Abstract
Background
Hibernation is an adaptation to extreme environments known to provide organ protection against ischemia-reperfusion (I/R) injury. An unbiased systems approach was utilized to investigate hibernation-induced changes characteristic of the hibernator cardioprotective phenotype, by comparing the myocardial proteome of winter hibernating arctic ground squirrels (HIB AGS), summer active (SA) AGS, and rats subjected to I/R, and further correlating with targeted metabolic changes.
Methods
In a well-defined rodent model of I/R by deep hypothermic circulatory arrest followed by 3h or 24h of reperfusion or sham, myocardial protein abundance in AGS (HIB, SA) and rats (n=4-5/group) was quantified by label-free proteomics (n=4-5/group), and correlated with metabolic changes.
Results
Compared to rats, HIB AGS displayed markedly reduced plasma levels of Troponin I, myocardial apoptosis, and left ventricular contractile dysfunction. Of the 1,320 rat and 1,478 AGS proteins identified, 545 were differentially expressed between HIB AGS and rat hearts (47% upregulated, 53% downregulated). Gene ontology analysis revealed downregulation in HIB AGS hearts of most proteins involved in mitochondrial energy transduction, including electron transport chain complexes, acetyl CoA biosynthesis, Krebs cycle, glycolysis and ketogenesis. Conversely, fatty acid oxidation enzymes and Sirtuin-3 were upregulated in HIB AGS, with preserved peroxisome proliferator activated receptor-α activity and reduced tissue levels of acylcarnitines and ceramides following I/R.
Conclusions
Natural cardioprotective adaptations in hibernators involve extensive metabolic remodeling, featuring increased expression of fatty acid metabolic proteins and reduced levels of toxic lipid metabolites. Robust upregulation of Sirtuin-3 suggests that post-translational modifications may underlie organ protection in hibernating mammals.
Introduction
Ischemia-reperfusion injury (I/R), a consequential component of cardiac surgery and organ transplantation, is a major determinant of morbidity and mortality. Cardioprotection after I/R remains an elusive target despite significant research and numerous compounds with promising preclinical data.1 Mammalian hibernation is a seasonal phenomenon characterized by prolonged bouts (days to weeks) of winter torpor, a state of decreased physiological activity, reduced body temperature, and metabolic rate, interspersed with brief (hours to a day) periods of arousal when metabolism and temperature temporarily return to normal.2 The entry into and exit from torpor is akin to I/R, in that blood flow is greatly reduced and then restored. During arousal from torpor, hibernators rewarm using a combination of shivering and non-shivering thermogenesis associated with peaks in oxygen consumption, while accelerating their heart rate 100-fold with corresponding increases in organ perfusion.3 Hibernating mammals like arctic ground squirrels (AGS) show natural resistance to experimental I/R in multiple organs,4-7 which remains yet uncharacterized for the heart. Moreover, although a suite of adaptations associated with the seasonal entry into torpor have been reported, including metabolic rate depression,8 increased reliance on lipid oxidation for energy production,3,9-11 and immune suppression,12 the mechanisms underlying this naturally evolved organ protection remain unknown.
In the present study, we hypothesized that experimental surgical I/R will result in reduced severity of myocardial injury and dysfunction in hibernating (HIB) AGS compared to a non-hibernator (rat); furthermore, we postulated that such hibernator cardioprotective phenotype will be attenuated in summer active (SA) AGS. As a first step to delineate the metabolic regulatory events involved, we employed an unbiased proteomics approach to identify and quantify differences in myocardial protein abundance between AGS and rats, and further correlate with targeted metabolic changes. The potential cardioprotective role of SIRT3 upregulation identified in HIB AGS hearts identified was further assessed in vitro using gain-of-function (rat) and loss-of-function (AGS) experiments in adult primary ventricular cardiomyocyte models of hypoxia-reoxygenation.
Materials and Methods
Animal Ischemia/Reperfusion (I/R) protocol – anesthetic, surgical and perfusion management
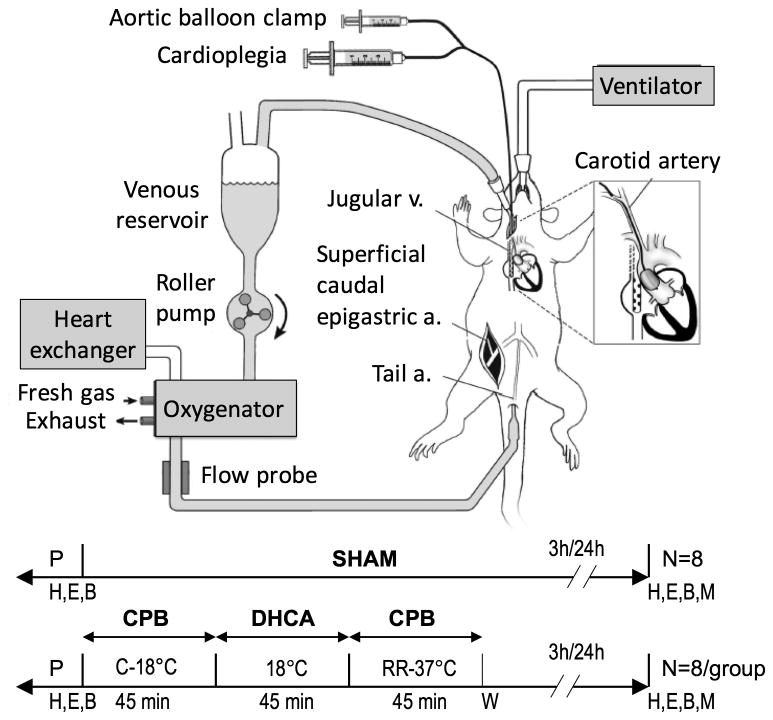
Adult male Brown Norway and Dahl/Salt Sensitive rat strains (Charles River, Wilmington, MA) were randomly assigned to three experimental groups: anesthetized and cannulated without undergoing cardiopulmonary bypass (CPB) (sham group); deep hypothermic circulatory arrest (45 min at 18°C), followed by either 3h (DHCA R3h group) or 24h (DHCA 24h group) of reperfusion (Figure 1).
Figure 1.
Experimental design and perfusion apparatus for deep hypothermic circulatory arrest (DHCA). P, surgical preparation; H, hemodynamic measurements; E, echocardiographic measurements; B, blood specimen; M, myocardial specimen collection; CPB, cardiopulmonary bypass; C, cooling; RR, rewarming and reperfusion; W, weaning from cardiopulmonary bypass.
Arctic ground squirrels (Urocitellus parryii) were trapped in July near the Toolik Field Station in northern Alaska and transported to the University of Alaska Fairbanks. In the fall, animals assigned to the HIB group were housed in temperature and photoperiod controlled facilities (5°C, photoperiod 4L:20D), after being implanted with intraperitoneal temperature-sensitive radio transmitters to monitor their body temperature and precisely determine stages of torpor and arousal, as described.13 Preoperatively, torpid animals (HIB AGS) were moved to a warm, lighted room, and aroused to euthermia before each experiment. Winter experiments were conducted in January-February, 2012 and 2013. Postreproductive summer euthermic animals were used as non-hibernating controls, with experiments conducted in August-September, 2012 and 2013. Summer animals were housed for at least one month to complete quarantine and infection and parasite testing. Summer animals had normal room temperature housing and light at least 12 hours/day. AGS included in these experimental groups were composed of N=16 males and N=7 females. As these animals are wild caught, exact age cannot be determined, however juvenile and aged animals were excluded.
Rats and AGS underwent an identical experimental protocol, as previously described.14,15 Briefly, animals were anesthetized with isoflurane, endotracheally intubated and mechanically ventilated; anesthesia was maintained with isoflurane, fentanyl and pancuronium. Surgical preparation involved cannulation of the ventral tail artery (rats) and right femoral artery (AGS) (20ga, arterial inflow), insertion of a multiorifice dual-stage cannula through the right internal jugular vein and advanced into the right atrium (4.5F, venous outflow), and insertion of a 3.5 mm balloon catheter (Sprinter OTW, Medtronic, Minneapolis, MN) via right carotid artery cutdown, positioned above the aortic valve under echocardiographic guidance. Electrocardiogram, pulse oximetry, end tidal capnography, peri-cranial and rectal temperatures, and invasive arterial blood pressure were continuously recorded. Arterial blood gases were monitored throughout the experimental protocol (GEM Premier 3000, Instrumentation Lab, Lexington, MA). Following systemic heparinization, cardiopulmonary bypass (CPB) was initiated and animals cooled to 18°C. Cardioplegic arrest was achieved by endo-aortic balloon inflation and antegrade administration of blood cardioplegia, with electromechanical arrest confirmed electro- and echocardiographically. DHCA was initiated for 45 min, at the conclusion of which the endoaortic balloon was deflated and CPB resumed. Animals were rewarmed to normothermia (1 hour), weaned from CPB, and recovered until the time of sacrifice, for either 3 hours (R3h) or 24 hours (R24h) post-reperfusion. CPB, ischemia, and rewarming times are identical between R3h and R24h groups, with only duration of reperfusion being different. All experiments were approved by the Institutional Animal Care and Use Committees of Duke University and University of Alaska Fairbanks, and compliant with the Guide for the Care and Use of Laboratory Animals.
