination (>20 unique colors) in amhc:CreER; priZm atria and pan-cardiomyocyte-labeled cmlc2:CreER; priZm atria at 7 dpf, indicating that identical analytical principles used in previous studies could be applied (Fig. S1E). At 7 dpf, cardiomyocyte clones, identifiable by colors distinguishable from their neighbors, comprised one or two cells, with two-cell clones constituting about 20% of all clones (20±2%, n=8 hemi-hearts) (Fig. S1F). This was verified by co-labeling of cardiomyocyte nuclei with transgene cmlc2:H2-EGFP containing an EGFP-tagged H2 histone core protein driven by the cmlc2 promoter (Fig. S1G). Thus, amhc:CreER; priZm animals provide a platform to trace and visualize the developmental contributions of individual 3 dpf atrial cardiomyocytes.
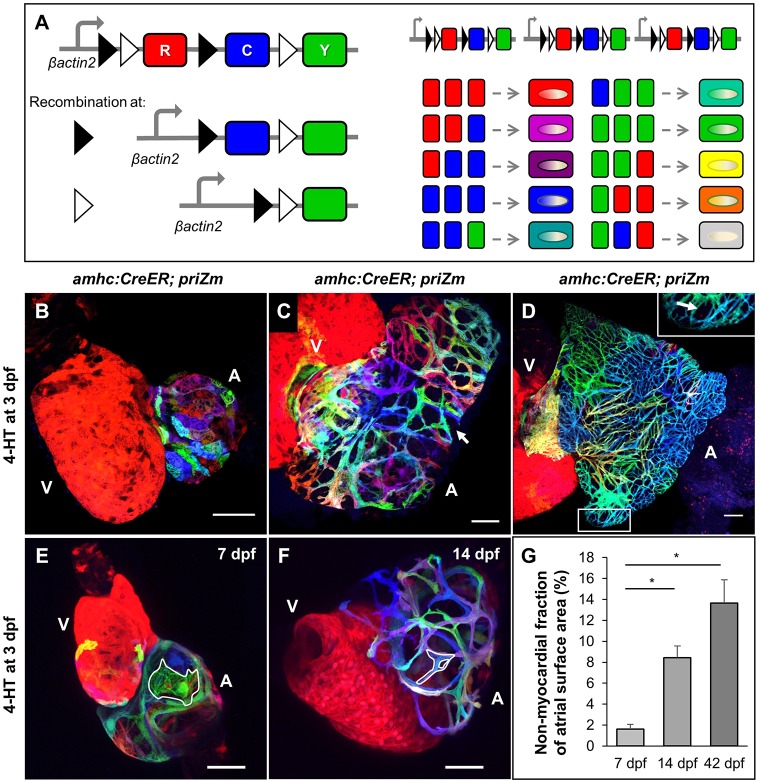
Fig. 1.
The larval zebrafish atrial wall undergoes permanent myocardial webbing. (A) Design of the priZm cassette and example spectrum of possible color combinations. (B-D) Surface myocardium of 7 dpf, 14 dpf and 42 dpf amhc:CreER; priZm hearts, respectively, after recombination with 4-HT at 3 dpf. V, ventricle; A, atrium. White arrows indicate gaps in myocardium. (E,F) 3D reconstructions of hearts from amhc:CreER; priZm animals treated with 4-HT at 3 dpf and collected at 7 dpf and 14 dpf, respectively. Examples of individual cardiomyocytes in the atrium (A) are outlined in white. Individual cardiomyocytes in the ventricle (V) are not distinguishable. (G) Percentage of total atrial surface area not covered by myocardium at 7 dpf, 14 dpf and 42 dpf (means±s.e.m.; n=7 hearts). Means are significantly heterogeneous (one-way ANOVA, P<0.001) and asterisks indicate means that are significantly different (Tukey–Kramer test, P<0.05). Scale bars: 50 µm in B,C,E,F and 100 µm in D.
The larval zebrafish atrial wall undergoes permanent myocardial webbing
Detailed atrial chamber and cardiomyocyte morphologies have not been described in developing zebrafish. Therefore, we assessed hearts from amhc:CreER; priZm zebrafish labeled at 3 dpf for any distinguishing atrial features at 7, 14 and 42 dpf (Fig. 1B-D). As previously reported (Chi et al., 2008), we noted a squamous morphology with rounded borders and broad cell-cell contacts in atrial cardiomyocytes up to 7 dpf (Fig. 1E; Movie 1). At this stage, atrial and ventricular wall cardiomyocytes are grossly identical in these features. Unexpectedly, by 14 dpf, atrial wall cardiomyocytes had undergone substantial morphologic changes that were not evident in the ventricle. Most 14 dpf atrial cardiomyocytes acquired a rod-shaped or branched morphology with limited areas of contact between adjacent cardiomyocytes, generating a webbed myocardial skeleton (Fig. 1F; Movie 2). Several myocardial markers and transgenes confirmed this structural change (Fig. S1H-I). By contrast, the epicardial cell layer and underlying extracellular matrix component Laminin had a contiguous presence enveloping the atrial wall myocardium (Fig. S1J-M). Thus, a complex morphological change occurs between 7 and 14 dpf that distinguishes atrial and ventricular wall morphogenesis.
We examined atria from larval (14 dpf) and juvenile (42 dpf) animals and noted that whereas the atrium grows markedly in post-larval animals, discontinuous myocardial regions persist and expand as a fraction of total atrial surface area (Fig. 1G). Even at adult stages, this webbed structure retains single-cardiomyocyte thickness and contains no coronary vasculature (as recently reported in Harrison et al., 2015). These characters sharply contrast with that of the ventricular wall, which develops a several-cardiomyocyte-thick, vascularized cortical layer to cover the single-cardiomyocyte-thick primordial layer during juvenile growth (Gupta and Poss, 2012).
Atrial pectinate muscle branches directly from wall cardiomyocytes
Recent lineage-tracing experiments identified a dynamic unexpected mechanism by which the major internal structure of the ventricle – trabecular muscle – arises. Starting around 3 dpf, a subpopulation of wall cardiomyocytes detaches from the ventricular surface without accompanying cell division, after which they typically seed trabecular myofibers distant from their wall origin. This results in trabecular cardiomyocytes that do not share a clonal relationship with the primordial wall cardiomyocytes to which they are attached (Liu et al., 2010; Gupta and Poss, 2012; Staudt et al., 2014).
