Abstract
The lack of software interoperability with respect to gating has traditionally been a bottleneck preventing the use of multiple analytical tools and reproducibility of flow cytometry data analysis by independent parties. To address this issue, ISAC developed Gating-ML, a computer file format to encode and interchange gates. Gating-ML 1.5 was adopted and published as an ISAC Candidate Recommendation in 2008. Feedback during the probationary period from implementors, including major commercial software companies, instrument vendors and the wider community, has led to a streamlined Gating-ML 2.0. Gating-ML has been significantly simplified and therefore easier to support by software tools. To aid developers, free, open source reference implementations, compliance tests and detailed examples are provided to stimulate further commercial adoption. ISAC has approved Gating-ML as a standard ready for deployment in the public domain and encourages its support within the community as it is at a mature stage of development having undergone extensive review and testing, under both theoretical and practical conditions.
Keywords: flow cytometry, bioinformatics, gating, data standard, file format
Introduction
The flow cytometry data file standard (FCS) format capturing the output of cytometers in an open manner was developed in 1984 and it has been continuously improved to keep up with technological advancements in the cytometry field ever since [1–3]. Through the adoption of the FCS standard by all analytical instrument vendors and software developers, the community can use third party software tools of their choice to analyze data acquired using different instruments. However, interoperability on the data analysis level has traditionally been a challenge in flow cytometry. Addressing this bottleneck is becoming increasingly important as both polychromatic and mass cytometry methods produce complex data sets with 20 and more distinct channels. The use of multiple software tools working in combination with manual analysis is becoming more commonplace as data complexity increases, and it is therefore imperative that additional data exchange solutions are developed.
Gating is the principal component of traditional analysis of flow cytometry data. Consequently, unambiguous description of gating, as addressed by Gating-ML, is critical for computational interchange of data analysis. Gating-ML 2.0 describes the mutual relations among all the different components and the workflow of the analysis. It is a significantly improved version of Gating-ML 1.5 that was adopted and published as an ISAC Candidate Recommendation in 2008 [4].
Materials and Methods
Feedback during the six years of Gating-ML's probationary period from implementors, including major commercial software companies and the wider community, has led to a streamlined Gating-ML 2.0. The complexity of the standard has been reduced, with components that were expected to be only very rarely used removed. As a result, the list of supported transformations has been reduced by half. Some new components have been added (e.g., Bounding Transformations) based on their usefulness as proposed by community members. The parameterizations of the nonlinear scaling transformations were modified and unified to make them more user friendly, and to allow programs substitute one scale for another with a relatively small effect on the populations defined by corresponding gates.
Results
Gating-ML files are Extensible Markup Language (XML) files that unambiguously describe gates and related data transformations in a way that is computationally reproducible and independent of any particular software tool. The text of the Gating-ML specification is available as Supplementary Information of this manuscript, and from http://flowcyt.sf.net/gating/latest.pdf, or from ISAC's web pages (https://isac-net.org/), where examples, additional documentation and extensive unit tests are also provided to aid in incorporation of the standard in third party software. When the specification is followed, Gating-ML gates applied on FCS data will result in the same sets of events (same cell populations) independently of which software tool is used.
Components of the Standard
The Gating-ML 2.0 specifications consist of several normative and informative parts. The normative parts are crucial for a compliant implementation. These consist of a detailed description of Gating-ML, and of XML schemas [5] formally defining the syntax of compliant files. Additional informative parts include examples of Gating-ML 2.0 files, HTML-based documentation of the XML schemas, detailed self-assessment compliance tests, and reference implementations of all the scaling transformations, which can be integrated into existing software tools. All these components are being shared in order to aid Gating-ML support in third-party software.
Improvements since Gating-ML 1.5
Gate types
Gating-ML 2.0 supports range, rectangular, quadrant, polygon, and ellipse gates. These gates may be ordered in hierarchical structures to describe gating strategies, or combined using Boolean collections (i.e., and, or, not) to define cell populations. Gating-ML 1.5 gate types that were deemed too complicated, too difficult to support or unlikely to be widely used are no longer supported. Specifically, decision tree gates were removed since the use of decision trees for gating proved to be nonintuitive and unnecessary as the same gates can be expressed by Boolean collections. Multidimensional polytope gates were also removed as their support is computationally difficult for most software tools and their use is currently limited to a handful of automated analysis algorithms.
