Abstract
The tumor suppressor PTEN restrains cell migration and invasion by a mechanism that is independent of inhibition of the PI3K pathway and decreased activation of the kinase AKT. PREX2, a widely distributed GEF that activates the GTPase RAC1, binds to and inhibits PTEN. We used mouse embryonic fibroblasts and breast cancer cell lines to show that PTEN suppresses cell migration and invasion by blocking PREX2 activity. In addition to metabolizing the phosphoinositide PIP3, PTEN inhibited PREX2-induced invasion by a mechanism that required the tail domain of PTEN, but not its lipid phosphatase activity. Fluorescent nucleotide exchange assays revealed that PTEN inhibited the GEF activity of PREX2 toward RAC1. PREX2 is a frequently mutated GEF in cancer, and examination of human tumor data showed that PREX2 mutation was associated with high PTEN expression. Therefore, we tested whether cancer-derived somatic PREX2 mutants, which accelerate tumor formation of immortalized melanocytes, were inhibited by PTEN. The three stably expressed, somatic PREX2 cancer mutants that we tested were resistant to PTEN-mediated inhibition of invasion but retained the ability to inhibit the lipid phosphatase activity of PTEN. In vitro analysis showed that PTEN did not block the GEF activity of two PREX2 cancer mutants and had a reduced binding affinity for the third. Thus, PTEN antagonized migration and invasion by restraining PREX2 GEF activity, and PREX2 mutants are likely selected in cancer to escape PTEN-mediated inhibition of invasion.
INTRODUCTION
The tumor suppressor PTEN (phosphatase and tensin homolog) is a lipid phosphatase that attenuates phosphatidylinositol 3-kinase (PI3K) signaling by dephosphorylating phosphatidylinositol-3,4,5-trisphosphate (PIP3). PTEN also has PI3K-independent functions, and many groups have reported that PTEN blocks cell movement and neurite branching by way of an unknown mechanism that is independent of its lipid phosphatase activity (1–5). Inhibition of cell migration and invasion and neurite branching by PTEN is an important, yet poorly understood, feature of its tumor suppressor activity.
Cell motility is largely regulated by the RHO guanosine triphosphatases (GTPases), which include RAC, RHO, and CDC42. This family of proteins links extracellular cues to changes in the actin cytoskeleton (6). RAC1 GTPase is ubiquitously distributed, and active RAC proteins promote membrane ruffling and the formation of lamellipodia at the leading edges of moving cell membranes. RAC1 cycles between an inactive guanosine diphosphate (GDP)–bound and an active GTP-bound state, and proper regulation of its activity is essential for many biological processes, including migration and neurite branching (6). RAC1 activation is controlled by guanine nucleotide exchange factors (GEFs). GEFs activate GTPases by catalyzing the release of GDP so that GTP, which is present at higher intracellular concentrations, can bind the GTPase.
The RAC-GEF PREX2 (phosphatidylinositol-3,4,5-trisphosphate–dependent Rac exchange factor 2) activates cell migration in response to PIP3 and Gβγ signaling (7–10). PREX2 interacts with PTEN and amplifies PI3K signaling by antagonizing PTEN-mediated catabolism of PIP3, which allows it to cooperate with an oncogenic mutant of PIK3CA to transform mammary cells (11). PREX2 is one of the most frequently mutated GEFs in cancer [about 9% of cell lines in the cancer cell line encyclopedia (CCLE) harbor mutations in PREX2] (12, 13), and PREX2 mutants identified in metastatic melanoma accelerate tumor formation of N-RAS–transformed melanocytes in vivo (14). The PREX2 homolog PREX1 is not commonly mutated in cancer, and unlike PREX2, PREX1 does not bind PTEN and does not inhibit PTEN activity (15). However, PREX1 has been implicated in tumor invasion and metastasis, and PREX1 overexpression promotes melanoma metastasis (16). The PREX2 gene, which is located on chromosome 8q13, maps to a region that is subject to recurrent amplification or copy number increases in many cancers, including breast, prostate, and colorectal cancer (17–19). PREX2 is overexpressed in all breast cancer subtypes, and its expression is associated with PTEN expression and activating PIK3CA mutations (11). PREX1, on the other hand, is overexpressed in luminal, but not basal-like, breast cancers (20).
Despite their potential importance in cancer, little is known about how PREX2 and other GEFs are inhibited. Here, we used cell biology, biochemistry, and genetic approaches to show that PTEN suppresses cell movement by blocking PREX2 GEF–catalyzed activation of the GTPase RAC1.
RESULTS
Pten loss–induced increases in MEF cell migration are dependent on Prex2
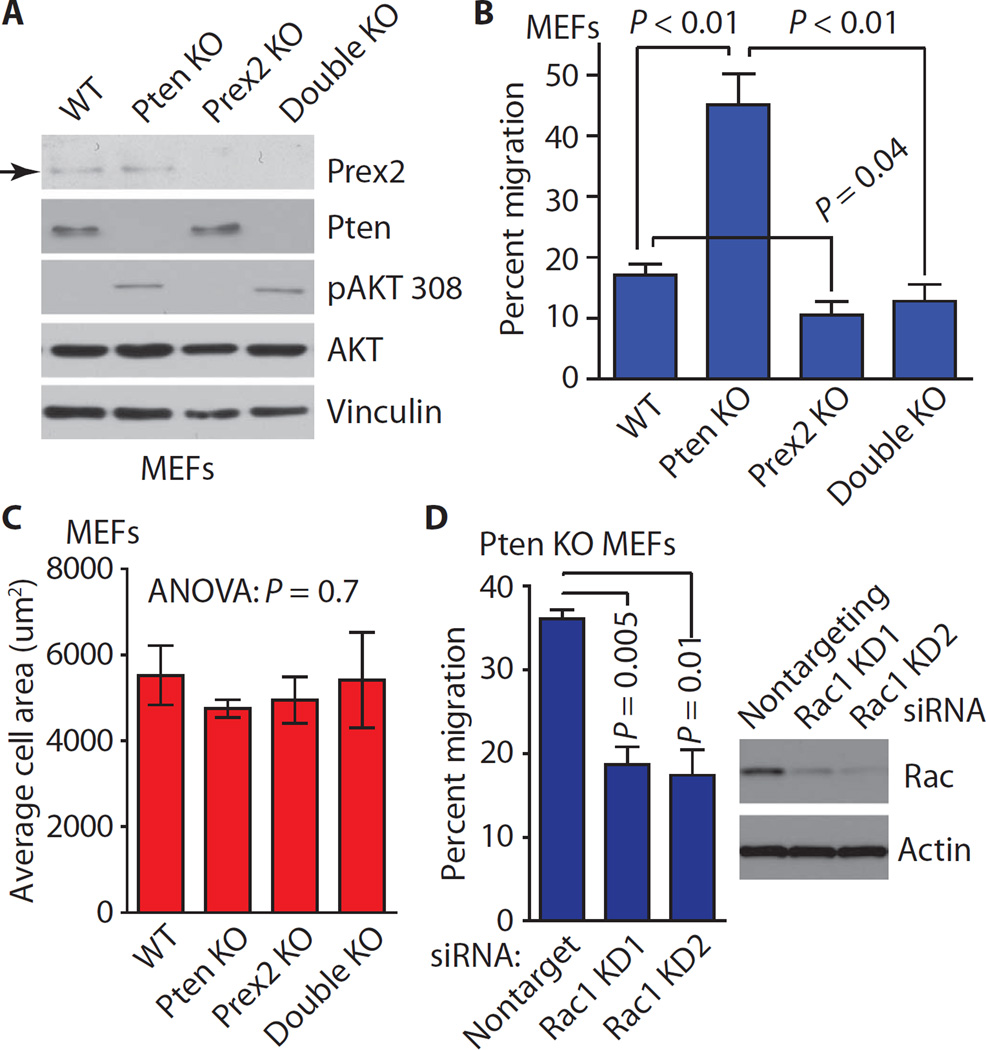
Because PTEN and PREX2 have opposing effects on cell migration and directly interact with each other, we hypothesized that PTEN restrains cell movement by inhibiting PREX2 function. To test whether PTEN and PREX2 act on the same pathway to control migration, we interbred Prex2+/− mice with Ptenflox/flox mice (15, 21) to generate Ptenflox/flox;Prex2+/+ and Ptenflox/flox;Prex2−/− mouse embryonic fibroblasts (MEFs) that were immortalized with dominant negative p53 (22) and infected with an adenovirus expressing either Cre recombinase or green fluorescent protein (GFP). Pten+/+Prex2+/+ and Pten−/−Prex2−/− MEFs will be referred to as “wild-type” and “double KO,” whereas Pten+/+Prex2−/− and Pten−/−Prex2+/+ MEFs will be referred to as “Prex2 KO” and “Pten KO.” As expected, Prex2 was present in wild-type and Pten KO MEFs but not in Prex2 KO or double KO MEFs (Fig. 1A). Under steady-state growth conditions, Pten KO and double KO MEFs had similar amounts of phospho-AKT (Thr308) (Fig. 1A). We next compared the ability of these MEFs to migrate toward a platelet-derived growth factor (PDGF) gradient (23). Pten KO MEFs displayed greatly increased migration relative to wild-type MEFs, whereas Prex2 KO MEFs exhibited reduced migration (Fig. 1B). Double KO MEFs did not display enhanced migration despite Pten loss and did not migrate significantly more than Prex2 KO MEFs (Fig. 1B). No differences in spreading were observed among the MEFs (Fig. 1C), suggesting that the migration differences are not due to changes in their abilities to attach to the substratum. In addition, Pten KO MEFs displayed significantly reduced migration after Rac1 knockdown (Fig. 1D). The observation that Pten KO–induced migration does not occur in the absence of Prex2 and can be suppressed by knockdown of Rac1 demonstrates that Pten and Prex2 operate on the same pathway to control cell migration and that Pten acts through Rac1 to suppress MEF movement. These findings suggest that the Pten loss–induced increases in MEF cell migration are primarily, if not solely, dependent on the presence of Prex2.
