Abstract
Ribosome assembly has been studied intensively using Förster resonance energy transfer (FRET) with fluorophore-labeled fragments of RNA produced by chemical synthesis. However, these studies are limited by the size of the accessible RNA fragments. We have developed a replicable unnatural base pair (UBP) formed between (d)5SICS and (d)MMO2 or (d)NaM, which efficiently directs the transcription of RNA containing unnatural nucleotides. We now report the synthesis and evaluation of several of the corresponding ribotriphosphates bearing linkers that enable the chemoselective attachment of different functionalities. We found that the RNA polymerase from T7 bacteriophage does not incorporate NaM derivatives, but does efficiently incorporate 5SICSCO, whose linker enables functional group conjugation via Click chemistry, and when combined with the previously identified MMO2A, whose amine side chains permits conjugation via NHS coupling chemistry, enables site-specific double labeling of transcribed RNA. To study ribosome assembly, we transcribed RNA corresponding to a 243-nt fragment of the central domain of Thermus thermophilus 16S ribosomal RNA containing 5SICSCO and MMO2A at defined locations, and then site-specifically attached the fluorophores Cy3 and Cy5. FRET was characterized using single-molecule total internal reflection fluorescence (smTIRF) microscopy in the presence of various combinations of added ribosomal proteins. We demonstrate that each of the fragment’s two three-helix junctions exist in open and closed states, with the latter favored by sequential protein binding. These results elucidate early and previously uncharacterized folding events underlying ribosome assembly, and demonstrate the applicability of UBPs for biochemical, structural, and functional studies of RNAs.
Graphical abstract
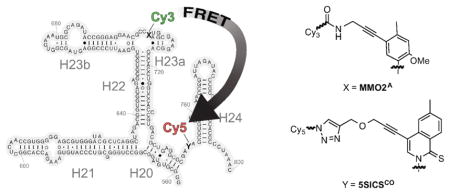
The ribosome is a large and intricate macromolecular machine that is highly conserved and responsible for protein synthesis in all cells. In bacteria, the ribosome consists of a 50S subunit that catalyzes the peptidyl transfer reaction and a 30S subunit that controls the decoding of mRNA by tRNAs. The 50S subunit is composed of the 23S and 5S RNAs (~2300 and 120 nts, respectively) and 31 proteins, and the 30S subunit is composed of the 16S RNA (1540 nts) and 20 proteins. The 30S subunit can be assembled in vitro from its purified components,1 and this remarkable property has fostered extensive biophysical characterizations of the assembly process over the past four decades.2 The initial Nomura assembly map defined the thermodynamic order of binding of the 20 ribosomal proteins to the 16S rRNA, and there was clear evidence for both sequential binding and parallel assembly processes.3 The assembly involves extensive RNA folding, and a series of RNA conformational changes that are accompanied by sequential binding of the ribosomal proteins, ultimately culminating in the fully assembled 30S subunit.
The hierarchical nature of the assembly has enabled the use of reduced subsystems that permit more detailed biophysical measurements to be made on the folding and binding reactions,4,5 and in particular, the central domain of the 30S subunit, which binds proteins S6, S8, S11, S15, and S21, has been subjected to a detailed cooperativity analysis for protein binding.6 The primary binding protein S15 binds to a three-helix RNA junction in the central domain consisting of helices 20, 21, and 22, stabilizing the native conformation of the junction over the unfolded form that predominates in free RNA.7 This conformational change can readily be monitored by attaching fluorophores to the ends of helices that rearrange upon S15 binding, and detecting changes in the spatial arrangement of the fluorophores via Förster resonance energy transfer (FRET).8–10 The thermodynamics and kinetics of both S15 binding and magnesium ion binding to an isolated three-helix RNA junction have been characterized extensively using single-molecule FRET.9 However, the detailed nature of the change(s) and their relationship to other parts of the central domain RNA or other ribosomal proteins, has not been defined.
Recently, the position-specific labeling of RNA (PLOR) method was reported which enables the synthesis of site-specifically modified RNAs using a biotin-immobilized DNA template and stepwise elongation by T7 RNA polymerase.11 Using this method, Cy3 and Cy5 dyes were site-specifically incorporated into a 71-nt riboswitch and used to monitor conformational changes. However, this method requires special and repeated manipulation of the reaction, and the size of RNAs accessible has yet to be determined. A procedure for the production of arbitrarily large labeled RNAs that is based on simple homogenous transcription would likely facilitate the characterization of many systems, including the ribosome.
