Abstract
Diamondback moth (DBM), Plutella xylostella (Linnaeus), is a notorious pest of brassica crops worldwide and is resistant to all groups of insecticides. The insect system harbors diverse groups of microbiota, which in turn helps in enzymatic degradation of xenobiotic-like insecticides. The present study aimed to determine the diversity of gut microflora in DBM, quantify esterase activity and elucidate their possible role in degradation of indoxacarb. We screened 11 geographic populations of DBM in India and analyzed them for bacterial diversity. The culturable gut bacterial flora underwent molecular characterization with 16S rRNA. We obtained 25 bacterial isolates from larvae (n = 13) and adults (n = 12) of DBM. In larval gut isolates, gammaproteobacteria was the most abundant (76%), followed by bacilli (15.4%). Molecular characterization placed adult gut bacterial strains into three major classes based on abundance: gammaproteobacteria (66%), bacilli (16.7%) and flavobacteria (16.7%). Esterase activity from 19 gut bacterial isolates ranged from 0.072 to 2.32 μmol/min/mg protein. Esterase bands were observed in 15 bacterial strains and the banding pattern differed in Bacillus cereus – KC985225 and Pantoea agglomerans – KC985229. The bands were characterized as carboxylesterase with profenofos used as an inhibitor. Minimal media study showed that B. cereus degraded indoxacarb up to 20%, so it could use indoxacarb for metabolism and growth. Furthermore, esterase activity was greater with minimal media than control media: 1.87 versus 0.26 μmol/min/mg protein. Apart from the insect esterases, bacterial carboxylesterase may aid in the degradation of insecticides in DBM.
Keywords: Plutella xylostella, Bacteria, 16S rRNA, Phylogeny, Carboxylesterase, Indoxacarb
Introduction
Diamondback moth (DBM), Plutella xylostella (Linnaeus), is a serious destructive pest of cruciferous crops worldwide.1 The cost associated with its management is estimated to be 4–5 US billion per year.2, 3 In India, DBM was identified as a major pest, with reported losses of up to 50%.4 DBM has become resistant to each new class of insecticide because of intensive and repeated use of insecticides. Insecticide resistance and absence of natural enemies are believed to explain the P. xylostella pest status worldwide.3, 5 Insects gain resistance to insecticides by increasing production of metabolic enzymes by esterases, glutathione-S-transferases and P450-dependent monooxygenases as well as non-metabolic mechanisms. Furthermore, an increased number of bacteria in the insect gut may produce detoxifying enzymes such as carboxylesterase and help in the detoxification of the xenobiotic compounds. DBM populations were found to harbor many gut bacteria.6
In recent decades, numerous investigations have aimed to understand the metabolic roles of associated microorganisms,7, 8 mainly bacteria, in gut microbiota.9, 10, 11, 12 However, despite the growing awareness of the key roles microbes play in metabolism, vitamin synthesis, pheromone production, pathogen prevention and pesticide degradation,13 the symbiotic relationship within the gut is far from being fully understood. Microscopy examination of the content of the alimentary canal of insects reveals a variety of microflora ranging from bacteria, yeast, molds and protozoa, but the interaction with host insects ranges from mutualistic symbiosis to pathogenesis. Insects with poor or restricted nutrient diet likely have primary endosymbionts, and none of these bacterial endosymbionts have been cultured.14
Degradation of insecticides usually combines a number of processes, including microbial degradation and chemical hydrolysis, which is also affected by some physiochemical properties such as pH, organic carbon and moisture content.15 However, degradation is the primary mechanism of insecticide breakdown and detoxification in insect systems; thus, microbes may have a major role in insecticide resistance. A significant number of microbial carboxylesterases have been discovered,16 but the role of bacterial carboxylesterase from insects in the degradation of insecticides is little known.
We aimed to determine whether gut microflora of DBM larvae play a role in insecticide degradation. We characterized and analyzed the diversity of gut bacteria of larvae and adults of DBM populations by biochemical and culture-dependent 16S rRNA gene-based molecular methods. Furthermore, we detected general esterase and carboxylesterase activity of bacterial strains by spectrophotometry and native PAGE assay, respectively, to examine the possible role of larval gut bacteria in insecticide degradation.
Materials and methods
Collection of DBM populations
Larvae and adults were collected from different geographical locations of India (Table 1S) and reared on mustard (Brassica juncia L.) seedlings in plastic cups (5.5 cm × 6.9 cm × 4.5 cm) containing moistened vermiculate. The individual cups were placed in plastic cages (24 cm × 24 cm × 24 cm) for adult emergence.
Culture-dependent isolation of bacterial flora and bacterial growth
Before dissection, 4th-instar larvae and 1-week-old DBM adults were surface-sterilized in sodium hypochlorite (0.1%) and ethanol (70%) for 5 s to remove the adhering contaminants, especially external microflora.17 The entire gut was removed under aseptic conditions from sterilized larvae and adults. Gut homogenates (100 μL) were plated on sterile LB-agar media (10 g tryptone, 5 g yeast extract, 10 g NaCl and 15 g agar) in three replicates and incubated at 30 °C for 48 h. The colonies were selected by morphological characteristics (shape, color, and elevation) from the three replicates and by Gram staining, then were streaked onto LB agar plates to obtain pure culture, which was inoculated on LB broth (10 g tryptone, 5 g yeast extract, 10 g NaCl) and incubated at 30 °C for 48–72 h for maximum growth.
Screening and identification based on biochemical and morphological characteristics
Preliminary identification of bacterial isolates was based on biochemical analysis, morphologic characteristics and Gram staining. All isolates underwent Voges–Proskauer and methyl red testing, with red indicating positive staining and yellow or brown negative staining.
