Abstract
Continued vulnerability to relapse during abstinence is characteristic of cocaine addiction and suggests that drug-induced neuroadaptations persist during abstinence. However, the precise cellular and molecular attributes of these adaptations remain equivocal. One possibility is that cocaine self-administration leads to enduring changes in DNA methylation. To address this possibility, we isolated neurons from medial prefrontal cortex and performed high throughput DNA sequencing to examine changes in DNA methylation following cocaine self-administration. Twenty-nine genomic regions became persistently differentially methylated during cocaine self-administration, and an additional 28 regions became selectively differentially methylated during abstinence. Altered DNA methylation was associated with isoform-specific changes in the expression of co-localizing genes. These results provide the first neuron-specific, genome-wide profile of changes in DNA methylation induced by cocaine self-administration and protracted abstinence. Moreover, our findings suggest that altered DNA methylation facilitates long-term behavioral adaptation in a manner that extends beyond the perpetuation of altered transcriptional states.
Keywords: DNA methylation, neuron, cocaine self-administration, relapse, next-generation sequencing, genome-wide, MBD
Introduction
A central factor in the development of cocaine addiction is the pathological overlearning of associations between cues in the drug use environment and the rewarding effects of the drug. Memories of these associations are extraordinarily persistent and readily reactivated following re-exposure to cocaine-paired cues, which can prompt powerful cravings for cocaine and relapse despite prolonged abstinence (Childress et al., 1999, O'brien et al., 1998). However, the molecular features that sustain cocaine-associated memories and enhanced vulnerability to relapse during abstinence remain enigmatic. Accumulating evidence suggests that persistent learning-induced epigenetic modifications support the maintenance of long-term memories (Miller et al., 2010, Mizuno et al., 2012) and long-term behavioral adaptation (Champagne & Curley, 2009, Weaver et al., 2005). In particular, relatively stable inducible epigenetic modifications, such as DNA methylation, may mediate the maintenance of memory and enduring behavioral propensities (Griffith & Mahler, 1969, Miller et al., 2010). Therefore, persistent learning-induced changes in DNA methylation could function as a conserved mechanism of memory maintenance and underlie both the persistence of cocaine-related memories and enduring cocaine-seeking behavior during abstinence.
Long-lasting changes in DNA methylation induced by cocaine self-administration may arise in a brain region- and cell type-specific manner. Enduring changes in DNA methylation likely occur throughout reward-related neural circuitry during drug abuse, yet those arising within the medial prefrontal cortex (mPFC) may be particularly relevant to the maintenance of enduring cocaine-related memories. In self-administering animals and human users, exposure to previously cocaine-paired cues is associated with hyperactivity within the mPFC (Childress et al., 1999, Ciccocioppo et al., 2001) and the mPFC is further required for the reinstatement of cocaine seeking during abstinence (Capriles et al., 2003, Di Pietro et al., 2006). Together, this suggests that lasting neuroadaptive changes in the mPFC underpin continued cocaine seeking during abstinence. Moreover, the enduring contribution of the mPFC to drug-seeking behaviour is echoed in steadfast transcriptional and proteomic changes that persist for up to 100 days of forced abstinence following cocaine self-administration (Freeman et al., 2010, Freeman et al., 2008, Lull et al. 2009). Finally, at the cellular level, the neuronal genome could represent a more suitable repository for persistent acquired epigenetic modifications, as mature neurons are not subject to cell division (Griffith & Mahler, 1969).
Recent evidence indicates that cocaine exposure can induce changes in DNA methylation (Anier et al., 2010, Barros et al., 2011, Carouge et al., 2010, Laplant et al., 2010, Massart et al., 2015, Pol Bodetto et al., 2013, Tian et al., 2012, Wright et al., 2015) and other base modifications (Feng et al., 2015); however, the majority of investigations fail to distinguish between modifications arising from simple cocaine exposure and those that are associated with learned cocaine-seeking. This distinction is important, as passive involuntary cocaine exposure is aversive (Twining et al., 2009) and fails to produce the persistent changes in long-term potentiation (Chen et al., 2008, Martin et al., 2006) and sharp rise in extracellular dopamine (Hemby et al., 1997) and acetylcholine (Mark et al., 1999) that may underlie overlearning and the extreme persistence of cocaine-associated memories as well as the development of addiction. We therefore employed a mouse model of intravenous cocaine self-administration (IVSA) that contrasts learned cocaine self-administration with passive cocaine exposure to identify modifications in DNA methylation that are uniquely associated with learned cocaine-seeking and enduring cocaine-seeking during abstinence.
Materials and Methods
Animals
Adult male C57BL/6J mice (8-9 weeks of age at the start of experiments, 20-24 g) were singly housed under a 12 h reverse light-dark cycle (lights off at 7 am) in standard housing conditions with ad libitum access to standard rodent chow and water unless otherwise specified. All experiments were performed with approval from the Animal Ethics Committee of the University of Melbourne in accordance with the Prevention of Cruelty to Animals Act (1986) and the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes.
Intravenous cocaine self-administration (IVSA)
Operant self-administration of cocaine (5 mg/kg/infusion) was performed with minor modifications to previously described protocols (Brown et al., 2009, Mcpherson et al., 2010). Briefly, animals were divided into three experimental subgroups (naïve: n=13; self-administering (IVSA), n= 33; yoked controls: n=7). Naïve animals remained in their home cages for the duration of operant conditioning. The instrumental response (lever press) was established in IVSA animals using 10% w/v oral sucrose under a FR1 (fixed ratio 1) schedule, as previously described (Brown et al., 2009). Yoked control animals received equivalent amounts of sucrose, contingent upon the responses of their paired self-administering counterparts. Following the acquisition of the instrumental response by IVSA animals, both yoked and self-administering animals were surgically implanted with an indwelling jugular catheter (Brown et al., 2009).
After a two-day recovery period, IVSA mice self-administered cocaine intravenously under an FR1 schedule, over 12 daily, 2 h sessions. Responses on the reward-paired lever resulted in the infusion of 0.5 mg/kg of cocaine hydrochloride (Sigma Aldrich, in physiological saline) over 1.7 s (infusion volume of 19 μl) and the concurrent presentation of a 5 s light cue. To minimize the risk of overdose, cocaine was unavailable during the ongoing presentation of the light cue (“time out” period, responses recorded) and a within-session maximum of 80 infusions was applied. Yoked animals received cocaine intravenously, concurrent with earned infusions delivered to paired self-administering mice. Following self-administration training, mice were assigned to groups (IVSA 1: 1 day of abstinence + relapse test, n = 18; IVSA 21 R: 21 days of abstinence + relapse test, n = 15; IVSA 21 NR: 21 days of abstinence + no relapse test, n=7) counterbalanced for cocaine-seeking behavior (determined by the average number of infusions received over the final 3 days of cocaine self-administration). Yoked animals were culled after either 1 (Yoked 1, n=3) or 21 days (Yoked 21, n=4) of forced abstinence. During the abstinence period all animals remained in their home cages.
Cue-induced cocaine seeking during abstinence
With the exception of IVSA 21 NR mice, self-administering mice were subject to a 1 h cocaine-seeking (relapse) test in the absence of cocaine after either 1 (IVSA 1) or 21 days (IVSA 21) of forced abstinence (Brown et al., 2009, Madsen et al., 2012). All cocaine-paired cues were present and cocaine-seeking behavior was measured as the sum of all responses on the previously cocaine-paired lever. Yoked animals underwent simple contextual re-exposure for 1 h. Animals were killed by cervical dislocation either immediately after testing (next-generation sequencing) or 2 h post-test (validation of sequencing and gene expression). Their brains were then removed, snap-frozen over liquid nitrogen and stored at −80°C.
Identification of genome-wide changes in DNA methylation by MBD Ultra-Seq
Individual mPFCs were isolated by Palkovits punch (Palkovits, 1973). Neuronal genomic DNA was isolated from the mPFC of individual animals, as previously described (Li et al., 2014). Next-generation sequencing was performed by Methyl CpG Binding Domain (MBD) Ultra-Seq (Li et al., 2014) with two minor modifications: 75 ng of genomic DNA from each animal was used for library preparation and the final pooled, amplified methyl-enriched library for each treatment group was loaded in duplicate, in different lanes of the flow cell, to prevent the production of artificial differences by inter-lane discrepancies. Regions of methylation enrichment (RMEs, or peaks) were identified using Model-based Analysis of ChIP-Seq (MACS) (Zhang et al., 2008) and previously established thresholds (Li et al., 2014).
