Abstract
Background and Purpose
Studies of brain tumors have identified altered tissue metabolism and water diffusion in MRI normal appearing tissue regions. In this retrospective study the relationship of these imaging measures with tumor grade in gliomas was investigated.
Methods
MR spectroscopic imaging of whole brain and mean diffusivity (MD) measurements were obtained in subjects with untreated glioma and from normal control subjects. Mean metabolite values for N-acetylaspartate (NAA), total creatine (Cre) and total choline (Cho) were obtained in grey- and white-matter regions for the hemisphere contralateral to the tumor location, and MD values were obtained from contralateral normal-appearing white matter. Analyses tested for differences in mean values between subject groups while accounting for age.
Results
Analysis demonstrated increased NAA/Cre and MD, and decreased Cho/NAA for all tumor grades relative to control values. Differences between tumor grades were also observed for NAA, NAA/Cre, and Cho/NAA. Abnormal values of water diffusion were also observed, but with only a weak association between alterations in diffusion and tissue metabolites.
Conclusions
This study supports previous observations of altered tissue metabolism and water diffusion in normal-appearing white matter while additionally finding differences of metabolite values in grey-matter and an association with tumor grade.
Keywords: 1H MRSI, Spectroscopic Imaging, MRI diffusion, brain, normal tissue, glioma
INTRODUCTION
Magnetic resonance spectroscopy (MRS) measurements of altered tissue metabolism in brain cancers provide a valuable biomarker that, together with structural MRI methods, can be used for diagnostic and treatment monitoring applications. In addition to detecting abnormal values within the tumor and infiltrated peritumoral regions, previous MRS studies have also reported that metabolic changes occur within tissue regions that appear normal with conventional MRI sequences. Findings include reductions of N-Acetylaspartate (NAA), a neuronal marker, and ratios of NAA to the combined signals from creatine and phosphocreatine (Cre) and mobile choline compounds (Cho) [1-4]; although these findings for NAA and Cho were not reproduced in another study of a group of subjects with glioblastoma multiforme that additionally found increased myo-Inositol and glutamine [5]. While several of these previous reports made measurements in normal-appearing white matter (NAWM) regions adjacent to the tumor, abnormal values have also been observed using single-voxel measurements in regions some distance away from the tumor [4, 5], suggesting that the observed metabolic changes are widespread throughout the brain. These previous reports have investigated several tumor types, but did not examine associations of the metabolic alterations with glioma grade.
Previous studies have also reported increases of water diffusivity, measured using diffusion tensor imaging (DTI) acquisitions, in NAWM regions [6], together with an association between whole-brain NAA and the mean diffusivity (MD) of tissue water [1]. Together with the MRS measures these results suggest the presence of a diffuse metabolic depression and edema throughout the brain of patients with a wide variety of brain tumor types, although the mechanisms for such changes remain unknown.
In this retrospective study MRS and DTI measurements were obtained from normal-appearing tissue regions in subjects with histologically-proven gliomas and associations with tumor grade evaluated. MRS data was obtained using a whole-brain MR spectroscopic imaging (MRSI) method that enabled data to be obtained over a wide region as well as separate measurements from grey-matter and white-matter. The associations between the MRS and MD measures were also evaluated.
METHODS
MRSI and DTI data were obtained at 3 T (General Electric Signa HDxt) from a group of 60 subjects with histologically-proven glioma of WHO Grade II, III, or IV, and aged 18 to 69 [7]. Studies were done prior to any treatment and the subjects were unaware of their diagnosis at the time of this initial diagnostic exam. Of these, one with a mixed histology grading was excluded and complete MRSI or DTI was not available for all subjects, resulting in 55 MRSI studies and 52 DTI studies. MRSI data from normal subjects was obtained from an existing database [7] acquired at a different site (Siemens Trio) but with the identical MRSI acquisition sequence, from which 174 studies were selected to cover the same age range as the glioma subject group. To confirm the suitability of the normative data this was compared with results from a group of seven control subjects obtained on the same scanner as the glioma subjects, and aged 19 to 33. A comparison between these two control groups indicated good agreement for the metabolite ratios between both groups with a difference between the means for an age-matched group of subjects of less than one standard deviation. Example results are mean NAA/Cr of 1.69±0.04 and 1.66±0.13 for the subject groups acquired on the Signa and Trio respectively. However, values of the individual metabolite values could not be compared due to differences in the intensity scaling on the different MR scanners. The control data was therefore only included in the analysis of the metabolite ratios. Additional DTI data was obtained from 54 control subjects using the same acquisition sequences. The number of subjects, age range, and gender distribution in each of the study groups are shown in Table 1.
