Abstract
Antibiotic resistance often evolves by mutations at conserved sites in essential genes, resulting in parallel molecular evolution between divergent bacterial strains and species. Whether these resistance mutations are having parallel effects on fitness across bacterial taxa, however, is unclear. This is an important point to address, because the fitness effects of resistance mutations play a key role in the spread and maintenance of resistance in pathogen populations. We address this idea by measuring the fitness effect of a collection of rifampicin resistance mutations in the β subunit of RNA polymerase (rpoB) across eight strains that span the diversity of the genus Pseudomonas. We find that almost 50% of rpoB mutations have background-dependent fitness costs, demonstrating that epistatic interactions between rpoB and the rest of the genome are common. Moreover, epistasis is typically strong, and it is the dominant genetic determinant of the cost of resistance mutations. To investigate the functional basis of epistasis, and because rpoB plays a central role in transcription, we measured the effects of common rpoB mutations on transcriptional efficiency across three strains of Pseudomonas. Transcriptional efficiency correlates strongly to fitness across strains, and epistasis arises because individual rpoB mutations have differential effects on transcriptional efficiency in different genetic backgrounds.
Keywords: antibiotic resistance, fitness costs, genetic background, Pseudomonas
1. Background
Antibiotic resistance is associated with pleiotropic costs that are expressed in terms of reduced competitive fitness in the absence of antibiotics [1–3]. Fitness costs play a key role in the dynamics of resistance as they generate selection against resistance in antibiotic-free conditions, such as when antibiotic use is discontinued, or during transmission between hosts. Understanding the factors that govern the cost of resistance is therefore crucial for predicting when resistance will persist in pathogen populations. Put simply, the larger the cost associated with resistance, the less likely the persistence of resistance in bacterial populations over the long term.
Most antibiotics target highly conserved domains in proteins that play pivotal roles in cell biology, such as DNA replication, cell-wall assembly and protein synthesis [4]. The most direct mechanism for bacteria to evolve resistance to antibiotics is by altering the structure of these targets [5]. Because the target sites of antibiotics are highly conserved, target site mutations show a strong tendency to evolve in parallel across species of bacteria [6–8]. This parallel molecular evolution implies that these mutations have some similar phenotypic effects in these differing bacterial species, in that these mutations increase resistance to antibiotics. However, the extent to which the fitness costs of resistance mutations are conserved across genetic backgrounds remains unclear (but see [6]). There is growing evidence that mutations often have epistatic effects on fitness in bacteria [9–12], suggesting that genetic background could play a key role in shaping the cost of resistance across species.
In this paper, we investigate the influence of genetic background on the fitness cost of antibiotic resistance, using a collection of rifampicin-resistant mutants from the genus Pseudomonas as a model system [7]. One advantage of working with Pseudomonas is that the genus is highly diverse, and yet it is still possible to culture most strains under a common set of laboratory conditions [13,14], making it possible to obtain equivalent measures of fitness in different species or strains of bacteria. Rifampicin is an antibiotic which binds to a highly conserved domain of RNA polymerase, preventing RNA-transcript elongation. Resistance to rifampicin evolves by mutations in rpoB that alter the structure of the rifampicin binding pocket. The fitness cost of rifampicin resistance has been measured across a wide range of bacteria, including Escherichia coli [15], Salmonella typhymurium [6], Pseudomonas aeruginosa [16], Mycobacterium tuberculosis [17] and Staphylococcus aureus [18]. However, previous studies have measured fitness using a range of techniques and under different environmental conditions [3,19]. As these variables can affect the estimate of the cost of resistance, it is questionable to solely rely on comparing fitness cost estimates from different studies.
To measure the overall contribution of genetic background to the cost of resistance, we estimated the competitive fitness of a collection of mutations across eight strains that span the diversity of Pseudomonas. We then did two further analyses to try to explain why certain mutations vary in their fitness effects between genetic backgrounds. First, we use comparative methods to test whether the effects of a mutation are more conserved between closely related strains. If genetic background is a significant but relatively minor determinant of the cost of a mutation, we would predict a greater probability of differing fitness effects with increasing genetic distance. Second, rifampicin resistance mutations are known to have a range of effects on transcription [15], and previous work has shown that the fitness effects of rifampicin resistance correlate with reductions in transcriptional efficiency [15,20]. Genetic background could influence the fitness effects of rifampicin resistance mutations by modulating the effect of altered RNA polymerase on transcriptional efficiency or by altering the strength of the correlation between transcriptional efficiency and fitness. In other words, genetic background can influence the fitness effects of mutations by altering the relationship between genotype and phenotype, or the relationship between phenotype and fitness. To address this issue, we used an inducible reporter construct to measure the transcriptional efficiency of a subset of rifampicin resistance mutations across three strains of Pseudomonas.
2. Material and methods
(a). Strains and culture conditions
Eight strains of bacteria from the genus Pseudomonas were used: Pseudomonas aeruginosa PAO1, P. stutzeri ATCC17588, P. mendocina CCUG7181, P fulva CCUG12573, P. putida KT2440, P. protegens PF5, P. fluorescens PF01 and P. fluorescens SBW25. Prior to experimentation, all strains were stored at −80°C in 25% glycerol. All culturing was performed at 30°C with constant shaking at 200 r.p.m., in King's B (KB) medium.
