Abstract
Strong lines of evidence have established a critical role for the homeodomain protein, HOXB7, in cancer. Specifically, molecular and cellular studies have demonstrated that HOXB7 is a master regulatory gene, capable of orchestrating a variety of target molecules, resulting in the activation of several oncogenic pathways. HOXB7 overexpression correlates with clinical progression and poor outcome of cancer patients. Specific inhibition of HOXB7 is particularly relevant in cancers still lacking effective therapies, such as tamoxifen-resistant breast cancer and melanoma. Mechanistic studies are providing additional targets of therapy, and biomarker studies are further establishing its importance in early diagnosis and prognosis.
Keywords: HOXB7, breast cancer, melanoma, ER, FGF-2
Introduction
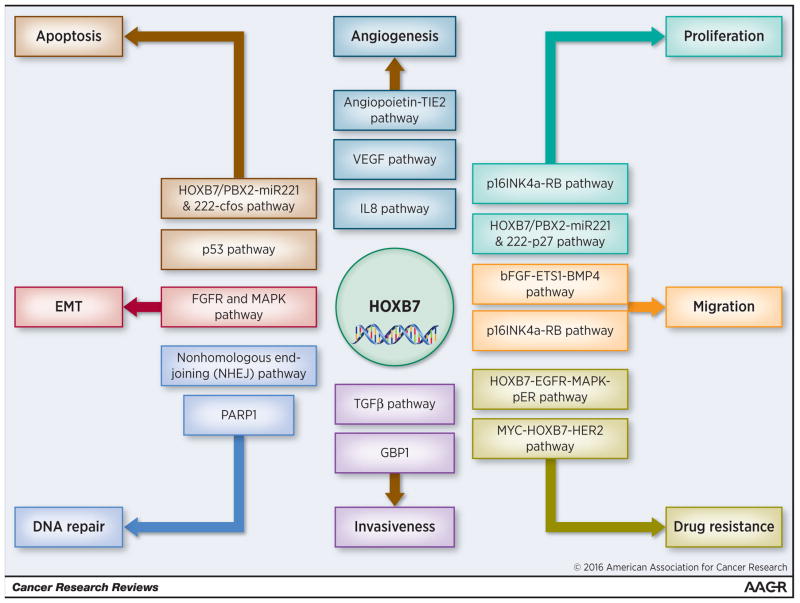
HOX genes encode a family of transcription factors that determines cellular identity during development (1). Although these proteins are generally silenced in adult cells, a substantial number of studies have reported their re-expression in a wide variety of neoplasia (2). The role of HOXB7 in cancer is not surprising given its involvement in all the major cellular processes where, through overexpression or amplification, it appears to act as a master regulatory gene capable of orchestrating a wide variety of target molecules in the oncogenic hierarchy (Figure 1) (2). Predictably, HOXB7 expression has been reported to be correlated with clinical progression and poor outcome of cancer patients, thus suggesting that this protein is a potential prognostic factor and therapeutic target (3).
Figure 1.
Involvement of HOXB7 in known pathways critical to initiation and progression of solid cancers
Although HOX gene clusters are a paradigm of genetic redundancy resulting from genome duplications, HOX proteins display a high level of regulatory specificity and stability of DNA-binding through cooperation with cofactors and other transcription factors. These cofactors are members of the three amino acid loop extension (TALE) family that includes PBX (PBX1, 2, 3 and 4), MEIS (MEIS1, 2, and 3) and PREP (PREP1 and 2) (4). An example of such cooperation is the HOXB7/PBX2-dependent up-regulation of miR-221 and -222 in human melanoma cell lines, where the requirement of PBX cofactors for HOXB7-dependent tumorigenesis was highlighted (5).
In addition, a number of epigenetic regulatory mechanisms have been reported for HOXB7, including changes in methylation status and histone posttranslational modifications, microRNA-based targeting and long non coding-RNAs (lncRNAs) modulation (6–9).
A functional role for overexpressed HOXB7 in solid tumors
A number of studies on HOXB7 have strongly established its critical role in different solid tumors (Figure 2). Here we highlight the growing body of knowledge on HOXB7 in a number of human cancers, including breast cancer, melanoma, colorectal cancer, pancreatic ductal adenocarcinoma, oral squamous cell carcinomas, epithelial ovarian carcinoma and lung cancer.