In agreement with previously published work (Hu et al., 2000), we found that the internal structure of the atrium – pectinate muscle – forms later in development than trabeculae, with the first fibers visible around 14 dpf (Fig. 2A,B). Pectinate morphologies include both the small networks of thin fibers near the atrial wall and also branches joining the primordial heart tube remnant that spans from the atrioventricular canal to the chamber inlet. In mature hearts, the pectinate muscle is composed of fibrous bands of cardiomyocytes that form a complex internal meshwork. By assessing amhc:CreER; priZm atria labeled with 4-HT at 3 dpf, we found that mature pectinate myofibers often present as fused bundles of multiple non-clonal partners. These myofibers are thickest in the interior of the chamber nearest the atrio-ventricular valve, becoming progressively thinner and composed of fewer clones as they approach the atrial wall (Fig. 2C). This suggests a more ramified structure than has been appreciated in the trabecular muscle of the ventricle.
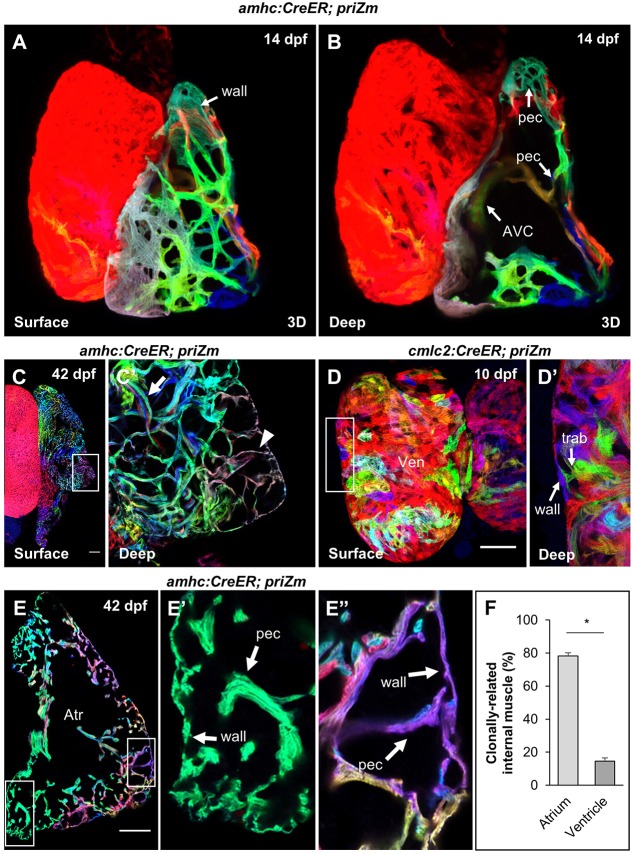
Fig. 2.
Atrial pectinate branches directly from wall cardiomyocytes. (A,B) 3D reconstructions of a 14 dpf amhc:CreER; priZm heart at the surface (A) and with the anterior hemi-section obscured to show interior structures (B). The atrio-ventricular canal (AVC) and newly formed pectinate fibers (pec) are indicated. (C) Surface and (C′) deep optical sections of a 42 dpf atrium. Arrowhead indicates monoclonal pectinate fiber contacting atrial wall. Arrow indicates thicker, polyclonal pectinate fiber deeper in the lumen. (D) Surface and (D′) deep optical sections of a 10 dpf cmlc2:CreER; priZm ventricle treated with 4-HT at 3 dpf. Arrows indicate a point of contact between a trabecular (trab) and ventricular wall cardiomyocyte (wall). Note that red cardiomyocytes, as the unrecombined color, are uninformative for clonal concordance assessment. (E) Cryosection of a 42 dpf amhc:CreER; priZm atrium exhibiting monoclonal, clonally concordant pectinate (pec) muscle (E′) and a polyclonal fiber containing clonally concordant muscle (E″); Atr, atrium. (F) Frequency with which pectinate and trabecular muscle share a clonal origin with adjacent cardiomyocytes of the atrial and ventricular wall, respectively [means±s.e.m.; n=5 (atria) and 11 (ventricles) from 16 hearts]. Asterisk indicates significantly different means (two-tailed Student's t-test, P<0.001). Scale bars: 100 µm in C,C′,E-E″ and 50 µm in D,D′.
To determine whether atrial pectinate fibers form by the same mechanism as ventricular trabeculae, we examined atria that had been labeled at 3 dpf and quantified the rate of clonal concordance between pectinate fibers and adjacent atrial wall cardiomyocytes. Importantly, 3 dpf ventricular cardiomyocytes do not give rise to clonally related areas of muscle spanning radially from wall into trabeculae (Fig. 2D). By contrast, individual 3 dpf atrial cardiomyocytes generate sizable, contiguous, clonally related areas of the juvenile and adult atrium that incorporate both wall and pectinate myocardium (Fig. 2E). Accordingly, we found that the rate of clonal concordance between atrial wall and pectinate cardiomyocytes was much higher than that between ventricular wall and trabecular cardiomyocytes (78±2%, n=25-54 pectinate fibers in 11 animals vs 15±2%; n=17-27 trabecular fibers in 5 animals; P<0.001) (Fig. 2F). Pectinate cardiomyocytes that did not match the adjoining wall cardiomyocyte color often formed a polyclonal, bundled wall attachment with pectinate cardiomyocytes that did match the wall color. This probably represents a secondary extension, in which a growing pectinate myofiber forms a second clonally discordant anchor site at the wall muscle by extending along an existing wall-concordant fiber. To assess whether the observed clonal concordance might represent atrial wall expansion after recombination but prior to pectinate formation, we treated animals at 12 dpf, shortly before the appearance of pectinate fibers. The majority of fiber anchor sites contained at least one colored myocardial cell that matched the adjacent wall cardiomyocyte (Fig. S2). These observations reveal that pectinate muscle fibers in the atrium arise in a mechanistically different manner than ventricular trabecular muscle. Although delamination with limited, local migration is possible, clonal analysis supports a dominant mechanism in which cardiomyocyte division in the atrial wall results in one wall daughter cardiomyocyte and one daughter pectinate cardiomyocyte.