(b). Isolation of rifampicin-resistant mutants
Rifampicin-resistant mutants were obtained from Vogwill et al. [7] where they were isolated by fluctuation tests on rifampicin agar. Briefly, for each strain, an overnight culture was diluted 1 million-fold and used to found 480 parallel cultures. These were grown for 48 h before being plated on agar containing either 30 or 60 mg ml−1 of rifampicin. After 48 h, mutants were isolated from 93 independent cultures and frozen in 25% glycerol at −80°C. We then sequenced the two regions of rpoB that can result in high-level rifampicin resistance. A single example of each mutation by strain combination was selected for further analysis.
(c). Measuring the cost of resistance
To facilitate the competition experiments, we transformed the ancestral genotype of each strain with a chromosomally integrated green fluorescent protein (GFP). These strains were generated by integrating a constitutively expressed GFP marker at the chromosomal tn7 insertion site using the methods of Choi & Schweizer [21]. Fitness costs were measured by competing rifampicin-resistant mutants against the appropriate GFP-tagged rifampicin-sensitive ancestral strain. Competitions took place in 200 µl of KB medium in a 96-well plate, incubated at 30°C with constant shaking at 250 r.p.m. Competitions lasted 24 h. Each competition was replicated six times, with the replicates of each competition spread across at least two separate occasions. For each competition, cells were grown overnight in KB medium. Each mutant was then mixed 50 : 50 by volume with the GFP-tagged ancestor, and this mixture was then diluted 10 000-fold. Initial and final ratios of GFP-tagged to untagged cells were determined using BD C6 flow cytometer. 10 000 cells per culture were counted and scored as either fluorescently tagged or not. Fitness was calculated at the ratio of the number of doublings of the rifampicin-resistant mutant compared with the GFP-tagged ancestral strain. To control for the cost of GFP-expression, fitness was standardized relative to the fitness of the unmarked ancestor in competition with the GFP-tagged ancestor.
(d). Correcting for phylogenetic distance
Using a selection of strains from across a genus results in potentially confounding any results with the effects of phylogeny. Specifically, the hierarchical nature of most phylogenies results in not all tips of a phylogeny being equally independent from each other. For example, in a phylogeny of three species, unless the phylogeny is a star configuration, two strains must be more closely related to each other than to the third strain. If a trait has any phylogenetic correlation or bias, then it would be pseudo-replication to consider all three strains equally independent. The method of Felsenstein [22] overcomes this issue by first comparing the phenotypes of the two closely related species, and then comparing the phenotype of the third species with the average of the first two species.
The analysis across phylogenies can be further conflated as not all evolutionary distances within a phylogeny are likely to be equal. This latter point could have important consequences for the influence of genetic background on the effects of a mutation, as it would seem intuitive that differing fitness effects are less likely between closely related strains. To control for this, we correct for the topological effects of phylogeny using the method of Felsenstein [22]. However, as we wished to test the effects of evolutionary distance on the effects of a mutation, we do not standardize our contrasts by evolutionary distance as in the conventional Felsenstein approach. We would therefore expect a positive correlation between evolutionary distance and the size of a contrast, assuming the probability of a mutation having conserved effects is greater between closely related strains.
The phylogeny of this study system has been previously determined (see [7] for full methods), based on 55 housekeeping genes taken from 28 fully sequenced Pseudomonas genomes. It is a maximum-likelihood phylogenetic tree assuming a GTR + G + I model in PHYML [23]. From this phylogeny, we trimmed it to just the eight strains used here, from which we extracted the relevant evolutionary distances for each pair of strains, or a strain and a node, or two nodes, as appropriate.
(e). Measuring gene expression per mutant
To estimate the effect of the rpoB mutations on transcriptional efficiency, we use the luciferase report gene system to measure gene expression [20]. In bacteria, transcription and translation are strongly coupled; therefore, gene expression should provide a good proxy for transcriptional efficiency. To measure the effects of rpoB mutations on transcriptional efficiency, strains were transformed with an IPTG-inducible luciferase reported gene (henceforth lux-transformed). The luciferase gene, which reacts with ATP to produce light, is commonly used to measure transcriptional activity by measuring the amount of light produced per cell. This was performed for the three most genetic tractable strains, specifically the ancestral strain for P. aeruginosa PAO1, P. putida KT2440 and P. fulva CCUG12573, as well as seven mutations common to these three strains. For each strain, overnight cultures of the lux-transformed ancestor and each of the lux-transformed mutants were grown in KB medium. Cultures were then diluted 10 000-fold into 200 µl of KB medium in black 96-well Costar microplates with clear bottom (Corning, USA). The medium was supplemented with 1 mM IPTG to induce luciferase production. The plate was then incubated in a Synergy 2 microplate reader (BioTek, USA) at 30°C, during which luminescence emission (RLU) per OD600 was measured every 20 min with shaking prior to each read. Each assay was replicated on four separate occasions.