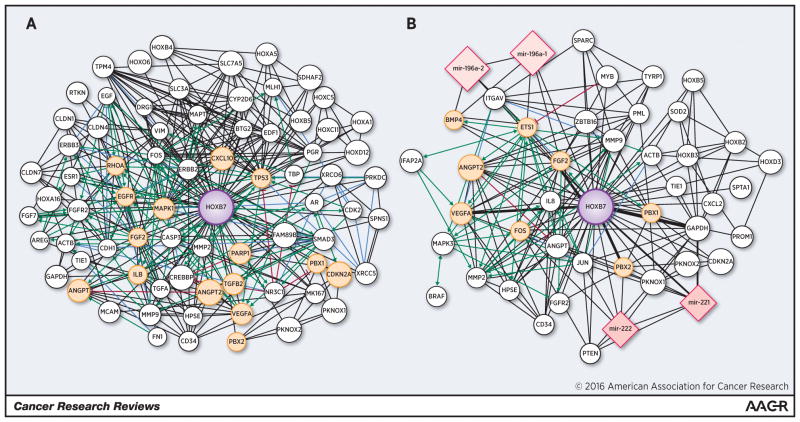
Figure 2. Network of proteins and microRNAs (nodes) linking HOXB7 described in (a) breast cancer and (b) melanoma.
Specifically the node size is proportional to the number of papers where these molecules have been studied, while the edge width is proportional to the number of documents where two molecules have been described together. The central and biggest node corresponds to HOXB7. Proteins and microRNAs described in this review are represented as yellow spheres and pink quadrangles, respectively. Protein pathways are visualized in green (activation), red (inhibition) or violet (binding information). The networks were generated using the web platform tool ProteinQuest (PQ) (http://www.proteinquest.com) and visualized using Cytoscape (42).
Breast Cancer
The study of cancer gene expression patterns has led to the identification of genes whose dysregulation, including recurrent DNA amplification, may contribute to tumorigenesis and tumor progression. Hyman and colleagues performed high resolution CGH analysis in breast tumors with accompanying cDNA microarray data, and showed that about half of the highly amplified genes were also overexpressed (10). Interestingly, they detected HOXB7 amplification in 10% of 363 primary breast cancers, and found a direct correlation between HOXB7 amplification and poor prognosis. In a different study, Sukumar’s group reported a significantly higher percentage of breast tumors that displayed HOXB7 overexpression (60%), indicating that mechanisms other than genomic amplification might be involved in HOXB7 deregulation (11). In addition, this group showed the ability of HOXB7 to drive epithelial cells towards a more mesenchymal phenotype (EMT) through the up-regulation of bFGF and the activation of the MAPK pathway (11). These studies supported the previously reported identification of bFGF as a direct target of HOXB7, both in melanoma (12) and in the SKBR3 breast adenocarcinoma cell line, where enforced expression of HOXB7 resulted in bFGF transcription and induction of a more aggressive phenotype (13,14). The high levels of expression of HOXB7 in lymph-node metastasis-positive ductal carcinomas strengthened its correlation with the malignant behavior of breast cancer cells (15). Interestingly, by using a double transgenic MMTV-HoxB7/HER2 mouse model of breast carcinogenesis, Chen and colleagues discovered dual roles for HoxB7, as HoxB7 inhibited the initial development of mammary tumors but later promoted faster tumor growth and dissemination to the lungs (16). Although previous reports showed that HOXB7 is capable of inducing angiogenesis in human breast cancer cell line xenografts by upregulating proangiogenic factors (VEGF, Ang2, GROα and IL-8) (13,14), no dramatic effects on angiogenesis were observed in the transgenic mice, likely due to innate differences in the immune-deficient transplanted versus immune-competent autochthonous tumor model systems (16). Recently, HoxB7-dependent activation of the TGFβ signaling pathway was also reported in the same model system. Specifically, HOXB7 overexpression induced TGFβ2, leading to increased cell motility and invasiveness as well as recruitment and activation of macrophages (17).