The atrial wall forms by heterogeneous expansion of clonally related muscle patches
To chart the proliferation dynamics responsible for constructing the atrial wall, we imaged and quantified the surface area of cardiomyocyte clones over the course of chamber growth and maturation. Three dpf amhc:CreER; priZm embryos were treated with 4-HT and collected at 7, 14 and 42 dpf in these experiments. We found that, like ventricular wall cardiomyocytes, atrial wall cardiomyocytes largely retain contacts with clonal partners during animal growth, resulting in a conglomeration of laterally expanding multicellular muscle patches.
To define the number and sizes of clonal muscle patches that contribute to the atrial wall, we used imaging software to quantify the number and areas of atrial surface muscle clones from whole-mounted hearts (Fig. 3A-C). The number of distinguishable clones remained relatively constant at 7, 14 and 42 dpf time points, ranging from 44 to 69 per atrium [7 dpf, 53±5; 14 dpf, 59±6; 42 dpf, 62±6; n=8] (Fig. 3D). Although it is possible that in rare events clones from two separate embryonic cardiomyocytes might have been counted as a single clone if they adopted the same color, we attempted to minimize this effect by analyzing hearts exhibiting a high degree of color diversity, as in our previous studies (Gupta and Poss, 2012; Gupta et al., 2013). A slight increase in the number of clones identified at later time points might be attributable to spatial fragmenting of like-colored patches such that they are counted as two separate clones; however, this increase was not statistically significant (one-way ANOVA, P>0.05). Our results suggest that ∼50-60 embryonic cardiomyocytes present at 3 dpf ultimately contribute to the atrial wall, very similar to the number of 3 dpf cardiomyocytes contributing to the juvenile ventricular wall (Gupta and Poss, 2012). This number is less than the total number of cardiomyocytes in 3 dpf larval atria, which we estimated as 81 per animal (81±10; n=4) (Fig. S3A,B). This disparity is probably explained in part by our observations that many amhc+ cardiomyocytes incorporate into the atrio-ventricular (AV) junction, which we have excluded from quantification because of its polyclonal composition of densely arranged cardiomyocytes (Fig. S3C).
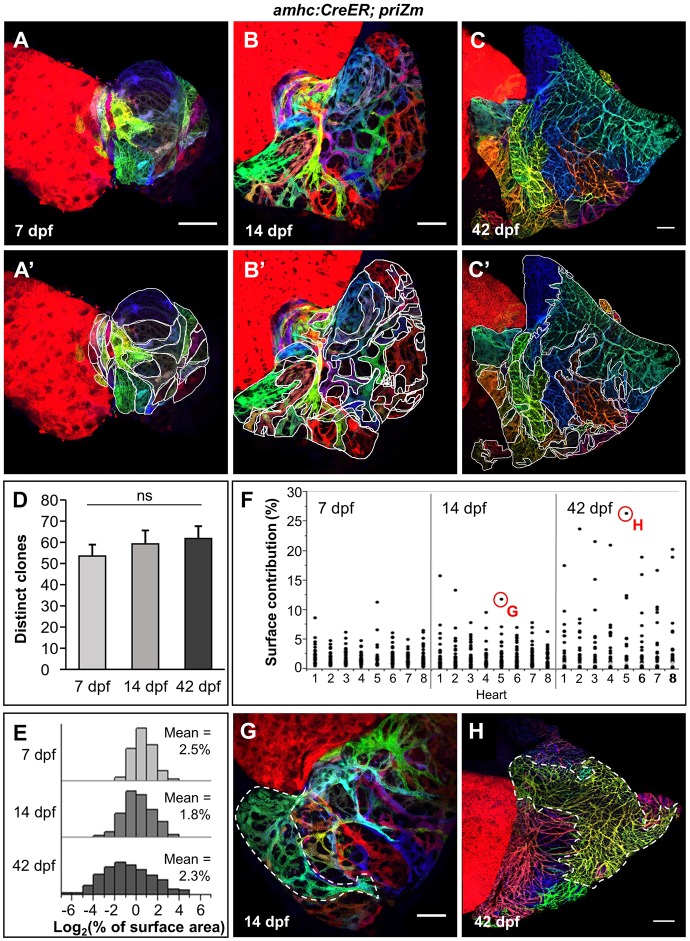
Fig. 3.
The atrial wall forms by heterogeneous expansion of clonally related muscle patches. (A-C) Surface myocardium of 7 dpf, 14 dpf and 42 dpf amhc:CreER; priZm hearts, respectively, with examples of traced clones outlined in white. (D) Number of distinct clones within 7 dpf, 14 dpf and 42 dpf amhc:CreER; priZm atria (means±s.e.m.; n=8 hearts each). The difference in the indicated means was not significant (ns) using one-way ANOVA (P>0.05). (E) Histograms of relative clone size distributions at 7 dpf, 14 dpf and 42 dpf. Data are aggregated from n=8 hearts per time point. The data are log-transformed to better show both tails of the distributions. (F-H) Distributions of relative clone sizes in individual hearts, grouped by age (n=8 at each time point), with indicated clones in F outlined in white from a 14 dpf (G) and a 42 dpf (H) heart. Scale bars: 50 µm in A,A′,B,B′,G and 100 µm in C,C′,H.
To assess clone size at each age, we examined the distribution of clone sizes relative to total atrial surface area (Fig. 3E). The distributions at each age were significantly different (two-sample Kolmogorov–Smirnov test, P<0.001 for each comparison). Whereas the mean relative clone size did not vary considerably, the number of very large and very small clones increased with age. This observation suggests heterogeneous rates of growth among clones. Examination of individual 7 dpf atria revealed no individual surface muscle clone occupying more than 12% of the total atrial wall surface area. By contrast, a majority of hearts contained at least one ‘dominant’ clone that contributed from 17 to 30% (19.1±7.2%, n=8) of the total wall surface area (Fig. 3F-H). We cannot exclude the possibility that a single ‘dominant’ clone resulted from the fusion of two identically colored clones during development; however, the consistency with which dominant clones are observed argues against this possibility. These dominant clones were present at several locations within the atrial wall in the population of 42 dpf animals we examined, most often incorporating a chamber appendage distant from the AV junction. Thus, the zebrafish atrium achieves its form by varied contributions of embryonic wall cardiomyocytes, including occasional cardiomyocytes with particularly high cardiogenic activity.