To estimate transcriptional efficiency, the mean light produced per bacterial cell was calculated at each time point by dividing the luminescence of each well (in RLU) by its absorbance (OD600). The maximum gradient of each RLU/OD600 curve was then calculated using nine consecutive data points during the early exponential phase. Within each assay, the relative lux expression rate in each rpoB mutant strain was obtained by normalizing each gradient to that of the ancestral strain control in the same plate. This provides a measure of the maximum rate of transcription per mutant.
3. Results
(a). Fitness costs of rifampicin resistance across species of Pseudomonas
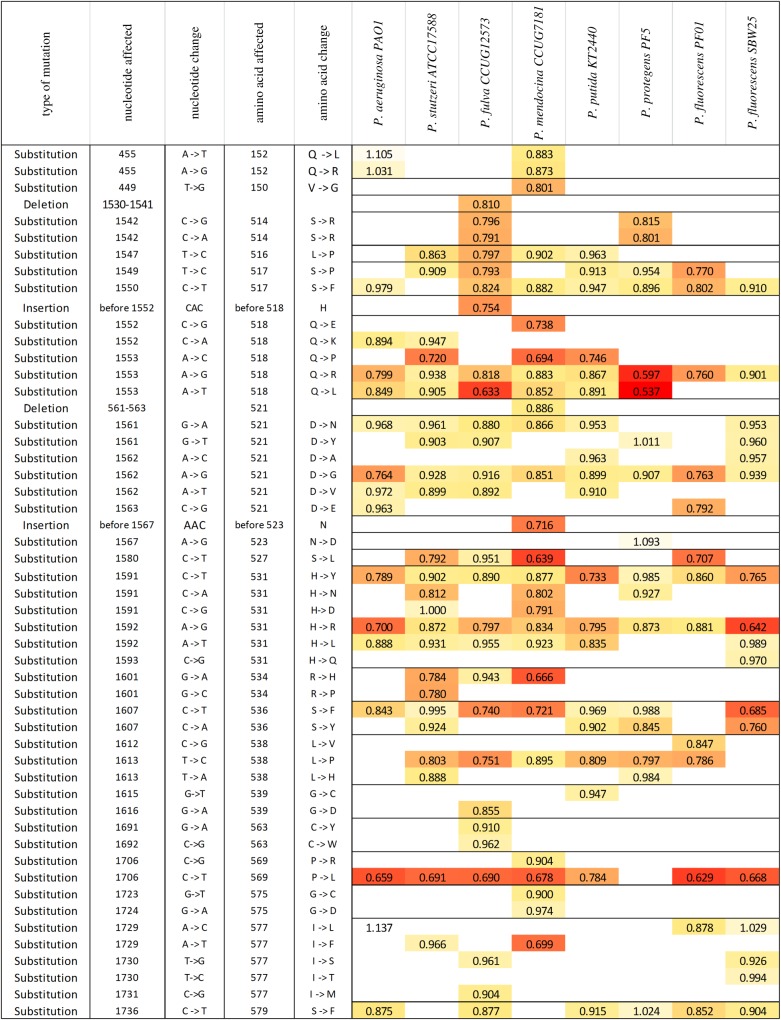
To estimate the relative importance of genetic background in determining the fitness cost of rifampicin resistance, we performed competitive fitness assays on a previously generated collection of rifampicin-resistant mutations from eight strains of Pseudomonas. Each of the competition experiments for the 161 unique strain-by-mutation combinations were replicated six times, giving 966 independent fitness measurements (figure 1). As shown, there is considerable variation in the cost of resistance. Owing to the low number of mutations common to all strains, as well as the high number of mutations found in only one strain, we analysed these data using a model consisting of just the main effects of strain and mutation. We find that although both factors significantly affect the cost of resistance (strain: F7,876 = 8.79, p < 0.001; mutation: F51,876 = 8.34, p < 0.001), mutation explains a greater proportion of the variation in fitness (mutation = 27.1% of variance; strain = 4.0% of variance). However, even if there is greater variance between mutations than strains, genetic background could still be affecting the cost of resistance. To test how commonly the cost of a mutation depended on its genetic background, we performed Bonferroni-corrected one-way ANOVAs on every mutation which arose in at least two backgrounds. Of these 33 comparisons, 14 (42.2%) are significant at the Bonferroni-corrected p-value of 0.0015, demonstrating that genetic background impacts fitness for almost half of these mutations under a conservative statistical test.
Figure 1.
Fitness effects of resistance mutations across species. Numbers show the fitness of rifampicin-resistant mutants relative to their respective ancestral strains in the absence of antibiotics, such that a value of 1 represents equal fitness. Each competition assay was carried out with sixfold replication and the colours highlight variation in fitness, ranging from red for very costly to white for beneficial. (Online version in colour; in printed version, intensity of shdading indicates the cost of mutation.)
