Abstract
The remodelling of organelle function is increasingly appreciated as a central driver of eukaryotic biodiversity and evolution. Kinetoplastids including Trypanosoma and Leishmania have evolved specialized peroxisomes, called glycosomes. Glycosomes uniquely contain a glycolytic pathway as well as other enzymes, which underpin the physiological flexibility of these major human pathogens. The sister group of kinetoplastids are the diplonemids, which are among the most abundant eukaryotes in marine plankton. Here we demonstrate the compartmentalization of gluconeogenesis, or glycolysis in reverse, in the peroxisomes of the free-living marine diplonemid, Diplonema papillatum. Our results suggest that peroxisome modification was already under way in the common ancestor of kinetoplastids and diplonemids, and raise the possibility that the central importance of gluconeogenesis to carbon metabolism in the heterotrophic free-living ancestor may have been an important selective driver. Our data indicate that peroxisome modification is not confined to the kinetoplastid lineage, but has also been a factor in the success of their free-living euglenozoan relatives.
Keywords: organelle evolution, metabolic compartmentalization, peroxisomes, glycolysis, diplonemids, kinetoplastids
1. Introduction
Glycolysis (the Embden–Meyerhof–Parnas pathway) and gluconeogenesis are common to all domains of life and play fundamental roles in essential cellular processes. Glycolysis consists of 10 enzymatic steps, and generates ATP, NADH and intermediate metabolites for a variety of metabolic processes, while gluconeogenesis produces glucose 6-phosphate (G6P) from other carbon sources including lipids and amino acids [1]. Glycolysis takes place in the cytosol, but glycolytic enzymes have also been associated with the plasma membrane of erythrocytes [2] and with mitochondria [3,4]. In addition, enzyme isoforms with non-glycolytic roles are found to locate to other compartments [5].
Relocation of the first 7 of 10 glycolytic enzymes to microbody-like organelles, called glycosomes, was first demonstrated in the kinetoplastid parasite, Trypanosoma brucei [6], and subsequently in other kinetoplastids [7]. So far, the kinetoplastids, which comprise bodonids and trypanosomatids, are the only organisms known to sequester a successive glycolytic cascade into organelles. These enzymes are targeted to glycosomes by a peroxisomal targeting signal (PTS), implying that glycosomes originated as modified peroxisomes. Glycosomes also contain enzymes of other pathways including gluconeogenesis, the pentose phosphate pathway, energy/redox metabolism and nucleotide synthesis [7]. Glycosomes are thus a classic example of how complex and physiologically important metabolic pathways can be relocated during evolution.
The relocation of entire metabolic pathways in eukaryotes is rarely observed, because isolation of the individual pathway enzymes by transfer from one compartment to another can abolish otherwise tightly coordinated enzymatic cascades. To overcome these difficulties, a ‘minor-mistargeting’ mechanism has been proposed to allow the dual location of pathways [8]. According to this hypothesis, the imperfect targeting of PTS-tagged enzymes to peroxisomes would allow the retention of functional glycolysis in the cytosol until the newly routed peroxisomal enzymes could reconstitute the pathway. This hypothesis provides a plausible mechanism for how functional retargeting could occur, but does not identify the physiological conditions or selective pressures that might drive the initial evolution of compartmentalization.
The phylum Euglenozoa currently contains the euglenoids, diplonemids and kinetoplastids, with diplonemids the sister group of kinetoplastids and euglenoids the outgroup to both [9]. In euglenoids, glycolysis occurs in the cytosol but some enzymes are also located to its secondary plastid to participate in the Calvin cycle, as in plants and green algae [10,11]. In the free-living marine diplonemid Diplonema papillatum, we have previously reported that fructose-1,6-biphosphate aldolase (FBPA), the fourth enzyme of glycolysis, has a type-2 PTS (PTS2) and is compartmentalized [12], suggesting that the transition to glycosomes may have already occurred in the common ancestor of diplonemids and kinetoplastids.
2. Material and methods
(a). Organisms
Diplonema papillatum (strain ATCC 50162) and the kinetoplastid Trypanosoma cruzi (Tulahuen stock) were grown axenically in ATCC 1532 and 1029 media, respectively. ATCC 1532 medium contains 0.1% (w/v) tryptone and 1% horse serum, corresponding to 10 mM amino acid ingredients and 40–60 µM glucose, respectively. ATCC 1029 LIT medium is a rich medium that contains 6 mM glucose and 0.9% (w/v) liver infusion broth/0.5% tryptone, equivalent to greater than 100 mM amino acid ingredients. All experiments were carried out using the standard medium unless otherwise stated.
(b). Draft genome sequencing of Diplonema papillatum
Diplonema papillatum genomic DNA was extracted using standard protocols, purified on agarose gel to remove mitochondrial DNA and used for genome sequencing. The D. papillatum genomic DNA was sequenced using a HiSeq 2000 apparatus (Illumina K.K., Tokyo, Japan) by Takara Bio Inc., Shiga, Japan and Eurofins Genomics K.K., Tokyo, Japan. Total reads were 21.5 Gb and the estimated genome size was 176.5 Mb. The D. papillatum open reading frames for genes of interest were identified using TBLASTN. We predicted PTS1 by the Prosite pattern PS00342 for the C-terminal tripeptides, [-(STAGCN)-(RKH)-(LIVMAFY)], and the PTS 1 Predictor (http://mendel.imp.ac.at/pts1/) [13]; we required a positive identification from both methods to diagnose the presence of a bona fide PTS. The presence of a PTS2 was manually inspected based on the N-terminal non-americ consensus sequence, (RK)-(LIV)-X5-(QH)-(LA) [14]. The T. brucei glycosomal proteins obtained by detailed proteomic analysis were used as references [15].
