Abstract
LASSO is a popular statistical tool often used in conjunction with generalized linear models that can simultaneously select variables and estimate parameters. When there are many variables of interest, as in current biological and biomedical studies, the power of LASSO can be limited. Fortunately, so much biological and biomedical data have been collected and they may contain useful information about the importance of certain variables. This paper proposes an extension of LASSO, namely, prior LASSO (pLASSO), to incorporate that prior information into penalized generalized linear models. The goal is achieved by adding in the LASSO criterion function an additional measure of the discrepancy between the prior information and the model. For linear regression, the whole solution path of the pLASSO estimator can be found with a procedure similar to the Least Angle Regression (LARS). Asymptotic theories and simulation results show that pLASSO provides significant improvement over LASSO when the prior information is relatively accurate. When the prior information is less reliable, pLASSO shows great robustness to the misspecification. We illustrate the application of pLASSO using a real data set from a genome-wide association study.
Keywords: Asymptotic efficiency, Oracle inequalities, Solution path, Weak oracle property
1 INTRODUCTION
In many scientific areas, it is common that experiments with similar goals are performed simultaneously or repetitively to establish a replicable conclusion. A typical example arises from the genome-wide association studies (GWAS). GWAS have become a common practice in exploring the genetic factors underlying complex diseases with the rapid advancement in high-throughput sequencing technology. It usually happens that multiple studies investigating the same question of interest (e.g., looking for the genetic variants associated with a particular human disease) have been carried out repetitively. This brings in the “prior” information for a current study. For example, previous studies provide us with some knowledge about possible effects of certain genes and/or single nucleotide polymorphisms (SNPs) on a disease, although the existing information may include both true and false positives.
Meanwhile, the massive data collected from current research are challenging the statistical analysis. Taking GWAS as an example again, it is well documented that the large number of genetic markers and the generally small size of their effects make it very unlikely for researchers to detect important genetic factors with a desired statistical significance in a single study. For example, hundreds of thousands of SNPs are typically genotyped, and yet their individual effect sizes are usually small. Therefore, it is imperative for us to utilize the existent biological and biomedical information. This prior information, if used appropriately, can alleviate the “curse of dimensionality” and hence improve the power of a study to discover important genetic factors.
Meta-analysis has actually provided effective means to combine different studies or their results to improve the statistical power, especially for genetic studies as well as biomedical studies broadly (Zeggini and Ioannidis, 2009 ; Cantor et al., 2010). Although meta-analysis has been proven successful in many studies (Zeggini et al., 2008), its applicability is restrictive for the present application. For example, the studies in a meta-analysis have to be similar in certain aspects such as the design of the experiments and the analytical methods. In addition, in genome-wide association studies, data are often collected using different genotyping platforms. As a consequence, the SNPs are different across studies, which makes it difficult to combine the results. While there exist methods to impute SNPs across platforms, the implication is not well understood between the results for genotyped and imputed SNPs. Most analytic methods treat the imputed SNPs naively as if they are genotyped SNPs.
We consider the situation where the data from the current study are analyzed using LASSO (Tibshirani, 1996) in conjunction with generalized linear models (McCullagh and Nelder, 1989). LASSO simultaneously selects a subset of explanatory variables and estimates their effects on the response. The goals of LASSO are achieved by balancing the model fitting and model complexity through a penalized likelihood approach. In general, LASSO considers all explanatory variables equally in terms of penalization. Suppose previous studies offered the plausible information about the importance of explanatory variables (e.g., genes or SNPs in GWAS). To incorporate the prior information, a seemingly natural method is to make use of the adaptive LASSO (Zou, 2006 ; Zhang and Lu, 2007), by assigning different weights of penalization to different variables to reflect various levels of prior evidence for significance. For example, variables with smaller p-values (or larger effect sizes) from previous studies should be penalized less such that they have a greater chance to be selected than the variables with larger p-values (or smaller effect sizes). However, this approach suffers from similar difficulties to those that we mentioned for meta-analysis methods. Moreover, this approach will perform poorly when the prior information contains a lot of false positives.
In this paper we propose a new solution named prior LASSO (pLASSO) to incorporate prior information into a LASSO problem. To utilize pLASSO, we only need to assume that the prior information can make certain prediction of the response. Then, pLASSO imposes an extra term using these predictive response values on the criterion function of LASSO to incorporate the prior information. In addition to the balance between model fitting and complexity in LASSO, pLASSO further balances the trade-off between the data and the prior information. It is noteworthy that pLASSO is a very flexible framework that can handle different types of prior information, including information presented as a set of significant explanatory variables (genes/SNPs), the effect sizes of some explanatory variables (genes/SNPs), or even the response values (disease status) given a set of explanatory variables (gene/SNP conditions).
Computationally, we investigate the solution path of pLASSO where the path reflects the change of belief in the prior information. Compared to the piecewise linear paths in the Least Angle Regression (LARS) of Efron et al. (2004) and in the similar work by Rosset and Zhu (2007), our path is piecewise smooth but not linear. Similar to the relationship between LARS and LASSO, our solution path fully characterizes the local effect of incorporating the prior information into a LASSO problem.
Theoretically, we compare the asymptotic properties of the LASSO and pLASSO estimators in three aspects: the asymptotic distribution in the low-dimensional setting, the weak oracle property in the high-dimensional setting, and the oracle inequalities for excess risk and estimation error in the high-dimensional setting. All three comparisons show that pLASSO results in a more efficient estimator than LASSO when the prior information is reasonably reliable. They also illustrate the trade-off between the variance reduction by incorporating the correct prior information and the additive bias due to the prior error.
Finally, numerically, the simulation results verify that pLASSO provides significant improvement over LASSO when the prior information is accurate. We will also see that when the prior information is less reliable, pLASSO shows great robustness to the misspecification. To illustrate the applicability of pLASSO to real data, we re-analyze a genetic data set on bipolar disorder using both LASSO and pLASSO and compare their results.
2 PRIOR LASSO
2.1 LASSO
Assume that we have a collection of independent observations
, where Yi’s are independently observed response values given the p-dimensional predictor vector Xi. Let Y = (Y1, …, Yn)T and
 be the design matrix with Xi as its i-th row. Assume that, with a canonical link, the conditional distribution of Yi given Xi belongs to the canonical exponential family with the following density function (ignoring a multiplier that depends on Yi only):
be the design matrix with Xi as its i-th row. Assume that, with a canonical link, the conditional distribution of Yi given Xi belongs to the canonical exponential family with the following density function (ignoring a multiplier that depends on Yi only):
| (1) |
where with . As is common in generalized linear models, b(θ) is assumed to be twice continuously differentiable with positive b″(θ). The loss function (the negative log-likelihood function) of a generalized linear model is given by
| (2) |
where β = (β0, β1, …, βp)T.
Traditionally, the maximum likelihood estimator (MLE) of β, denoted by β̂, is obtained by minimizing L(β;
 , Y). To select a subset of predictors, LASSO adds an L1-norm penalty term of (β1, …, βp)T to L(β;
, Y). To select a subset of predictors, LASSO adds an L1-norm penalty term of (β1, …, βp)T to L(β;
 , Y). Thus, the LASSO estimator β̂λ is calculated by minimizing the following L1-norm penalized loss function:
, Y). Thus, the LASSO estimator β̂λ is calculated by minimizing the following L1-norm penalized loss function:
| (3) |
where λ is a tuning parameter.
The following well-known observation about a generalized linear model provides a good intuition on how LASSO works. Note that the term in (2) is proportional to the Kullback-Leibler (KL) distance between the distribution in (1) and the following distribution:
| (4) |
Model (4) is the so-called saturated model which often serves as the reference model for evaluating the performance of a fitted model (Collett, 2003). In other words, a generalized linear model is a model reduction tool to approximate the saturated model. The idea of LASSO is to balance the trade-off between the approximation accuracy and the model complexity.
2.2 Prior LASSO
As discussed in the Introduction, we focus on the prior information about possible effects of certain predictors on the response. It can be a set of significant predictors, their effect sizes, or the predictive values given a set of predictors. Mathematically, these can be presented as some existing knowledge about the support of β, the values of β, or the predicted values of the response, etc. For the time being, let us assume that a preprocessing step can summarize the prior information into the predicted responses denoted by . Depending on the nature of the prior information, different methods may be used in this step, and we shall revisit this issue below.
To incorporate the prior information, we need to weigh between the prior information and the presently observed data. Then, in the same spirit of the loss function L(β;
 , Y), it is natural to add the KL distance between the distribution in (1) and the following distribution in Lλ(β;
, Y), it is natural to add the KL distance between the distribution in (1) and the following distribution in Lλ(β;
 , Y):
, Y):
| (5) |
Consequently, the pLASSO estimator β̂λ,η is defined to be the minimizer of the following criterion function:
| (6) |
where η is another tuning parameter playing in a similar role to λ. The parameter η balances the relative importance of the data themselves and the prior information. In the extreme case of η = 0, pLASSO is reduced to LASSO. If η = ∞, pLASSO will solely rely on the prior information to fit the model. Furthermore, the tuning parameter η has another appealing interpretation: it controls the variance of β in its prior distribution from a Bayesian viewpoint (see Section 2.3.1).
We can simplify Lλ,η(β;
 , Y, Ŷp) as follows:
, Y, Ŷp) as follows:
| (7) |
where Ỹ = (Y + ηŶp)/(1 + η) is the adjusted response values using the prior information. The pLASSO criterion function is in exactly the same form as the LASSO criterion function in (3). Essentially any LASSO fitting algorithm can be used to solve pLASSO.
As discussed above, a preprocessing step is needed to summarize the prior information into predictions Ŷp. For generalized linear models, we can obtain the predicted values of the response by plugging in a prior estimator β̂p of β and then calculate , i.e., the expectation of Yi given Xi and β̂p.
A common type of prior information is about the support of β: A subset of predictors, denoted by
 , have been identified to be associated with the response by previous studies. The following simple version of weighted LASSO can be employed to force the predictors
, have been identified to be associated with the response by previous studies. The following simple version of weighted LASSO can be employed to force the predictors
 to stay in the model,
to stay in the model,
| (8) |
The resultant estimator from (8) can be used as the prior estimator β̂p. In fact, minimizing (8) is to compute the predictions Ŷp given that we fully trust the previously identified signals in
 . Certainly, we also consider the variables not in
. Certainly, we also consider the variables not in
 but impose a L1 penalty on them. By doing so, both β̂p and Ŷp depend directly on the reliability of the prior information. Therefore, the pLASSO criterion function in (6) further balances the trade-off between the data and the prior information summarized in Ŷp from (8).
but impose a L1 penalty on them. By doing so, both β̂p and Ŷp depend directly on the reliability of the prior information. Therefore, the pLASSO criterion function in (6) further balances the trade-off between the data and the prior information summarized in Ŷp from (8).
2.3 Special Cases
In this subsection, we consider the application of pLASSO to linear regression and logistic regression—two simplest, yet most frequently used generalized linear models.
2.3.1 Linear regression
Assume both Y and
 are centralized, so we can set β0 = 0 and let β = (β1, …, βp)T without ambiguity. Setting b(θ) = θ
2/2 in (2) leads to the loss function for linear regression
are centralized, so we can set β0 = 0 and let β = (β1, …, βp)T without ambiguity. Setting b(θ) = θ
2/2 in (2) leads to the loss function for linear regression
| (9) |
Then the pLASSO criterion function for linear regression can be written as
| (10) |
| (11) |
where L is defined in (9). It is noteworthy that {Ỹi : i = 1, …, n} in Ỹ are usually not statistically independent, as { : i = 1, …, n} are in general not statistically independent.