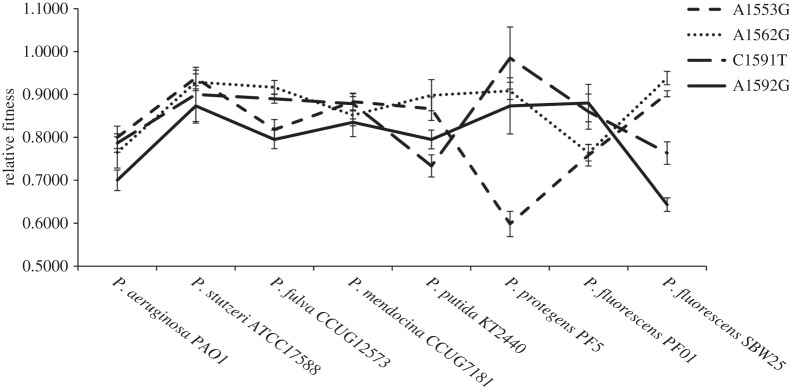
An alternative approach to estimate the effects of epistasis is to limit the analysis to just the four mutations common to all eight strains (figure 2). Although this limits the scope of the analysis, this approach makes it possible to estimate the contribution of the interaction between mutation and genetic background to the cost of a mutation. This analysis reveals significant effects of mutation (F3,158 = 7.53, p < 0.001) and genetic backgrounds (F7,158 = 7.13, p < 0.001) on fitness, and an interaction between mutation and genetic background (F21,158 = 7.37, p < 0.001). However, the proportion of variance explained by mutation is small (5.83%), whereas the explanatory power of genetic background is not much greater (12.88%). In contrast, the interaction term explains 39.92% of the variance. The implication of this result is that the interaction between a rifampicin resistance mutation and the genetic background plays a dominant role in determining the cost of resistance. Intriguingly, this epistatic interaction is driven by a complex interplay between mutations and genetic backgrounds, and not because all mutations are more costly in some strains than others.
Figure 2.
Epistastic effects of rifampicin resistance mutations on fitness across Pseudomonas. The fitness effect of the four common rifampicin resistance mutations across eight strains of Pseudomonas (±s.e.m; n = 6) is shown. The fitness of each resistant mutant was measured relative to its ancestor in the absence of antibiotics, such that a value of 1 represents equal fitness.
(b). Phylogeny and the costs of a mutation
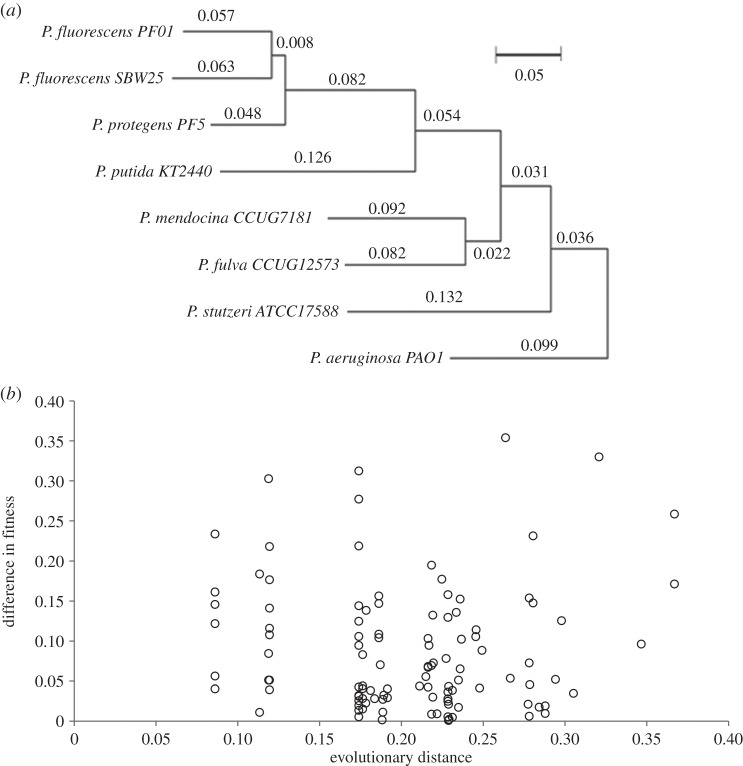
One important assumption of the above analyses is that they do not take phylogeny into account, and this may be an important bias. For example, intuition suggests that a mutation is more likely to have more similar effects on fitness in two closely related strains than in two distantly related strains. To test the hypothesis that phylogenetic distance drives epistasis, we tested for a positive correlation between genetic distance and epistasis using methods based on phylogenetically independent contrasts (PICs). As in PICs, we calculated the difference between neighbouring tips or nodes, thereby controlling for unequal relatedness between differing species (figure 3a). However, rather than standardize these contrasts by dividing by the underlying genetic distance, we looked for a relationship between genetic distance and the size of a contrast. If a mutation was found in all eight strains, it is possible to make seven contrasts. Because not all mutations were found in all strains, from the 33 mutations which were found in more than one strain it is possible to make 109 contrasts (figure 3b). We found no significant relationship between evolutionary distance and epistasis (Pearson's correlation: r = −0.480, d.f. = 107, p = 0.878). To see if evolutionary distance increases the probability of epistasis for any individual mutations, we re-analysed the data for the four mutations common to all strains, as these would have the most statistical power. None of the four show a significant correlation between evolutionary distance and epistasis (A1553G: r = −0.480, d.f. = 5, p = 0.275; A1562G: r = −0.379, d.f. = 5, p = 0.402; C1591T: r = −0.449, d.f. = 5, p = 0.312; A1592G: r = −0.619, d.f. = 5, p = 0.138). In short, mutations do not appear to have similar effects on fitness between more closely related strains. Therefore, at this scale, it appears that phylogenetic relatedness is not an important determinant of the probability of epistasis.