HOXB7 has been also reported to play a crucial role in DNA repair. Rubin and colleagues demonstrated that HOXB7-expressing breast cancer cell lines exhibited both a transformed phenotype and an enhanced survival after irradiation (IR) (18). Using a combination of in vitro and in vivo approaches, HOXB7’s ability to stimulate DNA double-strand breaks (DSB) repair was found to be mediated through its interaction with Ku70, Ku80 and DNA-PKcs, factors involved in the nonhomologous end-joining (NHEJ) pathway. Further, this effect was reversed by suppressing HOXB7 (18). The authors suggested that the enhanced resistance to IR observed in HOXB7-expressing cells could allow them to accumulate mutations that initiate tumorigenesis, thus reconciling HOXB7’s role in DNA repair with its involvement in neoplastic transformation (18).
Looking for a comprehensive profile of genes transcriptionally regulated by HOXB7, a recent study by Jin and colleagues, used bioinformatic analysis of combined ChIP-seq and gene expression datasets (ChIP-PED) (24) to identify more than 1,500 chromatin HOXB7-binding sites in the breast cancer cell line, MCF7. The genes located near the binding sites were potential targets of HOXB7 although, contrary to prediction, the authors demonstrated that HOXB7 preferentially bound to distal enhancer regions rather than to the promoters of its target genes (25). However, since HOXB7 up- or down-regulation per se might be insufficient to produce all the revealed effects, further studies are required to decipher the molecular and biological effects of HOXB7-mediated regulation.
Other Solid Cancers
Numerous independent studies unanimously reported HOXB7 deregulation in several other types of cancer, characterized by HOXB7 overexpression in tumor tissues in relation to their normal counterparts. Among these diseases, we will focus on gastrointestinal, gynecological tumors and oral carcinomas as well as lung carcinomas.
In colorectal cancer (CRC), one of the most prevalent and incident cancers worldwide, the first study reporting altered HOXB7 mRNA levels goes back to 1993 (26). Only recently, HOXB7 expression was shown to be significantly correlated with the invasive and aggressive characteristics of human CRC and therefore, associated with a poor clinical outcome (27).
Also, in pancreatic ductal adenocarcinoma (PDAC), one of the most lethal types of cancer with an overall 5-year survival below 4% (28), HOXB7 was found to be overexpressed and capable of augmenting invasiveness, correlating with regional lymph node dissemination and worse overall survival (29). Gene profiling analyses showed, among those genes modulated by HOXB7, the induction of GBP1, previously shown as a positive effector of tumor cell invasion in glioblastoma, and the decrease of CCBP2, a chemokine decoy receptor (29). Further insight into PDAC biology was provided by a study on microRNAs (miRs). Among miRs linked with tumor grade and venous invasion, miR-337 was found inversely correlated with HOXB7 in 44 cases of PDAC patients retrospectively examined, and in turn, with stage, lymph node status and prognosis, thus suggesting a tumor suppressor action played by miR-337 through the negative regulation of HOXB7 (9). But no mechanistic insights were offered. A miR-dependent regulation of HOXB7 was also reported in cervical cancer. Besides a number of deregulated miRs, miR-196b was downregulated in cervical cancer tissues and cell lines and was associated with worse DFS in patients treated with chemo-radiation (30). This study, and previous data reporting the HOXB7-based induction of VEGF in breast cancer (13) together highlight a novel miR-196b, HOXB7, VEGF axis, raising the speculation on the potential efficacy of an anti-angiogenic therapeutic strategy for cervical cancer.
In the oral cavity, HOXB7 expression was detected at levels significantly higher in oral squamous cell carcinomas (OSCC) than in normal mucosa. De Souza demonstrated the capability of HOXB7 to promote a proliferative phenotype correlating with features related to poor prognosis in OSCC patients (31). Finally, a recent study confirmed significant association of HOXB7 expression with regional lymph nodes involvement and clinical stage in oral and oropharyngeal squamous cell carcinoma (32). Additional information was presented by Di Pietro that demonstrated the correlation of HOXB7 expression in Barrett esophagus (BE), a metaplasia of the distal esophagus that predisposes to esophageal adenocarcinoma (EAC), with levels of the epigenetic histone marks H3K27me3 and AcH3 at gene regulatory regions (6). HOXB7 has been also included in a four-gene diagnostic signature (CLDN4, HOXB7, TMSB4 and TTR) that can distinguish liver tissues of cholangiocarcinoma (CCA) from hepatocellular carcinoma (HCC) and from benign biliary-liver diseases (6).