DNA extraction, PCR amplification and 16S rRNA gene sequence analysis
Culturable bacterial genomic DNA was isolated by SDS-lysis18 and the quantity of DNA was checked with 1% agarose gel. The 16S rRNA gene was amplified with the 16S primer (27F: AGAGTTTGATCMTGGCTCAG and 1492R: CGGTTACCTTGTTACGACTT). The PCR protocol was an initial denaturation at 95 °C for 1 min followed by 30 cycles of 95 °C (1 min), annealing 55 °C (1 min) and 72 °C (2 min) and a final extension step of 72 °C for 2 min (C1000 Thermal cycler). The amplified PCR product was verified with 1.5% agarose gel. The PCR product was purified by use of the Quagin PCR purification kit. Direct sequencing of the 16S rRNA gene for all bacterial isolates was performed at Eurofins analytical services (Bangalore, India).
Phylogenetics analysis
The NCBI database (http://www.ncbi.nlm.nih.gov) was BLAST searched for the 16S rRNA gene sequences, which were used to construct a phylogenetic tree by the character-based maximum-likelihood method based on the Tamura–Nei model19 with MEGA6 and 1000-iteration bootstrapping. The phylogenetic tree was visualized by using Tree View. Gamma distribution parameters and substitution rates were used in phylogenetic analysis.
Quantitative analysis of esterase and protein estimation
Bacterial cultures were transferred (1 mL) to Eppendorf tubes and cell suspensions were centrifuged at 8000 rpm for 10 min (4 °C). Pellets were re-suspended in 100 mM potassium phosphate buffer (pH 7.5) and DNase and centrifuged at 8000 rpm for 15 min (4 °C). Esterase activity was determined spectrophotometrically at 450 nm.20 An amount of 900 μL reaction mixture [α-naphthyl acetate + fast blue RR (4-benzoylamino-2,5-dimethoxyaniline)] and 4 mL potassium phosphate buffer was added to each 100 μL of enzyme extract. The initial spectrophotometric reading was at 450 nm. The mixture was incubated at room temperature (RT) for 10 min and absorbance was recorded at 450 nm. A standard curve was prepared with an identical procedure with a known amount of α-napthol. The rate of hydrolysis was expressed as micromoles of α-napthol produced per minute at RT and the specific activity was expressed as micromoles of α-napthol produced per minute per milligram protein at RT. The protein concentration of the sample was determined by the Lowry method21 with bovine serum albumin used as a standard.
Quantification of esterase in bacterial flora and detection of carboxylesterase
The carboxylesterase activity pattern in 25 bacterial isolates was determined in 8% polyacrylamide gel (pH 8.8) by a Biorad mini gel electrophoresis system.22 Esterase activity was estimated in 20 min at RT. Profenofos was used as a carboxylesterase inhibitor for detection.23 Native PAGE gel with the enzyme sample was incubated in the PAGE buffer containing 100 μL inhibitor solution for 30 min at RT and the control was no treatment. Gel was observed after staining with α-naphthyl acetate (0.4%) and Fast Blue B salt (0.1%) in 40 mM sodium phosphate buffer (pH 7.4) and compared with the control for carboxylesterase detection.
Screening of indoxacarb (92.02% purity) degrading bacteria (Bayer Crop Sciences, Mumbai, India)
Previously identified larval gut bacterial strains were screened for their ability to degrade indoxacarb. The bacterial culture was plated on agar media containing 5 mL indoxacarb (25, 50 and 100 ppm). The colonies able to grow on the agar plate with indoxacarb were selected and streaked on an LB agar plate to obtain pure culture. This procedure was repeated three times to ensure the purity of the isolated colonies. The pure culture was inoculated in LB broth and incubated at 30 °C for 48–72 h for maximum growth.
Indoxacarb degradation by B. cereus and gas chromatography–mass spectroscopy (GC–MS)
A flask (250 mL) was supplemented with indoxacarb as the only carbon source. In total, 50 mL minimal media and 1 mL inoculum were transferred to the flask and thoroughly homogenized. The final concentration of indoxacarb (5 mL) was 25, 50 and 100 ppm. Minimal media with inoculum and with indoxacarb without inoculum was a control. The pH of all media was adjusted to 7.0 and the culture was incubated at 37 °C without contact with the ambient atmosphere and kept in a shaker at 120 rpm for 6 days. Cell growth was measured spectrophotometrically by measuring absorbance at 660 nm. Growth rate was estimated by the slope of the line representing the linear fit of the increase in absorbance over time, during the exponential phase of culture growth.
The culture was extracted twice with an equal volume of ethyl acetate as an extracting reagent. The mixture was centrifuged at 8000 rpm for 10 min and ethyl acetate with residual indoxacarb was filtered. The filtrate was evaporated to dryness and re-dissolved in 50 μL high-performance liquid chromatography-grade dichloromethane for analysis by GC–MS. The operating conditions were injector temperature 245 °C, detector temperature 280 °C and carrier flow 2 mL/min. Minimal media and indoxacarb without bacteria and with inoculum and without indoxacarb were controls.
Quantification of esterase during indoxacarb degradation by B. cereus
The test sample (minimal media + indoxacarb + B. cereus) and control sample (minimal media + B. cereus) were assayed for esterase activity. The test sample was centrifuged to pellet out the cells (8000 rpm, 4 °C, and 15 min). Esterase activity was determined as described above.
Results
Biochemical characteristics, morphological characteristics and Gram staining properties of larval and adult gut bacterial isolates of DBM
Analysis of morphological characteristics revealed the same type of bacterial colonies in all three replicates. Therefore, a single colony was selected to maintain a pure culture. Among the 13 larval isolates, 9 were Gram-negative rods, 2 were Gram-positive rods and the remaining 2 were Gram-positive cocci. Among 12 adult isolates, 9 were Gram-negative rods and 3 were Gram-positive rods. Among 25 bacterial strains, 21 had negative results on methyl red testing, and 4 had positive results. Voges–Proskauer testing was positive for 12 isolates and negative for 13 (Tables 2S and 3S). For further confirmation and identification of isolates, molecular characterization was performed.