Figure 3.
Evolutionary divergence does not predict epistasis. (a) Phylogeny of the eight ancestral strains of Pseudomonas. Branch lengths are proportional to evolutionary distance. (b) The contrast in the fitness effect of rifampicin resistance mutations between tips or nodes on the phylogeny shown as a function of evolutionary distance. We find no evidence that rifampicin resistance mutations have more divergent effects on fitness between distantly related bacterial strains than between closely related strains.
(c). Molecular mechanisms underpinning cross-strain epistasis
Previous work has shown that compromised transcriptional efficiency explains much of the variation in the fitness cost of rifampicin resistance in P. aeruginosa [20], providing a potential molecular mechanism to explore the cost of resistance across strains. To link transcriptional efficiency with fitness, we transformed clones of P. aeruginosa PAO1, P. putida KT2440 and P. fulva CCUG12573 with a chromosomally integrated reporter that measures transcriptional efficiency. These strains were selected owing to their ease of genetic transformation, and the same seven rpoB mutations were transformed in each strain.
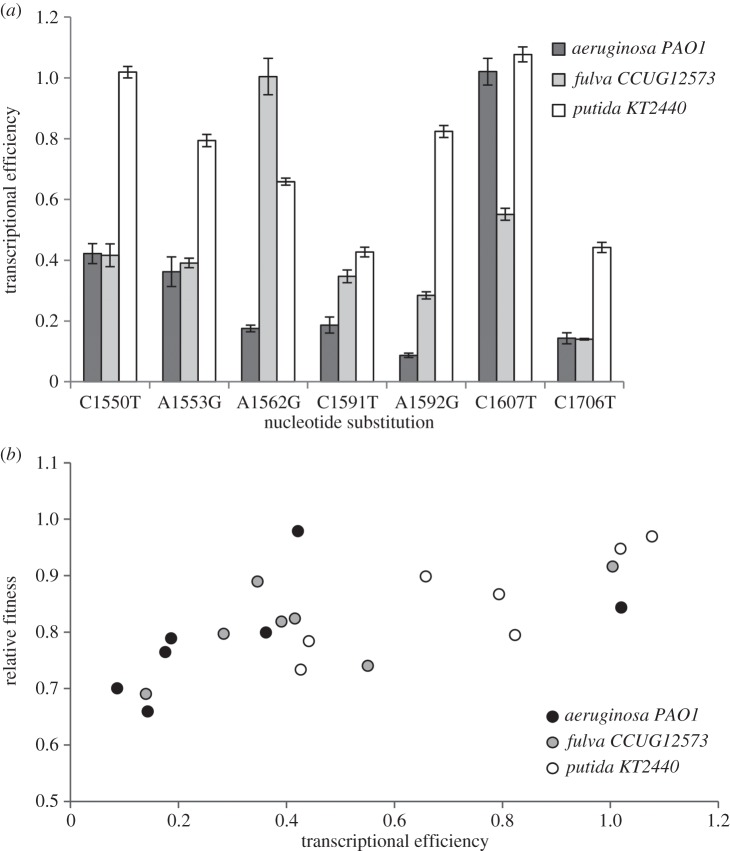
Given that transcriptional efficiency is likely to be an important determinant of fitness across strains, two mechanisms could explain why resistance mutations have background-dependent effects on fitness. First, it is possible that the same mutation has different effects on transcriptional efficiency in different strains. Consistent with this mechanism, we find transcriptional efficiency is significantly affected by mutation (figure 4a; F6,63 = 49.61, p < 0.001, variance explained = 37.5%), strain (figure 4a; F2,63 = 105.54, p < 0.001, variance explained = 26.6%) and, crucially, an interaction between strain and mutation (figure 4a; F12,63 = 18.48, p < 0.001, variance explained = 28.0%). In other words, the same rifampicin resistance mutation has different effects on transcriptional efficiency in different strains of Pseudomonas, resulting in epistatic interactions for fitness between resistance mutations and genetic background. Second, it is possible that even when resistance mutations have consistent effects on transcriptional efficiency across strains, compromised transcriptional efficiency carries a greater cost in some strains than others. To test if the relationship between transcription and fitness is strain-dependent, we analysed the fitness of these 21 transformed mutants using a general linear model with transcription rate fitted as a covariate and strain fitted as a fixed factor. As expected (figure 4b), we find that transcriptional efficiency significantly correlates with fitness (transcription: F1,15 = 12.3, p < 0.005, variance explained = 45.1%). This correlation is not significantly affected by strain (F2,15 = 0.45, p = 0.645, variance explained = 5.7%), nor by an interaction between strain and transcription rate (F2,15 = 0.37, p = 0.697, variance explained = 4.7%), demonstrating that the relationship between transcriptional efficiency and fitness is constant across strains.