HOXB7 was demonstrated to be epigenetically modulated by the long non-coding RNA (lncRNA) TUG1 in lung cancer, the most frequent cause of cancer-related death worldwide (8). Specifically, in non-small cell lung cancer TUG1 was reported to be downregulated and associated with a poor prognosis. Conversely, HOXB7 was found to be upregulated in NSCLC tissues, with this inverse correlation relying on TUG1 association with PRC2 protein complexes characterized by histone methyltransferase activity. The authors proposed a p53/TUG1/PRC2/HOXB7 pathway as a target for NSCLC diagnosis and therapy (8).
Cofactors that enhance HOXB7 activity
It is well known that HOX transcription factors require cofactors for strengthening the affinity and specificity of binding to the target sites. In this regard, in breast cancer and in melanoma, functional requirement of PBX cofactors for the oncogenic activity of HOXB7 has been demonstrated. In a breast cancer model the introduction of a dominant-negative PBX1 mutant, PBX1NT, interfered with the HOXB7-induced aggressive phenotype by increasing apoptosis, reducing cell cycling, and up-regulating p16 and p53 (19). HOX proteins achieve DNA sequence specificity by interacting with DNA-binding proteins acting as cofactors (4). Among these, the PBX family members appeared as preferential binding partners of HOXB7 in melanoma (19,20). Along these lines, Errico reported the activating role of the HOXB7/PBX2 complex on miR-221 and miR-222 transcription, supporting a model involving an intricate and reciprocal interplay between HOXB7, their cofactors and microRNAs (5). These two clustered microRNAs are highly upregulated in a variety of solid tumors. Specifically, in melanoma miR-221 and miR-222 enhance tumorigenicity by targeting, among many other molecules, p27Kip1, ETS-1, c-KIT receptor and c-FOS, thus leading to enhanced proliferation, dissemination, differentiation blockade and apoptosis reduction (5,21).
Another HOXB7-interacting partner that contributes to genomic stability is poly(ADP)ribose polymerase-1 (PARP-1), an enzyme that undergoes rapid activation in response to DNA damage (22). Indeed, Wu and colleagues further defined the protein/protein interaction of HOXB7 with PARP-1. Upon this binding, poly(ADP-ribosyl)ation of HOXB7 represses its transcriptional and DNA binding capabilities. In line with this, the inhibition of PARP-1 restored HOXB7 transactivation (22). PARP-1 was also reported to modulate the activity of Yin Yang 1 (YY1) (23), a transcription factor involved in the positive transcriptional regulation of HOXB7 gene by direct interaction with its promoter (23).
Clinical implications
Several findings have paved the way to investigations on HOXB7 as a potential druggable target. Despite major advances in breast cancer treatment and management, and successful treatment of ER-positive breast cancer with tamoxifen and aromatase inhibitors, one third of women with estrogen receptorα (ER)-positive breast cancer suffer from recurrent disease within fifteen years (33). As EMT has been observed in tamoxifen (TAM)-resistant MCF-7 breast cancer cells (30,34,35) and given the association between HOXB7 and EMT in breast cancer (11), HOXB7 protein might possibly play a role in tamoxifen resistance and ensuing tumor progression. Evidence supporting this hypothesis was elucidated by Jin and collaborators through work that demonstrated that HOXB7 overexpression conferred TAM-resistance to ER+ breast cancer cells through the HOXB7-dependent transcriptional induction of EGFR (3,11). In addition, the authors observed hyperphosphorylation of EGFR, activation of p44 MAPK and ER phosphorylation at Ser-118 (3,11). Further, in a recent paper Jin et al. demonstrated that the HOXB7/ER complex promoted the expression of ER-target genes including HER2 through recruitment of ER coactivators to their binding sites in TAM-resistant breast cancer cells (24). The upregulation of HOXB7 was attributed to direct repression of miR-196a by stabilized MYC in TAM-resistant cells. Upregulation of miR-196a or depletion of HOXB7 in TAM-resistant cells restored tamoxifen sensitivity by decreasing expression of HER2 and ER target genes. Also, inhibition of MYC synergized with HER2-targeted therapies (Trastuzumab) to achieve tumor regression (24). Furthermore, co-expression of HOXB7, HER2, and MYC was prognostic of poor overall and relapse-free survival in patients with ER-positive breast cancer who received endocrine therapy. These findings supported HOXB7’s role as an ER-cofactor and suggested that targeting the MYC–HOXB7–HER2 pathway may limit resistance to endocrine therapy in ER-positive breast cancer (24). In depth studies are needed to investigate whether other RTK-signaling pathways are involved in HOXB7-mediated resistance to TAM.