Diversity analysis of culturable bacteria
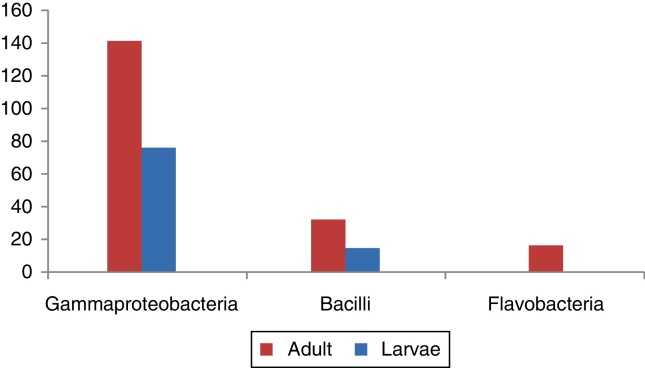
In total, 13 and 12 bacterial from larvae isolates (Table 1) and adults (Table 2), respectively, were identified and sequenced. 16S rRNA sequencing placed these 13 larval bacterial isolates into two major classes – gammaproteobacteria (76%) and bacilli (15.4%) and the 12 adult bacterial isolates into three major classes based on abundance – gammaproteobacteria (66%), bacilli (16.7%) and flavobacteria (16.7%).
Table 1.
Culturable larval gut bacteria isolated from diamondback moth (DBM) populations.
| Sample no. | Isolate | Bacteria | Accession no. |
|---|---|---|---|
| 1 | PX-1 | Enterococcus mundtii | KC985227 |
| 2 | PX-2 | Bacillus cereus | KC985225 |
| 3 | PX-3 | Enterobacter asburiae | KC410777 |
| 4 | PX-3 | Leclercia adecarboxylata | KC410778 |
| 5 | PX-3 | Enterobacteriaceae bacterium | KC410779 |
| 6 | PX-4 | Enterobacter sp. | KF468763 |
| 7 | PX-5 | Pseudomonas sp. | KF597541 |
| 8 | PX-6 | Bacillus cereus | KC139357 |
| 9 | PX-7 | Pseudomonas sp. | KF485084 |
| 10 | PX-8 | Enterococcus mundtii | KC985226 |
| 11 | PX-9 | Enterobacter sp. | KF485082 |
| 12 | PX-10 | Enterococcus mundtii | KF597539 |
| 13 | PX-11 | Enterobacter sp. | KC441058 |
Table 2.
Culturable adult gut bacteria isolated from DBM populations.
| Sample no. | Isolates | Bacteria | Accession no. |
|---|---|---|---|
| 1 | PX-1 | Chryseobacterium sp. | KF312303 |
| 2 | PX-2 | Chryseobacterium sp. | KC985228 |
| 3 | PX-3 | Enterobacter sp. | KC512248 |
| 4 | PX-4 | Enterobacter sp. | KF468764 |
| 5 | PX-5 | Enterococcus mundtii | KC512247 |
| 6 | PX-6 | Bacillus cereus | KC139358 |
| 7 | PX-6 | Weissella confusa | KC139359 |
| 8 | PX-7 | Pseudomonas sp. | KF597543 |
| 9 | PX-8 | Pantoea agglomerans | KC985229 |
| 10 | PX-9 | Morganella morganii | KF468765 |
| 11 | PX-10 | Pseudomonas sp. | KF597542 |
| 12 | PX-11 | Pseudomonas aeruginosa | KF485083 |
Molecular characterization and phylogeny analysis
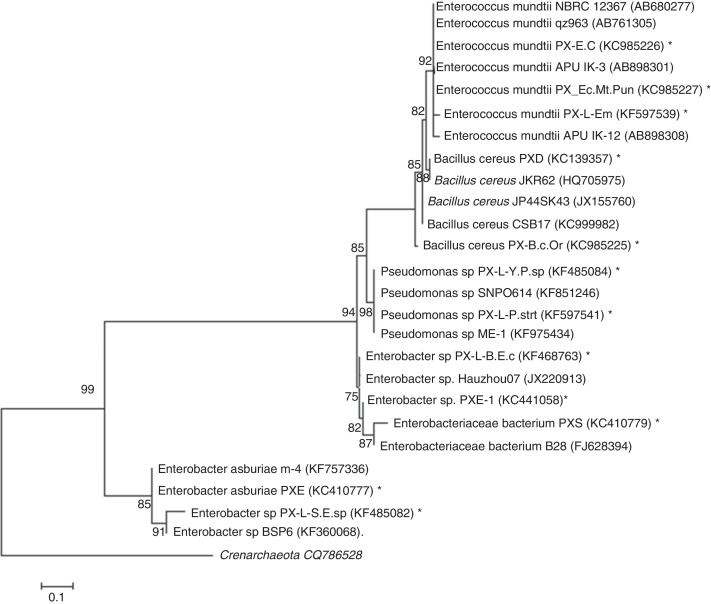
The phylogeny analysis of 16S rRNA gene from larval (Fig. 1) and adult (Fig. 2) bacterial isolates showed two clades. 16S rRNA gene sequencing was used to determine broad taxonomic groupings; the percentage distribution is in Fig. 3. For larval gut isolates, the phylogenetic tree showed two clades with 94% bootstrap value: clade 1, Enterococcus, Bacillus, Pseudomonas and Enterobacter, and clade 2, Enterobacter asburiae and Enterobacter sp. Furthermore, the first clade was divided into two branches: branch 1, Enterococcus mundtii and B. cereus, and branch 2, Pseudomonas and Enterobacter. In the second branch, Pseudomonas showed 98% similarity, Enterobacter sp. and Enterobacteriaceae bacterium were closely related, and the tree was rooted with Crenarchaeota.
Fig. 1.
Maximum-likelihood tree deduced from partial sequences of 16S rRNA gene of larval gut bacterial isolates of DBM. Entries from this work are represented with a star (*), accession numbers and phylotypes. Entries from the public database are represented by a generic name and accession numbers. Crenarchaeota was used as an out group.