Candidate DMR selection
To identify differentially methylated regions (DMRs), RMEs covered by a minimum of 5 normalized reads in at least one of the treatment groups were selected. From this selection, RMEs within 600 bp of each other were grouped and defined as a single RME. Student's t-tests were performed to compare the relative level of 5-methylcytosine (5mC) enrichment at each RME (as measured by the normalized read counts) between naïve and cocaine-treated animals (IVSA 1, IVSA 21, Yoked 1, Yoked 21). The Benjamini-Hochberg false discovery rate (FDR) correction was used to account for the effects of multiple testing and to calculate adjusted p-values, allowing for a false discovery rate of 10%. The level of 5mC enrichment was considered significantly different between groups if the FDR-adjusted p-value from the Student's t-test was less than 0.1 (Ellis et al., 2012, Non et al., 2014). Persistent DMRs arising from cocaine self-administration were those that were significantly different in the IVSA 1 and IVSA 21 groups, but not in the yoked cocaine controls, relative to naïve animals. Levels of 5mC enrichment at abstinence-associated DMRs were significantly different from those in naïve animals after 21 days of abstinence from cocaine self-administration, but not following 21 days of abstinence from yoked cocaine exposure. Furthermore, we verified that the genomic region (RME summit +/− 150 bp) surrounding each DMR of interest contained at least 5 CpG dinucleotides, as the MBD2b/3L1 complex captures fragments of DNA containing a minimum of 5 methylated CpGs (Active Motif). Finally, we confirmed that the 300 bp region overlapping each DMR corresponded to a unique location in the mouse reference genome (mm9) to avoid artefacts produced by poor mapping of repetitive regions. Associations between significant DMRs (persistent and abstinence-associated) and regulatory features of the genome were examined using EpiExplorer (Halachev et al., 2012), a web-based tool that identifies genes and regulatory features overlapped by user-defined genomic regions. DMRs were considered to co-localize with regulatory features if at least 10% of the DMR (peak summit +/− 150 bp) was shared by the regulatory feature annotation.
Validation of select DMRs by MBD quantitative PCR (MBD qPCR)
MBD pull-downs were performed using 150 ng of neuronal DNA derived from the mPFCs of individual animals from a second cohort of animals. Genomic DNA was extracted as previously described (Li et al., 2014), although two 250 μl aliquots were retained for the analysis of gene expression following the homogenization of the tissue. Briefly, genomic DNA was sheared by sonication in 130 μl of ultrapure H20 (Covaris S2, bath temperature: 4°C, duty: 10%, intensity: 6, cycle/burst: 100, time: 180 s) to fragments of ~300bp in length, as verified by Bioanalyzer HS (Agilent). Thirteen μl of fragmented DNA was diluted in a total volume of 80 μl of DNAse/RNase-free TE buffer (pH 8) and retained as input to control for slight variations in the amount of DNA used in each pull-down. MBD pull-downs were performed according to the manufacturer's protocol (Methylcollector Ultra, Active Motif). Captured methylated fragments were eluted in 60 μl of TE buffer (pH 8).
A 300 bp region surrounding each candidate DMR was retrieved (UCSC genome browser, mm9), and primers (Supplemental Table 1) were designed using Primer3 (Untergasser et al., 2012) to amplify 120-300 bp regions overlapping the peak summit of select candidate DMRs. qPCR was performed in duplicate using a Rotor gene Q (Qiagen) in 10 μl reactions (5 μl 2X SYBR-green master mix (Qiagen), 1 μl 10 μM primer (forward + reverse), 3 μl H2O, 1 μl MBD DNA/input DNA). Ct values for methyl-enriched DNA were normalized to input and the relative enrichment between groups was calculated using the ΔΔCT method, relative to naïve animals.
Analysis of gene expression
250 μl of whole mPFC homogenate was retained from each animal in the second biological cohort following homogenization of the brain tissue. RNA was extracted by Trizol LS (Invitrogen) according to the manufacturer's protocol and quantified by spectrophotometry (Nanodrop). 250 ng of RNA was reverse transcribed (QuantiTect Reverse Transcription, Qiagen). cDNA was diluted to approximately 100 ng/μl prior to qPCR (Nanodrop). Whole mPFC homogenate was used because it is not possible to obtain mRNA (which resides in the cytoplasm) from neuronal nuclei.
Primers (IDT) for gene expression analysis were designed using Primer3 (Untergasser et al., 2012) or AutoPrime (Wrobel et al., 2004) or obtained from PrimerBank (Wang et al., 2012) (Supplemental Table 2). In select cases, particularly when intragenic methylation was present, the expression of multiple isoforms of a gene was examined. qPCR was performed in triplicate using a Rotor gene Q (Qiagen) in 10 μl reactions (5 μl of 2X SYBR-Green master mix, 2 μl of 5 μM primer (forward+ reverse), 2 μl H2O, 1 μl 100 ng/μl cDNA). Relative expression was quantified using the ΔΔCT method relative to naïve animals. Within each animal, expression levels of genes of interest were normalized to the level of dynenin expression.
Results
Mice continue to seek cocaine during protracted abstinence
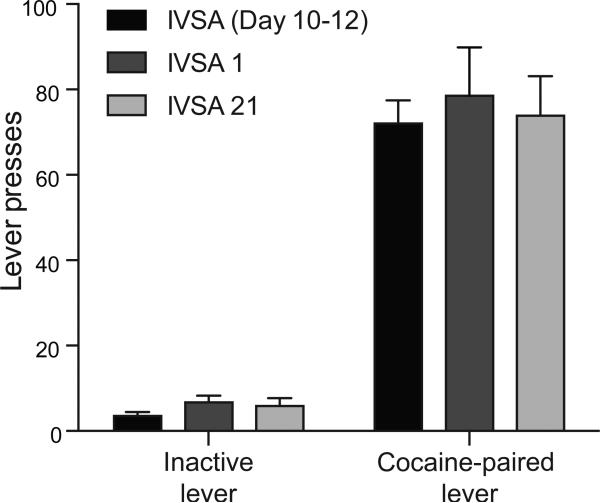
We first verified that mice continue to seek cocaine during abstinence, as continued cocaine-seeking behavior is indicative of the presence of long-lasting neuroadaptations. Continued cocaine-seeking behavior manifests as a sustained and significant preference for the previously cocaine-paired lever relative to the inactive lever. In our paradigm, mice trained to self-administer cocaine continued to seek cocaine during abstinence, with high discrimination for the cocaine-paired lever over the inactive lever during days 10-12 of intravenous self-administration and following 1 or 21 days of abstinence (F1,128 = 198.6, p<0.0001, Figure 1). Cocaine-seeking behavior did not differ across the end of self-administration training or throughout period of enforced abstinence (Holm-Sidak post hoc tests, IVSA (Day 10-12) vs. IVSA1, IVSA (Day 10-12) vs. IVSA 21 and IVSA 1 vs. IVSA 21, not significant). Therefore, this behavioral paradigm produces enduring cocaine-seeking behavior and is suitable for the examination of persistent neuroadaptations following cocaine-self administration.
Figure 1. Mice continue to seek cocaine during protracted abstinence.
A two-way ANOVA revealed that mice press the previously cocaine-paired lever significantly more than the inactive lever after 1 and 21 days of forced abstinence (F1,128 = 198.6, p<0.0001, IVSA (Day 10-12): n=34, IVSA 1: n=18 and IVSA 21: n=15). There was no difference in the number of cocaine-paired lever presses performed during the last 3 days of cocaine self-administration or after 1 or 21 days of forced abstinence (F2,128 = 0.40, not significant). Data are displayed as mean lever presses ± SEM; IVSA 1: mice tested after 1 day of abstinence, IVSA 21: mice tested after 21 days of abstinence.
