Abstract
Botrytis cinerea, a model necrotrophic fungal pathogen that causes gray mold as it infects different organs on more than 200 plant species, is a significant contributor to post-harvest rot in fresh fruit and vegetables, including tomatoes. By describing host and pathogen proteomes simultaneously in infected tissues, the plant proteins that provide resistance and allow susceptibility and the pathogen proteins that promote colonization and facilitate quiescence can be identified. This study characterizes fruit and fungal proteins solubilized in the B. cinerea-tomato interaction using shotgun proteomics. Mature green, red ripe wild type and ripening inhibited (rin) mutant tomato fruit were infected with B. cinerea B05.10 and the fruit and fungal proteomes were identified concurrently 3 days post-infection. One hundred and eighty-six tomato proteins were identified in common among red ripe and red ripe-equivalent ripening inhibited (rin) mutant tomato fruit infected by B. cinerea. However, the limited infections by B. cinerea of mature green wild type fruit resulted in 25 and 33% fewer defense-related tomato proteins than in red and rin fruit, respectively. In contrast, the ripening stage of genotype of the fruit infected, did not affected the secreted proteomes of B. cinerea. The composition of the collected proteins populations and the putative functions of the identified proteins argue for their role in plant-pathogen interactions.
Keywords: Botrytis cinerea, tomato, host-pathogen interaction, mass spectrometry
Graphical abstract
Table of Contents (TOC) Graphic & Synopsis: Characterization of fruit and fungal proteins solubilized into the microenvironment of the interaction sites of B. cinerea infection of tomato fruit using shotgun proteomics. This proteomic approach obtains sufficient information to identify both pathogen and host proteins from sites of infection and to describe the diverse classes of proteins present
Introduction
Botrytis cinerea (Pers.) [tel. Botryotinia fuckeliana] is a necrotrophic phytopathogenic fungus that grows particularly aggressively on the senescing aerial tissues of more than 200 plant species. B. cinerea causes gray mold on many economically important crops; pre- and post-harvest rotting infections by B. cinerea of fruits, vegetables, ornamental leaves and flowers cause significant losses.1 Because B. cinerea infects many types of tissues under a variety of conditions, the fungus is likely to have diverse infection and growth strategies.2 Furthermore, susceptibility to B. cinerea changes as the tissues develop and age. Green unripe fruit are largely resistant to rotting by B. cinerea, but ripe fruit are particularly susceptible, although B. cinerea is able to infect at least some non-ripening fruit.3-5
The interaction between plants and fungi includes communication that is undoubtedly in the form of proteins present in the microenvironment where the infections occur. The host-pathogen interaction is complex and dynamic, and is only partially elucidated by examining transcript abundances. Identifying which plant and which pathogenic proteins are present in infected sites may reveal some of the proteins that comprise the communication between the plant and the pathogen and may contribute to developing novel disease control strategies.
Limited proteomic information is available about plants interacting with pathogens. Proteomic work has cataloged plant, but not pathogen, proteomes in infected tissues and resulted in the identification of less than 100 proteins. A study of infections of pea with powdery mildew (Erisphye pisi) identified fewer than 100 pea proteins.6 Infection of Arabidopsis suspension cells with Pseudomonas syringae resulted in the secretion collection of 45 Arabidopsis secreted proteins,7 and similar sized collections of proteins were obtained from Xanthamonas campestris infection of Brassica oleracea although in this case most of the proteins identified were of bacterial origin.8 The proteome of tomato fruits (Lycopersicon esculentum) infected with TMV identified 16 proteins. including several pathogenesis-related (PR) proteins and antioxidant enzymes found that may be part of the plant resistance response to viral infection.9 In another Arabidopsis proteome study, it was reported that 62 apoplastic Arabidopsis proteins were identified after 2D gel electrophoresis of Arabidopsis treated with oligogalacturonides.10 Pathogen proteomics has focused on the on pathogen proteins obtained from cultures of fungal pathogens grown on synthetic and plant-derived sources such as glucose, cellulose, starch, pectin and partially purified total tomato fruit cell walls.11-16
In this study, we employed proteomic analysis of proteins released into the microenvironment of the infection sites of green and red tomato fruit with B. cinerea to identify the proteins produced by mature green (MG) and red ripe (RR) fruit in response to infection as well as proteins released by B. cinerea. We have identified proteins when only limited fungal growth is observed on MG fruit and when aggressive colonization is observed on ripened wildtype fruit and on non-ripening but susceptible ripening inhibited (rin) mutant fruit at the stage equivalent to ripe non-mutant fruit. Unlike previous studies of proteins secreted by B. cinerea which identified fungal proteins produced in cultures, this is the first analysis of the protein populations of both organisms simultaneously in a plant-fungal pathogen interaction.
Experimental Procedures
Fungal cultures
Botrytis cinerea (strain B05.10) conidia provided by Jan Van Kan (Wageningen University) were collected from sporulating cultures of the fungus grown on potato dextrose agar (Difco, Sparks, Maryland).
Inoculation
Mature green (MG, 34 days post anthesis) and red ripe (RR, 41 days post anthesis) tomato (Lycopersicon solanum cv. Ailsa Craig) fruit were harvested from plants grown in greenhouses in Davis, California. Fruit from plants with the ripening inhibited (rin) mutation in the Ailsa Craig background (provided by the Tomato Genome Resource Center, University of California, Davis) were grown at the same time in greenhouses and harvested at the RR-equivalent ripening stage (e.g. 41 days post anthesis). Six to ten fruit at each ripening stage were identified and collected from 3 to 4 plants of each genotype, sterilized by immersion in 10% bleach for 5 minutes followed by four rinses with distilled water.
Fruit inoculation was performed as described in Cantu et al. (2008).3 All fruit were inoculated on the day of harvest to minimize variation due to storage and handling. At the time of inoculation, the fruit were punctured at sites that were 2 mm deep and 1 mm in diameter and 5-6 mm apart covering the stylar hemisphere of the fruit; 25-50 wound sites, depending on the size of the fruit, were made per fruit. Ten microliters of an aqueous suspension containing 5000 conidia of B. cinerea were placed in each wound site of the 4-5 fruit at each ripening stage of each genotype and were identified as the infected samples. Negative control material was wounded and 10 μl of H2O was placed in the wound sites of similar numbers of fruit at the same ripening stages and genotypes from the same harvests used for the infected samples. Fruit were incubated at 20°C in high humidity chambers. Three days after infection by B. cinerea were visibly apparent on the surface of the infected RR AC and ripe equivalent rin fruit, but infections were not observed on the MG AC fruit (Figure 1). The infected fruit had no visible evidence of contaminating infections and the wounded fruit had no evidence of microbial infections. The 4-5 fruit from a particular ripening stage (MG or RR), genotype (AC or rin) and treatment (infected or wounded) were combined. Six collections of extracted proteins were, therefore, evaluated: Infected MG AC (MG), infected RR AC (RR), infected RR-equivalent rin (rin), wounded MG AC, wounded RR AC and wounded RR-equivalent rin. Wounded (negative control) samples were analyzed by LC-MS/MS and we were not able to identify any proteins with statistically significant scores in these samples. The fruit (ca. 200 gm total weight) from each ripening stage, genotype and treatment were placed in 500 ml of protein extraction buffer (50 mM HEPES, pH 7.4, containing 2 mM DTT, 3 mM NaHSO3, 2mM EDTA, 1.5 M NaCl) and gently shaken for 16 h at 4°C. The supernatants, after overnight extraction, were filtered through 2 layers of cheesecloth and then passed through 0.2 mm NYL membranes to remove all particulate material. The extraction buffer was dried at reduced pressure before the proteins in the extraction buffer were analyzed.
Figure 1.
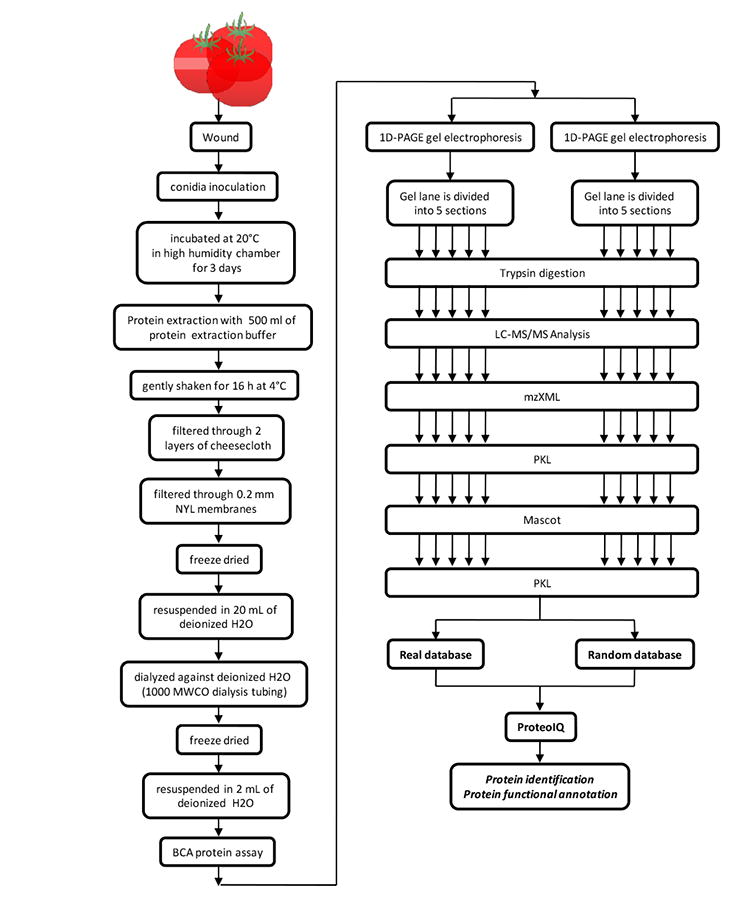
Schematic depiction of protein extraction and peptide preparation for proteomic analysis for RR, rin and MG tomato fruit infected with B. cinerea
Isolation and Separation of Secreted Proteins by 1D-SDS –PAGE
Each dried extractant was resuspended in 20 mL of deionized water and dialyzed against deionized water using a Spectra/Por CE 1000 MWCO dialysis tubing (Spectrum Labs, Rancho Dominguez, CA). The desalted protein solution was lyophylized and resuspended in 2 mL of deionized water. Approximately equal amounts of total protein based on BCA protein assay, were separated by 1D-PAGE. Protein solutions from MG, RR and rin tomato fruit infected with B. cinerea was mixed with Laemmli sample buffer (Invitrogen, Carlsbad, CA). The proteins were separated on a 4-12% polyacrylamide precast gradient gel (Invitrogen, Carlsbad, CA) at 150 Volts for 1.5 h. The gels were silver stained to visualize the protein bands,13 gels for trypsin digestion were not silver stained. All gels were done in duplicate; the duplicates were processed and analyzed separately as technical replicates.
In Gel Digestion
Gel lanes were divided into 5 sections of equal. Gel bands were then cut into smaller pieces (1 × 1 mm2), dried in a vacuum centrifuge, and the proteins reduced by submerging the gel pieces in a 100 mM ammonium bicarbonate solution containing 10 mM dithiotheritol for 1 h at 55°C. The dithiothreitol solution was then replaced by the same volume of a 55 mM iodoacetamide solution containing 100 mM ammonium bicarbonate and incubated for 45 min in the dark. After alkylation, the gel pieces were treated with 100 mM ammonium bicarbonate and acetonitrile sequentially and then dried under vacuum centrifugation. The dried gel pieces were submerged in a solution containing 2 mg of trypsin in 100 mM ammonium bicarbonate and digestion of the proteins was carried out at 37°C overnight. Peptides were eluted from the gel fragments by collecting five sequential washings of the gel fragments; the gel fragments were washed once with ammonium bicarbonate followed by acetonitrile, and twice with 5% formic acid followed by acetonitrile. The eluted peptides in the collected washings were dried and resuspended in a 0.1% formic acid solution for mass spectrometric analysis.
LC-MS/MS Analysis
Trypsin digested secreted proteins were analyzed in duplicate for each of the tomato fruit samples and analyzed in separate mass spectrometry runs. An Agilent 1100 capillary LC (Palo Alto, CA) was attached with a T splitter to deliver ηL flow rates into the mass spectrometer. Five μm diameter C18 beads (Rainin, Woburn, MA) were packed into a pulled fused silica capillary (10.5 cm × 100 μm ID) under 1000 psi pressure using nitrogen gas. Peptide samples were loaded onto the column for 45 min under the same pressure. The loaded column was then washed with 95% buffer A for 10 min prior to being interfaced with the mass spectrometer.
Mobile phase A was 0.1% formic acid in water, and phase B was 99.9% acetonitrile/0.1% formic acid. Peptides were eluted from the column with a 90 min linear gradient from 5 to 60% buffer B at a flow rate of ∼200 ηL/min and injected directly into a LTQ linear ion trap mass spectrometer through an electrospray source (Thermo Fisher, San Jose, CA) using a voltage of 2500 V.
The instrument was set to acquire MS/MS spectra on the 9 most abundant precursor ions from each MS scan with a repeat count set of 3 and a repeat duration of 5 sec. Dynamic exclusion was enabled for 160 sec. Raw tandem mass spectra were converted into a peak list using ReAdW followed by mzMXL2Other algorithms. The peak lists were then searched using Mascot 1.9 (Matrix Science, Boston, MA).