Figure 4.
Linking epistasis to transcriptional efficiency. (a) The transcriptional efficiency (±s.e.m; n = 6) of seven rifampicin resistance mutations in P. putida KT2440, P. aeruginosa PAO1 and P. fulva CCUG12573. (b) The correlation between relative fitness and transcriptional efficiency for the seven mutations common to these three strains. Transcriptional efficiency and fitness were independently assayed relative to the ancestral strains, such that a value of 1 denotes fitness or transcriptional efficiency equal to the ancestor.
4. Discussion
Although genetic background is increasingly being seen as an important influence on adaptation (reviewed in [24,25]), experimental estimates of its role remain rare. Here we demonstrate that epistatic interactions between antibiotic resistance mutations and the genetic backgrounds in which they evolve are key in determining the fitness costs of rifampicin resistance. Given that fitness costs have a central role in the long-term dynamics of resistance, genetic background is likely to play a key role in the evolution of resistance. Specifically, we find that the fitness effects of over 40% of rifampicin resistance mutations differ significantly between species of Pseudomonas. In agreement with previous works [15,20], we find that compromised transcriptional efficiency plays a key role in determining the cost of rifampicin resistance. Moreover, the relationship between transcriptional efficiency and fitness is constant across strains, and the epistatic effects of rifampicin resistance on mutations arise from the fact that mutations have background-specific effects on transcriptional efficiency. In other words, in this system the relationship between phenotype (transcriptional efficiency) and fitness is constant over the range of tested values, but the phenotypic effect of mutations is complex and strain-specific.
One feature of the design of our study is that we tested for background effects at quite a broad phylogenetic scale. Pseudomonas is a genus that is well known for its remarkable phenotypic and genetic diversity [14]. The strains used in this study were selected to sample a broad range of this diversity without sacrificing the experimental tractability that is required to accurately measure the fitness costs of resistance. From our data, it is clear that genetic background has a pervasive impact on the cost of resistance at this phylogenetic scale. However, our study does not provide any insights into whether this is likely to be the case at a finer phylogenetic scale, such as different clones from the same species. Although there is almost certainly less diversity within any one species than observed between our isolates from many different species, this is not to say there is considerable genomic diversity within species of bacteria [26–28]. Equally, the cost of resistance has been shown to be epistatic even when strains only vary by as little as a single mutation [11,12]. Taken together, there is no reason why genetic background effects cannot also be as pronounced as observed here at considerably smaller evolutionary scales.
The cost of antibiotic resistance is predicted to be crucial to the clinical evolutionary dynamics of resistance [1,2]. As genetic background strongly affects the cost of resistance, this may explain why certain lineages of pathogen species are more likely to be epidemic than others [29]. The explanation for these so-called dominant or epidemic strains is often the possession of specific traits, such as particular antibiotic resistance determinants or virulence factors. But this leads to the question of why these strains have successfully acquired these traits, but other clones have not. Although there are several potential reasons, one interpretation is that these genetic backgrounds are the ones where these traits possess the lowest cost. This raises the important point that the function and cost of clinically important traits measured on a particular (often laboratory-adapted) strain should be assumed to be identical for all strains and clones.
In our experiment, we investigated only the relationship between fitness and transcription in a single environment. However, not only do clinical populations of bacteria encounter many different environments, but the cost of antibiotic resistance has been shown to be environment-dependent [30]. It is unlikely that transcription will be equally correlated with fitness across all possible environments. Indeed, it has been previously shown that the costs of rpoB mutations are reduced when the need for transcription is artificially lowered [31]. Therefore, in some environments (or rather in some combinations of environments and genetic backgrounds) the relationship between transcription and fitness will disappear. However, the relationship between transcription and fitness in chronic Mycobacterium tuberculosis infections is likely to be strong. Using rifampicin to treat M. tuberculosis is rifampicin's most common clinical use, and consequently many M. tuberculosis populations evolve resistance to rifampicin via mutations in rpoB [6,32]. These populations will often subsequently adapt to the costs of rpoB mutations by fixing compensatory mutations [32], which in other systems have been shown to restore fitness by directly restoring transcriptional efficiency [15,33]. Therefore, it would appear that transcriptional efficiency is also likely to be tightly correlated with fitness in clinical M. tuberculosis infections.
Additionally, as genetic background can significantly affect the cost of resistance, this could have important consequences for adaptation to the cost of resistance. Compensatory adaptation to the cost of antibiotic resistance is predicted to be critical to its evolutionary dynamics [2], as it can slow or even prevent the extinction of resistant strains in the absence of selection for resistance. As the probability of compensatory adaptation should depend on the cost of a mutation, genetic background therefore has a central role in compensatory adaptation to the cost of resistance. Additional support for this comes from rifampicin-resistant M. tuberculosis, which is one of the best-characterized examples of compensatory adaptation [32]. It has been demonstrated that some strains can compensate very rapidly over the course of a few weeks. However, other strains, possessing the exact same mutation, took more than 2 years to compensate for this mutation, whereas yet other strains did not show compensatory dynamics at all [32]. Admittedly, there are other explanations for this besides genetic background, such as adaptation to the infection environment rather than to compensation per se. However, it could also be that compensation is more rapid on genetic backgrounds in which the mutation possesses the greatest cost.