Fig. 1. Pten KO–induced migration requires PREX2.
(A) Immunoblot of PI3K pathway components in immortalized wild-type (WT) or KO MEFs of indicated genotypes under steady-state growth conditions. Immunoblots are representative of three experiments. pAKT 308, phospho-AKT (Thr308). (B) MEFs were subjected to migration assays using collagen-coated membranes and a PDGF gradient. Error bars represent the mean ± SEM of three experiments using six pairs of MEFs from three different litters. (C) MEFs from each genotype were plated in serum-free medium, and cell spreading (cell area) was measured over three experiments. Spreading was measured for at least 50 MEFs from each genotype. (D) Pten KO MEFs were transfected with siRNAs against Rac1 (KD1 and KD2) or nontargeting siRNA and subjected to migration assays. Error bars represent the mean ± SEM of three experiments. All P values were calculated using two-tailed t tests. Experiments were performed using early-passage immortalized MEFs.
PTEN restrains invasion through PREX2 in breast cancer cells
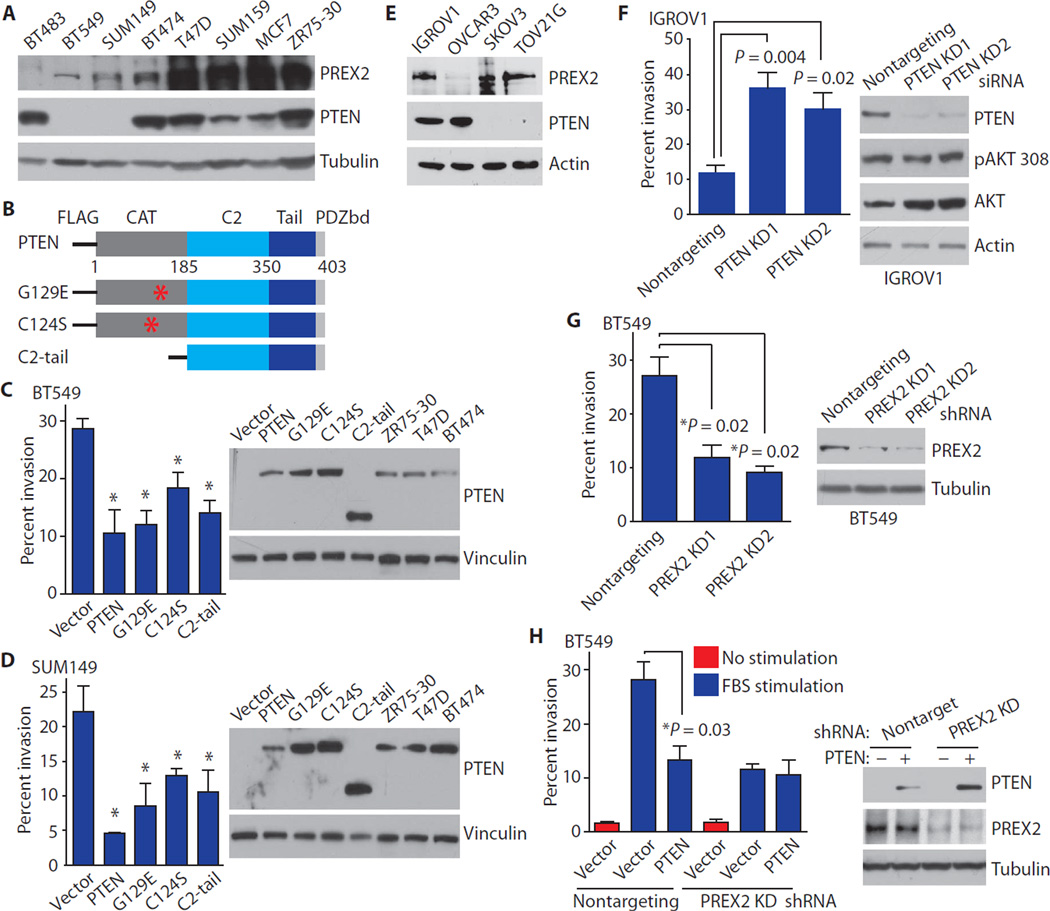
Having shown that Pten loss–induced migration requires Prex2 in MEFs, we explored whether a similar relationship exists between PTEN and PREX2 in breast cancer. PTEN loss occurs frequently in breast cancer (13) and is associated with decreased survival and increased metastasis (fig. S1, A and B). PREX2 mRNA is expressed in all breast cancer subtypes (fig. S1, C and D), and PREX2 protein is also generally present (fig. S1E) (24). Together, these observations raised the possibility that PREX2 stimulates cell migration and invasion in the setting of PTEN loss. Thus, we set out to test whether PTEN inhibits cell invasion through PREX2 in breast cancer cells.
BT549 and SUM149 breast cancer cells have varying amounts of PREX2 but do not have PTEN protein (Fig. 2A) (25, 26). We first tested whether reexpression of PTEN could suppress invasion in BT549 and SUM149 cells. Missense PTEN mutants that lack lipid phosphatase activity can inhibit migration and invasion (3, 4), and the PTEN C2 and tail domains are sufficient to block migration in glioma cells (1). We therefore tested whether expression of wild-type PTEN, G129E (a PTEN mutant without lipid phosphatase activity), PTEN C124S (a PTEN mutant without protein or lipid phosphatase activity), or the isolated PTEN C2-tail region (Fig. 2B) could decrease invasion in breast cancer cells. Wild-type PTEN, G129E, C124S, and C2-tail all decreased the ability of BT549 and SUM149 cells to invade toward a gradient of fetal bovine serum (FBS) (Fig. 2, C and D).
Fig. 2. PTEN does not suppress invasion in the absence of PREX2.
(A) PREX2 and PTEN proteins in breast cancer cell lines. (B) PTEN consists of a catalytic phosphatase domain (CAT), a C2 domain, and a tail with a PDZ binding domain. (C and D) WT PTEN, G129E, C124S, and C2-tail suppress invasion in (C) BT549 and (D) SUM149 cells. Cells were transfected and used in invasion assays with an FBS gradient. (E) PREX2 protein in ovarian cancer cell lines. (F) Knockdown (KD) of PTEN increases invasion in IGROV1 cells. (G) Knockdown of PREX2 reduces invasion in BT549 cells. (H) PTEN does not suppress invasion after PREX2 knockdown. BT549 cells stably expressing shRNA against PREX2 or nontargeting shRNA were transfected as indicated. Cells invaded toward an FBS gradient (blue) or no gradient (red). Error bars in (C), (D), and (F) to (H) represent the mean ± SEM of three experiments. P values were calculated using two-tailed t tests. Immunoblots in (A) and (C) to (H) are representative of at least two experiments.
Having shown that reexpression of catalytically dead PTEN suppresses invasion in PTEN null cancer cells, we tested whether cells endogenously expressing a PTEN mutant that lacks lipid phosphatase activity would be more invasive after PTEN knockdown. Indeed, knockdown of PTEN using two different small interfering RNAs (siRNAs) greatly increased the invasiveness of IGROV1 ovarian cancer cells, which are PREX2 wild-type and harbor an inactivating missense mutation in PTEN (Y155C) (Fig. 2, E and F) (12, 13, 27). The PTEN Y155C mutation in IGROV1 is heterozygous; however, this cell line also harbors a frameshift mutation in PTEN (V317fs) (13), which likely destabilizes PTEN. Knockdown of PTEN in IGROV1 did not result in increased phosphorylation of AKT at Thr308 (Fig. 2F), suggesting that the only intact PTEN protein in these cells is the phosphatase-dead Y155C mutant.
We next set out to test whether PTEN restrains invasion through PREX2 in breast cancer cells. We first depleted PREX2 from BT549 cells using short hairpin RNA (shRNA) and then overexpressed PTEN. PREX2 knockdown by two different shRNAs greatly reduced invasion (Fig. 2G). Expression of wild-type PTEN suppressed invasion in cells expressing non-targeting shRNA (Fig. 2H), but PTEN did not suppress invasion in cells in which PREX2 protein had been knocked down by shRNA (Fig. 2H).