Inhibition of the HOX/PBX interaction has been proposed as a therapeutic strategy. To this end, a small cell permeable peptide (HXR9) has been shown to trigger apoptosis in different cancer cells, including melanoma, antagonizing the interaction between HOX and PBX proteins, and disrupting the binding of these proteins to DNA consensus sites (36). In this regard, the study of Errico and collaborators suggest that the direct inhibition of miR-221 and miR-222 by antagomiR treatment and/or the disruption of the HOXB7/PBX2 dimers might represent innovative approaches for translation into the clinical setting (5).
Epithelial ovarian carcinoma is the fourth most common cause of cancer-related death in women in the developed world, where it is also the leading cause of death from gynecological malignancies (37). In the last two decades, advances in the understanding of ovarian cancer immunogenicity have opened the door to immunotherapeutic approaches to ovarian cancer treatment. The humoral immune response elicited by TAAs could also generate circulating autoantibodies (AAbs) possibly representing novel biomarkers for cancer diagnosis, prognosis, monitoring and prediction of response to chemotherapy. In normal individuals and in non-cancer conditions these antibodies are generally absent or present in very low titers; thus there is a growing interest in evaluating serum autoantibodies against TAAs as biomarkers in cancer immunodiagnosis (38). This idea was supported by Erkanli and colleagues who developed a serum assay evaluating the antibody response to a panel of TAAs for cancer diagnosis. Specifically, they demonstrated that measuring specific AAbs in a three-member panel of TAAs (p53, NY-CO-8 and HOXB7) in addition to serum CA-125, yielded a reasonable sensitivity and specificity in discriminating between epithelial ovarian carcinoma patients and healthy controls (39). Nonetheless, the effectiveness of HOXB7 as a TAA remains an unconfirmed finding.
Future perspectives
Although in recent times an enormous body of basic and translational research investigations has identified a large number of potential biomarkers, a number of cancers are diagnosed at advanced stages and patient survival has not significantly improved. The identification of additional, more informative biomarkers is, therefore, of outmost importance.
Over the last few decades a number of genes and noncoding RNAs in the HOX clusters have been found to be regulated in different tumor types. To therapeutically target HOX proteins has attractive clinical implications but it has been challenging to attain a high level of specificity due to their extreme homology with other HOX proteins. However, current advances in gene therapy and computational biology might contribute substantially in overcoming this restriction. By using siRNAs/shRNAs or RNA targeting tools (ribozymes, aptamers, ASOs, lncRNAs and microRNAs) delivered as nanoparticles, stable, selective and efficient knockdown or overexpression of genes can be achieved. In addition, pharmaceutically designing drugs that inhibit the interaction between HOX proteins and their partners is a promising approach to inhibit their function (5,40,41). As most of the data on HOXB7 expression and function were derived from retrospective analyses of patient tumors, sometimes without adequate treatment information, further prospective validation extended to larger, more controlled cohort of patients will be required. Nevertheless, there is strong evidence already that HOXB7 plays a dominant role in facilitating tumor progression in many solid tumors. If future work confirms its utility as a biomarker and its possible facile detection in easily accessible tissues (i.e. plasma, serum, sputum and urine), HOXB7 could eventually represent a fundamental tool in the oncologist’s armamentarium. The studies reviewed here raise hope regarding HOXB7’s potential role in early diagnosis, direct targeting and selection of targeted therapeutic approaches.