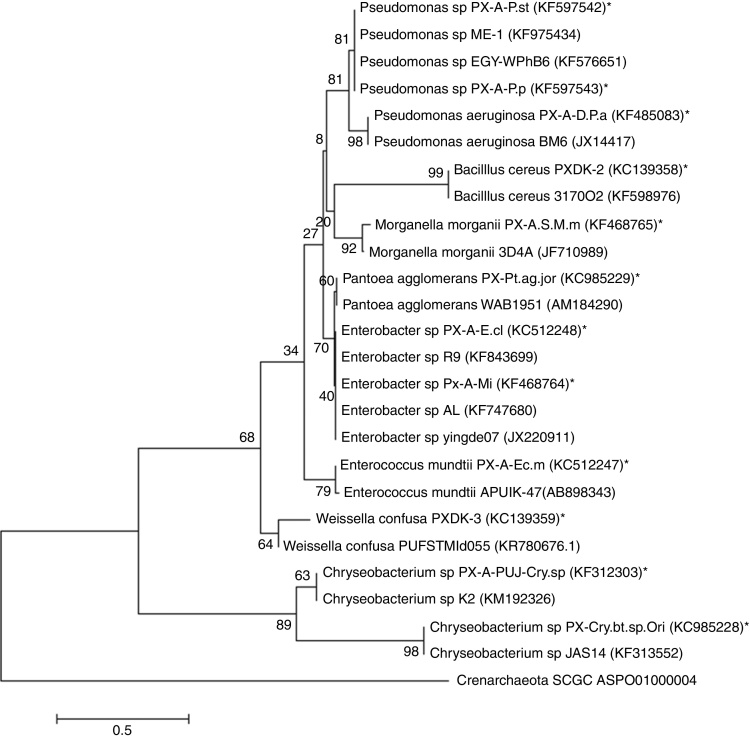
Fig. 2.
Maximum-likelihood tree constructed for partial 16S rRNA gene of adult gut bacterial isolates of DBM. Entries from this work are represented with a star (*), accession numbers and phylotypes. Entries from the public database are represented by a generic name and accession numbers. Crenarchaeota was used as an out group.
Fig. 3.
Percentage distribution of adult and larval gut isolates of DBM. Percentage distribution calculated on the basis of relative abundance.
The phylogenetic tree for adults showed two clades: clade 1, Pseudomonas, Enterococcus, Weissella, Pantoea, Enterobacter, Morganella and Bacillus, and clade 2, Chryseobacterium. The first clade was further divided into two branches; in the first branch, Pseudomonas sp. and P. aeruginosa were related to each other with 99% similarity. Enterobacter sp. was close to Pantoea agglomerans. The tree was rooted with Crenarchaeota. Enterobacter, Enterococcus, Pseudomonas and Bacillus were the most common in both larval and adult isolates.
Esterase analysis
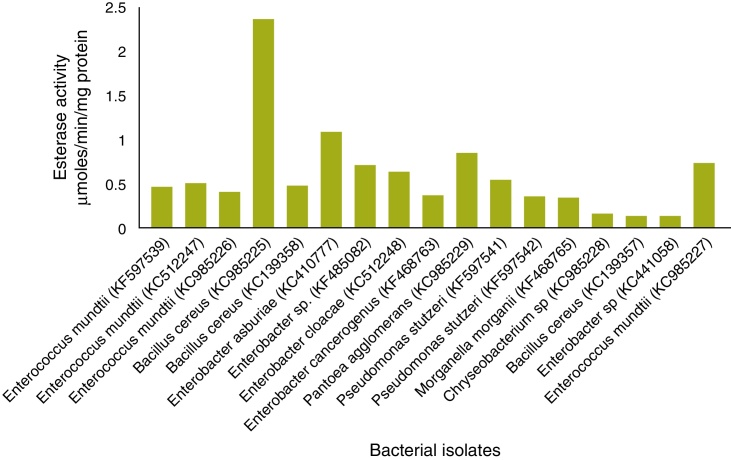
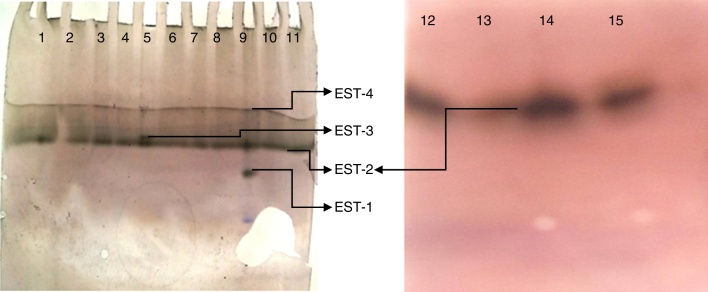
In total, 19 of 25 strains showed esterase activity, which ranged from 0.072 to 2.32 μmol/min/mg protein (Fig. 4). B. cereus (KC985225) showed high esterase activity. Esterase activity was observed in 15 of 25 bacterial strains: E. mundtii – KC985227, KF597539, KC512247, KC985226; B. cereus – KC985225, KC139358; E. asburiae – KC410777, Enterobacter sp. – KF485082; Enterobacter sp. – KC512248; E. cancerogenus – KF468763; P. agglomerans – KC985229; Pseudomonas sp. – KF597541, KF597542; Morganella morganii – KF468765 and Chryseobacterium sp. – KC985228. The esterase band patterns of the strains and the frequencies of each esterase band are in Fig. 5. Four esterase bands were detected, namely Est-1, Est-2, Est-3 and Est-4, was according to their relative mobility in the gel. Est-2 and Est-4 were common among all 15 strains, whereas Est-3 was observed in P. agglomerans – KC985229 and Est-1 in B. cereus – KC985225. The bands were scattered throughout three zones of the gel: fast-moving (Est-1), medium-moving (Est-2 and Est-3) and slow-moving (Est-4). All hydrolyzed the substrate α-naphthyl acetate and were classified as α-esterases.
Fig. 4.