(c). Phylogenetics
We used the D. papillatum sequences as queries in BLASTP searches against the other euglenozoan genomes and a broad selection of outgroups. The Euglena gracilis PFK sequence was obtained from http://euglenadb.org/. Sequences were aligned with Meta-Coffee [16]. Poorly aligning regions were masked using the ‘automated1’ option in trimAl [17]. Phylogenies were built using the LG [18], C20 and C60 [19] in PhyloBayes. The trees inferred under the three methods were very similar, with no differences impacting on our inferences of the relationships among the euglenozoans, and the consensus trees in figure 1; electronic supplementary material, figures S1–S5 and S8–S10 were inferred using the C60 model.
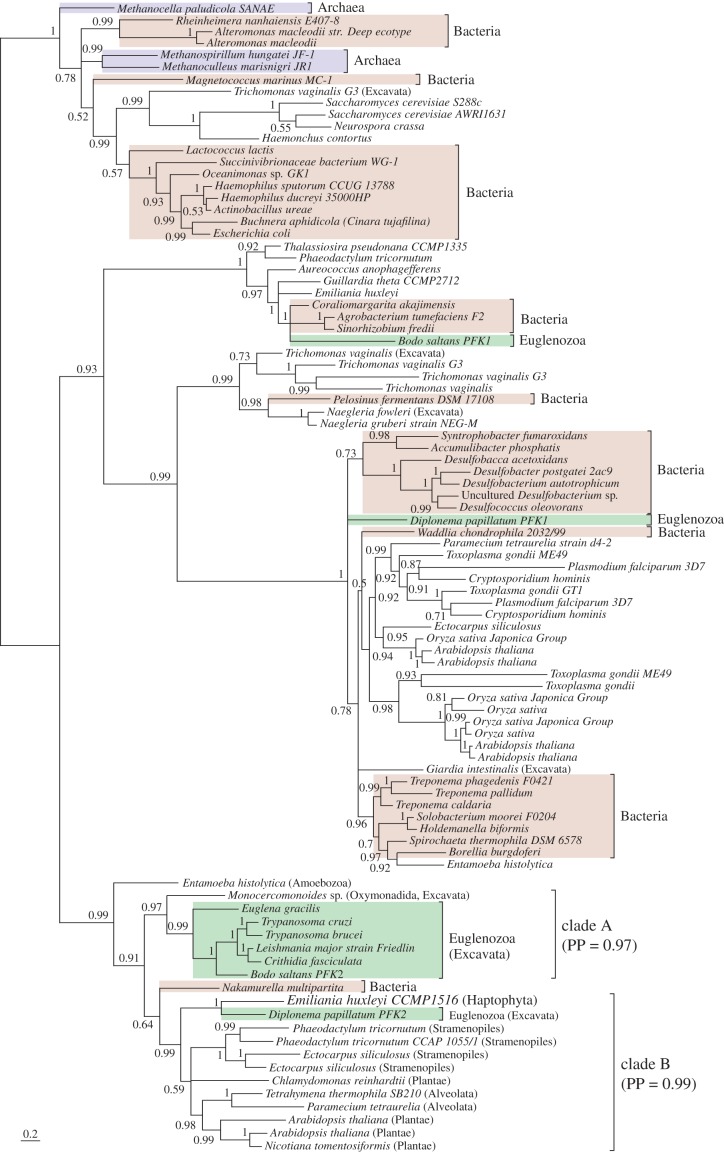
Figure 1.
Diplonema papillatum lacks an orthologue of the glycosome-targeted phosphofructokinase (PFK) gene found in other euglenozoans. A Bayesian consensus phylogeny of PFK was inferred under the C60 model in PhyloBayes. Euglena gracilis and kinetoplastid PFKs form a monophyletic group (clade A) with high posterior support (PP = 0.99). By contrast, D. papillatum PFK2 clusters with a PFK homologue from the haptophyte E. huxleyi (PP = 1) and is nested within the clade ‘B’ (PP = 0.99). Branch lengths are proportional to the expected number of substitutions.
(d). Indirect immunofluorescence analysis
Antisera to glycolytic enzymes were obtained as described in electronic supplementary material, supplementary materials and methods. Indirect immunofluorescence analysis (IFA) was performed as described [12]. Briefly, cells were fixed with 2% paraformaldehyde, probed with rabbit antisera to each glycolytic enzyme and mouse antisera to D. papillatum FBPA as a peroxisomal marker and then treated with anti-rabbit Alexa Fluor 568- and anti-mouse Alexa Fluor 488-conjugates (Life Technologies), respectively, followed by counterstaining of the nucleus with Hoechst 33342 dye. Fluorescence was observed using fluorescence microscopy (Axio Imager M2, Carl Zeiss Co., Tokyo, Japan). The reactions with the combination of the rabbit antisera and the 488-conjugate, without the anti-FBPA combination, showed the same patterns as observed in the co-localization experiments.
(e). Glucose and amino acid consumption
Diplonema papillatum cells of the mid-log phase were cultured in ATCC 1532 medium supplemented with 6 mM d-glucose, while T. cruzi cells were cultured in LIT medium. The glucose concentration of the culture supernatant was measured using a Glucose-Oxidase assay kit (Sigma-Aldrich). Amino acid consumption by cells was evaluated by the increase of NH4+ in the culture supernatant using the Ammonia assay kit (Sigma-Aldrich).