It is also interesting to interpret the above criterion function from the Bayesian viewpoint. The middle term in (10) can be considered from a prior distribution of the parameters β as p(β) ∝ exp{−η(Ŷp −
 β)T(Ŷp −
β)T(Ŷp −
 β)/(2n)}, provided that Ŷp is known. Assume that Ŷp is obtained from an estimator β̂p of β by Ŷp =
β)/(2n)}, provided that Ŷp is known. Assume that Ŷp is obtained from an estimator β̂p of β by Ŷp =
 β̂p. Then p(β) ∝ exp{−(β − β̂p)T(η
β̂p. Then p(β) ∝ exp{−(β − β̂p)T(η

 )(β − β̂p)/(2n)} is the density of a normal distribution with mean β̂p and a variance inversely proportional to η. Hence, the weight of the prior information increases as η increases. We shall revisit this observation in Section 3 when we investigate the pLASSO solution path with respect to η.
)(β − β̂p)/(2n)} is the density of a normal distribution with mean β̂p and a variance inversely proportional to η. Hence, the weight of the prior information increases as η increases. We shall revisit this observation in Section 3 when we investigate the pLASSO solution path with respect to η.
2.3.2 Logistic regression
Logistic regression fits each observation (Xi, Yi) by a Bernoulli distribution with probability p(Xi) = P(Yi = 1|Xi), where
| (12) |
The corresponding loss function is
| (13) |
which is obtained by setting b(θ) = log[1+exp(θ)] in (2). The prediction can be computed as . Following (7), the pLASSO criterion function for logistic regression is:
| (14) |
where L is defined in (13) and Ỹ = (Ỹ1, …, Ỹn)T.
Note that each term Yi log{p(Xi)} + (1 − Yi) log{1 − p(Xi)} in L(β;
 , Y) is the KL distance between two Bernoulli distributions—Bernoulli(Yi) and Bernoulli{p(Xi)}. Each term
in L(β;
, Y) is the KL distance between two Bernoulli distributions—Bernoulli(Yi) and Bernoulli{p(Xi)}. Each term
in L(β;
 , Ŷp) is the KL distance between
and Bernoulli{p(Xi)}. Alternatively, we can view the pLASSO criterion function as a LASSO criterion function with observations
. Here, Ỹi ∈ [0, 1] can be regarded as the probability of Yi = 1 obtained after adjusting the distribution Bernoulli(Yi) using the prior information.
, Ŷp) is the KL distance between
and Bernoulli{p(Xi)}. Alternatively, we can view the pLASSO criterion function as a LASSO criterion function with observations
. Here, Ỹi ∈ [0, 1] can be regarded as the probability of Yi = 1 obtained after adjusting the distribution Bernoulli(Yi) using the prior information.
We note that the same KL distance has been used in Schapire et al. (2005) to incorporate prior information into the boosting classification algorithm. It is not surprising that it also works for logistic regression given the close connection between logistic regression and boosting as shown in Friedman et al. (2000). However, the goal and development of our method are obviously different from those in Schapire et al. (2005).
3 SOLUTION PATH
For linear regression, the geometric interpretation of the LASSO estimator provides a useful insight into the mechanism of LASSO, leading to the development of the LARS algorithm (Efron et al., 2004 ). The pLASSO solution would enjoy the same property of the LASSO solution regarding the change of λ for a given η. In this section, we show that the pLASSO estimator of (10) has a similar solution path with respect to the change of η for a given λ. This result allows us to fully characterize the local effect of incorporating the prior information into a LASSO problem.
Let Gλ,η = n−1(1 + η)

 β̂λ,η − n−1
β̂λ,η − n−1
 (Y + ηŶp) be the gradient evaluated at β = β̂λ,η of the pLASSO criterion function in (10) excluding the L1 penalty. Similar to Efron et al. (2004), β̂λ,η and Gλ,η are continuous of both λ and η. Let
(Y + ηŶp) be the gradient evaluated at β = β̂λ,η of the pLASSO criterion function in (10) excluding the L1 penalty. Similar to Efron et al. (2004), β̂λ,η and Gλ,η are continuous of both λ and η. Let
 be the current active set, i.e.,
. The points of discontinuity of
be the current active set, i.e.,
. The points of discontinuity of
 are denoted by {η1, η2, η3, …} where 0 < η1
< ··· < ηk < ηk+1
< ··· < ∞. Therefore, within the interval of η ∈ (ηk, ηk+1),
are denoted by {η1, η2, η3, …} where 0 < η1
< ··· < ηk < ηk+1
< ··· < ∞. Therefore, within the interval of η ∈ (ηk, ηk+1),
 =
=
 is a constant set. Finally, define
is a constant set. Finally, define
 =
=
 to be the submatrix of
to be the submatrix of
 consisting of the columns in
consisting of the columns in
 , and similarly
to be the subvector of β consisting of the elements in
, and similarly
to be the subvector of β consisting of the elements in
 .
.
The necessary and sufficient conditions for β̂λ,η to be a pLASSO estimator are that
| (15) |
From condition (C1) in (15), we have that
| (16) |
Choose any η and η′ such that ηk < η′ < η < ηk+1. It follows that
from the continuity of β̂λ,η and the fact that
 does not change for η ∈ (ηk, ηk+1). Plugging η and η′ into (16) and then subtracting, we can solve the linear equation and obtain
does not change for η ∈ (ηk, ηk+1). Plugging η and η′ into (16) and then subtracting, we can solve the linear equation and obtain
Let us define
 as the vector that satisfies
and
. In fact,
as the vector that satisfies
and
. In fact,
 is the restricted least squares estimator with the response Ŷp and the design matrix restricted to the columns
is the restricted least squares estimator with the response Ŷp and the design matrix restricted to the columns
 . By the continuity of β̂λ,η, setting η′ → ηk leads to
. By the continuity of β̂λ,η, setting η′ → ηk leads to
| (17) |
Also note that . We have proved the following theorem.
Theorem 1
For η ∈ (ηk, ηk+1), the pLASSO solution of (10) satisfies that
Theorem 1 implies that the pLASSO solution follows a piecewise smooth path with respect to the change of η. A similar solution path can be found in the proof of Lemma 4 in Meinshausen and Yu (2009). Their solution path was obtained by moving the response variable along the direction to the noise, which modifies only the linear term of β in the loss function. In our case, the change to the loss function with respect to η is slightly more complicated than their situation because both linear and quadratic terms of β are involved. As a consequence, the path of pLASSO in Theorem 1 is smooth, but not linear as in Meinshausen and Yu (2009).
To understand the pLASSO solution path better, we need to derive how the active set evolves with respect to the change of η. In light of the conditions in (15), for any given λ and η, the elements of an estimator β̂λ,η can be partitioned into the following three non-overlapping groups,
| (18) |
| (19) |
| (20) |
Obviously, . The following theorem fully characterizes the evolvement of the active set with the change of η.
Theorem 2
Assume the “one at a time” condition in Efron et al. (2004). Given λ > 0 and the current active set
 ,
,
ηk+1 = η and
 =
=
 \ {j} if and only if j enters
from
at η;
\ {j} if and only if j enters
from
at η;ηk+1 = η and
 =
=
 ∪ {j} if and only if j enters
from
at η.
∪ {j} if and only if j enters
from
at η.
The active set
 can increase or decrease as η increases. “One at a time” means that the increases and decreases never involve more than a single index j. The proof of Theorem 2 can be found in the Appendix. Theorems 1 and 2 present the whole picture of the solution path of the pLASSO estimator. This piecewise smooth path is different from the piecewise linear path as in LARS or the similar work by Rosset and Zhu (2007). It is also different from the differential equation-based solution path in Wu (2011). This unique solution path deepens our understanding of how the prior information is incorporated into the original LASSO problem locally.
can increase or decrease as η increases. “One at a time” means that the increases and decreases never involve more than a single index j. The proof of Theorem 2 can be found in the Appendix. Theorems 1 and 2 present the whole picture of the solution path of the pLASSO estimator. This piecewise smooth path is different from the piecewise linear path as in LARS or the similar work by Rosset and Zhu (2007). It is also different from the differential equation-based solution path in Wu (2011). This unique solution path deepens our understanding of how the prior information is incorporated into the original LASSO problem locally.
Figure 1 shows an example of what the pLASSO solution path looks like. It is noteworthy that this solution path is limited to the linear regression setting. Also importantly, it is known that LARS, which is again a solution path algorithm, is not as efficient as some other algorithms such as the coordinate descent algorithm (Wu and Lange, 2008; Friedman et al., 2010). Hence, our presentation of the pLASSO solution path is purely to gain insight into how and why pLASSO works. In simulation and real data analysis, however, we use other algorithms that are more convenient.
Figure 1.
An Example of pLASSO Solution Path. This path is illustrated using the diabetes data in Efron et al. (2004), with η on the horizontal axis and coefficient estimates on the vertical axis. There are 10 predictors in the diabetes data, each of which is standardized to have a unit norm. We choose a prior set
 = {X5, X6, X7, X8}. λ is fixed at 316.07 with an initial set
= {X5, X6, X7, X8}. λ is fixed at 316.07 with an initial set
 = {3, 4, 9}. Each vertical line in the plot represents a change of the active set, with the inclusion/deletion of a variable noted at the top of the plot, and the corresponding η value noted at the bottom. The last vertical line is hypothetical to show the limit of coefficients when η → ∞.
= {3, 4, 9}. Each vertical line in the plot represents a change of the active set, with the inclusion/deletion of a variable noted at the top of the plot, and the corresponding η value noted at the bottom. The last vertical line is hypothetical to show the limit of coefficients when η → ∞.
4 THEORETICAL PROPERTIES
This section mainly deals with the theoretical properties of pLASSO. In Section 4.1, we compare the asymptotic distributions of the LASSO and pLASSO estimators when p is fixed. In Section 4.2, we study the weak oracle property of pLASSO when p → ∞ and compare it with LASSO. In Section 4.3, we establish the oracle inequalities for excess risk and estimation error of pLASSO, and again compare them with existing results of LASSO.
4.1 Low-Dimensional Asymptotic Distribution
Assuming that the dimension p is fixed, we derive the asymptotic distribution of the pLASSO estimator and compare it with that of the LASSO estimator. The asymptotic distribution of the LASSO estimator is studied thoroughly by Knight and Fu (2000) in the linear regression setting. A similar result can be obtained in the framework of generalized linear models.
Without loss of generality, let be the true values of the parameters β with nonzero parameters β1,0 = (β0,0, β1,0, …, βs,0)T and zero parameters β2,0 = (βs+1,0, …, βp,0)T = 0. Accordingly, write , where β1 = (β0, β1, …, βs)T and β2 = (βs+1, …, βp)T. Define
and F(β) =
 Σ(β)
Σ(β)
 with
with
 = (Z1, …, Zn)T. In the definition of μ(β) and Σ(β), for linear regression (9), b′(θ) = θ and b″(θ) ≡ 1; for logistic regression (13), b′(θ) = exp(θ)/[1 + exp(θ)] and b″(θ) = exp(θ)/[1 + exp(θ)]2.
= (Z1, …, Zn)T. In the definition of μ(β) and Σ(β), for linear regression (9), b′(θ) = θ and b″(θ) ≡ 1; for logistic regression (13), b′(θ) = exp(θ)/[1 + exp(θ)] and b″(θ) = exp(θ)/[1 + exp(θ)]2.
Theorem 3 provides the asymptotic distribution of the LASSO estimator β̂λ. This result is well known in the linear regression setting (Knight and Fu, 2000).