Cocaine self-administration produces distinct changes in DNA methylation
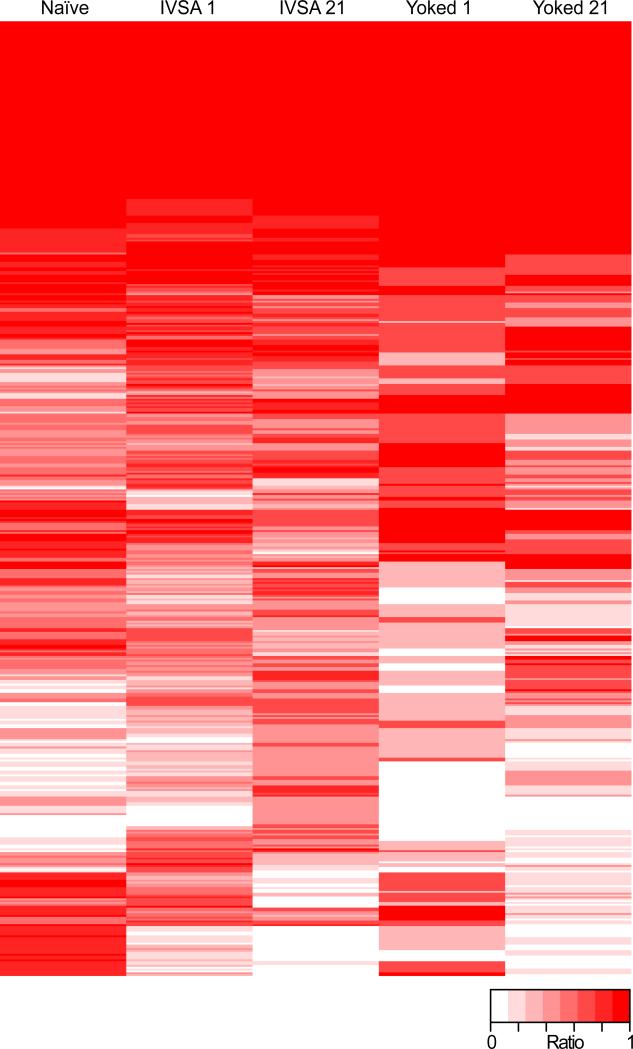
To identify changes in DNA methylation specific to self-administering animals, we performed genome-wide sequencing by MBD Ultra-Seq (Li et al., 2014) on neuronal DNA derived from the mPFCs of naïve animals (n=5), mice that had self-administered cocaine and been sacrificed after either 1 or 21 days of abstinence (n=7 and 6 respectively) and yoked cocaine controls sacrificed at the same time points (n=3 and 4 respectively). Comparisons between self-administering animals and yoked cocaine controls served to isolate modifications of DNA methylation produced by voluntary learned cocaine-seeking from those produced by passive cocaine exposure. MACS (Zhang et al., 2008) identified 46464 RMEs across the genome (Supplemental File 1). A representative heatmap of 5mC enrichment revealed that passive cocaine exposure and cocaine self-administration produced distinct patterns of 5mC enrichment (Figure 2).
Figure 2. Representative heatmap of 5mC enrichment.
All RMEs that were supported by enrichment (relative to background genomic coverage as identified by MACS) in at least 50% of biological replicates in naïve (n=5), IVSA 1 (n=7) or IVSA 21 (n=6) animals are plotted. White indicates that no biological replicates displayed enrichment for 5mC, while dark red indicates that all biological replicates had enrichment for 5mC at the given genomic locus. The ratio is indicative of the number of animals within a group that displayed an enrichment for 5mC at a given RME.
Persistent changes in DNA methylation
Patterns of DNA methylation that arise during cocaine self-administration and persist over time are consistent with the enduring nature of cocaine-seeking behavior and may mediate the maintenance of long-lasting cocaine-related memories. Twenty-nine genomic regions became differentially methylated in a persistent manner (relative to naïve animals) in response to cocaine self-administration but not passive cocaine exposure (p<0.1 in IVSA 1 and IVSA 21 animals, p>0.1 in Yoked 1 and Yoked 21 animals, FDR-adjusted p-values derived from Student's t-tests relative to naïve animals, Supplemental File 2). Five persistent DMRs became demethylated following cocaine self-administration, whereas the remainder showed an increase in DNA methylation. The genomic coordinates of the persistent DMRs and associated or proximal genes are listed in Supplemental Table 3.
EpiExplorer (Halachev et al., 2012) was used to identify associations between persistent IVSA-associated DMRs and genomic regulatory features. Fifteen of 29 DMRs were embedded within genes (coding and non-coding) and a further 5 were located distal (<10 kB) to a gene (NCBI Ensembl Build 37). A chief function of persistent IVSA-associated modifications of DNA methylation may therefore be the regulation of gene transcription. Moreover, several (13 of 29) persistent DMRs overlapped DNase I-hypersensitive sites (DNAse I HS sites). This further suggests that persistent changes in DNA methylation could regulate gene transcription, as DNAse I HS sites are classically associated with open chromatin and transcriptional activity (Weintraub & Groudine, 1976). Interestingly, gene-associated DMRs may regulate the expression of specific splice variants, as 12 of 15 persistent gene-associated DMRs were located within intronic regions or non-coding loci, whereas principal promoter regions (−5kb to 1kb from transcription start sites) and exons were relatively devoid of changes. Persistent IVSA-associated DMRs were often located within repetitive elements (19 of 29 DMRs overlapped repetitive elements identified by RepeatMasker); however, as repetitive elements tend to be heavily methylated, it is often easier to discern changes in DNA methylation at these loci when using enrichment-based methods to detect DNA methylation (Hardcastle, 2013). Nevertheless, cocaine exposure leads to repetitive element unsilencing (Maze et al., 2011), and it is therefore possible that persistent IVSA-associated modifications of DNA methylation regulate repetitive element activity.
Abstinence-associated changes in DNA methylation
Abstinence-associated changes in DNA methylation denotes changes in DNA methylation that were observed in animals subject to 21 days of forced abstinence following cocaine self-administration training, but not in those subject to 1 day of abstinence or in yoked cocaine controls (relative to naïve animals). Abstinence-associated DMRs may arise as a consequence of withdrawal, but could equally be pertinent to the maintenance of cocaine-related memories. During abstinence, interoceptive cues (such as withdrawal states) may prompt the retrieval and reconsolidation of cocaine-related memories. The retrieval and reconsolidation of memories could drive further changes in DNA methylation (Maddox & Schafe, 2011), which may be associated with an increased persistence of memory, as reconsolidation can strengthen memory (Tronson & Taylor, 2013). Abstinence-associated DNA modifications that are unique to self-administering animals could arise from the retrieval and reconsolidation of cocaine-related memories and therefore be relevant to the maintenance of these memories as well as enduring cocaine-seeking behavior. Twenty-eight genomic regions (Supplemental Table 4) became differentially methylated during abstinence from cocaine self-administration but not passive cocaine exposure (p<0.1 in IVSA 21 animals, p>0.1 in IVSA 1, Yoked 1 and Yoked 21 animals, FDR-adjusted p-values relative to naïve animals, p<0.05 IVSA 21 vs. IVSA 1 and Yoked 21, follow-up Student's t-tests, Supplemental File 3). Eight abstinence-associated DMRs became demethylated during prolonged abstinence whereas the remainder were methylated. Thirteen of 28 DMRs were located within genes and a further 3 were located within 10 kB of a gene. Again, gene-associated DMRs predominantly arose within introns or non-coding loci; only 5 of 19 gene-associated DMRs were located within exons or promoter regions. A greater percentage of abstinence-associated DMRs were found within 10-100 kB of annotated genes (39% of abstinence-associated DMRs as opposed to 14% of persistent DMRs) which may indicate a more pronounced role for abstinence-associated DMRs in the regulation of enhancer or insulator regions across the genome. Once more, abstinence-associated DMRs frequently occurred within repetitive elements (16 of 28 DMRs overlapped repetitive elements). Surprisingly, 24 of 28 abstinence-associated DMRs overlapped or were located proximal to nuclear lamina domains (13 overlapped and 11 were located within 1 kB of nuclear lamina domains). The unusual association of abstinence-associated DMRs with lamina domains suggests extensive repositioning of the genome during abstinence, potentially with profound transcriptional consequences.