Acknowledgments
Grant Support
This work was supported by grants from the Italian Ministry of Health (RF-2010-2310494) and the Italian Association for Cancer Research (AIRC IG13247) to A.C. and SKCCC Core grant P30 CA006973 to S.S.
Footnotes
Disclosure of Potential Conflicts of Interest
SS and KJ are named co-inventors in the U.S. Provisional Application 61/448,009, filed Mar. 01, 2012, named P11436-02 on the subject of “HOXB7 and tamoxifen resistance”. MCE and AC declare no potential conflicts of interest.
References
- 1.Duboule D. The rise and fall of Hox gene clusters. Development. 2007;134(14):2549–60. doi: 10.1242/dev.001065. [DOI] [PubMed] [Google Scholar]
- 2.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nature reviews Cancer. 2010;10(5):361–71. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 3.Jin K, Kong X, Shah T, Penet MF, Wildes F, Sgroi DC, et al. The HOXB7 protein renders breast cancer cells resistant to tamoxifen through activation of the EGFR pathway. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(8):2736–41. doi: 10.1073/pnas.1018859108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Longobardi E, Penkov D, Mateos D, De Florian G, Torres M, Blasi F. Biochemistry of the tale transcription factors PREP, MEIS, and PBX in vertebrates. Developmental dynamics : an official publication of the American Association of Anatomists. 2014;243(1):59–75. doi: 10.1002/dvdy.24016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Errico MC, Felicetti F, Bottero L, Mattia G, Boe A, Felli N, et al. The abrogation of the HOXB7/PBX2 complex induces apoptosis in melanoma through the miR-221&222-c-FOS pathway. International journal of cancer Journal international du cancer. 2013;133(4):879–92. doi: 10.1002/ijc.28097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.di Pietro M, Lao-Sirieix P, Boyle S, Cassidy A, Castillo D, Saadi A, et al. Evidence for a functional role of epigenetically regulated midcluster HOXB genes in the development of Barrett esophagus. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(23):9077–82. doi: 10.1073/pnas.1116933109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcinkiewicz KM, Gudas LJ. Altered epigenetic regulation of homeobox genes in human oral squamous cell carcinoma cells. Experimental cell research. 2014;320(1):128–43. doi: 10.1016/j.yexcr.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH, et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell death & disease. 2014;5:e1243. doi: 10.1038/cddis.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang R, Leng H, Huang J, Du Y, Wang Y, Zang W, et al. miR-337 regulates the proliferation and invasion in pancreatic ductal adenocarcinoma by targeting HOXB7. Diagnostic pathology. 2014;9:171. doi: 10.1186/s13000-014-0171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer research. 2002;62(21):6240–5. [PubMed] [Google Scholar]
- 11.Wu X, Chen H, Parker B, Rubin E, Zhu T, Lee JS, et al. HOXB7, a homeodomain protein, is overexpressed in breast cancer and confers epithelial-mesenchymal transition. Cancer research. 2006;66(19):9527–34. doi: 10.1158/0008-5472.CAN-05-4470. [DOI] [PubMed] [Google Scholar]
- 12.Care A, Silvani A, Meccia E, Mattia G, Stoppacciaro A, Parmiani G, et al. HOXB7 constitutively activates basic fibroblast growth factor in melanomas. Molecular and cellular biology. 1996;16(9):4842–51. doi: 10.1128/mcb.16.9.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Care A, Silvani A, Meccia E, Mattia G, Peschle C, Colombo MP. Transduction of the SkBr3 breast carcinoma cell line with the HOXB7 gene induces bFGF expression, increases cell proliferation and reduces growth factor dependence. Oncogene. 1998;16(25):3285–9. doi: 10.1038/sj.onc.1201875. [DOI] [PubMed] [Google Scholar]