Database Searching and Protein Identification
A target database was created by combining the B. cinerea BO5.10 from Broad Institute, MA (downloaded on 7/28/2009) and T4 databases from Genoscope, France (http://urgi.versailles.inra.fr/Species/Botrytis/Download; downloaded on 7/28/2009) with a tomato protein database from SOL Genomics Network, Cornell University, NY (ftp://ftp.sgn.cornell.edu/proteins/tomato_protein_with_hits.fasta) (released 7/5/2007). Combined databases are available for download as Supplemental Information Database).
A decoy database was constructed by reversing the sequences in the target database. Searches were performed against the target and decoy databases using the following parameters: (1) tryptic enzymatic cleavage with two possible missed cleavages; (2) peptide tolerance of 800 ppm; (3) fragment ion tolerance of 0.8 Da; and (4) variable modifications due to carboxyamidomethylation of cysteine residues (+ 57 Da) and deamidation of asparagine residues (+1 Da). Statistically significant proteins were determined for all of the samples at a 1% protein FDR using the ProValT algorithm as deployed in ProteoIQ (BIOINQUIRE, Athens, GA BioInquire is now NuSep, Bogart, GA).
Protein Functional Annotation
GOSlim terms were extracted from the AgBase web site (http://agbase.msstate.edu/cgi-bin/tools/GOanna.cgi)17 using GOSlim Viewer after summarizing GO data. For Botrytis proteins with no assigned function homology searches were performed using the BlastP program against all nonredundant protein sequences in the NCBI database released on September 2009 (http://blast.ncbi.nlm.nih.gov/). Protein alignments were considered significant if they were below an e-value threshold (≤ -50). Signal peptides in the deduced amino acid sequences were searched for using the SignalP web site.18 Annotation based on association rules between CAZY families and the pfam domains was done with the CAZYmes Analysis Toolkit (CAT) from the BESC KnowledgeBase website (http://bobcat.ornl.gov/besc/index.jsp).
Results and Discussion
This study was designed to provide a qualitative global proteomic analysis. To this end, a simple, non-destructive buffer extraction method was adapted to collect both tomato fruit proteins as well as B. cinerea proteins released from the fruit and by the pathogen as a consequence of this host-pathogen interaction. A shotgun LC-MS/MS approach was used to identify tryptic peptide fragments of the collected proteins and provide researchers with access to a descriptive proteomic analysis. Data were obtained that identified proteins constituting the proteomes of tomato and B. cinerea three days after introduction of ungerminated spores into small wound sites on the fruit epidermis, a time point at which very little fungal growth is visible on the MG fruit but robust fungal growth is visible on the RR and the ripe equivalent rin fruit (Figure 2). Due to differences in fruit cell wall degradation because of the extent of fungal growth following infections of MG, RR and rin fruit5 the greatest amount of tissue maceration occurs in infections of RR fruit. Less tissue maceration is observed in infections of ripe equivalent rin fruit and little maceration is seen in infections of MG fruit. Hence, the total amount of protein collected was significantly different from each type of infected fruit. A qualitative study was undertaken rather than attempting a semiquantitative or quantitative study due to the lack of information about the number of proteins involved in plant pathogen interactions, disparities in the amount of protein released in various infection conditions and the resulting impact of the abundance of particular proteins in the proteome pools.
Figure 2.
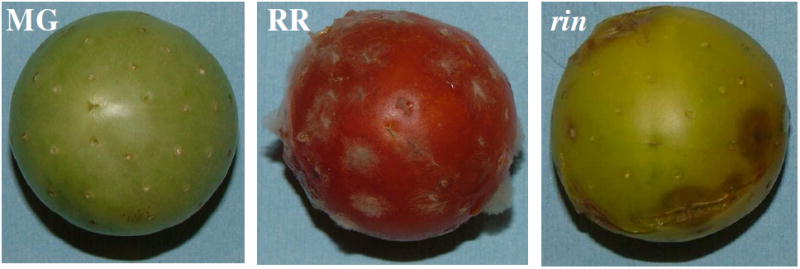
Tomato fruit infected by Botrytis cinerea at the MG (31 dpa), RR (42 dpa) stages of wild type AC and at 42 dpa of the rin non-ripening mutant.
Overview of the Proteomic Analysis of Tomato Fruit Infected with B. cinerea
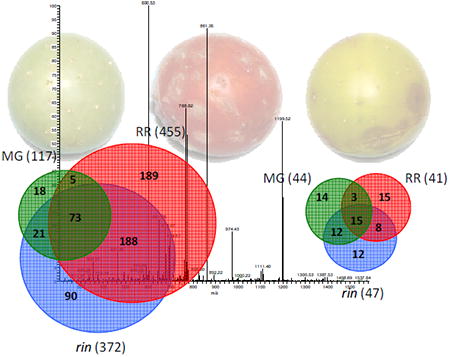
A total of 588 tomato proteins and 79 B. cinerea proteins were identified in the present study. Venn diagrams (Figure 3) depicts the number of proteins from tomato (Figure 3a) and B. cinerea (Figure 3b) that were found in common in the pairings as well as those which were unique to material from infected MG and RR wild type or rin mutant fruit. We identified 119, 456 and 374 tomato proteins from infected MG, RR and rin tomato fruit, respectively. Only 13% of the tomato proteins (74 tomato proteins) were in common among all collections, suggesting that while infections of unripe or ripe fruit released a common set of proteins, most of the proteins were unique and determined by the ripening stage or genotype of the host fruit tissue. In contrast, similar numbers of B. cinerea proteins were identified in all of the collections; 44, 41 and 47 fungal proteins were identified from infections of MG, RR and rin, respectively. Of these, 15 proteins (∼34%) of the B. cinerea proteins in each collection were common among all the collections. Supplemental Information Table 1 summarizes the spectra and proteins in each of the collections that were identified as tomato proteins and the number that were identified as B. cinerea proteins. The tomato proteins identified in MG, RR and rin fruit comprised 73, 92 and 89% respectively of the total proteins identified, and B. cinerea proteins comprised the remainder. Spectral analysis reveals an essentially identical profile.
Figure 3.
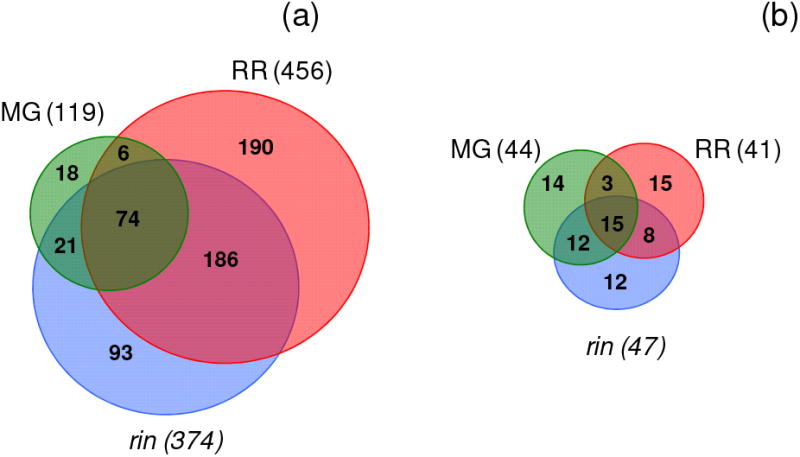
Area-proportional Venn diagrams showing overlap of tomato proteins (a) and Botrytis cinerea proteins (b) identified from RR, MG and rin tomatoes infected with Botrytis cinerea.
To obtain an overview of how proteins from the tomato-Botrytis interaction are associated with various molecular functions, Gene Ontology (GO) Slim terms were used to categorize the proteomes in the interaction. The functional assignments of the identified tomato and Botrytis proteins with molecular functions and biological processes in GOSlim terms based on the AgBase database are shown in Supplemental Information Figures 1-4.
Tomato Proteins
A total of 588 tomato proteins were identified from the proteins collected from Botrytis infections of MG and RR wild type and rin fruit (Supplemental Information Table 2 and 3). We focused on tomato defense related proteins for a more detailed analysis. Plants have evolved constitutive and inducible protective mechanisms of antimicrobial defense19 known as the defense or pathogen responses. Defense-related proteins20 are synthesized in response to biotic stress. Genes encoding pathogenesis-related proteins (PR proteins) are specifically induced in pathological or related situations and 17 families have been identified.21 Generally, these genes have been studied by transcript accumulation, which may or may not correlate with actual protein levels. The proteins identified in this work with putative biological or biochemical functions similar to the 17 PR protein families were grouped together and included as defense related proteins along with peroxidases, proteases and protease inhibiting proteins. The 114 defense related proteins were further classified into four sub-categories: PR proteins (43 proteins), proteases (43 proteins), peroxidases (13 proteins) and protease inhibiting proteins (15 proteins).
Among the 43 PR Proteins observed (Table 1), 12 chitinases were identified. Chitinases (EC 3.2.1.14) catalyze the hydrolysis of chitin, a linear homopolymer of β-1,4-linked N-acetylglucosamine (GlcNAc) residues. Chitinases constitute the second largest group of antifungal proteins,22 and a variety of chitinases have been identified during other plant-microbe interactions.9, 23-26 In addition, 10 1,3- and 1,4-β-endoglucanases were identified. The antifungal activity of these plant endoglucanases is thought to be a result of their hydrolysis of the structural 1,3-β-glucan present in the fungal cell wall, particularly at the hyphal apex of filamentous molds where glucan is most exposed, leading to cell lysis and cell death.27 Chitinases and β-1,3-glucanases are well-characterized classes of PR proteins and together can effectively inhibit fungal growth.28-30 Plant inhibitors of polysaccharide hydrolases produced by pathogens have been shown to provide reduced susceptibility to infections.4 A xyloglucan-specific fungal endoglucanase inhibitor protein31 was frequently detected in all protein collections and a polyglacturonase inhibiting protein (PGIP) whose expression has been shown to reduce susceptibility of tomato fruit to B. cinerea4 was identified less frequently but in all collections. Other proteins in the PR category include a disease response protein, an elicitor-inducible protein, a glycosyl hydrolase, three osmotin like proteins, and thaumatin.
Table 1. Identified tomato defense related proteins.