Myocardial injury phenotypes
Phenotypic characterization of myocardial injury severity was performed using biochemical, echocardiographic and histological measurements at baseline and at the 3h and 24h reperfusion time points. Levels of cardiac Troponin I (cTnI) were measured at each time point in ethylenediaminetetraacetic acid (EDTA) plasma using a multiplex immunoassay system (Meso Scale Discovery, Rockville, MD). Serial transthoracic echocardiographic measurements of LV systolic function were conducted using a Vivid i ultrasound system (GE Healthcare, Wauwatosa, WI) equipped with an M12L transducer, and analyzed using Xcelera R3.3 software (Phillips, Andover, MA). Left ventricular fractional area change (LV-FAC) was calculated as (LV end diastolic area – LV end systolic area)/LV end diastolic area x100. Percent change in LV-FAC at each reperfusion time point compared to baseline is presented for all groups. Myocardial apoptosis was assessed in LV tissue sections by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assays (Roche Diagnostics, Indianapoils, IN) and double immunofluorescence staining for activated caspase-3 and Troponin I, as well as by determining levels of cleaved caspase 3 in myocardial tissue (Western blot).
Sample preparation for mass spectrometry
At the time of sacrifice (3h or 24h reperfusion), animals were completely exsanguinated via the jugular venous cannula and perfused with 60mL warm PBS. The heart was rapidly removed, divided using a rat heart slicer matrix into axial sections for histology and snap frozen in liquid nitrogen. Details of sample preparation protocols for proteomic and metabolic profiling are presented in Supplementary Methods.
Proteomic data collection and processing
We employed a 5-fraction two dimensional liquid chromatography, tandem mass spectrometry (2D-LC/LC-MS/MS) method to identify and quantitate proteins expressed in LV myocardium of AGS (HIB, SA) and Brown Norway rats subjected to DHCA followed by 3h or 24h of reperfusion, or sham. Proteins extracted from LV homogenates were normalized, reduced, cysteine alkylated and trypsin digested, followed by label-free quantitative 2D-LC/LC-MS/MS. Robust peak detection and label-free alignment of individual peptides across all sample injections was performed using Rosetta Elucidator v3.3 (Rosetta Biosoftware, Seattle, WA) with PeakTeller algorithm. MS/MS data were searched against the NCBI RefSeq Rattus (for rat) and against a curated AGS database representing refined thirteen-lined ground squirrel (Ictidomys tridecemlineatus) and human protein sequences from Ensembl release 69, as described.10 After individual peptide scoring using the Peptide Prophet algorithm, the data was annotated at a <1% peptide false discovery rate. Relative peptide and protein abundance were calculated in Rosetta Elucidator. Details for mass spectrometry data collection, processing and quality control are presented in Supplementary Methods.
Arctic ground squirrel to rat orthology mapping
A key component of our analytical strategy to compare changes in myocardial protein expression across species involves the establishment of orthologous relationships among the proteins identified and quantified using label-free proteomic approaches in rat vs. AGS. Commonly used methods for orthology assessment typically use Basic Local Alignment Search Tool (BLAST) to search for pairs of reciprocal best hits in the genomes (or proteomes) being compared. However, these methods are easily misled when additional gene duplications occur after the species in question have diverged (“in-paralogs”), leading to the presence of co-orthologs in cross-genome comparisons. We have defined sets of putatively orthologous proteins among those identified within each species by 5-fraction 2D-LC/LC-MS/MS proteomics. These orthologies were established by adapting the EnsemblCompara GeneTrees resource developed by Vilella et al, a system that automates the clustering, multiple alignment, and phylogenetic analysis necessary to build gene trees for large numbers of gene families represented in the Ensembl project. Given the known one-to-one, one-to-many and many-to-many orthology relationships, we used the BioMart data-mining tool and Ensembl Genes Release 73 (Rnor_5.0 for rat, Spetri2 for squirrel) to conduct a bidirectional orthology matching using first the rat as the reference genome and then the AGS as the reference genome. The resulting orthology map - containing 702 unique rat GI protein IDs, 700 unique squirrel Ensembl Protein IDs, and 697 unique gene names – was used for subsequent cross-species comparative analyses of myocardial protein abundance. Details of the computational approach to establish the orthology map are presented in Supplementary Methods.
Mitochondrial DNA analysis
Total DNA (mitochondrial and nuclear) was extracted from rat and AGS LV myocardium samples using a QIAamp DNA Mini Kit (Qiagen, Valencia, CA). The content of mtDNA was calculated using quantitative real-time PCR by measuring the threshold cycle ratio (ΔCT) of a mitochondrial-encoded gene (Cytochrome c oxidase, subunit I, Mt-Co1) versus a nuclear-encoded gene (Cyclophilin A, Ppia) in a StepOnePlus RT-PCR System (Life Technologies, Carlsbad, CA) equipped with StepOne v2.2.3 software (see Supplemental Methods for details on primers used and RT-PCR protocol).
Western blot analysis
To validate mass spectrometry findings, Western blot imaging analysis was conducted for specific ETC proteins (NDUFS8, SDHA, UQCRQ, COX5A and ATP5A) and SIRT3, and all results normalized using ratios to either the constitutively expressed protein COXIV, HSP60, or β-actin depending on protein size (see Supplemental Methods for details).
Functional Sirtuin-3 (SIRT3) analyses
To characterize the functional relevance of differences in myocardial SIRT3 abundance identified by proteomic analysis, we conducted additional in vitro SIRT3 gain and loss-of-function studies in adult primary ventricular cardiomyocytes (APVC) isolated from rat and AGS,16 respectively, and assessed the effects on cell viability following simulated I/R. Rat APVCs were treated with increasing concentrations of resveratrol (Sigma-Aldrich, St. Louis, MO), an activator of SIRT3, or nicotinamide, an inhibitor of SIRT3 (control, 5μM, 10 μM) for 1h. AGS APVCs were transfected with SIRT3-siRNA (60pmol) or scrambled RNA control (Santa Cruz, Dallas, TX) overnight, according to manufacturer's protocols. Cells were subjected to 2h of oxygen and glucose deprivation followed by 24h of reoxygenation and determination of apoptosis (Cell Death Detection ELISA kit, Roche, Indianapolis, IN) and necrosis (cardiac Troponin I levels in supernatant, Life Diagnostics, West Chester, PA). SIRT3 expression (Western blot) and activity (fluorometric Sirt3 activity assay kit, Abcam, Cambridge, MA) were measured in APVC lysates. All experiments were run in triplicate.
PPAR-α activity assay
The nuclear fraction was isolated from cryopreserved myocardial samples using a commercial nuclear extraction kit (Panomics, Fremont, CA) according to manufacturer's specifications, and assayed for PPAR-α nuclear receptor activity by ELISA (Affymetrix, Santa Clara, CA) (see Supplemental Methods for details).
Targeted metabolic profiling
Cryopreserved myocardial samples were homogenized in 50% acetonitrile supplemented with 0.3% formic acid, and used to determine levels of acylcarnitines, organic acids, amino acids and ceramides via stable isotope dilution techniques, as described previously.17 Details of the metabolomic analysis are provided in Supplemental Methods.
Statistical analysis
Experimental sample sizes are based on previous reports of differences in protein abundance in hibernators, 3,10 comparative responses to I/R across species 18, as well as efforts to conserve scant resources (wild-caught arctic ground squirrels).
Principal component analysis (PCA) was performed to explore sources of variation in relative protein abundance across the experimental dataset, visualize whether biological replicate samples resemble each other, and identify any outliers. PCA uses orthogonal transformation to convert relative protein abundance into a set of linearly uncorrelated principal components.19 The top 3 principal components are presented as 3D scatter plots in Supplemental Results, with each point representing the proteomic data from one sample, and samples colored by species and hibernation state. Differentially expressed proteins in this multi-factor experiment were identified using mixed model ANOVA which included species, hibernation state (winter hibernating, summer active), reperfusion time (sham, reperfusion 3h, reperfusion 24h), and hibernation state * reperfusion time interaction (to test if seasonal hibernation states would result in temporal differences in myocardial protein abundance post-reperfusion). Differentially expressed proteins were defined based on a fold-change ≥ 1.5 or ≤ -1.5 and a Benjamini-Hochberg false discovery rate (FDR) adjusted p-value <0.05. For this exploratory analysis, we chose FDR to adjust for multiple comparisons over the more conservative family-wise error rate approach (e.g. Bonferroni correction), which would likely miss a number of potentially novel differences.20 Exploratory PCA analysis and differential protein expression analysis were conducted using Partek Genomics Suite v6.6 (Partek, St Louis, MO).
Differences in perioperative myocardial injury endpoints (biochemical, echocardiographic), as well as differences in abundance of individual proteins or metabolites between HIB AGS, SA AGS, and rats, or between timepoints were assessed by one-way ANOVA with Tukey's post hoc test for pairwise comparisons. Two-way ANOVA was further used to test whether differences in individual protein abundance between species and reperfusion timepoints were more than expected by chance. Mann-Whitney U test was used for two group comparisons of apoptosis and necrosis endpoints. Statistical analyses were conducted using GraphPad Prism v6 (GraphPad Software, Inc, La Jolla, CA).