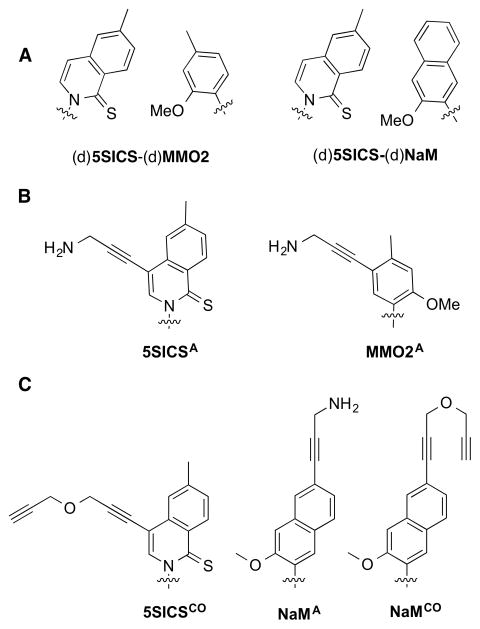
A novel approach to the preparation of site-specifically labeled DNA or RNA is based on the use of an unnatural base pair (UBP), such as that reported by Hirao and co-workers,12,13 or developed by our group and formed between (d)5SICS and (d)MMO2 or (d)NaM ((d)5SICS-(d)MMO2 or (d)5SICS-(d)NaM)14–16 (Figure 1A). To use the UBPs to site-specifically label DNA, we have explored the modification of the unnatural nucleobases with linkers that allow for the attachment of different cargos to the unnatural nucleoside triphosphates.17,18 Because some moieties could inhibit triphosphate incorporation,19,20 linkers that terminate with either a propinyl group or a protected amine were also developed, because their orthogonal reactivity (after amine deprotection) may be used to attach two different functional groups or labels once the corresponding triphosphates have been incorporated into DNA. To enable RNA labeling, a variety of similarly derivatized ribotriphosphates were synthesized and tested for incorporation into RNA during in vitro transcription (Figure 1B). We showed that dNaM efficiently directs bacteriophage T7 RNA polymerase (T7 RNAP) to specifically incorporate 5SICSATP into RNA.18 While we also showed that d5SICS directs T7 RNAP to incorporate MMO2ATP into RNA, pairs of 5SICS and NaM or MMO2 derivatives bearing linkers with orthogonal reactivity were not identified, precluding the double labeling of RNA with two different cargos.
Figure 1.
(A) Parental UBPs (d)5SICS-(d)MMO2) and (d)5SICS-(d)NaM. (B) Previously identified linker-derivatized unnatural nucleotides 5SICSATP and MMO2ATP. (C) Linker-derivatized analogs synthesized and characterized in this study, 5SICSCOTP, NaMATP, and NaMCOTP. Phosphates and ribose not shown.
Here we report the synthesis and evaluation of 5SICSCOTP, NaMATP, and NaMCOTP (Figure 1C). We find that the two NaM derivatives are not efficiently incorporated into RNA, but dNaM does direct the efficient synthesis of RNA containing 5SICSCO. Thus, for the first time, by incorporating MMO2ATP and 5SICSCOTP, and using a combination of copper-catalyzed azide-alkyne cycloaddition (Click) and NHS coupling chemistry, site specifically-labeled RNA with virtually any combination of two functional groups of interest may be prepared via straightforward enzymatic synthesis. To demonstrate the utility of the modified UBP, a 243-nt fragment of the central domain of Thermus thermophilus 16S RNA was transcribed with MMO2A and 5SICSCO replacing natural nucleotides, then the resulting RNA was site-specifically modified with the fluorophores Cy3 and Cy5, and the FRET properties of the double-labeled RNA were characterized in the presence and absence of several ribosomal proteins using smTIRF microscopy. We demonstrate that the RNA populates four distinct conformations, which are assigned to a combinatorial set arising from open and closed states of the two three-helix junctions in this fragment, and that the resulting conformational equilibrium is driven towards the doubly closed, most folded state, by sequential protein binding. These results shed light on early folding events in the central domain of 16S rRNA, and demonstrate the applicability of UBPs for biochemical, structural, and functional studies of RNAs.
RESULTS AND DISCUSSION
Unnatural nucleotides 5SICSCOTP, NaMATP, and NaMCOTP were synthesized as described in the Supporting Information. Briefly, 5SICSCO was generated from the previously reported iodo-5SICS intermediate,18 which was coupled to propargyl ether via a Sonogashira reaction. Phosphorylation under Ludwig conditions21 provided 5SICSCOTP, which was purified by anion exchange chromatography and HPLC. Toward NaMATP and NaMCOTP, the 6-(benzyloxy)-2-bromo-3-methoxynaphthalene nucleobase precursor22 was lithiated and coupled to a protected ribonolactone. The resulting intermediate was reduced to generate the corresponding C-nucleoside, and the ribose protecting groups were exchanged. The benzyl group was removed and the resulting 6-napthol was converted to the corresponding aryl triflate. Sonogashira coupling with either propargyl ether or DCA-protected propargylamine, followed by hydroxyl deprotection, yielded NaMCO and NaMPA respectively, which were phosphorylatyed and purified as described above for 5SICSCO. NaMPATP was treated with diluted sodium hydroxide affording NaMATP, which was further purified by HPLC.