Data transformations
Gating-ML 1.5 supported compound transformations, which could be created as an arbitrary combination of any of the supported transformation types. While this was a mathematically elegant way of describing any transformation workflow, the generic nature of this approach made it a challenge to implement and integrate in existing software tools. Considering that the gating workflow follows the same pattern in the vast majority of cases, Gating-ML 2.0 removes this flexibility and defines a fixed transformation template as follows:
Linearize the data if applicable (i.e., if stored on a log scale in the FCS data file). The linearization is defined by keywords in the FCS data file (e.g., $PnE) so it is not repeated in Gating-ML.
Compensate (unmix) the data if applicable. The compensation (spectrum) description can be encoded in the Gating-ML file or (newly in Gating-ML 2.0) a spillover matrix in the FCS data file can be referenced.
Apply channel specific scaling transformations (if applicable). Supported scales include linear, logarithmic, inverse hyperbolic sine, Hyperlog and Logicle. ISAC standards do not include any components that are believed to require the use of subject matter covered by patents or other intellectual property rights, or that may only be available under restrictive licensing conditions. Implementation of a standard shall be possible without charge and be non-restrictive. Consequently, the Logicle/Bi-exponential transformation [6] was not included in Gating-ML 1.5 as it was covered by U.S. patent 6954722 and was licensed under restrictive conditions. However, thanks to discussions of members of the ISAC Data Standards Task Force with the patent owner (Stanford University), this patent is no longer being asserted in the field of flow cytometry, which allowed for Logicle to be included as one of the scale transformations in Gating-ML 2.0 [7].
Apply gates.
A fixed workflow template significantly simplified the Gating-ML 2.0 language. It reduced the number of supported transformations by half, leaving only compensation (spectral unmixing), ratio, and five different scale types. Instead of chaining the transformation in a generic way, each gating dimension can now simply reference an FCS channel (or two FCS channels if ratio is used), a spillover matrix (optional), and a scaling transformation (optional). The order of applying these is fixed and implicit. This results in a much simpler XML construct and allows for the same scale definition to be reused for multiple channels and gates.
The parameterization of the scale transformations has been redesigned and unified to make these comprehensible and to ease the burden of implementation. They are defined in terms of a consistent parameterization that is closely related to the user experience. The top of scale value T is always mapped to the value 1. In addition, the logarithmic and log-like transforms are parameterized by M, the number of decades of data range mapped onto the unit display interval by the logarithmic transform. The log-like scales are also parameterized by W and A, which are commensurate with decades on the scale, although they do not represent ten fold changes in data values. W controls the degree of linearization for the Logicle and Hyperlog transforms. The parameter A specifies an additional range of negative data values that are to be brought on scale. For the Logicle and Hyperlog transforms, this is in addition to what is already brought on scale by W and generally should not be needed. The Logicle, Hyperlog, and parameterized inverse hyperbolic sine transforms with A = 0 all behave like the logarithmic transform with the same values of T and M for large data values. This choice of parameters also leads to a sensible fall back strategy when a software tool does not implement a particular transform. For example, if the Logicle transform is not available, then a Hyperlog transform with the same parameters should be a reasonable alternative, and vice versa, which allows programs to substitute one scale for another with a relatively small effect on the populations defined by corresponding gates. Figure 1 shows a comparison of the parameterized logarithmic and the supported log-like scale transformations with T = 1000, M = 4.5, W = 1 and A = 0 (W and A set where applicable). All transformations are very close to each other for large data values. In the low data range, the parameterized inverse hyperbolic sine, Logicle, and Hyperlog transforms show a linear-like behavior around zero and extend the scale to the negative data range. Unlike the parameterized inverse hyperbolic sine, the Logicle and Hyperlog transforms allow the width of the linearization region to be controlled independently from the logarithmic character for large data values.
Figure 1.