The C2-tail region of PTEN binds to PREX2 and is sufficient for antagonism of PREX2-driven invasion
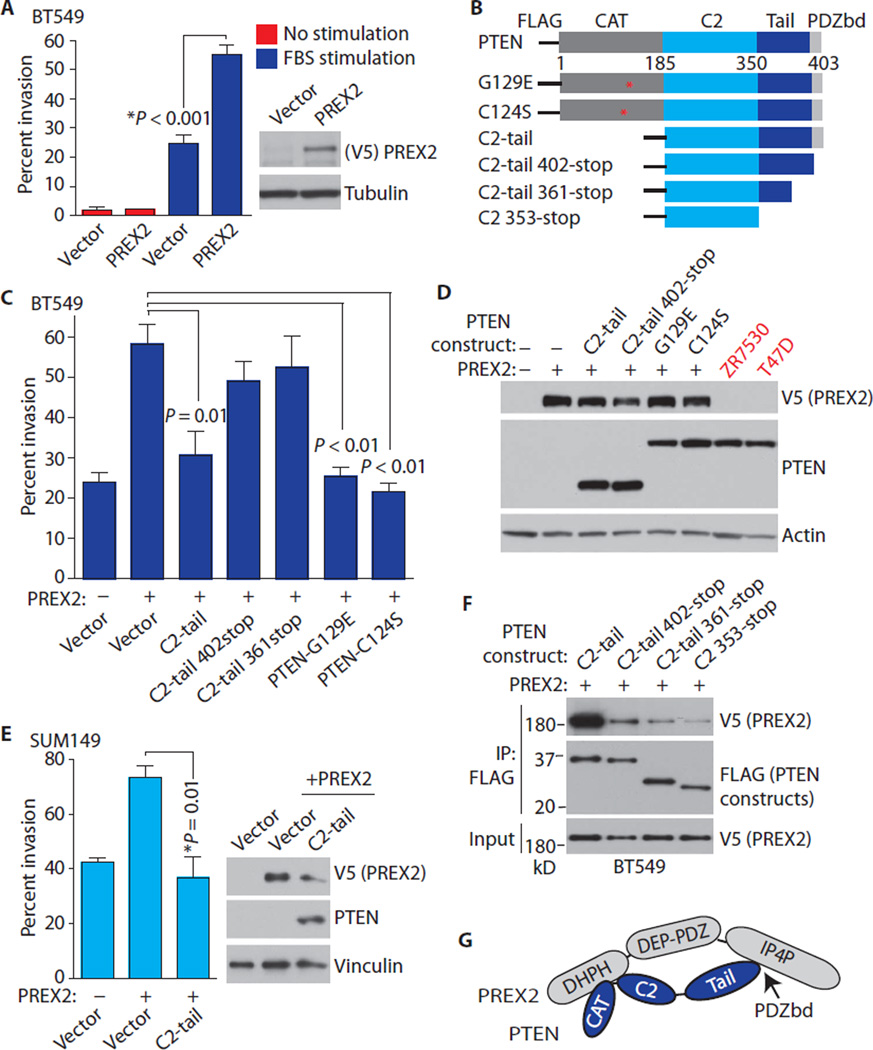
Because PREX2 knockdown suppresses invasion, we tested whether PREX2 overexpression, which frequently occurs in human breast cancer (fig. S1, C and E), stimulates invasion in breast cancer cells. Overexpression of PREX2 increased invasion in BT549 cells in response to an FBS gradient, but not in unstimulated cells (Fig. 3A). PREX2 overexpression also stimulated invasion in SUM149 cells (fig. S2A), albeit to a lesser extent than in BT549 cells, potentially because of the higher abundance of PREX2 in this cell line (Fig. 2A).
Fig. 3. PTEN’s intact PDZ binding domain is required for antagonism of PREX2-driven invasion, and the PTEN C2-tail is sufficient to suppress invasion.
(A) PREX2 drives invasion. BT549 cells were transfected as indicated and subjected to invasion assays using an FBS gradient or no gradient. (B) PTEN constructs used in invasion assays. (C) PTEN C2-tail, G129E, and C124S block PREX2-driven invasion. BT549 cells were transfected as indicated, and invasion experiments were performed using an FBS gradient. (D) Immunoblots showing PREX2 and PTEN from cells used in invasion assays. Lysates from breast cancer cell lines with endogenous PTEN are shown. (E) C2-tail antagonizesPREX2-driven invasion in SUM149 cells. SUM149 cells were transfectedas indicated, and invasion experiments were performed. (F) Coimmunoprecipitation of PTEN constructs and PREX2. BT549 cells were transfected with V5-PREX2 along with the indicated FLAG-PTEN constructs. (G) Schematic showing the binding of the PTEN PDZ binding domain to the IP4P domain of PREX2 and the binding of the C2 domain of PTEN to the Dbl homology–pleckstrin homology (DHPH) domain of PREX2. Error bars in (A), (C), and (E) represent the mean ± SEM of three experiments. P values were calculated using two-tailed t tests. Immunoblots in (D) to (F) are representative of two experiments.
Having observed that transfected PREX2 could stimulate invasion in breast cancer cells, we set out to determine the domains of PTEN that might be required for antagonism of PREX2-stimulated invasion. PTEN contains a catalytic N-terminal phosphatase domain, a lipid-binding C2 domain, and a C-terminal tail that contains a PDZ-interacting motif (Fig. 3B). We therefore cotransfected PTEN deletion constructs (Fig. 3B) along with full-length PREX2 and performed immunoprecipitation experiments and invasion studies. The isolated PTEN C2-tail, which has an intact PDZ binding domain, suppressed PREX2-driven invasion in BT549 breast cancer cells to a similar degree as PTEN G129E (Fig. 3, C and D, and fig. S2B) without affecting AKT signaling (fig. S2C). The PTEN C2-tail also blocked PREX2-driven invasion in SUM149 breast cancer cells (Fig. 3E). In contrast, PTEN constructs that lacked an intact PDZ binding domain (C2-tail 402-stop and C2-tail 361-stop) did not antagonize PREX2-driven invasion (Fig. 3, C and D). These results agreed with the data from our binding experiments in BT549 cells, which showed that the PTEN C2-tail bound to PREX2 more strongly than did C2-tail 402-stop and C2-tail 361-stop (Fig. 3F). We next tested whether the PTEN tail, including the PDZ domain, is sufficient for inhibition of invasion. Because the isolated PTEN tail is unstable, we used a GFP-tagged tail construct (amino acids 353 to 403). The GFP-tail suppressed PREX2-driven invasion in BT549 and SUM149 cells, suggesting that the PTEN tail is sufficient for antagonism of PREX2-driven invasion (fig. S2, D and E).
Our binding data are in line with previously published data showing that PTEN interacts with PREX2 through two interfaces in human embryonic kidney (HEK) 293 cells: the PDZ binding domain within the PTEN tail binds strongly to the IP4P domain of PREX2, and the catalytic and C2 domains of PTEN interact with the PH domain of PREX2, albeit more weakly (Fig. 3G) (15). PTEN switches between open and closed states, and dephosphorylation of residues within the PTEN tail promotes an open PTEN conformation that enables association with the membrane and promotes phosphatase activity (28). Because the PTEN C2-tail bound to PREX2 more strongly than did G129E in unstimulated transfected cells (fig. S2F), it is likely that C2-tail, but not G129E, is in an open PTEN state under these conditions. However, PTEN G129E inhibits invasion as efficiently as C2-tail in invasion experiments (Fig. 3C), which were done in a setting of FBS stimulation. Thus, serum stimulation potentially regulates the protein conformation of PTEN G129E and its interaction with PREX2.
Collectively, results from our binding and invasion studies indicate that an intact PDZ binding domain is required for strong binding to PREX2 and for antagonism of PREX2-driven invasion. Our data suggest that the PTEN tail is sufficient for inhibition of PREX2; however, we cannot rule out a role for the PTEN C2 domain in enhancing tail function and further suppressing PREX2 activity. Notably, PTEN mutations that result in deletion of the PTEN tail or PDZ binding domain have been identified in tumors (12, 13). In addition, neither wild-type PTEN nor the isolated PTEN C2-tail interacted with PREX1, and the PTEN C2-tail did not inhibit PREX1-stimulated invasion (fig. S2, G and H), which suggests that PI3K-independent inhibition of cell invasion by PTEN may be specific to PREX2.
PTEN inhibits the GEF activity of PREX2 and blocks the activation of the GTPase RAC1
Given that the PTEN C2-tail is sufficient for antagonism of invasion, we wondered how PTEN functioned to inhibit PREX2. Previously published data indicate that PTEN colocalizes with PREX2 at the cell membrane (11), suggesting that PTEN acts at the cell membrane to inhibit PREX2 activity. To rule out the possibility that C2-tail inhibits invasion by preventing PREX2 membrane localization, we transfected HCC1937 cells with C2-tail and PREX2 and examined their localization by immunofluorescence. PREX2 localization was not altered by coexpression of C2-tail, because PREX2 showed both cytoplasmic and membrane localization when transfected alone or along with C2-tail (fig. S3).