To begin to explore the transcription of DNA containing the linker-derivatized unnatural nucleotides, DNA templates T85 and T86 were produced by standard oligonucleotide synthesis and PCR assembly (see Supporting Information). Template T85 includes the T7 promoter and three GC base pairs, resulting in a 5’-GGG initiation sequence, and the sequence corresponding to nucleotides C719 to G783 of the T. thermophilus 16S central domain with A750 replaced with d5SICS. Template T86 directs the transcription of RNA with a 5’GG and the sequence corresponding to nucleotides G668 to U734 of the T. thermophilus 16S central domain, with G704 replaced by dNaM. In vitro transcription reactions were performed for 1 h at 37 °C in 20 μL volumes of 100 nM DNA (2 pmol), 1× Takara buffer, 16 mM MgCl2, 5 mM DTT, 0.01% Triton X-100, 2 mM NTPs, [γ– 32P]GTP, 2.5 unit/μL T7 RNAP, and 1 mM of an unnatural ribotriphosphate. In all cases, both full-length product and product corresponding to truncation at the position of the unnatural nucleotide in the template were observed (Figure S23). The ratio of full-length to truncated product varied from 81% – 87% for NaMTP, 5SICSTP, and 5SICSATP, to 70% – 75% for 5SICSCOTP and MMO2ATP, and to 55% for NaMCOTP, and 35% for NaMATP (in each case the incorporation of the unnatural triphosphate was directed by its cognate unnatural nucleotide, d5SICS or dNaM, in the template). Importantly, the relative efficiency with which dNaM directed the incorporation of 5SICSCOTP, with its Click chemistry compatible side chain, into RNA provides the first feasible route to post-transcription double labeling of RNA when used in conjunction with the primary amine-derivatized MMO2ATP.
To explore the labeling of RNA with two different fluorophores, we constructed the transcription template T135. Template T135 includes two GC base pairs inserted between the T7 promoter and the sequence corresponding to nucleotides G668 to G783 of the T. thermophilus 16S central domain, with G704 replaced with dNaM, and A750 replaced with d5SICS. Positions 704 and 750 were selected for these exploratory studies, because based on the reported structure of the 30S subunit,23 they were expected to allow for conformational changes of major helices 20 and 23a to be monitored without disruption of base pairing. Transcription reactions were performed as described above, except that reactions were run for 5 h. Transcription of T135 with 5SICSCOTP and MMO2ATP was as efficient as with 5SICSTP and MMO2TP, further illustrating that the addition of linkers is not significantly deleterious. The T135 transcript was then incubated with azide-Cy5-sulfo, to react with the alkyne of 5SICSCO, and NHS-Cy3-sulfo, to react with the primary amine of MMO2A, and the UV/vis spectra of the purified product demonstrated that a significant percentage of the transcripts had been double labeled (Figures S24 and S25).
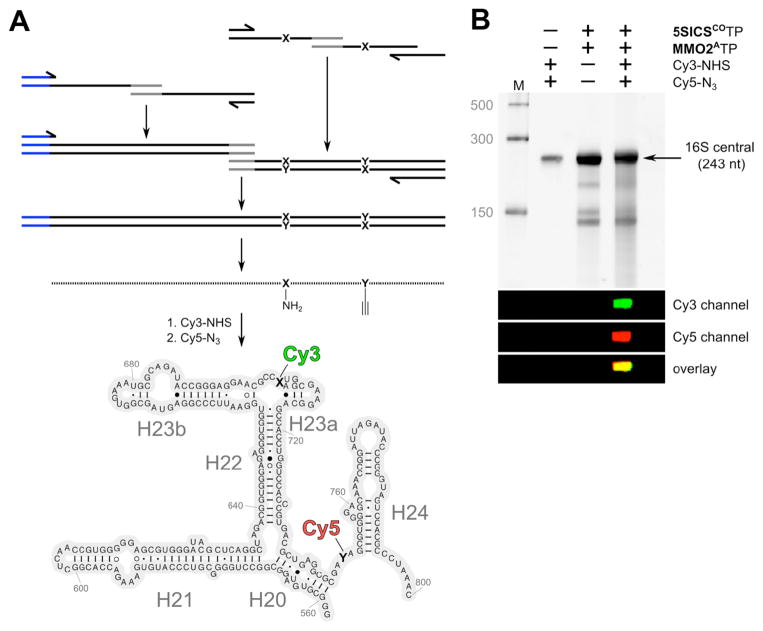
With validation of double labeling, we next assembled template T260 via overlap extension PCR (Figure 2A). T260 directs the transcription of residues G558 to C800 of the central domain, with dNaM and d5SICS incorporated at the same positions as with T135. Transcription and labeling were performed as described above for template T135, and analysis revealed a single product band for which both Cy3 and Cy5 emission was detected by gel electrophoresis (Figures 2B and S26). The UV/vis absorption spectrum again indicated that a significant fraction of the RNA was double labeled (Figure S27). In contrast, the transcription product of a fully natural analog of T260 showed no labeling after reaction with azide-Cy5-sulfo and NHS-Cy3-sulfo (Figure S28), confirming that labeling is mediated by the unnatural nucleotides.