Validation of changes in DNA methylation
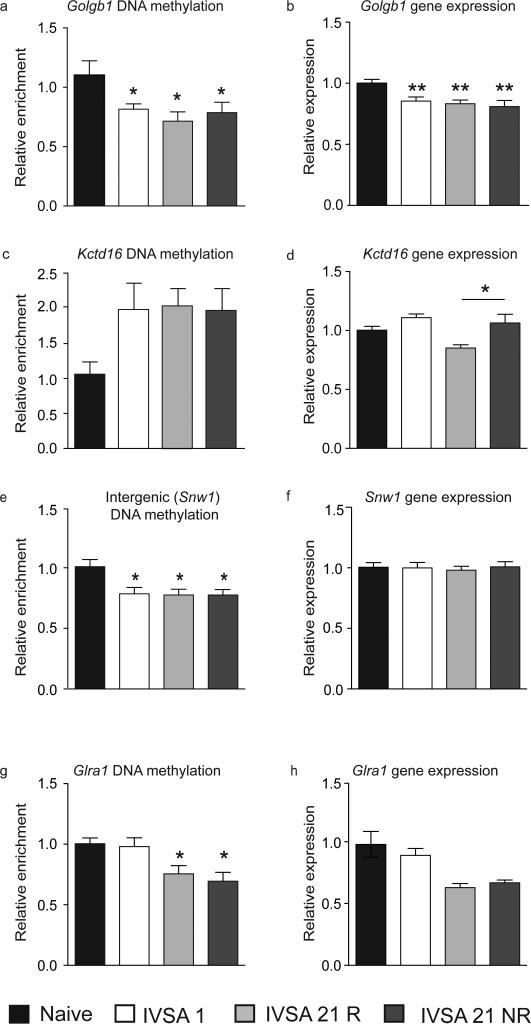
A number of DMRs were validated by MBD qPCR in an independent cohort of animals. An additional group that did not undergo relapse testing after 21 days of abstinence (IVSA 21 NR) was included to ensure that abstinence-associated changes in DNA methylation were not simply the product of the cocaine-seeking (relapse) test. Select DMRs included three regions that were persistently differentially methylated following cocaine self-administration and one that became differentially methylated during abstinence. The mean normalized read distribution at each candidate DMR is provided in Supplemental Figure 1. All candidate DMRs selected for validation by MBD qPCR validated reliably (Figure 3), as was an additional candidate DMR within Cdh13 (Supplemental Figure 2, this candidate did not meet the final statistical criteria for selection), indicating that MBD Ultra-Seq and the statistical limitations employed reliably identify changes in DNA methylation induced by cocaine self-administration.
Figure 3. Validation of candidate DMRs and expression of co-localizing genes.
In an independent cohort of animals, MBD qPCR was performed to assess the relative levels of 5mC enrichment at select DMRs and qPCR was used to examine expression of co-localizing genes. (a) In accordance with sequencing results, the locus within Golgb1 was demethylated following cocaine self-administration; F3,17= 3.57, p<0.05. (b) The decreased DNA methylation was correlated with a significant reduction in the expression of Golgb1 (all isoforms) at all time points regardless of relapse testing; F3,26 = 5.42, p<0.01. (c) The DMR located within Kctd16 displayed a near-significant trend towards persistent methylation in all treatment groups following cocaine self-administration; F3,16 = 2.73, p=0.07. (d) Interestingly, a trend towards a significant decrease in Kctd16 gene expression was observed only after 21 days of abstinence and relapse testing; F3,26 = 6.55, p<0.01, Holm-Sidak post hoc test IVSA 21 R vs. Naïve, p= 0.07. After 21 days of abstinence, Kctd16 expression was decreased in animals that underwent relapse testing (IVSA 21 R) compared to those that were simply sacrificed (IVSA 21 NR), p<0.05 (Holm-Sidak post hoc test), which suggests that the relapse test may influence the relationship between altered DNA methylation and gene expression. (e) Demethylation was also replicated at the intergenic locus located proximal to Snw1; F3,17= 4.35, p<0.05. (f) Decreased DNA methylation was not associated with a significant change in the expression of Snw1; F3,26 = 0.07. (g) Finally, demethylation of the Glra1-associated DMR was reproduced following 21 days of abstinence regardless of relapse testing; F3,17 = 5.59, p<0.01. (h) Demethylation of the Glra1-associated DMR was associated with a trend towards a significant reduction in the expression of Glra1 (all isoforms) after 21 days of abstinence; Welch's F3,13.36 = 7.52, p<0.01, Games-Howell post hoc tests. All data are displayed as mean ± SEM, and p-values are derived from Holm-Sidak post hoc tests relative to naïve animals, except where specifically indicated. * p<0.05, ** p<0.01.
Altered DNA methylation is associated with changes in gene expression
In addition to propagating enduring changes in gene transcription, persistent experience-induced modifications of DNA methylation could act as silent signatures of cocaine-related learning and prime the transcription of the affected locus upon reactivation of cocaine-related memories, representing a form of genomic metaplasticity (Baker-Andresen et al., 2012). To examine this possibility, we examined gene expression in animals that were subject to a relapse test after 21 days of abstinence (IVSA21 R) and others that were simply sacrificed at the same time point (IVSA21 NR). The relapse test served to explicitly reactivate cocaine-associated memories, whereas these memories would have remained relatively dormant in animals that were not subject to a relapse test. Golgb1 and Glra1 expression were decreased regardless of whether or not relapse occurred (Golgb1: F3,26 = 5.42, p<0.01, Holm-Sidak post hoc test, naïve vs. IVSA 1, IVSA 21 R and IVSA 21 NR all p<0.01, GLRa1: Welch's F3,13.36 = 7.52, p<0.01, Games-Howell post hoc test, naïve vs IVSA 21 R and IVSA 21 NR, p<0.1) (Figure 3b and 3h respectively). Conversely, Kctd16 expression was altered only in animals that underwent a relapse test (F3,26 = 6.55, p<0.01, Holm-Sidak post hoc test, naïve vs. IVSA 21 R, p<0.1, IVSA 21 R vs. IVSA 21 NR, p<0.05) (Figure 3d), despite increased DNA methylation across all groups following cocaine self-administration (Figure 3c). The intergenic change in DNA methylation located distal to Snw1 (Figure 3e) had no effect on its expression (F3,26 = 0.07, not significant; Figure 3f). The change in DNA methylation within Cdh13 was associated with a trend towards reduction in the expression of this gene (F3,26 =2.53, p=0.07; Supplemental Figure 2) regardless of relapse testing. Therefore, IVSA-related changes in DNA methylation are associated with altered expression of co-localizing genes, although in some instances this association is modulated by the reactivation state of the cocaine-related memories.
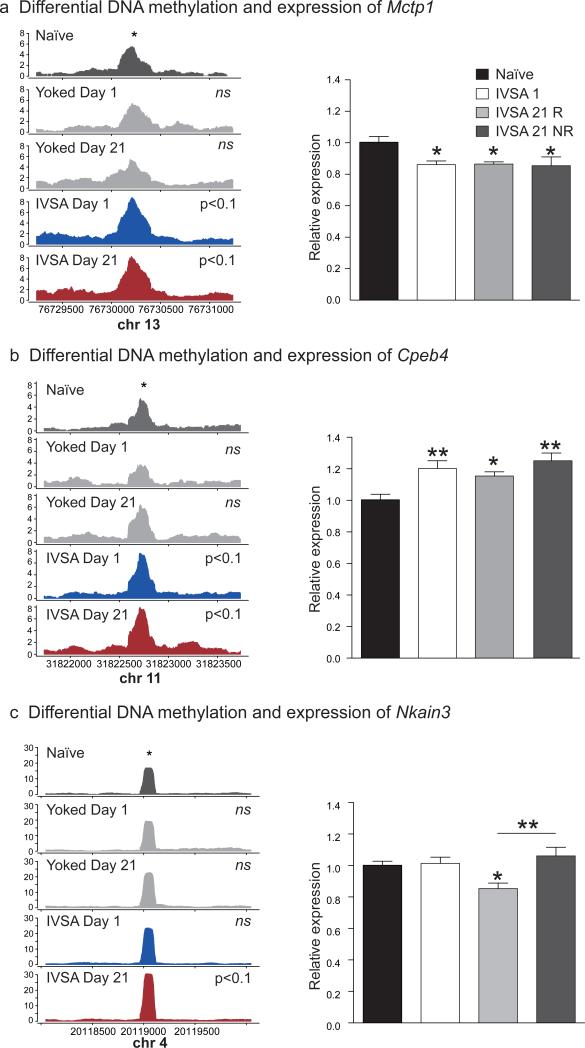
A number of significant DMRs were located within lengthy repetitive elements and unsuited to validation by MBD qPCR. However, these DMRs merit mention, as they are located within genes that have been implicated in addiction, including Mctp1 (multiple C2 domains, transmembrane 1), Cpeb4 (cytoplasmic polyadenylation element binding protein 4) and Nkain3 (Na+/K+ transporting ATPase interacting 3). Mctp1- and Cpeb4- associated DMRs were persistently methylated following cocaine self-administration (Figure 4a,b), whereas the DMR within Nkain3 became methylated during prolonged abstinence from cocaine self-administration (Figure 4c). The persistent increase in DNA methylation within Mctp1 was associated with an enduring and significant decrease in the expression of Mctp1 (−001 isoform; F3,26 = 4.07, p<0.05, Holm-Sidak post hoc test, naïve vs. IVSA 1, IVSA 21 R and IVSA 21 NR all p<0.05) while a similar increase in DNA methylation within Cpeb4 was associated with a significant increase in the expression of this gene (−001 isoform; F3,26 = 5.96, p<0.01, Holm-Sidak post hoc test, naïve vs. IVSA 1 and 21 NR, p<0.01, naïve vs. IVSA 21 R, p<0.05; Figure 4 a,b). Interestingly, the expression of Nkain3 was exclusively decreased in mice that had undergone relapse testing after 21 days of abstinence (F3,26 =4.56, p<0.05, Holm-Sidak post hoc test, naïve vs. IVSA 21 R, p<0.05, IVSA 21 R vs. IVSA 21 NR, p<0.01; Figure 4c), which provides further preliminary evidence of a complex relationship between learning-induced changes in DNA methylation, gene expression and the reactivation state of the memory.