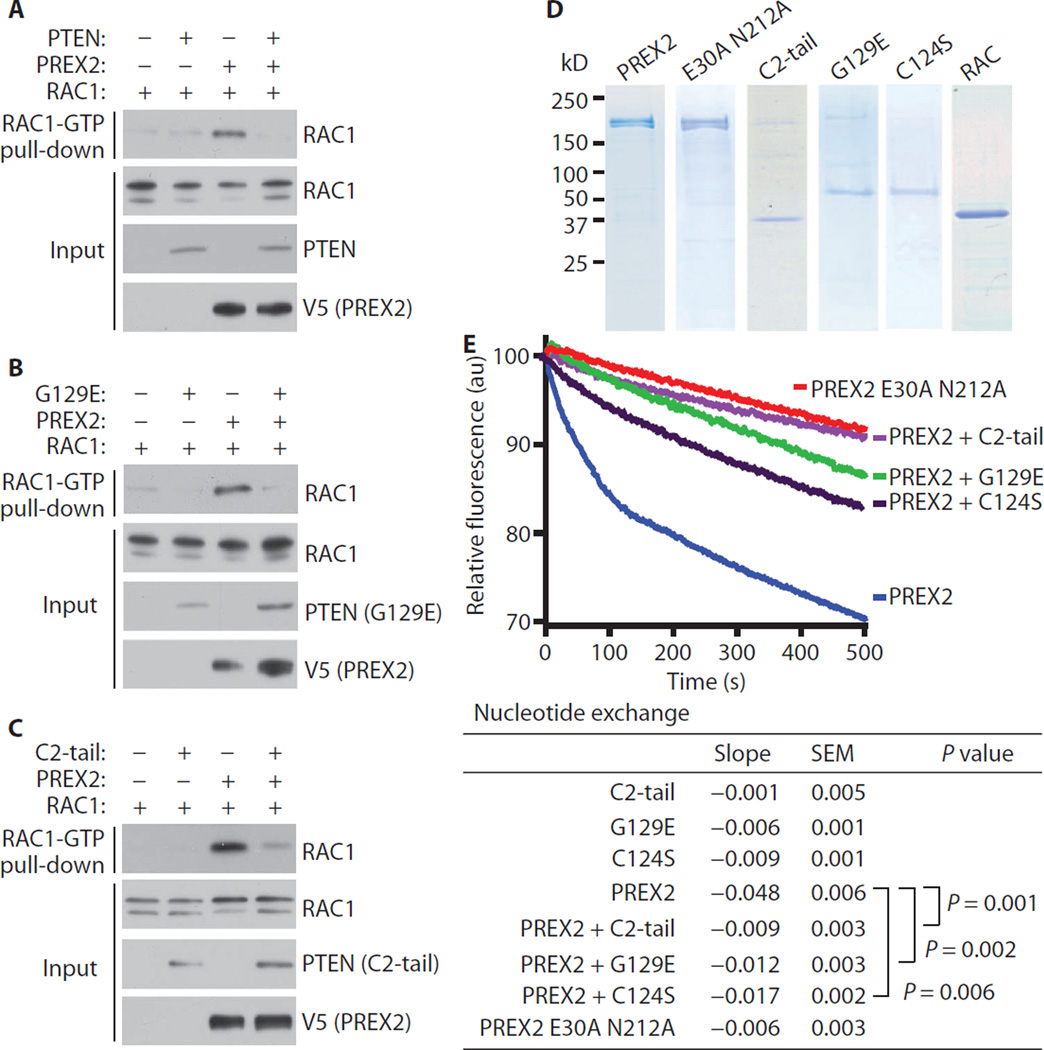
To test whether PTEN impeded RAC1 activation in cells, we cotransfected HEK-293 cells with RAC1, PREX2, and either empty vector, wild-type PTEN, G129E, C2-tail, or GFP-tail. We then used a cellular RAC activation assay to pull down GTP-bound and activated RAC1 (29). PREX2 stimulated RAC1 binding to GTP, whereas wild-type PTEN, G129E, C2-tail, and GFP-tail all decreased RAC1-GTP amounts to a similar extent (Fig. 4, A to C, and fig. S4A). In addition, wild-type PTEN, G129E, and C2-tail all decreased the activation of endogenous RAC1 in BT549 cells transfected with PREX2 (fig. S4B).
Fig. 4. PTEN inhibits PREX2 GEF activity.
(A to C) PTEN suppresses RAC activation. HEK-293 cells were cotransfected with RAC and PREX2 plus either (A) WT PTEN, (B) G129E, or (C) C2-tail. Cells were starved overnight, and RAC activation was measured by pull-down of RAC-GTP. Immunoblots in (A) to (C) are representative of at least two experiments. (D) Coomassie stains of purified PREX2, PTEN, and RAC1 proteins. (E) Effect of PREX2 and PTEN on the kinetics of mant-GDP dissociation from RAC as measured by a fluorimeter. RAC, PTEN (G129E, C124S, and C2-tail), and PREX2 (WT and GEF-dead double mutant E30A N212A) were present in exchange reactions at concentrations of about 20, 10, and 5 nM, respectively. One thousand–fold molar excess of GTP was added to the reaction to initiate dissociation of mant-GDP from RAC. Lines represent the average of at least three independent runs, and the slopes of the linear phases of nucleotide exchange were calculated. Two-tailed Mann-Whitney nonparametric tests were used to compare slopes of nucleotide exchange experiments. au, arbitrary units.
Next, we used a fluorescent nucleotide exchange assay to examine more directly the effect of PTEN on PREX2 GEF activity. We purified wild-type PREX2 protein and PTEN protein constructs (G129E, C124S, and C2-tail) from HEK-293 cells, and we purified RAC1 protein from Escherichia coli (Fig. 4D). We also purified PREX2 ΔDHPH and GEF-dead PREX2 mutants (N212A and E30A/N212A single and double mutants) from HEK-293 cells for use as negative controls (Fig. 4D and fig. S4C). GEF-dead PREX2 mutants were generated on the basis of published GEF-dead PREX1 mutants (30). Wild-type PREX2 greatly increased the exchange of GDP for GTP on RAC1 (Fig. 4E). In contrast, neither the GEF-dead PREX2 mutants nor PREX2 ΔDHPH catalyzed nucleotide exchange (Fig. 4E and fig. S4, C and D). Addition of PTEN G129E, C124S, or C2-tail to the reaction inhibited PREX2 GEF activity toward RAC1 (Fig. 4E and fig. S4E), indicating that PTEN binds PREX2 and directly inhibits GEF activity.
PREX2 mutation is associated with high PTEN expression, and PREX2 cancer mutants are resistant to antagonism by PTEN
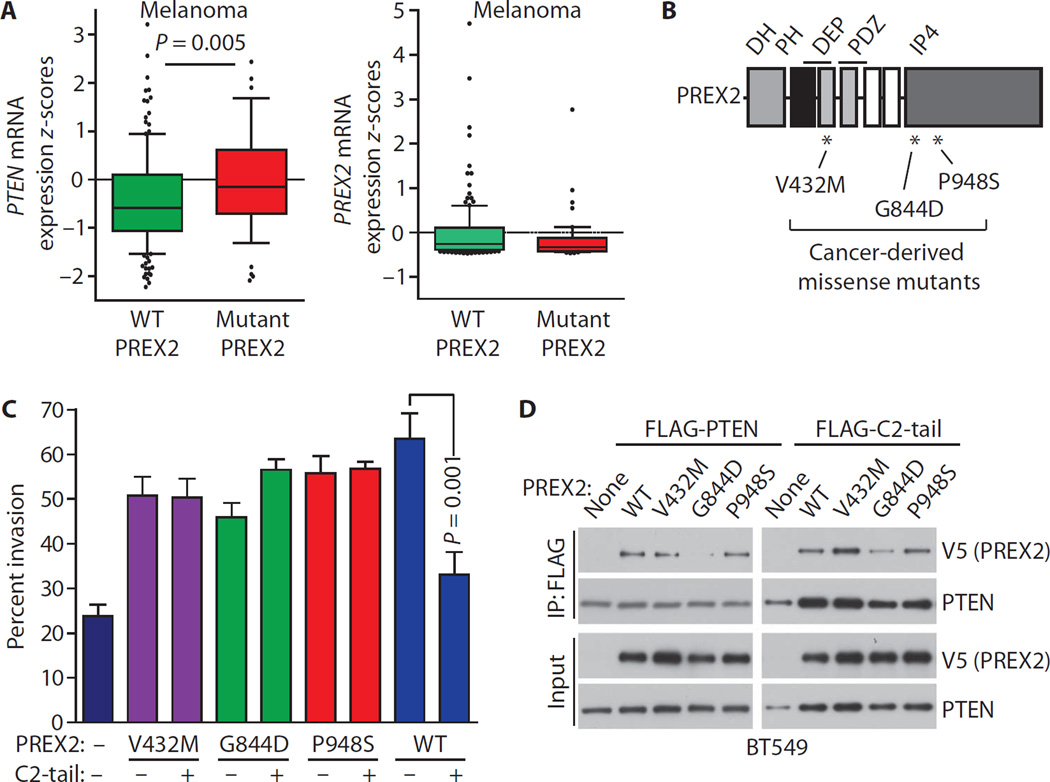
Somatic PREX2 mutations have been reported in many cancers. PREX2 mutation is particularly common in melanoma, where about 14% of tumors harbor PREX2 mutations (13, 14). More than 10% of stomach, colorectal, and lung cancers have PREX2 mutations as well (13). PREX2 mutation has functional consequences because the expression of cancer-derived PREX2 mutants in immortalized melanocytes accelerates tumor formation in mice (14). Thus, we decided to explore the relationships between PREX2 and PTEN in the TCGA using cBioPortal (13). In melanoma, PREX2 mutation was associated with significantly higher PTEN expression (Fig. 5A). Such data suggest that PREX2 mutation is selected for under conditions of high PTEN expression, or that loss of PTEN is no longer selected for after PREX2 mutation.
Fig. 5. Cancer-derived PREX2 mutants evade inhibition by PTEN.