Given that rpoB is highly conserved, it is surprising that the same mutation has different effects on transcriptional efficiency in different strains of Pseudomonas. However, this is overlooking the many different and often less conserved cellular systems involved in transcriptional efficiency. In support of this argument, previous work has shown that rpoB mutations can have highly pleiotropic effects [34–36], including altered expression of rpoC [20], another subunit of RNA polymerase. Therefore, alterations to the expression of other genes involved in transcription may explain the variable impacts of these mutations on transcriptional efficiency, assuming the regulation of these genes is less conserved between species than the structure of rpoB. However, we stress that this is quite a speculative point, and further molecular work beyond the scope of this study would be required to test this hypothesis rigorously.
In summary, we present evidence that genetic background is a key determinant of the fitness costs of antibiotic resistance. As such, the effect of genetic background should not be ignored when studying the evolution and epidemiology of bacterial pathogens.
Data accessibility
Data are available from Dryad (doi:10.5061/dryad.qn370).
Authors' contributions
Conceived and designed the experiment: T.V., M.K., R.C.M.; performed the experimental work: T.V., M.K.; analysed the experiment: T.V.; wrote the paper: T.V., R.C.M.; commented on the paper: T.V., M.K., R.C.M.
Competing interests
We declare we have no competing interests.
Funding
The research leading to these results has received funding from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007–2013)/ERC grant agreement no. 281591 and from the Royal Society.
References
- 1.Andersson DI. 2006. The biological cost of mutational antibiotic resistance: any practical conclusions? Curr. Opin. Microbiol. 9, 461–465. ( 10.1016/j.mib.2006.07.002) [DOI] [PubMed] [Google Scholar]
- 2.Andersson DI, Hughes D. 2010. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 8, 260–271. ( 10.1038/nrmicro2319) [DOI] [PubMed] [Google Scholar]
- 3.Vogwill T, MacLean RC. 2015. The genetic basis of the fitness costs of antimicrobial resistance: a meta-analysis approach. Evol. Appl. 8, 284–295. ( 10.1111/eva.12202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat. Rev. Microbiol. 8, 423–435. ( 10.1038/nrmicro2333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV. 2015. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51. ( 10.1038/nrmicro3380) [DOI] [PubMed] [Google Scholar]
- 6.Brandis G, Pietsch F, Alemayehu R, Hughes D. 2015. Comprehensive phenotypic characterization of rifampicin resistance mutations in Salmonella provides insight into the evolution of resistance in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 70, 680–685. ( 10.1093/jac/dku434) [DOI] [PubMed] [Google Scholar]
- 7.Vogwill T, Kojadinovic M, Furió V, MacLean RC. 2014. Testing the role of genetic background in parallel evolution using the comparative experimental evolution of antibiotic resistance. Mol. Biol. Evol. 31, 3314–3323. ( 10.1093/molbev/msu262) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weigel LM, Steward CD, Tenover FC. 1998. gyrA mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae. Antimicrob. Agents Chemother. 42, 2661–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breen MS, Kemena C, Vlasov PK, Notredame C, Kondrashov FA. 2012. Epistasis as the primary factor in molecular evolution. Nature 490, 535–538. ( 10.1038/nature11510) [DOI] [PubMed] [Google Scholar]
- 10.Costanzo M, et al. 2010. The genetic landscape of a cell. Science 327, 425–431. ( 10.1126/science.1180823) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacLean RC, Perron GG, Gardner A. 2010. Diminishing returns from beneficial mutations and pervasive epistasis shape the fitness landscape for rifampicin resistance in Pseudomonas aeruginosa. Genetics 186, 1345–1354. ( 10.1534/genetics.110.123083) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ward H, Perron GG, Maclean RC. 2009. The cost of multiple drug resistance in Pseudomonas aeruginosa. J. Evol. Biol. 22, 997–1003. ( 10.1111/j.1420-9101.2009.01712.x) [DOI] [PubMed] [Google Scholar]
- 13.Silby MW, Winstanley C, Godfrey SAC, Levy SB, Jackson RW. 2011. Pseudomonas genomes: diverse and adaptable. FEMS Microbiol. Rev. 35, 652–680. ( 10.1111/j.1574-6976.2011.00269.x) [DOI] [PubMed] [Google Scholar]
- 14.Spiers AJ, Buckling A, Rainey PB. 2000. The causes of Pseudomonas diversity. Microbiology 146, 2345–2350. ( 10.1099/00221287-146-10-2345) [DOI] [PubMed] [Google Scholar]
- 15.Reynolds MG. 2000. Compensatory evolution in rifampin-resistant Escherichia coli. Genetics 156, 1471–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall AR, MacLean RC. 2011. Epistasis buffers the fitness effects of rifampicin-resistance mutations in Pseudomonas aeruginosa. Evolution 65, 2370–2379. ( 10.1111/j.1558-5646.2011.01302.x) [DOI] [PubMed] [Google Scholar]