| Sub-category | Gene Id | Putative Function | SignalP | MG | RR | rin | CAZy Families | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Peptide | Spectra | Peptide | Spectra | Peptide | Spectra | |||||
|
| ||||||||||
| Pathogenesis related proteins | SGN-U312944 | beta-1,3-Endoglucanase | 0.999 Y | 21 | 73 | 22 | 56 | 32 | 167 | GH17 |
| SGN-U312558 | Basic Endochitinase | 0.001 N | 20 | 68 | 21 | 66 | 27 | 110 | GH19|CBM18 | |
| SGN-U312562 | Basic Endochitinase | 0.113 N | 17 | 74 | 23 | 110 | 33 | 195 | GH19|CBM18 | |
| SGN-U314071 | Xyloglucan-specific fungal Endoglucanase inhibitor protein | 0.998 Y | 17 | 43 | 20 | 83 | 30 | 129 | - | |
| SGN-U312653 | Thaumatin | 0.994 Y | 17 | 44 | 15 | 52 | 14 | 39 | - | |
| SGN-U342004 | beta 1,3 glucanase | 0.803 Y | 17 | 29 | 12 | 14 | 13 | 24 | GH17 | |
| SGN-U314797 | P14 | 1.000 Y | 16 | 79 | 6 | 20 | 14 | 51 | - | |
| SGN-U316008 | P2 | 0.012 N | 15 | 64 | 14 | 79 | 18 | 82 | - | |
| SGN-U313265 | Basic Endochitinase | 0.000 N | 14 | 33 | 8 | 17 | 18 | 52 | GH19 | |
| SGN-U312654 | Osmotin like | 0.656 Y | 12 | 30 | 12 | 37 | 13 | 35 | - | |
| SGN-U312559 | Basic Endochitinase | 0.966 Y | 11 | 57 | 11 | 54 | 14 | 79 | GH19|CBM18 | |
| SGN-U312561 | Basic Endochitinase | 0.008 N | 8 | 40 | 9 | 36 | 12 | 64 | GH19|CBM18 | |
| SGN-U312560 | Basic Endochitinase | 0.017 N | 7 | 15 | 7 | 16 | 11 | 23 | GH19|CBM18 | |
| SGN-U312367 | PR-1 precursor | 0.999 Y | 6 | 20 | 8 | 33 | 11 | 21 | - | |
| SGN-U333664 | Basic Endochitinase | 0.000 N | 6 | 10 | 5 | 8 | 8 | 17 | GH19|CBM18 | |
| SGN-U314382 | beta 1,3 glucanase | 0.001 N | 6 | 10 | 2 | 4 | 3 | 7 | GH17 | |
| SGN-U313266 | Basic Endochitinase | 0.000 N | 4 | 6 | 4 | 4 | 1 | 1 | GH19 | |
| SGN-U313763 | PR protein | 0.880 Y | 4 | 14 | 3 | 8 | 3 | 11 | CBM18 | |
| SGN-U315737 | PR protein | 0.000 N | 4 | 5 | 1 | 3 | 3 | 3 | - | |
| SGN-U312368 | PR protein | 0.000 N | 3 | 6 | 7 | 26 | 4 | 9 | - | |
| SGN-U322252 | Chitinase | 0.001 N | 3 | 7 | 3 | 3 | 4 | 6 | GH18 | |
| SGN-U316224 | Chitinase | 0.959 Y | 3 | 4 | 3 | 4 | 0 | 0 | GH19|CBM18 | |
| SGN-U312323 | Osmotin like protein | 0.999 Y | 2 | 3 | 0 | 0 | 0 | 0 | - | |
| SGN-U315727 | PGIP1 | 0.000 N | 1 | 2 | 5 | 8 | 5 | 16 | - | |
| SGN-U324260 | Endo beta 1,4 glucanase | 0.958 Y | 1 | 1 | 1 | 2 | 3 | 5 | GH9|GH19|CBM18 | |
| SGN-U312369 | PR protein | 0.000 N | 1 | 1 | 1 | 1 | 0 | 0 | - | |
| SGN-U312981 | PR protein | 0.004 N | 0 | 0 | 13 | 215 | 10 | 87 | - | |
| SGN-U317470 | PR protein | 0.000 N | 0 | 0 | 6 | 46 | 3 | 4 | - | |
| SGN-U317219 | beta 1,3 glucanase | 0.000 N | 0 | 0 | 2 | 2 | 1 | 1 | - | |
| SGN-U319531 | Basic Endochitinase | 0.998 Y | 0 | 0 | 1 | 1 | 1 | 1 | - | |
| SGN-U316555 | beta 1,3 glucanase | 0.004 N | 0 | 0 | 1 | 1 | 0 | 0 | GH17|CBM43 | |
| SGN-U322253 | Chitinase | 0.985 Y | 0 | 0 | 1 | 1 | 0 | 0 | GH18 | |
| SGN-U316817 | Endo 1,3 beta glucosidase | 0.998 Y | 0 | 0 | 1 | 1 | 0 | 0 | GH17|CBM43 | |
| SGN-U345256 | Endo-1,4-glucanase | 0.651 Y | 0 | 0 | 1 | 1 | 1 | 2 | - | |
| SGN-U318558 | Osmotin like protein | 1.000 Y | 0 | 0 | 1 | 1 | 1 | 3 | - | |
| SGN-U319298 | PR protein | 0.000 N | 0 | 0 | 1 | 1 | 2 | 2 | - | |
| SGN-U317352 | Disease resistance-responsive protein | 0.991 Y | 0 | 0 | 0 | 0 | 1 | 1 | - | |
| SGN-U317182 | Elicitor-inducible protein | 0.020 N | 0 | 0 | 0 | 0 | 1 | 1 | - | |
| SGN-U315668 | Endo 1,3 beta glucosidase | 0.076 N | 0 | 0 | 0 | 0 | 1 | 1 | GH17 | |
| SGN-U321793 | Endo 1,3 beta glucosidase | 0.000 N | 0 | 0 | 0 | 0 | 1 | 1 | GH17|CBM43 | |
| SGN-U340225 | Glycosyl hydrolase | 0.000 N | 0 | 0 | 0 | 0 | 1 | 1 | GH17|CBM43 | |
| SGN-U315428 | PR protein | 1.000 Y | 0 | 0 | 0 | 0 | 2 | 4 | - | |
| SGN-U330712 | PR protein | 0.000 N | 0 | 0 | 0 | 0 | 2 | 2 | - | |
|
| ||||||||||
| Proteases | SGN-U313379 | Aminopeptidase | 0.001 N | 0 | 0 | 6 | 7 | 2 | 2 | - |
| SGN-U317362 | Aminopeptidase | 0.009 N | 0 | 0 | 1 | 1 | 0 | 0 | - | |
| SGN-U312783 | ATP-dependent protease | 0.016 N | 0 | 0 | 2 | 2 | 0 | 0 | - | |
| SGN-U322932 | Carboxypeptidase | 0.000 N | 0 | 0 | 3 | 3 | 0 | 0 | - | |
| SGN-U322736 | Nucellin | 0.057 N | 0 | 0 | 0 | 0 | 1 | 1 | - | |
| SGN-U313835 | Peptidase | 0.000 N | 11 | 23 | 15 | 42 | 15 | 37 | - | |
| SGN-U334617 | Peptidase | 0.000 N | 4 | 6 | 5 | 12 | 7 | 9 | - | |
| SGN-U319568 | Peptidase | 0.999 Y | 1 | 1 | 0 | 0 | 3 | 3 | - | |
| SGN-U321102 | Peptidase | 0.000 N | 0 | 0 | 2 | 2 | 3 | 3 | - | |
| SGN-U324600 | Peptidase | 0.987 Y | 1 | 1 | 0 | 0 | 2 | 3 | - | |
| SGN-U316263 | Peptidase | 0.000 N | 0 | 0 | 0 | 0 | 1 | 1 | - | |
| SGN-U317724 | Protease | 0.000 N | 3 | 5 | 7 | 9 | 23 | 43 | - | |
| SGN-U322187 | Protease | 0.002 N | 2 | 4 | 14 | 29 | 14 | 33 | - | |
| SGN-U314479 | Protease | 0.999 Y | 2 | 3 | 6 | 9 | 15 | 30 | - | |
| SGN-U318298 | Protease | 0.991 Y | 1 | 1 | 4 | 5 | 11 | 16 | - | |
| SGN-U313775 | Protease | 0.994 Y | 1 | 2 | 0 | 0 | 6 | 10 | - | |
| SGN-U315460 | Protease | 0.715 Y | 0 | 0 | 6 | 11 | 8 | 13 | - | |
| SGN-U312375 | Protease | 0.000 N | 0 | 0 | 14 | 22 | 4 | 4 | - | |
| SGN-U315473 | Protease | 0.001 N | 0 | 0 | 7 | 12 | 7 | 9 | - | |
| SGN-U312919 | Protease | 0.971 Y | 0 | 0 | 8 | 11 | 4 | 4 | - | |
| SGN-U316289 | Protease | 0.000 N | 0 | 0 | 0 | 0 | 4 | 7 | - | |
| SGN-U313378 | Protease | 0.001 N | 0 | 0 | 9 | 10 | 3 | 3 | - | |
| SGN-U313997 | Protease | 1.000 Y | 3 | 4 | 13 | 39 | 3 | 8 | - | |
| SGN-U315317 | Protease | 0.996 Y | 0 | 0 | 2 | 2 | 2 | 4 | - | |
| SGN-U313773 | Protease | 0.002 N | 2 | 2 | 0 | 0 | 1 | 1 | - | |
| SGN-U314619 | Protease | 0.995 Y | 0 | 0 | 3 | 6 | 3 | 6 | - | |
| SGN-U316051 | Protease | 0.000 N | 0 | 0 | 7 | 9 | 2 | 2 | - | |
| SGN-U322487 | Protease | 0.000 N | 0 | 0 | 1 | 1 | 2 | 3 | CBM5 | |
| SGN-U315577 | Protease | 0.997 Y | 0 | 0 | 2 | 4 | 2 | 4 | - | |
| SGN-U320558 | Protease | 0.000 N | 0 | 0 | 1 | 1 | 0 | 0 | - | |
| SGN-U316057 | Protease | 0.650 Y | 0 | 0 | 0 | 0 | 1 | 1 | - | |
| SGN-U312592 | Protease | 0.999 Y | 2 | 3 | 1 | 2 | 1 | 1 | - | |
| SGN-U326813 | Protease | 0.791 Y | 0 | 0 | 2 | 2 | 0 | 0 | - | |
| SGN-U317291 | Protease | 0.002 N | 0 | 0 | 1 | 1 | 0 | 0 | - | |
| SGN-U337147 | Protease | 0.002 N | 0 | 0 | 1 | 2 | 0 | 0 | - | |
| SGN-U321739 | Protease | 0.008 N | 1 | 1 | 0 | 0 | 0 | 0 | - | |
| SGN-U312589 | Protease | 0.953 Y | 2 | 2 | 0 | 0 | 4 | 5 | - | |
| SGN-U312588 | Protease | 0.233 N | 1 | 1 | 0 | 0 | 2 | 2 | - | |
| SGN-U313739 | Serine carboxypeptidase | 0.999 Y | 0 | 0 | 17 | 27 | 5 | 6 | - | |
| SGN-U323007 | Serine carboxypeptidase | 0.000 N | 0 | 0 | 6 | 9 | 2 | 2 | - | |
| SGN-U316728 | Serine carboxypeptidase | 0.098 N | 0 | 0 | 1 | 2 | 1 | 1 | - | |
| SGN-U322415 | Serine carboxypeptidase | 0.987 Y | 0 | 0 0 | 2 | 2 | 0 | 00 | - | |
| SGN-U315356 | Ubiquitin specific protease | 0.000 N | 0 | 0 | 1 | 1 | 0 | 0 | - | |
|
| ||||||||||
| Peroxidases | SGN-U315418 | Peroxidase | 0.000 N | 7 | 18 | 1 | 1 | 12 | 26 | - |
| SGN-U313411 | Peroxidase | 0.997 Y | 9 | 26 | 10 | 18 | 11 | 27 | - | |
| SGN-U321125 | Peroxidase | 0.000 N | 7 | 11 | 0 | 0 | 4 | 4 | - | |
| SGN-U321126 | Peroxidase | 0.032 N | 8 | 16 | 0 | 0 | 4 | 6 | - | |
| SGN-U323529 | Peroxidase | 0.000 N | 18 | 61 | 2 | 2 | 3 | 4 | - | |
| SGN-U315693 | Peroxidase | 0.957 Y | 4 | 20 | 3 | 12 | 3 | 6 | - | |
| SGN-U316807 | Peroxidase | 0.989 Y | 5 | 12 | 3 | 3 | 2 | 6 | - | |
| SGN-U315420 | Peroxidase | 0.957 Y | 0 | 0 | 0 | 0 | 1 | 1 | - | |
| SGN-U315694 | Peroxidase | 0.992 Y | 1 | 2 | 0 | 0 | 0 | 0 | - | |
| SGN-U314196 | Peroxidase | 0.999 Y | 9 | 22 | 1 | 1 | 1 | 1 | - | |
| SGN-U327467 | Peroxidase | 0.000 N | 2 | 2 | 0 | 0 | 0 | 0 | - | |
| SGN-U314124 | Peroxidase | 0.994 Y | 3 | 4 | 0 | 0 | 0 | 0 | - | |
| SGN-U346809 | Peroxidase | 0.599 Y | 1 | 1 | 0 | 0 | 0 | 0 | - | |
|
| ||||||||||
| Protease inhibiting proteins | SGN-U315605 | Endoglucanase inhibitor protein | 0.998 Y | 0 | 0 | 2 | 2 | 3 | 3 | - |
| SGN-U317739 | Protease inhibitor | 0.006 N | 8 | 16 | 7 | 19 | 8 | 30 | - | |
| SGN-U312820 | Protease inhibitor | 0.966 Y | 0 | 0 | 5 | 14 | 5 | 22 | - | |
| SGN-U320265 | Protease inhibitor | 0.829 Y | 3 | 4 | 4 | 8 | 6 | 9 | - | |
| SGN-U313384 | Protease inhibitor | 0.998 Y | 2 | 7 | 4 | 36 | 6 | 13 | - | |
| SGN-U316036 | Protease inhibitor | 0.001 N | 1 | 1 | 3 | 3 | 5 | 8 | - | |
| SGN-U312826 | Protease inhibitor | 0.996 Y | 2 | 2 | 4 | 4 | 5 | 10 | - | |
| SGN-U312623 | Protease inhibitor | 0.000 N | 2 | 2 | 0 | 0 | 5 | 6 | - | |
| SGN-U315288 | Protease inhibitor | 0.916 Y | 2 | 3 | 4 | 4 | 1 | 1 | - | |
| SGN-U315879 | Protease inhibitor | 0.986 Y | 4 | 7 | 4 | 7 | 4 | 7 | - | |
| SGN-U322543 | Protease inhibitor | 0.000 N | 5 | 12 | 7 | 19 | 1 | 3 | - | |
| SGN-U320540 | Protease inhibitor | 0.003 N | 0 | 0 | 1 | 1 | 2 | 2 | - | |
| SGN-U312825 | Protease inhibitor | 0.996 Y | 1 | 1 | 0 | 0 | 1 | 1 | - | |
| SGN-U318506 | Protease inhibitor | 0.000 N | 0 | 0 | 0 | 0 | 1 | 1 | - | |
| SGN-U312828 | Protease inhibitor | 0.963 Y | 0 | 0 | 0 | 0 | 1 | 2 | - | |
Gene Id: identification number provided by the tomato protein database (ftp.sgn.cornell.edu/proteins/)
Putative function: was assigned based on sequence similarity when blasted using NCBI non redundant database
Signal P prediction value using algorithm SignalP3.0 server (http://www.cbs.dtu.dk/services/SignalP/)
Spectra: Total number of spectra matched to proteins in all replicates in an infected tomato fruit
Peptide: Total number of non-redundant peptides above a threshold score of 54 matched to the tomato protein in that infected tomato fruit
CAZy families: Predicted with CAZYmes Analysis Toolkit (CAT) from the BESC KnowledgeBase website (http://bobcat.ornl.gov/besc/index.jsp)
Forty three proteases were a second subcategory of defense related proteins. Plants have a large arsenal of proteolytic enzymes that regulates the fate of proteins. These proteins are proposed to have primarily a housekeeping role. However, proteases also play a key role in the regulation of biological processes, such as the recognition of pathogens and pests and the induction of effective defense responses.32-34
Thirteen peroxidases were identified among the 114 defense related proteins. In the course of pathogen–plant interaction, peroxidases are key enzymes in the detoxification systems involved in scavenging reactive oxygen forms, whose increased generation is closely associated with the induction of plant defense reactions.35 In tomato it has been shown that heat treatment after inoculation induces peroxidases that prevent B. cinerea development.36
We identified 15 protease inhibiting (pin) proteins in the 114 defense proteins and, more than 60% belong to the Type I class of pin proteins. Pin proteins are among the defense proteins in plant tissues that are both developmentally regulated and induced in response to insect and pathogen attack.37 Type I pin proteins are best known in potatoes but are found in other plant species, for example, barley endosperm chymotrypsin inhibitor is a Type I pin.38 It has been shown that protease inhibitors isolated from healthy bean and tomato plants reduced the activities of proteases from Fusarium solani and C. lindemuthianum.39, 40 In tomatoes, serine protease inhibitors I and II accumulate in endosperm cell walls and in secretory cells of root cap, and are secreted into the milieu.41
Fruit cell wall related proteins
Forty-four proteins from the three collections were assigned to the cell wall related categories (Table 2). Fruit cell wall related proteins include subcategories of cell wall (structural) proteins and proteins thought to cause cell wall modifications (both catabolic and anabolic).