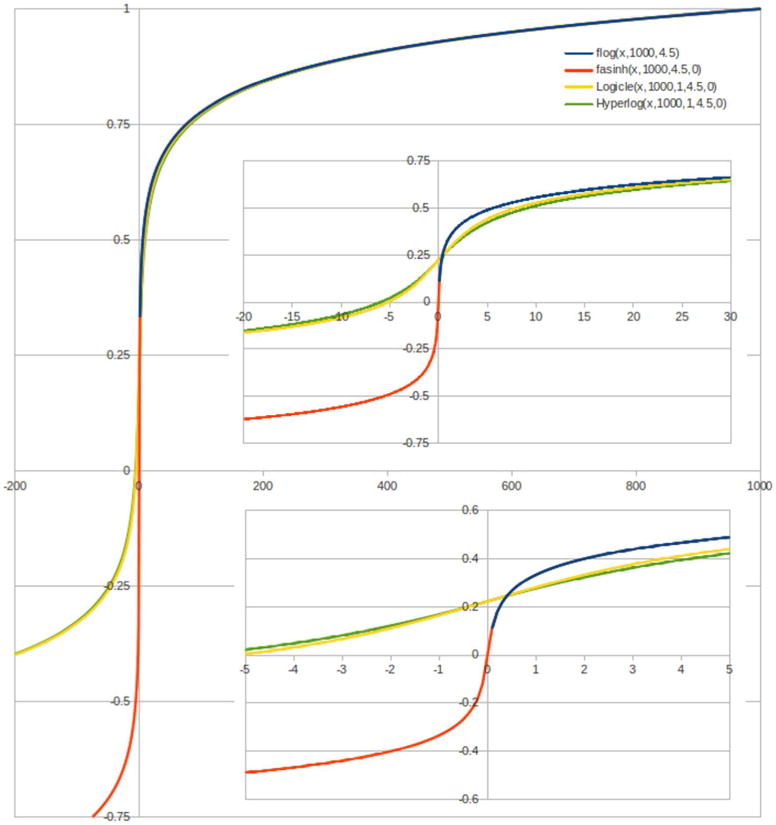
Comparison of non-linear scale transformations. All transformations are very similar in the far positive range, where they approximate the logarithmic scale. In the low data range (zoomed-in in the two sub figures), the parameterized inverse hyperbolic sine, Logicle, and Hyperlog transforms show a linear-like behavior around zero and extend the scale to the negative data range. The similarity of these transformations guides a sensible fall back strategy when software does not implement a particular scale.
Finally, an optional boundary may be defined for any transformation. A boundary restricts the result of a transformation to a predefined interval. Using a boundary allows for simple unambiguous encoding of gating performed by software tools that pile off-scale events on the graph axes. In these cases, if the selected visualization is not appropriate, some events could fall outside of the display area. Some analysis tools shift these to a predefined minimum or maximum (usually the graph boundary), which alters the gate membership of these events. A Gating-ML boundary transformation may be used in order to mimic such behavior and encode these gates in a reproducible manner that addresses this use case.
Other changes
A few additional changes have been implemented in response to comments from the early implementors. For example, similar to the FCS data file standard, custom and vendor specific information may now be included in a standardized way. Vendors can associate additional information with the whole Gating-ML file, or with specific sections, such as particular scale transformations or gate definitions. The elimination of the in-line definition option of Boolean operands represents an example of a change that simplifies the XML parsing while retaining the power of the Gating-ML language. The operand gates may still be defined outside of the Boolean gate definition and referenced from any Boolean gate. In addition, operands may also be used as complements, which allows for a simple creation of expressions in the form of “A AND NOT(B)” (e.g., “Lymphocytes but not T-cells”).