- 17.Mariam DH, Mengistu Y, Hoffner SE, Andersson DI. 2004. Effect of rpoB mutations conferring rifampin resistance on fitness of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 48, 1289–1294. ( 10.1128/AAC.48.4.1289-1294.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wichelhaus TA, Böddinghaus B, Besier S, Schäfer V, Brade V, Ludwig A. 2002. Biological cost of rifampin resistance from the perspective of Staphylococcus aureus. Antimicrob. Agents Chemother. 46, 3381–3385. ( 10.1128/AAC.46.11.3381-3385.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melnyk AH, Wong A, Kassen R. 2015. The fitness costs of antibiotic resistance mutations. Evol. Appl. 8, 273–283. ( 10.1111/eva.12196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi Q, Preston GM, MacLean RC. 2014. Linking system-wide impacts of RNA polymerase mutations to the fitness cost of rifampin resistance in Pseudomonas aeruginosa. mBio 5, e01562-14. ( 10.1128/mBio.01562-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi K-H, Schweizer HP. 2006. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc. 1, 153–161. ( 10.1038/nprot.2006.24) [DOI] [PubMed] [Google Scholar]
- 22.Felsenstein J. 1985. Phylogenies and the comparative method. Am. Nat. 125, 1–15. ( 10.1086/284325) [DOI] [Google Scholar]
- 23.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. ( 10.1093/sysbio/syq010) [DOI] [PubMed] [Google Scholar]
- 24.Chandler CH, Chari S, Dworkin I. 2013. Does your gene need a background check? How genetic background impacts the analysis of mutations, genes, and evolution. Trends Genet. 29, 358–366. ( 10.1016/j.tig.2013.01.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandler CH, Chari S, Tack D, Dworkin I. 2014. Causes and consequences of genetic background effects illuminated by integrative genomic analysis. Genetics 196, 1321–1336. ( 10.1534/genetics.113.159426) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergthorsson U, Ochman H. 1998. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol. Biol. Evol. 15, 6–16. ( 10.1093/oxfordjournals.molbev.a025847) [DOI] [PubMed] [Google Scholar]
- 27.Loper JE, et al. 2012. Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 8, e1002784 ( 10.1371/journal.pgen.1002784) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson JR, Pacocha S, Pharino C, Klepac-Ceraj V, Hunt DE, Benoit J, Sarma-Rupavtarm R, Distel DL, Polz MF. 2005. Genotypic diversity within a natural coastal bacterioplankton population. Science 307, 1311–1313. ( 10.1126/science.1106028) [DOI] [PubMed] [Google Scholar]
- 29.Hallin M, Denis O, Deplano A, De Ryck R, Crèvecoeur S, Rottiers S, de Mendonça R, Struelens MJ. 2008. Evolutionary relationships between sporadic and epidemic strains of healthcare-associated methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 14, 659–669. ( 10.1111/j.1469-0691.2008.02015.x) [DOI] [PubMed] [Google Scholar]
- 30.Björkman J, Nagaev I, Berg OG, Hughes D, Andersson DI. 2000. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science 287, 1479–1482. ( 10.1126/science.287.5457.1479) [DOI] [PubMed] [Google Scholar]
- 31.Hall AR, Iles JC, MacLean RC. 2011. The fitness cost of rifampicin resistance in pseudomonas aeruginosa depends on demand for RNA polymerase. Genetics 187, 817–822. ( 10.1534/genetics.110.124628) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Comas I, Borrell S, Roetzer A, Rose G, Malla B, Kato-Maeda M, Galagan J, Niemann S, Gagneux S. 2012. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat. Genet. 44, 106–110. ( 10.1038/ng.1038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi Q, Toll-Riera M, Heilbron K, Preston GM, MacLean RC. 2016. The genomic basis of adaptation to the fitness cost of rifampicin resistance in Pseudomonas aeruginosa. Proc. R. Soc. B 283, 20152452 ( 10.1098/rspb.2015.2452) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao W, et al. 2013. The RpoB H481Y rifampicin resistance mutation and an active stringent response reduce virulence and increase resistance to innate immune responses in Staphylococcus aureus. J. Infect. Dis. 207, 929–939. ( 10.1093/infdis/jis772) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez-Verdugo A, Gaut B, Tenaillon O. 2013. Evolution of Escherichia coli rifampicin resistance in an antibiotic-free environment during thermal stress. BMC Evol. Biol. 13, 50 ( 10.1186/1471-2148-13-50) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu J, Tozawa Y, Lai C, Hayashi H, Ochi K. 2002. A rifampicin resistance mutation in the rpoB gene confers ppGpp-independent antibiotic production in Streptomyces coelicolor A3(2). Mol. Gen. Genomics 268, 179–189. ( 10.1007/s00438-002-0730-1) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available from Dryad (doi:10.5061/dryad.qn370).