Table 2. Identified tomato cell wall-related proteins.
| Sub categorie | Gene Id | Putative Function | SignalP | MG | RR | rin | CAZy Families | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Peptide | Spectra | Peptide | Spectra | Peptide | Spectra | |||||
|
| ||||||||||
| Cell wall related proteins | SGN-U320073 | alpha-galactosidase | 1.000 Y | - | - | 1 | 1 | - | - | GH27 |
| SGN-U319599 | alpha-galactosidase | 0.003 N | - | - | 3 | 5 | 3 | 4 | - | |
| SGN-U321036 | alpha-glucosidase | 0.000 N | - | - | 2 | 3 | 3 | 4 | GH31 | |
| SGN-U319244 | alpha-xylosidase | 0.000 N | - | - | 3 | 8 | 6 | 14 | GH31 | |
| SGN-U320295 | alpha-xylosidase | 0.001 N | 1 | 1 | - | - | 7 | 10 | GH31 | |
| SGN-U320011 | alpha-xylosidase | 0.000 N | - | - | 7 | 10 | 4 | 8 | - | |
| SGN-U336993 | alpha-xylosidase | 0.000 N | 1 | 1 | 4 | 5 | 3 | 4 | - | |
| SGN-U312614 | arabinogalactan protein | 1.000 Y | 2 | 8 | 4 | 24 | 7 | 24 | - | |
| SGN-U322781 | arabinogalactan protein | 0.785 Y | 3 | 7 | 4 | 8 | 10 | 11 | - | |
| SGN-U320398 | arabinogalactan protein | 0.999 Y | 3 | 7 | 3 | 3 | 4 | 5 | - | |
| SGN-U316315 | arabinogalactan protein | 1.000 Y | - | - | - | - | 1 | 1 | GH18 | |
| SGN-U319146 | beta-D-glucan exohydrolase | 0.000 N | - | - | - | - | 1 | 1 | GH3 | |
| SGN-U345075 | beta-D-glucan exohydrolase | 0.015 N | - | - | - | - | 1 | 1 | - | |
| SGN-U314662 | beta-fructosidase | 0.000 N | 10 | 14 | 26 | 97 | 14 | 21 | - | |
| SGN-U319861 | beta-fructosidase | 0.681 Y | - | - | - | - | 5 | 11 | GH32 | |
| SGN-U319056 | beta-galactosidase | 0.020 N | - | - | 4 | 4 | - | - | - | |
| SGN-U315107 | beta-galactosidase | 0.013 N | - | - | 2 | 3 | - | - | - | |
| SGN-U322726 | beta-galactosidase | 0.362 N | - | - | 2 | 2 | - | - | - | |
| SGN-U328864 | beta-galactosidase | 0.000 N | - | - | 1 | 1 | 1 | 1 | - | |
| SGN-U316373 | beta-Ig-H3 domain-containing protein unknown | 0.811 Y | - | - | 1 | 2 | 2 | 4 | - | |
| SGN-U313742 | endo-beta-mannanase | 0.019 N | - | - | 7 | 14 | 2 | 3 | - | |
| SGN-U312953 | expansin 1 | 0.998 Y | - | - | 2 | 3 | - | - | - | |
| SGN-U312510 | expansin 10 | 1.000 Y | - | - | - | - | 2 | 2 | - | |
| SGN-U317347 | expansin-like | 1.000 Y | - | - | 2 | 3 | - | - | - | |
| SGN-U313100 | extensin | 0.000 N | - | - | 6 | 8 | 2 | 4 | - | |
| SGN-U322174 | germin-like protein | 0.999 Y | 1 | 1 | - | - | - | - | - | |
| SGN-U318102 | germin-like protein | 0.982 Y | 1 | 1 | 4 | 6 | 1 | 2 | - | |
| SGN-U314144 | glycosyl hydrolase | 0.000 N | 1 | 1 | 2 | 3 | 7 | 8 | - | |
| SGN-U316037 | glycosyl hydrolase | 0.000 N | - | - | 1 | 1 | 2 | 2 | GH3 | |
| SGN-U326255 | hydroxyproline-rich glycoprotein | 0.932 Y | - | - | 1 | 1 | - | - | - | |
| SGN-U318294 | pectin acetylesterase | 0.998 Y | 1 | 2 | 8 | 26 | 11 | 28 | - | |
| SGN-U312758 | pectin esterase | 0.002 N | - | - | 21 | 114 | 14 | 68 | CE8 | |
| SGN-U313648 | pectin esterase | 0.001 N | - | - | 5 | 6 | 9 | 17 | - | |
| SGN-U318232 | pectin esterase | 0.252 N | - | - | 4 | 10 | 4 | 10 | - | |
| SGN-U313531 | pectin esterase | 0.992 Y | - | - | 3 | 4 | 1 | 1 | - | |
| SGN-U314637 | pectin methylesterase | 0.986 Y | - | - | 3 | 5 | - | - | - | |
| SGN-U312757 | pectin methylesterase | 0.966 Y | 7 | 17 | 63 | 1039 | 55 | 861 | CE8 | |
| SGN-U313725 | pectin methylesterase | 0.009 N | 2 | 2 | 14 | 66 | 11 | 87 | - | |
| SGN-U313233 | pectin methylesterase | 0.156 N | - | - | - | - | 1 | 1 | CE8 | |
| SGN-U330767 | pectinacetylesterase | 0.846 Y | - | - | 1 | 1 | - | - | CE13 | |
| SGN-U320881 | pectinacetylesterase | 0.964 Y | - | - | 1 | 1 | 1 | 1 | CE13 | |
| SGN-U324003 | PG 2A precursor | 0.999 Y | 2 | 3 | 1 | 2 | 3 | 4 | GH28 | |
| SGN-U321477 | polygalacturonase | 0.000 N | 1 | 1 | - | - | - | - | - | |
| SGN-U312703 | polygalacturonase | 0.992 Y | 8 | 10 | 63 | 435 | 23 | 59 | GH28 | |
Gene Id: identification number provided by the tomato protein database (ftp.sgn.cornell.edu/proteins/)
Putative function: was assigned based on sequence similarity when blasted using NCBI non redundant database
Signal P: prediction value using algorithm SignalP3.0 server (http://www.cbs.dtu.dk/services/SignalP/)
Spectra: Total number of spectra matched to proteins in all replicates in an infected tomato fruit
Peptide: Total number of non-redundant peptides above a threshold score of 54 matched to the tomato protein in that infected tomato fruit
CAZy families: Predicted with CAZYmes Analysis Toolkit (CAT) from the BESC KnowledgeBase website (http://bobcat.ornl.gov/besc/index.jsp)
Other tomato proteins
The other categories of tomato proteins identified in the collections include sub-classes that have not been implicated directly in host-pathogen interaction. However, such a role for these proteins cannot be disregarded a priori. The presence of these proteins is most likely due to cell lysis during the infection, thus they should be considered an indirect response to the presence of the pathogen.
Carbohydrate-active enzymes from tomato
Families of structurally-related catalytic and carbohydrate-binding modules (CBMs) for 93 tomato proteins (Supplemental Information Table 2, Supplemental Information Figure 5) were assigned based on CAZYmes Analysis Toolkit (CAT) (http://bobcat.ornl.gov/besc/index.jsp); this represents 16% of the 586 tomato proteins identified. One major class of CAZy identified for tomato proteins was the glycoside hydrolases (GH) with 45 members. CBM, carbohydrate esterase (CE) and glucosyl transferase (GT) families had 24, 18 and 6 members, respectively.
Tomato transcriptome and proteome analysis
To gain insights into the accumulation of specific proteins, we compared the tomato protein data with microarray transcriptome data from Cantu et al (2009) 5. The changes in expression from the transcriptome analysis that were also observed in the proteome analysis are listed in Supplemental Information Table 4. Of the 186 identified tomato proteins, 32 had transcript probes represented on the microarray.
In general, there was no significant up or down regulation of any transcripts when MG or RR fruit were inoculated or wounded; transcript abundance of these 32 genes stayed remarkably constant, in both ripening stages and in healthy, wounded or infected tissues. Transcriptional changes observed in both MG and RR fruit in response to B. cinerea suggested pathways transcriptionally activated by B. cinerea regardless of the ripening stage of the fruit. In most tomato-botrytis combinations expression of an embryo-abundant protein-related, a saposin B domain-containing protein, a putative pectinesterase, and two FLA1 (Fasciclin-Like Arabinogalactan 1) transcripts was activated by the fungus and proteins from these genes also accumulated.
In infected rin fruit only a copper ion binding / oxidoreductase (SGN-U314232; BG735509) was identified from both transcriptome and proteome analysis. Fourteen probe sets were identified in proteins unique to RR fruit and 3 out of 14 are involved in defense mechanisms (SGN-U317362, SGN-U317362 and SGN-U316817). Between RR and rin fruit, nine probe sets were linked to 9 proteins. Three out of the 9 are cell wall defense related proteins (SGN-U318232, SGN-U318232 and SGN-U318232) and 2 proteins were involved in defense mechanisms (SGN-U315473 and SGN-U315473). Four probe sets matched two proteins found in common in MG, RR and rin fruit (SGN-U320398 and SGN-U322781). Overall, no close correlation was found comparing results of the proteome and transcriptome. The low correlations between the presence of a transcript and the presence of its corresponding protein suggest that post-transcriptional regulatory mechanisms play an important role in the fruit-pathogen interaction and has not been described previously. Osorio et al., (2011) reported also a low correlation between transcriptome and proteome data, especially in MG fruit.42
Botrytis cinerea Proteins
Seventy-nine proteins encoded by B. cinerea genes during the infection of tomato fruit were identified in the collections from MG, RR and rin infected tomatoes (Table 3, Supplemental Information Figure 6). In order to identify secretory signal peptides in B. cinerea proteins, we used the SignalP 3.0 algorithm (Table 3). Using this algorithm, 58 out of 79 fungal proteins were found to have a secretory signal peptide, representing 73% of the total fungal proteins, while the remaining 21 (27%) proteins did not contain an identifiable signal peptide. However, use of the WoLF PSORT algorithm (http://wolfpsort.seq.cbrc.jp) indicated an additional seven proteins out of the remaining 21 were categorized as extracellular because of possible signal peptides. Four of the proteins were found to be intracellular when B. cinerea was grown in 1% carboxymethylcellulose,11 raising the possibility that the presence of these proteins in our study is due to lysis of B. cinerea cells. Further studies will determine whether these proteins are truly secreted or are present as the result of either autolysis or lysis occurring as a host's defense response to the pathogen.