(A) PREX2 mutation is associated with high PTEN expression in melanoma. mRNA z-scores for PTEN and PREX2 expression in melanomas with WT PREX2 (n =160) or with PREX2 mutations (n = 49). Whiskers extend from the 10th to 90th percentiles, and black dots denote values outside this range. Boxes delineate the first to third quartiles, and the central bar represents the median. P values were calculated using the Mann-Whitney test. Data were obtained from cBioPortal (13). Eighteen samples had PTEN mutations and were excluded. (B) PREX2 consists of DH, PH, DEP, PDZ, and IP4P domains. *, locations of cancer-derived PREX2 mutations used in this study. (C) The PTEN C2-tail is unable to inhibit invasion driven by PREX2 cancer mutants. BT549 cells were transfected as indicated, and invasion assays were performed. C2-tail and WT PREX2 are shown for comparison. Error bars represent the mean ± SEM of at least three experiments. (D) Coimmunoprecipitation of PREX2 mutants along with full-length PTEN or C2-tail. Immunoblots are representative of three experiments.
To examine how PREX2 mutants function in the presence of PTEN, we tested whether PREX2 mutants were sensitive to PTEN-mediated inhibition of invasion. We generated forms of PREX2 with mutations identified in pancreatic cancer (V432M) and melanoma (G844D and P948S) (Fig. 5B), two cancer types in which PREX2 protein is present (fig. S5, A and B). These PREX2 mutants (V432M, G844D, and P948S) were stable and were expressed at amounts similar to that of wild-type PREX2 (fig. S5C). All three of these PREX2 mutations occurred in PTEN wild-type tumors (12, 14, 31, 32). PTEN C2-tail did not suppress invasion driven by any of the PREX2 cancer mutants (Fig. 5C). We next tested whether PREX2 mutations affect PTEN binding. PREX2 V432M and P948S interacted with C2-tail and full-length PTEN to a degree similar to wild-type PREX2 (Fig. 5D). On the other hand, the interaction between PREX2 G844D and C2-tail or full-length PTEN was much reduced (Fig. 5D). The G844D mutation occurs in the IP4P domain of PREX2, which is the site of interaction with the PTEN PDZ binding domain (15), so the decreased binding is likely a result of a disrupted interaction between these regions.
Cancer-derived PREX2 mutants escape PTEN-mediated inhibition of GEF activity and activate PI3K signaling
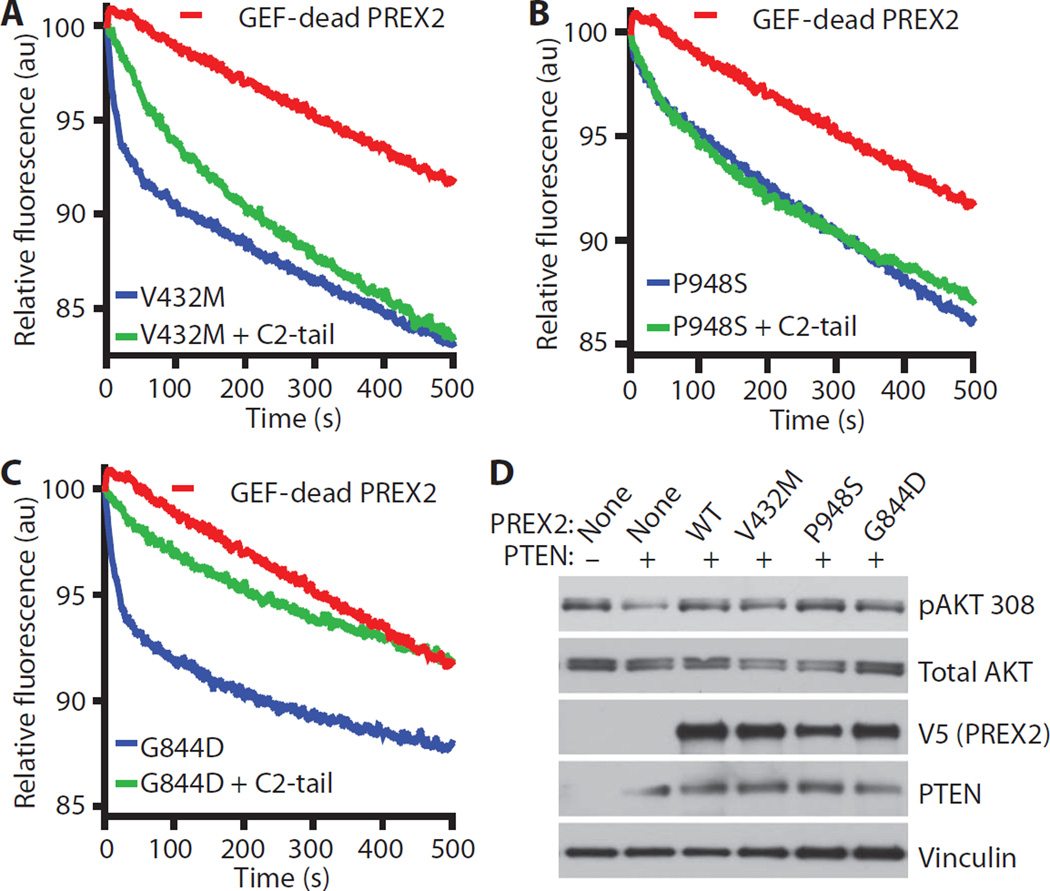
Having observed that PREX2 cancer mutants are resistant to antagonism by PTEN in invasion assays, we purified PREX2 cancer mutant proteins (fig. S6A) and tested whether their GEF activity could be inhibited by PTEN C2-tail in the fluorescent nucleotide exchange assay. In this experimental setup, we used the GEF activity of the purified PREX2 mutants as an internal control for ensuring the purification of properly folded, functional proteins. PREX2 V432M, G844D, and P948S all catalyzed nucleotide exchange on RAC1 (Fig. 6, A to C). V432M and P948S were resistant to GEF inhibition by C2-tail, whereas G844D, which interacts with PTEN relatively weakly inside cells, was more sensitive to inhibition of GEF activity in vitro (Fig. 6, A to C, and fig. S6B). Together, these data suggest that PREX2 mutations are selected in tumors that highly express PTEN as a means of escaping PTEN-mediated inhibition of PREX2. Wild-type PREX2 inhibits the lipid phosphatase activity of PTEN to stimulate AKT (11), and we wondered whether cancer-derived PREX2 mutants were also capable of antagonizing PTEN. Expression of PTEN in PTEN-deficient U87-MG cells resulted in decreased phosphorylation of AKT at Thr308, but coexpression of wild-type PREX2 or cancer-derived PREX2 mutants restored phosphorylation at this site in PTEN-transfected cells (Fig. 6D), suggesting that these PREX2 mutants retain the ability to inhibit PTEN signaling activity.
Fig. 6. PREX2 cancer mutants are resistant to PTEN C2-tail–mediated inhibition of RAC-GEF activity.
(A to C) Effect of PTEN C2-tail on PREX2 mutant-induced mant-GDP dissociation from RAC. Nucleotide exchange catalyzed by a GEF-dead PREX2 double mutant (E30A N212A) is also shown. Each line represents the average of at least three experiments. Slopes of nucleotide exchange were quantified (fig. S6B). (D) Cancer-derived PREX2 mutants retain the ability to inhibit PTEN signaling activity. U87-MG cells were transfected as indicated, and lysates were collected from cells growing under steady-state conditions. Immunoblots were probed as indicated. Immunoblots are representative of two experiments.
DISCUSSION
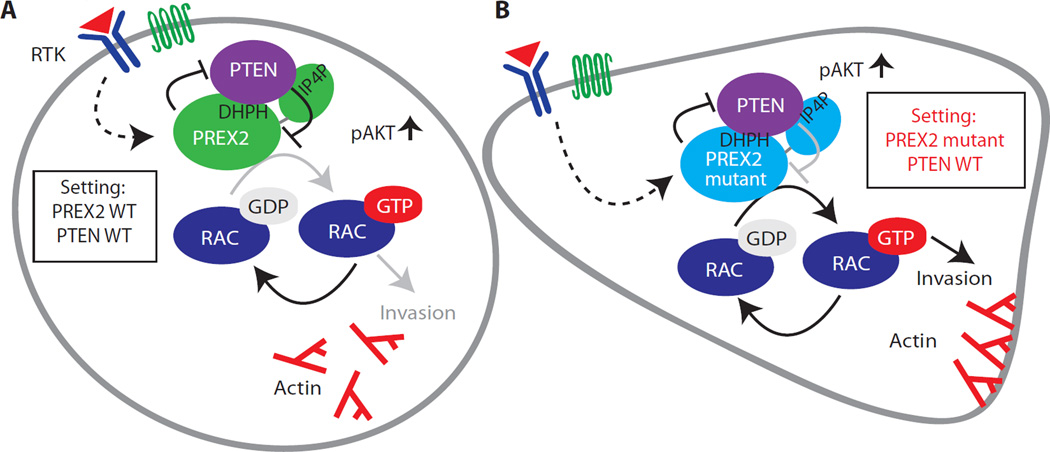
Here, we show that PTEN suppresses cell migration and invasion by directly inhibiting PREX2 function. Our data indicate that the PTEN C2 and tail domains are sufficient to inhibit PREX2 GEF–catalyzed RAC1 activation and suppress cell migration and invasion (Fig. 7A). Data from our Pten and Prex2KOMEFs demonstrate that Pten loss–induced cell migration is primarily dependent on the presence of Prex2 (Fig. 1B), and our studies in breast cancer cells demonstrate that PREX2 makes a substantial contribution to their invasiveness, which can be suppressed by wild-type PTEN, G129E, C124S, and the isolated PTEN C2-tail (Fig. 2, C and D). Evidence from invasion studies (Fig. 3C), binding experiments (Fig. 3F), and nucleotide exchange assays (Fig. 4E) supports a model in which the PDZ binding domain within the PTEN tail is important for docking to PREX2 and is sufficient for inhibition of PREX2 function. Although our results suggest that the PTEN tail is sufficient for inhibition of PREX2, we cannot rule out a role for the C2 domain. We suspect that the C2 domain may act in concert with the PTEN tail to restrain PREX2 GEF activity because it is the C2 domain that binds to the catalytic GEF domain of PREX2 (namely, the DHPH domain). Direct inhibition of PREX2-catalyzed nucleotide exchange by PTEN represents a unique mechanism of GEF inhibition, and our data are consistent with previous studies that have demonstrated the importance of the PTEN C2 and tail domains in suppressing migration, invasion, and neurite branching (1, 4, 5, 33).
