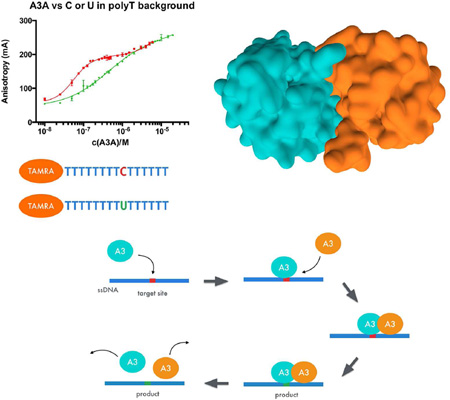
Abstract
Deaminase activity mediated by the human APOBEC3 family of proteins contributes to genomic instability and cancer. APOBEC3A is by far the most active in this family and can cause rapid cell death when overexpressed, but in general how the activity of APOBEC3s is regulated on a molecular level is unclear. In this study the biochemical and structural basis of APOBEC3A substrate binding and specificity is elucidated. We find that specific binding of single-stranded DNA is regulated by the cooperative dimerization of APOBEC3A. The crystal structure elucidates this homo-dimer as a symmetric domain swap of the N-terminal residues. This dimer interface provides insights into how cooperative protein-protein interactions may impact function in the APOBEC3 enzymes, and provides a potential scaffold for strategies aimed at reducing their mutation load.
Graphical Abstract
E-TOC
Human APOBEC3A is a strongly mutagenic ssDNA single domain cytosine deaminase. Bohn et al. demonstrate that potent cooperative binding occurs to substrate, while lost for efficient product release. The crystal structure revealed a homodimer, which mutational analysis confirmed is required for the cooperativity, providing a model for substrate recognition.
Introduction
Several exogenous and endogenous factors act as mutagens, contributing to carcinogenesis. The APOBEC3 proteins have been described as a major endogenous source for mutations in various types of cancer. Acting on chromosomal DNA, the APOBEC3 family of cytidine deaminases can introduce G-to-A hypermutations, as observed in clusters of APOBEC3-mediated mutational signatures found in breast cancer genomes (Nik-Zainal et al., 2012). APOBEC3B (A3B) was recently identified as a direct enzymatic source for this type of clustered mutations (Burns et al., 2013a). In addition to breast cancer, several other cancers such as bladder cancer, head and neck cancer, cervical cancer and lung cancer exhibit a similar genomic mutation pattern (Burns et al., 2013b; Roberts et al., 2013). Urothelial bladder cancer exhibits the most pronounced contribution of APOBEC3-mediated hypermutations to the overall mutation load (Cancer Genome Atlas Research Network, 2014). In lung cancer, APOBEC3-induced genomic instability appears to increase over time as the tumor progresses (de Bruin et al., 2014). APOBEC3A (A3A) shares the same genomic locus as A3B but is much more catalytically active and potentially linked to breast cancer (Caval et al., 2014; Nik-Zainal et al., 2014).
APOBEC3 proteins belong to a superfamily of deaminases and catalyze a cytidine to uridine zinc-dependent deamination reaction (Betts et al., 1994; Wilson et al., 1991). Common ancestry links the seven proteins of the contiguous human APOBEC3 locus (Wedekind et al., 2003) and allows classification based on phylogeny (LaRue et al., 2009). A3A, A3C and A3H comprise a single, catalytically active deaminase domain, whereas A3B, A3D, A3F and A3G are two-domain proteins with an N-terminal pseudocatalytic deaminase domain (NTD) and a C-terminal catalytic domain (CTD). Spatial extent of the substrate accommodating active-site region appears to be a determinant of whether a deaminase domain exhibits catalytic activity or not (Shandilya et al., 2014). The APOBEC3 proteins act on ssDNA to introduce strand-coordinated G-to-A point mutations. These mutations not only compromise the informational integrity of DNA but may also lead to double strand breaks (Burns et al., 2013a; Landry et al., 2011) contributing to genomic damage observed in the cancer genomes (Roberts et al., 2012; Sakofsky et al., 2014).
Four members of the APOBEC3 family (A3D, A3F, A3G and A3H) apply strong selective pressure on HIV-1 in the absence of Vif (Bishop et al., 2004; Dang et al., 2006; Harari et al., 2009; Harris et al., 2003; Hultquist et al., 2011; Lecossier et al., 2003; Liddament et al., 2004; Mangeat et al., 2003; OhAinle et al., 2008; Sheehy et al., 2002; Wiegand et al., 2004; Zhang et al., 2003; Zheng et al., 2004). These proteins are incorporated into budding virions and upon subsequent infection of a target cell, introduce point mutations in the newly reverse-transcribed viral genomic ssDNA, leading to direct degradation of the highly mutated product (Weil et al., 2013) or detrimental G-to-A mutations (Harris et al., 2003; Loeb et al., 1999).
A3G and A3F form high molecular mass complexes with polynucleotides that are relevant for biological function (Wang et al., 2007; Wedekind et al., 2006). The four antiretroviral APOBEC3 proteins were recently demonstrated to form multimeric complexes in living cells (Li et al., 2014). Over the last few years, Atomic Force Microscopy (AFM) studies have provided insights into the mechanistic details influencing this complex formation (Shlyakhtenko et al., 2011; 2014; 2013). The crystal structures of A3C (Kitamura et al., 2011), A3F-CTD (Bohn et al., 2013) and A3G-CTD (Holden et al., 2008; Li et al., 2012; Shandilya et al., 2010), and the NMR structures of A3G-CTD (Chen et al., 2008; Harjes et al., 2009) and A3A (Byeon et al., 2013) have provided further insights into the structural factors influencing this activity. However, significant details are still missing due to the lack of APOBEC3-ssDNA complex structures that could illuminate the molecular basis of complex formation.
The functional oligomerization state of A3 enzymes and whether or not cooperativity contributes to DNA binding affinity are not clear. The two-domain A3G binds ssDNA both as a monomer and dimer, but the dimerization is not induced by ssDNA binding (Shlyakhtenko et al., 2013), which is consistent with the observation that A3G binds its substrate non-cooperatively (Chelico et al., 2006; 2008). Substrate is being bound with high affinity (KD ~ 50–75 nM) (Chelico et al., 2006; Iwatani et al., 2006; Yu et al., 2004).
The single domain A3A has been reported to be monomeric in vitro, in solution, as detected by AFM (Shlyakhtenko et al., 2014) as well as in living cells monitored by Fluorescence Fluctuation Spectroscopy (FFS) (Li et al., 2014). The binding affinity of catalytically active A3A to ssDNA substrate, as compared to A3G, is reported to be lower by 100-fold (Byeon et al., 2013; Mitra et al., 2014) or 50-fold (Love et al., 2012), depending on the chosen experimental conditions. These reports are further complicated by the observation that catalytically active A3A is the most potent enzyme of the A3 family, with deamination rates up to 10-fold above those of APOBEC3G (Carpenter et al., 2012). Catalytically inactive A3A, on the other hand, dimerizes readily on the target ssDNA substrate (Shlyakhtenko et al., 2014). However, catalytically-inactive A3A can bind substrates with similar affinity as A3G (Logue et al., 2014). The fundamental reasons for these apparent discrepancies are not well understood.