Figure 4. Expression of genes co-localizing with DMRs that cannot be validated by MBD qPCR.
(a) Cocaine self-administration yielded a persistent increase in DNA methylation at a locus within Mctp1 (b) Increased DNA methylation correlated with a concomitant decrease in the expression of Mctp1 (−001 isoform) across treatment groups; F3,26 = 4.07, p<0.05. (c) Cocaine self-administration resulted in a persistent increase in DNA methylation within an intragenic region of Cpeb4. (d) Increased DNA methylation was associated with a long-lasting upregulation of Cpeb4 (001/201 isoform) expression; F3,26 = 5.96, p<0.01. (e) A significant increase in DNA methylation within an intron of Nkain3 was observed after 21 days of abstinence (f) The increase in DNA methylation was associated with a decrease in the expression of Nkain3 (all isoforms) after 21 days of abstinence and relapse testing, but this decrease was not observed when the relapse test did not occur; F3,26 =4.56, p<0.05, naïve vs. IVSA 21 R, p<0.05, IVSA 21 R vs. IVSA 21 NR, p<0.01. Data are displayed as mean ± SEM, all p-values are derived from Holm-Sidak post hoc tests relative to naïve animals, * p<0.05 ** p<0.01.
Differential DNA methylation regulates the expression of specific splice variants
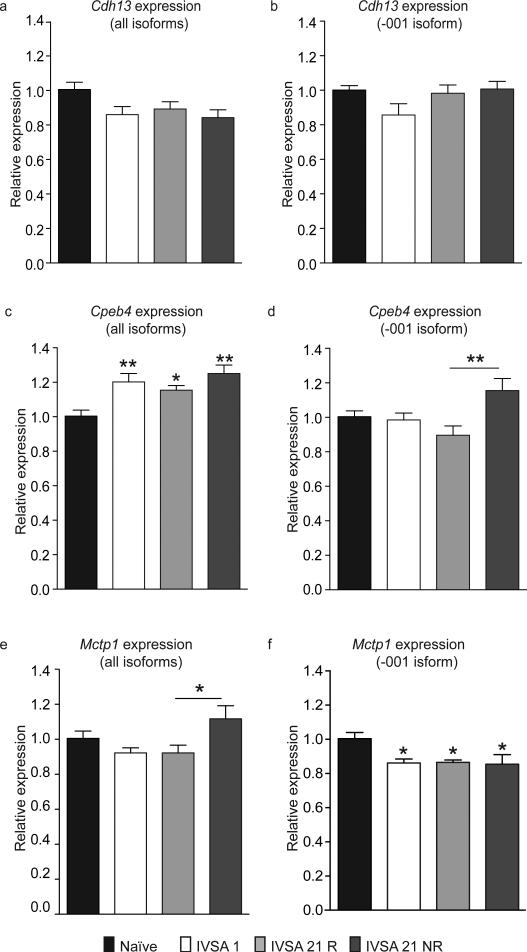
A principal function of intragenic methylation is the regulation of alternative splicing (Oberdoerffer, 2012). Accordingly, DMRs located within Cdh13, Cpeb4 and Mctp1 were associated with changes in the expression of specific isoforms of each gene (Figure 5). There was a trend towards the overall decreased expression of Cdh13 (F3,26 =2.53, p=0.07; Figure 5a), however, this is likely due to the regulation of non-coding transcripts of Cdh13, as the expression of the protein-coding transcript (Cdh13-001) was not significantly altered following cocaine self-administration (F3,26 = 1.35, not significant; Figure 5b). The overall expression of Cpeb4 (all isoforms) increased following cocaine self-administration (F3,26 = 5.96, p<0.01; Figure 5c), yet the principal isoform (Cpeb4-001, Ensembl 37) exhibited a different pattern of expression (F3,26 = 4.32, p<0.01; Figure 5d). The persistent increase in methylation within Mctp1 was also associated with an enduring decrease in the expression of Mctp1-001 (F3,26 = 4.07, p<0.05; Figure 5f), but the overall expression of all protein-coding transcripts of this gene was only significantly different between animals that underwent relapse testing after 21 days of abstinence (IVSA 21 R) and those that were simply sacrificed (IVSA 21 NR) (F3,26 = 3.62, p<0.05, Holm-Sidak post hoc test, IVSA 21R vs. IVSA 21 NR; p<0.05; Figure 5e). Together, these data suggest that the intragenic changes in DNA methylation produced as a result of cocaine self-administration may regulate the expression of associated genes in an isoform-specific manner.
Figure 5. Regulation of alternative splicing and isoform-specific expression by intragenic DNA methylation.
Intragenic modifications of DNA methylation arising during cocaine self-administration may contribute to the differential regulation of select splice variants. The isoforms selected include the major protein-coding isoform of the select gene and isoforms transcribed from the genomic region proximal to the DMR of interest. (a) There was a trend towards the overall decreased expression of Cdh13, F3,26 =2.53, p=0.07. (b) However, this is likely due to the regulation of non-coding transcripts of Cdh13, as the expression of the protein-coding transcript was not significantly altered following cocaine self-administration; F3,26 = 1.35, not significant. (c). Subsequent to self-administration, overall Cpeb4 expression increased in all treatment groups; F3,26 = 5.96, p<0.01. (d) Nevertheless, the expression of the common isoform, Cpeb4-001, displayed a different pattern of expression relative to that in naïve animals, with the sole significant difference in expression being between animals subject to relapse testing at 21 days of abstinence compared to those that were not (F3,26 = 4.32, p<0.01, IVSA 21 R vs. IVSA 21 NR, p<0.01, Holm-Sidak post hoc test). (e) When the collective expression of all protein-coding isoforms of Mctp1 was explored, the sole significant difference was again between animals subject to relapse testing at 21 days and those that were not (F3,26 = 3.62, p<0.05, Holm-Sidak post hoc, IVSA 21 R vs. IVSA 21 NR, p<0.05). (f) However, when explored individually, the expression of one protein-coding isoform (Mctp1-001, Ensembl 37) was persistently decreased at all time points relative to naïve animals; F3,26= 4.07, p<0.05. Data are displayed as mean ± SEM; all p-values are derived from Holm-Sidak post hoc tests relative to naïve animals except where indicated, * p<0.05, ** p<0.01.
Discussion
Taken together, the findings of this study reveal that cocaine self-administration produces distinct and enduring DNA methylation states within neurons of the mPFC. Functionally, IVSA-associated changes in DNA methylation are associated with concomitant isoform-specific modifications of gene expression. In select cases the association between IVSA-associated DMRs and transcription is only evident following reactivation of cocaine-associated memories, which provides a putative demonstration of experience-dependent genomic metaplasticity in the brain, and suggests that persistent changes in DNA methylation could contribute to memory maintenance in ways that extend beyond the perpetuation of altered patterns of gene expression.