Theorem 3
Suppose , F(β0)/n → Ω with a positive definite matrix Ω, and for any δ > 0. Let u ~ N(0, Ω), then the LASSO estimator β̂λ follows that , where
| (21) |
In (21), Q and Pλ0 are defined as
In parallel, we will show that a similar result to the above theorem exists for pLASSO. To proceed, we need to be slightly more specific about how the prior information is summarized. As in Section 2.2, we use a prior set
 to indicate the set of those identified variables of potential importance. Then, a prior estimator β̂p is obtained using (8), and the prior information is summarized into
. Theorem 4 provides the asymptotic distribution of the pLASSO estimator β̂λ,η when n → ∞.
to indicate the set of those identified variables of potential importance. Then, a prior estimator β̂p is obtained using (8), and the prior information is summarized into
. Theorem 4 provides the asymptotic distribution of the pLASSO estimator β̂λ,η when n → ∞.
Theorem 4
Suppose , F(β0)/n → Ω with a positive definite matrix Ω, and for any δ > 0. Then, the pLASSO estimator β̂λ,η follows that , where
| (22) |
when η → η0 ≥ 0, and
| (23) |
when η → ∞. In (22) and (23), Q and Pλ0 are defined as in Theorem 3, and
 is defined as
is defined as
The influence of incorporating the prior information can be revealed by comparing the asymptotic distributions of the LASSO and pLASSO estimators. In the extreme case when η → ∞, the prior information dominates, and it is not surprising to observe that the asymptotic distribution of the pLASSO estimator is the same as the prior estimator as in (23). In the other extreme case when η → 0, the contribution of the prior information vanishes eventually so that the asymptotic distribution of the pLASSO estimator simplifies to that of the LASSO estimator.
The non-trivial situation arises when η → η0 > 0. For simplicity, let’s assume λ0 = 0, which implies that the L1 penalization has a negligible effect asymptotically. This is a fair assumption as both (21) and (22) have the same term Pλ0(ϕ), which leads to
| (24) |
| (25) |
Compared with the asymptotic form arg min(Q) of the LASSO estimator, the pLASSO estimator yields a sum of arg min(Q) and argmin(Q +
 ) weighted by the tuning parameter η0. Certainly, whether arg min(Q +
) weighted by the tuning parameter η0. Certainly, whether arg min(Q +
 ) is an improvement or a degradation of arg min(Q) depends on the quality of the prior set
) is an improvement or a degradation of arg min(Q) depends on the quality of the prior set
 as well as the extent (reflected by the parameter κ0) to which we penalize the variables in (
as well as the extent (reflected by the parameter κ0) to which we penalize the variables in (
 )c when constructing Ŷp.
)c when constructing Ŷp.
Writing
by splitting the subscripts {0, 1, …, p} into {0, 1 …, s} and {s + 1, …, p}, we use the following two extreme cases for illustration. On the one hand, suppose that we happen to have the best prior set
 = {X1, …, Xs} and we believe they are the only nonzero parameters by setting κ0 large enough. It leads to that
with
. On the other hand, suppose that we have the worst prior set
= {X1, …, Xs} and we believe they are the only nonzero parameters by setting κ0 large enough. It leads to that
with
. On the other hand, suppose that we have the worst prior set
 = {Xs+1, …, Xp}. This yields arg min(Q +
= {Xs+1, …, Xp}. This yields arg min(Q +
 ) = Ω−1[u − κ0sign(b)] with b = (0, β1,0, …, βs,0, 0, …, 0)T and sign(0) = 0. These are the best and worst cases when we incorporate the prior information, while it is common to have an intermediate case in practice.
) = Ω−1[u − κ0sign(b)] with b = (0, β1,0, …, βs,0, 0, …, 0)T and sign(0) = 0. These are the best and worst cases when we incorporate the prior information, while it is common to have an intermediate case in practice.
In the above two cases, the asymptotic distribution in (25) is respectively simplified into
| (26) |
| (27) |
with . In the best case (26), Ω − Ω̃ is non-negative definite because is non-negative definite. Thus, the pLASSO estimator has a smaller asymptotic variance than the LASSO estimator by comparing (26) and (24). Both estimators are asymptotically unbiased. In the worst case (27), the LASSO and pLASSO estimators have the same asymptotic variance Ω−1 but the pLASSO estimator becomes asymptotically biased. This result clearly illustrates the trade-off between variance reduction and increased bias depending on the quality of the prior information. It is also apparent from the asymptotic distributions in (26)–(27) that the theoretical choice of η depends on the quality of the prior information.
4.2 High-Dimensional Weak Oracle Property
Statistical properties of LASSO for high-dimensional data (p ≫ n) have been studied by many works, including Zhao and Yu (2006) for variable selection consistency, Meinshausen and Yu (2009) for estimation consistency, and Fan and Lv (2011) for weak oracle property. We focus on the weak oracle property of pLASSO in this subsection.
To provide a coherent comparison with the existing results, we adopt the same notation as in Fan and Lv (2011). Therefore, we do not include the intercept β0 and set β = (β1, …, βp)T as in Fan and Lv (2011). Slightly different from Section 4.1, β1 = (β1, …, βs)T and β2 = (βs+1, …, βp)T. This setting results in the following LASSO and pLASSO criterion functions:
| (28) |
| (29) |
We review in the following the weak oracle property of LASSO in Fan and Lv (2011). Let
 and
and
 respectively be the submatrices of the design matrix
respectively be the submatrices of the design matrix
 = (x1, …, xp) formed by columns in the nonzero positions of β0 and the complement. Without loss of generality we assume that each covariate xj has been standardized so that
. Let dn = 2−1 min{|βj,0| : βj,0 ≠ 0}. Similar to Section 4.1, define
= (x1, …, xp) formed by columns in the nonzero positions of β0 and the complement. Without loss of generality we assume that each covariate xj has been standardized so that
. Let dn = 2−1 min{|βj,0| : βj,0 ≠ 0}. Similar to Section 4.1, define
Condition A: The design matrix
 satisfies that
satisfies that
where the L∞ norm of a matrix is the maximum of the L1 norm of each row, bs is a diverging sequence of positive numbers depending on s,
 = {β1 ∈ ℝs : ||β1 − β1,0||∞ ≤ dn}, and ∘ denotes the Hadamard product.
= {β1 ∈ ℝs : ||β1 − β1,0||∞ ≤ dn}, and ∘ denotes the Hadamard product.
Condition B: Assume that dn ≥ n−γ log n and , for some γ ∈ (0, 1/2]. In addition, assume that λ satisfies and λ ≫ n−α(log n)2 with , s = O(nα0), and α0 < 1, and that if the responses are unbounded.
Condition C: With a constant c1 > 0, P(|aTY − aT μ(β0)| > ||a||2ε) ≤ 2e−c1ε2 where ε ∈ (0, ∞) if the responses are bounded and ε ∈ (0, ||a||2/||a||∞] if the responses are unbounded.
Conditions A–C are rewritten from Conditions 1–3 and the exponential bound (22) in Fan and Lv (2011), specifically for the LASSO situation. The following theorem is rewritten from Theorem 2 in that paper for the LASSO estimator.
Theorem 5
Assume that Conditions A–C are satisfied and log p = O(n1−2α). Then, there exists a LASSO estimator β̂λ such that with probability tending to one,
;
, with γ defined in Condition B.
In comparison, we present a parallel result of the pLASSO estimator. Similar to the previous subsection, we assume that the prior information in pLASSO is captured by a prior estimator β̂p so that . We show that the weak oracle property can be achieved by imposing some extra conditions on the prior estimator (information).
Condition D: The prior estimator β̂p satisfies that
and that
where is the line segment between β0 and β̂p.
Theorem 6
Assume that Conditions A–D are satisfied and log p = O(n1−2α). Then, there exists a pLASSO estimator β̂λ,η such that with probability tending to one,
;
, with γ defined in Condition B.
It is obvious that η plays an important role in Condition D. All the conditions involved with η in Condition D have the same explanation: how much we should believe in the prior information (measured by η) depends on how well the prior estimator β̂p performs. When η = 0, those conditions are automatically satisfied and the conclusion of Theorem 6 becomes identical to that of Theorem 5.
Theorem 6 implies that, if the prior information is reasonably reliable, the pLASSO estimator can achieve the same property of variable selection consistency as the LASSO estimator. In addition, the pLASSO possesses a lower L∞-norm loss for the nonzero coefficients compared to the LASSO estimator, similar to what we concluded in Section 4.1 when the dimension p is fixed and low. Both theoretical conclusions show the improvement of efficiency of pLASSO over LASSO with reliable prior information.
4.3 Oracle Inequalities
In addition to statistical properties on variable selection and asymptotic distribution, a great deal of attention has been devoted to the oracle inequalities for the excess risk (prediction error) and/or estimation error of the LASSO estimator. In linear models, these results include Bunea et al. (2007) for prediction error of LASSO, Bickel et al. (2009) for prediction error of both LASSO and Dantzig selector as well as bounds on estimation error, and Koltchinskii et al. (2011) for a sharp oracle inequality of prediction error. Some parallel results have been developed for generalized linear models, including but not limited to, Van de Geer (2008), Bach (2010) and Kwemou (2012). In this subsection, we establish the oracle inequalities for pLASSO in the framework of generalized linear models, including its excess risk and estimation error.
We utilize the same notation as in Section 4.2. Specifically, the intercept β0 is omitted, β0 denotes the true values of the parameters β, and the pLASSO estimator β̂λ,η is defined to be the minimizer of the criterion function (29). J(β) = {j ∈ {1, …, p} : βj ≠ 0} denotes the subscript set of nonzero components of β and |J(β)| denotes the cardinality of J(β). Thus, from Section 4.2, J(β0) = {1, …, s} and |J(β0)| = s. In addition, we introduce the following notation:
| (30) |
where as in Sections 4.1–4.2.
Following Van de Geer (2008), the excess risk of an estimator β̂ in a generalized linear model is defined as
| (31) |
Before presenting the main result, let us review the restricted eigenvalue condition in Bickel et al. (2009) as follows.
Condition RE(r, c0): For some integer r such that 1 ≤ r ≤ p and a positive number c0, the following condition holds:
Moreover, we impose two other regularity conditions.
Condition E: Assume that supn≥1,1≤i≤n ||Xi||∞ < ∞. Moreover, there exists a constant M > 0 such that supn≥1,1≤i≤n E(|εi|m) ≤ m!Mm for m = 2, 3, 4, ….
Condition F: The parameter space under consideration is restricted to for a constant δ > 0.
The first part of Condition E was employed by Kwemou (2012) when they derived oracle inequalities for LASSO. The second part of Condition E, justified by Jiang and Zhang (2013), is used here to control the probability with which the oracle inequalities hold, by applying Bernstein’s inequality [e.g., Lemma 2.2.11 in van der Vaart and Wellner (1996)]. It is noteworthy that this condition is automatically satisfied in logistic regression. Condition F essentially imposes a uniform lower bound on the function b″(·), and it depends on the function b. For linear regression, the lower bound is automatically satisfied as b″(·) ≡ 1 so
 = ℝp when we choose any δ ≤ 1/2. For logistic regression, b″(θ) = exp(θ)/[1 + exp(θ)]2. Intuitively, Condition F enforces that the probabilities of the Bernoulli variables are uniformly bounded away from 0 and 1. Rigorously speaking, Condition F requires that δ < 1/8, and
and
are uniformly bounded by the interval
for 1 ≤ i ≤ n. When δ → 0, this interval tends to ℝ so
= ℝp when we choose any δ ≤ 1/2. For logistic regression, b″(θ) = exp(θ)/[1 + exp(θ)]2. Intuitively, Condition F enforces that the probabilities of the Bernoulli variables are uniformly bounded away from 0 and 1. Rigorously speaking, Condition F requires that δ < 1/8, and
and
are uniformly bounded by the interval
for 1 ≤ i ≤ n. When δ → 0, this interval tends to ℝ so
 → ℝp. For Poisson regression, b″(θ) = exp(θ), and Condition F is satisfied as long as
and
for 1 ≤ i ≤ n. Again, when δ → 0, log(2δ) → − ∞ and thus
→ ℝp. For Poisson regression, b″(θ) = exp(θ), and Condition F is satisfied as long as
and
for 1 ≤ i ≤ n. Again, when δ → 0, log(2δ) → − ∞ and thus
 → ℝp.