In this study we explore the structural and biochemical basis underlying A3A ssDNA binding activity and the direct functional impact of cooperative dimerization on binding affinity. We developed a novel fluorescence anisotropy based high-throughput binding assay, solved a high-resolution crystal structure and generated a range of A3A mutants to demonstrate that catalytically-inactive A3A binds ssDNA with high affinity and specificity while exhibiting a high degree of cooperativity. Cooperative dimerization of APOBEC3A provides fundamental insights into the function of the entire APOBEC3 family of proteins and their respective roles in anti-retroviral and anti-cancer therapies.
Results
APOBEC3A preferentially binds the TTC trinucleotide sequence
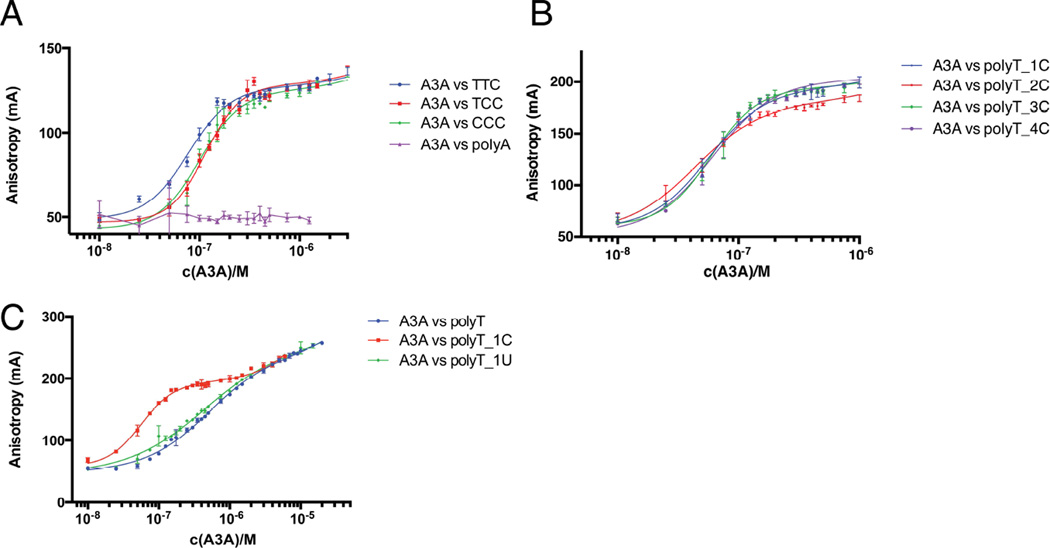
APOBEC3 enzymes recognize target deamination site cytidines within the context of specific trinucleotide motifs. A binding assay was developed using short, fluorescently labeled 15-mer oligonucleotides with single target sites for deamination to quantitatively assess motif specificity. A catalytically inactive mutant of A3A (E72A/C171A) (Figure 1a, Table 1) was used and the dissociation constant determined for several trinucleotide deamination motifs. This variant of A3A binds the target trinucleotide motif (5’-TTC-3’) with high affinity (KD = 77 ± 3 nM) and strong specificity as compared to an adenine polynucleotide. This affinity is comparable to that reported for A3G (Chelico et al., 2006). Other cytidine containing trinucleotide motifs known to be substrates for A3A are bound with similar affinities, KD = 114 ± 4 nM for 5’-TCC-3’ and KD = 100 ± 4 nM for 5’-CCC-3’. These data are consistent with previously reported values for trinucleotide motifs (Love et al., 2012), but in our observations 5-TTC-3’ is slightly more preferred with a ~20–30 nM difference in KD.
Figure 1. APOBEC3A binding specifically to trinucleotide deamination motifs.
(A) Fluorescence anisotropy measurements show how introduction of a single TTC motif in a poly A background leads to high affinity binding. Other motifs have a similar effect, but TTC appears to be the preferred substrate (B) APOBEC3A binding to an ideal substrate consisting of a polyT oligomer containing a single cytidine residue. Varying the number of cytidines does not have any pronounced effect on APOBEC3A affinity. (C) Difference in affinity to the target deamination site versus a poly T oligomer. Binding is cooperative with higher affinity to the oligomer containing the TTC motif (polyT_1C). APOBEC3A has the same affinity to the deaminated base (polyT_1U) as to the polyT background. Binding is specific to the substrate and not to the product. See also Supplemental Figures 2 and 3.
Table 1.
ssDNA binding affinity and cooperativity of APOBEC3A and interface mutants. The hill coefficient (nH) is expected to be 1 for non-cooperative binding and 2 for cooperative binding by a dimer. Note: A3A refers to the inactive variant E72A. See also Supplemental Table 1 for oligomer sequences.
| KD [nM] | KD relative to polyT_1C |
nH | |
|---|---|---|---|
| A3A vs TTC | 77 ± 3 | 1.4 | 2.1 ± 0.1 |
| A3A vs TCC | 114 ± 4 | 2.0 | 2.5 ± 0.2 |
| A3A vs CCC | 100 ± 4 | 1.8 | 2.1 ± 0.2 |
| A3A vs polyT_1C | 56 ± 2 | 1.0 | 1.8 ± 0.1 |
| A3A vs polyT_2C | 44 ± 2 | 0.8 | 1.6 ± 0.1 |
| A3A vs polyT_3C | 59 ± 2 | 1.1 | 2.2 ± 0.1 |
| A3A vs polyT_4C | 61 ± 3 | 1.1 | 1.7 ± 0.1 |
| A3A vs polyT | 502 ± 27 | 9.0 | 0.96 ± 0.04 |
| A3A vs polyT_1U | 434 ± 35 | 7.8 | 0.80 ± 0.04 |
| A3A H11A vs polyT_1C | 855 ± 311 | 15.3 | 0.7 ± 0.1 |
| A3A H16A vs polyT_1C | 584 ± 92 | 10.4 | 1.2 ± 0.2 |
| A3A K30E vs polyT_1C | 284 ± 20 | 5.1 | 1.2 ± 0.1 |
| A3A H56A vs polyT_1C | 86 ± 13 | 1.5 | 0.9 ± 0.1 |
| A3A E72Q vs polyT_1C | 26 ± 2 | 0.5 | 1.8 ± 0.1 |
| A3A E72Q vs polyT_1U | 296 ± 125 | 5.3 | 1.0 ± 0.2 |
| A3A E72Q vs polyT | 205 ± 20 | 3.7 | 1.4 ± 0.2 |
Using substrates based on the high affinity trinucleotide (5’-TTC-3’) a poly-thymidine oligomer containing two cytidine bases was identified as the tightest binding partner for A3A (Figure 1b, Table 1) with KD = 44 ± 2 nM. Varying the number of consecutive cytidines, ranging from one to four, did not significantly alter substrate affinity. An oligonucleotide consisting solely of thymidine, lacking cytidine, binds to the enzyme (Figure 1c) but the affinity is an order of magnitude weaker than in the presence of a single cytidine (KD = 502 ± 27 nM).