A growing body of evidence indicates that cocaine interacts with components of the DNA methylation machinery to give rise to brain region-specific changes in DNA methylation (Anier et al., 2010, Laplant et al., 2010, Pol Bodetto et al., 2013, Tian et al., 2012). However, few of the aforementioned reports distinguished between epigenetic modifications associated with voluntary cocaine-seeking and self-administration and those that are simply induced by passive cocaine exposure. Although examining the epigenetic consequences of passive cocaine exposure can yield important insight into the pharmacological effects of cocaine, this approach cannot be used to determine the epigenetic changes underlying the cognitive adaptations that give rise to addiction-like behavior.
We found that the majority of IVSA-related modifications of DNA methylation were located within, or proximal to, genes (both coding and non-coding) and predominantly within introns, which is consistent with the observation that the majority of neuronal activity-induced changes in DNA methylation arise within introns (Guo et al., 2011). Equally predictable was the relative enrichment of DMRs within repetitive elements; approximately 42% of the mouse genome corresponds to repetitive elements (Church et al., 2009) and changes in DNA methylation within these elements are often more readily detected due to high levels of basal DNA methylation. However, the myriad of abstinence-associated DMRs within or proximal to nuclear lamina-associated domains was surprising. The nuclear lamina is a protein framework located beneath the nuclear envelope, to which heterochromatin is often tethered. Approximately 35-40% of the genome comprises lamina-associated domains, which are key determinants of higher-order chromatin architecture and are classically transcriptionally silent (Peric-Hupkes & Van Steensel, 2010). Importantly, the association of the genome with the nuclear lamina is altered following neuronal activity (Walczak et al., 2013) and dysregulation of genome-lamina associations has been implicated in the etiology of neuropsychiatric disorders (Ito et al., 2014, Wilczynski, 2014). Moreover, DNA methylation directly influences the association between genomic regions and the nuclear lamina, as the methyl-CpG binding protein MeCP2 interacts with inner nuclear lamina-associated proteins (Guarda et al., 2009). Therefore, abstinence-associated changes in DNA methylation may alter genome-lamina associations and heterochromatin stability, although an explanation for the specific enrichment of abstinence-associated DMRs within lamina-associated domains remains elusive.
A number of gene-associated DMRs were located within genes that have been implicated in learning, memory and addiction, and their potential biological functions merit discussion. KCTD16 can act as an auxiliary subunit of the GABAB receptor, enhancing its sensitivity and accelerating responses to agonists (Schwenk et al., 2010). Activation of the GABAB receptor, which is equally subject to persistent changes in methylation following cocaine self-administration (data not shown), is associated with a reduction in cocaine self-administration (Roberts & Brebner, 2000). By altering the kinetics and sensitivity of this receptor, the differential methylation within Kctd16 and Gabab may therefore contribute to the modulation of cocaine seeking behavior. CDH13 is a particularly striking candidate, as polymorphisms within this gene have been implicated in substance dependence (Uhl et al., 2008), vulnerability to addiction (Johnson et al., 2011), and disorders of impulse control (Arias-Vasquez et al., 2011), although no studies have explored the epigenetic regulation of this gene in cocaine seeking and addiction. Polymorphisms within Mctp1 are associated with bipolar disorder (Scott et al., 2009), and the expression of Mctp1 is altered during abstinence from several drugs of abuse (Le Merrer et al., 2012), though the cellular changes that give rise to this alteration remain to be determined. Together with the other DMRs identified, these changes in DNA methylation have a clear potential to contribute to the development of addiction and, given the malleable nature of epigenetic modifications, may represent promising targets for pharmacological interventions in the treatment of this condition. Nevertheless, it will be necessary to establish that these DMRs regulate addiction and not simply cocaine self-administration. Moreover, as many DMRs are located within repetitive regions, further verification will require the application of novel techniques that currently in development, such as SMRT sequencing (PacBio). Future experiments will focus on epigenetic changes in neurons selectively engaged by cocaine-related memories, using inducible neuron-specific activity-driven reporter proteins (i.e. E-SARE-driven GFP (Kawashima et al., 2013) and fluorescence activated cell sorting). Excitingly, with emergent technologies, it may be possible to directly edit or reverse locus-specific changes in DNA methylation incurred by self-administration in recently activated neurons and directly demonstrate their functional role in behavior.
Repeated exposure to cocaine can generate ‘silent’ glutamatergic synapses that do not influence the basal efficacy of synaptic transmission but are pronounced sites of plasticity in response to subsequent stimulation (Lee & Dong, 2011). This investigation provides preliminary evidence of an analogous phenomenon within the genome, whereby persistent IVSA-induced modifications of DNA methylation do not necessarily produce long-lasting changes in the transcription of co-localizing genes, but instead prime transcription in response to subsequent neuronal and memory reactivation. In a subset of genes (Kctd16, Cpeb4 and Nkain3), an association between differential methylation and altered gene transcription was only evident following the explicit re-activation of cocaine-related memories through relapse testing, which suggests that a number of IVSA-associated changes in DNA methylation represent a form of genomic metaplasticity (Baker-Andresen et al., 2012). The Kctd16-associated data are particularly persuasive as there was no difference in methylation in animals that underwent relapse testing after 21 days of abstinence compared to those that were simply sacrificed. However, caution must be used when interpreting the Nkain3 and Cpeb4-related data, as it is possible that changes in DNA methylation were produced by the relapse test, resulting in the differences in expression between animals that underwent testing and those that did not. Additional experiments will extend these findings and conclusively demonstrate that the observed metaplastic priming of gene transcription is a direct consequence of memory reactivation and DNA methylation at a specific locus.
Finally, IVSA-associated changes in intragenic DNA methylation are associated with the regulation of alternative splicing and the expression of specific isoforms of co-localizing genes. Importantly, we demonstrated that the absence of an overall change in the expression of a gene does not preclude the possibility that the expression of individual splice variants is altered. Although the overall expression of Mctp1 appeared unchanged, the expression of a single isoform (Mctp1-001) was differentially regulated in this paradigm. In principle, opposing changes in the expression of specific isoforms of genes could obscure global changes in the expression of a given gene. Importantly, each isoform of a gene may produce proteins with vastly different functional capabilities and it is therefore critical that the expression of specific splice variants of candidate genes is determined and acknowledged. At the level of individual DMRs, exploring the expression of each splice variant of co-localizing genes may be too time-consuming and expensive; it would therefore be preferable to simultaneously perform whole-transcriptome sequencing to broadly explore the regulation of the expression of alternative splice variants.
In summary, this investigation has revealed persistent and abstinence-associated changes in DNA methylation in neurons of the mPFC that are specific to voluntary cocaine self-administration. These long-lasting changes in DNA methylation could in turn underlie the maintenance of cocaine-related memories and continued cocaine seeking during abstinence. Moreover, this study provides preliminary evidence of memory-related genomic metaplasticity: the priming of transcription in response to memory reactivation by enduring epigenetic modifications. Future experiments will functionally establish the role of these time-dependent changes in DNA methylation in cocaine seeking and memory maintenance and explore their contribution to the long-term regulation of transcriptional changes.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge grant support from the National Health and Medical Research Council (APP1023127), and the Australian Research Council (DP1096148). A postgraduate award from the National Sciences and Engineering Research Council of Canada supports DBA, and XL is supported by a University of Queensland postgraduate scholarship. The authors would also like to thank Ms. Virginia Nink for assistance with FACS, Dr. Robyn Brown and Ms. Nicola Chen for assistance with the behavioural paradigm and Ms. Rowan Tweedale for helpful editing of the manuscript.