Figure 2.
Design and assembly of the double-labeled 16S ribosomal RNA fragment. (A) T260 DNA template containing T7 promoter (shown in blue) with MMO2A704 (X) and 5SICSCO750 (Y) was assembled by PCR from smaller DNA fragments that were obtained by solid-phase DNA synthesis followed by transcription of the 243 nt fragment of rRNA and labeling with Cy3 and Cy5 dyes. (B) PAGE analysis of T260 transcription product labeling with NHS-Cy3-sulfo and/or azide-Cy5-sulfo.
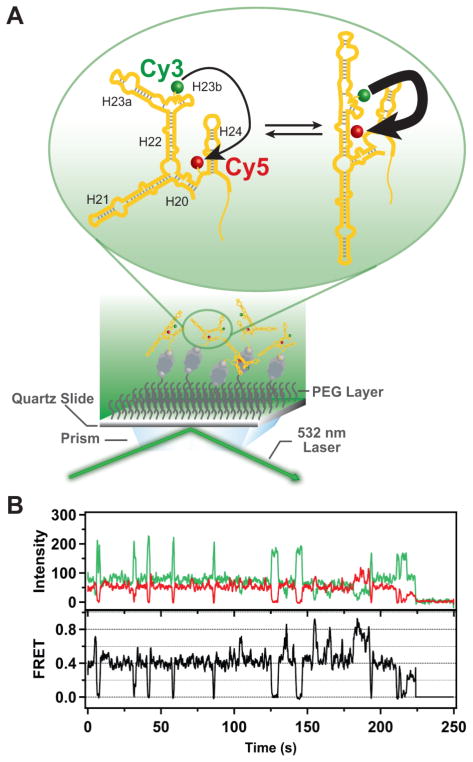
To explore the use of the Cy3/Cy5-labeled central domain RNA fragment to study 30S assembly, the RNA was immobilized on a quartz surface passivated with polyethylene glycol by means of biotin-streptavidin attachment (Figure S29), to enable characterization using TIRF microscopy (Figure 3A). We first estimated the extent of double labeling by simultaneously illuminating the surface-immobilized RNA molecules with green (532 nm, to excite Cy3) and red (643 nm, to excite Cy5) lasers and recording separate TIRF images of the resulting Cy3 and Cy5 emission. Typical images are shown in Fig. S30, where each of the spots in either the green or red images represents an immobilized RNA molecule. By counting the number of spots in the green image that have a corresponding spot in the red image, and comparing this to the total number of immobilized RNA molecules in the field of view, we estimated that 32% of the RNA molecules contained both the Cy3 and Cy5 labels. For smFRET measurements, we illuminated the surface with the green laser only and then selected for analysis only those individual RNA molecules that exhibited both green and red emission. The upper panel of Figure 3B shows a representative pair of time traces of the Cy3 (donor, green) and Cy5 (acceptor, red) emission from such a selected RNA molecule in the presence of protein S15. The donor and acceptor emissions exhibit anti-correlated fluctuations, characteristic of FRET, and the calculated FRET efficiency is shown in the lower panel of Figure 3B. The abrupt and irreversible loss of the donor and acceptor emission at ~ 225 s, due to a single-step photobleaching transition of the donor, confirms that a single RNA molecule is being observed. All smFRET data were compiled from single RNA molecules immobilized and selected in an identical fashion.
Figure 3.