A second parameter undergoing change upon introducing a target site is the hill coefficient of the binding curve. When the substrate contains a cytidine, the Hill coefficient is ~ 2 (Figures 1a and 1b, Table 1), whereas the Hill coefficient is ~1 in the absence of a cytidine. This difference in hill coefficient may imply that APOBEC3A is in a monomeric “low affinity” state in the absence of a target cytidine but assumes a dimeric “high affinity” state when encountering a deamination substrate. Such a binding mechanism would predict that the affinity to the product of the deamination reaction would likely also be weaker. To test this hypothesis we replaced the cytidine base with a uracil, in which case both the affinity and Hill coefficient are similar to those for the all-thymidine oligomer. (1U KD = 434 ± 35 nM, h = 0.80 ± 0.04; TTT KD = 502 ± 27 nM, h = 0.96 ± 0.04) (Figure 1c, Table 1).
Crystal structure of APOBEC3A
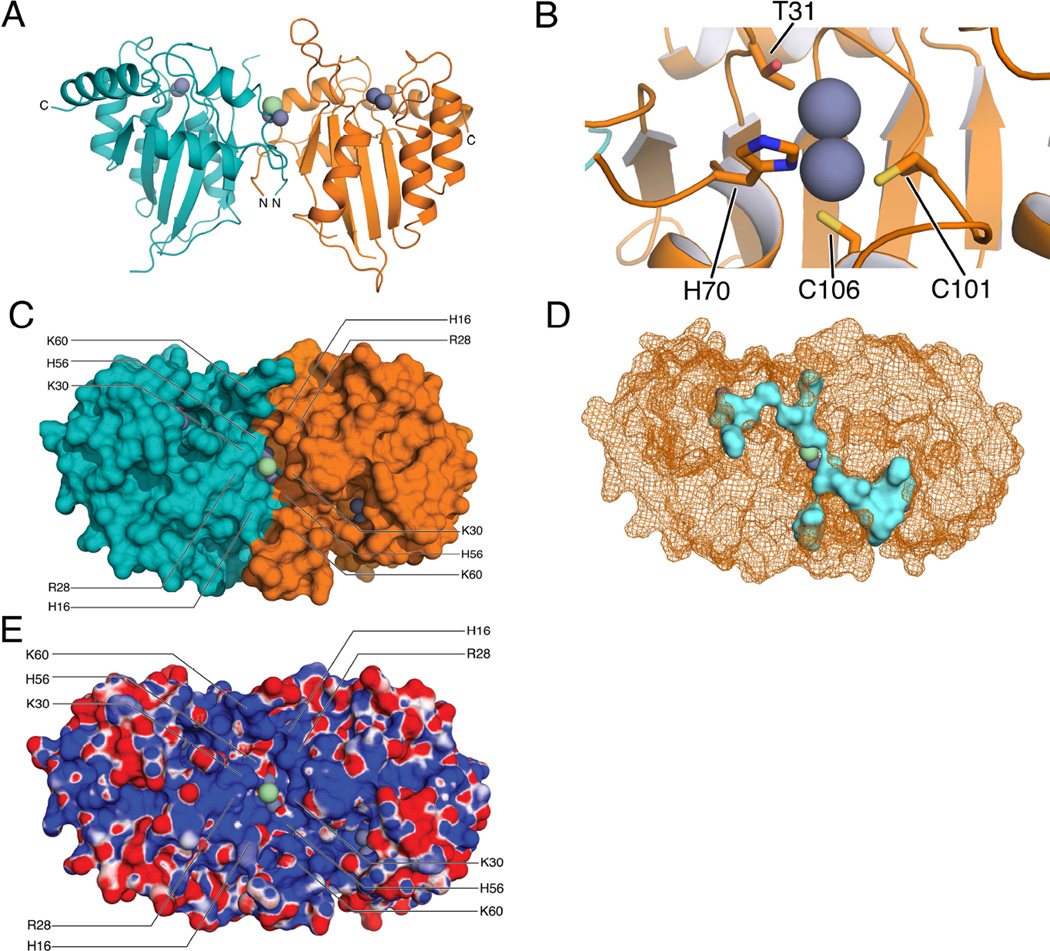
The crystal structure of A3A-E72A-C171A was determined to 2.85 Å resolution with two molecules per asymmetric unit (Figure 2a) forming a homodimer. The A3A dimer crystallized in spacegroup P6522 and refined with good statistics (Table 2).
Figure 2. Crystal structure of APOBEC3A.
(A) Crystallographic dimer of APOBEC3A. The two monomers are in orange and cyan, with the metal ions at the active sites depicted as steel grey spheres and chloride ions at the dimerization interface shown as green spheres. (B) Close-up view of the active site. Recruitment of a second metal via a threonine residue (T31) protects active site geometry in the presence of a mutation of the catalytic glutamate residue. (C) Surface representation of the dimeric structure in A reveals a groove connecting both active sites via the dimer interface. (D) SiteMap prediction (Halgren, 2009) of putative binding sites (blue) on a wireframe representation of the surface in C matches the groove connecting both active sites. (E) Electrostatic potential map, same orientation as A and C. Positive (blue) and negative (red) charges are indicated on the surface. The groove connecting the two active sites is mainly positively charged. See also Supplemental Figure 1 for a superposition of both monomers.
Table 2.
Crystallographic statistics for APOBEC3A structure.
| resolution | 2.85 Å |
| temperature | cryogenic (−180 °C) |
| spacegroup | P6522 |
| Cell dimensions | |
| a | 94.75 Å |
| b | 94.75 Å |
| c | 213.96 Å |
| α | 90.00° |
| β | 90.00° |
| γ | 120.00° |
| molecules in AU | 2 |
| completeness | 99.7% |
| total reflections | 153753 |
| unique reflections | 14142 |
| I/sigI (mean) | 7.0 |
| I/sigI (outer shell) | 0.6 |
| av redundancy | 11.0 |
| rmerge (linear) | 0.239 |
| rmeas | 0.251 |
| rpim | 0.091 |
| CC1/2 (outer shell) | 0.665 |
| CC* (outer shell) | 0.894 |
| rmsd in: | |
| bonds | 0.002 Å |
| angles | 0.59° |
| Rfactor | 0.21 |
| Rfree | 0.25 |
As was observed in the NMR solution structure (Byeon et al., 2013), A3A has a canonical DNA cytosine deaminase fold, composed of five β-strands, six α-helices and the catalytic zinc-binding site. The zinc atom is coordinated by direct interactions with H70, C101, C106 and, as the catalytic glutamic acid (E72A) is inactive, to what appears to be a second zinc ion which is 3.3 Å apart from and appears to stabilize the geometry around the catalytic zinc (Figure 2b). This type of site with two zinc ions in close proximity resembles cocatalytic zinc sites which can be found in class III hydrolases (Auld, 2001). Comparing with the NMR ensemble of catalytically active A3A (Byeon et al., 2013) shows that the loop connecting the zinc-coordinating cysteine residues has moved, which is necessary to conserve the active site geometry in the presence of the E72A mutation and allows recruitment of T31 to coordinate the second ion. This loop contains 104-WG-105, an insertion unique to A3A and the closely related C-terminal domain of A3B. All zinc-coordinating residues are located in helices α2 and α3 that also provide a structural backbone for the catalytic pocket.