→ ℝp.
Theorem 7
Fix a constant φ > 0 and an integer 1 ≤ r ≤ p. Let Condition RE(r, 3+4/φ) be satisfied. Assume that log p = o(n). Suppose b(·) in (29) is twice continuously differentiable with b″(·) > 0. Let
for a large enough constant A > 0 and Bε̂p = n−1/2||
 ε̂p||∞. Under Conditions E and F, with probability tending to one, the pLASSO estimator β̂λ,η satisfies
ε̂p||∞. Under Conditions E and F, with probability tending to one, the pLASSO estimator β̂λ,η satisfies
| (32) |
with δ defined in Condition F and C(φ) = (2 + φ)2/[φ(1 + φ)].
Theorem 7 provides a general oracle inequality for the excess risk of the pLASSO estimator. This result is parallel to those for LASSO, such as Theorem 2.1 in Van de Geer (2008). Compared to the well known oracle rate rLASSO = |J(β)|log p/n for the LASSO estimator (Van de Geer, 2008; Bickel et al., 2009 ), the rate of pLASSO’s excess risk has two components (by omitting the foregoing constants):
rpLASSO,1 is a fraction of rLASSO, and rpLASSO,2 depends on the prediction error of the prior estimator ε̂p = Ŷp − μ(β0).
Clearly, the result illustrates the trade-off between the variance reduction by incorporating the correct prior information and the additive bias due to the prior error, exactly in the same spirit as in Sections 4.1–4.2. On the one hand, let’s consider the situation when the prior information has a high quality. Take linear regression as an example where ε1, …, εn independently follow an identical normal distribution. Suppose we know the support of β0, then the best prior estimator is the oracle estimator with . In this case, , so we have
Firstly, the Bernstein’s inequality [Lemma 2.2.11 in van der Vaart and Wellner (1996)] leads to . Secondly, noticing that aTε follows a normal distribution for any vector a satisfying ||a||2 = 1, it is easy to verify that
where
. Therefore,
can have a much smaller order than log p with a large probability [depending on the maximum L2 norm of the projections of xj, j ∉ ∈ J(β0), on the space spanned by
 ], then the pLASSO excess risk rate is only a fraction of the LASSO rate. On the other hand, if the prior information is not reliable, a small choice of η will control the additional excess risk caused by the wrong information.
], then the pLASSO excess risk rate is only a fraction of the LASSO rate. On the other hand, if the prior information is not reliable, a small choice of η will control the additional excess risk caused by the wrong information.
Following Theorem 7, Corollary 1 provides some specific oracle inequalities for excess risk and estimation bias for the pLASSO estimator. Except the constants in the bounds, the rates are essentially the same as in (32). Therefore, Corollary 1 provides the same insight for us to better understand the pLASSO method.
Corollary 1
Assume the same conditions as in Theorem 7, except that Condition RE(r, 3+ 4/φ) is replaced by Condition RE(s, 3). Then, with probability tending to one, the pLASSO estimator β̂λ,η satisfies
5 SIMULATION
5.1 Unweighted Penalization Methods
In this subsection, we perform simulation studies to compare the following three estimators: the LASSO estimator β̂λ without using any prior information as in (3), the prior estimator β̂p which fully trusts the prior information, and our proposed pLASSO estimator β̂λ,η which incorporates the prior information into LASSO as in (6). All three methods use L1-norm penalty, so we refer to them as unweighted penalization methods in contrast to those in Section 5.2.
As in Section 2.3, we utilize both linear regression and logistic regression in our simulations. For linear regression, the training data are generated from the following model:
| (33) |
where Σ = (σij) with σij = ρ|i−j|, i, j = 1, …, p. In (33), we fix ρ = 0.5, σ = 1, and β0 = (−0.5, −2, 0.5, 2, −1.5, 1, 2, −1.5, 2, −2, 1, 1.5, −2, 1, 1.5, 0, …, 0)T, with X1, …, X15 relevant to the response.
With the same β0 and Σ, the training data are generated from the following model for logistic regression:
| (34) |
It is noteworthy that the above design matrix X has the so-called “power decay correlation.” It has been shown in Corollary 3 of Zhao and Yu (2006) that X satisfies the strong irrepresentable condition in linear regression, which is our main condition for the weak oracle property (the second condition of Condition A in Section 4.2). Moreover, X also satisfies the restricted eigenvalue condition in Bickel et al. (2009) with a large probability, which is our main condition for the oracle inequalities [Condition RE(r, c0) in Section 4.3 ]. For details, we refer to Example 1 in Raskutti et al. (2010).
In the above models, we set n = 200, p = 1000 in (33), and n = 200, p = 400 in (34). We adopt the form of prior information introduced in Section 2.2. A subset
 of explanatory variables is provided and the criterion function in (8) is used to estimate β̂p. In our computation, twelve different sets
of explanatory variables is provided and the criterion function in (8) is used to estimate β̂p. In our computation, twelve different sets
 are used for comparison, classified in four groups:
are used for comparison, classified in four groups:
The sets
 in the first and second groups (
in the first and second groups (
 and
and
 ) respectively consist of all relevant and irrelevant predictors only. The sets in the third and fourth groups (
) respectively consist of all relevant and irrelevant predictors only. The sets in the third and fourth groups (
 and
and
 ) are mixed with relevant and irrelevant variables. The relevant predictors dominate the sets in the third group
) are mixed with relevant and irrelevant variables. The relevant predictors dominate the sets in the third group
 while the irrelevant predictors dominate the sets in the fourth group
while the irrelevant predictors dominate the sets in the fourth group
 .
.
We use the coordinate descent algorithm developed by Friedman et al. (2010) and the algorithm from Meier et al. (2008) for our linear regression setting and logistic regression setting, respectively. After an estimator is obtained, it is further refitted using only those selected predictors to improve the estimation/prediction accuracy. The tuning parameters λ and η are selected by a 3-fold cross validation to maximize the average empirical likelihood. Specifically, for the selection of λ, we adopt the “one standard error rule” (Breiman et al., 1984) to achieve a more parsimonious model. That is, the parameter λ is selected to be the largest possible which gives an average empirical likelihood within one standard error from the highest average empirical likelihood. The standard error is estimated from the three folds in cross validation. With an independently simulated test set of the same size as the training set, the following criteria are used to assess the performance of an estimator in the setting of linear regression:
#CNZ, #INZ: the number of variables that are correctly and incorrectly selected, respectively;
Bias: the L1 norm bias of an estimator for the nonzero coefficients;
-
MSR: the mean squared residuals of an estimator evaluated on the test data.
In addition to (a) and (b), the following criteria are used to assess the performance of an estimator in the setting of logistic regression:
ME: the model error defined as EX[{logit−1(XTβ̂) − logit−1(XTβ0)}2]. RME: the relative model error as the ratio of the model error to that of the LASSO estimator. In our simulation, the model error is approximated by the empirical average using the test set;
MR: the misclassification rate of the classifier induced by an estimator, evaluated on the test data.
Therefore, criteria (a)–(c) apply to linear regression, while criteria (a), (b), (d) and (e) apply to logistic regression. In summary, the criteria in (a) consider the variable selection property of an estimator; criteria (b) and (d) take into account its estimation property; and criteria (c) and (e) evaluate its prediction performance on the test data. For each choice of
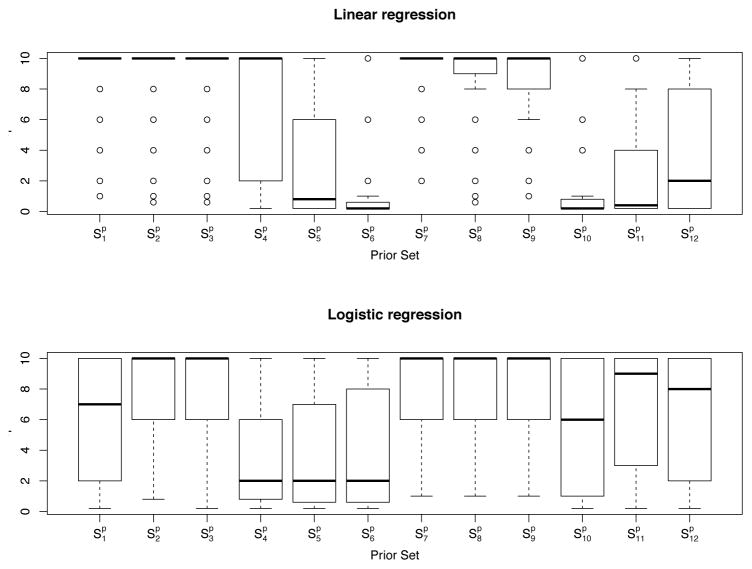
 , 100 training/test data sets are generated from each model. Figures 2–3 provide the boxplots of the criteria (a)–(c) for linear regression from the 100 runs; Figures 4–5 provide the boxplots of the criteria (a), (b), (d) and (e) for logistic regression. In the figures, we denote the three methods by LASSO, p, and pLASSO, respectively. In addition, we report the optimal selection of η by pLASSO from the 100 runs in Figure 6.
, 100 training/test data sets are generated from each model. Figures 2–3 provide the boxplots of the criteria (a)–(c) for linear regression from the 100 runs; Figures 4–5 provide the boxplots of the criteria (a), (b), (d) and (e) for logistic regression. In the figures, we denote the three methods by LASSO, p, and pLASSO, respectively. In addition, we report the optimal selection of η by pLASSO from the 100 runs in Figure 6.
Figure 2.
Simulation results of unweighted penalization methods for linear regression. Rows 1–3 correspond to prior sets
in
 respectively, and rows 4–6 correspond to prior sets
in
respectively, and rows 4–6 correspond to prior sets
in
 respectively. Columns 1–4 correspond to #CNZ, #INZ, Bias and MSR, respectively.
respectively. Columns 1–4 correspond to #CNZ, #INZ, Bias and MSR, respectively.
Figure 3.
Simulation results of unweighted penalization methods for linear regression. Rows 1–3 correspond to prior sets
in
 respectively, and rows 4–6 correspond to prior sets
in
respectively, and rows 4–6 correspond to prior sets
in
 respectively. Columns 1–4 correspond to #CNZ, #INZ, Bias and MSR, respectively.
respectively. Columns 1–4 correspond to #CNZ, #INZ, Bias and MSR, respectively.
Figure 4.
Simulation results of unweighted penalization methods for logistic regression. Rows 1–3 correspond to prior sets
in
 respectively, and rows 4–6 correspond to prior sets
in
respectively, and rows 4–6 correspond to prior sets
in
 respectively. Columns 1–5 correspond to #CNZ, #INZ, Bias, RME and MR, respectively.
respectively. Columns 1–5 correspond to #CNZ, #INZ, Bias, RME and MR, respectively.
Figure 5.