Table 3. Indentified Botrytis cinerea proteins.
| Sequence Id | Putative Function | Singal P | MG | RR | rin | Predicted CAZy Class | ||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Pep | Spec | Pep | Spec | Pep | Spec | |||||
|
| ||||||||||
| BC1G_07319.1 | 1,3-beta glucanaseCWDE | 0.999 | Y | - | - | 3 | 3 | - | - | - |
| BC1G_10455.1 | 1,3-beta-glucanosyltransferase Gel1CWDE | 0.998 | Y | - | - | 4 | 19 | 3 | 4 | GH72 |
| BC1G_02623.1 | alpha-amylase | 0.994 | Y | - | - | 2 | 2 | 4 | 5 | GH13 |
| BC1G_08975.1 | alpha-fucosidase | 0.962 | Y | 3 | 3 | 2 | 2 | 4 | 4 | - |
| BC1G_12859.1 | alpha-glucosidase precursorCWDE | 1 | Y | - | - | 2 | 2 | 2 | 2 | - |
| BC1G_08372.1 | alpha-L-arabinofuranosidase ACWDE | 0.995 | Y | 3 | 3 | - | - | - | - | - |
| BC1G_04994.1 | alpha-L-arabinofuranosidaseCWDE | 0.999 | Y | - | - | - | - | 2 | 2 | GH54|CBM42 |
| BofuT4_P078550.1 | alpha-L-rhamnosidase BCWDE | 0.072 | N | - | - | - | - | 1 | 1 | GH78 |
| BC1G_03983.1 | beta-1,3 exoglucanaseCWDE | 0.27 | N | - | - | 2 | 2 | - | - | - |
| BC1G_14030.1 | beta-1-3-glucanosyltransferase | 1 | Y | 3 | 4 | - | - | 3 | 4 | GH72|CBM43 |
| BC1G_03567.1 | beta-galactosidaseCWDE | 0.998 | Y | 9 | 17 | 15 | 20 | 22 | 33 | GH35 |
| BC1G_10221.1 | beta-glucosidase 1 precursorCWDE | 0.816 | Y | 2 | 2 | - | - | - | - | GH3 |
| BC1G_02364.1 | beta-glucosidase, putativeCWDE | 0.991 | Y | 5 | 7 | - | - | - | - | GH3 |
| BC1G_05538.1 | beta-xylosidaseCWDE | 0.978 | Y | 1 | 1 | - | - | - | - | GH3 |
| BC1G_09079.1 | cell wall beta-1,3-endoglucanaseCWDE | 0.999 | Y | 3 | 3 | 4 | 6 | 8 | 11 | - |
| BC1G_14702.1 | cellobiohydrolase 1 catalytic domainCWDE | 1 | Y | 3 | 4 | - | - | 1 | 1 | - |
| BC1G_06035.1 | cellobiohydrolaseCWDE | 0.999 | Y | 12 | 21 | - | - | - | - | GH7 |
| BC1G_03188.1 | cellobiose dehydrogenaseCWDE | 0 | N | - | - | - | - | 7 | 8 | CBM1 |
| BC1G_10880.1 | cellobiose dehydrogenaseCWDE | 0.992 | Y | 17 | 57 | - | - | 12 | 19 | GH7 |
| BC1G_14012.1 | choline dehydrogenase, putative | 0.998 | Y | - | - | 2 | 2 | - | - | CBM1 |
| BC1G_08529.1 | cytochrome c | 0 | N | - | - | - | - | 2 | 2 | - |
| BC1G_00576.1 | endo-1,4 beta-D-xylanaseCWDE | 1 | Y | 2 | 2 | 2 | 3 | - | - | GH10 |
| BofuT4_P061530.1 | endo-1,4-beta-mannosidaseCWDE | 0.998 | Y | 1 | 1 | - | - | - | - | - |
| BofuT4_P124780.1 | endo-glucanase, putativeCWDE | 0.999 | Y | 1 | 3 | - | - | - | - | GH12 |
| BC1G_11143.1 | endo-polygalacturonase precursorCWDE | 1 | Y | 15 | 57 | 13 | 87 | 25 | 161 | GH28 |
| BofuT4_P024480.1 | esterase | 0.025 | N | - | - | 2 | 2 | 1 | 1 | - |
| BC1G_07822.1 | extracellular endo-glucanaseCWDE | 0.999 | Y | 3 | 3 | - | - | - | - | GH5 |
| BC1G_07496.1 | extracellular exo-polygalacturonase, putativeCWDE | 0 | N | 2 | 3 | - | - | - | - | GH28 |
| BC1G_00448.1 | extracellular serine-rich protein | 0.983 | Y | 3 | 3 | 2 | 2 | - | - | - |
| BofuT4_P151530.1 | formate dehydrogenase | 0.659 | Y | - | - | 1 | 1 | 3 | 4 | - |
| BC1G_04836.1 | fructose-bisphosphate aldolase | 0 | N | 4 | 4 | - | - | - | - | - |
| BofuT4_P119480.1 | GDSL lipase/acylhydrolase family protein | 0.988 | Y | 2 | 4 | - | - | - | - | - |
| BC1G_04151.1 | glucan 1,4-alpha-glucosidase | 0.912 | Y | 5 | 7 | 11 | 15 | 20 | 84 | CBM20|GH15 |
| BC1G_10788.1 | glucose oxidase | 0.997 | Y | 1 | 1 | - | - | - | - | - |
| BC1G_11898.1 | glucosidaseCWDE | 0.999 | Y | 3 | 5 | 5 | 11 | 4 | 5 | GH17 |
| BofuT4_P033100.1 | glutaminase GtaA | 0.992 | Y | - | - | 3 | 3 | - | - | - |
| BC1G_01204.1 | glyoxal oxidaseCWDE | 0.983 | Y | - | - | - | - | 4 | 4 | CBM18 |
| BC1G_01393.1 | hypothetical protein | 1 | Y | - | - | 2 | 2 | - | - | - |
| BC1G_02060.1 | hypothetical protein | 0.999 | Y | 1 | 2 | 2 | 3 | - | - | - |
| BC1G_02930.1 | hypothetical protein | 0 | N | - | - | 7 | 11 | - | - | - |
| BC1G_03976.1 | hypothetical protein | 0.959 | Y | - | - | - | - | 7 | 7 | - |
| BC1G_05298.1 | hypothetical protein | 0 | N | - | - | 2 | 2 | - | - | - |
| BC1G_05503.1 | hypothetical protein | 0 | N | 2 | 6 | - | - | 2 | 2 | - |
| BC1G_05523.1 | hypothetical protein | 0 | N | - | - | - | - | 5 | 5 | - |
| BC1G_05980.1 | hypothetical protein | 0.004 | N | - | - | 2 | 4 | - | - | - |
| BC1G_07073.1 | hypothetical protein | 1 | Y | - | - | 2 | 2 | - | - | - |
| BC1G_07275.1 | hypothetical protein | 1 | Y | - | - | - | - | 2 | 2 | - |
| BC1G_07903.1 | hypothetical protein | 0.001 | N | - | - | 3 | 3 | 2 | 2 | - |
| BC1G_08393.1 | hypothetical protein | 0.999 | Y | 3 | 5 | - | - | 2 | 4 | - |
| BC1G_08615.1 | hypothetical protein | 0.962 | Y | - | - | - | - | 2 | 2 | - |
| BC1G_08719.1 | hypothetical protein | 0.019 | N | 1 | 1 | - | - | 1 | 2 | - |
| BC1G_10630.1 | hypothetical protein | 1 | Y | 3 | 4 | - | - | - | - | - |
| BC1G_12374.1 | hypothetical protein | 0.999 | Y | 5 | 40 | 9 | 287 | 6 | 42 | - |
| BC1G_15041.1 | hypothetical protein | 0.988 | Y | 4 | 5 | - | - | 4 | 5 | - |
| BC1G_16040.1 | hypothetical protein | 0 | N | - | - | - | - | 1 | 1 | - |
| BofuT4_P129550.1 | hypothetical protein | 0.998 | Y | - | - | 2 | 2 | - | - | - |
| BC1G_12487.1 | hypothetical protein | 0 | N | - | - | 3 | 5 | 3 | 3 | - |
| BC1G_14129.1 | hypothetical protein | 0.997 | Y | 2 | 4 | 2 | 2 | 2 | 4 | - |
| BC1G_08553.1 | laccase 2CWDE | 0.999 | Y | 4 | 5 | 2 | 2 | 9 | 16 | - |
| BC1G_10329.1 | laccaseCWDE | 0.989 | Y | - | - | 4 | 6 | - | - | - |
| BofuT4_P040250.1 | L-PSP endoribonuclease family protein, putative | 0 | N | - | - | 2 | 2 | - | - | - |
| BC1G_00455.1 | mannosyl-oligosaccharide alpha-1,2-mannosidase precursorCWDE | 0.998 | Y | 4 | 5 | - | - | 2 | 2 | GH47 |
| BofuT4_P108020.1 | nucleoside diphosphate kinase | 0 | N | 1 | 6 | 1 | 3 | 2 | 3 | - |
| BC1G_12017.1 | pectin lyase A precursorCWDE | 0.998 | Y | - | - | 3 | 8 | 3 | 3 | PL1 |
| BofuT4_P131540.1 | pectin methyl esterase 1CWDE | 1 | Y | 5 | 9 | 4 | 6 | 6 | 12 | CE8 |
| BC1G_00617.1 | pectin methyl esteraseCWDE | 1 | Y | 2 | 4 | - | - | 2 | 5 | CE8 |
| BC1G_11144.1 | pectin methylesterase, putativeCWDE | 1 | Y | 2 | 4 | - | - | - | - | CE8 |
| BC1G_13367.1 | polygalacturonase 2 CWDE | 0.999 | Y | - | - | - | - | 2 | 3 | GH28 |
| BC1G_00896.1 | predicted protein | 1 | Y | 5 | 39 | 5 | 44 | 6 | 87 | - |
| BC1G_09084.1 | predicted protein | 1 | Y | - | - | 1 | 1 | - | - | - |
| BC1G_14009.1 | rhamnogalacturonan acetylesteraseCWDE | 0.983 | Y | - | - | 3 | 10 | - | - | - |
| BofuT4_P124300.1 | rhamnogalacturonan hydrolase CWDE | 1 | Y | 1 | 1 | - | - | 1 | 1 | GH28 |
| BC1G_06836.1 | serine protease | 0.999 | Y | - | - | 3 | 4 | - | - | - |
| BC1G_02163.1 | SnodProt1CWDE | 0.996 | Y | 13 | 65 | 27 | 253 | 19 | 154 | - |
| BC1G_14403.1 | thioredoxin | 0 | N | 2 | 2 | 3 | 3 | 2 | 3 | - |
| BofuT4_P021640.1 | transaldolase 1 | 0 | N | 2 | 2 | 2 | 2 | 1 | 5 | - |
| BC1G_00979.1 | tripeptidyl-peptidase 1 precursor | 0.015 | N | 1 | 1 | - | - | 1 | 2 | - |
| BC1G_02944.1 | tripeptidyl-peptidase 1 precursor | 0.999 | Y | - | - | - | - | 2 | 2 | - |
| BC1G_10797.1 | xylosidase : arabinofuranosidaseCWDE | 0.997 | Y | 2 | 2 | - | - | 2 | 2 | - |
Sequence id: Identification number provided by the Broad Institute BO5.10 database
Putative function: was assigned based on sequence similarity whne blasted using NCBI non redundant database
Signal P: prediction value using algoritm SignalP3.0 server (http://www.cbs.dtu.dk/services/SignalP/); Y, signal peptide predicted. N,no signal peptide predicted.
Peptides: Total number of non-redundant peptides above a threshold score of 54 matched to the fungal protein in that infected tomato fruit
Spectra: Total number of spectra matched to proteins in all replicates in that infected tomato fruit
CAZy families: Predicted with CAZYmes Analysis Toolkit (CAT) from the BESC KnowledgeBase website (http://bobcat.ornl.gov/besc/index.jsp)
Plant cell wall degrading enzymes
To accomplish successful penetration and colonization of a plant, pathogenic fungi must break the cell wall. Degradation of plant cell wall compounds by plant pathogens is a complex process involving the synergistic action of a large number of extracellular cell wall degrading enzymes and proteins that help pathogens penetrate and colonize plant tissues. Enzymatic cleavage of the plant cell wall releases carbohydrates, which can become a carbon source for the pathogen. Of the 79 B. cinerea proteins detected, 36 (46%) were classified as cell wall degrading proteins are listed in Table 4. BofuT4_P131540.1 with homology to a PME-1 was identified in MG, RR and rin tomato. Other PMEs identified included BC1G_00617.1 in MG and rin tomato and BC1G_11144.1 in MG tomato. Interruption of Bcpme1 reduces pathogenicity on several plant hosts,42 indicating that pectin methyl esterase contributes to B. cinerea pathogenicity.
Table 4. Common Botrytis cinerea proteins in secretome studies.