(f). Metabolic labelling
Cells in the mid-log phase of growth were starved in artificial seawater (ATCC 1405 HESNW medium) for 2 h and then incubated with 6 mM 13C6 d-glucose or 6 mM 13C5 l-glutamine (Sigma-Aldrich) in artificial seawater for 0, 4 and 8 h or with 30 mM 13C6 d-glucose in ATCC 1532 medium for 72 h without starvation. The detailed experimental procedures for extraction of metabolites and analysis by CE-Q-TOFMS and LC-MS were described in the electronic supplementary material, supplementary materials and methods.
3. Results
(a). Identification of peroxisome targeting signals in the glycolytic enzymes of Diplonema papillatum
To investigate glycolytic compartmentalization in diplonemids, we sequenced the genome of D. papillatum and identified all of the genes for glycolysis (table 1). We identified putative PTS for two types of hexose kinase, namely a high-affinity type hexokinase (HK) and a low-affinity type glucokinase (GLK), and for phosphoglucose isomerase (PGI) and phosphoglycerate kinase (PGK), similar to kinetoplastid homologues. We identified two homologues of phosphofructokinase (PFK), PFK1 and PFK2, in the genome assembly. PFK1 possesses a predicted PTS1 but is probably a pseudogene or a bacterial contaminant (see below). We also found a PTS for triose-phosphate isomerase (TIM), in spite of the absence of an apparent PTS in the kinetoplastid TIM. By contrast, putative PTS were absent from the predicted coding sequences of PFK2, phosphoglycerate mutase (PGAM, 2,3-bisphosphoglycerate-independent type), enolase (ENO), pyruvate kinase (PK) and two isoforms of glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Table 1.
Characteristics of glycolytic enzymes of Diplonema papillatum. n.d., not detected.
| PTS |
||||
|---|---|---|---|---|
| enzyme | D. papillatum | T. cruzi | monophylya | activityb |
| 1. HK | PTS2 | PTS2 | yes (0.85) | 0.92c |
| 1. GLK | PTS1 | PTS1 | yes (0.96) | |
| 2. PGI | PTS1 | PTS1 | yes (0.93) | 39.1 |
| 3. PFK | PTS1/n.d. | PTS1 | no (0.99) | n.d. |
| 3. FBPase (gluconeogenic) | PTS1 | PTS1 | no (1.00) | 4.7 |
| 4. FBPA | PTS2 | PTS2 | yes (1.00)d | 14.2 |
| 5. TIM | PTS1 | n.d. | unresolved | 123.2 |
| 6. GAPDH | n.d. | PTS1/n.d. | no (0.90) | 48.5 |
| 7. PGK | PTS1 | PTS1/n.d. | yes (0.81) | 103.7 |
| 8. PGAM | n.d. | n.d. | no (1.00) | 2100 |
| 9. ENO | n.d. | n.d. | yes (0.99) | 3.7 |
| 10. PK | n.d. | n.d. | yes (0.98) | 15.1 |
aMonophyly of D. papillatum and kinetoplastids with posterior probability in parenthesis based on the relevant protein phylogeny (figure 1; electronic supplementary material, figures).
bnmol min−1 mg−1 protein of the D. papillatum cell extract (see also electronic supplementary material).
cThe activities of HK (high-affinity type) and GLK (low-affinity type) are indistinguishable.
dRef [12].
Phylogenetic analysis demonstrated that most of these enzymes (HK, GLK, PGI, PGK, ENO and PK) were present in the common ancestor of D. papillatum and kinetoplastids (electronic supplementary material, figures S1–S3). The phylogeny for TIM was weakly supported and hence cannot reject the hypothesis of a single common origin for TIM (electronic supplementary material, figure S1d). By contrast, three glycolytic enzymes—PFK1 and PFK2, the two isoforms of GAPDH, and PGAM—appear to have separate evolutionary origins for D. papillatum and the kinetoplastids (figure 1; electronic supplementary material, figure S1e and S4).
The protein phylogeny for PFK revealed that E. gracilis and kinetoplastids are monophyletic with high posterior support (PP = 0.99), suggesting their shared evolutionary origin (clade A, figure 1). By contrast, D. papillatum PFK2 clusters with a PFK homologue from the haptophyte Emiliania huxleyi (PP = 1) and is nested within a different eukaryotic clade (clade B, PP = 0.99), suggesting replacement of the ancestral euglenozoan PFK by lateral gene transfer. Diplonema papillatum PFK1 detected in our genome assembly is probably a pseudogene or a bacterial contaminant: it does not group with other eukaryotic PFKs in the tree; the putative pfk1 gene lacks a splice acceptor site at its 5′-untranslated region, which is essential for trans-splicing in euglenozoans; and we were unable to detect any PFK activity in the D. papillatum lysate (table 1). In summary, our data suggest that PTS-driven relocation of most, but not all, glycolytic enzymes had already occurred in the common ancestor of diplonemids and kinetoplastids.
We also investigated the presence of a PTS in the non-glycolytic enzymes of D. papillatum. Among 18 PTS-harbouring enzymes specific to kinetoplastid glycosomes, only three of these enzymes were also predicted to have PTS in D. papillatum, which participate in gluconeogenesis and energy metabolism (table 1; electronic supplementary material, table S1). These include the gluconeogenic fructose-1,6-biphosphatase (FBPase) that catalyses the reverse reaction to PFK, ATP-producing glycerol kinase (GK) and pyruvate phosphate dikinase (PPDK). None of the enzymes identified for the Diplonema pentose phosphate pathway (PPP), pyrimidine biosynthesis or purine salvage were predicted to locate to peroxisomes. Notably, despite of the presence of PTS1, the protein phylogeny of FBPase revealed different evolutionary origins between D. papillatum and kinetoplastids, implying independent LGT events and convergent PTS-driven relocation of FBPase in D. papillatum and kinetoplastids (table 1; electronic supplementary material, figure S5). Our data suggest that metabolic compartmentalization originated in association with glycolysis, gluconeogenesis and energy metabolism in the common ancestor of D. papillatum and kinetoplastids.