Specific activity of bacterial esterase isolated from larval and adult gut of DBM.
Fig. 5.
Esterase isozyme pattern from bacterial isolates of DBM.
Detection of carboxylesterase
The bands inhibited by profenofos were classified as carboxylesterases. Incubation of the gel in profenofos completely inhibited Est-2 in all bacterial strains, Est-4 in all strains except P. agglomerans – KC985229 and Est-1 in B. cereus – KC985225, whereas Est-3 and Est-4 were not inhibited by profenofos in P. agglomerans – KC985229. According to this classification, Est-2, Est-3 and Est-4 in the remaining strains were found to be carboxylesterase.
Screening of indoxacarb-degrading bacteria
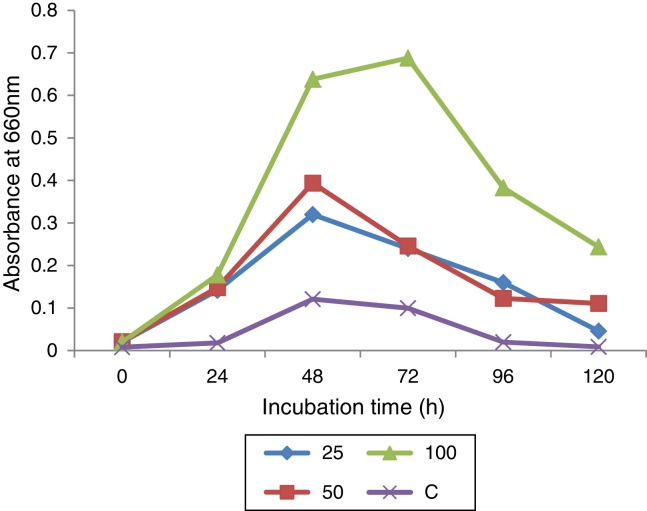
Of 13 larval gut bacterial isolates, only B. cereus – KC985225 was able to grow in the agar medium containing 100 ppm indoxacarb. B. cereus grown on minimal media lacking carbon and supplemented with indoxacarb showed enhanced growth rate, at 660 nm, whereas B. cereus grown on minimal media without indoxacarb showed retarded growth (Fig. 6), which confirms that growth was limited by depletion of indoxacarb as an available carbon source. The maximum growth was observed in medium supplemented with 100 ppm indoxacarb.
Fig. 6.
Effect of indoxacarb on Bacillus cereus growth.
Quantification of esterase activity during indoxacarb degradation by B. cereus
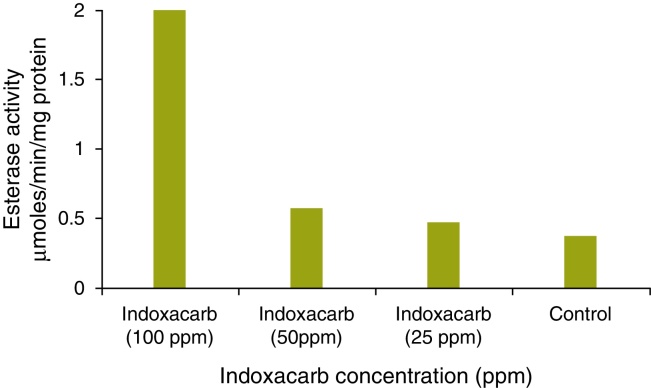
The esterase activity in the test sample (minimal media + indoxacarb + B. cereus) was 1.87, 0.46 and 0.35 μmol/min/mg protein for 100, 50 and 25 ppm respectively, and the same in the control (minimal media + B. cereus) was 0.26 μmol/min/mg protein (Fig. 7).
Fig. 7.
Quantification of esterases from Bacillus cereus in minimal media with indoxacarb.
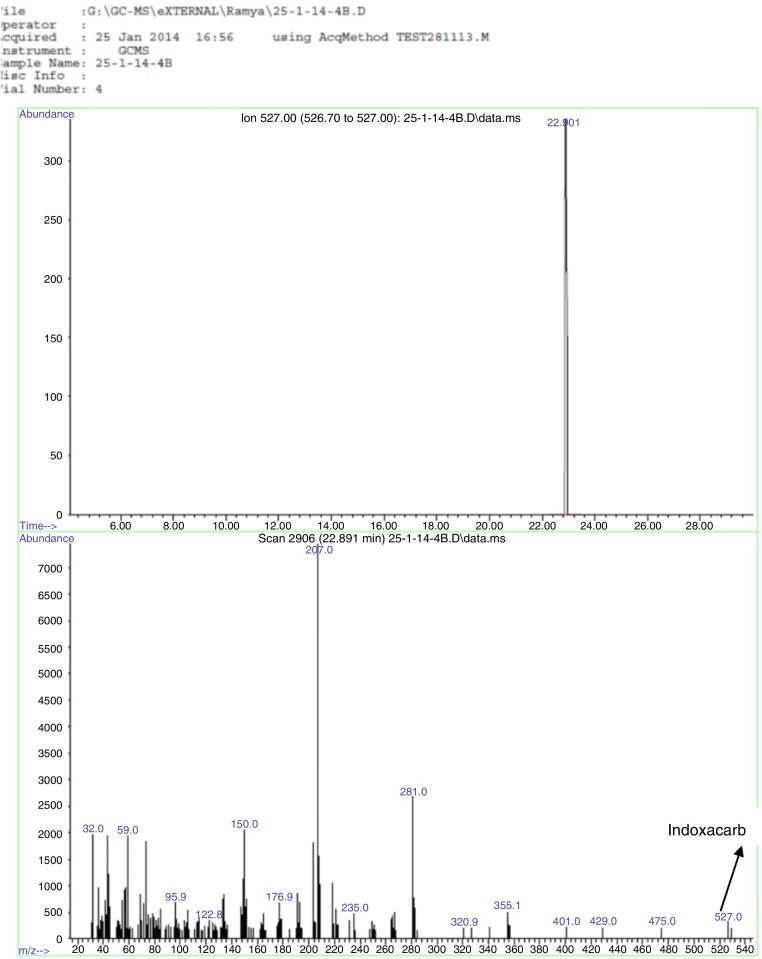
GC–MS analysis
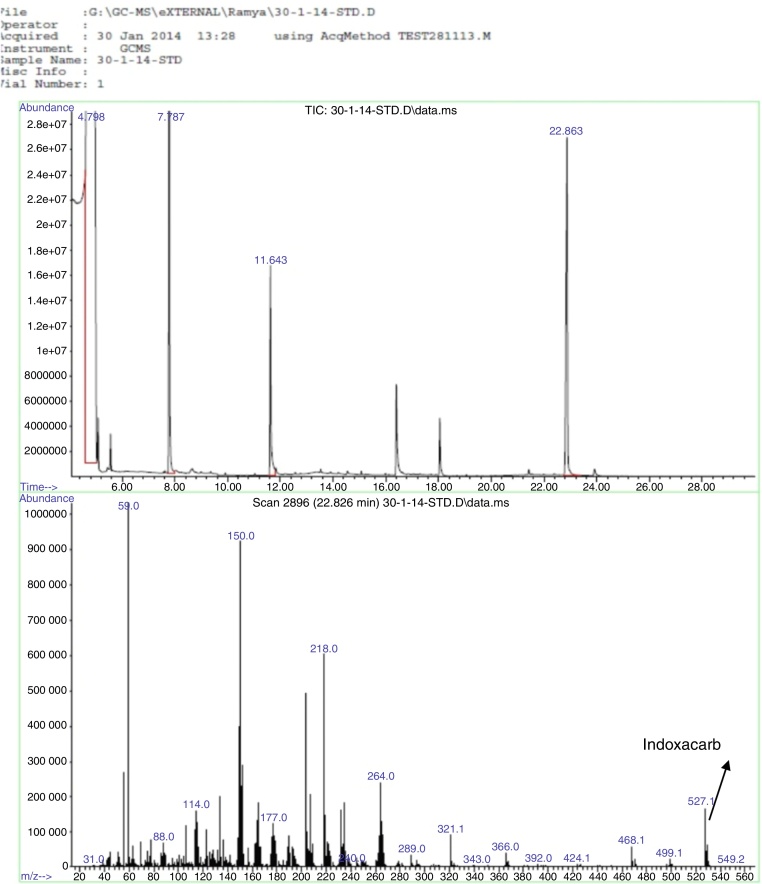