Results
The hibernator cardioprotective phenotype
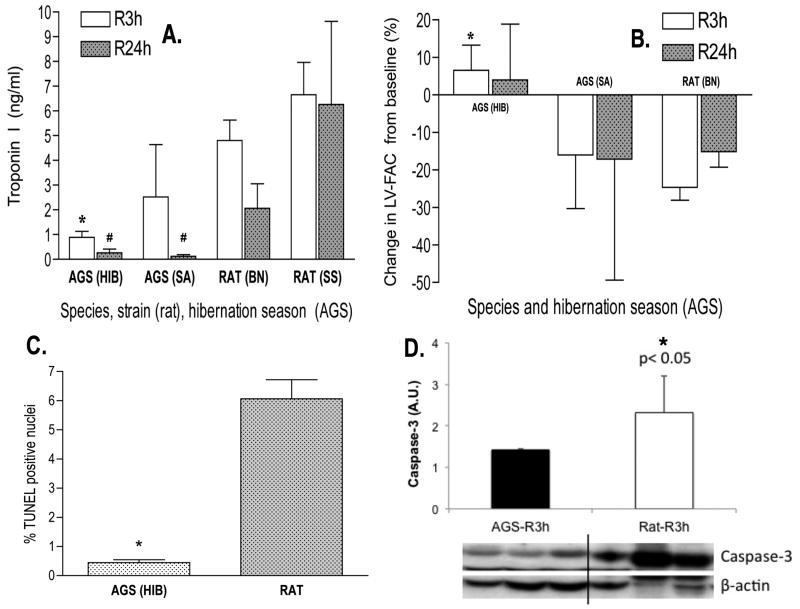
Compared to rats, AGS exhibited significantly reduced myocardial injury at both reperfusion time points (mean ± SD of plasma Troponin I concentrations following 3h of reperfusion were 0.9±0.2, 2.5±2.1, 4.8±0.8, and 6.7±1.3, and following 24h of reperfusion were 0.2±0.1, 0.1±0.1, 2.1±0.9, and 6.3±3.3 for HIB AGS, SA AGS, Brown Norway rats and Dahl-Salt Sensitive rats, respectively; Figure 2A). Reflecting known differences in susceptibility to myocardial I/R between the rat strains used as non-hibernator experimental model organisms, severity of myocardial injury in HIB AGS was ∼5-fold lower compared to Brown Norway rats (a strain characterized by resistance against myocardial I/R), and ∼8-fold lower than in Dahl/Salt Sensitive rats (a strain more susceptible to myocardial I/R). HIB AGS displayed the most robust cardioprotection, with SA AGS demonstrating an intermediate level of protection (Figure 2A). The biochemical results were corroborated echocardiographically, with HIB AGS showing preserved left ventricular systolic function following I/R, whereas LV-FAC declined in both SA AGS and rats (mean ± SD of percent changes in LV-FAC compared to preoperative baseline following 3h of reperfusion were +7±6, -16±14, and -25±3, and following 24h of reperfusion were 4±14, -17±32, and -15±4 for HIB AGS, SA AGS and rats, respectively; Figure 2B). Myocardial apoptosis was attenuated in HIB AGS compared to rats following I/R, as evidenced by lower fraction of TUNEL-positive nuclei (Figure 2C), and reduced tissue levels of active caspase-3 (Figure 2D).
Figure 2.
The cardioprotective hibernator phenotype. Winter (HIB) and Summer Active (SA) AGS exhibit reduced myocardial injury following I/R compared to Brown Norway (BN) or Dahl Salt Sensitive (SS) rats, as evidenced by reduced plasma markers of myonecrosis at both the 3h (R3h) and 24h (R24h) reperfusion time points (#, p<0.01, *, p<0.05 by ANOVA with Tukey's post-hoc correction) (A). Winter (HIB) AGS further demonstrated preserved echocardiographic left ventricular systolic function (LV-FAC) after I/R compared to SA AGS or rats (*, p<0.05, ANOVA with Tukey's post-hoc correction) (B) and reduced myocardial apoptosis as evidenced by Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining (C) corroborated by cleaved caspase-3 levels (D) at the 3h reperfusion timepoint (*, p<0.05, Mann-Whitney U test). Data presented as mean ± SD, N=8/group.
Cross-species quantitative LV myocardial proteomic analyses
Following quality control, the final quantitative dataset for rat consisted of 8482 peptides/1320 proteins (786 proteins contained at least 2 unique peptides), and 9292 peptides/1478 proteins (806 proteins contained at least 2 unique peptides) for AGS. Differential expression analysis across species was conducted based on an orthology map consisting of 697 proteins. Principal component analysis was performed to identify high-level differences between sample groups (Supplemental Figure S3). Signal vs. noise across all orthologous proteins for each of the factors and interactions in the ANOVA model identified that all factors contributed significant variation to the data across the set, with species (rat vs AGS) representing the dominant source of variation (Supplemental Figure S4).
A total of 545 proteins were differentially expressed between HIB AGS and rat based on a fold change ≥1.5 or ≤-1.5 and an FDR adjusted p-value <0.05, with 255 upregulated and 290 downregulated proteins (Supplemental Figure S5). Of these, 191 proteins had more than a ±6 fold change between HIB AGS and rat, and 498 had a Bonferroni-adjusted p-value <0.05. Using pathway analysis these proteins were mapped onto 77 canonical pathways. Top canonical pathways differentially expressed between HIB and rat included oxidative phosphorylation (p=1.1e-20), protein ubiquitination (1.4e-13), tricarboxylic acid cycle (6e-9), branched chain amino acid metabolism (7.3e-7) and cytoskeletal remodeling (7.8e-7).
Electron transport chain protein downregulation
The most prominent proteomic finding is a general downregulation of electron transport chain proteins in the hibernator hearts (Table 1). Significant reductions in levels of subunits of all five electron transport chain (ETC) complexes were found in hibernating AGS compared to rats, with only complex V showing increased expression of some subunits after I/R. Complex I subunits NADH-ubiquinone oxidoreductase (NDUFs) 1-13 were reduced 1.14-17.68 fold. Complex II subunits Succinate Dehydrogenase Complex (SDH) A, SDHB, and SDHC declined 2.47-5.1 fold. Complex III subunits Ubiquinol Cytochrome-C Reductase (UQCR)Q, UQCRFS1, UQCRC2, and UQCR10 declined 2.04-3.57 fold. Complex IV subunit Cytochrome C oxidase (COX) 5A declined 1.23 fold. Complex V subunits ATP synthase mitochondrial F1/F0 complex (ATP) AF2, ATP5F1, and ATP5B declined 1.75-19.11 fold; however subunits ATP5A1-ATP5J2 increased in abundance 1.41-15.94 fold (Table 1).
Table 1. Expression differences in hibernating AGS vs rat hearts for members of the electron transport chain.
| Protein symbol | Protein name | Fold Change HIB vs Rat | FDR adjusted p-value |
|---|---|---|---|
| Complex I (NADH-ubiquinone reductase) | |||
| Ndufa10 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex 10-like protein | -1.14 | 1.61E-02 |
| Ndufa13 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13 | -2.76 | 7.76E-10 |
| Ndufa5 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 | -5.26 | 4.19E-07 |
| Ndufab1 | NADH dehydrogenase [ubiquinone] 1, alpha/beta subcomplex, 1 | -16.51 | 1.06E-12 |
| Ndufb10 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10 | -1.87 | 2.60E-11 |
| Ndufb5 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex, 5 | -1.49 | 1.68E-05 |
| Ndufb6 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex, 6 | -6.28 | 6.15E-13 |
| Ndufs1 | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial | -3.62 | 1.27E-12 |
| Ndufs2 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 2, mitochondrial | -2.18 | 4.12E-11 |
| Ndufs3 | NADH dehydrogenase [ubiquinone] Fe-S protein 3 | -1.49 | 6.15E-05 |
| Ndufs4 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial | -1.20 | 1.88E-02 |
| Ndufs5 | NADH dehydrogenase [ubiquinone] Fe-S protein 5b, 15kDa (NADH-coenzyme Q reductase) | -3.31 | 1.58E-16 |
| Ndufs6 | NADH dehydrogenase [ubiquinone] Fe-S protein 6 | -1.20 | 1.58E-02 |
| Ndufs7 | NADH dehydrogenase [ubiquinone] Fe-S protein 7 | -7.11 | 5.89E-25 |
| Ndufs8 | NADH dehydrogenase [ubiquinone] Fe-S protein 8 | -17.68 | 7.93E-12 |
| Ndufv2 | NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial | -1.37 | 1.16E-05 |
| Ndufv3 | NADH dehydrogenase [ubiquinone] flavoprotein 3, mitochondrial | -2.69 | 9.07E-12 |
| Complex II (Succinate dehydrogenase-CoQ reductase) | |||
| Sdha | Succinate dehydrogenase complex subunit A | -5.10 | 3.20E-16 |
| Sdhb | Succinate dehydrogenase complex subunit B | -2.60 | 3.05E-11 |
| Sdhc | Succinate dehydrogenase complex subunit C | -2.47 | 3.91E-08 |
| Complex III (Cytochrome reductase) | |||
| Uqcrc2 | Cytochrome b-c1 complex subunit 2, mitochondrial | -2.48 | 8.02E-08 |
| Uqcrfs1 | Cytochrome b-c1 complex subunit Rieske, mitochondrial | -2.92 | 4.79E-21 |
| Uqcrq | Cytochrome b-c1 complex subunit 8 | -3.57 | 7.23E-12 |
| Uqcr10 | Ubiquinol-cytochrome c reductase, complex III subunit X | -2.04 | 3.75E-06 |
| Complex IV (Cytochrome c oxidase) | |||
| Cox5a | Cytochrome c oxidase subunit 5A, mitochondrial | -1.23 | 1.21E-03 |
| Complex V (ATP synthase) | |||
| Atp5b | ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide | -19.11 | 3.66E-23 |
| Atp5f1 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit B1 | -3.52 | 1.51E-09 |
| Atpaf2 | ATP synthase mitochondrial F1 complex assembly factor 2 | -1.75 | 4.79E-04 |
| Atp5a1 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle | 2.24 | 9.07E-10 |
| Atp5c1 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | 1.41 | 1.49E-06 |
| Atp5d | ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit | 1.59 | 4.02E-11 |
| Atp5e | ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit | 3.19 | 9.70E-13 |
| Atp5h | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit d | 2.17 | 8.17E-10 |
| Atp5i | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E | 2.02 | 1.07E-07 |
| Atp5j2 | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F2 | 3.16 | 5.43E-05 |
| Atp5l | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G | 15.94 | 1.02E-18 |
| Atp5o | ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit | 2.46 | 1.07E-10 |
| Atpif1 | ATPase inhibitory factor 1 | 1.50 | 8.26E-04 |
| Other | |||
| Etfa | Electron transfer flavoprotein, alpha polypeptide | -1.83 | 3.15E-12 |
| Etfdh | Electron transfer flavoprotein dehydrogenase | -1.62 | 1.06E-05 |
Abbreviations: AGS- arctic ground squirrel
ATP- adenosine triphosphate
CoQ- coenzyme Q10
FDR- false discovery rate
HIB- hibernating arctic ground squirrel
NADH- nicotinamide adenine dinucleotide, reduced form
To ensure that lower quantities of ETC proteins detected in AGS were not secondary to lower mitochondrial density in the myocardium, mtDNA quantity was calculated as the ratio of COX1 to cyclophillin A DNA levels determined by RT-PCR, and found to be higher for AGS compared to rat (Supplemental Figure S10), consistent with an increase in mitochondrial biogenesis in the hibernator heart. A decrease in mtDNA copy number, however, was seen in both species under the R3h condition compared to sham, coincident with peak reperfusion injury (Figure S10).