(b). Localization of peroxisomal targeting signal-harbouring glycolytic enzymes in the peroxisomes of Diplonema papillatum
To investigate the expression and confirm the cellular location of the D. papillatum glycolytic enzymes, we raised antisera to 11 glycolytic enzymes including GLK. Western blot analysis of the D. papillatum extracts showed the expected band for all enzymes tested, with the exception of the two homologues of PFK and HK (electronic supplementary material, figure S6a). The absence of the signals for both PFK1 and PFK2 suggests that neither of the two PFK homologues is expressed or functional in the culture condition of D. papillatum, consistent with the lack of PFK activity. To confirm the absence of HK, we measured hexose kinase activity in D. papillatum cell extracts and found that the Km value for glucose of the native enzyme harbouring hexose kinase activity was 0.33 mM, whereas the Km values of the recombinant HK and GLK were 0.064 and 0.66 mM, respectively. These results suggest that the native hexose kinase activity is probably attributable to GLK in D. papillatum under the experimental conditions.
The IFA was performed to investigate the cellular location of the glycolytic enzymes expressed in D. papillatum. Our previous study showed that our mouse antiserum to D. papillatum FBPA detected a single band on Western blots of the membrane-rich fraction (10 500g sediment) from the D. papillatum extract and also demonstrated the punctate pattern in the cell by IFA [12]. Immuno-electron microscopy using the same antiserum also detected labelling of FBPA inside single-membrane compartments with the characteristic morphology of peroxisomes (electronic supplementary material, figure S6b). Therefore, we used FBPA as a peroxisomal marker. The signals for GLK, PGI, TIM and PGK co-localized with that of FBPA, indicating their shared peroxisomal location (figure 2). The signal for the anti-GAPDH1 antibody gave a patchy distribution in the cytosol but did not co-localize with FBPA, consistent with the absence of a PTS. PTS-lacking PGAM, ENO and PK were also exclusively detected in the cytosol of D. papillatum, similar to their kinetoplastid PTS-lacking homologues.
Figure 2.
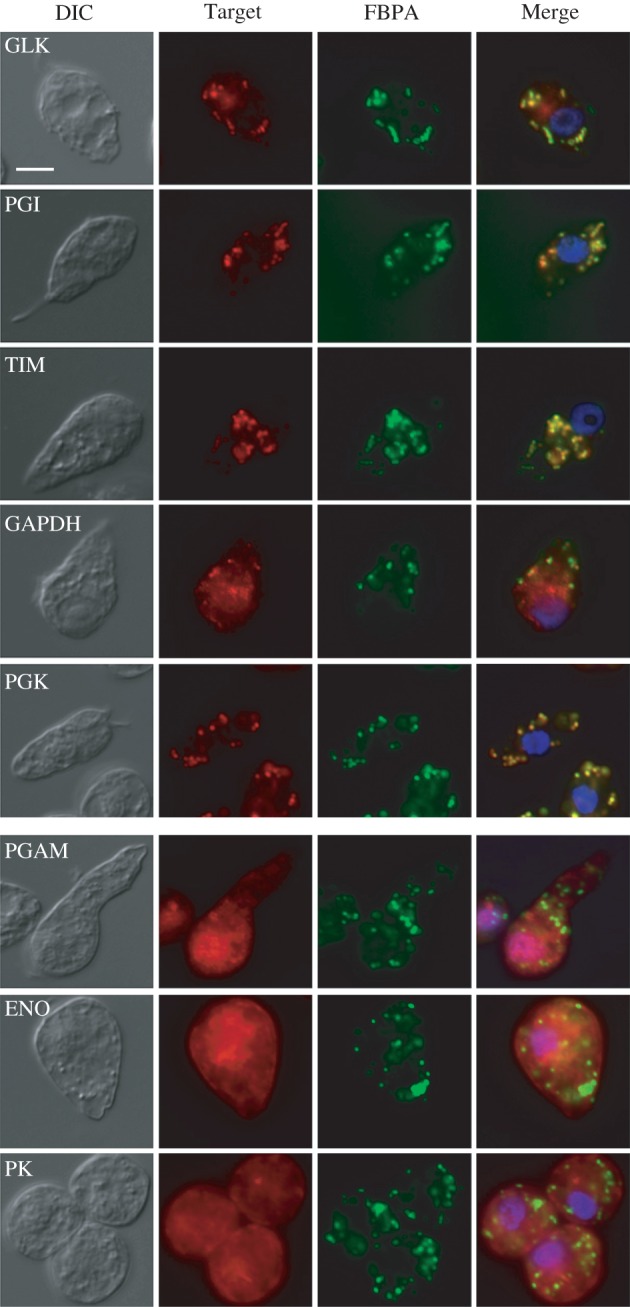
Cellular localization of glycolytic enzymes in Diplonema papillatum using immunofluorescence microscopy. Cells were labelled with polyclonal antisera to GLK, PGI, TIM, GAPDH, PGK, PGAM, ENO or PK (Target, red) and with antisera to D. papillatum FBPA as a peroxisomal marker (FBPA, green), and the reactions were visualized using Alexa Fluor 568- and 488-conjugates, respectively. Images of the same cells were merged together with the labels of Hoechst 33342 (blue) to visualize the nucleus (Merge). DIC, differential interference contrast image; Merge, merged image. Scale bar, 10 µm.