Initial Software Implementations
A fully compliant, free and open source reference implementation of all the components of the standard is available as an R/BioConducor package (flowUtils [8]). Gating-ML 2.0 is also supported by FlowRepository (as described in documentation [9]), and Cytobank (CytoBank Inc.). These tools can interchange gating details as they both read and write Gating-ML 2.0 compatible files. A free open source Gating-ML 2.0 reader is available in MATLAB from MATLAB CENTRAL [10]. GenePattern module ApplyGatingML2 [11] can also read Gating-ML 2.0 files and apply these on FCS data in order to extract all cell populations defined in the Gating-ML file. While FlowJo's Gating-ML 2.0 support is not well documented, users can drag and drop a Gating-ML 2.0 file onto their samples, and FlowJo X will extract and apply the gates. In order to export Gating-ML 2.0 files from FlowJo, users must save the workspace as an Archival Cytometry Standard file [12] (ACS; currently under development by ISAC) and then extract the Gating-ML 2.0 file from the archive by changing the archive file extension to “.zip” and extracting the ZIP file. However, FlowJo X (version 10.0.7) does not yet properly support all the scale transformations, it does not save scale definitions as part of the Gating-ML output, and the output is not fully compatible.
Discussion
Gating-ML has been well tested by several different implementations (FlowRepository, Cytobank, R/BioConductor, GenePattern, MATLAB and FlowJo) and ISAC has approved it as a Recommendation (i.e., it is a specification that, after extensive consensus-building, has been adopted as the version that ISAC recommends for wide deployment). Its design is a compromise providing enough expressive power to describe gating and traditional two-dimensional analyses that are common in existing software, while keeping the format simple and therefore appealing to implement by third party software vendors. It also attempts to accommodate innovations in automated gating and clustering by incorporating multidimensional gate types whenever such extensions are simple and inexpensive to calculate, such as hyper rectangular gates, ellipsoids and hyper-ellipsoids. This is implemented in a consistent manner on both, syntactic and semantic level.
The mathematical accuracy of the Gating-ML specification ensures that gates described in Gating-ML are exchangeable among compliant software applications and will provide the same subsets of events (cells) when applied on the same data using different tools. Gating-ML 2.0 is an open standard developed and maintained via a collaborative and consensus driven process under the auspice of ISAC. Its adoption is expected to enable the exchange of analysis between software tools, enabling advanced analyses and accelerating flow cytometry discovery. With Gating-ML 2.0, it is possible to use a combination of tools for their different strengths, such as a friendly gating interface, statistical analysis capabilities, or the power of automated computational approaches.
Gating-ML files can be used in conjunctions with CLR files [13] to exchange the results of gating by enumerating which cells (events) belong to which of the defined populations. CLR can communicate the results even in cases where gates (boundaries) are unknown, which is the case when certain computational methods identify cell populations of interest. Finally, the ACS specification (in development) is intended to tie all the components (files) together into a single archive (ZIP) with additional semantical information.
ISAC encourages adoption of Gating-ML to support the exchange of open analysis with the expectation that its adoption will aid members of the society in the same way that the widespread adoption of FCS file standard has done.
Supplementary Material
Acknowledgments
This work was supported by funding by National Institute of Health [R01 EB008400], Natural Sciences and Engineering Research Council of Canada, ISAC and the Wallace H. Coulter Foundation.
Footnotes
Disclosures of Potential Conflicts of Interest: ISAC Data Standards Task Force (DSTF) includes members of companies that make flow cytometry hardware or software. ISAC undertakes to establish standards (i.e. ISAC Recommendations) that do not require the purchase of patent licenses for compliance. For that reason, DSTF members must immediately disclose any patents or patent applications held by them, and which they know, or have reason to believe, have a likelihood of being infringed by compliance with any draft or approved standard. Users and reviewers are also encouraged to disclose any procedures that require a user to purchase a patent license. In addition, by participating in any Task Force, members affirm that, with respect to any patent that may be infringed by compliance with any draft or approved standard and is not disclosed, such patent shall either (1) not be enforced with respect to such compliance, or (2) be freely licensed to all users on a fair, reasonable and non-discriminatory basis without imposing a licensing fee or other charge of any kind. Every Task Force member has pledged to act in good faith and in an open and honest manner at all times.