Atrial-specified cardiomyocytes contribute a portion of the ventricular apex
We noticed that amhc:CreER animals treated with 4-HT at 3 dpf consistently displayed a low percentage of labeled (non-red) cardiomyocytes in the ventricle, in either the priZm or β-actin2:RSG background (Fig. 4A, Fig. S4A). The proportion of amhc-labeled ventricular cardiomyocytes per animal was independent of the duration of 4-HT treatment at 3 dpf (Fig. S4B) and ventricular labeling consistently occurred in the apex and regions spanning the apex to AV junction. These results suggest that amhc:CreER-mediated recombination in ventricular cardiomyocytes does not arise from leaky stochastic expression of the amhc:CreER transgene, but might instead reflect endogenous amhc expression. To test this, we examined the published amhc:EGFP reporter line (Zhang et al., 2013) and generated a new amhc:EGFP reporter construct using the same 4.5 kb upstream regulatory sequence employed for the amhc:CreER transgene (-4.5amhc:EGFP). In contrast to previously reported results (Zhang et al., 2013), we observed faint but consistent EGFP fluorescence in the ventricular apex and junctional regions of amhc:EGFP hearts up to 3 dpf (Fig. 4B). Hearts collected from 30 embryos injected with the -4.5amhc:EGFP construct exhibited mosaic EGFP fluorescence in similar domains (Fig. S4F-H), indicating a consistent gene regulatory domain that is not dependent on the transgene integration site.
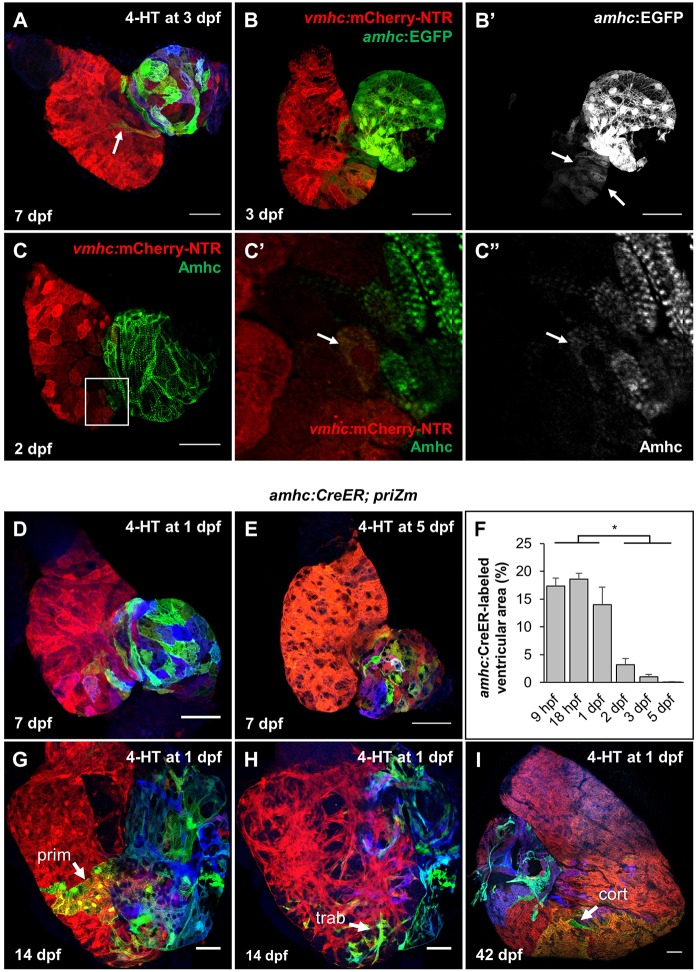
Fig. 4.
Transiently amhc+ cardiomyocytes populate the ventricular apex. (A) Surface myocardium of a 7 dpf heart from an amhc:CreER; priZm animal treated with 4-HT at 3 dpf, displaying a non-red cardiomyocyte (white arrow) on the ventricular side of the AV junction (dashed line). (B) Surface myocardium of a 3 dpf heart from a vmhc:mCherry-NTR and amhc:EGFP animal. (C) Surface myocardium of a 2 dpf heart expressing vmhc:mCherry-NTR and immunostained for Amhc. Magnified region shows area of EGFP expression within the ventricle (V) near the AV junction (dashed line). Arrow indicates an Amhc+ cardiomyocyte in the ventricle. (D,E) Surface myocardium of 7 dpf amhc:CreER; priZm hearts from animals treated with 4-HT at 1 dpf or 5 dpf, respectively. (F) Percentage of ventricular surface area labeled in 7 dpf amhc:CreER; priZm hearts treated at 9 hpf (n=7), 18 hpf (n=8), 1 dpf (n=7), 2 dpf (n=7), 3 dpf (n=8) or 5 dpf (n=7) (means±s.e.m.). Asterisk indicates that the means are significantly heterogeneous (one-way ANOVA, P<0.05) and a significant difference exists between the indicated groupings (Tukey–Kramer test, P<0.05). (G,H) Surface and deep myocardium, respectively, of a 14 dpf amhc:CreER; priZm heart treated with 4-HT at 1 dpf. Cardiomyocytes of the primordial ventricular wall (prim) and trabecular muscle (trab) are indicated with an arrow. (I) Surface myocardium of a 42 dpf amhc:CreER; priZm heart treated with 4-HT at 1 dpf. A labeled region of the ventricular cortical myocardium (cort) is indicated with an arrow. Scale bars: 100 µm in I; 50 µm in A,B,D,E,G,H; 25 µm in C-C′.
To confirm whether Amhc protein is endogenously produced by ventricular cardiomyocytes, we performed direct immunofluorescence. To assess whether amhc-expressing ventricular cardiomyocytes co-expressed ventricular markers, we incorporated a vmhc:mCherry-NTR transgenic line into our experiments (Zhang et al., 2013). At 2 dpf, faint Amhc signals were detectable in ventricular cardiomyocytes adjacent to the AV junction, whereas such signals were undetectable by 5 dpf (Fig. 4C, Fig. S4C). Interestingly, a small number of Amhc+; vmhc:mCherry-NTR+ cardiomyocytes with quantifiably lower levels of each marker were detectable, suggesting a transition from amhc to vmhc expression that possibly reflects a sub-lineage switch from an atrial to ventricular pattern of gene expression (Fig. S4D,E).