(c). Preference of amino acids as carbon source in Diplonema papillatum
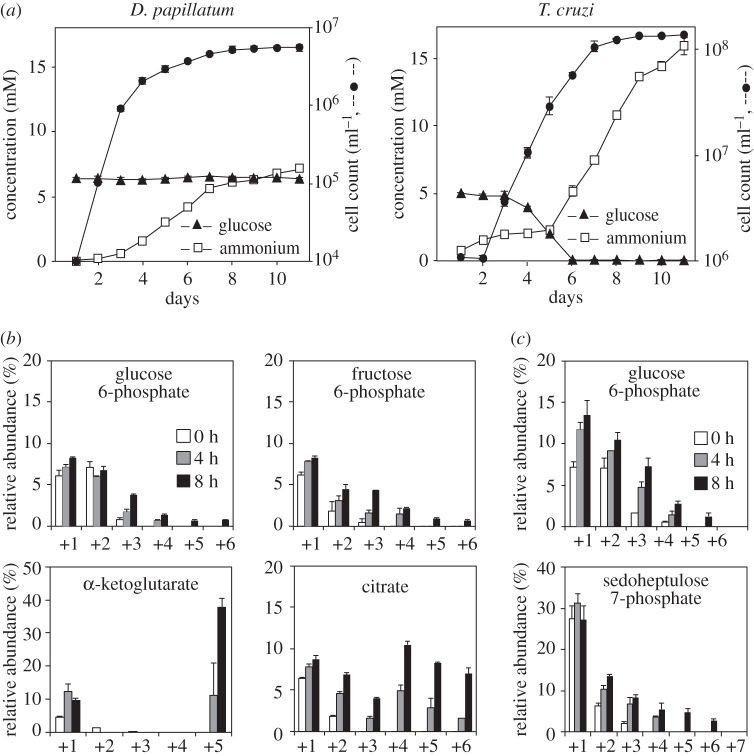
The failure to detect PFK activity in D. papillatum suggests that the glycolytic enzymes together with FBPase play a replenishing role for gluconeogenesis in this organism. Notably, D. papillatum can grow with a trace level of glucose. The modified ATCC 1532 medium supplemented with 1% dialysed fetal bovine serum (Thermo Fisher Scientific Inc.), which corresponds to less than 2.7 µM glucose, can also support the normal growth of D. papillatum (data not shown). However, the growth of D. papillatum even in the absence of glucose does not necessarily indicate the loss of glucose uptake and glycolytic activity. Therefore, we investigated firstly the relative preference of carbon sources in D. papillatum. Consistent with our hypothesis, D. papillatum does not significantly consume glucose (6 mM), but preferentially uses amino acids (figure 3a, left). This is demonstrated by the significant production of ammonium in the presence of glucose during early exponential growth. By contrast, epimastigotes (an insect form) of the parasitic kinetoplastid T. cruzi prefer glucose to amino acids, as previously reported [20]. In experiments with T. cruzi (figure 3a, right), the levels of glucose decreased significantly at day 3 to 6 along with exponential growth and became undetectable (less than 10 µM) at day 7, while the levels of ammonium remained constantly by day 5 and then increased significantly. Because amino acids are the major nutritional components in the diet of phagocytic heterotrophs like D. papillatum, this preference probably reflects nutritional adaptation.
Figure 3.
Carbon metabolism in Diplonema papillatum. (a) Consumption of glucose and amino acids in the culture media of D. papillatum (left) and Trypanosoma cruzi epimastigotes (right). Production of ammonium, a catabolite of amino acids, denotes the consumption of amino acids. (b,c) Metabolome analysis of D. papillatum. Diplonema papillatum cells were incubated in conditioned marine water for 2 h and then supplemented with 13C5-l-glutamine (b) or 13C6-d-glucose (c) for 0, 4 and 8 h. The y-axis indicates the relative abundance (%) of each isotopomer of the relevant metabolite. The x-axis represents the number of 13C in the isotopomers. Data for all metabolites in these experiments are shown in electronic supplementary material, figures S7a and S7b, respectively. Data points and error bars represent the mean ± s.d. of three independent experiments.
(d). Gluconeogenesis is the central carbon metabolism in Diplonema papillatum
To investigate the metabolic fate of amino acids, we performed tracer experiments using 6 mM 13C-uniformly labelled (13C5, +5 carbons) l-glutamine, and D. papillatum cells which had been starved in artificial seawater for 2 h prior to labelling. We detected specific enrichment with +5 carbon in the α-ketoglutarate fraction, indicating that glutamine is readily catabolized and incorporated into metabolites of the TCA cycle of D. papillatum (figure 3b; electronic supplementary material, figure S7a). The presence of +5 and +6 carbons in the citrate fraction also indicated an active TCA cycle and the recycling of labelled oxaloacetate. The time-dependent increase of 13C-labelling with +1 to +6 carbons for hexose phosphates provided evidence of active gluconeogenesis. In addition, the extensive metabolic dilution of 13C-isotopologues in the hexose 6-phosphate fractions indicated the presence of an internal carbon source. Quantitative metabolomics demonstrated that D. papillatum contains higher amounts of free amino acids when compared with T. cruzi, suggesting that they may serve as a carbon reservoir (electronic supplementary material, table S2). We conclude that amino acids are the preferred carbon/energy source for D. papillatum and that they are catabolized via the TCA cycle and can subsequently be anabolized via gluconeogenesis.