| Sequence Id | Putative Function | MG | RR | rin | O-glycosilation | N-glycosilation | Secretomes Studies |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Spectra | Spectra | Spectra | |||||
| BC1G_12374.1 | hypothetical protein | 40 | 287 | 42 | none | Yes | This study, Sha et al 2009a, Sha et al 2009b, Espino et al., 2010, Fernández-Acero et al., 2010 |
| BC1G_02163.1 | SnodProt1 | 65 | 253 | 154 | none | none | This study, Sha et al 2009a, Sha et al 2009b, Espino et al., 2010 |
| BC1G_ 11143.1 | endo-polygalacturonase precursor | 57 | 87 | 161 | none | none | This study, Sha et al 2009a |
| BC1G_00896.1 | predicted protein | 39 | 44 | 87 | Yes | none | This study, Sha et al 2009a, Sha et al 2009b, Fernández-Acero et al., 2010 |
| BC1G_03567.1 | beta-galactosidase | 17 | 20 | 33 | none | Yes | This study, Sha et al 2009b, Espino et al., 2010 |
| BC1G_04151.1 | glucoamylase precursor | 7 | 15 | 84 | Yes | Yes | This study, Sha et al 2009a, Fernández-Acero et al., 2010 |
| BC1G_11898.1 | glucosidase | 5 | 11 | 5 | none | none | This study, Sha et al 2009a, Espino et al., 2010, Fernández-Acero et al., 2010 |
| BC1G_09079.1 | GPI-anchored cell wall beta-1,3-endoglucanase | 3 | 6 | 11 | Yes | Yes | This study, Sha et al 2009a, Sha et al 2009b, Espino et al., 2010 |
| BofuT4_P131540.1 | pectin methyl esterase | 9 | 6 | 12 | none | Yes | This study, Espino et al., 2010, Fernández-Acero et al., 2010 |
| BC1G_ 14403.1 | thioredoxin | 2 | 3 | 3 | none | Yes | This study, Sha et al 2009a, Sha et al 2009b |
| BofuT4_P108020.1 | nucleoside diphosphate kinase | 6 | 3 | 3 | none | none | This study |
| BC1G_08553.1 | laccase 2 | 5 | 2 | 16 | none | Yes | This study, Espino et al., 2010 |
| BC1G_08975.1 | alpha-fucosidase | 3 | 2 | 4 | Yes | Yes | This study, Sha et al 2009b |
| BC1G_14129.1 | hypothetical protein | 4 | 2 | 4 | none | Yes | This study |
| BofuT4_P021640.1 | transaldolase 1 | 2 | 2 | 5 | none | Yes | This study |
Sequence id: Identification number provided by the Broad Institute BO5.10 database
Putative function: was assigned based on sequence similarity when blasted using NCBI non redundant database
Spectra: Total number of spectra matched to proteins in all replicates in that infected tomato fruit
O-glycosylation: Predicted with NetOGlyc 3.1 Server (http://www.cbs.dtu.dk/services/NetOGlyc/)
N-glycosilation: Predicted with NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/)
Endo-PGs were also identified in the present study, including some endo-PGs that had been previously cloned and identified in the B. cinerea genomes.43 Endo-PG activity is regulated by at least six endo-PG genes, and B. cinerea mutants of endo-BcPG1 and endo-BcPG2 have reduced virulence on tomato and other hosts.44, 45 Peptides from BcPG1, BcPG2 and an exo-PG were identified in the present study. Although this study was designed to provide a qualitative global proteomic analysis, some proteins showed high spectral counts and we consider that is important to mention them because they are likely to be particularly abundant. For example, endo-BcPG1 (BC1G_11143.1) was the most abundantly detected B. cinerea protein in all collections (89, 126 and 305 spectra) from infections of MG, RR and rin tomatoes, respectively; while BcPG2 was identified in the rin mutant with three spectral counts and exo-PG was identified in MG tomato, also with three spectral counts. The expression of exo-PGs has been previously identified in cucumber leaves inoculated with spores of B. cinerea.46 This expression either increased with time of culture (exo-PG I) or was transiently expressed soon after the start of culture (exo-PGII), suggesting that the exo-PGs play an important role in pathogenesis at an early stage of infection as well as in tissue maceration of host plants.46
β-Galactosidase (BC1G_03567.1) is another fungal enzyme identified in MG, RR and rin tomato. Plants also express β-galactosidases, which degrade plant cell wall pectins during cell wall loosening that occurs prior to cell elongation.47 Presumably B. cinerea uses β-galactosidases to facilitate the breakdown of fruit cell walls, but its spectral abundance from infected fruit suggests that it may have a more significant role than had been previously assumed.
B. cinerea cellobiohydrolases were only identified in collections from MG and/or rin fruit but not in RR tomato fruit. In MG tomato fruit infected by B. cinerea, three putative cellobiohydrolase or cellulase proteins (BC1G_14702.1, BC1G_06035.1, and BC1G_10880.1), all with a putative cellobiohydrolase or cellulase function, were identified. In infected rin fruit, BC1G_14702.1, BCG_03188.1 and BC1G_10880.1 were also identified. Because transcripts of the B. cinerea cellobiohydrolase gene are absent in infected tomato leaves, it has been suggested that cellobiohydrolase does not play an important role in infection of leaves, but its role in infection of fruit is not known.48 Cellulase enzyme activity from Trichoderma reesie produces a soluble inducer from insoluble cellulose,49, 50 which triggers the expression of cellobiohydrolase in T. reesie.51 Claviceps purpurea cellulolytic enzymes are expressed during the infection process of rye.52 These findings suggest that pathogen cellulases could be involved in the penetration and degradation of host cell walls, thus playing a role in the infection process.
Lacasses are another of the classes of B. cinerea proteins identified, and are present in either all the collections (BC1G_08553.1) or in proteins from infected RR fruit only (BC1G_12487.1). During infection, Botrytis cleaves the polysaccharides within the host cell wall, releasing plant wall fragments that in some hosts may result in the production of phytoalexins, a class of compounds which act as antifungal agents involved in the host response to pathogen attack.53, 54 Hydrogen peroxide is one of the forms of active oxygen species (AOS), which can arise from the response of plant tissues to infection by pathogens.55 Therefore, it may not be unexpected that Botrytis would produce laccases which are capable of inactivating these compounds.
Five B. cinerea glucosidases were identified in the present study (BC1G_12859.1, BC1G_10221.1, BC1G_02364.1, BC1G_04151.1 and BC1G_11898.1). The importance of glucosidases was demonstrated when a positive correlation was found between the β-glucosidase activity of B. cinerea in liquid culture and pathogenicity, as expressed in the area of lesion caused by B. cinerea in different hosts.56
Pathogenicity-related proteins
The category of B. cinerea pathogenicity-related proteins identified contained a homologue of Snodprot (BC1G_02163.1) with a large number of spectral counts in all samples (Table 3) as also seen in previous studies (Table 4). The Botrytis Snodprot homologue (BcSPL1) has a similar sequence to cerato-platanin, and is induced by plant ethylene production at an early stage of infection, and has been reported as an elicitor of pathogen responses and mutants have reduced pathogenicity.57 Elevated BcSPL1 expression has been observed during the first 72 hours after infection. Tomato plants contain a saponin called α-tomatine that has been proposed to kill sensitive cells by binding to cell membranes resulting in leakage of cell components.58 It has been shown that α-tomatine kills a broad range of fungi in vitro and functions as a resistance substance against phytopathogens in tomato.59 As is expected, fungi that invade saponin-containing plants are likely to have a strategy to protect themselves from these saponins and B. cinerea appears not to be the exception, as it produces tomatinase, an enzyme in the GH10 family of β-glucosyl hydrolases that degrades α-tomatine.60 In the NCBI protein database there are 75 tomatinase amino acid sequences.61 A B. cinerea xylosyl hydrolase that degrades α-tomatine has also been purified,61 but its relationship to other fungal glycosyl hydrolases is unclear as the protein has not been sequenced nor has the gene encoding the enzyme been identified.
Carbohydrate-active enzymes from Botrytis
Families of structurally-related catalytic and carbohydrate-binding modules for 28 Botrytis proteins (Table 3, Supplemental Information Figure 6) were assigned based on the CAZYmes Analysis Toolkit (CAT). One major class of CAZy was identified for Botrytis proteins: glycoside hydrolases (GH) with 21 members. Three minor groups, CBMs, CEs and polysaccharide lyases (PLs), were identified with 6, 3 and 1 members, respectively.
Common Botrytis proteins
Fifteen proteins from Botrytis were identified in common in MG, RR and rin infected tomato (Table 4). Of these, 12 Botrytis proteins had been reported in at least one other secretome study12-15 and 3 were identified as unique to this study. A few proteins (BofuT4_P108020.1, BC1G_14129.1, BC1G_12374.1 and BCIG_00896.1) appear worthy of special comment.
BofuT4_P108020 has homology to a nucleoside diphosphate kinase (NDPK) which has been identified in other wound-response studies.62 In addition, it has been shown that NDK in Neurospora crassa together with a catalase plays an important role in supporting the survival of conidia under oxidative and light-induced stress including singlet oxygen.63
Three proteins are present in all samples and their function could not be identified after a Blastp analysis. BC1G_12374.1 and BCIG_00896.1 were previously identified in other secretome studies, and BC1G_14129.1 has not been reported previously. Their presence in all three studies (Table 4) suggests that these proteins could be involved in pathogenicity. It would be of interest to isolate the genes, over-express them, and characterize the resultant proteins to evaluate their role in pathogenicity.
Another feature interesting to highlight from these common proteins is their glycosylation. Many proteins can present with either O-linked or N-linked glycosylation or both (Table 4). Filamentous fungi are known to carry high-mannose N-glycans.64 O-Mannosylation is found in glycoproteins of many higher eukaryotes as well as in most fungi,65 including the filamentous fungi.66 Cell wall degrading enzymes, especially PGs from a single strain of fungus, may exist in a variety of isoforms.67-69 The PG isoforms may each exist as a series of glycoforms, and may vary in their mode of action as well as in their ability to interact with their plant target (i.e., specific pectin classes of PGIPs).67, 70 In addition, the PGIPs of a single plant species may be present as a set of isoforms, each of which exists as a series of glycoforms.71 Protein glycosylation has proven to be important in maintaining protein structure and function and can play a key role in protein-protein interactions.72, 73 In the case of the PG-PGIP complex, this structural variation provides the potential for a wide range of specificity in their interactions within any plant–pathogen pairing. Whether glycosylation of proteins might alter the proteolytic stability of a given protein or its ability to interact with its plant target needs to be investigated, since it could be one of the critical factors in determining whether the fungus is a viable pathogen.
Conclusion
When pathogens encounter plant tissues, proteins from both the plant and the pathogen enable infections and form responses to the interaction. The proteins produced by the pathogen include those that facilitate the pathogen's penetration and growth on the plant tissue, those that repress resistance responses by the plant, and those that allow the pathogen to utilize the nutrient resources within the plant. The proteins produced by the plant include those that limit pathogenic infection, those that facilitate growth of the infecting organism, and those that protect the plant tissue from additional damage. By documenting simultaneously the proteomes of the pathogen and host in the microenvironment of infection sites, the plant proteins that provide resistance and facilitate susceptibility and the pathogen proteins that promote colonization and quiescence can be identified.
In order to grow, a plant pathogenic fungus must secure a carbon source from the plant. A complex cocktail of extracellular enzymes is secreted from the pathogen to fragment polymers, such as cellulose, lignin, proteins and lipids, and then deliver the resulting simple sugars, amino acids, and fatty acids to fungal hyphae, allowing the pathogen to develop and fulfill its life cycle. In response, the host will produce a battery of defense mechanisms to protect itself from the intruder (Figure 4).
Figure 4.
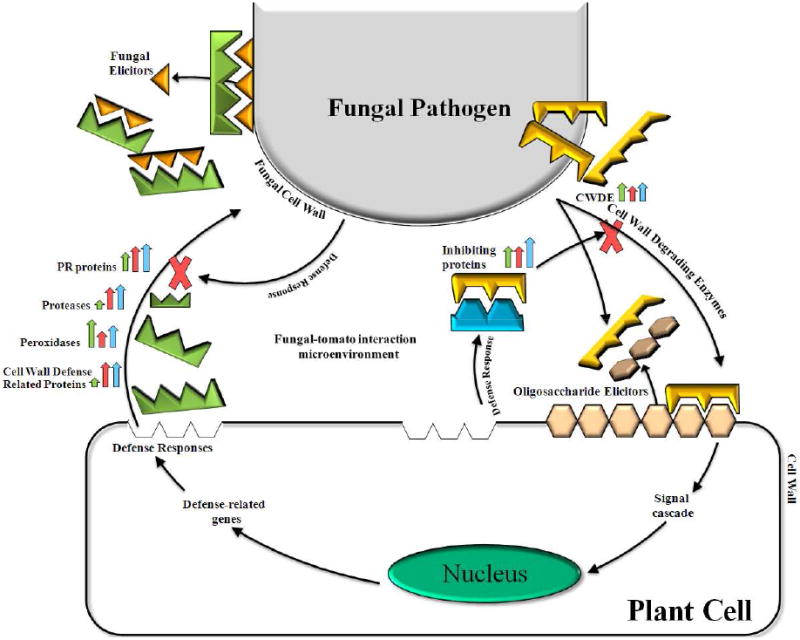
Hypothetical model representing the proteins secreted during a plant-fungal interaction. Pathogenic fungi facilitate their colonization of plant tissue by producing Cell Wall Degrading Enzymes (CWDEs) that fragment plant cell wall polysaccharides. The oligosaccharides that are generated by these glycanases provide the fungus with a carbon source but are also perceived by and elicit defense responses in the host plant. As part of that response, plants produce proteins that inhibit fungal glycanases and thereby increase the life time of the biologically active oligosaccharides. The fragmentation of the fungal cell wall by plant defense related proteins also generates oligosaccharides that induce plant defense responses. The fungus itself may in turn produce defensive proteins to prevent the degradation of its own cell wall and limit its perception by the plant. Thus, the interplay between fungal and plant glycanases and their respective inhibitors may in large part determine the outcome of attempted pathogenesis. The height of the colored arrows indicates the relative abundance of that particular protein group identified when MG (green arrows), RR (red arrows) and rin (blue arrows) tomatoes are infected with B. cinerea.
The proteome analysis described in this work allowed simultaneous identification of B. cinerea and tomato fruit proteins released into the microenvironment of fruit tissue infected by B. cinerea. These results show that this approach is feasible, both for obtaining sufficient information to identify both pathogen and host proteins from sites of infection and to describe the diverse classes of proteins. The results obtained show concurrent information about the host and the pathogen proteomes, identifies significant number of diverse proteins involved in pathogenicity and proteins involved in protection against the oxidative stress response by the host. The identification of proteins with unknown function opens new research avenues, possibly identifying new biological functions that may play important roles in the plant-pathogen interaction.