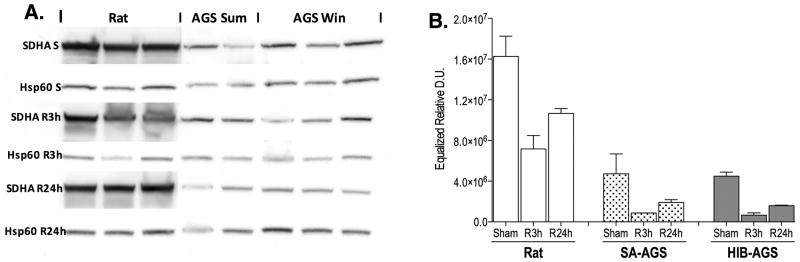
Western blot analyses confirmed mass spectrometry findings in select ETC proteins of interest. Downregulation of Complex II protein succinate dehydrogenase A (SDHA) in HIB AGS and SA AGS compared with rat at all timepoints is shown in figure 3. Results for Complex I protein NDUFS8, Complex III protein UQCRQ, Complex IV protein COX5A, and Complex V protein ATP5A are shown in figure S11 and table S7.
Figure 3.
Western blot analysis for succinate dehydrogenase A (SDHA) a protein in ETC complex II. Blots are shown for rat, summer active arctic ground squirrel (SA-AGS), and hibernating arctic ground squirrel (HIB-AGS) at sham (S), 3 hour (R3h), and 24 hour of reperfusion (R24h) time points. Heat-shock protein 60 (HSP60) is shown as a mitochondrial protein loading control (A). Densitometric analysis of HSP60 normalized SDHA bands demonstrating downregulation in SA and HIB-AGS compared to rat under sham, R3h, and R24h conditions, consistent with the mass spectrometry findings (B). Results displayed as mean ± standard error of the mean (SEM). Two-way ANOVA analysis identified both species and timepoint as being significant factors (p<0.0001) affecting SDHA expression.
Increased myocardial Sirtuin-3 levels and the cardioprotective hibernator phenotype
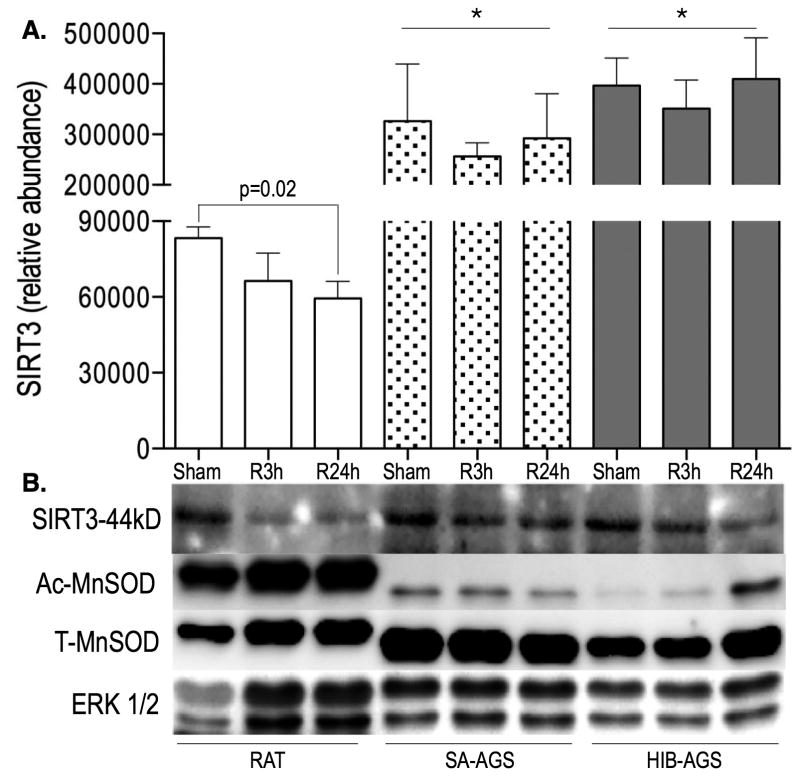
Sirtuin-3 (SIRT3), the major mitochondrial protein deacetylase with multiple cardioprotective targets including ETC proteins, is significantly upregulated in both HIB and SA AGS compared to rat by proteomic profiling (Figure 4A). Species differences also exist in the dynamic expression changes following I/R – unlike rats, AGS recover their baseline SIRT3 levels by R24h (Figure 4A), Western blot analysis shows a SIRT3 expression pattern consistent with the proteomic findings (Figure 4B). We also corroborated the SIRT3 result with changes in target proteins known to be regulated by SIRT3, including an upregulation in AGS hearts of manganese superoxide dismutase (total-MnSOD)21 accompanied by post-translational deacetylation of MnSOD,22 (Figure 4B) known to increase its ROS scavenging activity.23
Figure 4.
Proteomic analyses identified a significant increase in expression of myocardial Sirtuin-3 (SIRT3) in arctic ground squirrel (AGS) (both summer active [SA] and hibernating [HIB]) compared to rat, at every timepoint. Species differences also exist in the dynamic expression changes following ischemia reperfusion injury (I/R): unlike rats, AGS recover their baseline SIRT3 levels by R24h. Data shown as mean ± standard deviation (SD), N=6-8/group (A). Results were validated by Western blot, and were corroborated with changes in proteins known to be regulated by SIRT3 – upregulation of manganese superoxide dismutase (MnSOD, T-total levels increased in AGS), and deacetylation of MnSOD (Ac-MnSOD levels decreased in AGS), which increases its ROS scavenging activity. Extracellular signal-regulated kinase (ERK)1/2 used as loading control (B). *, p<0.05 by analysis of variance (ANOVA) with Tukey's post-hoc correction, relative to respective timepoint in the rat.
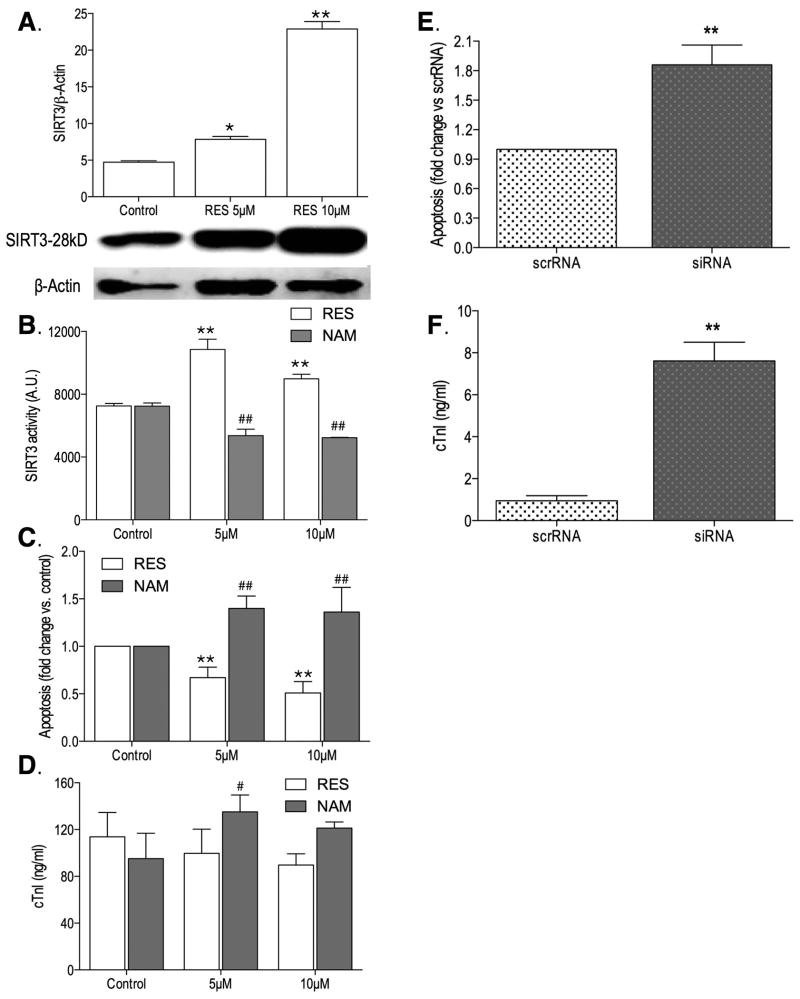
To further explore the potential mechanistic links between SIRT3 expression, activity, and the hibernator cardioprotective phenotype, we conducted comparative gain- and loss-of-function experiments in adult primary ventricular cardiomyocyte cell models of hypoxia-reoxygenation. In rat APVCs, resveratrol (RES) increased SIRT3 expression and activity at both 5μM and 10μM doses (Figure 5A,B), which was associated with reduced apoptotic and necrotic cell death (Figure 5C,D). Conversely, nicotinamide (NAM) reduced SIRT3 activity at both 5μM and 10μM (Figure 5B), and was associated with increased cardiomyocyte death (Figure 5C,D) On the other hand, siRNA SIRT3 silencing in AGS cardiomyocytes resulted in significant increases of apoptotic and necrotic death compared to scrambled RNA treated cells (Figure 5E,F). Furthermore, the observed increase in AGS cardiomyocyte viability following hypoxia-reoxygenation stress (Figure S12) is accompanied by reduced acetylation of mitochondrial proteins compared to the rat (Figure S13).