Table 1.
Number of subjects and age ranges used for the analysis.
| Age | ||||
|---|---|---|---|---|
| MRSI Analysis |
Number | Minimum | Maximum | Median |
| Control 1 | 174 | 19 | 68 | 36 |
| Grade II | 20 | 18 | 45 | 33.5 |
| Grade III | 12 | 28 | 55 | 45.5 |
| Grade IV | 23 | 30 | 69 | 47 |
| DTI Analysis | ||||
| Control 2 | 54 | 15 | 60 | 29 |
| Grade II | 19 | 18 | 45 | 34 |
| Grade III | 11 | 28 | 53 | 46 |
| Grade IV | 22 | 30 | 69 | 46.5 |
Subjects underwent a MRI study that included a whole-brain MRSI, DTI, T2-weighted MRI, and pre- and post-contrast T1-weighted MRI. The MRSI acquisition used a volumetric Echo-Planar acquisition that obtained data from a 135-mm slab covering the cerebrum and resulted in an effective voxel volume of approximately 1 mL. The sequence used spin-echo excitation with TE=70 ms and inversion-nulling to suppress subcutaneous lipid signals. Processing was carried out using the MIDAS package [7, 8] and details of the acquisition and processing have been previously reported [8, 9]. An automated spectral analysis procedure resulted in maps of NAA, Cre, Cho, and lactate, from which results for NAA/Cre, Cho/Cre, and Cho/NAA were also derived. Additional maps used in the analysis included the spectral linewidth and the tissue fraction in each SI voxel as described below. The individual metabolite measures were normalized using tissue water content, which was obtained using a water-reference SI that was acquired in an interleaved fashion with the water suppressed metabolite acquisition. The resultant metabolite values were not corrected for relaxation effects and results are expressed in institutional units. Analyses were based on both metabolite ratios and individual metabolite values. The ratio measurements have the advantage of excluding changes of tissue water as a variable and enabled comparisons with previous studies, but since these results remain ambiguous as to which value was altered the individual metabolite values were also examined.
Mean metabolite values for the normal-appearing grey- and white-matter tissue were derived using the following procedure. The tumor and surrounding edema were first manually outlined using the coregistered T1- and T2-weighted MRIs to create a mask that was identified as an “other” tissue category. This mask was incorporated into a tissue segmentation procedure so as to exclude this region from the T1-weighted MRI that was used to derive the maps for CSF, grey-matter (GM), and white-matter (WM) in the normal tissue region. These tissue maps were then resampled to correspond to the image resolution and spatial response function of the MRSI. All metabolite and tissue maps were then spatially transformed into a standard reference space at 2 mm isotropic resolution that was associated with a brain atlas that defined the left and right frontal, temporal, parietal, and occipital lobes [8]. Voxels within each lobar region with a fractional tissue content of GM+WM of greater than 80% were then selected, and a linear regression was carried out for each metabolite measure against the WM fraction to derive the mean value corresponding to 100% GM and WM. The single metabolite measures were also corrected for the CSF fractional volume contribution. Voxels were excluded if the spectral fitting reported a linewidth of greater than 12 Hz or had outlying values based on three times the standard deviation away from the mean value. An initial analysis based on the mean values from all subjects with each tumor grade found no differences in the associations of the metabolite values from each brain lobe within the contralateral hemisphere with tumor grade. Therefore, results from all lobes were combined and the average values for GM and WM within the hemisphere contralateral to the tumor were then obtained. In two subjects (Grade III) where the tumor occupied parts of both hemispheres the results from all unaffected lobes were summed.