Simulation results of unweighted penalization methods for logistic regression. Rows 1–3 correspond to prior sets
in
 respectively, and rows 4–6 correspond to prior sets
in
respectively, and rows 4–6 correspond to prior sets
in
 respectively. Columns 1–5 correspond to #CNZ, #INZ, Bias, RME and MR, respectively.
respectively. Columns 1–5 correspond to #CNZ, #INZ, Bias, RME and MR, respectively.
Figure 6.
Optimal selection of the tuning parameter η by pLASSO in linear regression (upper panel) and logistic regression (lower panel).
In the case where
 includes only the relevant variables (group
includes only the relevant variables (group
 ), the prior estimator performs the best among the three methods: it selects the most variables correctly and the fewest variables incorrectly; it also yields the lowest estimation bias (and/or smallest model error) and the smallest mean squared residuals (or misclassification rate). Moreover, pLASSO behaves better than LASSO in most criteria we have examined. However, pLASSO includes a similar (or even a larger) number of irrelevant variables to LASSO. For more discussion about this, we refer to Section 5.2 and the Discussion section.
), the prior estimator performs the best among the three methods: it selects the most variables correctly and the fewest variables incorrectly; it also yields the lowest estimation bias (and/or smallest model error) and the smallest mean squared residuals (or misclassification rate). Moreover, pLASSO behaves better than LASSO in most criteria we have examined. However, pLASSO includes a similar (or even a larger) number of irrelevant variables to LASSO. For more discussion about this, we refer to Section 5.2 and the Discussion section.
In the other case where
 includes only irrelevant variables (group
includes only irrelevant variables (group
 ), the prior estimator performs the worst, especially since it selects all those wrong variables in the prior set. pLASSO and LASSO are also better in identifying relevant variables. In terms of estimation bias and prediction accuracy, pLASSO performs comparably with LASSO, with a clear advantage over the prior estimator.
), the prior estimator performs the worst, especially since it selects all those wrong variables in the prior set. pLASSO and LASSO are also better in identifying relevant variables. In terms of estimation bias and prediction accuracy, pLASSO performs comparably with LASSO, with a clear advantage over the prior estimator.
When the prior set includes a mixture of relevant variables and irrelevant variables (groups
 and
and
 ), the numerical performance depends on the “quality” of the prior set. On the one hand, benefitted from the higher-quality prior information in
), the numerical performance depends on the “quality” of the prior set. On the one hand, benefitted from the higher-quality prior information in
 , the prior estimator and pLASSO reduce the estimation bias (and/or model error) and yield a better prediction on the test data. They also correctly include more relevant variables in the model. On the other hand, the lower-quality prior information in
, the prior estimator and pLASSO reduce the estimation bias (and/or model error) and yield a better prediction on the test data. They also correctly include more relevant variables in the model. On the other hand, the lower-quality prior information in
 forces the prior estimator to select all irrelevant variables in the prior set, not as appealing as LASSO or pLASSO. LASSO and pLASSO perform similarly with a lower estimation bias and more accurate prediction than the prior estimator.
forces the prior estimator to select all irrelevant variables in the prior set, not as appealing as LASSO or pLASSO. LASSO and pLASSO perform similarly with a lower estimation bias and more accurate prediction than the prior estimator.
In terms of the tuning parameter η, it is clearly seen from Figure 6 that the optimal selection of η conform with our intuition. In group
 , the tuning parameter η is usually selected at the highest value we set; while the optimal η is much smaller in group
, the tuning parameter η is usually selected at the highest value we set; while the optimal η is much smaller in group
 . For groups that have both relevant and irrelevant variables, η is selected at higher values in
. For groups that have both relevant and irrelevant variables, η is selected at higher values in
 than in
than in
 , since there are more relevant variables in
, since there are more relevant variables in
 and more irrelevant variables in
and more irrelevant variables in
 .
.
In summary, the prior and LASSO estimators are the preferred choices when the prior information is of high and low quality, respectively. However, they are not very robust and can perform poorly in some other situations. The pLASSO method is a more robust choice regardless of the quality of the prior information.
5.2 Weighted Penalization Methods
Now, we include the adaptive LASSO method (Zou, 2006) in our simulation studies. Adaptive LASSO is a weighted penalization method which uses an initial estimator βinitial to weight the L1 penalties. In our context, the adaptive LASSO estimator minimizes the following criterion function:
| (35) |
where λ and τ are tuning parameters. A referee suggested we investigate the case where the initial estimator is the prior estimator β̂p.
For a more comprehensive and fair comparison, we also include the cases where the initial estimator is chosen to be the LASSO and pLASSO estimator. Altogether, we have three weighted penalization methods to compare, which respectively use β̂λ, β̂p and β̂λ,η as initial estimators. For convenience, we refer to them as LASSO-A, p-A and pLASSO-A.
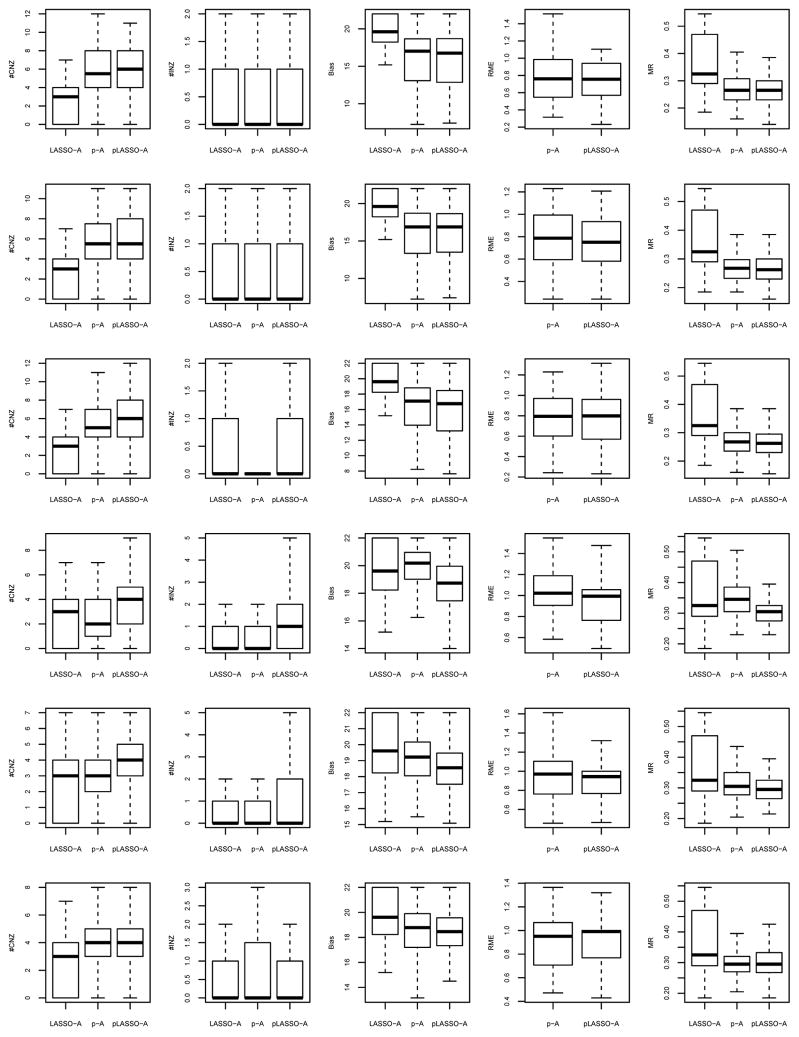
The simulation settings and computational algorithms utilized in this subsection are almost the same as in Section 5.1. Thus, we only point out the differences here. The tuning parameters (including λ and τ) are still optimized using 3-fold cross validation. The relative model error (RME) of the p-A/pLASSO-A estimator is defined to be the ratio of the model error to that of the LASSO-A estimator. Except this minor difference, the criteria used to compare among methods are exactly the same as those in Section 5.1. Figures 7–8 provide the boxplots of the criteria (a) – (c) for linear regression from the 100 runs; Figures 9–10 provide the boxplots of the criteria (a), (b), (d) and (e) for logistic regression.
Figure 7.
Simulation results of weighted penalization methods for linear regression. Rows 1–3 correspond to prior sets
in
 respectively, and rows 4–6 correspond to prior sets
in
respectively, and rows 4–6 correspond to prior sets
in
 respectively. Columns 1–4 correspond to #CNZ, #INZ, Bias and MSR, respectively.
respectively. Columns 1–4 correspond to #CNZ, #INZ, Bias and MSR, respectively.
Figure 8.
Simulation results of weighted penalization methods for linear regression. Rows 1–3 correspond to prior sets
in
 respectively, and rows 4–6 correspond to prior sets
in
respectively, and rows 4–6 correspond to prior sets
in
 respectively. Columns 1–4 correspond to #CNZ, #INZ, Bias and MSR, respectively.
respectively. Columns 1–4 correspond to #CNZ, #INZ, Bias and MSR, respectively.
Figure 9.
Simulation results of weighted penalization methods for logistic regression. Rows 1–3 correspond to prior sets
in
 respectively, and rows 4–6 correspond to prior sets
in
respectively, and rows 4–6 correspond to prior sets
in
 respectively. Columns 1–5 correspond to #CNZ, #INZ, Bias, RME and MR, respectively.
respectively. Columns 1–5 correspond to #CNZ, #INZ, Bias, RME and MR, respectively.
Figure 10.
Simulation results of weighted penalization methods for logistic regression. Rows 1–3 correspond to prior sets
in
 respectively, and rows 4–6 correspond to prior sets
in
respectively, and rows 4–6 correspond to prior sets
in
 respectively. Columns 1–5 correspond to #CNZ, #INZ, Bias, RME and MR, respectively.
respectively. Columns 1–5 correspond to #CNZ, #INZ, Bias, RME and MR, respectively.
Overall, the weighted penalization methods perform better than the unweighted penalization methods in the corresponding settings. The weighted L1 penalty improves all unweighted penalization methods in terms of variable selection, as well as estimation and prediction. It is noteworthy that pLASSO-A performs much better than pLASSO for eliminating irrelevant variables. As follows, we focus on the comparison within three weighted penalization methods. Their relative performance follows similar trends to their unweighted counterparts, although the differences are less obvious.
When
 includes only the relevant variables (group
includes only the relevant variables (group
 ), p-A and pLASSO-A outperform LASSO-A in all criteria including variable selection, estimation and prediction. The difference gets larger when the prior set size gets larger.
), p-A and pLASSO-A outperform LASSO-A in all criteria including variable selection, estimation and prediction. The difference gets larger when the prior set size gets larger.
When
 includes only the irrelevant variables (group
includes only the irrelevant variables (group
 ), p-A is not forced to include all the irrelevant variables in
), p-A is not forced to include all the irrelevant variables in
 . However, its performance can still be affected by the wrong information, especially when there are many irrelevant variables in
. However, its performance can still be affected by the wrong information, especially when there are many irrelevant variables in
 (e.g., rows 5–6 in Figure 7). In linear regression, LASSO-A and pLASSO-A exclude more irrelevant variables than p-A (e.g., rows 5–6 in Figure 7); while in logistic regression, the difference is not obvious (rows 4–6 in Figure 9). pLASSO-A also identifies more relevant variables than p-A. Moreover, LASSO-A and pLASSO-A outperform p-A in terms of the precision of the estimation and prediction.
(e.g., rows 5–6 in Figure 7). In linear regression, LASSO-A and pLASSO-A exclude more irrelevant variables than p-A (e.g., rows 5–6 in Figure 7); while in logistic regression, the difference is not obvious (rows 4–6 in Figure 9). pLASSO-A also identifies more relevant variables than p-A. Moreover, LASSO-A and pLASSO-A outperform p-A in terms of the precision of the estimation and prediction.