To define a possible developmental window for cardiomyocyte gene expression plasticity, we treated amhc:CreER; priZm embryos with 4-HT at several different ages before assessment of ventricular labeling. Embryos treated at 1 dpf or earlier exhibited large areas (∼15-20%) of labeled ventricular surface myocardium by 7 dpf. By contrast, hearts from embryos treated on or after 5 dpf later contained few or no labeled ventricular cardiomyocytes (Fig. 4D-F). The contributions of these early labeled clones to 14 dpf and 42 dpf ventricles included primordial, trabecular and cortical muscle (Fig. 4G-I). Notably, embryos treated with 4-HT for 24 h immediately after fertilization showed labeling in a consistent fraction and location of the ventricle, comprising on average 14±4% of the ventricle, predominantly in the apex and near the AV junction (Fig. S4I-Q). This supports the notion that a defined subpopulation of ventricular cardiomyocytes expresses amhc during an early window of cardiogenesis. Taken together, our experiments reveal that a substantial number of cardiomyocytes that express amhc early in development subsequently contribute to ventricular, rather than atrial, myocardium and that this subpopulation turns off amhc expression and adopts a ventricular gene expression signature after the first few days of life.
DISCUSSION
Multicolor fate-mapping technology is a powerful approach to dissect the proliferation dynamics of organogenesis; yet, due to technical challenges it has been sparingly employed. Here, to complement our analyses of the zebrafish ventricle, we have dissected the morphological and proliferative dynamics of atrial cardiomyocytes.
We found that the zebrafish atrial myocardium adopts a webbed structure during larval development. This transformation, which is not observed in the ventricle, is driven by changes in the morphology of individual atrial cardiomyocytes to a mature, branched form and results in a major expansion of atrial surface area. Accordingly, the endocardium, extracellular matrix and epicardium are in many cases the only barrier between the circulation and pericardial cavity environment. The physiological significance of this morphological change remains unclear, but it might reflect a preference to prioritize atrial volume over contractility. In addition to altered myocardial structure, chamber expansion is driven by cardiomyocyte proliferation, which we have characterized by tracking clonal populations of atrial cardiomyocytes at successive stages of development. Our results demonstrate that, as in adult ventricular cortical muscle, the heterogeneous proliferation of muscle clones contributes to atrial wall growth and this contribution can be dominated by a small number of expansive clones. These findings do not rule out the possibility that there are additional contributions to heterogeneity in clone size by cell hypertrophy. How this clonal expansion is regulated is a challenging question, which is likely to require the innovation of longitudinal cardiac imaging in individual developing animals to begin to resolve. Also crucial will be similar multicolor clonal analyses of atrial cardiomyocytes in higher vertebrates.
The inner mesh of the atrium, which starts to develop by 14 dpf, forms in a different method than that of the ventricular chamber. Our data reveal that atrial pectinate cardiomyocytes typically share a clonal relationship with the adjacent atrial wall, with large regions of clonally related atrial muscle spanning from wall to lumen in final form. This suggests that the pectinate arises through a simple mechanism of division, budding and branching. This mechanism contrasts with the delamination behavior of ventricular cardiomyocytes, which happens much earlier at 3 dpf. We suspect that there must be distinguishing influences, whether mechanical or chemical, that are responsible for these distinct behaviors. The development of live-imaging techniques to study pectinate formation in a range of conditions will assist the understanding of such influences. We have generated evidence that mechanical strain is a stimulus for cortical muscle formation in the zebrafish ventricle (Gupta et al., 2013); thus, the lower pressures in the atrium might account for its lack of a cortical layer. From our cardiomyocyte fate-mapping studies, one can visualize how a single cell type – the cardiac muscle cell – can display markedly different proliferative behaviors that ultimately drive chamber-specific morphogenesis and functional specialization. We anticipate that these chamber-specific principles of cardiomyocyte proliferation dynamics will provide new resolution to the roles of molecular factors disrupted in genetic mutant strains.
Unexpectedly, our investigation of atrial fate-mapping indicated that some cardiomyocytes expressing amhc in the early stages of cardiogenesis are present within and near the ventricular apex and subsequently switch to a ventricular phenotype. This finding is consistent with previous observations in mouse and rat (De Groot et al., 1989; Moorman and Lamers, 1994); however, the amhc-expressing component of the zebrafish ventricle has a more stereotyped localization than that reported for other animal models. One possible source of these cells is the early atrioventricular junction myocardium, which has been shown in mouse and chick to express Amhc and to contribute cardiomyocytes to both the atrium and the ventricle (Aanhaanen et al., 2009). Alternatively, this developmental phenomenon might be analogous to what has been observed in the setting of ventricular injury, in which atrial amhc+ cardiomyocytes were reported to reprogram to ventricular cardiomyocytes during regeneration (Zhang et al., 2013). The authors proposed this as a regeneration-specific event, as they could not detect the ventricular expression of amhc:EGFP or ventricular labeling using amhc:CreER; βactin2:RSG transgenes in uninjured larvae that we observe and report here. Because of these endogenous ventricular amhc expression domains, one cannot exclude that the regeneration after larval ventricular damage is mediated in some part by proliferation of amhc+ ventricular cardiomyocytes, rather than injury-induced reprogramming of atrial cardiomyocytes. To address this issue, it will be important to identify new markers that are expressed in a chamber-specific or chamber-preferential manner. The next key steps relevant to cardiac chamber specification and heart regeneration are to examine whether amhc-derived ventricular cardiomyocytes are physiologically distinct from other ventricular cardiomyocytes and how amhc+ to amhc+vmhc+ to vmhc+ transitions occur at the genetic and epigenetic levels.