(e). The pentose phosphate pathway complements PFK-deficiency in the glycolytic process
Metabolic labelling using 6 mM 13C6-d-glucose as a sole external carbon source showed enrichment with +1 to +5 carbons in the hexose phosphate fractions, indicating the occurrence of glucose catabolism in D. papillatum at a very low rate (figure 3c; electronic supplementary material, figure S7b). Importantly, sedoheptulose 7-phosphate (S7P), an intermediate metabolite of PPP, was significantly labelled with +1 to +6 carbons (figure 3c). Glucose uptake by D. papillatum was further investigated by cells incubated for 72 h in normal medium containing 30 mM 13C6-glucose. Under these conditions, the levels of +6 carbon were significantly lower than that of +3 carbon in the hexose phosphate fractions (electronic supplementary material, figure S7c). These data suggest that G6P is catabolized via the PPP to produce triose phosphates and their subsequent catabolites, which can then be recycled to form hexose phosphate species. Quantitative determination of the glycolytic metabolites also showed a relative accumulation of hexose phosphates in D. papillatum compared with T. cruzi, regardless of the nutritional conditions (electronic supplementary material, table S2), suggesting the predominance of anabolism towards the production of hexose phosphates. Taken together, these data demonstrate that glycolysis does not significantly contribute to central carbon metabolism in D. papillatum and that gluconeogenesis from amino acids is the dominant process.
(f). Retargeting of non-glycolytic pathway enzymes to peroxisomes occurred in the common ancestor of kinetoplastids
To investigate the evolution of kinetoplastid glycosomes after the split from the diplonemid lineage, we inferred additional phylogenies for PTS-harbouring enzymes specific to kinetoplastids. Both the phylogeny of adenylate kinase—which catalyses the reversible interconversion of 2 ADP into ATP plus AMP—and of inosine 5-monophosphate dehydrogenase—a purine salvage enzyme—revealed that the glycosomal forms of these enzymes have been acquired laterally from bacteria in kinetoplastids (electronic supplementary material, figures S8 and S9). Thus, kinetoplastids encode two copies of these genes: an ancestral copy shared with D. papillatum, and a glycosomal copy obtained by LGT. The phylogeny of a PPP enzyme, transketolase, showed monophyly of D. papillatum and kinetoplastids within the eukaryotic clade, indicating that the ancestral forms of this enzyme were retargeted to the glycosomes in the lineage leading to kinetoplastids (electronic supplementary material, figure S10). The phylogeny of other PTS-targeted enzymes was ambiguous, largely due to a lack of resolution commonly encountered in single-gene trees. We conclude that the transfer to the glycosomes of non-glycolytic enzymes, including those for pyrimidine biosynthesis [21], occurred on the branch leading to the kinetoplastids after their split from the diplonemid lineage (figure 4).
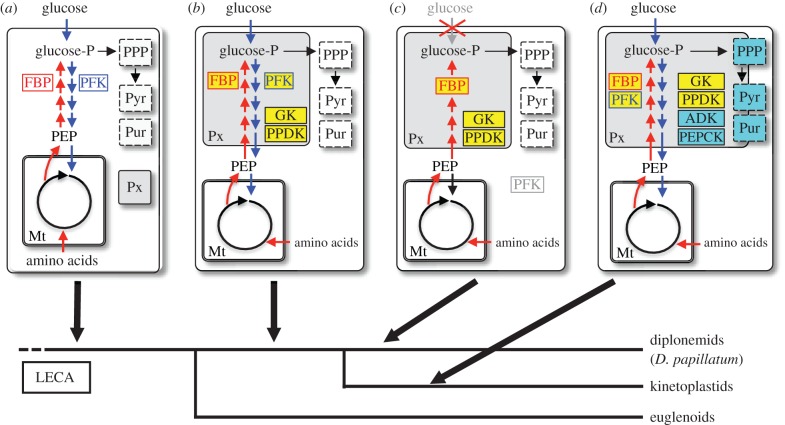
Figure 4.
A proposed metabolic model and peroxisome remodelling in Euglenozoa. (a) Peroxisomes (Px) and mitochondria (Mt) have been present in the last eukaryotic common ancestor (LECA), with cytosolic location of glycolysis (blue arrows), gluconeogenesis (red arrows), pentose phosphate pathway (PPP), pyrimidine biosynthesis (Pyr) and purine salvage (Pur). PFK and FBPase (FBP) are critical allosteric enzymes in glycolysis and gluconeogenesis, respectively. (b) After the divergence of the lineage leading to Euglenoids, the peroxisomes of the common ancestor of diplonemids and kinetoplastids were remodelled through the sequestration of a subset of glycolytic enzymes, possibly FBPase and others (boxed in yellow). In these remodelled compartments, both glycolysis and gluconeogenesis were functional. (c) The ancestral diplonemids or Diplonema lost glycolysis accompanied by the loss of PFK activity as suggested in our analysis. (d) The ancestral kinetoplastid further sequestered into peroxisomes a part of enzymes for PPP, Pyr, Pur and others such as PEPCK and NADH-fumarate reductase (boxed in blue).
4. Discussion
Recent advances in our sampling of microbial eukaryotic diversity have begun to shed light on the unique biological features of diplonemids, not only as a relative of important human pathogens but also as a major protistan group in ocean plankton [22,23]. The ecological success of diplonemids as marine plankton [23] suggests that independence from glucose accompanied by peroxisome remodelling is a viable strategy for heterotrophic life. Our draft genome sequencing of Diplonema provides an important source of genomic data for this group.
The phylogeny of PFK revealed that kinetoplastid and E. gracilis PFKs share the same evolutionary origin. Therefore, it is likely that, as well as euglenoids, a common ancestor of diplonemids and kinetoplastids used PFK to perform glycolysis. Whether this ancestral PFK had already relocated to peroxisomes in their common ancestor is, as yet, unclear. By contrast, D. papillatum PFK2 shares the same origin with a PFK homologue of the haptophyte Emiliania huxleyi, which is the most prominent marine cosmopolitan phytoplankton and often causes coccolithophore blooms [24]. It is possible that marine heterotrophs like Diplonema feed on haptophytes, which may have been an evolutionary source of LGT; alternatively, both E. huxleyi and D. papillatum may have obtained these PFK genes by LGT from other members of clade B.