The data obtained reveal that unsuccessful infections of green fruit result in the identification of 25 and 33% fewer defense related proteins encoded by the host plant as compared to infections of red and non-ripening rin tomato fruit, and that regardless of whether ripe red or non-ripening rin tomato fruit are successfully infected by B. cinerea, but differences in the plant proteomes could be due to the extent of tissue maceration or differences in response by the fruit. In contrast, irrespective of whether B. cinerea is able to infect MG, RR or rin tomato fruit, the proteins coming from the fungus in the infection microenvironment are similar. As noted above, however, shotgun studies cannot detect isoforms or post-translationally modified forms of the proteins, which may prove to be important factors in the change in host-pathogen interactions that lead to susceptibility vs resistance.
This proteome analysis identifies both plant and pathogen proteins with as yet unknown functions which may play a crucial role in plant-pathogen interactions. Proteomics analysis at different times during infection, along with the use of comparative and quantitative techniques, may allow identification of key proteins for recognition and early defense or attack during the interaction. Plant-pathogen proteomics is still in its early stage, but will likely become an active field with a large impact on plant biology. The rapid development of new proteomic analysis techniques is revolutionizing the study of plant-pathogen interactions and shedding more light on the complex network of signaling cascades involved in plant defense responses. Finally, a plant-pathogenic fungus proteome database would be of help to the scientific community studying either the pathogen or the host, and will assist in identifying those genes that are related to different infection and developmental stages as well as growth conditions.
Supplementary Material
Supplemental Information Database. Fasta sequence for tomato and Botrytis cinerea proteins
Supplemental Information Figure 1-2. Percentage of (Fig 1.) molecular function and (Fig 2.) biological process classification of the tomato proteins identified when MG (green bars), RR (red bars) and rin (blue bars) tomatoes are infected with B. cinerea. Identified proteins were classified by function according to GOSlim Viewer from AgBase datase (http://agbase.msstate.edu/cgi-bin/tools/GOanna.cgi).
Supplemental Information Figure 3-4. Percentage of (Fig. 3) molecular function and (Fig. 4) biological process classification of the B. cinerea proteins identified when MG (green bars), RR (red bars) and rin (blue bars) tomatoes are infected with B. cinerea. Identified proteins were classified by function according to GOSlim Viewer from AgBase datase (http://agbase.msstate.edu/cgi-bin/tools/GOanna.cgi).
Supplemental Information Figure 5-6. Number of families of structurally-related catalytic and carbohydrate-binding modules (CBMs) identified from (Fig. 5) tomato and (Fig. 6) Botrytis cinerea. Carbohydrate binding module (CBM), carbohydrate esterase (CE), glycoside hydrolases (GH) and glucosyl transferase (GT) and polysaccharide lyases (PL).
Supplemental Information Table 1. Summary of spectra and proteins in each of the protein collection that were identified as tomato and the number that were identified as B. cinerea proteins.
Supplemental Information Table 2. Tomato proteins identified from MG, RR and rin infected fruit by B. cinerea.
Supplemental Information Table 3. Summary of all tomato and Botrytis cinerea proteins identified from RR, MG and rin infected tomatoes
Supplemental Information Table 4. Fold changes of selected genes
Acknowledgments
This work was supported in part by NSF “Systems Biology Approach to Tomato Fruit Susceptibility to a Necrotrophic Fungus” (IOB-0544504), DOE “Structures and Functions of Oligosaccharins” (DE-FG02-96ER20221), the DOE-funded Center for Plant and Microbial Complex Carbohydrates (DE-FG05-93ER20097), and the NIH/NCRR Integrated Technology Resource for Biomedical Glycomics (P41 RR018502). We thank A. Martinez-Espinoza and L. Wells for critical reading of the manuscript.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Elad Y, Williamson B, Tudzynski P, Delen N. Botrytis spp. and diseases they cause in agricultural systems - an introduction. In: Elad Y, Williamson B, Tudzynski P, Delen N, editors. Botrytis: Biology, Pathology and Control. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2004. pp. 1–8. [Google Scholar]
- 2.Van Kan J. Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 2006;11:247–253. doi: 10.1016/j.tplants.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Cantu D, Vicente AR, Greve LC, Dewey FM, Bennett AB, Labavitch JM, Powell AL. The intersection between cell wall disassembly, ripening, and fruit susceptibility to Botrytis cinerea. Proc Natl Acad Sci USA. 2008;105:859–864. doi: 10.1073/pnas.0709813105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Powell AL, Van Kan J, ten Have A, Visser J, Greve LC, Bennett AB, Labavitch JM. Transgenic expression of Pear PGIP in tomato limits fungal colonization. Mol Plant Microbe Interact. 2000;13:942–950. doi: 10.1094/MPMI.2000.13.9.942. [DOI] [PubMed] [Google Scholar]
- 5.Cantu D, Blanco-Ulate B, Yang L, Labavitch JM, Bennett AB, Powell AL. Ripening regulated susceptibility of tomato fruit to Botrytis cinerea requires NOR but not RIN or ethylene. Plant Physiol. 2009;150:1434–1449. doi: 10.1104/pp.109.138701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curto M, Camafeita E, Lopez JA, Maldonado AM, Rubiales D, Jorrin JV. A proteomic approach to study pea (Pisum sativum) responses to powdery mildew. (Erysiphe pisi) Proteomics. 2006;6:S163–S174. doi: 10.1002/pmic.200500396. [DOI] [PubMed] [Google Scholar]
- 7.Kaffarnik FAR, Jones AME, Rathjen JP, Peck SC. Effector proteins of the bacterial pathogen Pseudemonas syringae alter the extracellular proteome of the host plant, Arabidopsis thaliana. Mol Cell Proteom. 2009;8:145–156. doi: 10.1074/mcp.M800043-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Andrade AE, Silva LP, Pereira JL, Noronha EF, Reis FB, Jr, Bloch C, Jr, dos Santos MF, Domont GB, Franco OL, Mehta A. In vivo proteome analysis of Xanthomonas campestris pv. campestris in the interaction with the host plant Brassica oleracea. FEMS Microbiol Lett. 2008;281:167–174. doi: 10.1111/j.1574-6968.2008.01090.x. [DOI] [PubMed] [Google Scholar]
- 9.Casado-Vela J, Selles S, Martinez RB. Proteomic analysis of tobacco mosaic virus-infected tomato (Lycopersicon esculentum M.) fruits and detection of viral coat protein. Proteomics. 2006;6:S196–S206. doi: 10.1002/pmic.200500317. [DOI] [PubMed] [Google Scholar]
- 10.Casasoli M, Spadoni S, Lilley KS, Cervone F, De Lorenzo G, Mattei B. Identification by 2-D DIGE of apoplastic proteins regulated by oligogalacturonides in Arabidopsis thaliana. Proteomics. 2008;8:1042–1054. doi: 10.1002/pmic.200700523. [DOI] [PubMed] [Google Scholar]
- 11.Fernández-Acero FJ, Jorge I, Calvo E, Vallejo I, Carbú M, Camafeita E, López JA, Cantoral JM, Jorrín J. Two-dimensional electrophoresis protein profile of the phytopathogenic fungus Botrytis cinerea. Proteomics. 2006;6:S88–S96. doi: 10.1002/pmic.200500436. [DOI] [PubMed] [Google Scholar]
- 12.Fernández-Acero FJ, Colby T, Harzen A, Cantoral JM, Schmidt J. Proteomic analysis of the phytopathogenic fungus Botrytis cinerea during cellulose degradation. Proteomics. 2009;9:1–11. doi: 10.1002/pmic.200800540. [DOI] [PubMed] [Google Scholar]
- 13.Shah P, Atwood JAI, Orlando R, El Mubarek H, Podila GK, Davis MR. Comparative proteomic analysis of Botrytis cinerea secretome. J Proteome Res. 2009;8:1123–1130. doi: 10.1021/pr8003002. [DOI] [PubMed] [Google Scholar]
- 14.Shah P, Gutierrez-Sanchez G, Orlando R, Bergmann C. A proteomic study of pectin degrading enzymes secreted by Botrytis cinerea grown in liquid culture. Proteomics. 2009;9:3126–3135. doi: 10.1002/pmic.200800933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernández-Acero FJ, Colby T, Harzen A, Carbú M, Wieneke U, Cantoral JM, Schmidt J. 2-DE proteomic approach to the Botrytis cinerea secretome induced with different carbon sources and plant-based elicitors. Proteomics. 2010;10:2270–2280. doi: 10.1002/pmic.200900408. [DOI] [PubMed] [Google Scholar]
- 16.Espino JJ, Gutierrez-Sanchez G, Brito N, Shah P, Orlando R. The Botrytis cinerea early secretome. Proteomics. 2010;16:3020–3034. doi: 10.1002/pmic.201000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCarthy FM, Wang N, Magee GB, Nanduri B, Lawrence ML, Camon EB, Barrell DG, Hill DP, Dolan ME, Williams WP, Luthe DS, Bridges SM, Burgess SC. AgBase: A functional genomics resource for agriculture. BMC Genomics. 2006;7:229. doi: 10.1186/1471-2164-7-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 19.Fritig B, Heitz T, Legrand M. Antimicrobial proteins in induced plant defense. Curr Opin Immunol. 1998;10:16–22. doi: 10.1016/s0952-7915(98)80025-3. [DOI] [PubMed] [Google Scholar]
- 20.Bowles DJ. Defense-related proteins in higher plants. Annu Rev Biochem. 1990;59:873–907. doi: 10.1146/annurev.bi.59.070190.004301. [DOI] [PubMed] [Google Scholar]
- 21.van Loon LC, Rep M, Pietersen CMJ. Significance of inducible defense-related proteins in infected plants. Ann Rev Phytopathol. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- 22.Ferreira RB, Monteiro SARA, Freitas R, Santos CN, Chen Z, Batista LM, Duarte JOAO, Borges A, Teixeira AR. The role of plant defense proteins in fungal pathogenesis. Mol Plant Pathol. 2007;8:677–700. doi: 10.1111/j.1364-3703.2007.00419.x. [DOI] [PubMed] [Google Scholar]
- 23.Coaker GL, Willard B, Kinter M, Stockinger EJ, Francis DM. Proteomic analysis of resistance mediated by Rcm 2.0 and Rcm 5.1, two loci controlling resistance to bacterial canker of tomato. Mol Plant Microbe Interact. 2004;17:1019–1028. doi: 10.1094/MPMI.2004.17.9.1019. [DOI] [PubMed] [Google Scholar]
- 24.Rep M, Dekker HL, Vossen JH, de Boer AD, Houterman PM, Speijer D, Back JW, de Koster CG, Cornelissen BJ. Mass spectrometric identification of isoforms of PR proteins in xylem sap of fungus-infected tomato. Plant Physiol. 2002;130:904–917. doi: 10.1104/pp.007427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim ST, Kim SG, Hwang DH, Kang SY, Kim HJ, Lee BH, Lee JJ, Kang KY. Proteomic analysis of pathogen responsive proteins from rice leaves induced by rice blast fungus, Magnaporthe grisea. Proteomics. 2004;4:3569–3578. doi: 10.1002/pmic.200400999. [DOI] [PubMed] [Google Scholar]
- 26.Elvira MI, Galdeano MM, Gilardi, Garcia-Luque I, Serra MT. Proteomic analysis of pathogenesis-related proteins (PRs) induced by compatible and incompatible interactions of pepper mild mottle virus (PMMoV) in Capsicum chinense L3 plants. J Exp Bot. 2008;59:1253–1265. doi: 10.1093/jxb/ern032. [DOI] [PubMed] [Google Scholar]
- 27.Selitrennikoff CP. Antifungal proteins. Appl Environ Microbiol. 2001;67:2883–2894. doi: 10.1128/AEM.67.7.2883-2894.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mauch F, Macuch-Mani B, Boller T. Antifungal hydrolases in pea tissue: II. Inhibition of fungal growth by combinations of chitinase and β-1,3-glucanase. Plant Physiol. 1988;88:936–942. doi: 10.1104/pp.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sela-Buurlage MB, Ponstein AS, Bres-Vloemans SA, Melcchers LS, van den Elzen PJM, Cornelissen BJC. Only specific tobacco (Nicotiana tabacum) chitinases and β-1,3-glucanases exhibit antifungal activity. Plant Physiol. 1993;101:857–863. doi: 10.1104/pp.101.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stintzi A, Heitz T, Prasad V, Wiedemann-Merdinoglu S, Kauffmann S, Geoffroy P, Legrand M, Fritig B. Plant “pathogenesis-related” proteins and their role in defense against pathogens. Biochimie. 1993;75:687–706. doi: 10.1016/0300-9084(93)90100-7. [DOI] [PubMed] [Google Scholar]
- 31.Qin Q, Bergmann CW, Rose JKC, Saladie M, Kolli VS, Albersheim P, Darvill AG, York WS. Characterization of a tomato protein that inhibits a xyloglucan-specific endoglucanase. Plant J. 2003;34:327–338. doi: 10.1046/j.1365-313x.2003.01726.x. [DOI] [PubMed] [Google Scholar]