Fig. 7. Model of PTEN-mediated inhibition of PREX2 GEF activity.
(A) PTEN binds PREX2 and inhibits its GEF activity, blocking RAC1 activation and suppressing invasion. WT PREX2 inhibits PTEN activity and amplifies PI3K signaling. (B) Cancer-derived PREX2 mutants are resistant to inhibition by PTEN. PREX2 cancer mutants stimulate invasion and maintain the ability to antagonize PTEN activity and amplify PI3K signaling. Arrows and blunted lines denote activation and inhibition, respectively. Black and gray lines denote full and reduced activity, respectively. PREX2 is activated downstream of receptor tyrosine kinases (RTK) or G protein (heterotrimeric guanine nucleotide–binding protein)–coupled receptors (GPCRs).
Whereas our data suggest that PTEN’s catalytic activity is not required for direct inhibition of PREX2 GEF function, PTEN’s lipid phosphatase activity provides another important layer of GEF inhibition. By reducing PIP3 concentrations, PTEN can indirectly reduce the activity of PIP3-responsive GEFs and thereby inhibit cell migration and invasion. Because PREX2-mediated inhibition of PTEN phosphatase activity leads to increased PIP3 concentrations (11), which in turn leads to increased PREX2 activity (9), PTEN’s ability to antagonize PREX2 function through a PIP3-independent mechanism may have evolved as a way to prevent a short circuit in which PREX2 is always active. Such a mechanism would allow PTEN to “turn off ” PREX2 downstream of PIP3 and would prevent PREX2 from constantly increasing its own activity through inhibition of PTEN. Given that PTEN-mediated inhibition of PREX2 GEF activity is independent of PTEN phosphatase activity, catalytically inactive PTEN mutants could still antagonize PREX2, making PREX2 mutation important even in cells harboring such PTEN mutations. This may explain why PREX2 and PTEN catalytic domain mutations sometimes co-occur in tumors.
PREX2 mutations are associated with high PTEN expression (Fig. 5A), suggesting that PREX2 mutation provides a mechanism for overcoming inhibition by PTEN. Indeed, we find that PREX2 mutants from melanoma (G844D and P948S) and pancreatic cancer (V432M) drive cancer cell invasion and escape antagonism by PTEN (Fig. 7B). Berger et al. (14) have shown that PREX2 G844D, but not P948S, cooperates with N-RAS to enhance tumor growth and decrease survival in mice. Whereas these two PREX2 mutants behave similarly in our in vitro experiments, the differences observed between G844D and P948S in tumor xenograft studies suggest that they are functionally distinct in vivo. Such functional differences could be explained by PTEN-independent effects or by differences in the mutants’ abilities to inhibit PTEN or catalyze nucleotide exchange in vivo.
Collectively, our findings point to a new mechanism of invasion suppression by PTEN and suggest that PTEN restrains cell movement by blocking the GEF activity of PREX2. It will be important for future studies to further examine the mechanisms by which PREX2 is deregulated in cancer because aberrant PREX2 activation may promote tumor development and drive metastasis. Selection of PREX2 cancer mutations for the ability to escape antagonism by PTEN is especially intriguing because it appears to be distinct from the selection of gain-of-function mutations.
MATERIALS AND METHODS
Antibodies
The following antibodies were used: a rabbit polyclonal antibody to amino acids 960 to 973 (VQLDSRKHNSHKE) of PREX2 (Zymed Laboratories), anti-V5 (Invitrogen), anti–FLAG M2 (Sigma), anti-RAC1 (Millipore), and antibodies against PTEN 138G6 (9559L), AKT (9272S), and phospho-AKT (Thr308) (244F9, Cell Signaling). A mouse monoclonal antibody against amino acids 1341 to 1375 of human PREX1 (clone 6F12) was used as previously described (10).
Plasmids and constructs
PREX2 was cloned into a pcDNA3.1V5/His vector as previously described (11). PREX2 cancer mutants were made by using PREX2 pcDNA3.1V5/His as a template. FLAG-PTEN pIRES has been previously described (11). C2-tail deletion constructs were made by using pIRES C2-tail as a template. A construct containing dominant negative p53 (pBABE-hygro p53DD) (22) was used for making retrovirus to immortalize MEFs.
Mouse embryonic fibroblasts
Ptenflox/flox mice (21) were crossed with Prex2−/− mice (15). Ptenflox/+Prex2−/+ mice in a SV129/C57B1/6 background were interbred, and MEFs were harvested. MEFs were immortalized by infection with a retrovirus expressing dominant negative p53. For deletion of Pten, MEFs were infected with adenovirus expressing Cre recombinase (Vector Biolabs). GFP adenovirus (Vector Biolabs) was used as a control. MEFs were used for experiments only between passages 2 and 5 after immortalization.
Cell spreading/area assays
Spreading assays were done according to previously published methods (34) using at least three MEFs for each genotype. Coverslips were coated overnight in six-well plates at 4°C with collagen (10 µg/ml), blocked in 1% bovine serum albumin (BSA) for 1 hour, and then washed with phosphate-buffered saline (PBS). About 50,000 MEFs were plated per coverslip and then placed at 37°C for 1 hour. MEFs were stained with CellMask Green membrane stain (Life Technologies) for 5 min, and coverslips were washed once in PBS. Coverslips were then mounted onto glass slides with Vectashield mounting medium. Ten 10× images of cells were taken using a Nikon fluorescence microscope, and ImageJ software was used to calculate cell area for 50 cells for each genotype.
Migration and invasion assays
Transwell inserts (BD Biosciences) were coated with either collagen I (MEF experiments) or Matrigel (Trevigen) (cell line experiments), and assays were performed as described (23). PDGF (20 ng/ml) (Sigma) and 10% FBS in medium were used as chemoattractants for MEFs and cancer cell lines, respectively. Three independent Transwell inserts were used per experiment. At least six independent fields of cells were counted for each experimental condition. For MEF experiments, about 30,000 cells were plated per well, and cells were allowed to migrate for 16 hours. For BT549 and SUM149 experiments measuring PREX2-driven invasion, cells were transfected with a 1:3 ratio of PTEN/PREX2 (that is, 6 µg of PTEN and 18 µg of PREX2). For invasion experiments with endogenous PREX2, cells were transfected with 15 µg of PTEN, G129E, or C2-tail. About 30,000 cells were plated per well, and cell invasion was allowed for 3 hours. For IGROV1 experiments, 50,000 cells were plated per well, and cell invasion was allowed for 24 hours. Percent invasion and percent migration were calculated by dividing the number of invaded cells by the total number of cells plated per membrane.
Nucleotide exchange
Exchange assays were performed at room temperature, using an LS-55 fluorescence spectrometer (Perkin Elmer) according to published methods (35). RAC1 was preloaded with mant-GDP (Invitrogen) in exchange buffer [20 mM tris (pH 7.4), 4 mM EDTA, and 50 mM NaCl] by incubating end over end at room temperature for 10 min. Next, reactions were supplemented with 15 mM MgCl2, incubated end over end for 10 min at room temperature, and then kept on ice. Mant-GDP–loaded RAC was transferred to a cuvette. V5-tagged PREX2 in PB buffer [100 mM NaCl, 25 mM tris (pH 7.4), 15 mM MgCl2] or FLAG-tagged C2-tail in tris-buffered saline (TBS) [150 mM NaCl, 10 mM tris (pH 7.4), 15 mM MgCl2] was added to the reaction. For exchange reactions containing both C2-tail and PREX2, C2-tail and PREX2 were preincubated for 10 min. For exchange reactions containing only PREX2 or C2-tail, the reaction was balanced by adding PB buffer or TBS buffer in place of PREX2 or C2-tail, respectively. Nucleotide exchange was initiated by adding GTP (Sigma) in 1000-fold molar excess relative to mant-GDP. After addition of GTP, fluorescence (emission, 440 nm; excitation, 360 nm) was monitored for 10 min. The final reaction volume for nucleotide exchange assays was 440 µl. RAC1 was present in exchange reactions at 20 nM. PREX2 and C2-tail were present in exchange reactions at 5 and 10 nM, respectively.
RAC activation assay
HEK-293 cells were transfected with RAC1, PREX2, and PTEN. Twenty-four hours after transfection, cells were washed in serum-free medium and then starved overnight. Glutathione S-transferase–CRIB resin was prepared as described previously (11), and RAC activation assays were done as described previously (11).