References
- Anier K, Malinovskaja K, Aonurm-Helm A, Zharkovsky A, Kalda A. DNA methylation regulates cocaine-induced behavioral sensitization in mice. Neuropsychopharm. 2010;35:2450–2461. doi: 10.1038/npp.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias-Vasquez A, Altink ME, Rommelse NN, Slaats-Willemse DI, Buschgens CJ, Fliers EA, Faraone SV, Sergeant JA, Oosterlaan J, Franke B, Buitelaar JK. CDH13 is associated with working memory performance in attention deficit/hyperactivity disorder. Genes Brain Behav. 2011;10:844–851. doi: 10.1111/j.1601-183X.2011.00724.x. [DOI] [PubMed] [Google Scholar]
- Baker-Andresen D, Ratnu VS, Bredy TW. Dynamic DNA methylation: a prime candidate for genomic metaplasticity and behavioral adaptation. Trends Neurosci. 2012;36:3–13. doi: 10.1016/j.tins.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Barros M, Dempster EL, Illott N, Chabrawi S, Maior RS, Tomaz C, De Souza Silva MA, Huston JP, Mill J, Muller CP. Decreased methylation of the NK3 receptor coding gene (TACR3) after cocaine-induced place preference in marmoset monkeys. Addict. Biol. 2011;18:452–454. doi: 10.1111/j.1369-1600.2011.00409.x. [DOI] [PubMed] [Google Scholar]
- Brown RM, Short JL, Cowen MS, Ledent C, Lawrence AJ. A differential role for the adenosine A2A receptor in opiate reinforcement vs opiate-seeking behavior. Neuropsychopharm. 2009;34:844–856. doi: 10.1038/npp.2008.72. [DOI] [PubMed] [Google Scholar]
- Capriles N, Rodaros D, Sorge RE, Stewart J. A role for the prefrontal cortex in stress- and cocaine-induced reinstatement of cocaine seeking in rats. Psychopharm. 2003;168:66–74. doi: 10.1007/s00213-002-1283-z. [DOI] [PubMed] [Google Scholar]
- Carouge D, Host L, Aunis D, Zwiller J, Anglard P. CDKL5 is a brain MeCP2 target gene regulated by DNA methylation. Neurobiol. Dis. 2010;38:414–424. doi: 10.1016/j.nbd.2010.02.014. [DOI] [PubMed] [Google Scholar]
- Champagne FA, Curley JP. Epigenetic mechanisms mediating the long-term effects of maternal care on development. Neurosci. Biobehav. Rev. 2009;33:593–600. doi: 10.1016/j.neubiorev.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Chen BT, Bowers MS, Martin M, Hopf FW, Guillory AM, Carelli RM, Chou JK, Bonci A. Cocaine but not natural reward self-administration nor passive cocaine infusion produces persistent LTP in the VTA. Neuron. 2008;59:288–297. doi: 10.1016/j.neuron.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childress AR, Mozley PD, McElgin W, Fitzgerald J, Reivich M, O'Brien CP. Limbic activation during cue-induced cocaine craving. Am. J. Psych. 1999;156:11–18. doi: 10.1176/ajp.156.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church DM, Goodstadt L, Hillier LW, Zody MC, Goldstein S, She X, Bult CJ, Agarwala R, Cherry JL, DiCuccio M, Hlavina W, Kapustin Y, Meric P, Maglott D, Birtle Z, Marques AC, Graves T, Zhou S, Teague B, Potamousis K, Churas C, Place M, Herschleb J, Runnheim R, Forrest D, Amos-Landgraf J, Schwartz DC, Cheng Z, Lindblad-Toh K, Eichler EE, Ponting CP, Mouse Genome Sequencing C. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 2009;7:e1000112. doi: 10.1371/journal.pbio.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccocioppo R, Sanna PP, Weiss F. Cocaine-predictive stimulus induces drug-seeking behavior and neural activation in limbic brain regions after multiple months of abstinence: reversal by D(1) antagonists. PNAS. 2001;98:1976–1981. doi: 10.1073/pnas.98.4.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro NC, Black YD, Kantak KM. Context-dependent prefrontal cortex regulation of cocaine self-administration and reinstatement behaviors in rats. Euro. J. Neurosci. 2006;24:3285–3298. doi: 10.1111/j.1460-9568.2006.05193.x. [DOI] [PubMed] [Google Scholar]
- Ellis JA, Munro JE, Chavez RA, Gordon L, Joo JE, Akikusa JD, Allen RC, Ponsonby AL, Craig JM, Saffery R. Genome-scale case-control analysis of CD4+ T-cell DNA methylation in juvenile idiopathic arthritis reveals potential targets involved in disease. Clin Epigenetics. 2012;4:20. doi: 10.1186/1868-7083-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Shao N, Szulwach KE, Vialou V, Huynh J, Zhong C, Le T, Ferguson D, Cahill ME, Li Y, Koo JW, Ribeiro E, Labonte B, Laitman BM, Estey D, Stockman V, Kennedy P, Courousse T, Mensah I, Turecki G, Faull KF, Ming GL, Song H, Fan G, Casaccia P, Shen L, Jin P, Nestler EJ. Role of Tet1 and 5-hydroxymethylcytosine in cocaine action. Nat. Neurosci. 2015;18:536–544. doi: 10.1038/nn.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman WM, Lull ME, Patel KM, Brucklacher RM, Morgan D, Roberts DC, Vrana KE. Gene expression changes in the medial prefrontal cortex and nucleus accumbens following abstinence from cocaine self-administration. BMC Neurosci. 2010;11:29. doi: 10.1186/1471-2202-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman WM, Patel KM, Brucklacher RM, Lull ME, Erwin M, Morgan D, Roberts DC, Vrana KE. Persistent alterations in mesolimbic gene expression with abstinence from cocaine self-administration. Neuropsychopharm. 2008;33:1807–1817. doi: 10.1038/sj.npp.1301577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JS, Mahler HR. DNA ticketing theory of memory. Nature. 1969;223:580–582. doi: 10.1038/223580a0. [DOI] [PubMed] [Google Scholar]
- Guarda A, Bolognese F, Bonapace IM, Badaracco G. Interaction between the inner nuclear membrane lamin B receptor and the heterochromatic methyl binding protein, MeCP2. Exp. Cell Res. 2009;315:1895–1903. doi: 10.1016/j.yexcr.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, Zhang K, Ming GL, Gao Y, Song H. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci. 2011;13:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halachev K, Bast H, Albrecht F, Lengauer T, Bock C. EpiExplorer: live exploration and global analysis of large epigenomic datasets. Genome Biol. 2012;13:R96. doi: 10.1186/gb-2012-13-10-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle TJ. High-throughput sequencing of cytosine methylation in plant DNA. Plant methods. 2013;9:16. doi: 10.1186/1746-4811-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemby SE, Co C, Koves TR, Smith JE, Dworkin SI. Differences in extracellular dopamine concentrations in the nucleus accumbens during response-dependent and response-independent cocaine administration in the rat. Psychopharm. 1997;133:7–16. doi: 10.1007/s002130050365. [DOI] [PubMed] [Google Scholar]
- Ito S, Magalska A, Alcaraz-Iborra M, Lopez-Atalaya JP, Rovira V, Contreras-Moreira B, Lipinski M, Olivares R, Martinez-Hernandez J, Ruszczycki B, Lujan R, Geijo-Barrientos E, Wilczynski GM, Barco A. Loss of neuronal 3D chromatin organization causes transcriptional and behavioural deficits related to serotonergic dysfunction. Nat. Commun. 2014;5:4450. doi: 10.1038/ncomms5450. [DOI] [PubMed] [Google Scholar]
- Johnson C, Drgon T, Walther D, Uhl GR. Genomic regions identified by overlapping clusters of nominally-positive SNPs from genome-wide studies of alcohol and illegal substance dependence. PLoS One. 2011;6:e19210. doi: 10.1371/journal.pone.0019210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T, Kitamura K, Suzuki K, Nonaka M, Kamijo S, Takemoto-Kimura S, Kano M, Okuno H, Ohki K, Bito H. Functional labeling of neurons and their projections using the synthetic activity-dependent promoter E-SARE. Nat. Methods. 2013;10:889–895. doi: 10.1038/nmeth.2559. [DOI] [PubMed] [Google Scholar]
- LaPlant Q, Vialou V, Covington HE, 3rd, Dumitriu D, Feng J, Warren BL, Maze I, Dietz DM, Watts EL, Iniguez SD, Koo JW, Mouzon E, Renthal W, Hollis F, Wang H, Noonan MA, Ren Y, Eisch AJ, Bolanos CA, Kabbaj M, Xiao G, Neve RL, Hurd YL, Oosting RS, Fan G, Morrison JH, Nestler EJ. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat. Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Merrer J, Befort K, Gardon O, Filliol D, Darcq E, Dembele D, Becker JA, Kieffer BL. Protracted abstinence from distinct drugs of abuse shows regulation of a common gene network. Addict. Biol. 2012;17:1–12. doi: 10.1111/j.1369-1600.2011.00365.x. [DOI] [PubMed] [Google Scholar]