MATERIALS AND METHODS
Zebrafish
Male and female wild-type or transgenic zebrafish of the EK/AB strain of the indicated ages were used for all experiments. All transgenic strains were analyzed as hemizygotes. Published transgenic lines used in this study were: priZm [Tg(bactin2:Brainbow1.0L)pd49] (Gupta and Poss, 2012); βactin2:RSG [Tg(bactin2:loxP-DsRed-STOP-loxP-eGFP)s928] (Kikuchi et al., 2010); cmlc2:CreER [Tg(cryaa:DsRed,-5.1myl7:CreERT2)pd13] (Gupta et al., 2013); amhc:CreER [Tg(amhc:CreERT2)sd20] (Zhang et al., 2013); vmhc:mCherry-NTR [Tg(vmhc:mCherry-Eco.NfsB)] (Zhang et al., 2013); tcf21:dsRed [TgBAC(tcf21:DsRed2)pd37] (Kikuchi et al., 2011) and βactin2:tagBFP [Tg(βactin2:loxP-mTagBFP-STOP-loxP-Neuregulin1)pd107] (Gemberling et al., 2015). All cmlc2:CreER; priZm and amhc:CreER; priZm animals were labeled with 4-hydroxytamoxifen (4-HT) at the indicated ages. Embryos labeled at 1 dpf were treated with 5 μM 4-HT in egg water for 4 h from a 1 mM stock made in 100% ethanol. Embryos treated after 1 dpf were treated with 10 μM 4-HT for 4 h. After treatment, embryos were washed thoroughly and returned to egg water. Work with zebrafish was performed according to an approved institutional animal care and use committee (IACUC) protocol (A100-12-04) at Duke University.
Embryo injection and mosaic analysis
A 4.5 kb segment of the amhc promoter (Zhang et al., 2013) was cloned into a plasmid containing a downstream EGFP sequence and flanking I-SceI meganuclease sites. The plasmid was linearized with meganuclease prior to injection to facilitate integration into the genome. EGFP+ embryos were identified at 2 dpf using a Zeiss AxioZoom dissection microscope. EGFP+ embryos were fixed in 4% paraformaldehyde overnight at 2 dpf or 3 dpf and prepared for confocal microscopy as described below.
Construction of -4.5amhc:EGFP transgenic zebrafish
The construct to create -4.5amhc:EGFP was made by cloning a 4.5 kb amhc promoter [forward primer (KpnI): 5′-CAGATGGTACCCCTGGAACTCAACAATGCTC-3′ and reverse primer (BamHI): 5′-CAGATGGATCCGATCTGGATCTCTCCTCAGTTG-3′] into the pCS2 vector and EGFP was cloned downstream of the promoter. The entire construct was flanked with I-SceI sites to facilitate transgenesis. Animals mosaic for this construct were analyzed in this study and one transgenic line was saved. The full name of this transgenic line is Tg(-4.5amhc:EGFP)pd116.
Construction of cmlc2:H2A F/Z-EGFP transgenic zebrafish
For the transgenic line referred to as cmlc2:H2-EGFP, the ORF sequence of zebrafish H2A F/Z (Pauls et al., 2001) was amplified using primers: H2A-F (EcoRI): CGGAATTCATGGCAGGTGGAAAAGC and H2A-R: (BamHI): CGGGATCCCCTGCGGTTTTCTGCTGG. The sequence was inserted into the EcoRI-BamHI sites of pEGFP-N1 to make a pH2A-EGFP construct. The H2A-EGFP-SV40 PolyA cassette was then digested by AflII, blunted with Klenow fragment, digested with EcoRI and cloned downstream of the 1.6 kb cmlc2 promoter (Huang et al., 2003) in a pBluescript backbone. The entire construct was flanked with I-SceI sites to facilitate transgenesis. The full name of this transgenic line is Tg(cmlc2:H2A F/Z-EGFP)pd115.
Construction of cmlc2:EGFP-CAAX transgenic zebrafish
cmlc2:EGFP-CAAX was generated by subcloning an EGFP-CAAX cassette that had been amplified from Tol2kit plasmid 384 (Kwan et al., 2007) downstream of a published 5.1 kb promoter sequence of zebrafish cmlc2 (Kikuchi et al., 2010). The full name of this transgenic line is Tg(cmlc2:EGFP-CAAX)pd117.
Tissue collection
To facilitate confocal imaging of the atrium, hearts were collected from fish after fixation of the whole organism. Fish were anesthetized completely in Tricaine and immersed in 4% paraformaldehyde for 1 day (fish 14 dpf or younger) or 2 days (fish older than 14 dpf) at 4°C. Whole hearts were then manually dissected and washed in PBS prior to mounting.
Immunofluorescence
Whole mount immunofluorescence was performed on hearts collected as described above. Primary antibodies used in this study were anti-laminin (rabbit, Sigma L9393) at 1:50 and S46 (anti-Myh6; mouse, Developmental Studies Hybridoma Bank) at 1:20 (Miller et al., 1985). Secondary antibodies used in this study were Alexa Fluor 488 goat anti-rabbit (1:200) and Alexa Fluor 488 goat anti-mouse (1:200). Antibody staining was performed as described previously (Yang and Xu, 2012).
Imaging
Fluorescent images from all samples were acquired using a Leica SP8 or a Zeiss LSM 700 microscope. For imaging of the cardiac surface, fixed hearts were placed in Fluoromount G (Southern Biotech) and compressed between two coverslips to allow imaging of both sides. Confocal slices through whole-mounted hearts were acquired by adjusting the Z-position until trabecular and pectinate muscle could be visualized. Images from priZm samples were acquired as described previously using laser lines 458 nm, 515 nm and 561 nm to excite CFP, YFP and RFP, respectively (Gupta and Poss, 2012). When indicated, laser line 488 nm was also used to excite GFP. Channels were acquired sequentially, overlaid and imported into ImageJ (NIH), where uniform adjustments to brightness and contrast were made.
Image analysis
To calculate the non-myocardial fraction of the atrial surface area, regions within the atrial wall of 7 dpf and 14 dpf priZm hearts that did not express any fluorescent protein were measured manually in ImageJ (http://imagej.nih.gov/ij/). For 42 dpf hearts, the thresholding function of ImageJ was manually calibrated to isolate and measure non-labeled regions within the atrial wall in µm2. The total area of non-myocardial surface was divided by the total atrial wall surface area to obtain the non-myocardial fraction.