ISAC Data Standards Task Force: Ryan Brinkman (BC Cancer Agency, ISAC DSTF chair), Jay Almarode (FlowJo, LLC), Ernie Anderson (Beckman Coulter Life Sciences), Kim Blenman (Yale University), Chris Bray (Verity Software House), Martin Büscher (Miltenyi Biotechnology), James Cavenaugh, Michael Goldberg (BD Biosciences), Bill Hyun (University of California, San Francisco), David Kripal (Cytek Development, Inc.), Kevin Krouse (Labkey, Inc.), Robert Leif (Newport Instruments), Wayne Moore (Stanford University), David Novo (De Novo Software), David Parks (Stanford University), Josef Spidlen (BC Cancer Agency), Adam Treister (Fluourish, Inc.), James Wood (Wake Forest University), Michael Zordan (Sony Biotechnology)
References
- 1.Data File Standards Committee of the Society for Analytical Cytology. Data file standard for flow cytometry. Cytometry. 1990;11(3):323–332. doi: 10.1002/cyto.990110303. [DOI] [PubMed] [Google Scholar]
- 2.Seamer LC, Bagwell CB, Barden L, Redelman D, Salzman GC, Wood JC, Murphy RF. Proposed new data file standard for flow cytometry, version FCS 3.0. Cytometry. 1997;28(2):118–122. doi: 10.1002/(sici)1097-0320(19970601)28:2<118::aid-cyto3>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 3.Spidlen J, Moore W, Parks D, Goldberg M, Bray C, Bierre P, Gorombey P, Hyun B, Hubbard M, Lange S, Lefebvre R, Leif R, Novo D, Ostruszka L, Treister A, Wood J, Murphy RF, Roederer M, Sudar D, Zigon R, Brinkman RR. Data File Standard for Flow Cytometry, version FCS 3.1. Cytometry Part A. 2010;77(1):97–100. doi: 10.1002/cyto.a.20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spidlen J, Leif RC, Moore W, Roederer M, Brinkman RR for the Advancement of Cytometry Data Standards Task Force IS. Gating-ML: XML-based gating descriptions in flow cytometry. Cytometry Part A. 2008;73(12):1151–1157. doi: 10.1002/cyto.a.20637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Wide Web Consortium (W3C) [Accessed June 10, 2014];XML Schema 1.0 (W3C Recommendation) 2004 URL: http://www.w3.org/TR/xmlschema-1/
- 6.Parks DR, Roederer M, Moore WA. A new “Logicle” display method avoids deceptive effects of logarithmic scaling for low signals and compensated data. Cytometry Part A. 2006;69(6):541–551. doi: 10.1002/cyto.a.20258. [DOI] [PubMed] [Google Scholar]
- 7.Stanford University – Office of Technology Licensing. [Accessed June 10, 2014];An Improved Method for Visualization of Multidimensional Data (“Logicle”) 2004 URL: http://techfinder.stanford.edu/technology_detail.php?ID=23438.
- 8.Spidlen J, Gopalakrishnan N, Hahne F, Ellis B, Gentleman R, Dalphin M, LeMeur N, Purcell B. [Accessed October 30, 2014];flowUtils – Utilities for flow cytometry. 2014 URL: http://www.bioconductor.org/packages/devel/bioc/html/flowUtils.html.
- 9.FlowRepository Development Team. [Accessed June 10, 2014];Gating-ML 2.0 support in FlowRepository. 2014 URL: http://flowrepository.org/gating-ml-2-support.
- 10.Finck R. [Accessed June 10, 2014];Gating-ML support in MATLAB. 2014 URL: http://www.mathworks.com/matlabcentral/fileexchange/46106-gating-ml.
- 11.Spidlen J. [Accessed June 10, 2014];GPArc – GenePattern Archive, ApplyGatingML2 module. 2014 URL: http://tinyurl.com/GatingML2GPArc.
- 12.International Society for Advancement of Cytometry Data Standards Task Force. [Accessed June 10, 2014];Archival Cytometry Standard (ACS) 2010 URL: http://flowcyt.sf.net/acs/ACS.v1.0.101013.pdf.
- 13.Spidlen J, Bray C, Brinkman R. ISAC's Classification Results File Format (CLR) [accepted on October 14, 2014];Cytometry A. 2014 doi: 10.1002/cyto.a.22586. Manuscript 14-093.R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