- 32.Estelle M. Proteases and cellular regulation in plants. Curr Opin Plant Biol. 2001;4:254–260. doi: 10.1016/s1369-5266(00)00169-2. [DOI] [PubMed] [Google Scholar]
- 33.Pechan T, Ye L, Chang Y, Mitra A, Lin L, Davis FM, Williams WP, Luthe DS. A unique 33-kD cysteine proteinase accumulates in response to larval feeding in maize genotypes resistant to fall armyworm and other Lepidoptera. Plant Cell. 2000;12:1031–1040. doi: 10.1105/tpc.12.7.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LeMosy EK, Tan YQ, Hashimoto C. Activation of a protease cascade involved in patterning the Drosophila embryo. Proc Natl Acad Sci USA. 2001;98:5055–5060. doi: 10.1073/pnas.081026598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gay PA, Tuzun S. Temporal and spatial assessment of defense responses in resistant and susceptible cabbage varieties during infection with Xanthomonas campestris pv. Campestris. Physiol Mol Plant Pathol. 2000;57:201–210. [Google Scholar]
- 36.Lurie S, Fallik E, Handros A, Shapira R. The possible involvement of peroxidase in resistance to Botrytis cinerea in heat treated tomato fruit. Physiol Mol Plant Pathol. 1997;50:141–149. [Google Scholar]
- 37.Ryan CA. Proteinase inhibitor gene families: Strategies for transformation to improve plant defenses against herbivores. Bioessays. 1989;1:20–24. doi: 10.1002/bies.950100106. [DOI] [PubMed] [Google Scholar]
- 38.Williamson MS, Forde J, Buxton B, Kreis M. Nucleotide sequence of barley chymotrypsin inhibitor-2 (CI-2) and its expression in normal and high-lysine barley. Eur J Biochem. 1987;165:99–106. doi: 10.1111/j.1432-1033.1987.tb11199.x. [DOI] [PubMed] [Google Scholar]
- 39.Mosolov VV, Loginova MD, Fedurkina NV, Benken II. The biological significance of proteinase inhibitors in plants. Plant Sci Lett. 1976;7:77–80. [Google Scholar]
- 40.Mosolov VV, Loginova MD, Malova EL, Benken II. A specific inhibitor of Collectotrichum lindemunthianum protease from kidney beans (Phaseolus vulgaris) seeds. Planta. 1979;144:265–269. doi: 10.1007/BF00388768. [DOI] [PubMed] [Google Scholar]
- 41.Narváez-Vásquez J, Franceschi VR, Ryan CA. Proteinase-inhibitor synthesis in tomato plants: Evidence for extracellular deposition in roots through the secretory pathway. Planta. 1993;189:257–266. [Google Scholar]
- 42.Valette-Collet O, Cimerman A, Reignault P, Levis C, Boccara M. Disruption of Botrytis cinerea pectin methylesterase gene Bcpme 1 reduces virulence on several host plants. Mol Plant Microbe Interact. 2003;16:360–367. doi: 10.1094/MPMI.2003.16.4.360. [DOI] [PubMed] [Google Scholar]
- 43.Wubben JP, Mulder W, ten Have A, Van Kan J, Visser J. Cloning and partial characterization of endopolygalacturonase genes from Botrytis cinerea. Appl Environ Microbiol. 1999;65:596–1602. doi: 10.1128/aem.65.4.1596-1602.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kars I, Krooshof G, Wagemakers L, Joosten R, Benen J, Van Kan J. Necrotizing activity of five Botrytis conerea endopolygalacturonases produced in Pichia pastoris. Plant J. 2005;43:213–225. doi: 10.1111/j.1365-313X.2005.02436.x. [DOI] [PubMed] [Google Scholar]
- 45.ten Have A, Mulder W, Visser J, Van Kan J. The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol Plant Microbe Interact. 1998;11:1009–1016. doi: 10.1094/MPMI.1998.11.10.1009. [DOI] [PubMed] [Google Scholar]
- 46.Rha R, Park HJ, Kim MO, Chung YR, Lee CW, Kim JW. Expression of exopolygalacturonase in Botrytis cinerea. FEMS Microbiol Lett. 2001;210:105–109. doi: 10.1111/j.1574-6968.2001.tb10740.x. [DOI] [PubMed] [Google Scholar]
- 47.Martin I, Dopico B, Munoz FJ, Esteban R, Oomen R, Driouich A, Vincken JP, Visser R, Labrado E. In vivo expression of a Cicer arietinum β-galactosidase in potato tubers leads to a reduction of the galactan side-chains in cell wall pectin. Plant Cell Physiol. 2005;46:1613–1622. doi: 10.1093/pcp/pci177. [DOI] [PubMed] [Google Scholar]
- 48.Cole L, Dewery FM, Hawes CR. Immunochemical studies of the infection mechanisms of Botrytis fabae: II. Host cell wall breakdown. New Phytol. 1998;139:611–622. [Google Scholar]
- 49.Carle-Urioste JC, Escobar-Vera J, El-Gogary S, Henrique-Silva F, Torigoi E, Crivellaro O, Herrera-Estrella A, El-Dorry H. Cellulase induction in Trichoderma reesei by cellulose requires its own basal expression. J Biol Chem. 1997;272:10169–10174. doi: 10.1074/jbc.272.15.10169. [DOI] [PubMed] [Google Scholar]
- 50.Tweddell RJ, Jabaji-Hare SH, Charest PM. Production of chitinases and β-1,3-glucanses by Stachybotrys elegansa mycoparasite of Rhizoctonia solani. Appl Environ Microbiol. 1994;60:489–495. doi: 10.1128/aem.60.2.489-495.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.el-Gogary S, Leite A, Crivellaro O, Eveleigh DE, el-Dorry H. Mechanism by which cellulose triggers cellobiohydrolase I gene expression in Trichoderma reesei. Proc Natl Acad Sci USA. 1989;86:6138–6141. doi: 10.1073/pnas.86.16.6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muller U, Tenberge KB, Oeser B, Tudzynski P. Cel1, probably encoding a cellobiohydrolase lacking the substrate binding domain, is expressed in the initial infection phase of Claviceps purpurea on Secale cereale. Mol Plant Microbe Interact. 1997;10:268–279. doi: 10.1094/MPMI.1997.10.2.268. [DOI] [PubMed] [Google Scholar]
- 53.Darvill AG, Albersheim P. Phytoalexins and their elicitors - a defense against microbial infection in plants. Annu Rev Plant Physiol. 1984;35:243–275. [Google Scholar]
- 54.Mansfield JW. Mechanisms of resistance to ‘Botryti’. In: Coley-Smith JR, Verhoeff K, Jarvis WR, editors. The Biology of Botrytis. Academic Press; New York: 1980. pp. 181–218. [Google Scholar]
- 55.Lamb C, Dixon RA. The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Mol Biol. 1997;48:251–275. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- 56.Gueguen Y, Chemardin P, Arnaud A, Galzy P. Purification and characterization of an intracellular β-glucosidase from Botrytis cinerea. Enzyme Microb Technol. 1995;17:900–906. [Google Scholar]
- 57.Chague V, Levanoni-Visel D, Siewers V, Schulze Gronover C, Tudzynski P, Tudzynski B, Sharon A. Ethylene sensing and gene activation in Botrytis cinerea: A missing link in ethylene regulation of fungus-plant interaction? Mol Plant Microbe Interact. 2006;19:33–42. doi: 10.1094/MPMI-19-0033. [DOI] [PubMed] [Google Scholar]
- 58.Ito S, Ihara T, Tamura H, Tanaka S, Ikeda T, Kajihara H, Dissanayake C, Abdel-Motaal FF, El-Sayed MA. α-Tomatine, the major saponin in tomato, induces programmed cell death mediated by reactive oxygen species in the fungal pathogen Fusarium oxysporum. FEBS Lett. 2007;581:3217–3222. doi: 10.1016/j.febslet.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 59.Sandrock RW, VanEtten HD. Fungal sensitivity to and enzymatic degradation of the phytoanticipin α-tomatine. Phytopathology. 1998;88:137–143. doi: 10.1094/PHYTO.1998.88.2.137. [DOI] [PubMed] [Google Scholar]
- 60.Osbourn AE, Bowyer P, Lunness P, Clarke B, Daniels M. Fungal pathogens of oat roots and tomato leaves employ closely related enzymes to detoxify host plant saponins. Mol Plant Microbe Interact. 1995;8:971–978. doi: 10.1094/mpmi-8-0971. [DOI] [PubMed] [Google Scholar]
- 61.Quidde T, Osbourn A, Tudzynski P. Detoxification of α-tomatine by Botrytis cinerea. Physiol Mol Plant Pathol. 1998;52:151–165. [Google Scholar]
- 62.Bergey DR, Howe GA, Ryan CA. Polypeptide signaling for plant defensive genes exhibits analogies to defense signaling in animals. Proc Natl Acad Sci USA. 1996;93:12053–12058. doi: 10.1073/pnas.93.22.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang N, Yoshida Y, Hasunuma K. Loss of catalase-1 (CAT-1) results in decreased conidial viability enhanced by exposure to light in Neurospora crassa. Mol Gen Genom. 2007;277:13–22. doi: 10.1007/s00438-006-0170-4. [DOI] [PubMed] [Google Scholar]
- 64.Maras M, van Die I, Contreras R, Van Den Hondel CA. Filamentous fungi as production organism for glycoproteins of bio-medical interest. Glycoconj J. 1999;16:99–107. doi: 10.1023/a:1026436424881. [DOI] [PubMed] [Google Scholar]
- 65.Strahl-Bolsinger S, Gentzsch M, Tanner W. Protein O-mannosylation. Biochim Biophys Acta. 1999;1426:297–307. doi: 10.1016/s0304-4165(98)00131-7. [DOI] [PubMed] [Google Scholar]
- 66.Goto M. Protein O-glycosylation in fungi: Diverse structures and multiple functions. Biosci Biotechnol Biochem. 2007;71:1415–1427. doi: 10.1271/bbb.70080. [DOI] [PubMed] [Google Scholar]
- 67.Cook BJ, Clay RP, Bergmann CW, Albersheim P, Darvill AG. Fungal polygalacturonases exhibit different substrate degradation patterns and differ in their susceptibilities to polygalacturonase inhibiting proteins. Mol Plant Microbe Interact. 1999;12:703–711. doi: 10.1094/MPMI.1999.12.8.703. [DOI] [PubMed] [Google Scholar]
- 68.Cervone F, De Lorenzo G, Salvi G, Bergmann C, Hahn MG, Ito Y, Darvill A, Albersheim P. Release of phytoalexin elicitor-active oligogalacturonides by microbial pectic enzymes. In: Lugtenberg BJJ, editor. Signal Molecules in Plants and Plant-Microbe Interactions. H36. Springer Verlag; Heidelberg: 1989. NATO ASI Series. [Google Scholar]
- 69.Centis S, Guillas I, Séjalon N, Esquerré-Tugayé MT, Dumas B. Endopolygalacturonase genes from Colletotrichum lindemuthianum: Cloning of CLPG2 and comparison of its expression to that of CLPG1 during saprophytic and parasitic growth of the fungus. Mol Plant Microbe Interact. 1997;10:769–775. doi: 10.1094/MPMI.1997.10.6.769. [DOI] [PubMed] [Google Scholar]
- 70.Stotz HU, Bishop J, Bergmann CW, Koch M, Albersheim P, Darvill AG, Labavitch JM. Identification of target amino acids that affect interactions of fungal polygalaturonases and their inhibitors. Physiol Mol Plant Pathol. 2000;56:117–130. [Google Scholar]
- 71.Lim JM, Aoki K, Angel PM, Garrison D, King D, Tiemeyer M, Bergmann C, Wells L. Mapping glycans onto specific N-linked glycosylation sites of Pyrus communis PGIP redefines the interface for EPG-PGIP interactions. J Proteome Res. 2009;8:673–680. doi: 10.1021/pr800855f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haltiwanger RS, Lowe JB. Role of glycosylation in development. Annu Rev Biochem. 2004;73:491–537. doi: 10.1146/annurev.biochem.73.011303.074043. [DOI] [PubMed] [Google Scholar]
- 73.Martin PT. Dystroglycan glycosylation and its role in matrix binding in skeletal muscle. Glycobiology. 2003;13:55R–66R. doi: 10.1093/glycob/cwg076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information Database. Fasta sequence for tomato and Botrytis cinerea proteins
Supplemental Information Figure 1-2. Percentage of (Fig 1.) molecular function and (Fig 2.) biological process classification of the tomato proteins identified when MG (green bars), RR (red bars) and rin (blue bars) tomatoes are infected with B. cinerea. Identified proteins were classified by function according to GOSlim Viewer from AgBase datase (http://agbase.msstate.edu/cgi-bin/tools/GOanna.cgi).
Supplemental Information Figure 3-4. Percentage of (Fig. 3) molecular function and (Fig. 4) biological process classification of the B. cinerea proteins identified when MG (green bars), RR (red bars) and rin (blue bars) tomatoes are infected with B. cinerea. Identified proteins were classified by function according to GOSlim Viewer from AgBase datase (http://agbase.msstate.edu/cgi-bin/tools/GOanna.cgi).
Supplemental Information Figure 5-6. Number of families of structurally-related catalytic and carbohydrate-binding modules (CBMs) identified from (Fig. 5) tomato and (Fig. 6) Botrytis cinerea. Carbohydrate binding module (CBM), carbohydrate esterase (CE), glycoside hydrolases (GH) and glucosyl transferase (GT) and polysaccharide lyases (PL).
Supplemental Information Table 1. Summary of spectra and proteins in each of the protein collection that were identified as tomato and the number that were identified as B. cinerea proteins.
Supplemental Information Table 2. Tomato proteins identified from MG, RR and rin infected fruit by B. cinerea.
Supplemental Information Table 3. Summary of all tomato and Botrytis cinerea proteins identified from RR, MG and rin infected tomatoes
Supplemental Information Table 4. Fold changes of selected genes