For clonal analysis, individual clones were distinguished by color from their neighbors and traced in ImageJ and quantified in µm2. Areas of red myocardium were aggregated and divided by the average clone size within the same heart in order to estimate the number of unlabeled clones. The percentage area occupied by a clone was calculated by dividing its measured area by the total surface area of both sides of the atrium. For counting the number of cells per clone, nuclei were identified by the density of fluorescent signal; these densities consistently colocalized with a known nuclear reporter (cmlc2:H2-GFP). The characteristics of the priZm cassette, including distribution of observed colors and other details, were published previously (Gupta and Poss, 2012; Gupta et al., 2013).
3D reconstructions
For 3D reconstructions, 7 dpf and 14 dpf hearts were fixed in the organism and manually dissected. Hearts were then washed and transferred into 1% low-melting point agarose in a Petri dish that was then filled with water. An upright Leica SP8 with a 20× water immersion objective was used to obtain images in a Z-stack with 1.5 µm optical sections. Multicolor imaging was performed as described above. 3D reconstruction was performed using Imaris (Bitplane).
Statistical analysis
Statistical analysis was performed using JMP11 (SAS). Sample sizes were determined based on prior clonal analysis in the ventricle (Gupta and Poss, 2012). According to pre-established criteria, specimens were excluded only if there was no detectable non-red fluorescence, which was presumed to be due to genotyping error or if damaged during isolation. Treatment with 4-HT was done by clutch and animals were selected from clutch-matched tanks at random for each time point. For all experiments comparing the means of a set of percentages, an arcsine transformation was applied to the data. In experiments comparing the means of two distributions, Student's t-test was used with a threshold for significance at P<0.05. For experiments comparing the means of more than two groups, one-way ANOVA was used to detect significant differences in the group means (P<0.05) with a post hoc application of the Tukey–Kramer test to show subgroups with significantly different means. To analyze the relative clone sizes in hearts of different ages, we used the non-parametric two-sample Kolmogorov–Smirnov test to determine the probability that the distributions arose from identical populations. A probability of P<0.05 was used as a threshold for significance. All statistics reported within the text are means±s.e.m.
Acknowledgements
We thank V. Gupta for generating priZm animals; N. Chi for providing the amhc:CreER and vmhc:mCherry-NTR strains; J. Burris, N. Lee, T. Hume Thoren and N. Benkaci for zebrafish care; M. Bagnat, D. Fox and Poss laboratory members for comments on the manuscript; and B. Carlson for imaging advice.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
M.J.F. and K.D.P. designed the experimental strategy, analyzed data, and prepared the manuscript. J.C. and V.A.T. generated transgenic lines. M.J.F. performed all of the experiments.
Funding
M.J.F. was supported by a National Heart, Lung, and Blood Institute (NHLBI) pre-doctoral fellowship [F30 HL126487] and a Medical Scientist Training Program Training Grant [T32 GM007171]. J.C. was supported by a post-doctoral fellowship from the American Heart Association. V.A.T. was supported by a Graduate Research Fellowship [1106401] from the National Science Foundation. This work was supported by grants from the March of Dimes Foundation (FY14-205) and NHLBI [HL081674] to K.D.P. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.136606/-/DC1
References
- Aanhaanen W. T. J., Brons J. F., Domínguez J. N., Rana M. S., Norden J., Airik R., Wakker V., de Gier-de Vries C., Brown N. A., Kispert A. et al. (2009). The Tbx2+ primary myocardium of the atrioventricular canal forms the atrioventricular node and the base of the left ventricle. Circ. Res. 104, 1267-1274. 10.1161/CIRCRESAHA.108.192450 [DOI] [PubMed] [Google Scholar]
- Abu-Issa R. and Kirby M. L. (2008). Patterning of the heart field in the chick. Dev. Biol. 319, 223-233. 10.1016/j.ydbio.2008.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkers J. (2011). Zebrafish as a model to study cardiac development and human cardiac disease. Circ. Res. 91, 279-288. 10.1093/cvr/cvr098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdougo E., Coleman H., Lee D. H., Stainier D. Y. R. and Yelon D. (2003). Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development 130, 6121-6129. 10.1242/dev.00838 [DOI] [PubMed] [Google Scholar]
- Bruneau B. G. (2008). The developmental genetics of congenital heart disease. Nature 451, 943-948. 10.1038/nature06801 [DOI] [PubMed] [Google Scholar]
- Chen J. N. and Fishman M. C. (1996). Zebrafish tinman homolog demarcates the heart field and initiates myocardial differentiation. Development 122, 3809-3816. [DOI] [PubMed] [Google Scholar]
- Chi N. C., Shaw R. M., Jungblut B., Huisken J., Ferrer T., Arnaout R., Scott I., Beis D., Xiao T., Baier H. et al. (2008). Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol. 6, e109 10.1371/journal.pbio.0060109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels V. M., Habets P. E. M. H., Franco D., Campione M., de Jong F., Lamers W. H., Bao Z.-Z., Palmer S., Biben C., Harvey R. P. et al. (2000). Chamber formation and morphogenesis in the developing mammalian heart. Dev. Biol. 223, 266-278. 10.1006/dbio.2000.9753 [DOI] [PubMed] [Google Scholar]
- De Groot I. J. M., Lamers W. H. and Moorman A. F. M. (1989). Isomyosin expression patterns during rat heart morphogenesis: an immunohistochemical study. Anat. Rec. 224, 365-373. 10.1002/ar.1092240305 [DOI] [PubMed] [Google Scholar]