When
 includes a mixture of relevant variables and irrelevant variables (groups
includes a mixture of relevant variables and irrelevant variables (groups
 and
and
 ), the numerical performance depends again on the quality of the prior set. The results from prior set groups
), the numerical performance depends again on the quality of the prior set. The results from prior set groups
 and
and
 are similar to those from groups
are similar to those from groups
 and
and
 , respectively. This is consistent to our observation in Section 5.1 for unweighted penalization methods.
, respectively. This is consistent to our observation in Section 5.1 for unweighted penalization methods.
6 REAL DATA ANALYSIS
In this section, we re-analyze the data from a study of bipolar disorder to illustrate how to carry out a real data analysis with pLASSO. Bipolar disorder is a serious and potentially life-threatening mood disorder (Merikangas et al., 2007). It is known that bipolar disorder has a substantial genetic component (Craddock and Forty, 2006). However, the underlying genetic mechanism of bipolar disorder remains elusive, despite significant research effort. There have been at least six genome-wide association studies reported in the literature so far (WTCCC, 2007; Baum et al., 2007; Ferreira et al., 2008; Scott et al., 2009; Sklar et al., 2008; Smith et al., 2009). While these studies revealed promising association signals, top findings from various studies do not show obvious overlap (Baum et al., 2008; Ollila et al., 2009).
In this work, we use the data collected in the Wellcome Trust Case Control Consortium (WTCCC) study as the primary data set. Then we choose the top findings from the other studies as prior information. Among the five prior studies, both Ferreira et al. (2008) and Scott et al. (2009) used meta-analysis by jointly analyzing an independently collected data set and the WTCCC data. Another study, Smith et al. (2009), focused on analyzing the data from two independent populations—individuals of European ancestry and individuals of African ancestry. Since we use the WTCCC data as the primary data and this data set includes only individuals of European ancestry, we do not utilize the results from the above three studies. Therefore, the other two studies—Baum et al. (2007) and Sklar et al. (2008)— serve as “previous” studies which provide us with prior information of potentially important variants for bipolar disorder. The following genes have been reported as top findings in these two studies: MYO5B, TSPAN8, EGFR, SORCS2, DFNB31, DGKH, VGCNL1, and NXN. In our context, we regard the SNPs in those genes as a previously identified set of variants whose information we can borrow in our current study. GeneALaCart (http://www.genecards.org/BatchQueries/index.php), the batch-querying platform of the GeneCards database, is used to extract the SNPs within these eight genes.
Certain quality control and prescreening procedures are performed before we carry out the association study for the WTCCC data. All SNPs with Minor Allele Frequency (MAF) less than 0.05 or failing the Hardy-Weinberg equilibrium test at p-value 0.0001 are excluded from further analysis. To reduce the computational burden, a univariate SNP analysis was performed to select a smaller set of SNPs that we are going to analyze with a regression model. Specifically, a covariate-adjusted (adjusted by age and gender) regression model using bipolar disorder as the response variable is run separately for each SNP after quality control. A SNP is included in the final analysis if either of the following two conditions is satisfied: (a) the p-value of the SNP from the logistic regression is smaller than 0.0001; (b) the SNP belongs to one of the eight previously identified genes and its logistic regression p-value is smaller than 0.1. With these selection criteria, the final data set in our analysis includes 916 SNPs in total, among which 174 SNPs compose the prior set
 as in Section 2.2. The tuning parameters λ and η in both LASSO and pLASSO are selected by a 3-fold cross validation, together with the “one standard error rule” for λ. The final model is fitted using the optimal choice of tuning parameters. The algorithm in Meier et al. (2008) is employed in this example.
as in Section 2.2. The tuning parameters λ and η in both LASSO and pLASSO are selected by a 3-fold cross validation, together with the “one standard error rule” for λ. The final model is fitted using the optimal choice of tuning parameters. The algorithm in Meier et al. (2008) is employed in this example.
From our final results, the optimal choice of η is selected to be 0.2 in pLASSO. Compared with the weight of 1 for the data part in (6), it indicates that pLASSO imposes a mild belief on the prior information by balancing the trade-off between the data and the prior information. The relatively small value of η partially supports the findings that various studies might not agree with each other in detecting the genetic factors underlying bipolar disorder (Baum et al., 2008; Ollila et al., 2009). However, it also gives us some evidence that the data can be better fitted by incorporating prior information to some extent. Therefore, compared with LASSO based on the data themselves, pLASSO has the flexibility of deciding whether or not and to what extent we should incorporate the previous findings in a current study. In addition, both pLASSO and LASSO select a small value of λ, resulting in a large final model for this data set. This observation supports the common belief that bipolar disorder is a complex disease with multiple genetic variants of small effect sizes.
In GWAS, researchers are often looking for a list of top SNPs ranked by statistical significance, which deserves further investigation. With penalized regression, researchers are using different criteria to rank the SNPs, including effect size (Cho et al., 2009), self-developed significance (Wu et al., 2009), etc. To keep our situation simple, we provide a list of top SNPs sorted by effect size from pLASSO and LASSO separately. We will compare how two methods affect the list. Table 1 provides the top twenty SNPs with the largest effect sizes, along with the gene in which each SNP is located or its nearest gene within a distance of 60 kb (Johnson and O’Donnell, 2009).
Table 1.
Real Data Results. Top 20 SNPs with the largest effect size from LASSO and pLASSO separately.
| LASSO | pLASSO | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| SNP | Chr. | Gene | OR | SNP | Chr. | Gene | OR |
|
|
|
||||||
| Matched | |||||||
| rs2226284 | 1 | LRRC7 | 1.51 | rs2226284 | 1 | LRRC7 | 1.30 |
| rs6691577* | 1 | LRRC7 | 0.67 | rs6691577* | 1 | LRRC7 | 0.77 |
| rs4407218* | 2 | - | 1.25 | rs4407218* | 2 | - | 1.18 |
| rs10496366 | 2 | - | 0.81 | rs10496366 | 2 | - | 0.85 |
| rs12640371 | 4 | - | 1.26 | rs12640371 | 4 | - | 1.20 |
| rs4451017 | 5 | - | 1.28 | rs4451017 | 5 | - | 1.22 |
| rs10041150 | 5 | - | 1.24 | rs10041150 | 5 | - | 1.17 |
| rs2609653* | 8 | - | 1.24 | rs2609653* | 8 | - | 1.17 |
| rs884672 | 10 | SORCS3 | 0.78 | rs884672 | 10 | SORCS3 | 0.84 |
| rs10994548 | 10 | RHOBTB1 | 0.81 | rs10994548 | 10 | RHOBTB1 | 0.85 |
| rs659991 | 13 | DGKH | 1.28 | rs659991 | 13 | DGKH | 1.32 |
| rs7159947 | 14 | PARP2 | 1.22 | rs7159947 | 14 | PARP2 | 1.17 |
| rs7176592 | 15 | C15orf23 | 1.24 | rs7176592 | 15 | C15orf23 | 1.18 |
| rs12938916 | 17 | MRPS23 | 1.36 | rs12938916 | 17 | MRPS23 | 1.28 |
| rs2837588 | 21 | DSCAM | 1.24 | rs2837588 | 21 | DSCAM | 1.18 |
|
| |||||||
| Unmatched | |||||||
| rs6668411 | 1 | DAB1 | 0.82 | rs12472797 | 2 | - | 0.86 |
| rs6895541 | 5 | FBXL21 | 1.22 | rs11103407 | 9 | COL5A1 | 1.17 |
| rs17053171 | 6 | - | 0.81 | rs10982246* | 9 | DFNB31 | 0.86 |
| rs6987445 | 8 | - | 1.22 | rs17600642 | 10 | ADAMTS14 | 1.17 |
| rs4984405 | 15 | - | 1.22 | rs6574988 | 14 | - | 1.17 |
The SNPs/genes in the prior set are in bold characters. Chr.: chromosome; Gene: within or nearby gene; OR: odds ratio estimate;
identified in WTCCC (2007) with a strong or moderate association with bipolar disorder.
It is seen that 15 SNPs are overlapping between these two lists, and they possess similar odds ratio estimates. This is due to the small choice of η from our calculation. In other words, two methods partially agree with each other when they are applied to the WTCCC data. It is also noteworthy that rs659991, a SNP in both two top lists, belongs to the gene DGKH which is one of the potential gene associated with bipolar disorder by Baum et al. (2007). This provides us with some evidence for the common genetic variants from the two studies (Baum et al., 2007; WTCCC, 2007).
There are 5 SNPs, however, in each list that are not matched between the two lists. This illustrates the effect of pLASSO compared with LASSO. In particular, one of the 5 unmatched SNPs selected by pLASSO, rs10982246, belongs to the gene DFNB31 as the previously identified information (Baum et al., 2007). Without considering the prior information, we would have missed this gene in the top findings solely by LASSO. Therefore, pLASSO provides a flexible framework to make analysis using information from multiple data sets.
It is also interesting to compare the results from multivariate analysis (LASSO/pLASSO) with univariate analysis in the original paper WTCCC (2007). As indicated in Table 1, some of our top SNPs/genes were identified to possess a strong or moderate association with bipolar disorder in WTCCC (2007). In summary, this example illustrates the applicability of pLASSO to, but not limited to, genetic studies. Certainly, our findings warrant further investigation, but we will leave this to our future work.
7 DISCUSSION
7.1 Summary
This paper presents pLASSO to incorporate prior information into penalized generalized linear models. pLASSO summarizes the prior information into an additional loss function term to achieve a balance between the prior information and the data, where the balance is controlled by a tuning parameter η. Notice that this is different from the elastic net method where the balance is between the L1 and L2 penalties (Zou and Hastie, 2005; Bunea, 2008). However, a shared feature is that neither the L2 penalty nor our additional loss is meant to be standalone. Instead, they provide additional balance to improve the overall performance of the estimation process. Specifically, the L2 penalty stabilizes the estimator and our additional loss increases its efficiency if the prior information is reliable.
Although we keep the settings simple, this represents the first attempt to examine such a scenario, to the best of our knowledge. A few comments are noteworthy. Firstly, the generalized linear models under consideration are coupled with the canonical link functions. Secondly, we keep the L1 penalty as in LASSO and mainly compare pLASSO to LASSO but not other variable selection methods. We study both theoretically and empirically the effect of incorporating prior information by altering the loss function part. Both theoretical and numerical results show that pLASSO is more efficient when the prior information is of high quality, and is robust to the low quality prior information with a good choice of the tuning parameter η.
7.2 Tuning Parameters
For the choice of the tuning parameter η, we mainly investigate the performance of cross validation in our simulations. If incorporating the prior information improves the estimator’s performance, cross validation will pick a relatively large tuning parameter η to improve the efficiency of the resultant estimator. Otherwise, cross validation will select a small tuning parameter to maintain the estimator’s robustness. Simulation results support the effectiveness of cross validation for our purpose. Depending on the quality, pLASSO automatically incorporates the prior information to a proper extent and achieves estimation efficiency and prediction accuracy.
We should note that practical tuning methods, such as the cross validation used in our numerical studies, may not satisfy the conditions imposed on λ or η in our theory. Theoretical justification of practical tuning methods is known to be very challenging and remains to be an open problem, especially for high-dimensional data. We utilize a commonly-used practical tuning approach, and recognize that it is important to investigate this problem and pursue alternative approaches to selecting the tuning parameters. Methods that are computationally more efficient than cross-validation grid search are preferred and need to be pursued, e.g., the bisection method in Bunea and Barbu (2009).