All spatial transformations were visually checked to confirm appropriate registration of the interhemispheric fissure. Since the voxel selection procedure ensured that voxels from the tumor region and ventricles were not included and only mean metabolite values over the contralateral hemisphere was obtained, it was not necessary to achieve accurate spatial normalization of individual brain structures, which is problematic when space-occupying lesions are present, and precludes the use of atlas-based analysis using smaller brain regions.
DTI data were acquired using a single-shot echo-planar sequence, with TR=10 s, TE=100 ms, 46 slices of thickness=3mm and interslice gap=0, FOV= 240 mm, image matrix = 128*128, and diffusion weighting b-factor of 1000 s/mm2 applied in 12 directions in addition to the reference measurement with b=0 s/mm2. DTI data was processed using in-house developed software to compute the MD and FA for each voxel. Mean values of the MD and FA were obtained from a region of interest placed in supraventricular white matter in the hemisphere contralateral to the tumor using between 200 and 400 voxels. A region contralateral to the tumor was not used since this would cover different brain regions and different grey- and white-matter distributions.
Analysis of covariance (ANCOVA) was used to test for between-group differences of the metabolite measures in GM and WM and the WM MD of the contralateral hemisphere, while controlling for subject age. For the single metabolite values this was carried out for the three glioma subject groups, while the metabolite ratio and MD measures included the corresponding control group. Additional regression analyses were carried out to test for associations of the imaging measures with tumor volume and subject age, and to examine associations between metabolite and MD measures.
RESULTS
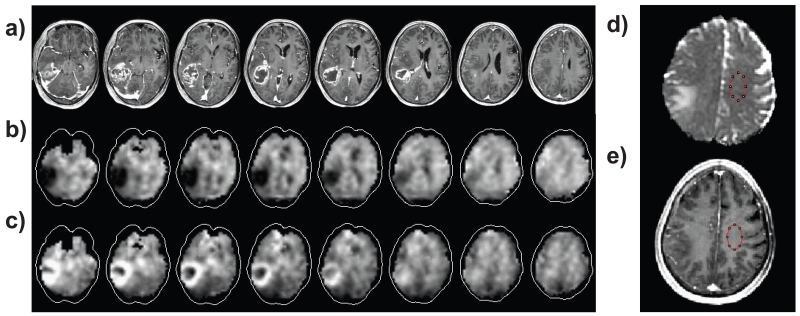
An example MRSI result together with the post-contrast T1-weighted MRI is shown in Figure 1, for the NAA and Cho maps of a 76 year old subject with a Grade IV glioblastoma. Also illustrated in Figs. 1d and 1e is the region selection used for the NAWM MD measurement.
Figure 1.
Example images showing selected slices from the multi-planar MRSI and typical ROI selection used for the MD measurement, with a) post-contrast T1-weighted MRI at slices corresponding to the center of the MRSI slices; b) NAA map; c) Cho map; d) MD map with ROI selection; and e) T1 MRI with the corresponding ROI.