Schematic illustration of the single-molecule fluorescence measurements and representative experimental data. (A) RNA labeled with Cy3 (donor) and Cy5 (acceptor) probes is immobilized on a quartz surface by biotin-streptavidin attachment (biotin is shown as a gray sphere and streptavidin as a dark grey rectangle with a biotin binding site in each corner). The surface is passivated with polyethylene glycol (PEG) molecules (wavy lines) to suppress non-specific adsorption of proteins. Upon optical excitation of the Cy3 dye by the evanescent field of the totally internally reflected green laser beam, FRET can occur from Cy3 to Cy5 (indicated by the arrows). The RNA is depicted as exchanging between two distinct conformations, with corresponding differences in FRET efficiency (indicated by the weights of the arrows). (B) Representative time traces of the Cy3 (green) and Cy5 (red) emission from a single RNA molecule immobilized on the surface. Anti-correlated fluctuations of the donor and acceptor emission are evident, indicating that FRET is occurring. The corresponding FRET intensity trajectory is shown by the black trace. In the example shown, the protein S15 is also present. A single-step donor photobleaching event is observed at ~225 s, resulting in the abrupt loss of both donor and acceptor emission.
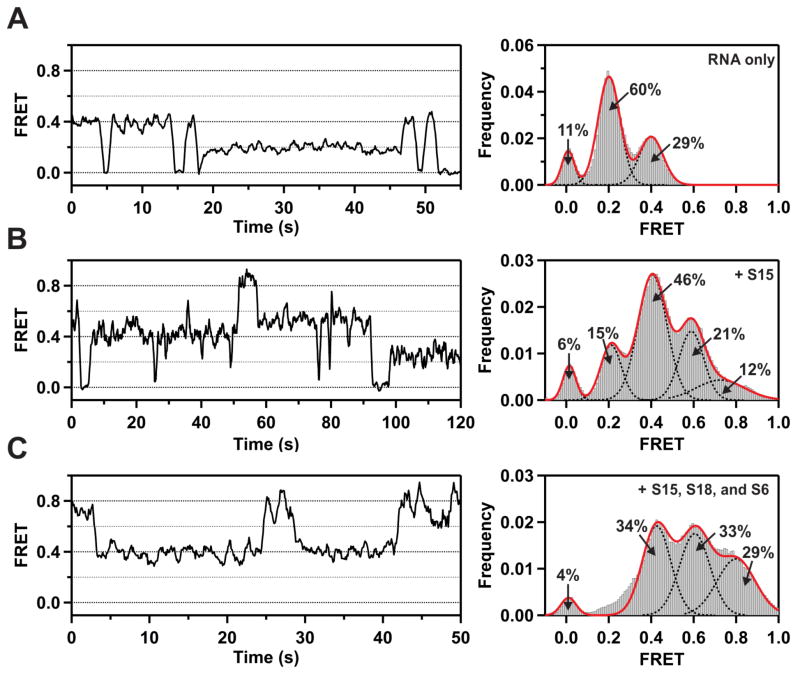
In the absence of any added proteins, individual RNA molecules populate two states with FRET efficiencies of ~0.2 and ~0.4, interspersed with brief periods in a state of zero FRET efficiency (Figure 4A, left panel). The latter state could arise from transient blinking of the Cy5 acceptor. To assess this possibility, we examined RNA molecules labeled with only the Cy5 acceptor and observed that 7% of these exhibited transient blinking when directly excited at 643 nm. A histogram of FRET efficiencies compiled from 111 individual RNA molecules reveals that the state with 0.2 FRET efficiency is dominant, followed by the state with 0.4 FRET efficiency (Figure 4A, right panel), while the population of the zero efficiency state (11%) is similar to the percentage of Cy5 acceptors that exhibit transient blinking upon direct excitation (7%), confirming the origin of this state. Thus, the free RNA exists in two distinct, at least partially folded conformations, and is capable of spontaneously exchanging between them. We attribute the major 0.2 FRET species and the minor 0.4 FRET species to open and closed conformations, respectively, of the lower three-way junction between helices 20, 21 and 22. Previous smFRET studies of a small RNA fragment consisting of only this three-helix junction show that the RNA naturally partitions between open (major) and closed (minor) conformations in the absence of any ribosomal proteins.8 In the open conformation, helices 20, 21 and 22 are extended in opposite directions, which would result in a large distance between the Cy3 donor and Cy5 acceptor in the present double-labeled RNA. In contrast, in the closed conformation, helices 21 and 22 are coaxially stacked, with helix 20 forming an acute angle with helix 22.24 This folded conformation would result in a shorter donor-acceptor distance and a higher FRET efficiency, as observed in the double-labeled RNA.
Figure 4.