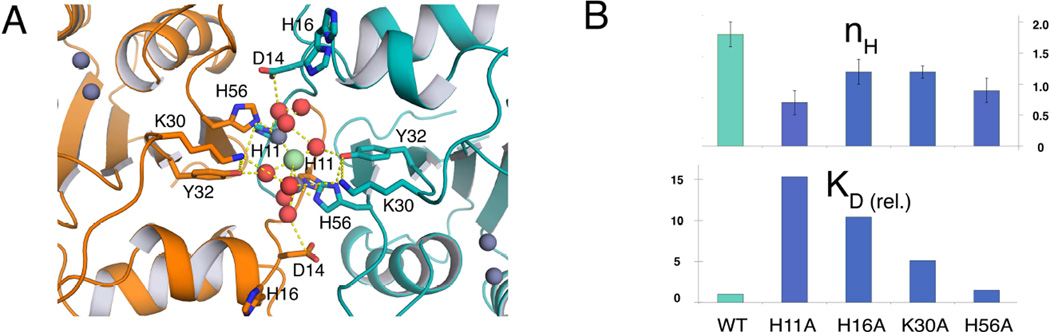
The crystal structure reveals that A3A forms a dimer that defines the asymmetric unit. This dimer is formed via a symmetric domain swap between two A3A molecules. The RMSD between the two molecules is 0.64 Å with the largest deviations in residues 25–27, 50 and 86–87 with Cα-Cα distances of 1.3 Å (Supplemental Figure 1). 17 residues that form the N-terminal loop regions of both molecules form an intimate handshake (Figure 2a) burying >1000 Å2 acting as the dimerization interface. The dimerization interface is away from the active sites and coordinates three metal ions by residues H11 and H56 from both chains and a network of water molecules. In addition, the side chains of residues K30 are coordinated via a water molecule at the interface bridging the two molecules (Figure 3a).
Figure 3. Residues contributing to interface formation are determinants for cooperativity and affinity.
(A) The dimer interface in the APOBEC3A crystal structure. Residues forming the dimerization interface are shown as sticks and zinc (grey), chloride (green) and water (red) as spheres. Side-chain oxygen and nitrogen atoms are colored red and green, respectively. (B) Bar graphs show how point mutations at the highlighted sites affect KD and hill coefficient. WT represents data collected for A3A-E72A-C171A. KD (rel.) represents the fold change in KD relative to A3A-E72A-C171A binding polyT_1C.
On the surface of the dimer (Figure 2c), a symmetric groove of 36 Å length connects the active site regions of the two molecules via the dimerization interface. This groove has been identified as a potential ligand binding site with a calculated (Halgren, 2009) pocket volume of ~800 Å3 (Figure 2d). Based on surface electrostatic potential the groove is mostly positively charged (Figure 2e). Residues H16, R28, K30, H56 and K60 are prominently contributing to both the accessible pocket volume and charge.
Assessing the functional significance of the crystallographic dimer
The crystal structure allowed us to identify and test the potential determinants for substrate recognition and binding. Based on the crystallographic dimer we engineered a series of mutant constructs to probe whether the observed dimer might play a role in substrate binding. If the crystallographic dimer indeed corresponds to a biochemically relevant structure then the designed mutations should affect and provide insights into substrate recognition.
Since the crystallographic interface brings the N-terminal 17 residues in close proximity, we tested an N-terminally truncated version of A3A to investigate the potential role of the crystallographic dimer as the structure responsible for the observed cooperativity. Under the same experimental conditions as used above, the truncated protein lost the high affinity to the ideal substrate (Supplemental Figure 2), and protein expression yields and the construct’s solubility were severely compromised.
In order to confirm that the active site rearrangement caused by the E72A mutation did not cause the observed differential substrate recognition we repeated the binding experiment presented in Figure 1c with an E72Q variant A3A E72Q exhibits the same kind of marked increase in affinity and cooperativity upon encountering the target cytidine (Supplemental Figure 3, Table 1).
The crystal structure was further used to narrow down the determinants for dimerization and identify sites amenable to single amino acid substitutions based on the charge distribution and interatomic distances described above. A series of mutant proteins were then engineered to measure the contribution of these individual amino acids to cooperativity and affinity (Figure 3). From the structure two pairs of residues were identified that might contribute to dimer formation. H11 and H56 are the first set, which forms the base of the dimerization interface (Figure 3a, blue sticks) not only disrupt the protein-protein interface but also severely affect cooperativity of binding H11A (KD = 855 ± 311 nM, h = 0.7 ± 0.1) and H56A (KD = 86 ± 13 nM, h = 0.9 ± 0.1). A second set of ionic residues H16 and K30 closer to the surface (Figure 3a, purple sticks) of the groove also reduce cooperativity markedly H16A (KD = 584 ± 92 nM, h = 1.2 ± 0.2) and K30E (KD = 284 ± 20 nM, h = 1.2 ± 0.1) and greatly compromise affinity. H56A was the only mutation drastically reducing cooperativity while maintaining substrate affinity. Together, the results show that both sets of interactions predicted by the dimerization interface observed in the crystal structure are critical to cooperative DNA recognition in solution.
Discussion
In this study we characterized the cooperativity in specific binding of A3A to ssDNA substrate, determined the crystal structure of the A3A dimer, and engineered mutations that interrogated the functional implications of this dimer interface. We found that A3A recognizes substrate cooperatively and with high affinity and specificity. Key to this recognition is the A3A dimer that forms an extensive positively charged groove connecting the active sites of both monomers. A3A exists as a monomer and as dimer both in solution and bound to substrate (Logue et al., 2014; Shlyakhtenko et al., 2014) yet a functional implication for transition between those states was missing. The identification of this substrate-binding groove and mutational analysis provide key insights into structural basis of A3A substrate specificity, and helps explain the previously reported apparent discrepancies on A3A function.