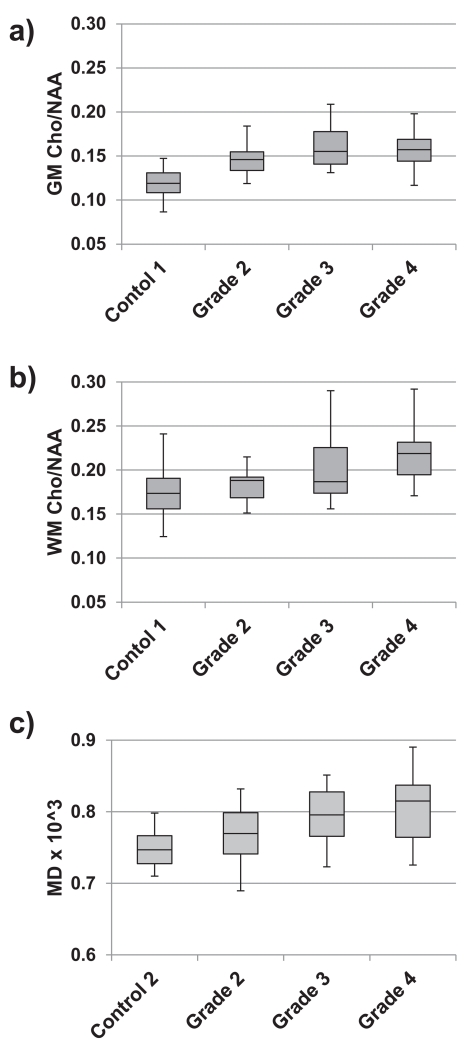
In Figs. 2a and 2b are shown examples of the distributions of Cho/NAA values within each subject group for the NAWM GM and WM. Note that data from all subjects are included, which results in a larger spread of the data values from the control group due to wide range of subject age in this group, notably for WM that has a greater association with age than GM [7]. Shown in Fig. 2c are the corresponding values for MD.
Figure 2.
Example distributions of imaging measures for all subjects within each of the subject groups. Box-and-whisker plots are shown for a) grey-matter Cho/NAA, b) white-matter Cho/NAA, and c) MD. The plots show the minimum, the first quartile, the median, the third quartile, and the maximum value within the group.
Results of the ANCOVA analyses for WM are shown in Table 2 for those measures for which significant differences (for p<0.05) were found. No significant differences were found for Cho or Cre. The corresponding results for GM showed significant decreases of NAA/Cre and increases of Cho/NAA for all tumor grades against control values (p<0.001), but no significant differences between grades for any of the metabolite measures. These results indicate highly significant differences between all tumor groups and controls for NAA/Cre, Cho/NAA, and MD. The absence of significant differences for the individual Cre and Cho measures would suggest that the abnormal metabolite ratio measures are being driven by decreased NAA in all tumor subject groups. Additional findings are significant differences between the grade II and III groups with the grade IV group for NAA, and differences between grade II and grade IV for NAA/Cre and Cho/NAA.
Table 2.
Summary of p-values for the difference in imaging measures in white matter between subject groups. Only measures for which significant differences were noted are shown. Significant difference for p < 0.05 are highlighted in bold. The NAA values are shown in institutional units and MD in mm2/s ×10−3.
| Mean | Between-Group p-Values | ||||
|---|---|---|---|---|---|
| Ctrl | Grade II | Grade II | |||
| NAA | Grade II | 1.680E+04 | |||
| Grade III | 1.685E+04 | - | 1.000 | ||
| Grade IV | 1.451E+04 | - | 0.011 | 0.033 | |
| NAA/Cre | Control 1 | 1.682 | |||
| Grade II | 1.54 | 0.000 | |||
| Grade III | 1.521 | 0.000 | 1.000 | ||
| Grade IV | 1.427 | 0.000 | 0.007 | 0.114 | |
| Cho/NAA | Control 1 | 0.175 | |||
| Grade II | 0.186 | 0.453 | |||
| Grade III | 0.203 | 0.001 | 0.243 | ||
| Grade IV | 0.219 | 0.000 | 0.000 | 0.153 | |
| MD | Control 2 | 0.745 | |||
| Grade II | 0.768 | 0.000 | |||
| Grade III | 0.802 | 0.000 | 0.076 | ||
| Grade IV | 0.804 | 0.000 | 0.084 | 1.000 | |
Since the ANCOVA analysis indicated that Cho/NAA had the strongest associations with tumor grade, the possible associations of this parameter with the total volume of the tumor and edema was examined. Regression analysis of Cho/NAA in contralateral WM and GM with tumor+edema volume showed no significant findings.