Single-molecule FRET data obtained under various experimental conditions. (A) Representative FRET trajectory of an individual RNA molecule in the absence of any proteins (left). Histogram of FRET efficiencies compiled from 111 individual RNA molecules (right). The dashed black lines are Gaussian fits to individual peaks, with percentage populations as indicated, and the solid red line is the composite fit to the overall histogram. (B) Representative FRET trajectory (left) and histogram of FRET efficiencies compiled from 145 individual RNA molecules (right) in the presence of ribosomal protein S15 (20 nM). Other details are as in panel (A). (C) Representative FRET trajectory (left) and histogram of FRET efficiencies compiled from 152 individual RNA molecules (right) in the presence of ribosomal proteins S15 (20 nM), S16 (30 nM) and S18 (30 nM). Other details are as in panel (A).
In the presence of saturating concentrations of the primary ribosomal protein S15, the same three states are again observed, and in addition, two new states with ~0.6 and ~0.8 FRET efficiencies are populated (Figure 4B, left panel). A histogram of FRET efficiencies compiled from 145 individual RNA molecules reveals that the dominant state is now that with 0.4 FRET efficiency (Figure 4B, right panel). Control experiments confirmed that S15 has no direct effect on the emission intensity of either the Cy3 donor or Cy5 acceptor (Figure S31). The increase in the fractional population of the 0.4 FRET state is consistent with previous studies showing that S15 favors the closed conformation of the lower junction.24 The appearance of two new states at higher FRET efficiency is likely due to a conformational change of the upper three-helix junction formed by helices 22, 23a and 23b induced by S15. Previous biochemical studies have also shown that S15 binding induces a conformational change in the upper three-helix junction25 and there is evidence from crystallographic studies that S15 stabilizes the tertiary fold of the upper junction.26 Our results suggest that the upper three-helix junction can adopt two distinct conformations, which we term “open” and “closed”.
The upper three-helix junction is the binding site for proteins S6 and S18, which form a heterodimer and bind cooperatively to RNA with protein S15.5,6 In the presence of saturating concentrations of S15 and the S6/S18 heterodimer, the fractional populations of the 0.6 and 0.8 FRET efficiency states increase markedly (Figure 4C). A previous crystallographic study has shown that binding of the S6/S18 heterodimer stabilizes an upper junction conformation in which helix 23b stacks with helix 22, while helix 23a rotates towards the lower junction and packs alongside helix 22.26 This conformation of the upper junction is expected to further shorten the donor-acceptor distance and is likely responsible for the 0.8 FRET state.
In contrast to the changes observed upon addition of S15, S6, and S18, the addition of ribosomal protein S8 alone or in combination with S15 has no effect on the FRET distributions, as can be seen by comparing Figures 4A and S32A or Figures 4B and S32B, respectively. Similarly, the FRET distributions are insensitive to the concentration of S8 (Figure S32C and D). The S8 protein is known to bind near the center of helix 21 and is not expected to induce any change in the relative orientations of the helices flanking the lower junction. This is consistent with the absence of any observed effect, and demonstrates that the effects observed with S15, S6, and S18 arise from specific protein-RNA interactions.
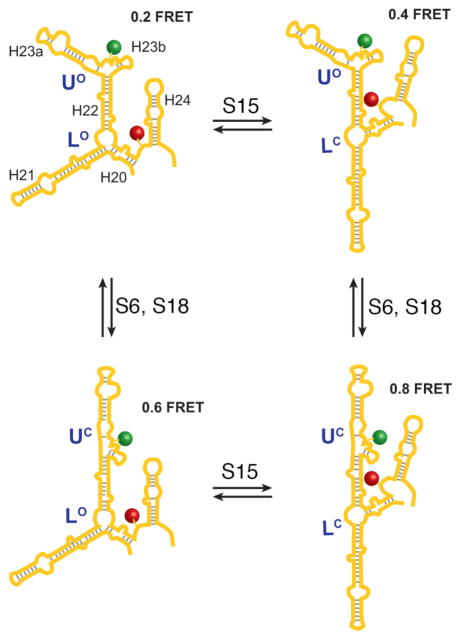
The large double-labeled RNA construct examined in this study reveals the conformational preferences and folding behavior of a major portion of the central domain of 16S rRNA. Our results suggest that the upper and lower three-helix junctions can each exist in two conformations, giving rise to a total of four distinct conformations for the RNA as a whole (Figure 5). We tentatively assign these as LOUO (0.2 FRET state), LCUO (0.4 FRET state), LOUC (0.6 FRET state) and LCUC (0.8 FRET state), where “L” and “U” refer to the lower and upper junctions, respectively, and the superscripts “O” and “C” refer to open and closed conformations, respectively. In the absence of any ribosomal proteins, the RNA populates the LOUO (major) and LCUO (minor) conformations. In the presence of S15 alone, the distribution shifts in favor of the LCUO conformer and the LOUC and LCUC conformers are also populated to some extent. When the S6/S18 heterodimer is also present, the LOUC and LCUC conformers are further stabilized. Interestingly, the presence of a very high concentration of Mg2+ ions (100 mM) also stabilizes the LOUC and LCUC conformers, although the ions are much less effective at stabilizing these states than the ribosomal proteins (Figure S33). Additional smFRET experiments utilizing different donor and acceptor labeling sites and/or RNA mutants that disrupt specific tertiary interactions will be required to unambiguously determine the precise nature of the four distinct RNA conformations. Based on the results of this initial study, it is evident that a wealth of information on RNA conformation and dynamics can be obtained through the combination of site-specific labeling and smFRET methods described here.