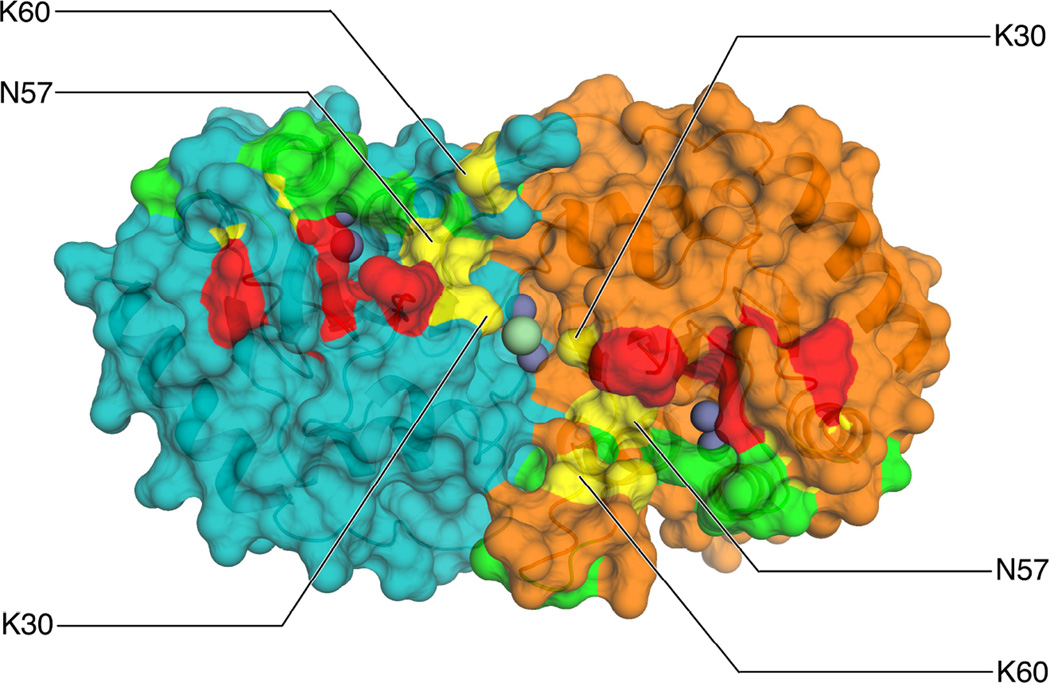
The contiguous and positively charged groove on the A3A surface is consistent with DNA recognition and binding. In fact, many residues around the active site region appear to be involved in substrate binding. Our structure unifies many of the residues previously associated with ssDNA binding to A3A (Bulliard et al., 2011; Byeon et al., 2013; Mitra et al., 2014) by mapping them to a contiguous band on the molecular surface crossing the dimer interface (Figure 4, Supplemental Figure 4). This bridges the data from the two previous studies where only relatively few residues identified were overlapping (Figure 4, yellow), most likely experimental conditions account for the differences (Figure 4, green (Mitra et al., 2014) and red (Bulliard et al., 2011)). Our crystal structure demonstrates the consistency of both studies in that all residues identified by either study lie within the groove. In fact two residues outside the active site both studies identified, K30 and K60 that bridge the dimer interface and contribute to the grooves charge, respectively, impact substrate affinity and deamination (Bulliard et al., 2011; Mitra et al., 2014). The H56A mutant, which has a significant impact on dimerization and a much smaller effect on substrate affinity, seems to allow a separation of function and shows that high affinity can be achieved in the monomeric state as well.
Figure 4. Residues implicated in deamination activity.
Shown are independently determined substrate binding surfaces by enzymatic activity (red) (Bulliard et al., 2011) or chemical shift perturbation accompanied by enzymatic activity (green) (Mitra et al., 2014). Residues identified in both studies are colored yellow. See also Supplemental Figure 4.
Although A3A can and does exist in monomeric form in solution and in the cell (Li et al., 2014; Logue et al., 2014; Shlyakhtenko et al., 2014), our analysis strongly implicates the high-affinity DNA-binding functional form is a homodimer formed by swapping the N-terminal loop. A naturally occurring isoform of A3A, which lacks the initial 12 residues (Stenglein et al., 2010; Thielen et al., 2010) was described to be 5-fold less active compared to the full length enzyme (Carpenter et al., 2012) in an in vitro deamination assay. This only modest reduction in activity could arise from key residues of the interface lying outside of the N-terminus. Dimerization via the N-terminus could be communicated via K30 and H56, which are involved in interface formation, to neighboring residues T31 and N57 forming the pocket containing the catalytic site. Residues H56 and K30, which form the top of the groove of the dimerization interface, are also positioned to favorably interact with the negatively charged phosphates of the substrate and thus can not only communicate dimerization, but, more specifically, dimerization on the substrate to the active site region.
The necessity for A3A to form a cooperative dimer for high affinity binding effectively explains the apparent discrepancy between high enzymatic activity and the great variation in reported substrate affinities (Byeon et al., 2013; Love et al., 2012; Mitra et al., 2014; Pham et al., 2013). As we show, affinity drops by an order of magnitude and the hill coefficient drops dramatically when binding to product is compared to substrate binding. Since most binding experiments to determine affinity are conducted at equilibrium and A3A has very fast deamination rates, experiments done with active enzyme will observe binding to the reaction product instead of the substrate. Weaker and monomeric binding to product corroborates our results (Shlyakhtenko et al., 2014) in which, after incubation with substrate, active A3A was observed in a predominantly monomeric form with whereas inactive A3A would form a dimer on substrate. A recent study in which the catalytically deficient E72A mutant was used was also able to measure a similarly high affinity to the ssDNA substrate (Logue et al., 2014). In the presence of active A3A, substrate would quickly have been turned over to product and therefore appear to bind with low affinity. Taken together the differences in binding product and substrate explain why binding with high affinity and in a dimeric state could only be observed for catalytically inactive enzyme.
A cooperative model of APOBEC3 activity also explains the ability to achieve the required fidelity of substrate recognition despite high deamination activity. Being the most active deaminase of the APOBEC3 family, A3A also serves as the most effective restrictor of foreign DNA (Stenglein et al., 2010). Less discriminatory than other APOBEC3 enzymes and the only known family member reported to deaminate modified cytidine residues (Carpenter et al., 2012; Suspène et al., 2013; Wijesinghe and Bhagwat, 2012) A3A can also be implied as an agent in demethylation pathways (Franchini et al., 2012; Guo et al., 2011). At the same time, the random nature of mutations introduced by A3A can be very detrimental to cell viability (Burns et al., 2013a; Stenglein et al., 2010) or can be the source for mutations in cancer, as has been shown for the close relative A3B, the catalytic domain of which shares 97% similarity with A3A. The cooperative model of substrate interaction for A3A can have implications on how the mutation load inflicted by A3A is regulated in vivo. At low concentrations and in the dense milieu of the cell the enzyme would encounter a short, exposed ssDNA substrate mostly as monomer with only modest affinity. At higher concentrations and in the presence of longer stretches of foreign ssDNA A3A can act with very high specificity and affinity on a target sequence, leading to the observed high rates of deamination. Also, the change in binding affinity towards a stretch of thymidine bases (KD = 502 ± 27 nM) upon introduction of a single cytidine (KD = 56 ± 2) could suggest a possible mechanism of substrate binding wherein A3A initially binds the thymidine bases with low affinity, followed by identifying the target cytidine and binding more tightly.