7.3 pLASSO, LASSO, and adaptive LASSO
The distinction between pLASSO and LASSO is the additional loss function which measures the goodness of fit to the prior information (instead of the fit to the data in the usual loss function). Incorporating high-quality prior information will result in a more efficient estimator than LASSO, which is theoretically supported by a smaller asymptotic variance (Section 4.1), a smaller L∞ loss (Section 4.2), and a lower excess risk rate (Section 4.3). It is also practically verified by simulations (Section 5.1). However, incorporating low-quality prior information will lead to the asymptotic bias (Section 4.1) or a higher excess risk rate (Section 4.3). Fortunately, we can adaptively select the tuning parameter η, making pLASSO robust to the low-quality prior information (Section 5.1).
Here, pLASSO only alters the loss function part but keeps the L1 penalty exactly the same as in LASSO. In our numerical studies, pLASSO and adaptive LASSO does not dominate each other and have different advantages under different settings, and hence complement each other. Adaptive LASSO can incorporate the prior information into the penalty part through its adaptive weights, e.g., as in (35). It is interesting to explore in the future whether we can incorporate prior information into both loss and penalty so that estimation efficiency and variable selection advantage can be simultaneously achieved. We have made preliminary progress in investigating the combination of pLASSO and adaptive LASSO (i.e., pLASSOA) in our simulation studies (Section 5.2), and demonstrated its advantage over adaptive LASSO. However, this issue warrants further studies.
Acknowledgments
The authors thank the editor, the associate editor, and two anonymous referees for their comments and suggestions that led to considerable improvements of the paper. This research is supported in part by grants R01 DA016750 and R01 DA029081 from the National Institutes of Health (NIH). The dataset used for the analyses described in this manuscript was obtained from the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113 and 085475.
8 APPENDIX
8.1 Proof of Theorem 2
By the continuity of β̂λ,η and Gλ,ηof η, it is impossible for the two groups
and
to exchange elements directly. Hence, any change of
 can only happen if
is not empty, and that single element in
will be the one either entering or deleted from
can only happen if
is not empty, and that single element in
will be the one either entering or deleted from
 . Therefore, the “only if” parts for (a) and (b) are straightforward. We focus on the “if” parts as follows.
. Therefore, the “only if” parts for (a) and (b) are straightforward. We focus on the “if” parts as follows.
For (a), it is seen from (17) that
So
is a monotone function of η ∈ (ηk, ηk+1) for any j ∈
 . Then if some j enters
from
at η, i.e.,
becomes 0 at η for some j ∈
. Then if some j enters
from
at η, i.e.,
becomes 0 at η for some j ∈
 , η must be a discontinuity point of
, η must be a discontinuity point of
 . That is, ηk+1 = η. If not, η < ηk+1 and
. That is, ηk+1 = η. If not, η < ηk+1 and
 stays as
stays as
 . By the monotonicity,
will change its sign, which is contradictive to the continuity of Gλ,η and the fact that
for any j ∈
. By the monotonicity,
will change its sign, which is contradictive to the continuity of Gλ,η and the fact that
for any j ∈
 . Moreover, since ηk+1 = η is proved,
. Moreover, since ηk+1 = η is proved,
 =
=
 \{j} is the only allowed change of
\{j} is the only allowed change of
 .
.
For (b), by Theorem 1, we have that when η ∈ (ηk, ηk+1),
Hence
is a monotone function of η ∈ (ηk, ηk+1) for any
. Then if j enters
from
at η, i.e., |
| becomes λ for some
, η must be a discontinuity point of
 . That is, ηk+1 = η. If not, η <ηk+1 and
. That is, ηk+1 = η. If not, η <ηk+1 and
 stays as
stays as
 . By the monotonicity, |
| will be larger than λ which is impossible. Moreover, since ηk+1 = η is proved,
. By the monotonicity, |
| will be larger than λ which is impossible. Moreover, since ηk+1 = η is proved,
 =
=
 ∪ {j} is the only allowed change of
∪ {j} is the only allowed change of
 .
.
8.2 Proof of Theorem 3
Let . In the LASSO criterion function (3), using Taylor’s expansion,
| (A.1) |
| (A.2) |
where ≃ stands for the equality if ignoring an additive term independent with ϕ. (A.2) follows from Lemma 1 in Fahrmeir and Kaufmann (1985) by verifying their conditions (D) and (N). Furthermore, since ,
Thus, the proof is finished using the same argument as in the proof of Theorem 2 in Knight and Fu (2000).
8.3 Proof of Theorem 4
We briefly outline the proof. As the pLASSO criterion function (6) includes the two loss functions L(β;
 , Y) and L(β;
, Y) and L(β;
 , Ŷp), we need to evaluate them jointly. To do so, we first write L(β;
, Ŷp), we need to evaluate them jointly. To do so, we first write L(β;
 , Y) in terms of β̂, the maximum likelihood estimator of β, i.e., the minimizer of the criterion function (2); and also write L(β;
, Y) in terms of β̂, the maximum likelihood estimator of β, i.e., the minimizer of the criterion function (2); and also write L(β;
 , Ŷp) in terms of β̂p, the prior estimator of β. Then, the two loss functions can be jointly evaluated by studying the joint distribution of β̂ and β̂p. In another word, the relationship between the observed response Y and the prior response Ŷp can be investigated through jointly studying β̂ and β̂p, which is feasible because the two estimators minimize (2) and (8) separately but based on the same data set. This is a key step in the proof. The detailed proof follows.
, Ŷp) in terms of β̂p, the prior estimator of β. Then, the two loss functions can be jointly evaluated by studying the joint distribution of β̂ and β̂p. In another word, the relationship between the observed response Y and the prior response Ŷp can be investigated through jointly studying β̂ and β̂p, which is feasible because the two estimators minimize (2) and (8) separately but based on the same data set. This is a key step in the proof. The detailed proof follows.
8.3.1 Case 1: η → η0 ≥ 0
Let . In the pLASSO criterion function (6), similar to (A.1),
| (A.3) |
First, we write the first terms in (A.1) and (A.3) in terms of β̂ and β̂p respectively as follows:
| (A.4) |
| (A.5) |
By verifying conditions (D) and (N) in Fahrmeir and Kaufmann (1985), (A.4) is a direct conclusion from Theorems 3 therein and also (3.13) in its proof. For (A.5),
with β̃ between β0 and β̂p. The above equalities hold due to the fact that , which can be proved similarly to Theorem 3 since .
Next, we evaluate the asymptotic joint distribution of β̂ and β̂p. It is observed that
and a similar argument to (A.1) and (A.2) leads to
Similar to Theorem 2 in Knight and Fu (2000), the above observations imply that
Finally, from (A.1), (A.3)–(A.5), and the above asymptotic joint distribution,
Then the proof is finished similar to that of Theorem 3. F
8.3.2 Case 2: η → ∞
Let . Similar to the proof in Case 1,
ignoring an additive term independent with ϕ. So the conclusion is proved.
8.4 Proof of Theorem 6
Similar to Theorem 1 in Fan and Lv (2011), the Karush-Kuhn-Tucker (KKT) sufficient conditions for (29) are
| (A.6) |
| (A.7) |
where f(u) = [f(u1), …, f(un)]T for any univariate function f and vector u = (u1, …, un)T for simplicity of notation hereafter. (A.6) and (A.7) imply that is a pLASSO estimator.
8.4.1 Step 1: Existence of a solution to (A.6)
We prove that (A.6) has a solution β̂1 inside the hypercube
For any β1 ∈
 , since dn ≥ n−γ log n, we have sign(β1) = sign(β1,0) and
, since dn ≥ n−γ log n, we have sign(β1) = sign(β1,0) and
With a slight abuse of notation, let γ(β) =
 b′(
b′(
 β), γ(β1) =
β), γ(β1) =
 b′(
b′(
 β1), ξ =
β1), ξ =
 [Y − b′(
[Y − b′(
 β0)] and ξp =
β0)] and ξp =
 [Ŷp − b′(
[Ŷp − b′(
 β0)], then (A.6) is equivalent to Ψ1(β̂1) = 0 where
β0)], then (A.6) is equivalent to Ψ1(β̂1) = 0 where
Similar to the proof of Theorem 2 in Fan and Lv (2011), , and
where r1 = (r1, …, rs)T and for each j = 1, …, s,
with β̃1j lying on the line segment between β1 and β1,0. Therefore,
where
In addition, we have similarly that
where and for each j = 1, …, s,
with β̃j lying between β̂p and β0.
This leads to u = u1 + u2 + u3 with
Using the similar arguments as in the proof of Theorem 2 in Fan and Lv (2011),
It is also seen that
As ||β1 − β1,0||∞ = n−γ log n/(1 + η) for β1 ∈
 , it is already proved that ||u1||∞ = oP [(1 + η)||β1 − β1,0||∞] by Condition B. In addition, under Condition D, it is verified that ||u2||∞ = oP [(1 + η)||β1 − β1,0||∞] and ||u3||∞ = oP [(1 + η)||β1 − β1,0||∞] for β1 ∈
, it is already proved that ||u1||∞ = oP [(1 + η)||β1 − β1,0||∞] by Condition B. In addition, under Condition D, it is verified that ||u2||∞ = oP [(1 + η)||β1 − β1,0||∞] and ||u3||∞ = oP [(1 + η)||β1 − β1,0||∞] for β1 ∈
 . Therefore, the existence of β̂1 inside
. Therefore, the existence of β̂1 inside
 satisfying Ψ1(β̂1) = 0 is implied by the Miranda’s existence theorem as similarly argued by Fan and Lv (2011).
satisfying Ψ1(β̂1) = 0 is implied by the Miranda’s existence theorem as similarly argued by Fan and Lv (2011).
8.4.2 Step 2: Verification of condition (A.7)
Let β̂1 be the solution of (A.6). Note that (A.7) is equivalent to ||Ψ2||∞ < nλ, where
Similar to Fan and Lv (2011), , and
where r2 = (rs+1, …, rp)T and for each j = s + 1, …, p,
with β̃1j lying between β̂1 and β̂1,0. It follows from Fan and Lv (2011) that
In addition, by Step 1,
where and ||r1||∞ = O[sn1−2γ(log n)2/(1+η)2] from Step 1. Moreover,
with from Step 1. Therefore,
Similar to in Step 1,
where and for each j = s + 1, …, p,
with β̃j lying between β̂p and β0. Thus,
It is implied that ||ξ2||∞ = o(nλ) from Condition B and from Condition D. In addition, ||(1 + η)[γ2(β̂1) − γ2(β1,0)]||∞ ≤ Cnλ + oP (nλ) from Conditions B and D. Therefore, with probability tending to one, (A.7) holds with the solution β̂1 in Step 1.
8.5 Proof of Theorem 7
Recall from Condition F that the parameter space under consideration is restricted to
 . As β̂λ,η minimizes (29) for β ∈
. As β̂λ,η minimizes (29) for β ∈
 , the following inequality holds for any β ∈
, the following inequality holds for any β ∈
 ,
,
| (A.8) |
With R(β) defined in (31), by noticing that
(A.8) can be rewritten as
| (A.9) |
where
with εi and defined in (30). By direct calculations,
Recall that
where Bε̂p = n−1/2||
 ε̂p||∞. Consider the following two events,
ε̂p||∞. Consider the following two events,
Our derivation will be restricted on
 ∩
∩
 hereafter, and we will bound the probability of (
hereafter, and we will bound the probability of (
 ∩
∩
 )c at the end of the proof. On
)c at the end of the proof. On
 ∩
∩
 ,
,
Then (A.9) leads to
| (A.10) |
When j ∉ J(β), βj = 0, then . Then, (A.10) leads to
| (A.11) |
Define another event
for the constant φ > 0 defined in Theorem 7. On the one hand, on event
 , (A.11) implies that
, (A.11) implies that
Thus, the inequality (32) in Theorem 7 is proved on event
 .