- Devine W. P., Wythe J. D., George M., Koshiba-Takeuchi K. and Bruneau B. G. (2014). Early patterning and specification of cardiac progenitors in gastrulating mesoderm. Elife 3, e03848 10.7554/eLife.03848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemberling M., Karra R., Dickson A. L. and Poss K. D. (2015). Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. Elife 4, e05871 10.7554/eLife.05871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V. and Poss K. D. (2012). Clonally dominant cardiomyocytes direct heart morphogenesis. Nature 484, 479-484. 10.1038/nature11045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V., Gemberling M., Karra R., Rosenfeld G. E., Evans T. and Poss K. D. (2013). An injury-responsive gata4 program shapes the zebrafish cardiac ventricle. Curr. Biol. 23, 1221-1227. 10.1016/j.cub.2013.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M. R. M., Bussmann J., Huang Y., Zhao L., Osorio A., Burns C. G., Burns C. E., Sucov H. M., Siekmann A. F. and Lien C.-L. (2015). Chemokine-guided angiogenesis directs coronary vasculature formation in zebrafish. Dev. Cell 33, 442-454. 10.1016/j.devcel.2015.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N., Sedmera D., Yost H. J. and Clark E. B. (2000). Structure and function of the developing zebrafish heart. Anat. Rec. 260, 148-157. [DOI] [PubMed] [Google Scholar]
- Huang C.-J., Tu C.-T., Hsiao C.-D., Hsieh F.-J. and Tsai H.-J. (2003). Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev. Dyn. 228, 30-40. 10.1002/dvdy.10356 [DOI] [PubMed] [Google Scholar]
- Keegan B. R., Meyer D. and Yelon D. (2004). Organization of cardiac chamber progenitors in the zebrafish blastula. Development 131, 3081-3091. 10.1242/dev.01185 [DOI] [PubMed] [Google Scholar]
- Kikuchi K., Holdway J. E., Werdich A. A., Anderson R. M., Fang Y., Egnaczyk G. F., Evans T., MacRae C. A., Stainier D. Y. R. and Poss K. D. (2010). Primary contribution to zebrafish heart regeneration by gata4+ cardiomyocytes. Nature 464, 601-605. 10.1038/nature08804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Gupta V., Wang J., Holdway J. E., Wills A. A., Fang Y. and Poss K. D. (2011). tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development 138, 2895-2902. 10.1242/dev.067041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan K. M. Fujimoto E., Grabher C., Mangum B. D, Hardy M. E., Campbell D. S., Parant J. M., Yost H. J., Kanki J. P. and Chien C.-B. (2007). The Tol2kit: a multisite Gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088-3099. 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- Lescroart F., Chabab S., Lin X., Rulands S., Paulissen C., Rodolosse A., Auer H., Achouri Y., Dubois C., Bondue A. et al. (2014). Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nat. Cell Biol. 16, 829-840. 10.1038/ncb3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Bressan M., Hassel D., Huisken J., Staudt D., Kikuchi K., Poss K. D., Mikawa T. and Stainier D. Y. R. (2010). A dual role for ErbB2 signaling in cardiac trabeculation. Development 137, 3867-3875. 10.1242/dev.053736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livet J., Weissman T. A., Kang H., Draft R. W., Lu J., Bennis R. A., Sanes J. R. and Lichtman J. W. (2007). Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56-62. 10.1038/nature06293 [DOI] [PubMed] [Google Scholar]
- Meilhac S. M., Esner M., Kelly R. G., Nicolas J.-F. and Buckingham M. E. (2004). The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Dev. Cell 6, 685-698. 10.1016/S1534-5807(04)00133-9 [DOI] [PubMed] [Google Scholar]
- Mikawa T., Borisov A., Brown A. M. C. and Fischman D. A. (1992). Clonal analysis of cardiac morphogenesis in the chicken embryo using a replication-defective retrovirus: I. Formation of the ventricular myocardium. Dev. Dyn. 193, 11-23. 10.1002/aja.1001930104 [DOI] [PubMed] [Google Scholar]
- Miller J. B., Crow M. T. and Stockdale F. E. (1985). Slow and fast myosin heavy chain content defines three types of myotubes in early muscle cell cultures. J. Cell. Biol. 101, 1643-1650. 10.1083/jcb.101.5.1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman A. F. M. and Lamers W. H. (1994). Molecular anatomy of the developing heart. Trends Cardiovasc. Med. 4, 257-264. 10.1016/1050-1738(94)90029-9 [DOI] [PubMed] [Google Scholar]
- Pauls S., Geldmacher-Voss B. and Campos-Ortega J. A. (2001). A zebrafish histone variant H2A. F/Z and a transgenic H2A. F/Z: GFP fusion protein for in vivo studies of embryonic development. Dev. Genes Evol. 211, 603-610. 10.1007/s00427-001-0196-x [DOI] [PubMed] [Google Scholar]
- Rana M. S., Christoffels V. M. and Moorman A. F. M. (2013). A molecular and genetic outline of cardiac morphogenesis. Acta Physiol. 207, 588-615. 10.1111/apha.12061 [DOI] [PubMed] [Google Scholar]
- Stainier D., Lee R. K. and Fishman M. C. (1993). Cardiovascular development in the zebrafish. I. Myocardial fate map and heart tube formation. Development 119, 31-40. [DOI] [PubMed] [Google Scholar]
- Stainier D. Y., Fouquet B., Chen J. N., Warren K. S., Weinstein B. M., Meiler S. E., Mohideen M. A., Neuhauss S. C., Solnica-Krezel L., Schier A. F. et al. (1996). Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development 123, 285-292. [DOI] [PubMed] [Google Scholar]
- Staudt D. W., Liu J., Thorn K. S., Stuurman N., Liebling M. and Stainier D. Y. (2014). High-resolution imaging of cardiomyocyte behavior reveals two distinct steps in ventricular trabeculation. Development 141, 585-593. 10.1242/dev.098632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targoff K. L. Colombo S., George V., Schell T., Kim S.-H., Solnica-Krezel L. and Yelon D. (2013). Nkx genes are essential for maintenance of ventricular identity. Development 140, 4203-4213. 10.1242/dev.095562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bom T., Zomer A. C., Zwinderman A. H., Meijboom F. J., Bouma B. J. and Mulder B. J. (2011). The changing epidemiology of congenital heart disease. Nat. Rev. Cardiol. 8, 50-60. 10.1038/nrcardio.2010.166 [DOI] [PubMed] [Google Scholar]
- van der Linde D., Konings E. E. M., Slager M. A., Witsenburg M., Helbing W. A., Takkenberg J. J. M. and Roos-Hesselink J. W. (2011). Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J. Am. Coll. Cardiol. 58, 2241-2247. 10.1016/j.jacc.2011.08.025 [DOI] [PubMed] [Google Scholar]
- Yang J. and Xu X. (2012). Immunostaining of dissected zebrafish embryonic heart. J. Vis. Exp. 59, e3510 10.3791/3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Han P., Yang H., Ouyang K., Lee D., Lin Y.-F., Ocorr K., Kang G., Chen J., Stainier D. Y. R. et al. (2013). In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature 498, 497-501. 10.1038/nature12322 [DOI] [PMC free article] [PubMed] [Google Scholar]