The cooperative binding model may also provide insights into the evolution of APOBEC3 domain structure. Four of the seven members of the human APOBEC3 protein family (A3B, A3D, A3F and A3G) comprise two cytidine deaminase domains connected via a short linker. This repertoire of double-domain APOBEC3 proteins likely evolved during a series of gene duplication events from single domain precursors (Jarmuz et al., 2002; Wedekind et al., 2003). The evolutionary linkage between the members of the modern primate APOBEC3 locus can be understood from a prototypical set of single domain APOBEC3 proteins, one for each Z-domain subtype (LaRue et al., 2008). In primates A3A is the sole member comprising a single Z1 domain, sharing phylogenetic origin with the catalytic domains of A3B and A3G. Both of these enzymes possess a pseudocatalytic N-terminal Z2 domain which is required for efficient substrate binding but does not catalyze a deamination reaction (Chelico et al., 2010; Hache et al., 2005). The functional A3A dimer identified here might provide a reason why APOBEC3’s might form two domain fusions. With a single target site, one of the A3A monomers does not act on the target cytidine but is involved in substrate binding. The evolution of double-domain enzymes appears to have allowed for a separation of function between binding and catalysis leading to less active proteins that became more specific to their target. The interdomain linker region has recently been shown to have the determinants for processivity in A3G and A3F and alterations in the linker can impair enzyme function (Ara et al., 2014).
Damage in ssDNA was shown to be a major source for mutation clusters in cancer (Roberts et al., 2012; Sakofsky et al., 2014) and can also contribute to the diversity of viral genomes (Kim et al., 2010; Sadler et al., 2010). Targeting the activity of deaminases may have implications for novel strategies in treatment of infectious diseases and cancer therapies and the insights into the structural mechanism of substrate binding described in this study could help guide efforts to alleviate detrimental mutagenic activity of cellular deaminases.
Materials and Methods
Expression and Purification of APOBEC3A-E72A-C171A
E. coli BL21 DE3 Star (Stratagene) cells were transformed with a pColdIII vector (Takara Biosciences) encoding a Glutathione-S-transferase (GST)-based construct. The E72A mutation was chosen to render the protein inactive and C171A to increase solubility. Expression occurred at 16 °C for 22 h in Lysogeny Broth medium containing 1 mM IPTG and 100 µg/mL ampicillin. Cells were pelleted, resuspended in purification buffer (50 mM Tris-HCl pH 8.0, 300 mM NaCl, 1 mM DTT) and disrupted through sonication. Cellular debris was separated by centrifugation (45,000 g, 30 min, 4 °C). The fusion protein was separated using Glutathione Sepharose (GE Healthcare). The GST-tag was removed by means of a PreScission protease digest over night at 4 °C. Size-exclusion chromatography using a HiLoad 16/60 Superdex 75 column (GE Healthcare) was used as a final purification step.
Crystallization and Structural data analysis of APOBEC3A-E72A-C171A
The protein solution was concentrated to 19.5 mg/mL in crystallization buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM DTT, 50 µM ZnCl) and crystals were grown at 4 °C in crystallization solution (100 mM Sodium-Cacodylate pH 6.0, 40% MPD, 8% PEG8000) by sitting-drop vapor diffusion over 3 years.
Diffraction experiments were conducted using a rotating anode x-ray source (Rigaku Micromax-007 HF) and CCD detector (Rigaku Saturn 944) at 100 K.
Data were indexed and scaled using the software HKL2000 (Otwinowski and Minor, 1997). CC1/2 and CC* were used to determine the data cutoff. The molecular replacement solution was calculated by PHASER (McCoy, 2007) using PDB ID: 3V4K as a search model (Li et al., 2012). The structure was rebuilt using phenix.autobuild (Zwart et al., 2008). An automated pipeline (REdiii) was used for processing data from subsequent diffraction experiments (Frederick-Bohn, unpublished). Multiple crystals were diffracted with exposure times between 2–4 min per oscillation image; only one led to diffraction spots beyond 3 Å. Refinement was carried out using Coot (Emsley and Cowtan, 2004) and phenix.refine (Zwart et al., 2008). Molecular graphics images were generated using PyMOL (Schrödinger LLC) (DeLano, 2002). SiteMap (Halgren, 2009) was used to identify and evaluate volumes of binding sites, using a fine grid to search around the Zn2+ atoms in the dimerization interface.
High-throughput DNA binding assay
Carboxytetramethylrhodamine (5’-TAMRA) labeled ssDNA (IDT) served as substrate (sequences are listed in Supplemental Table 1). 10 nM of substrate was added to A3A-E72A-C171A in concentrations varying between 10 nM and 20 µM, and to a control without protein. The A3A protein concentrations were 0, 10, 25, 50, 75, 100, 125, 150, 175, 200, 250, 300, 350, 400, 450, 500, and 750 nM; 1, 1.25, 1.5, 2, 3, 4, 5, 6, 7, 8, 9, 10, 15 and 20 µM. The mixtures were incubated for 1 h at room temperature in non-binding 96-well plates (Greiner) in 50 mM MES pH 6.0, 100 mM NaCl, 2 mM TCEP in a total reaction volume of 150 µL per well.. Fluorescence anisotropy was measured for triplicate experiments using an EnVision plate reader (PerkinElmer) equipped with the Optimized Tamra Acyclo Prime SNP Label detection kit, allowing excitation at 531 nm and detection of polarized emission at 579 nm wavelength. Data analysis was performed using Prism (GraphPad) performing least-square fitting of the measured fluorescence anisotropy values (Y) at different protein concentrations (X) with a single-site binding curve with hill slope, a non-specific linear term and a constant background using the equation Y=(Bmax*X^h)/(Kd^h + X^h)+NS*X+Background, where Kd is the equilibrium dissociation constant, h is the hill coefficient, and Bmax is the extrapolated maximum anisotropy at complete binding. Standard deviation was calculated for each measurement point from the three independent repeats, and is shown as error bars in the corresponding data figures.
Supplementary Material
Highlights.
Single domain APOBEC3A binds cooperatively to ssDNA substrate but not product
Crystal structure of APOBEC3A reveals a homodimer coordinated by Zinc
Mutational interrogation of dimer interface disrupts cooperatively
Single domain A3A homodimer provides an APOBEC3 cooperativity and regulation model
Acknowledgments
This work was supported by the National Institute of General Medical Sciences P01 GM091743. TVS was supported by NIH P01 GM091743-03S1. We would like to thank Dr Chojiro Kojima for the gift of the pCold vector used in this research and Dr. Hiroshi Matsuo for plasmids.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The Protein Data Bank accession number for the structure factors and coordinates for A3A-E72A-C171A reported in this paper is 4XXO.