- Lee BR, Dong Y. Cocaine-induced metaplasticity in the nucleus accumbens: Silent synapse and beyond. Neuropharm. 2011;61:1060–1069. doi: 10.1016/j.neuropharm.2010.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Baker-Andresen D, Zhao Q, Marshall V, Bredy TW. Methyl CpG Binding Domain Ultra-Sequencing: a novel method for identifying inter-individual and cell-type-specific variation in DNA methylation. Genes, brain, and behav. 2014;13:721–731. doi: 10.1111/gbb.12150. [DOI] [PubMed] [Google Scholar]
- Lull ME, Erwin MS, Morgan D, Roberts DC, Vrana KE, Freeman WM. Persistent proteomic alterations in the medial prefrontal cortex with abstinence from cocaine self-administration. Proteomics. Clin. App. 2009;3:462–472. doi: 10.1002/prca.200800055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox SA, Schafe GE. Epigenetic alterations in the lateral amygdala are required for reconsolidation of a Pavlovian fear memory. Learn Mem. 2011;18:579–593. doi: 10.1101/lm.2243411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen HB, Brown RM, Short JL, Lawrence AJ. Investigation of the neuroanatomical substrates of reward seeking following protracted abstinence in mice. J Physiol. 2012;590:2427–2442. doi: 10.1113/jphysiol.2011.225219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark GP, Hajnal A, Kinney AE, Keys AS. Self-administration of cocaine increases the release of acetylcholine to a greater extent than response-independent cocaine in the nucleus accumbens of rats. Psychopharm. 1999;143:47–53. doi: 10.1007/s002130050918. [DOI] [PubMed] [Google Scholar]
- Martin M, Chen BT, Hopf FW, Bowers MS, Bonci A. Cocaine self-administration selectively abolishes LTD in the core of the nucleus accumbens. Nat. Neurosci. 2006;9:868–869. doi: 10.1038/nn1713. [DOI] [PubMed] [Google Scholar]
- Massart R, Barnea R, Dikshtein Y, Suderman M, Meir O, Hallett M, Kennedy P, Nestler EJ, Szyf M, Yadid G. Role of DNA methylation in the nucleus accumbens in incubation of cocaine craving. J. Neurosci. 2015;35:8042–8058. doi: 10.1523/JNEUROSCI.3053-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Feng J, Wilkinson MB, Sun H, Shen L, Nestler EJ. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. PNAS. 2011;108:3035–3040. doi: 10.1073/pnas.1015483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson CS, Mantamadiotis T, Tan SS, Lawrence AJ. Deletion of CREB1 from the dorsal telencephalon reduces motivational properties of cocaine. Cereb Cortex. 2010;20:941–952. doi: 10.1093/cercor/bhp159. [DOI] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, Rivera IM, Rubio MD, Rumbaugh G, Sweatt JD. Cortical DNA methylation maintains remote memory. Nat. Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Dempster E, Mill J, Giese KP. Long-lasting regulation of hippocampal Bdnf gene transcription after contextual fear conditioning. Genes, brain, and behav. 2012;11:651–659. doi: 10.1111/j.1601-183X.2012.00805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Non AL, Binder AM, Kubzansky LD, Michels KB. Genome-wide DNA methylation in neonates exposed to maternal depression, anxiety, or SSRI medication during pregnancy. Epigenetics. 2014;9:964–972. doi: 10.4161/epi.28853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien CP, Childress AR, Ehrman R, Robbins SJ. Conditioning factors in drug abuse: can they explain compulsion? J. Psychopharmacol. 1998;12:15–22. doi: 10.1177/026988119801200103. [DOI] [PubMed] [Google Scholar]
- Oberdoerffer S. A conserved role for intragenic DNA methylation in alternative premRNA splicing. Epigenetics. 2012;3:1–3. doi: 10.4161/trns.19816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palkovits M. Isolated removal of hypothalamic or other brain nuclei of the rat. Brain Res. 1973;59:449–450. doi: 10.1016/0006-8993(73)90290-4. [DOI] [PubMed] [Google Scholar]
- Peric-Hupkes D, van Steensel B. Role of the nuclear lamina in genome organization and gene expression. Cold Spring Harb Symp Quant Biol. 2010;75:517–524. doi: 10.1101/sqb.2010.75.014. [DOI] [PubMed] [Google Scholar]
- Pol Bodetto S, Carouge D, Fonteneau M, Dietrich JB, Zwiller J, Anglard P. Cocaine represses protein phosphatase-1Cbeta through DNA methylation and Methyl-CpG Binding Protein-2 recruitment in adult rat brain. Neuropharm. 2013;73:31–40. doi: 10.1016/j.neuropharm.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Roberts DC, Brebner K. GABA modulation of cocaine self-administration. Ann. N. Y. Acad. Sci. 2000;909:145–158. doi: 10.1111/j.1749-6632.2000.tb06680.x. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Metz M, Zolles G, Turecek R, Fritzius T, Bildl W, Tarusawa E, Kulik A, Unger A, Ivankova K, Seddik R, Tiao JY, Rajalu M, Trojanova J, Rohde V, Gassmann M, Schulte U, Fakler B, Bettler B. Native GABA(B) receptors are heteromultimers with a family of auxiliary subunits. Nature. 2010;465:231–235. doi: 10.1038/nature08964. [DOI] [PubMed] [Google Scholar]
- Scott LJ, Muglia P, Kong XQ, Guan W, Flickinger M, Upmanyu R, Tozzi F, Li JZ, Burmeister M, Absher D, Thompson RC, Francks C, Meng F, Antoniades A, Southwick AM, Schatzberg AF, Bunney WE, Barchas JD, Jones EG, Day R, Matthews K, McGuffin P, Strauss JS, Kennedy JL, Middleton L, Roses AD, Watson SJ, Vincent JB, Myers RM, Farmer AE, Akil H, Burns DK, Boehnke M. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. PNAS. 2009;106:7501–7506. doi: 10.1073/pnas.0813386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Zhao M, Li M, Song T, Zhang M, Quan L, Li S, Sun ZS. Reversal of cocaine-conditioned place preference through methyl supplementation in mice: altering global DNA methylation in the prefrontal cortex. PLoS One. 2012;7:e33435. doi: 10.1371/journal.pone.0033435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronson NC, Taylor JR. Addiction: a drug-induced disorder of memory reconsolidation. Curr. Opin. Neurobiol. 2013;23:573–580. doi: 10.1016/j.conb.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twining RC, Bolan M, Grigson PS. Yoked delivery of cocaine is aversive and protects against the motivation for drug in rats. Behav. Neurosci. 2009;123:913–925. doi: 10.1037/a0016498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl GR, Drgon T, Liu QR, Johnson C, Walther D, Komiyama T, Harano M, Sekine Y, Inada T, Ozaki N, Iyo M, Iwata N, Yamada M, Sora I, Chen CK, Liu HC, Ujike H, Lin SK. Genome-wide association for methamphetamine dependence: convergent results from 2 samples. Arch. Gen. Psychiatry. 2008;65:345–355. doi: 10.1001/archpsyc.65.3.345. [DOI] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak A, Szczepankiewicz AA, Ruszczycki B, Magalska A, Zamlynska K, Dzwonek J, Wilczek E, Zybura-Broda K, Rylski M, Malinowska M, Dabrowski M, Szczepinska T, Pawlowski K, Pyskaty M, Wlodarczyk J, Szczerbal I, Switonski M, Cremer M, Wilczynski GM. Novel higher-order epigenetic regulation of the Bdnf gene upon seizures. J. Neurosci. 2013;33:2507–2511. doi: 10.1523/JNEUROSCI.1085-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Spandidos A, Wang H, Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 2012;40:D1144–1149. doi: 10.1093/nar/gkr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Champagne FA, Brown SE, Dymov S, Sharma S, Meaney MJ, Szyf M. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J. Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976;193:848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Wilczynski GM. Significance of higher-order chromatin architecture for neuronal function and dysfunction. Neuropharm. 2014;80:28–33. doi: 10.1016/j.neuropharm.2014.01.016. [DOI] [PubMed] [Google Scholar]
- Wright KN, Hollis F, Duclot F, Dossat AM, Strong CE, Francis TC, Mercer R, Feng J, Dietz DM, Lobo MK, Nestler EJ, Kabbaj M. Methyl supplementation attenuates cocaine-seeking behaviors and cocaine-induced c-Fos activation in a DNA methylation-dependent manner. J. Neurosci. 2015;35:8948–8958. doi: 10.1523/JNEUROSCI.5227-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel G, Kokocinski F, Lichter P. AutoPrime: selecting primers for expressed sequences. Genome Biol. 2004;5:P11. [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.