Transfection and infection
Transfections were done using Lipofectamine 2000 (Invitrogen). Retrovirus was produced by transfection of pBabe constructs into Phoenix HEK-293 packaging cell line (36). Lentiviral shRNA vectors (Sigma Mission) targeting PREX2 (target sequence 5′-CCGGCCCTGGACAGTGCATTATCAACTCGAGTTGATAATGCACTGTCCAGGGTTTTTTG-3′ and target sequence 5′-CCGGGCCCAAAGGTTTCTTCAGCTTCTCGAGAAGCTGAAGAAACCTTTGGGCTTTTTTG-3′) were used. shRNA knockdown lentiviruses were produced in HEK-293T cells (37). For PTEN knockdown experiments, cells were transfected with PTEN siRNA (GS5728, Qiagen). Cells were used in invasion assays 36 hours after transfection.
Immunoprecipitation, immunoblotting, and immunofluorescence
Cells were washed in cold PBS and lysed in cold BC-200 buffer [200 mM KCl, 1 mM EDTA, 20 mM tris (pH 7.5), 0.2% Triton X, 10% glycerol, and protease inhibitor cocktail]. Lysates were centrifuged at 16,000g for 30 min and then precleared. Complexes were precipitated overnight at 4°C using anti–FLAG (M2)–linked agarose or anti-V5 agarose. Complexes were washed and eluted into 2× Laemmli sample buffer. Elutions were resolved by SDS–polyacrylamide gel electrophoresis (SDS-PAGE), and proteins were transferred onto polyvinylidene difluoride membranes using wet transfer methods. Immunofluorescence was performed as previously described (11).
Human breast samples
Deidentified human breast samples were obtained from the Columbia University tumor bank. Samples were lysed in cold radioimmunoprecipitation assay buffer [50 mM tris-HCl (pH 7.4), 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA] containing a protease inhibitor cocktail (Sigma), sonicated for 15 s, and centrifuged at 20,000g for 1 hour. Protein concentrations were measured using the BCA protein assay (Pierce). Equal amounts of protein were suspended in 2× Laemmli sample buffer, loaded on SDS-PAGE gels, and immunoblotted.
Quantitative polymerase chain reaction
Quantitative polymerase chain reaction was performed in an ABI 7500 machine, using Fast SYBR Green (Invitrogen) according to the manufacturer’s instructions. The primer sequences were as follows: PREX2 forward, 5′-AACCATGAGAAGGCACAAAAA-3′, and reverse, 5′-CTTGCATATTCTTTGTATTGGTGT-3′. Samples were run in triplicate. The efficiencies of PREX2 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers were measured by using standard curves and were calculated to be 2.3 and 2.0, respectively. Quantitative analysis was performed on the basis of the ΔΔCt method using the housekeeping gene GAPDH. The GAPDH primer sequences were as follows: forward, 5′-GAAGGTGAAGGTCGGAGTCAAC-3′, and reverse, 5′-CAGAGTTAAAAGCAGCCCTGGT-3′.
Protein purification
V5-tagged PREX2 was purified as previously described (11); for purification of FLAG–C2-tail, a slightly modified procedure was used with FLAG agarose. HEK-293 cells were transfected with PREX2 or C2-tail. Forty-eight hours after transfection, cells were lysed in Triton X radioimmunoassay (TRIPA) buffer [500 mM NaCl, 50 mM tris (pH 7.5), 0.1% Triton X, and protease inhibitor cocktail]. Lysates were sonicated for 20 s (5-s pulses) at 4°C and then centrifuged at 25,000g for 90 min, filtered, and precleared. Precleared lysates were incubated with anti-V5 agarose or anti-FLAG resin overnight at 4°C. Resins were transferred to 10-ml Poly-Prep columns (Bio-Rad) and washed four times with TRIPA and four times with PB [100 mM NaCl, 25 mM tris (pH 7.4), 15 mM MgCl2] for V5-tagged proteins or with TBS [150 mM NaCl, 10 mM tris (pH 7.4), 15 mM MgCl2] for FLAG-tagged proteins. Proteins were eluted from beads using V5 or FLAG peptides in PB or TBS supplemented with 1 mM dithiothreitol. For purification of RAC1, BL21 (DE3) pLysE cells (Invitrogen) were transformed with a plasmid encoding RAC1 tagged with a 6His-Sumo N-terminal tag (38). Bacterial cultures were induced with 0.1 mMisopropyl-β-d-thiogalactopyranoside (Sigma) and grown for 16 hours at 21°C. Bacteria were centrifuged, and pellets were resuspended in PB, sonicated, centrifuged for 1 hour at 25,000g, and filtered. For AKTA fast protein liquid chromatography, HisTrap and desalting columns were used to isolate His-RAC1. Purified proteins were quantified by Coomassie with known BSA standards.
Mutagenesis
The QuikChange II XL Site-Directed Mutagenesis Kit (Agilent) was used. Primers were designed using the Agilent Web site for site-directed mutagenesis. Clones were sequenced to confirm the presence of the engineered mutations and the absence of secondary mutations.
Statistical analysis
Thresholds for statistical significance were set at P < 0.05. All migration data are presented as means ± SEM. Box and whisker plots show data from human cancer databases. Whiskers denote the 10th to 90th percentiles for each group. Boxes delineate the first to third quartiles of expression, and the central bar represents the median. The one-tailed Mann-Whitney nonparametric test was used to compare unpaired groups of mRNA expression z-scores from cancer data sets. The nonparametric Spearman correlation value (R) was calculated for mRNA z-scores, and P values were calculated using two-tailed tests. F tests were used to compare variances. Two-tailed Mann-Whitney nonparametric tests were used to compare slopes of nucleotide exchange experiments.
Supplementary Material
Acknowledgments
We thank all the members of the Parsons laboratory and M. Maurer for reading the manuscript. We thank Y. Song and B. Zheng for help with immunoblots from skin, ovary, and pancreatic cancer cell lines. We also thank M. Horton for help with statistics. We thank N. Leslie for providing the PTEN GFP-tail vector. Funding: This work was supported by the National Cancer Institute (R01 CA155117) and the Komen Leadership Grant (SAC110028). Additional support was given to S.M.M. from the Department of Defense Breast Cancer Research Program (BC096345 and CA078207).
Footnotes
www.sciencesignaling.org/cgi/content/full/8/370/ra32/DC1
Fig. S1. PTEN loss is associated with poor prognosis in breast cancer; PREX2 is expressed in breast cancer.
Fig. S2. PTEN C2-tail suppresses invasion in BT549 and SUM149 cells; C2-tail does not antagonize PREX1-driven invasion.
Fig. S3. PTEN C2-tail does not alter PREX2 localization.
Fig. S4. PTEN inhibits PREX2-catalyzed RAC activation.
Fig. S5. PREX2 abundance in melanoma and pancreatic cancer cells and expression of PREX2 cancer mutants used in invasion studies.
Fig. S6. Purification of PREX2 mutants for use in nucleotide exchange assays.