Figure 5.
Model for protein binding-induced folding of the junctions formed by G558 to C800 of the T. thermophilus 16S central domain. FRET efficiencies for each state are indicated and each helix is indicated in the upper left while the positions of fluorophore labeling are indicated with stars. The designations “L” and “U” refer to the lower and upper junctions, respectively, while the “O” and “C” superscripts denote open and closed conformations, respectively. The ribosomal proteins are indicated where they exert their primary effect (see text for details).
These smFRET measurements reveal previously unknown complexity in the series of RNA conformational changes necessary for assembly of the central domain of the 30S ribosomal subunit. For the first time, it has been possible to observe the conformations of two three-helix junctions separated by a common helix independently, and to measure the changes in conformational equilibria that accompany protein binding. Four distinct conformations can be identified, and the populations are shifted toward more folded states upon ordered protein binding. The results imply that there is a gradual and sequential adoption of more native-like structure as sequential protein binding proceeds, until the final native 30S subunit is formed.
This is also the first reported method for labeling a large RNA molecule with two different fluorophores at defined locations using in vitro transcription. Two 5SICS-MMO2 UBPs were used, with the d5SICS of one directing the incorporation of MMO2A into the RNA and the dMMO2 of the other directing the incorporation 5SICSCO. The orthogonal linkers were then selectively derivatized using Click chemistry and amine-reactive chemistry, respectively. While fluorophore-labeled RNAs were synthesized for the present study, there are few limitations to the functional groups that could be introduced. Thus, the approach based on an in vitro expanded genetic alphabet should be broadly applicable to biochemical, structural and functional studies of many different RNAs.
METHODS
Full details for all experimental methods are provided in the Supporting Information.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (GM060005 to F.E.R.; GM044060 to D.P.M.).
Footnotes
Figures, tables, and additional text describing detailed experimental procedures, synthesis and characterization of compounds, and additional experimental results. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Mizushima S, Nomura M. Assembly mapping of 30S ribosomal proteins from E coli. Nature. 1970;226:1214–1218. doi: 10.1038/2261214a0. [DOI] [PubMed] [Google Scholar]
- 2.Shajani Z, Sykes MT, Williamson JR. Assembly of Bacterial Ribosomes. Annu Rev Biochem. 2011;80:501–526. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- 3.Held WA, Ballou B, Mizushima S, Nomura M. Assembly mapping of 30 S ribosomal proteins from Escherichia coli Further Studies. J Biol Chem. 1974;249:3103–3111. [PubMed] [Google Scholar]
- 4.Adilakshmi T, Bellur DL, Woodson SA. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature. 2008;455:1268–1272. doi: 10.1038/nature07298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agalarov SC, Williamson JR. A hierarchy of RNA subdomains in assembly of the central domain of the 30 S ribosomal subunit. RNA. 2000;6:402–408. doi: 10.1017/s1355838200991945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Recht MI, Williamson JR. RNA tertiary structure and cooperative assembly of a large ribonucleoprotein complex. J Mol Biol. 2004;344:395–407. doi: 10.1016/j.jmb.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Batey RT, Williamson JR. Interaction of the Bacillus stearothermophilus ribosomal protein S15 with 16 S rRNA. 1. Defining the minimal RNA site. J Mol Biol. 1996;261:536–549. doi: 10.1006/jmbi.1996.0481. [DOI] [PubMed] [Google Scholar]