Regression analyses of individual metabolite measures with MD indicated no significant associations of WM NAA, Cre, Cho and NAA/Cre with MD, although there was a trend to a negative association of MD with NAA (R2=0.02, p=0.29) and a positive association of MD with Cho (R2=0.01, p=0.38) leading to a significant association of increasing MD with the value of mean WM Cho/NAA with R2=0.11 and p=0.021.
DISCUSSION
This study confirms previous observations of reduced NAA and NAA/Cr, relative to normal control values, in normal appearing WM [3, 4] remote from the location of a tumor in subjects with histologically-proven gliomas, while additionally finding increased Cho/NAA in WM and reduced NAA/Cre and increased Cho/NAA in normal-appearing GM. A new finding of this study is that these metabolic alterations are increased for tumors of a higher grade of malignancy. While these findings are statistically significant for the mean values for each study group there remains considerable overlap between subjects with glioma and normal control values, and between the glioma grades, indicating that the measurements are not sufficiently robust to be considered as a diagnostic marker.
This study also found an association of increasing MD values within NAWM with increasing tumor grade. This finding is consistent with the report of Provenzale et al. [6] that found a 6% increase of MD in contralateral NAWM for a group of 17 grade II and IV glioma subjects relative to normal control values, while this study additionally found MD to be increased by 3.1%, 7.7% and 7.9% for tumor grades II, III, and IV respectively, although the differences between tumor grade were not statistically significant.
This study did not reproduce the observation of Inglese et al. [1] in a group of 10 subjects with glioma grades II to IV that found a strong negative association of whole-brain NAA with increasing NAWM MD values, although the observed association of increasing Cho/NAA with MD is consistent with this previous report. The R2 values found in this study, of 0.01 and 0.11 for NAA and Cho/NAA, indicate a much weaker association than the value of R2 = 0.77 reported by Inglese et al. for whole brain NAA. A likely reason for this difference is that this study excluded the influence of the tumor, edema, and CSF volume contribution on the NAA measurement, whereas that of Inglese et al. did not. This observation is consistent with the findings of Wagnerova et al. [10] that indicate strong associations of NAA and Cho with MD across the tumor, though with no association indicated for voxels in the NAWM immediately adjacent to the tumor. The findings of this study suggest only a weak association between the microstructural changes responsible for increased water diffusivity and altered metabolite levels in the NAWM.
Limitations of this study include that signal normalization for the individual metabolite values used tissue water as an internal reference, with an assumption of normal values for GM and WM. However, the strongest finding, that of Cho/NAA, would not be affected by any alteration of water concentration relative to normal values. An additional limitation is that different control groups were used for the MRS and DTI studies, with the result that the normal association between these measures could not be evaluated. Finally, an alternative factor that could affect brain metabolites in the NAWM could be that these are related to decreased neurological function; however, measures such as neuropsychological evaluations or the Karnofsky Performance Scale were not available for this study.
In conclusion, this study confirms findings from a small number of reports of altered brain metabolism and cellular structure, and extends these findings to tissue type and tumor grade. These findings suggest the presence of widespread metabolic alteration and inflammation that increases with the degree of malignancy of the tumor. Diffuse infiltration of glioma cells into the healthy tissue has been proposed as a mechanism for such changes [1, 4], although appears to be less likely for changes in the hemisphere contralateral to the tumor. A systemic metabolic depression caused by release of compounds from the tumor, including cytokines or tumor-necrosis factor has also been proposed, which is supported by an association between the cytokine TNF-α and decreased NAA reported in lung cancer patients prior to treatment [11].
ACKNOWLEDGEMENTS
This work was supported by NIH grant R01EB000822 and by Indo-US Science & Technology Forum award #20-2009. Bhaswati Roy received financial assistance from University Grant Commission, New Delhi, India. Rishi Awasthi received financial assistance from Indian Council of Medical Research, New Delhi, India.