Figure 5.
Functional Sirtuin-3 (SIRT3) analyses in adult primary ventricular cardiomyocytes isolated from rat and arctic ground squirrel (AGS) and subjected to 2h of oxygen and glucose deprivation followed by 24h of reoxygenation. In rat cardiomyocytes, resveratrol (RES), an activator of SIRT3 (A,B), was associated with reduced apoptosis (C) and a trend towards reduced necrosis (D) (*, p<0.05, **, p<0.01, analysis of variance (ANOVA) with Tukey's post-hoc correction), whereas nicotinamide (NAM), an inhibitor of SIRT3 (B), had the opposite effects (C,D, #, p<0.05, ##, p<0.01, ANOVA with Tukey's post-hoc correction). Two-way ANOVA analyses identified significant (p<0.0001) treatment (RES versus NAM) and concentration (5μM, 10μM) effects on SIRT3 activity (B) and cardiomyocyte apoptosis (C). Conversely, in AGS cardiomyocytes, short inhibitory RNA (siRNA) mediated SIRT3 knockdown was associated with increased apoptosis, but not scrambled RNA (scrRNA). (E) and necrotic cell death (F) following simulated ischemia reperfusion injury (I/R) (**, p<0.01, Mann-Whitney U test). Data presented as mean ± standard deviation (SD), all experiments conducted in triplicate.
Metabolic correlates of the hibernator cardioprotective phenotype
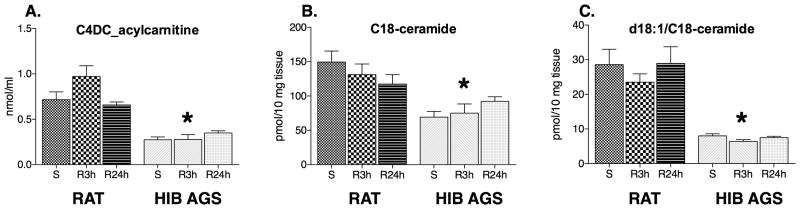
The proteomic analyses revealed a sharp contrast between a general downregulation of key metabolic enzymes associated with glycolysis, TCA cycle, ketolysis, amino acid catabolism and ETC, whereas a significant proportion of proteins involved in fatty acid uptake, transport and catabolism were upregulated in the hibernator hearts (Table 2). Accordingly, we used mass spectrometry-based targeted metabolomic profiling to provide additional insight into the energy metabolic changes occurring in the hibernator heart during I/R stress. The most striking distinguishing metabolite signature in the AGS compared to rat myocardium was exhibited by the acylcarnitine profile. Accummulation of toxic intermediates of lipid metabolism (acylcarnitines and ceramides) was found in LV myocardial tissue homogenates from rat compared to HIB AGS at both reperfusion timepoints (Figure 6).
Table 2. Changes in protein abundance for key regulators of intermediary metabolism.
| Protein symbol | Protein name | Fold Change HIB vs Rat | FDR adjusted p-value |
|---|---|---|---|
| Acetyl CoA biosynthesis - PDH complex | |||
| Pdha1 | pyruvate dehydrogenase (lipoamide) alpha 1 | -2.91 | 6.31E-19 |
| Pdhb | pyruvate dehydrogenase (lipoamide) beta | -4.58 | 1.02E-14 |
| Dlat | dihydrolipoamide S-acetyltransferase | -15.01 | 1.55E-16 |
| Dld | dihydrolipoamide dehydrogenase | -2.04 | 2.88E-14 |
| Ketogenesis – Ketolysis | |||
| Acat1 | acetyl-CoA acetyltransferase 1 | -3.12 | 9.54E-14 |
| Bdh1 | 3-hydroxybutyrate dehydrogenase, type 1 | -1.46 | 1.60E-04 |
| Hadhb | hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit | -1.74 | 7.89E-06 |
| Oxct1 | 3-oxoacid CoA transferase 1 | -2.57 | 3.93E-15 |
| Branched-chain Amino Acid Metabolism | |||
| Acat1 | Acetyl-CoA acetyltransferase, mitochondrial | -3.12 | 9.54E-14 |
| Aldh6a1 | Methylmalonate-semialdehyde dehydrogenase [acylating], mitochondrial | -5.55 | 4.17E-16 |
| Bckdha | branched chain ketoacid dehydrogenase E1, alpha polypeptide | -2.84 | 2.93E-19 |
| Hadhb | Trifunctional enzyme subunit beta, mitochondrial | -1.74 | 7.89E-06 |
| Hibch | 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial | -1.27 | 4.94E-04 |
| Ivd | Isovaleryl-CoA dehydrogenase, mitochondrial | -4.60 | 1.90E-20 |
| Mccc2 | Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial | -185.75 | 3.58E-14 |
| Oxct1 | Succinyl-CoA:3-ketoacid coenzyme A transferase 1, mitochondrial | -2.57 | 3.93E-15 |
| Pcca | Propionyl-CoA carboxylase alpha chain, mitochondrial | -12.07 | 6.07E-15 |
| Pccb | Propionyl-CoA carboxylase beta chain, mitochondrial | -5.40 | 5.11E-18 |
| TCA Cycle | |||
| Aco2 | Aconitate hydratase, mitochondrial | -1.63 | 4.00E-07 |
| Cs | Citrate synthase, mitochondrial | -1.56 | 7.66E-04 |
| Dld | Dihydrolipoyl dehydrogenase, mitochondrial | -2.04 | 2.88E-14 |
| Dlst | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | -1.75 | 1.57E-06 |
| Idh2 | Isocitrate dehydrogenase [NADP], mitochondrial | -5.69 | 4.07E-20 |
| Idh3B | Isocitrate dehydrogenase [NAD] subunit beta, mitochondrial | -2.06 | 1.17E-11 |
| Sdha | Succinate dehydrogenase complex, subunit A | -5.10 | 3.20E-16 |
| Sdhb | Succinate dehydrogenase complex, subunit B | -2.60 | 3.05E-11 |
| Sdhc | Succinate dehydrogenase complex, subunit C | -2.47 | 3.91E-08 |
| Sucla2 | succinate-Coenzyme A ligase, ADP-forming, beta subunit | -1.50 | 1.27E-08 |
| Suclg1 | Succinyl-CoA ligase [ADP/GDP-forming] subunit alpha, mitochondrial | -4.52 | 1.54E-19 |
| Glycolysis and Gluconeogenesis | |||
| Aldoa | Fructose-bisphosphate aldolase A | -2.62 | 5.97E-12 |
| Eno1 | Alpha-enolase | -40.94 | 9.81E-18 |
| Eno3 | Beta-enolase | -31.90 | 2.67E-18 |
| Gapdhs | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific | -38.11 | 3.14E-17 |
| Mdh1 | Malate dehydrogenase, cytoplasmic | -1.94 | 1.35E-11 |
| Pgam1 | Phosphoglycerate mutase 1 | -2.41 | 1.83E-11 |
| Pgk1 | Phosphoglycerate kinase 1 | -1.52 | 5.83E-08 |
| Pgk2 | phosphoglycerate kinase 2 | -5.66 | 5.32E-04 |
| Fatty Acid Metabolism | |||
| Acaa2 | acetyl-CoA acyltransferase 2 | 1.20 | 3.97E-02 |
| Acadsb | acyl-CoA dehydrogenase, short/branched chain | 1.45 | 4.04E-02 |
| Adh5 | alcohol dehydrogenase 5 (class III), chi polypeptide | 695.23 | 3.41E-19 |
| Aldh2 | aldehyde dehydrogenase 1 family, member A1 | 1.79 | 4.63E-07 |
| Aldh5a1 | aldehyde dehydrogenase 4 family, member A1 | 1.93 | 1.52E-03 |
| Cd36 | fatty acid translocase | 5.08 | 1.82E-09 |
| Cpt1b | carnitine palmitoyltransferase 1b, muscle | 4.88 | 3.71E-10 |
| Dhrs4 | dehydrogenase/reductase (SDR family) member 4 | 2.78 | 2.04E-11 |
| Echs1 | enoyl CoA hydratase, short chain, 1, mitochondrial | 1.36 | 8.39E-06 |
| Fabp4 | fatty acid binding protein 4, adipocyte | 1.78 | 5.72E-11 |
| Hadha | hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit | 2.49 | 7.40E-11 |
| Hsd17b8 | hydroxysteroid (17-beta) dehydrogenase 8 | 5.39 | 7.01E-17 |
| Slc25a3 | solute carrier family 25 (mitochondrial carrier, phosphate) carrier), member 3 | 21.60 | 3.02E-26 |
| Slc25a4 | solute carrier family 25 (mitochondrial carrier; adenine) nucleotide translocator), member 4 | 4.13 | 1.01E-14 |
Abbreviations: ADP- adenosine diphosphate, AGS- arctic ground squirrel, GDP- guanosine diphosphate,
CoA- coenzyme A, FDR- false discovery rate, HIB- hibernating arctic ground squirrel,
NAD- nicotinamide adenine dinucleotide, oxidized form,
NADP- nicotinamide adenine dinucleotide phosphate, oxidized form
PDH- pyruvate dehydrogenase
SDR- short-chain dehydrogenases/reductases
TCA- tricarboxylic acid
Figure 6.