- 8.Ha T, Zhuang XW, Kim HD, Orr JW, Williamson JR, Chu S. Ligand-induced conformational changes observed in single RNA molecules. Proc Natl Acad Sci USA. 1999;96:9077–9082. doi: 10.1073/pnas.96.16.9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim HD, Nienhaus GU, Ha T, Orr JW, Williamson JR, Chu S. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc Natl Acad Sci USA. 2002;99:4284–4289. doi: 10.1073/pnas.032077799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klostermeier D, Sears P, Wong CH, Millar DP, Williamson JR. A three-fluorophore FRET assay for high-throughput screening of small-molecule inhibitors of ribosome assembly. Nucleic Acids Res. 2004;32:2707–2715. doi: 10.1093/nar/gkh588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Holmstrom E, Zhang J, Yu P, Wang J, Dyba MA, Chen D, Ying J, Lockett S, Nesbitt DJ, Ferre-D'Amare AR, Sousa R, Stagno JR, Wang YX. Synthesis and applications of RNAs with position-selective labelling and mosaic composition. Nature. 2015;522:368–372. doi: 10.1038/nature14352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishizuka T, Kimoto M, Sato A, Hirao I. Site-specific functionalization of RNA molecules by an unnatural base pair transcription system via click chemistry. Chem Commun. 2012;48:10835–10837. doi: 10.1039/c2cc36293g. [DOI] [PubMed] [Google Scholar]
- 13.Someya T, Ando A, Kimoto M, Hirao I. Site-specific labeling of RNA by combining genetic alphabet expansion transcription and copper-free click chemistry. Nucleic Acids Res. 2015;43:6665–6676. doi: 10.1093/nar/gkv638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavergne T, Malyshev DA, Romesberg FE. Major groove substituents and polymerase recognition of a class of predominantly hydrophobic unnatural base pairs. Chem Eur J. 2012;18:1231–1239. doi: 10.1002/chem.201102066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leconte AM, Hwang GT, Matsuda S, Capek P, Hari Y, Romesberg FE. Discovery, characterization, and optimization of an unnatural base pair for expansion of the genetic alphabet. J Am Chem Soc. 2008;130:2336–2343. doi: 10.1021/ja078223d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malyshev DA, Dhami K, Lavergne T, Chen T, Dai N, Foster JM, Correa IR, Jr, Romesberg FE. A semi-synthetic organism with an expanded genetic alphabet. Nature. 2014;509:385–388. doi: 10.1038/nature13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, Lavergne T, Malyshev DA, Zimmermann J, Adhikary R, Dhami K, Ordoukhanian P, Sun Z, Xiang J, Romesberg FE. Site-specifically arraying small molecules or proteins on DNA using an expanded genetic alphabet. Chem Eur J. 2013;19:14205–14209. doi: 10.1002/chem.201302496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seo YJ, Malyshev DA, Lavergne T, Ordoukhanian P, Romesberg FE. Site-specific labeling of DNA and RNA using an efficiently replicated and transcribed class of unnatural base pairs. J Am Chem Soc. 2011;133:19878–19888. doi: 10.1021/ja207907d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimoto M, Sato A, Kawai R, Yokoyama S, Hirao I. Site-specific incorporation of functional components into RNA by transcription using unnatural base pair systems. Nucleic Acids Symp Ser. 2009;53:73–74. doi: 10.1093/nass/nrp037. [DOI] [PubMed] [Google Scholar]
- 20.Morohashi N, Kimoto M, Sato A, Kawai R, Hirao I. Site-specific incorporation of functional components into RNA by an unnatural base pair transcription system. Molecules. 2012;17:2855–2876. doi: 10.3390/molecules17032855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ludwig J, Eckstein F. Rapid and efficient synthesis of nucleoside 5'-0-(1-thiotriphosphates), 5'-triphosphates and 2',3'-cyclophosphorothioates using 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one. J Org Chem. 1989;54:631–635. [Google Scholar]
- 22.Anderson S, Neidlein U, Diederich F, Gramlich V. Eine neue Klasse chiraler, von 1,1'-Binaphthyl abgeleiteter Cyclophan-Rezeptoren: Komplexierung von Pyranosiden. Angew Chem. 1995;107:1722–1725. [Google Scholar]
- 23.Clemons WM, Jr, Brodersen DE, McCutcheon JP, May JL, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: purification, crystallization and structure determination. J Mol Biol. 2001;310:827–843. doi: 10.1006/jmbi.2001.4778. [DOI] [PubMed] [Google Scholar]
- 24.Orr JW, Hagerman PJ, Williamson JR. Protein and Mg2+-induced conformational changes in the S15 binding site of 16 S ribosomal RNA. J Mol Biol. 1998;275:453–464. doi: 10.1006/jmbi.1997.1489. [DOI] [PubMed] [Google Scholar]
- 25.Svensson P, Changchien LM, Craven GR, Noller HF. Interaction of ribosomal proteins, S6, S8, S15 and S18 with the central domain of 16 S ribosomal RNA. J Mol Biol. 1988;200:301–308. doi: 10.1016/0022-2836(88)90242-2. [DOI] [PubMed] [Google Scholar]
- 26.Agalarov SC, Sridhar Prasad G, Funke PM, Stout CD, Williamson JR. Structure of the S15,S6,S18-rRNA complex: assembly of the 30S ribosome central domain. Science. 2000;288:107–113. doi: 10.1126/science.288.5463.107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.