REFERENCES
- [1].Inglese M, Brown S, Johnson G, Law M, Knopp E, Gonen O. Whole-brain N-acetylaspartate spectroscopy and diffusion tensor imaging in patients with newly diagnosed gliomas: a preliminary study. AJNR Am J Neuroradiol. 2006;27:2137–40. [PMC free article] [PubMed] [Google Scholar]
- [2].Cohen BA, Knopp EA, Rusinek H, Babb JS, Zagzag D, Gonen O. Assessing global invasion of newly diagnosed glial tumors with whole-brain proton MR spectroscopy. AJNR Am J Neuroradiol. 2005;26:2170–7. [PMC free article] [PubMed] [Google Scholar]
- [3].Goebell E, Fiehler J, Ding XQ, Paustenbach S, Nietz S, Heese O, Kucinski T, Hagel C, Westphal M, Zeumer H. Disarrangement of fiber tracts and decline of neuronal density correlate in glioma patients--a combined diffusion tensor imaging and 1H-MR spectroscopy study. AJNR Am J Neuroradiol. 2006;27:1426–31. [PMC free article] [PubMed] [Google Scholar]
- [4].Busch M, Liebenrodt K, Gottfried S, Weiland E, Vollmann W, Mateiescu S, Winter S, Lange S, Sahinbas H, Baier J, van Leeuwen P, Gronemeyer D. Influence of brain tumors on the MR spectra of healthy brain tissue. Magn Reson Med. 2011;65:18–27. doi: 10.1002/mrm.22612. [DOI] [PubMed] [Google Scholar]
- [5].Kallenberg K, Bock HC, Helms G, Jung K, Wrede A, Buhk JH, Giese A, Frahm J, Strik H, Dechent P, Knauth M. Untreated glioblastoma multiforme: increased myo-inositol and glutamine levels in the contralateral cerebral hemisphere at proton MR spectroscopy. Radiology. 2009;253:805–12. doi: 10.1148/radiol.2533071654. [DOI] [PubMed] [Google Scholar]
- [6].Provenzale JM, McGraw P, Mhatre P, Guo AC, Delong D. Peritumoral brain regions in gliomas and meningiomas: investigation with isotropic diffusion-weighted MR imaging and diffusion-tensor MR imaging. Radiology. 2004;232:451–60. doi: 10.1148/radiol.2322030959. [DOI] [PubMed] [Google Scholar]
- [7].Maudsley AA, Domenig C, Govind V, Darkazanli A, Studholme C, Arheart K, Bloomer C. Mapping of brain metabolite distributions by volumetric proton MR spectroscopic imaging (MRSI) Magn Reson Med. 2009;61:548–59. doi: 10.1002/mrm.21875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Maudsley AA, Darkazanli A, Alger JR, Hall LO, Schuff N, Studholme C, Yu Y, Ebel A, Frew A, Goldgof D, Gu Y, Pagare R, Rousseau F, Sivasankaran K, Soher BJ, Weber P, Young K, Zhu X. Comprehensive processing, display and analysis for in vivo MR spectroscopic imaging. NMR Biomed. 2006;19:492–503. doi: 10.1002/nbm.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Roy B, Gupta RK, Maudsley AA, Awasthi R, Sheriff S, Gu M, Husain N, Mohakun S, Benhari S, Pandey CM, Spielman DM, Alger JR. Utility of multiparametric 3T MRI for glioma characterization. Neuroradiology. 2013 doi: 10.1007/s00234-013-1145-x. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wagnerova D, Herynek V, Malucelli A, Dezortova M, Vymazal J, Urgosik D, Syrucek M, Jiru F, Skoch A, Bartos R, Sames M, Hajek M. Quantitative MR imaging and spectroscopy of brain tumours: a step forward? Eur Radiol. 2012;22:2307–18. doi: 10.1007/s00330-012-2502-6. [DOI] [PubMed] [Google Scholar]
- [11].Benveniste H, Zhang S, Reinsel RA, Li H, Lee H, Rebecchi M, Moore W, Johansen C, Rothman DL, Bilfinger TV. Brain metabolomic profiles of lung cancer patients prior to treatment characterized by proton magnetic resonance spectroscopy. Int J Clin Exp Med. 2012;5:154–64. [PMC free article] [PubMed] [Google Scholar]