- 14.Care A, Felicetti F, Meccia E, Bottero L, Parenza M, Stoppacciaro A, et al. HOXB7: a key factor for tumor-associated angiogenic switch. Cancer research. 2001;61(17):6532–9. [PubMed] [Google Scholar]
- 15.Makiyama K, Hamada J, Takada M, Murakawa K, Takahashi Y, Tada M, et al. Aberrant expression of HOX genes in human invasive breast carcinoma. Oncology reports. 2005;13(4):673–9. [PubMed] [Google Scholar]
- 16.Chen H, Lee JS, Liang X, Zhang H, Zhu T, Zhang Z, et al. Hoxb7 inhibits transgenic HER-2/neu-induced mouse mammary tumor onset but promotes progression and lung metastasis. Cancer research. 2008;68(10):3637–44. doi: 10.1158/0008-5472.CAN-07-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S, Jin K, Hui Y, Fu J, Jie C, Feng S, et al. HOXB7 promotes malignant progression by activating the TGFbeta signaling pathway. Cancer research. 2015;75(4):709–19. doi: 10.1158/0008-5472.CAN-14-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubin E, Wu X, Zhu T, Cheung JC, Chen H, Lorincz A, et al. A role for the HOXB7 homeodomain protein in DNA repair. Cancer research. 2007;67(4):1527–35. doi: 10.1158/0008-5472.CAN-06-4283. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez LC, Errico MC, Bottero L, Penkov D, Resnati M, Blasi F, et al. Oncogenic HoxB7 requires TALE cofactors and is inactivated by a dominant-negative Pbx1 mutant in a cell-specific manner. Cancer letters. 2008;266(2):144–55. doi: 10.1016/j.canlet.2008.02.042. [DOI] [PubMed] [Google Scholar]
- 20.Shiraishi K, Yamasaki K, Nanba D, Inoue H, Hanakawa Y, Shirakata Y, et al. Pre-B-cell leukemia transcription factor 1 is a major target of promyelocytic leukemia zinc-finger-mediated melanoma cell growth suppression. Oncogene. 2007;26(3):339–48. doi: 10.1038/sj.onc.1209800. [DOI] [PubMed] [Google Scholar]
- 21.Felicetti F, Errico MC, Bottero L, Segnalini P, Stoppacciaro A, Biffoni M, et al. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer research. 2008;68(8):2745–54. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 22.Wu X, Ellmann S, Rubin E, Gil M, Jin K, Han L, et al. ADP ribosylation by PARP-1 suppresses HOXB7 transcriptional activity. PloS one. 2012;7(7):e40644. doi: 10.1371/journal.pone.0040644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oei SL, Shi Y. Poly(ADP-ribosyl)ation of transcription factor Yin Yang 1 under conditions of DNA damage. Biochemical and biophysical research communications. 2001;285(1):27–31. doi: 10.1006/bbrc.2001.5115. [DOI] [PubMed] [Google Scholar]
- 24.Jin K, Park S, Teo WW, Korangath P, Cho SS, Yoshida T, et al. HOXB7 Is an ERalpha Cofactor in the Activation of HER2 and Multiple ER Target Genes Leading to Endocrine Resistance. Cancer discovery. 2015;5(9):944–59. doi: 10.1158/2159-8290.CD-15-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heinonen H, Lepikhova T, Sahu B, Pehkonen H, Pihlajamaa P, Louhimo R, et al. Identification of several potential chromatin binding sites of HOXB7 and its downstream target genes in breast cancer. International journal of cancer Journal international du cancer. 2015;137(10):2374–83. doi: 10.1002/ijc.29616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Vita G, Barba P, Odartchenko N, Givel JC, Freschi G, Bucciarelli G, et al. Expression of homeobox-containing genes in primary and metastatic colorectal cancer. European journal of cancer. 1993;29A(6):887–93. doi: 10.1016/s0959-8049(05)80432-0. [DOI] [PubMed] [Google Scholar]
- 27.Liao WT, Jiang D, Yuan J, Cui YM, Shi XW, Chen CM, et al. HOXB7 as a prognostic factor and mediator of colorectal cancer progression. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17(11):3569–78. doi: 10.1158/1078-0432.CCR-10-2533. [DOI] [PubMed] [Google Scholar]