References
- Ara A, Love RP, Chelico L. Different Mutagenic Potential of HIV-1 Restriction Factors APOBEC3G and APOBEC3F Is Determined by Distinct Single-Stranded DNA Scanning Mechanisms. PLoS Pathog. 2014;10:e1004024. doi: 10.1371/journal.ppat.1004024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auld DS. Zinc coordination sphere in biochemical zinc sites. Biometals. 2001;14:271–313. doi: 10.1023/a:1012976615056. [DOI] [PubMed] [Google Scholar]
- Betts L, Xiang S, Short SA, Wolfenden R, Carter CW. Cytidine deaminase. The 2.3 A crystal structure of an enzyme: transition-state analog complex. Journal of Molecular Biology. 1994;235:635–656. doi: 10.1006/jmbi.1994.1018. [DOI] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho S-J, Malim MH. Cytidine Deamination of Retroviral DNA by Diverse APOBEC Proteins. Current Biology. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Bohn M-F, Shandilya SMD, Albin JS, Kouno T, Anderson BD, McDougle RM, Carpenter MA, Rathore A, Evans L, Davis AN, et al. Crystal Structure of the DNA Cytosine Deaminase APOBEC3F: The Catalytically Active and HIV-1 Vif-Binding Domain. Structure. 2013;21:1042–1050. doi: 10.1016/j.str.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulliard Y, Narvaiza I, Bertero A, Peddi S, Röhrig UF, Ortiz M, Zoete V, Castro-Díaz N, Turelli P, Telenti A, et al. Structure-function analyses point to a polynucleotide-accommodating groove essential for APOBEC3A restriction activities. J Virol. 2011;85:1765–1776. doi: 10.1128/JVI.01651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013a;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet. 2013b;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byeon I-JL, Ahn J, Mitra M, Byeon C-H, Hercík K, Hritz J, Charlton LM, Levin JG, Gronenborn AM. NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity. Nat Commun. 2013;4:1890. doi: 10.1038/ncomms2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter MA, Li M, Rathore A, Lackey L, Law EK, Land AM, Leonard B, Shandilya SMD, Bohn M-F, Schiffer CA, et al. Methylcytosine and Normal Cytosine Deamination by the Foreign DNA Restriction Enzyme APOBEC3A. J. Biol. Chem. 2012;287:34801–34808. doi: 10.1074/jbc.M112.385161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caval V, Suspène R, Shapira M, Vartanian J-P, Wain-Hobson S. A prevalent cancer susceptibility APOBEC3A hybrid allele bearing APOBEC3B 3’UTR enhances chromosomal DNA damage. Nat Commun. 2014;5:5129. doi: 10.1038/ncomms6129. [DOI] [PubMed] [Google Scholar]
- Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3” --> 5” on single-stranded DNA. Nat Struct Mol Biol. 2006;13:392–399. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- Chelico L, Sacho EJ, Erie DA, Goodman MF. A model for oligomeric regulation of APOBEC3G cytosine deaminase-dependent restriction of HIV. J Biol Chem. 2008;283:13780–13791. doi: 10.1074/jbc.M801004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelico L, Prochnow C, Erie DA, Chen XS, Goodman MF. Structural model for deoxycytidine deamination mechanisms of the HIV-1 inactivation enzyme APOBEC3G. J. Biol. Chem. 2010;285:16195–16205. doi: 10.1074/jbc.M110.107987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K-MM, Harjes E, Gross PJ, Fahmy A, Lu Y, Shindo K, Harris RS, Matsuo H. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–119. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- Dang Y, Wang X, Esselman WJ, Zheng Y-H. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J Virol. 2006;80:10522–10533. doi: 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, Jamal-Hanjani M, Shafi S, Murugaesu N, Rowan AJ, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL2. The PyMOL User’s Manual. San Carlos, CA, USA: DeLano Scientific LLC; 2002. [Google Scholar]
- Emsley P, Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallographica. Section D: Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Franchini D-M, Schmitz K-M, Petersen-Mahrt SK. 5-Methylcytosine DNA Demethylation: More Than Losing a Methyl Group. Annu. Rev. Genet. 2012;46:419–441. doi: 10.1146/annurev-genet-110711-155451. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming G-L, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hache G, Liddament MT, Harris RS. The Retroviral Hypermutation Specificity of APOBEC3F and APOBEC3G is Governed by the C-terminal DNA Cytosine Deaminase Domain. J Biol Chem. 2005;280:10920–10924. doi: 10.1074/jbc.M500382200. [DOI] [PubMed] [Google Scholar]
- Halgren TA. Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model. 2009;49:377–389. doi: 10.1021/ci800324m. [DOI] [PubMed] [Google Scholar]
- Harari A, Ooms M, Mulder LCF, Simon V. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J Virol. 2009;83:295–303. doi: 10.1128/JVI.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harjes E, Gross PJ, Chen K-MM, Lu Y, Shindo K, Nowarski R, Gross JD, Kotler M, Harris RS, Matsuo H. An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model. Journal of Molecular Biology. 2009;389:819–832. doi: 10.1016/j.jmb.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- Holden LG, Prochnow C, Chang YP, Bransteitter R, Chelico L, Sen U, Stevens RC, Goodman MF, Chen XS. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008;456:121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultquist JF, Lengyel JA, Refsland EW, Larue RS, Lackey L, Brown WL, Harris RS. Human and Rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H Demonstrate a Conserved Capacity to Restrict Vif-deficient HIV-1. J Virol. 2011;85:1765–1776. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani Y, Takeuchi H, Strebel K, Levin JG. Biochemical Activities of Highly Purified, Catalytically Active Human APOBEC3G: Correlation with Antiviral Effect. J Virol. 2006;80:5992–6002. doi: 10.1128/JVI.02680-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- Kim E-Y, Bhattacharya T, Kunstman K, Swantek P, Koning FA, Malim MH, Wolinsky SM. Human APOBEC3G–mediated editing can promote HIV-1 sequence diversification and accelerate adaptation to selective pressure. J Virol. 2010;84:10402–10405. doi: 10.1128/JVI.01223-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura S, Ode H, Iwatani Y. Structural Features of Antiviral APOBEC3 Proteins are Linked to Their Functional Activities. Front Microbiol. 2011;2:258. doi: 10.3389/fmicb.2011.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry S, Narvaiza I, Linfesty DC, Weitzman MD. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 2011;12:444–450. doi: 10.1038/embor.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRue RS, Andresdottir V, Blanchard Y, Conticello SG, Derse D, Emerman M, Greene WC, Jonsson SR, Landau NR, Lochelt M, et al. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol. 2009;83:494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRue RS, Jónsson SR, Silverstein KAT, Lajoie M, Bertrand D, El-Mabrouk N, Hötzel I, Andrésdóttir V, Smith TPL, Harris RS. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol Biol. 2008;9:104. doi: 10.1186/1471-2199-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecossier D, Bouchonnet F, Clavel F, Hance AJ. Hypermutation of HIV-1 DNA in the Absence of the Vif Protein. Science. 2003;300:1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- Li J, Chen Y, Li M, Carpenter MA, McDougle RM, Luengas EM, Macdonald PJ, Harris RS, Mueller JD. APOBEC3 multimerization correlates with HIV-1 packaging and restriction activity in living cells. Journal of Molecular Biology. 