Author contributions: S.M.M. designed experiments, performed experiments, analyzed data, and wrote the paper. D.B., N.S., C.H., W.S., B.F., D.S., and B.D.H. performed experiments. T.S. and H.H. provided reagents and helped design the study. R.P. designed the studies, analyzed data, and wrote the paper.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Raftopoulou M, Etienne-Manneville S, Self A, Nicholls S, Hall A. Regulation of cell migration by the C2 domain of the tumor suppressor PTEN. Science. 2004;303:1179–1181. doi: 10.1126/science.1092089. [DOI] [PubMed] [Google Scholar]
- 2.Davidson L, Maccario H, Perera NM, Yang X, Spinelli L, Tibarewal P, Glancy B, Gray A, Weijer CJ, Downes CP, Leslie NR. Suppression of cellular proliferation and invasion by the concerted lipid and protein phosphatase activities of PTEN. Oncogene. 2010;29:687–697. doi: 10.1038/onc.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada KM. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science. 1998;280:1614–1617. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- 4.Leslie NR, Yang X, Downes CP, Weijer CJ. PtdIns(3,4,55)P3-dependent and -independent roles for PTEN in the control of cell migration. Curr. Biol. 2007;17:115–125. doi: 10.1016/j.cub.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang XC, Piccini A, Myers MP, Van Aelst L, Tonks NK. Functional analysis of the protein phosphatase activity of PTEN. Biochem. J. 2012;444:457–464. doi: 10.1042/BJ20120098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cain RJ, Ridley AJ. Phosphoinositide 3-kinases in cell migration. Biol. Cell. 2009;101:13–29. doi: 10.1042/BC20080079. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, Paik JH, Wang Z, Hla T, Wu D. Role of guanine nucleotide exchange factor P-Rex-2b in sphingosine 1-phosphate-induced Rac1 activation and cell migration in endothelial cells. Prostaglandins Other Lipid Mediat. 2005;76:95–104. doi: 10.1016/j.prostaglandins.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Welch HC, Coadwell WJ, Ellson CD, Ferguson GJ, Andrews SR, Erdjument-Bromage H, Tempst P, Hawkins PT, Stephens LR. P-Rex1, a PtdIns(3,4,5)P3- and Gβγ-regulated guanine-nucleotide exchange factor for Rac. Cell. 2002;108:809–821. doi: 10.1016/s0092-8674(02)00663-3. [DOI] [PubMed] [Google Scholar]
- 9.Donald S, Hill K, Lecureuil C, Barnouin R, Krugmann S, John Coadwell W, Andrews SR, Walker SA, Hawkins PT, Stephens LR, Welch HC. P-Rex2, a new guanine-nucleotide exchange factor for Rac. FEBS Lett. 2004;572:172–176. doi: 10.1016/j.febslet.2004.06.096. [DOI] [PubMed] [Google Scholar]
- 10.Welch HC, Condliffe AM, Milne LJ, Ferguson GJ, Hill K, Webb LM, Okkenhaug K, Coadwell WJ, Andrews SR, Thelen M, Jones GE, Hawkins PT, Stephens LR. P-Rex1 regulates neutrophil function. Curr. Biol. 2005;15:1867–1873. doi: 10.1016/j.cub.2005.09.050. [DOI] [PubMed] [Google Scholar]
- 11.Fine B, Hodakoski C, Koujak S, Su T, Saal LH, Maurer M, Hopkins B, Keniry M, Sulis ML, Mense S, Hibshoosh H, Parsons R. Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science. 2009;325:1261–1265. doi: 10.1126/science.1173569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, Teague JW, Campbell PJ, Stratton MR, Futreal PA. COS-MIC: Mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, Ivanova E, Watson IR, Nickerson E, Ghosh P, Zhang H, Zeid R, Ren X, Cibulskis K, Sivachenko AY, Wagle N, Sucker A, Sougnez C, Onofrio R, Ambrogio L, Auclair D, Fennell T, Carter SL, Drier Y, Stojanov P, Singer MA, Voet D, Jing R, Saksena G, Barretina J, Ramos AH, Pugh TJ, Stransky N, Parkin M, Winckler W, Mahan S, Ardlie K, Baldwin J, Wargo J, Schadendorf D, Meyerson M, Gabriel SB, Golub TR, Wagner SN, Lander ES, Getz G, Chin L, Garraway LA. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodakoski C, Hopkins BD, Barrows D, Mense SM, Keniry M, Anderson KE, Kern PA, Hawkins PT, Stephens LR, Parsons R. Regulation of PTEN inhibition by the pleckstrin homology domain of P-REX2 during insulin signaling and glucose homeostasis. Proc. Natl. Acad. Sci. U.S.A. 2014;111:155–160. doi: 10.1073/pnas.1213773111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindsay CR, Lawn S, Campbell AD, Faller WJ, Rambow F, Mort RL, Timpson P, Li A, Cammareri P, Ridgway RA, Morton JP, Doyle B, Hegarty S, Rafferty M, Murphy IG, McDermott EW, Sheahan K, Pedone K, Finn AJ, Groben PA, Thomas NE, Hao H, Carson C, Norman JC, Machesky LM, Gallagher WM, Jackson IJ, Van Kempen L, Beermann F, Der C, Larue L, Welch HC, Ozanne BW, Sansom OJ. P-Rex1 is required for efficient melanoblast migration and melanoma metastasis. Nat. Commun. 2011;2:555. doi: 10.1038/ncomms1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun J, Liu W, Adams TS, Li X, Turner AR, Chang B, Kim JW, Zheng SL, Isaacs WB, Xu J. DNA copy number alterations in prostate cancers: A combined analysis of published CGH studies. Prostate. 2007;67:692–700. doi: 10.1002/pros.20543. [DOI] [PubMed] [Google Scholar]
- 18.Fejzo MS, Godfrey T, Chen C, Waldman F, Gray JW. Molecular cytogenetic analysis of consistent abnormalities at 8q12-q22 in breast cancer. Genes Chromosomes Cancer. 1998;22:105–113. doi: 10.1002/(sici)1098-2264(199806)22:2<105::aid-gcc4>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 19.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Mc Henry KT, Pinchback RM, Ligon AH, Cho YJ, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA, Tanaka KE, Chiang DY, Bass AJ, Loo A, Hoffman C, Prensner J, Liefeld T, Gao Q, Yecies D, Signoretti S, Maher E, Kaye FJ, Sasaki H, Tepper JE, Fletcher JA, Tabernero J, Baselga J, Tsao MS, Demichelis F, Rubin MA, Janne PA, Daly MJ, Nucera C, Levine RL, Ebert BL, Gabriel S, Rustgi AK, Antonescu CR, Ladanyi M, Letai A, Garraway LA, Loda M, Beer DG, True LD, Okamoto A, Pomeroy SL, Singer S, Golub TR, Lander ES, Getz G, Sellers WR, Meyerson M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sosa MS, Lopez-Haber C, Yang C, Wang H, Lemmon MA, Busillo JM, Luo J, Benovic JL, Klein-Szanto A, Yagi H, Gutkind JS, Parsons RE, Kazanietz MG. Identification of the Rac-GEF P-Rex1 as an essential mediator of ErbB signaling in breast cancer. Mol. Cell. 2010;40:877–892. doi: 10.1016/j.molcel.2010.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lesche R, Groszer M, Gao J, Wang Y, Messing A, Sun H, Liu X, Wu H. Cre/loxP-mediated inactivation of the murine Pten tumor suppressor gene. Genesis. 2002;32:148–149. doi: 10.1002/gene.10036. [DOI] [PubMed] [Google Scholar]
- 22.Bowman T, Symonds H, Gu L, Yin C, Oren M, Van Dyke T. Tissue-specific inactivation of p53 tumor suppression in the mouse. Genes Dev. 1996;10:826–835. doi: 10.1101/gad.10.7.826. [DOI] [PubMed] [Google Scholar]
- 23.Marshall J. Transwell® invasion assays. Methods Mol. Biol. 2011;769:97–110. doi: 10.1007/978-1-61779-207-6_8. [DOI] [PubMed] [Google Scholar]
- 24.Ringner M, Fredlund E, Hakkinen J, Borg A, Staaf J. GOBO: Gene expression-based outcome for breast cancer online. PLOS One. 2011;6:e17911. doi: 10.1371/journal.pone.0017911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Simpson L, Takahashi M, Miliaresis C, Myers MP, Tonks N, Parsons R. The PTEN/MMAC1 tumor suppressor induces cell death that is rescued by the AKT/protein kinase B oncogene. Cancer Res. 1998;58:5667–5672. [PubMed] [Google Scholar]
- 26.Saal LH, Gruvberger-Saal SK, Persson C, Lovgren K, Jumppanen M, Staaf J, Jonsson G, Pires MM, Maurer M, Holm K, Koujak S, Subramaniyam S, Vallon-Christersson J, Olsson H, Su T, Memeo L, Ludwig T, Ethier SP, Krogh M, Szabolcs M, Murty VV, Isola J, Hibshoosh H, Parsons R, Borg A. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat. Genet. 2008;40:102–107. doi: 10.1038/ng.2007.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han SY, Kato H, Kato S, Suzuki T, Shibata H, Ishii S, Shiiba K, Matsuno S, Kanamaru R, Ishioka C. Functional evaluation of PTEN missense mutations using in vitro phosphoinositide phosphatase assay. Cancer Res. 2000;60:3147–3151. [PubMed] [Google Scholar]
- 28.Rahdar M, Inoue T, Meyer T, Zhang J, Vazquez F, Devreotes PN. A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proc. Natl. Acad. Sci. U.S.A. 2009;106:480–485. doi: 10.1073/pnas.0811212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benard V, Bokoch GM. Assay of Cdc42, Rac, and Rho GTPase activation by affinity methods. Methods Enzymol. 2002;345:349–359. doi: 10.1016/s0076-6879(02)45028-8. [DOI] [PubMed] [Google Scholar]
- 30.Hill K, Krugmann S, Andrews SR, Coadwell WJ, Finan P, Welch HC, Hawkins PT, Stephens LR. Regulation of P-Rex1 by phosphatidylinositol (3,4,5)-trisphosphate and Gβγ subunits. J Biol. Chem. 2005;280:4166–4173. doi: 10.1074/jbc.M411262200. [DOI] [PubMed] [Google Scholar]
- 31.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, Dicara D, Ramos AH, Lawrence MS, Cibulskis K, Sivachenko A, Voet D, Saksena G, Stransky N, Onofrio RC, Winckler W, Ardlie K, Wagle N, Wargo J, Chong K, Morton DL, Stemke-Hale K, Chen G, Noble M, Meyerson M, Ladbury JE, Davies MA, Gershenwald JE, Wagner SN, Hoon DS, Schadendorf D, Lander ES, Gabriel SB, Getz G, Garraway LA, Chin L. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tibarewal P, Zilidis G, Spinelli L, Schurch N, Maccario H, Gray A, Perera NM, Davidson L, Barton GJ, Leslie NR. PTEN protein phosphatase activity correlates with control of gene expression and invasion, a tumor-suppressing phenotype, but not with AKT activity. Sci. Signal. 2012;5:ra18. doi: 10.1126/scisignal.2002138. [DOI] [PubMed] [Google Scholar]
- 34.Yamada KM. Cell adhesion. Curr. Protoc. Cell Biol. 2003;(Suppl 18):9.0.1–9.0.9. [Google Scholar]
- 35.Yang MH, Nickerson S, Kim ET, Liot C, Laurent G, Spang R, Philips MR, Shan Y, Shaw DE, Bar-Sagi D, Haigis MC, Haigis KM. Regulation of RAS oncogenicity by acetylation. Proc. Natl. Acad. Sci. U.S.A. 2012;109:10843–10848. doi: 10.1073/pnas.1201487109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swift S, Lorens J, Achacoso P, Nolan GP. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell–based systems. Curr. Protoc. Immunol. 2001;Chapter 10(Unit 10.17C) doi: 10.1002/0471142735.im1017cs31. [DOI] [PubMed] [Google Scholar]
- 37.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 38.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.