.
On the other hand, on event
 , (A.11) leads to
, (A.11) leads to
| (A.12) |
Denoted by δ = β̂λ,η − β, (A.12) implies that
Under Condition RE(r, 3 + 4/φ), for any β satisfying |J(β)| ≤ r,
| (A.13) |
From (A.11) and (A.13), we have that
| (A.14) |
Furthermore, Lemma 1 shows that, for β ∈
 ,
,
Then, (A.14) becomes
| (A.15) |
where we have applied the inequality 2uv ≤ bu2 + v2/b for b > 0. With b > 1/(1 + η), (A.15) implies
Setting (b + bη + 1)/(b + bη − 1) = 1 + φ leads to b = (2 + φ)/[φ (1 + η)] and therefore,
for any β ∈
 satisfying that J(β) ≤ r. Therefore, the inequality (32) in Theorem 7 is proved on event
satisfying that J(β) ≤ r. Therefore, the inequality (32) in Theorem 7 is proved on event
 .
.
The last part of the proof is to bound the probability of (
 ∩
∩
 )c. Recall that
. With the same notation as in Condition E, define M′ = max[M supn,i ||Xi||∞, 2(M supn,i ||Xi||∞)2] < ∞. For
)c. Recall that
. With the same notation as in Condition E, define M′ = max[M supn,i ||Xi||∞, 2(M supn,i ||Xi||∞)2] < ∞. For
 , applying Bernstein’s inequality (Lemma 2.2.11 in van der Vaart and Wellner (1996)) leads to
, applying Bernstein’s inequality (Lemma 2.2.11 in van der Vaart and Wellner (1996)) leads to
It is seen that
when A > 0 is large enough and log p = o(n). In addition, for
 ,
,
as long as A ≥ 4. Then
. Therefore, P[(
 ∩
∩
 )c] → 0.
)c] → 0.
8.6 Proof of Corollary 1
In the proof of Theorem 7, setting β = β0 in (A.11) leads to
| (A.16) |
where δ0 = β̂λ,η − β0, J0 = J(β0) with |J0| = s. Thus, Condition RE(r, 3 + 4/φ) becomes RE(s, 3) and κ(r, 3+4/φ) becomes κ(s, 3) in the proof. Also
 = ∅ when β = β0. Meanwhile, setting β = β0 in (A.14) we have that
= ∅ when β = β0. Meanwhile, setting β = β0 in (A.14) we have that
In addition, by Lemma 1,
Furthermore, (A.16) implies that . Therefore, under Condition RE(s, 3),
Therefore, all results in Corollary 1 are proved by noticing that
8.7 A Lemma
Lemma 1
Under Condition F, for any β ∈
 ,
,
Proof
For t ∈ [0, 1], consider the following function
Clearly, g is twice continuously differentiable. By Taylor’s expansion,
Note that the left-hand side of the above equation is just
 (β), and the right-hand side is
(β), and the right-hand side is
under Condition F. The proof is completed.
Contributor Information
Yuan Jiang, Email: yuan.jiang@stat.oregonstate.edu.
Yunxiao He, Email: yunxiaohe@gmail.com.
Heping Zhang, Email: heping.zhang@yale.edu.
References
- Bach F. Self-concordant analysis for logistic regression. Electronic Journal of Statistics. 2010;4:384–414. [Google Scholar]
- Baum A, Akula N, Cabanero M, Cardona I, Corona W, Klemens B, Schulze T, Cichon S, Rietschel M, Nöthen M, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Molecular Psychiatry. 2007;13:197–207. doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A, Hamshere M, Green E, Cichon S, Rietschel M, Noethen M, Craddock N, McMahon F. Meta-analysis of two genome-wide association studies of bipolar disorder reveals important points of agreement. Molecular Psychiatry. 2008;13:466–467. doi: 10.1038/mp.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel PJ, Ritov Y, Tsybakov AB. Simultaneous analysis of Lasso and Dantzig selector. The Annals of Statistics. 2009;37:1705–1732. [Google Scholar]
- Breiman L, Friedman J, Olshen R, Stone C. Classification and Regression Trees. Wadsworth International Group; 1984. [Google Scholar]
- Bunea F. Honest variable selection in linear and logistic regression models via ℓ1 and ℓ1 + ℓ2 penalization. Electronic Journal of Statistics. 2008;2:1153–1194. [Google Scholar]
- Bunea F, Barbu A. Dimension reduction and variable selection in case control studies via regularized likelihood optimization. Electronic Journal of Statistics. 2009;3:1257–1287. [Google Scholar]
- Bunea F, Tsybakov A, Wegkamp M. Sparsity oracle inequalities for the Lasso. Electronic Journal of Statistics. 2007;1:169–194. [Google Scholar]
- Cantor R, Lange K, Sinsheimer J. Prioritizing GWAS results: A review of statistical methods and recommendations for their application. The American Journal of Human Genetics. 2010;86:6–22. doi: 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Kim H, Oh S, Kim K, Park T. Elastic-net regularization approaches for genome-wide association studies of rheumatoid arthritis. BMC proceedings. 2009;3:S25. doi: 10.1186/1753-6561-3-s7-s25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett D. Modelling Binary Data. CRC Press; 2003. [Google Scholar]
- Craddock N, Forty L. Genetics of affective (mood) disorders. European Journal of Human Genetics. 2006;14:660–668. doi: 10.1038/sj.ejhg.5201549. [DOI] [PubMed] [Google Scholar]
- Efron B, Hastie T, Johnstone I, Tibshirani R. Least Angle Regression. Annals of Statistics. 2004;32:407–451. [Google Scholar]
- Fahrmeir L, Kaufmann H. Consistency and Asymptotic Normality of the Maximum Likelihood Estimator in Generalized Linear Models. Annals of Statistics. 1985;13:342–368. [Google Scholar]
- Fan J, Lv J. Non-concave penalized likelihood with NP-dimensionality. IEEE Transactions on Information Theory. 2011;57:5467–5484. doi: 10.1109/TIT.2011.2158486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira M, O’Donovan M, Meng Y, Jones I, Ruderfer D, Jones L, Fan J, Kirov G, Perlis R, Green E, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nature Genetics. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Special invited paper. Additive logistic regression: A statistical view of boosting. Annals of Statistics. 2000;28:337–374. [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. Journal of Statistical Software. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Zhang CM. High-dimensional regression and classification under a class of convex loss functions. Statistics and Its Interface. 2013;6:285–299. [Google Scholar]
- Johnson A, O’Donnell C. An open access database of genome-wide association results. BMC medical genetics. 2009;10:6. doi: 10.1186/1471-2350-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight K, Fu W. Asymptotics for lasso-type estimators. The Annals of Statistics. 2000;28:1356–1378. [Google Scholar]
- Koltchinskii V, Lounici K, Tsybakov AB. Nuclear-norm penalization and optimal rates for noisy low-rank matrix completion. The Annals of Statistics. 2011;39:2302–2329. [Google Scholar]
- Kwemou M. Non-asymptotic Oracle Inequalities for the Lasso and Group Lasso in high dimensional logistic model. 2012 arXiv preprint arXiv:1206.0710. [Google Scholar]
- McCullagh P, Nelder J. Generalized Linear Models. Chapman & Hall/CRC; 1989. [Google Scholar]
- Meier L, van der Geer S, Bühlmann P. The group lasso for logistic regression. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2008;70:53–71. [Google Scholar]
- Meinshausen N, Yu B. Lasso-type recovery of sparse representations for high-dimensional data. Annals of Statistics. 2009;37:246–270. [Google Scholar]
- Merikangas K, Akiskal H, Angst J, Greenberg P, Hirschfeld R, Petukhova M, Kessler R. Lifetime and 12-month prevalence of bipolar spectrum disorder in the National Comorbidity Survey replication. Archives of general psychiatry. 2007;64:543–552. doi: 10.1001/archpsyc.64.5.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollila H, Soronen P, Silander K, Palo O, Kieseppä T, Kaunisto M, Lönnqvist J, Peltonen L, Partonen T, Paunio T. Findings from bipolar disorder genome-wide association studies replicate in a Finnish bipolar family-cohort. Molecular Psychiatry. 2009;14:351–353. doi: 10.1038/mp.2008.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskutti G, Wainwright MJ, Yu B. Restricted eigenvalue properties for correlated Gaussian designs. The Journal of Machine Learning Research. 2010;11:2241–2259. [Google Scholar]
- Rosset S, Zhu J. Piecewise linear regularized solution paths. Annals of Statistics. 2007;35:1012–1030. [Google Scholar]
- Schapire R, Rochery M, Rahim M, Gupta N. Boosting with prior knowledge for call classification. Speech and Audio Processing, IEEE Transactions on. 2005;13:174–181. [Google Scholar]
- Scott L, Muglia P, Kong X, Guan W, Flickinger M, Upmanyu R, Tozzi F, Li J, Burmeister M, Absher D, et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proceedings of the National Academy of Sciences. 2009;106:7501–7506. doi: 10.1073/pnas.0813386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P, Smoller J, Fan J, Ferreira M, Perlis R, Chambert K, Nimgaonkar V, McQueen M, Faraone S, Kirby A, et al. Whole-genome association study of bipolar disorder. Molecular Psychiatry. 2008;13:558–569. doi: 10.1038/sj.mp.4002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Bloss C, Badner J, Barrett T, Belmonte P, Berrettini W, Byerley W, Coryell W, Craig D, Edenberg H, et al. Genome-wide association study of bipolar disorder in European American and African American individuals. Molecular Psychiatry. 2009;14:755–763. doi: 10.1038/mp.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 1996;58:267–288. [Google Scholar]
- Van de Geer SA. High-dimensional generalized linear models and the Lasso. The Annals of Statistics. 2008;36:614–645. [Google Scholar]
- van der Vaart AW, Wellner JA. Weak Convergence and Empirical Processes: With Applications to Statistics. New York: Springer-Verlag; 1996. [Google Scholar]
- WTCCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Chen Y, Hastie T, Sobel E, Lange K. Genome-wide association analysis by lasso penalized logistic regression. Bioinformatics. 2009;25:714–721. doi: 10.1093/bioinformatics/btp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Lange K. Coordinate descent algorithms for lasso penalized regression. Annals of Applied Statistics. 2008;2:224–244. [Google Scholar]
- Wu Y. An ordinary differential equation-based solution path algorithm. Journal of nonparametric statistics. 2011;23:185–199. doi: 10.1080/10485252.2010.490584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E, Ioannidis J. Meta-analysis in genome-wide association studies. Pharmacogenomics. 2009;10:191–201. doi: 10.2217/14622416.10.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E, Scott L, Saxena R, Voight B, Marchini J, Hu T, de Bakker P, Abecasis G, Almgren P, Andersen G, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nature Genetics. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lu W. Adaptive Lasso for Cox’s proportional hazards model. Biometrika. 2007;94:691–703. [Google Scholar]
- Zhao P, Yu B. On model selection consistency of Lasso. Journal of Machine Learning Research. 2006;7:2541–2563. [Google Scholar]
- Zou H. The adaptive lasso and its oracle properties. Journal of the American Statistical Association. 2006;101:1418–1429. [Google Scholar]
- Zou H, Hastie T. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2005;67:301–320. [Google Scholar]