The sample with B. cereus + indoxacarb + minimal media showed a single peak at retention time 22.9 min in the chromatogram, and MS analysis determined that it had an identical spectrum of indoxacarb (Fig. 8). The same spectrum of indoxacarb was observed at retention time 22.86 min when analyzing the control sample without B. cereus (Fig. 9), but the total area of the indoxacarb (Fig. 10) peak was reduced in the test sample. B. cereus showed partial degradation of indoxacarb (20%).
Fig. 8.
Chromatogram and mass spectra of indoxacarb degradation in control sample.
Fig. 9.
Chromatogram and mass spectrum of indoxacarb degradation by B. cereus in test sample.
Fig. 10.
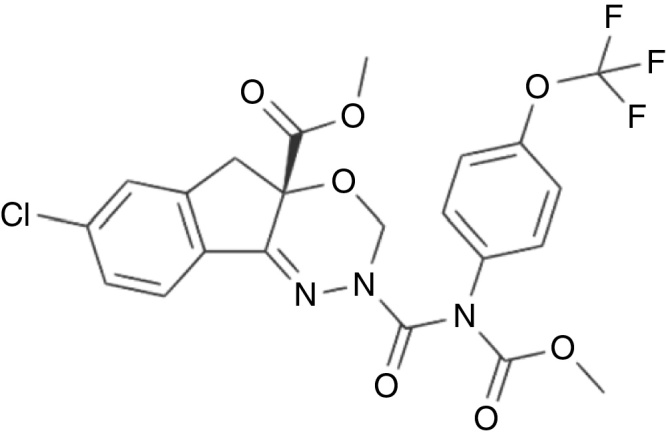
Structure of indoxacarb.
Discussion
In the present study, we isolated various bacteria associated with different field populations of DBM, which led to the identification of 13 culturable isolates from larvae gut and 12 from adult gut. We found E. mundtii, B. cereus, E. asburiae, Enterobacter sp., Pseudomonas sp., P. aeruginosa and Bacillus sp. as the major bacterial isolates from larval and adult gut. Bacillus sp., Pseudomonas sp. has been isolated from the gut of the Discladispa armigera (Olivier).24 The Pseudomonas sp. and Stenotrophomonas sp. were reported in the larval gut of DBM.6 Presence of P. aeruginosa, Pseudomonas sp. and Enterococcus sp. was reported in the larval gut of H. armigera.25 Therefore, Bacillus sp., Psuedomonas and Enterobacter occur commonly in different groups of insects.
At the genus level, Enterobacter (38.46%) and Pseudomonas (25%) dominated in larval and adult DBM gut, respectively. The adult gut showed more diverse bacterial communities as compared with larvae. The gut during the larvae to adult transition is believed to undergo a sterilization process and adults recruit new microbiota.26 B. cereus was exclusively retained by both larvae and adults in the PX-6 strain. The differences in midgut bacterial composition between larvae and adults were large and could be due to different food habits and environmental conditions. Here, the larval gut bacterial associations are more important because they involve detoxification of insecticide and may confer resistance to DBM.