Metabolic correlates of hibernator cardioprotective phenotype. We used mass-spectrometry-based metabolomics to quantify ischemia reperfusion injury (I/R) induced changes in targeted metabolites between hibernating arctic ground squirrels (HIB AGS) and rats. Accumulation of toxic intermediates of lipid metabolism, acylcarnitine (C4DC acylcarnitine) (A) and ceramide species (C18-ceramide and d18:1/C18-ceramide) (B-C), was found in left ventricular myocardial tissue homogenates from rats compared to arctic ground squirrels (AGS), at both reperfusion 3 hour (R3h) and 24 hour (R24h) time points, sham surgery (S) is shown for comparison (*, p<0.05, ANOVA with Tukey's post-hoc correction). Data presented as mean ± standard deviation, N=6/group.
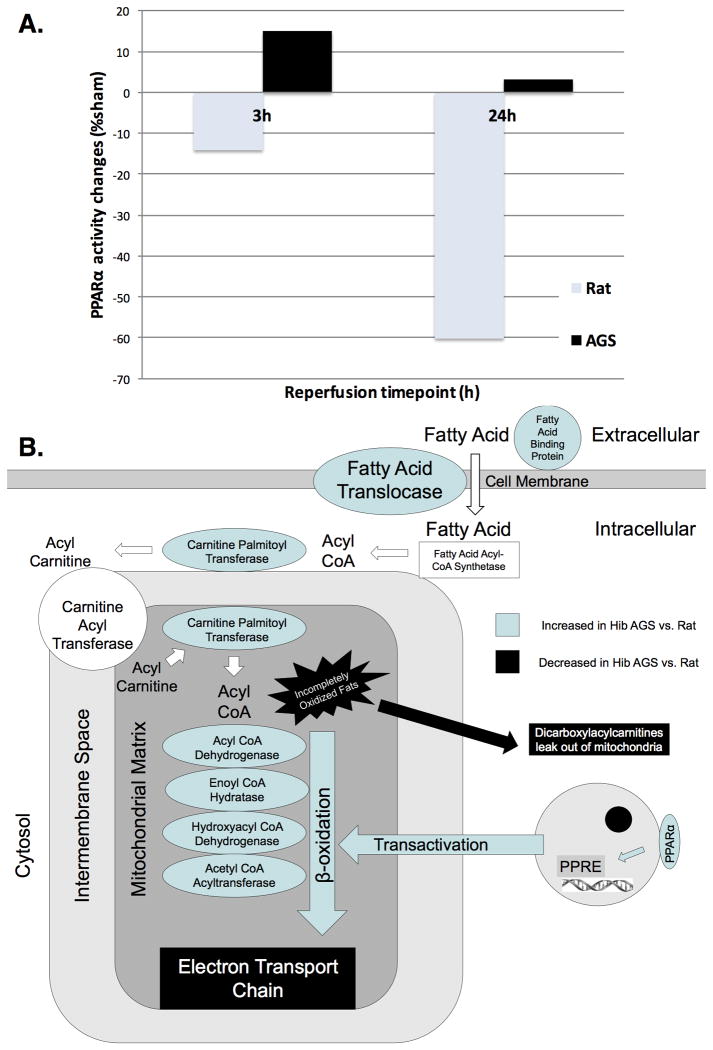
While the prime regulatory mechanism underlying the hibernator phenotype remains unclear, key molecules known to play a role in lipid metabolism and energy flux are significantly different between rat and HIB AGS in our analyses. We therefore conducted comparative analyses of peroxisome proliferator activated receptor-alpha (PPARα) activity, a master regulator of mitochondrial fatty acid utilization, in the hearts of rats and HIB AGS in response to I/R. While PPARα activity levels significantly declined with I/R in rats, HIB AGS were able to maintain baseline levels of PPARα activity despite I/R, and may be involved in the upregulation of fatty acid oxidation enzymes observed in AGS through its trans-activation effects (Figure 7).
Figure 7.
The hibernator cardioprotective phenotype is associated with preservation of peroxisome proliferator activated receptor alpha (PPARα) activity by transcription factor binding assay in hibernating arctic ground squirrel (AGS) compared to rat, at both reperfusion time points (A). A diagram of lipid metabolism focused on lipid transport into the mitochondrion is given in (B). Increased expression of multiple fatty acid transporters and enzymes of fatty acid metabolism may contribute to increased metabolic efficiency in hibernating AGS hearts. PPARα-regulated transactivation effects maintain lipid metabolism under conditions of physiologic stress.
Discussion
Here we report for the first time a cross-species analysis demonstrating cardioprotection in hibernating AGS compared with summer AGS or rat. Following a clinically relevant I/R model and employing robust endpoint measures, hibernating AGS displayed significantly attenuated myocardial injury, as assessed by plasma levels of troponin I, myocardial apoptosis, and preservation of LV contractile function. Our comparative analyses employed two different strains of rat with known differences in susceptibility to I/R (Brown Norway and Dahl Salt Sensitive), and revealed a further significant reduction in susceptibility in the hibernator heart. Moreover, we were able to demonstrate a seasonal difference in responses to I/R in AGS, with increased protection in HIB AGS. While AGS as a species seem to be resistant to I/R, many of the adaptations that allow maximum protection appear to maximize in the winter.24 Our experimental model of I/R injury employs standardized temperature (18°C), anesthetic, and cannulation strategies between rat, summer AGS, and hibernating AGS, and therefore the observed cardioprotection in HIB AGS can be ascribed to adaptations that accompany the hibernator state, not to protective effects of hypothermia.
We undertook an unbiased approach to identify changes in metabolic enzymes and metabolite pools between species and hibernation states in response to I/R stress that may shed light onto the mechanisms underlying cardioprotection in hibernators. The most striking difference is the profound decline in expression of ETC proteins in hibernating AGS compared to rat, a pattern also associated with the most significant cardioprotection of all three groups. Recent work by an independent laboratory has characterized proteomic differences in adaptation to myocardial ischemia among non-hibernating mammals. Cabrera et al, examined the proteomic signatures of myocardial hibernation and ischemic preconditioning in swine.25 Hibernating myocardium, an adaptation to chronic ischemia in which the myocardium decreases its oxygen consumption by adopting a quiescent state26 (and a confusing term for hibernation biologists), was found to have decreased quantities of all five ETC complex proteins - including NADH-ubiquinone oxidoreductase (NDUFs), succinate dehydrogenase (SDH), ubiquinol cytochrome-c reductase (UQCR), cytochrome c oxidase (COX), and ATP synthase mitochondrial F1/F0 complex subunits. This quiescent state allows for the preservation of viable myocardium that can regain function if blood supply is restored.27 Ironically, our data demonstrate that the cardioprotective strategy adopted by hibernating mammals in response to I/R closely approximates the proteomic features of hibernating myocardium in non-hibernators, suggesting that at some level the regulatory components of the hibernator cardioprotective phenotype may elicit a conserved response to chronic ischemia in all mammals. However, hibernating AGS hearts also downregulate the molecular machinery involved in glycolysis, TCA cycle, and the metabolism of ketones and amino acids. This is in contrast to what is known about hibernating myocardium in mammals that do not seasonally hibernate such as rats or humans, which is more reliant on glycolysis.28 Our comparative analyses revealed that rats suffer more myocardial injury, and have a proteomic profile characterized by maintenance of ETC protein abundance. This more closely approximates the adaptations seen in ischemic preconditioning. This occurs when the myocardium is exposed to brief non-lethal periods of ischemia and becomes resistant to further ischemic insults,29-31 yet it does not significantly reduce ETC proteins.25 In our model, rats demonstrate inferior cardioprotection to AGS associated with a pattern of maintained expression of ETC proteins and increased glycolysis, ketolysis, and amino acid metabolism. Consistent with these findings, reversible pharmacological blockade of electron transport at the onset of reperfusion was recently reported to decrease cardiac injury in aged hearts by improving the inner mitochondrial membrane potential.32
The cardioprotective adaptations in hibernators were associated with robust upregulation of Sirtuin-3 (SIRT3) in AGS hearts, the main mitochondrial protein deacetylase and key regulator of metabolic (including fatty acid oxidation) stress-response pathways in the heart.33 Increasing evidence also suggests Sirtuin involvement in regulating the hypometabolic states associated with hibernation. During entrance into torpor, reductions in metabolism precede reductions in body temperature (even when thermogenesis is not active) suggesting active mechanisms regulating metabolic suppression rather than passive thermal effects.8,34 Mitochondrial respiration decreases quickly during entrance into torpor when body temperature is high (e.g. by 70% between 37°C during interbout arousal and 30°C during torpor entrance).34 This occurs faster than transcriptional or translational changes; moreover, peptide elongation in hibernators ceases below 18°C,35 yet in early arousal (when body temperature is much lower), mitochondrial respiration increases significantly. This pattern suggests that body temperature- sensitive, enzyme-mediated post-translational modifications of oxidative phosphorylation complexes like differential acetylation could be involved. Although the heart accounts for <0.5% of basal metabolic rate, cardiac mitochondria from AGS display 60% reductions in respiration rates during torpor.36 Inhibition of succinate-fueled state 3 mitochondrial respiration has been observed in three different tissues (heart, skeletal muscle, and liver), appearing to be a hallmark of hibernation in AGS. The underlying mechanisms remain unclear, but do not appear to involve inhibitions of either succinate oxidation (SHD activity)37 or transport (dicarboxylate transporter). Nonetheless, the report of SIRT3-mediated deacetylation of SDHA regulating ETC complex II activity38 is of particular relevance to hibernators, given the profound suppression of succinate oxidation during torpor. Although we did not specifically assess acetylation of ETC complexes in our study, increased myocardial expression of SIRT3 in AGS was associated with activation of key cardioprotective targets like MnSOD and consequently reduced oxidative stress. We are providing a preliminary characterization of SIRT3 involvement in the hibernator cardioprotective phenotype, by reporting in cardiomyocyte models of I/R the association between SIRT3 gain- and loss-of-function and changes in cell survival. Furthermore, consistent with the differences in SIRT3 expression, rat cardiomyocytes had hyperacetylated mitochondrial proteins compared to AGS, which was further increased following I/R.