Our data demonstrate that the remodelling of peroxisome function already occurred in the common ancestor of diplonemids and kinetoplastids, and involved the retargeting of at least a subset of glycolytic enzymes and GK and PPDK. It is unclear whether FBPase also relocated to peroxisomes in their common ancestor, because both FBPases have different origins. Retargeting to peroxisomes of ATP-producing GK and PPDK may be related to the requirement of ATP hydrolysis in the first seven enzymatic steps of glycolysis, in terms of the maintenance of ATP homeostasis inside the peroxisomes. That is, the maintenance of the ATP levels by the action of GK, acting in the reverse direction, and PPDK in the peroxisomes might have played an important role in maintaining the efficiency of glycolysis and gluconeogenesis in these organisms.
As with enzymatic composition of the ancestral glycosomes, the primary metabolic status in the common ancestor of diplonemids and kinetoplastids is still unclear. Our metabolome data indicate that D. papillatum lacks a fully functional glycolytic pathway. By contrast, our phylogenetic analyses suggest that the ancestor of diplonemids and kinetoplastids likely retained glycolysis. A possible explanation is that the gluconeogenic state of Diplonema may not be an ancestral, but is instead a derived feature, which was accompanied by nutritional adaptations to heterotrophic life followed by the loss of expression and activity of PFK.
From a metabolic viewpoint, it seems rational that ancestral glycosomes harboured the first seven enzymes of glycolysis and the corresponding enzymes of gluconeogenesis for completion of a contiguous enzymatic cascade (figure 4). Diplonema papillatum may represent an interesting case whereby nutritional adaptations leading to glucose-independent metabolism have evolved, resulting in peroxisomes housing gluconeogenesis (gluconeosomes). At the same time, preservation of glycolytic activity in kinetoplastids might have potentiated adaptation to a different mode of life: parasitism. Parasitism has developed repeatedly among kinetoplastids, and this adaptability is made possible by the compartmentalization into glycosomes of metabolic processes that have to undergo drastic and rapid changes during some transitions in the complex life cycles [25]. Hence, the sequestering of core metabolic pathways into peroxisomes, begun in the common ancestor of diplonemids and kinetoplastids, may have laid the foundations for the success of a major clade of medically and economically important parasites. Indeed, high levels of glucose (in the millimolar range) are present in the blood and body fluids of vertebrates, which can be readily used by haemoflagellates as carbon source.
Supplementary Material
Acknowledgements
We thank Paul Michels for critical reading and the Euglena gracilis nucleotide sequencing consortium (http://euglenadb.org/) for E. gracilis PFK sequence.
Data accessibility
The D. papillatum genome was deposited in DDBJ/EMBL/GenBank under the accession LMZG00000000. Diplonema papillatum sequences for relevant enzymes reported here have been deposited individually at DDBJ (accession numbers AB970479–AB970481, AB970483–AB970501, LC127115, LC127507). The datasets supporting this article have been uploaded as part of the electronic supplementary material.
Authors' contributions
T.N. designed research; J.M., M.H., T.A.W., H.H.-M., A.T., N.K., H.T., T.F., M.K. and T.N. performed research; J.M., T.A.W. and T.N. analysed data; F.B. and J.L.C. provided materials; T.A.W., T.Ma., T.Mi., F.B., J.L.C. and T.H. helped to draft manuscript; T.A.W. and T.M.E edited the paper; J.M. and T.N. wrote the paper.
Competing interests
We have no competing interests.
Funding
T.N. acknowledges support from grants-in-aid for scientific research (24390102 and 25670205) and from the Foundation of Strategic Research Projects in Private Universities (S1201013) from the Ministry of Education, Culture, Sport, Science and Technology, Japan (MEXT). T.M.E. acknowledges support from the Wellcome Trust and the European Advanced Investigator Programme. T.A.W. is supported by a Royal Society University Research Fellowship.