- 28.Lowe AW, Olsen M, Hao Y, Lee SP, Taek Lee K, Chen X, et al. Gene expression patterns in pancreatic tumors, cells and tissues. PloS one. 2007;2(3):e323. doi: 10.1371/journal.pone.0000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen Kovochich A, Arensman M, Lay AR, Rao NP, Donahue T, Li X, et al. HOXB7 promotes invasion and predicts survival in pancreatic adenocarcinoma. Cancer. 2013;119(3):529–39. doi: 10.1002/cncr.27725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.How C, Hui AB, Alajez NM, Shi W, Boutros PC, Clarke BA, et al. MicroRNA-196b regulates the homeobox B7-vascular endothelial growth factor axis in cervical cancer. PloS one. 2013;8(7):e67846. doi: 10.1371/journal.pone.0067846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Souza Setubal Destro MF, Bitu CC, Zecchin KG, Graner E, Lopes MA, Kowalski LP, et al. Overexpression of HOXB7 homeobox gene in oral cancer induces cellular proliferation and is associated with poor prognosis. International journal of oncology. 2010;36(1):141–9. [PubMed] [Google Scholar]
- 32.Rivera C, Gonzalez-Arriagada WA, Loyola-Brambilla M, de Almeida OP, Coletta RD, Venegas B. Clinicopathological and immunohistochemical evaluation of oral and oropharyngeal squamous cell carcinoma in Chilean population. International journal of clinical and experimental pathology. 2014;7(9):5968–77. [PMC free article] [PubMed] [Google Scholar]
- 33.Brown RJ, Davidson NE. Adjuvant hormonal therapy for premenopausal women with breast cancer. Seminars in oncology. 2006;33(6):657–63. doi: 10.1053/j.seminoncol.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 34.Yuan J, Liu M, Yang L, Tu G, Zhu Q, Chen M, et al. Acquisition of epithelial-mesenchymal transition phenotype in the tamoxifen-resistant breast cancer cell: a new role for G protein-coupled estrogen receptor in mediating tamoxifen resistance through cancer-associated fibroblast-derived fibronectin and beta1-integrin signaling pathway in tumor cells. Breast Cancer Res. 2015;17:69. doi: 10.1186/s13058-015-0579-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward A, Balwierz A, Zhang JD, Kublbeck M, Pawitan Y, Hielscher T, et al. Re-expression of microRNA-375 reverses both tamoxifen resistance and accompanying EMT-like properties in breast cancer. Oncogene. 2013;32(9):1173–82. doi: 10.1038/onc.2012.128. [DOI] [PubMed] [Google Scholar]
- 36.Morgan R, Boxall A, Harrington KJ, Simpson GR, Gillett C, Michael A, et al. Targeting the HOX/PBX dimer in breast cancer. Breast cancer research and treatment. 2012;136(2):389–98. doi: 10.1007/s10549-012-2259-2. [DOI] [PubMed] [Google Scholar]
- 37.Chester C, Dorigo O, Berek JS, Kohrt H. Immunotherapeutic approaches to ovarian cancer treatment. Journal for immunotherapy of cancer. 2015;3:7. doi: 10.1186/s40425-015-0051-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Q, Liu M, Dai L, Ying X, Ye H, Zhou Y, et al. Using immunoproteomics to identify tumor-associated antigens (TAAs) as biomarkers in cancer immunodiagnosis. Autoimmunity reviews. 2013;12(12):1123–8. doi: 10.1016/j.autrev.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Erkanli A, Taylor DD, Dean D, Eksir F, Egger D, Geyer J, et al. Application of Bayesian modeling of autologous antibody responses against ovarian tumor-associated antigens to cancer detection. Cancer research. 2006;66(3):1792–8. doi: 10.1158/0008-5472.CAN-05-0669. [DOI] [PubMed] [Google Scholar]
- 40.Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. The international journal of biochemistry & cell biology. 2013;45(8):1895–910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 41.Brock A, Krause S, Li H, Kowalski M, Goldberg MS, Collins JJ, et al. Silencing HoxA1 by intraductal injection of siRNA lipidoid nanoparticles prevents mammary tumor progression in mice. Science translational medicine. 2014;6(217):217ra2. doi: 10.1126/scitranslmed.3007048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]