Bacteria associated with Lepidoptera are little known, and studies related to gut microbiota suggest that microorganisms play a role in host digestion, provide essential nutrients and help in detoxification of pesticides.27 Microorganisms are known to metabolize several insecticides. Insecticides with aliphatic, aromatic and heterocyclic compounds were completely mineralized by various microbes. Catabolism is a type of degradation whereby an organic chemical is completely degraded, and the energy or nutrient gained used for the cell growth and co-metabolism is another method of degradation with no net benefit to the organism.28 Three classes of bacterial enzymes are known to degrade organophosphate compounds, namely, phosphotriesterases (EC 3.1.8.1), methyl parathion hydrolases (MPH, EC 3.1.8.1) and organophosphorus acid anhydrolases (EC 3.1.8.2).29 Our study revealed esterase in the gut bacteria of DBM larvae. The role of insect gut bacterial esterases in degradation of insecticides has not been studied extensively; however, the role played by soil bacterial esterases in insecticide degradation has been reported.30
Native PAGE is an important technique in the study of bacterial esterases. The characteristics of each esterase isoenzyme can be determined by the use of specific inhibitors. Carboxylesterases (EC 3.1.1.1) from the thermophillic bacterium Alicyclobacillus tengchongensis completely degraded malathion.31 In our study, esterases (EC 3.1.1) from B. cereus degraded indoxacarb up to 20%. From the preliminary characterization of insect esterases from DBM, a high level of resistance was found associated with slow-moving esterases, which are more important in resistance monitoring than fast-moving esterases.32 In the present study, among 25 bacterial strains tested, 15 showed esterase bands, whereas 19 showed esterase activity when quantified spectrophotometrically. All 15 strains showed slow-moving esterase and were characterized as carboxylesterases, except in one strain. Therefore, the presence of slow-moving carboxylesterase in the bacterial strains may play a role in insecticide detoxification and may confer resistance to DBM. Studies of the use of microbial carboxylesterase for insecticide degradation are limited. Indoxacarb is one of the most commonly used insecticides belonging to the oxadiazine group worldwide and until now, we lack published information on microbial degradation of indoxacarb. Our study reveals that bacteria (B. cereus) in DBM could utilize indoxacarb for metabolism and growth. GC–MS analysis of the extracts of the minimal media study with B. cereus revealed its indoxacarb-degrading capability. B. cereus was also found to degrade fenvalerate, cypermethrin, and chlorpyrifos in soil.30, 33, 34 In the present study, B. cereus may have degraded indoxacarb by enhanced esterase activity. Quantification of esterase in the minimal media clearly showed the role of esterase in degradation of indoxacarb. From the results of this study, we cannot predict whether this hydrolysis results in complete mineralization or if the resulting metabolite remains in the medium. Bacteria isolated from the insect gut exposed to insecticides generate enzymes, thereby resulting in a new metabolic pathway of insecticide degradation. Our study revealed the role of esterase in the partial degradation of indoxacarb. We have insufficient evidence to state the symbiotic role of these bacteria with DBM. The present results open an avenue for the identification of novel enzyme carboxylesterase and its role in the degradation of indoxacarb. Furthermore, intensive work on understanding the pathways and amino acids involved may lead to the synthesis of specific insecticides with a new mode of action for the management of DBM.
Conclusions
We isolated 13 and 12 bacterial isolates from larval and adult gut of DBM, respectively. The gut bacterial association differed between adults and larvae. In larval gut isolates, gammaproteobacteria was the most abundant (76%), followed by Bacilli (15.4%). Esterase bands were increased in 15 bacterial strains. B. cereus degraded indoxacarb up to 20%, which showed its ability to utilize indoxacarb for metabolism and growth. Besides the insect esterases, bacterial carboxylesterase may aid in the degradation of insecticides in DBM.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgement
This is a part of Ph.D. work of the first author.
Associate Editor: Valeria Maia de Oliveira
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bjm.2016.01.012.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Shelton A.M. The management of diamondback moth and other crucifer pests. Proceedings of the Fourth International Workshop on Diamondback Moth; Melbourne, Australia; 2004. pp. 3–8. [Google Scholar]
- 2.Zalucki M.P., Shabbir A., Silva R., Adamson D., Shu-Sheng L. Estimating the economic cost of one of the world's major insect pests, Plutella xylostella (Lepidoptera: Plutellidae): just how long is a piece of string? J Econ Entomol. 2012;105:1115–1129. doi: 10.1603/EC12107. [DOI] [PubMed] [Google Scholar]
- 3.Furlong M.J., Wright D.J., Dosdall L.M. Diamondback moth ecology and management: problems, progress and prospects. Annu Rev Entomol. 2013;58:517–541. doi: 10.1146/annurev-ento-120811-153605. [DOI] [PubMed] [Google Scholar]
- 4.Sandur S. Consultant Report for the Centre for Environmental Stress and Adaptation Research. La Trobe University, Victoria; Australia: 2004. Implications of diamondback moth control for Indian farmers; p. 31. [Google Scholar]
- 5.Talekar N.S., Shelton A.M. Biology ecology and management of the diamondback moth. Annu Rev Entomol. 1993;38:275–301. [Google Scholar]
- 6.Indiraghandhi P., Anandham R., Madhaiyan M., Poonguzhali S., Lim G.H., Saravanan V.S. Cultivable bacteria associated with larval gut of prothiofos-resistant, prothiofos susceptible and field caught populations of diamondback moth, Plutella xylostella and their potential for anatagonism towards entomopathogenic fungi and host insect nutrition. J Appl Microbiol. 2007;103:2664–2675. doi: 10.1111/j.1365-2672.2007.03506.x. [DOI] [PubMed] [Google Scholar]