2014;426:1296–1307. doi: 10.1016/j.jmb.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Shandilya SMD, Carpenter MA, Rathore A, Brown WL, Perkins AL, Harki DA, Solberg J, Hook DJ, Pandey KK, et al. First-in-class small molecule inhibitors of the single-strand DNA cytosine deaminase APOBEC3G. ACS Chemical Biology. 2012;7:506–517. doi: 10.1021/cb200440y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddament MT, Brown WL, Schumacher AJ, Harris RS. APOBEC3F Properties and Hypermutation Preferences Indicate Activity against HIV-1 in Vivo. Current Biology. 2004;14:1385–1391. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- Loeb LA, Essigmann JM, Kazazi F, Zhang J, Rose KD, Mullins JI. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc. Natl. Acad. Sci. U.S.a. 1999;96:1492–1497. doi: 10.1073/pnas.96.4.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue EC, Bloch N, Dhuey E, Zhang R, Cao P, Herate C, Chauveau L, Hubbard SR, Landau NR. A DNA Sequence Recognition Loop on APOBEC3A Controls Substrate Specificity. PLOS One. 2014;9:e97062. doi: 10.1371/journal.pone.0097062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love RP, Xu H, Chelico L. Biochemical Analysis of Hypermutation by the Deoxycytidine Deaminase APOBEC3A. J. Biol. Chem. 2012;287:30812–30822. doi: 10.1074/jbc.M112.393181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallographica. Section D: Biological Crystallography. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra M, Hercík K, Byeon I-JL, Ahn J, Hill S, Hinchee-Rodriguez K, Singer D, Byeon C-H, Charlton LM, Nam G, et al. Structural determinants of human APOBEC3A enzymatic and nucleic acid binding properties. Nucleic Acids Res. 2014;42:1095–1110. doi: 10.1093/nar/gkt945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Wedge DC, Alexandrov LB, Petljak M, Butler AP, Bolli N, Davies HR, Knappskog S, Martin S, Papaemmanuil E, et al. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat. Genet. 2014;46:487–491. doi: 10.1038/ng.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OhAinle M, Kerns JA, Li MM, Malik HS, Emerman M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe. 2008;4:249–259. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pham P, Landolph A, Mendez C, Li N, Goodman MF. A Biochemical Analysis Linking APOBEC3A to Disparate HIV-1 Restriction and Skin Cancer. J. Biol. Chem. 2013 doi: 10.1074/jbc.M113.504175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol. Cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler HA, Stenglein MD, Harris RS, Mansky LM. APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis. J Virol. 2010;84:7396–7404. doi: 10.1128/JVI.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakofsky CJ, Roberts SA, Malc E, Mieczkowski PA, Resnick MA, Gordenin DA, Malkova A. Break-induced replication is a source of mutation clusters underlying kataegis. Cell Rep. 2014;7:1640–1648. doi: 10.1016/j.celrep.2014.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shandilya SMD, Bohn M-F, Schiffer CA. A computational analysis of the structural determinants of APOBEC3’s catalytic activity and vulnerability to HIV-1 Vif. Virology. 2014;471–473:105–116. doi: 10.1016/j.virol.2014.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shandilya SMD, Nalam MNL, Nalivaika EA, Gross PJ, Valesano JC, Shindo K, Li M, Munson M, Royer WE, Harjes E, et al. Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure. 2010;18:28–38. doi: 10.1016/j.str.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Lushnikov AY, Li M, Lackey L, Harris RS, Lyubchenko YL. Atomic force microscopy studies provide direct evidence for dimerization of the HIV restriction factor APOBEC3G. J. Biol. Chem. 2011;286:3387–3395. doi: 10.1074/jbc.M110.195685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Lushnikov AJ, Li M, Harris RS, Lyubchenko YL. Interaction of APOBEC3A with DNA Assessed by Atomic Force Microscopy. PLOS One. 2014;9:e99354. doi: 10.1371/journal.pone.0099354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Lushnikov AY, Miyagi A, Li M, Harris RS, Lyubchenko YL. Atomic force microscopy studies of APOBEC3G oligomerization and dynamics. Journal of Structural Biology. 2013;184:217–225. doi: 10.1016/j.jsb.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nature Structural & Molecular Biology. 2010;17:222–229. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suspène R, Aynaud M-M, Vartanian J-P, Wain-Hobson S. Efficient deamination of 5-methylcytidine and 5-substituted cytidine residues in DNA by human APOBEC3A cytidine deaminase. PLOS One. 2013;8:e63461. doi: 10.1371/journal.pone.0063461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thielen BK, McNevin JP, McElrath MJ, Hunt BVS, Klein KC, Lingappa JR. Innate immune signaling induces high levels of TC-specific deaminase activity in primary monocyte-derived cells through expression of APOBEC3A isoforms. J. Biol. Chem. 2010;285:27753–27766. doi: 10.1074/jbc.M110.102822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Tian C, Zhang W, Luo K, Sarkis PTN, Yu L, Liu B, Yu Y, Yu X-F. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J Virol. 2007;81:13112–13124. doi: 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedekind JE, Dance GSC, Sowden MP, Smith HC. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends in Genetics. 2003;19:207–216. doi: 10.1016/S0168-9525(03)00054-4. [DOI] [PubMed] [Google Scholar]
- Wedekind JE, Gillilan R, Janda A, Krucinska J, Salter JD, Bennett RP, Raina J, Smith HC. Nanostructures of APOBEC3G support a hierarchical assembly model of high molecular mass ribonucleoprotein particles from dimeric subunits. J Biol Chem. 2006;281:38122–38126. doi: 10.1074/jbc.C600253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil AF, Ghosh D, Zhou Y, Seiple L, McMahon MA, Spivak AM, Siliciano RF, Stivers JT. Uracil DNA glycosylase initiates degradation of HIV-1 cDNA containing misincorporated dUTP and prevents viral integration. Proc. Natl. Acad. Sci. U.S.a. 2013;110:E448–E457. doi: 10.1073/pnas.1219702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. Embo J. 2004;23:2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijesinghe P, Bhagwat AS. Efficient deamination of 5-methylcytosines in DNA by human APOBEC3A, but not by AID or APOBEC3G. Nucleic Acids Res. 2012;40:9206–9217. doi: 10.1093/nar/gks685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DK, Rudolph FB, Quiocho FA. Atomic structure of adenosine deaminase complexed with a transition-state analog: understanding catalysis and immunodeficiency mutations. Science. 1991;252:1278–1284. doi: 10.1126/science.1925539. [DOI] [PubMed] [Google Scholar]
- Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nature Structural & Molecular Biology. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- Zhang H, Pomerantz RJ, Zhang C, Arunachalam SC, Yang B, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y-H, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM. Human APOBEC3F Is Another Host Factor That Blocks Human Immunodeficiency Virus Type 1 Replication. J Virol. 2004;78:6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, et al. Automated structure solution with the PHENIX suite. Methods in Molecular Biology. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.