References
- 1.Gruning NM, Rinnerthaler M, Bluemlein K, Mulleder M, Wamelink MM, Lehrach H, Jakobs C, Breitenbach M, Ralser M. 2011. Pyruvate kinase triggers a metabolic feedback loop that controls redox metabolism in respiring cells. Cell Metab. 14, 415–427. ( 10.1016/j.cmet.2011.06.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campanella ME, Chu H, Low PS. 2005. Assembly and regulation of a glycolytic enzyme complex on the human erythrocyte membrane. Proc. Natl Acad. Sci. USA 102, 2402–2407. ( 10.1073/pnas.0409741102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giegé P, Heazlewood JL, Roessner-Tunali U, Millar AH, Fernie AR, Leaver CJ, Sweetlove LJ. 2003. Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell 15, 2140–2151. ( 10.1105/tpc.012500) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith DG, Gawryluk RM, Spencer DF, Pearlman RE, Siu KW, Gray MW. 2007. Exploring the mitochondrial proteome of the ciliate protozoon Tetrahymena thermophila: direct analysis by tandem mass spectrometry. J. Mol. Biol. 374, 837–863. ( 10.1016/j.jmb.2007.09.051) [DOI] [PubMed] [Google Scholar]
- 5.Freitag J, Ast J, Bölker M. 2012. Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi. Nature 485, 522–525. ( 10.1038/nature11051) [DOI] [PubMed] [Google Scholar]
- 6.Opperdoes FR, Borst P. 1977. Localization of nine glycolytic enzymes in a microbody-like organelle in Trypanosoma brucei: the glycosome. FEBS Lett. 80, 360–364. ( 10.1016/0014-5793(77)80476-6) [DOI] [PubMed] [Google Scholar]
- 7.Gualdrón-López M, Brennand A, Hannaert V, Quiñones W, Cáceres AJ, Bringaud F, Concepción JL, Michels PA. 2012. When, how and why glycolysis became compartmentalised in the Kinetoplastea: a new look at an ancient organelle. Int. J. Parasitol. 42, 1–20. ( 10.1016/j.ijpara.2011.10.007) [DOI] [PubMed] [Google Scholar]
- 8.Martin W. 2010. Evolutionary origins of metabolic compartmentalization in eukaryotes. Phil. Trans. R. Soc. B 365, 847–855. ( 10.1098/rstb.2009.0252) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simpson AG, Roger AJ. 2004. Protein phylogenies robustly resolve the deep-level relationships within Euglenozoa. Mol. Phylogenet. Evol. 30, 201–212. ( 10.1016/S1055-7903(03)00177-5) [DOI] [PubMed] [Google Scholar]
- 10.Hannaert V et al. 2000. Enolase from Trypanosoma brucei, from the amitochondriate protist Mastigamoeba balamuthi, and from the chloroplast and cytosol of Euglena gracilis: pieces in the evolutionary puzzle of the eukaryotic glycolytic pathway. Mol. Biol. Evol. 17, 989–1000. ( 10.1093/oxfordjournals.molbev.a026395) [DOI] [PubMed] [Google Scholar]
- 11.Plaxton WC. 1996. The organization and regulation of plant glycolysis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 185–214. ( 10.1146/annurev.arplant.47.1.185) [DOI] [PubMed] [Google Scholar]
- 12.Makiuchi T, Annoura T, Hashimoto M, Hashimoto T, Aoki T, Nara T. 2011. Compartmentalization of a glycolytic enzyme in Diplonema, a non-kinetoplastid euglenozoan. Protist 162, 482–489. ( 10.1016/j.protis.2010.11.003) [DOI] [PubMed] [Google Scholar]
- 13.Neuberger G, Maurer-Stroh S, Eisenhaber B, Hartig A, Eisenhaber F. 2003. Prediction of peroxisomal targeting signal 1 containing proteins from amino acid sequence. J. Mol. Biol. 328, 581–592. ( 10.1016/S0022-2836(03)00319-X) [DOI] [PubMed] [Google Scholar]
- 14.Lazarow PB. 2006. The import receptor Pex7p and the PTS2 targeting sequence. Biochim. Biophys. Acta 1763, 1599–1604. ( 10.1016/j.bbamcr.2006.08.011) [DOI] [PubMed] [Google Scholar]
- 15.Güther ML, Urbaniak MD, Tavendale A, Prescott A, Ferguson MA. 2014. High-confidence glycosome proteome for procyclic form Trypanosoma brucei by epitope-tag organelle enrichment and SILAC proteomics. J. Proteome. Res. 13, 2796–2806. ( 10.1021/pr401209w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moretti S, Armougom F, Wallace IM, Higgins DG, Jongeneel CV, Notredame C. 2007. The M-Coffee web server: a meta-method for computing multiple sequence alignments by combining alternative alignment methods. Nucleic. Acids. Res. 35, W645–W648. ( 10.1093/nar/gkm333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. ( 10.1093/bioinformatics/btp348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. ( 10.1093/molbev/msn067) [DOI] [PubMed] [Google Scholar]
- 19.Quang le S, Gascuel O, Lartillot N. 2008. Empirical profile mixture models for phylogenetic reconstruction. Bioinformatics 24, 2317–2323. ( 10.1093/bioinformatics/btn445) [DOI] [PubMed] [Google Scholar]
- 20.Bringaud F, Riviere L, Coustou V. 2006. Energy metabolism of trypanosomatids: adaptation to available carbon sources. Mol. Biochem. Parasitol. 149, 1–9. ( 10.1016/j.molbiopara.2006.03.017) [DOI] [PubMed] [Google Scholar]
- 21.Makiuchi T, Annoura T, Hashimoto T, Murata E, Aoki T, Nara T. 2008. Evolutionary analysis of synteny and gene fusion for pyrimidine biosynthetic enzymes in Euglenozoa: an extraordinary gap between kinetoplastids and diplonemids. Protist 159, 459–470. ( 10.1016/j.protis.2008.02.002) [DOI] [PubMed] [Google Scholar]
- 22.Lukeš J, Flegontova O, Horák A. 2015. Diplonemids. Curr. Biol. 25, R702–R704. ( 10.1016/j.cub.2015.04.052) [DOI] [PubMed] [Google Scholar]
- 23.de Vargas C, et al. 2015. Ocean plankton: eukaryotic plankton diversity in the sunlit ocean. Science 348, 1261605 ( 10.1126/science.1261605) [DOI] [PubMed] [Google Scholar]
- 24.Read BA, et al. 2013. Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 499, 209–213. ( 10.1038/nature12221) [DOI] [PubMed] [Google Scholar]
- 25.Szöör B, Haanstra JR, Gualdrón-López M, Michels PA. 2014. Evolution, dynamics and specialized functions of glycosomes in metabolism and development of trypanosomatids. Curr. Opin. Microbiol. 22C, 79–87. ( 10.1016/j.mib.2014.09.006) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The D. papillatum genome was deposited in DDBJ/EMBL/GenBank under the accession LMZG00000000. Diplonema papillatum sequences for relevant enzymes reported here have been deposited individually at DDBJ (accession numbers AB970479–AB970481, AB970483–AB970501, LC127115, LC127507). The datasets supporting this article have been uploaded as part of the electronic supplementary material.