A precisely controlled fuel shift from myocardial carbohydrate to fatty acid metabolism in hibernating AGS appears to be orchestrated through downregulation of key metabolic enzymes associated with glycolysis, TCA cycle, ketolysis, and branched-chain amino acid catabolism, while increasing enzymes involved in fatty acid catabolism. This represents something of a contradiction to classic teaching for myocardial substrate utilization during ischemic stress, when the heart's ability to process a wide variety of substrates as fuel would appear to be an advantage; moreover, oxidation of carbohydrate, ketone, and amino acid fuel sources requires less oxygen that the catabolism of fatty acids.39 However, increased expression of enzymes involved in fatty acid catabolism may help explain why hibernating AGS are able to catabolize fats without suffering ill-effects, even with the addition of I/R injury. We show that increased expression levels of fatty acid oxidation enzymes is associated with preserved activity of the PPAR-α transcription factor following I/R in AGS but not in rats. The mechanism of increased myocardial injury in non-hibernators exposed to high fat may in part be due to incomplete oxidation of fatty acids. Accumulation of partially oxidized lipids and sphingolipids can give rise to lipotoxicity as these reactive intermediates accumulate in the mitochondria of organisms unable to metabolize a large fat load.40 To investigate the role of lipotoxicity in our model we used targeted metabolomic profiling to measure levels of partially oxidized lipid intermediates previously implicated as deleterious in I/R injury, like dicarboxylacylcarnitines and ceramides.41-43 Compared to rats, winter AGS have significantly lower myocardial levels of C4-dicarboxylacylcarnitine and C-18 ceramide (Figure 6). During early reperfusion (R3h), rats experience a rise in C4-dicarboxylacylcarnitines indicative of mitochondrial dysfunction, whereas winter AGS maintain their baseline levels.
In an attempt to employ an unbiased analysis of differences in myocardial proteins between hibernators vs. non-hibernators, we have utilized a label-free proteomic profiling method. This technique has the limitation of detecting high abundance proteins over rare ones. One shortcoming of our study is that regulatory proteins may not be adequately represented. Also, in this experiment we have focused on the identification and abundance of proteins as a first step, many regulatory steps are undoubtedly controlled by post-translational modifications including: phosphorylation, acetylation, Neddylation, SUMOylation, and ubiquitinylation. These modifications may exert effects by modifying protein function or simply marking a protein for degradation. Oxidation and partial degradation of protein might also not be adequately detected by our technique. While we have been able to demonstrate quantitative changes in ETC proteins in whole myocardial tissue, we did not perform the analysis in isolated mitochondria, and thus cannot speak to changes in protein abundance within the mitochondria specifically. Our data relate to proteomic changes within a specific quantity of myocardial protein. We did however demonstrate that the ratio of mitochondrial to nuclear DNA is in fact higher in AGS than rat, thus eliminating the possibility that the decline in ETC proteins is secondary to falling numbers of mitochondria.
In summary, we present the first comparative proteomics study of myocardial protein expression changes following experimental I/R in hibernating versus non-hibernating mammals. We show that several prominent features of myocardial I/R injury like myocardial necrosis, apoptosis and mechanical stunning were significantly reduced in the hibernator heart, and accompanied by differential expression of proteins mainly involved in regulation of mitochondrial fuel and energy metabolism. Our data support the idea that mammals have a dichotomous response to I/R injury, one pathway leading towards the development of hibernating myocardium, the other to ischemic preconditioning. Ironically, hibernating animals exhibit a seasonal cardioprotective phenotype that resembles hibernating myocardium with respect to the decline of ETC protein abundance. In contrast, rats maintain levels of ETC proteins consistent with what has been observed in models of ischemic preconditioning. Enhanced lipid metabolism possibly regulated by SIRT3, PPARs and other master regulators of metabolism may be key to the metabolic reprogramming associated with cardioprotection in hibernators. Further understanding this unique model of extreme metabolic plasticity and developing strategies to ‘switch’ myocardial metabolism to resemble that naturally occurring in mammalian hibernators represent a transformative approach that could ultimately have a positive impact in patients undergoing cardiac surgery and transplantation, victims of cardiac arrest, trauma and hypothermia, in addition to fundamentally advancing cardiovascular biology.
Supplementary Material
Acknowledgments
We would like to thank Jeanette Moore, Ph.D., Franziska Kohl, MSc, and Lori Bogren, Ph.D. (University of Alaska Fairbanks) for technical assistance with DHCA experiments in Alaska; Jun Yan, Ph.D. (Chinese Academy of Sciences-German Max Planck Society, Shanghai, China) for help in building phylogenetic orthology maps for cross-species analyses; and G. Burkhard Mackensen, M.D., Ph.D. (University of Washington, Department of Anesthesiology, Seattle, WA) for his role in developing the DHCA model in rodents.
Funding Sources: Foundation for Anesthesia Education and Research, Research Fellowship Grant, Duke University Department of Anesthesiology, PI: Quintin Quinones, Mentor: Mihai Podgoreanu;
NIGMS T32 GM08600, Duke University Medical Center, Fellow: Quintin Quinones;
NIH R01-HL092071, Duke Innovation Grant Award, Duke University Medical Center, PI: Mihai Podgoreanu
American Heart Association, 11BGIN7620055 Beginning Grant in Aid, Duke University Medical Center, PI: Zhiquan Zhang
Footnotes
Disclosures: None
References
- 1.Schwartz Longacre L, Kloner RA, Arai AE, Baines CP, Bolli R, Braunwald E, Downey J, Gibbons RJ, Gottlieb RA, Heusch G, Jennings RB, Lefer DJ, Mentzer RM, Murphy E, Ovize M, Ping P, Przyklenk K, Sack MN, Vander Heide RS, Vinten-Johansen J, Yellon DM. New horizons in cardioprotection: recommendations from the 2010 National Heart, Lung, and Blood Institute Workshop. Circulation. 2011;124(10):1172–1179. doi: 10.1161/CIRCULATIONAHA.111.032698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carey HV, Andrews MT, Martin SL. Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiological reviews. 2003;83(4):1153–1181. doi: 10.1152/physrev.00008.2003. [DOI] [PubMed] [Google Scholar]
- 3.Grabek KR, Karimpour-Fard A, Epperson LE, Hindle A, Hunter LE, Martin SL. Multistate proteomics analysis reveals novel strategies used by a hibernator to precondition the heart and conserve ATP for winter heterothermy. Physiological genomics. 2011;43(22):1263–1275. doi: 10.1152/physiolgenomics.00125.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurtz CC, Lindell SL, Mangino MJ, Carey HV. Hibernation confers resistance to intestinal ischemia-reperfusion injury. American journal of physiology Gastrointestinal and liver physiology. 2006;291(5):G895–901. doi: 10.1152/ajpgi.00155.2006. [DOI] [PubMed] [Google Scholar]
- 5.Lindell SL, Klahn SL, Piazza TM, Mangino MJ, Torrealba JR, Southard JH, Carey HV. Natural resistance to liver cold ischemia-reperfusion injury associated with the hibernation phenotype. American journal of physiology Gastrointestinal and liver physiology. 2005;288(3):G473–480. doi: 10.1152/ajpgi.00223.2004. [DOI] [PubMed] [Google Scholar]
- 6.Dave KR, Anthony Defazio R, Raval AP, Dashkin O, Saul I, Iceman KE, Perez-Pinzon MA, Drew KL. Protein kinase C epsilon activation delays neuronal depolarization during cardiac arrest in the euthermic arctic ground squirrel. Journal of neurochemistry. 2009;110(4):1170–1179. doi: 10.1111/j.1471-4159.2009.06196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dave KR, Prado R, Raval AP, Drew KL, Perez-Pinzon MA. The arctic ground squirrel brain is resistant to injury from cardiac arrest during euthermia. Stroke; a journal of cerebral circulation. 2006;37(5):1261–1265. doi: 10.1161/01.STR.0000217409.60731.38. [DOI] [PubMed] [Google Scholar]
- 8.Heldmaier G, Ortmann S, Elvert R. Natural hypometabolism during hibernation and daily torpor in mammals. Respiratory physiology & neurobiology. 2004;141(3):317–329. doi: 10.1016/j.resp.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 9.Frank CL, Karpovich S, Barnes BM. Dietary fatty acid composition and the hibernation patterns in free-ranging arctic ground squirrels. Physiological and biochemical zoology : PBZ. 2008;81(4):486–495. doi: 10.1086/589107. [DOI] [PubMed] [Google Scholar]
- 10.Shao C, Liu Y, Ruan H, Li Y, Wang H, Kohl F, Goropashnaya AV, Fedorov VB, Zeng R, Barnes BM, Yan J. Shotgun proteomics analysis of hibernating arctic ground squirrels. Molecular & cellular proteomics : MCP. 2010;9(2):313–326. doi: 10.1074/mcp.M900260-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams DR, Epperson LE, Li W, Hughes MA, Taylor R, Rogers J, Martin SL, Cossins AR, Gracey AY. Seasonally hibernating phenotype assessed through transcript screening. Physiological genomics. 2005;24(1):13–22. doi: 10.1152/physiolgenomics.00301.2004. [DOI] [PubMed] [Google Scholar]