- 7.Douglas A.E. The microbial dimension in insect nutritional ecology. Funct Ecol. 2009;23:38–47. [Google Scholar]
- 8.Moran N.A. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc Natl Acad Sci U S A. 2007;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dillon R.J., Dillon V.M. The gut microbiota of insects: nonpathogenic interactions. Annu Rev Entomol. 2003;49:71–92. doi: 10.1146/annurev.ento.49.061802.123416. [DOI] [PubMed] [Google Scholar]
- 10.Nicholson J.K., Holmes E., Kinross J. Host–gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 11.Ryu J.H., Kim S.H., Lee H.Y. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- 12.Storelli G., Defaye A., Erkosar B., Hols P., Royet J., Leulier F. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 2011;14:403–414. doi: 10.1016/j.cmet.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 13.Dillon R.J., Dillon V.M. The gut bacteria of insects: nonpathogenic interactions. Annu Rev Entomol. 2004;49:71–92. doi: 10.1146/annurev.ento.49.061802.123416. [DOI] [PubMed] [Google Scholar]
- 14.Frederick B.A., Caesar A.J. Analysis of bacterial communities associated with insect biological control agents using molecular techniques. In: Spencer N.R., editor. Proceedings of the X International Symposium on Biological Control of Weeds. Montana State University, Bozeman; Montana, USA: 2000. pp. 261–267. [Google Scholar]
- 15.Gunther A.F. Residues of pesticides and other contaminants in the total environment. Residue Rev. 1974:51. [PubMed] [Google Scholar]
- 16.Bomscheur T.U. Microbial carboxylesterases: classification, properties and application in biocatalysis. FEMS Microbiol Rev. 2006;26:73–81. doi: 10.1111/j.1574-6976.2002.tb00599.x. [DOI] [PubMed] [Google Scholar]
- 17.Gebbardi K., Schimana J., Muller J., Krantal P., Zeeck A., Vater I. Screening for biologically active metabolites with endosymbiotic bacilli isolated from arthropods. FEMS Microbiol Lett. 2001;217:199–205. doi: 10.1111/j.1574-6968.2002.tb11475.x. [DOI] [PubMed] [Google Scholar]
- 18.Syn C.K., Swarup S. A scalable protocol for the isolation of large-sized genomic DNA within an hour from several bacteria. Anal Biochem. 2000;278:86–90. doi: 10.1006/abio.1999.4410. [DOI] [PubMed] [Google Scholar]
- 19.Tamura K., Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 20.Miller R.B., Karan R.C. A rapid spectrophotometric method for the determination of esterase activity. J Biochem Biophys Methods. 1980;3:345–354. doi: 10.1016/0165-022x(80)90043-3. [DOI] [PubMed] [Google Scholar]
- 21.Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with Folin phenol reagent. J Biol Chem. 1951;93:265–275. [PubMed] [Google Scholar]
- 22.Sousa-Polezzi R.D., Bicudo H. Genetic variation along time in a Brazilian population of Aedes aegypti (Diptera: Culicidae) detected by changes in the esterase patterns. Genetica. 2005;125:43–53. doi: 10.1007/s10709-005-4915-5. [DOI] [PubMed] [Google Scholar]
- 23.Eman M.R. Esterase activity and detection of carboxylesterase and phosphotriesterase in female desert locust Schistocerca gregaria (Forskal) in relation to tissues and ages. Egypt Acad J Biol Sci. 2008;1:135–143. [Google Scholar]
- 24.Thakur D., Bhuyan M., Majumdar S. Isolation, characterization, in-vitro antibiotic susceptibility and pesticide tolerance of gut bacteria from rice hispa, Dicladispa armigea (Olivier) Indian J Microbiol. 2005;45:217–221. [Google Scholar]
- 25.Madhusudan S., Jalali S.K., Venkatesan T., Lalitha Y., Prasannasrinivas R. 16S rRNA gene based identification of gut bacteria from laboratory and wild larvae of Helicoverpa armigera (Lepidoptera: Noctuidae) from tomato farm. The Bioscan. 2011;6:175–183. [Google Scholar]
- 26.Asha R., Anil S., Raman R., Tridibesh A., Raj K.B. Bacterial diversity analysis of larvae and adult midgut microflora using culture-dependent and culture-independent methods in lab-reared and field-collected Anopheles stephensi – an Asian malarial vector. BMC Microbiol. 2009;9:96. doi: 10.1186/1471-2180-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Broderick N.A., Raffa K.F., Goodman R.M., Handelsman J. Census of bacterial community of gypsy moth larval mid gut by using culturing and culture independent methods. Appl Environ Microbiol. 2004;70:290–300. doi: 10.1128/AEM.70.1.293-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Racke K.D. Environmental fate of chlorpyrifos. Rev Environ Contam Toxicol. 1993;131:1–151. doi: 10.1007/978-1-4612-4362-5_1. [DOI] [PubMed] [Google Scholar]
- 29.Munnecke D.M., Hsieh D.P. Pathways of microbial metabolism of parathion. Appl Environ Microbiol. 1976;31:63–69. doi: 10.1128/aem.31.1.63-69.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selvam A., Gnana D., Thatheyu A.J., Vidhya R. Biodegradation of the synthetic pyrethroid, fenvalerate by Bacillus cereus Mtcc 1305. World J Environ Eng. 2013;2:21–26. [Google Scholar]
- 31.Zhenrong X., Bo X., Junmei D., Lingyun L., Junjun L., Zunxi H. Heterologous expression and characterization of a malathion-hydrolysing carboxylesterase from a thermophilic bacterium, Alicyclobacillus tengchongensis. Biotechnol Lett. 2013;35:1283–1289. doi: 10.1007/s10529-013-1195-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maa W.C., Lee S., Guh S.H., Wang C.M., Hou J.W. Esterase zymograms as an assay for detection of population of diamondback moth. In: Talekar N.S., editor. Proceedings of the 2nd International Workshop. Asian Vegetable Research and Development Center; Shanhua, Taiwan: 1990. [Google Scholar]
- 33.Shaohua C., Jianjun L., Meiying L., Peng G., Huasheng H. Enhancement of cypermethrin degradation by a co-culture of Bacillus cereus ZH-3 and Streptomyces aureus HP-S-01. Bioresource Technol. 2012;110:97–104. doi: 10.1016/j.biortech.2012.01.106. [DOI] [PubMed] [Google Scholar]
- 34.Zhiyuan L., Xin C., Yi S., Zhen C. Bacterial degradation of chloropyrifos by Bacillus cereus. Adv Mater Res. 2012:356–360. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.