- 12.Bouma HR, Carey HV, Kroese FG. Hibernation: the immune system at rest? Journal of leukocyte biology. 2010;88(4):619–624. doi: 10.1189/jlb.0310174. [DOI] [PubMed] [Google Scholar]
- 13.Yan J, Barnes BM, Kohl F, Marr TG. Modulation of gene expression in hibernating arctic ground squirrels. Physiological genomics. 2008;32(2):170–181. doi: 10.1152/physiolgenomics.00075.2007. [DOI] [PubMed] [Google Scholar]
- 14.de Lange F, Yoshitani K, Podgoreanu MV, Grocott HP, Mackensen GB. A novel survival model of cardioplegic arrest and cardiopulmonary bypass in rats: a methodology paper. Journal of cardiothoracic surgery. 2008;3:51. doi: 10.1186/1749-8090-3-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jungwirth B, Mackensen GB, Blobner M, Neff F, Reichart B, Kochs EF, Nollert G. Neurologic outcome after cardiopulmonary bypass with deep hypothermic circulatory arrest in rats: description of a new model. The Journal of thoracic and cardiovascular surgery. 2006;131(4):805–812. doi: 10.1016/j.jtcvs.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 16.Li XC, Wei L, Zhang GQ, Bai ZL, Hu YY, Zhou P, Bai SH, Chai Z, Lakatta EG, Hao XM, Wang SQ. Ca2+ cycling in heart cells from ground squirrels: adaptive strategies for intracellular Ca2+ homeostasis. PloS one. 2011;6(9):e24787. doi: 10.1371/journal.pone.0024787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turer AT, Stevens RD, Bain JR, Muehlbauer MJ, van der Westhuizen J, Mathew JP, Schwinn DA, Glower DD, Newgard CB, Podgoreanu MV. Metabolomic profiling reveals distinct patterns of myocardial substrate use in humans with coronary artery disease or left ventricular dysfunction during surgical ischemia/reperfusion. Circulation. 2009;119(13):1736–1746. doi: 10.1161/CIRCULATIONAHA.108.816116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutchison JS, Frndova H, Lo TY, Guerguerian AM, Hypothermia Pediatric Head Injury Trial I, Canadian Critical Care Trials G Impact of hypotension and low cerebral perfusion pressure on outcomes in children treated with hypothermia therapy following severe traumatic brain injury: a post hoc analysis of the Hypothermia Pediatric Head Injury Trial. Developmental neuroscience. 2010;32(5-6):406–412. doi: 10.1159/000323260. [DOI] [PubMed] [Google Scholar]
- 19.Sydenham E, Roberts I, Alderson P. Hypothermia for traumatic head injury. The Cochrane database of systematic reviews. 2009;(2):CD001048. doi: 10.1002/14651858.CD001048.pub3. [DOI] [PubMed] [Google Scholar]
- 20.Glickman ME, Rao SR, Schultz MR. False discovery rate control is a recommended alternative to Bonferroni-type adjustments in health studies. Journal of clinical epidemiology. 2014;67(8):850–857. doi: 10.1016/j.jclinepi.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 21.Kozinski M, Pstragowski K, Kubica JM, Fabiszak T, Kasprzak M, Kuffel B, Paciorek P, Navarese EP, Grzesk G, Kubica J. ACS network-based implementation of therapeutic hypothermia for the treatment of comatose out-of-hospital cardiac arrest survivors improves clinical outcomes: the first European experience. Scandinavian journal of trauma, resuscitation and emergency medicine. 2013;21:22. doi: 10.1186/1757-7241-21-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernard SA, Gray TW, Buist MD, Jones BM, Silvester W, Gutteridge G, Smith K. Treatment of comatose survivors of out-of-hospital cardiac arrest with induced hypothermia. The New England journal of medicine. 2002;346(8):557–563. doi: 10.1056/NEJMoa003289. [DOI] [PubMed] [Google Scholar]
- 23.Engler RL. Lidoflazine in the treatment of comatose survivors of cardiac arrest. The New England journal of medicine. 1991;325(14):1046–1047. doi: 10.1056/nejm199110033251416. [DOI] [PubMed] [Google Scholar]
- 24.Barnes BM. Freeze avoidance in a mammal: body temperatures below 0 degree C in an Arctic hibernator. Science. 1989;244(4912):1593–1595. doi: 10.1126/science.2740905. [DOI] [PubMed] [Google Scholar]
- 25.Cabrera JA, Butterick TA, Long EK, Ziemba EA, Anderson LB, Duffy CM, Sluiter W, Duncker DJ, Zhang J, Chen Y, Ward HB, Kelly RF, McFalls EO. Reduced expression of mitochondrial electron transport chain proteins from hibernating hearts relative to ischemic preconditioned hearts in the second window of protection. Journal of molecular and cellular cardiology. 2013;60:90–96. doi: 10.1016/j.yjmcc.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 26.Rahimtoola SH. The hibernating myocardium. American heart journal. 1989;117(1):211–221. doi: 10.1016/0002-8703(89)90685-6. [DOI] [PubMed] [Google Scholar]
- 27.Ceconi C, La Canna G, Alfieri O, Cargnoni A, Coletti G, Curello S, Zogno M, Parrinello G, Rahimtoola SH, Ferrari R. Revascularization of hibernating myocardium: rate of metabolic and functional recovery and occurrence of oxidative stress. European heart journal. 2002;23(23):1877–1885. doi: 10.1053/euhj.2002.3364. [DOI] [PubMed] [Google Scholar]
- 28.Vogt AM, Elsasser A, Nef H, Bode C, Kubler W, Schaper J. Increased glycolysis as protective adaptation of energy depleted, degenerating human hibernating myocardium. Molecular and cellular biochemistry. 2003;242(1-2):101–107. [PubMed] [Google Scholar]
- 29.Sumeray MS, Yellon DM. Ischaemic preconditioning reduces infarct size following global ischaemia in the murine myocardium. Basic research in cardiology. 1998;93(5):384–390. doi: 10.1007/s003950050106. [DOI] [PubMed] [Google Scholar]
- 30.Sack S, Mohri M, Arras M, Schwarz ER, Schaper W. Ischaemic preconditioning--time course of renewal in the pig. Cardiovascular research. 1993;27(4):551–555. doi: 10.1093/cvr/27.4.551. [DOI] [PubMed] [Google Scholar]
- 31.Schott RJ, Rohmann S, Braun ER, Schaper W. Ischemic preconditioning reduces infarct size in swine myocardium. Circulation research. 1990;66(4):1133–1142. doi: 10.1161/01.res.66.4.1133. [DOI] [PubMed] [Google Scholar]
- 32.Chen Q, Ross T, Hu Y, Lesnefsky EJ. Blockade of electron transport at the onset of reperfusion decreases cardiac injury in aged hearts by protecting the inner mitochondrial membrane. Journal of aging research. 2012;2012:753949. doi: 10.1155/2012/753949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr, Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464(7285):121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toien O, Blake J, Edgar DM, Grahn DA, Heller HC, Barnes BM. Hibernation in black bears: independence of metabolic suppression from body temperature. Science. 2011;331(6019):906–909. doi: 10.1126/science.1199435. [DOI] [PubMed] [Google Scholar]
- 35.van Breukelen F, Martin SL. Translational initiation is uncoupled from elongation at 18 degrees C during mammalian hibernation. American journal of physiology Regulatory, integrative and comparative physiology. 2001;281(5):R1374–1379. doi: 10.1152/ajpregu.2001.281.5.R1374. [DOI] [PubMed] [Google Scholar]
- 36.Chung D, Lloyd GP, Thomas RH, Guglielmo CG, Staples JF. Mitochondrial respiration and succinate dehydrogenase are suppressed early during entrance into a hibernation bout, but membrane remodeling is only transient. Journal of comparative physiology B, Biochemical, systemic, and environmental physiology. 2011;181(5):699–711. doi: 10.1007/s00360-010-0547-x. [DOI] [PubMed] [Google Scholar]
- 37.Brown JC, Staples JF. Substrate-specific changes in mitochondrial respiration in skeletal and cardiac muscle of hibernating thirteen-lined ground squirrels. Journal of comparative physiology B, Biochemical, systemic, and environmental physiology. 2014;184(3):401–414. doi: 10.1007/s00360-013-0799-3. [DOI] [PubMed] [Google Scholar]
- 38.Cimen H, Han MJ, Yang Y, Tong Q, Koc H, Koc EC. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry. 2010;49(2):304–311. doi: 10.1021/bi901627u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grynberg A, Demaison L. Fatty acid oxidation in the heart. Journal of cardiovascular pharmacology. 1996;28(Suppl 1):S11–17. doi: 10.1097/00005344-199600003-00003. [DOI] [PubMed] [Google Scholar]
- 40.Drosatos K, Schulze PC. Cardiac lipotoxicity: molecular pathways and therapeutic implications. Current heart failure reports. 2013;10(2):109–121. doi: 10.1007/s11897-013-0133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park TS, Hu Y, Noh HL, Drosatos K, Okajima K, Buchanan J, Tuinei J, Homma S, Jiang XC, Abel ED, Goldberg IJ. Ceramide is a cardiotoxin in lipotoxic cardiomyopathy. Journal of lipid research. 2008;49(10):2101–2112. doi: 10.1194/jlr.M800147-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park TS, Goldberg IJ. Sphingolipids, lipotoxic cardiomyopathy, and cardiac failure. Heart failure clinics. 2012;8(4):633–641. doi: 10.1016/j.hfc.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shah SH, Sun JL, Stevens RD, Bain JR, Muehlbauer MJ, Pieper KS, Haynes C, Hauser ER, Kraus WE, Granger CB, Newgard CB, Califf RM, Newby LK. Baseline metabolomic profiles predict cardiovascular events in patients at risk for coronary artery disease. American heart journal. 2012;163(5):844–850. doi: 10.1016/j.ahj.2012.02.005. e841. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.