Abstract
Colorectal cancer (CRC) is a genetic disease, due to progressive accumulation of mutations in oncogenes and tumor suppressor genes. Large scale genomic sequencing projects revealed >100 mutations in any individual CRC. Many of these mutations are likely passenger mutations and fewer are driver mutations. Of these, activating mutations in RAS proteins are essential for cancer initiation, progression, and/or resistance to therapy. There has been significant interest in developing drugs targeting mutated cancer gene products or downstream signaling pathways. Due to the number of mutations involved and inherent redundancy in intracellular signaling, drugs targeting one mutation or pathway have been either ineffective or led to rapid resistance. We have devised a strategy whereby multiple cancer pathways may be simultaneously targeted for drug discovery. For proof-of-concept, we targeted the oncogenic KRAS, and HIF pathways, since oncogenic KRAS has been shown to be required for cancer initiation and progression, and HIF-1α and HIF-2α are induced by the majority of mutated oncogenes and tumor suppressor genes in CRC. We have generated isogenic cell lines defective in either oncogenic KRAS or both HIF-1α and HIF-2α, and subjected them to multiplex genomic, siRNA, and high-throughput small molecule screening. We have identified potential drug targets and compounds for preclinical and clinical development. Screening of our marine natural product library led to the rediscovery of the microtubule agent dolastatin 10 and the class I histone deacetylase (HDAC) inhibitor largazole to inhibit oncogenic KRAS and HIF pathways. Largazole was further validated as an anti-angiogenic agent in a HIF-dependent manner in human cells and in vivo in zebrafish using a genetic model with activated HIF. Our general strategy, coupling functional genomics with drug susceptibility or chemical-genetic interaction screens, enables the identification of potential drug targets and candidates with requisite selectivity. Molecules prioritized in this manner can easily be validated in suitable zebrafish models due to the genetic tractability of the system. Our multidimensional platform with cellular and organismal components can be extended to larger scale multiplex screens that include other mutations and pathways.
Oncogenic RAS mutations including HRAS, KRAS, and NRAS are found in approximately 30% of all human tumors, with KRAS being the most prevalent1,2. KRAS mutations are most prevalent in pancreatic (72–90%), thyroid (55%), colorectal (32–57%), and lung cancers (15–50%). Activating KRAS mutations are important for cancer initiation and progression, and cause primary resistance to therapy targeting EGFR. Signaling downstream of oncogenic KRAS turns on genes that promote cell proliferation, obstruct cell death, and induce angiogenesis and metabolic adaptation.
The hypoxia-inducible factors-1α and -2α (HIF-1α and HIF-2α) are transcription factors that are overexpressed in cancer and often linked to cancer progression3. HIF-1α and HIF-2α overexpression is driven by intratumoral hypoxia and genetic mutations in oncogenes and tumor suppressor genes3, and their target genes important for tumor angiogenesis, cell growth and survival, and metastasis. MAPK and mTOR/AKT signaling downstream of RAS has been shown to lead to the transcriptional activation of HIF-1α by HIF-1α phosphorylation and induction of HIF-1α expression, respectively4. To evaluate the relationship between oncogenic KRAS, HIF-1α, and HIF-2α, we generated isogenic cell lines from HCT116 human colorectal cell lines, containing both a wildtype (WT) KRAS allele and an oncogenic KRAS allele. Using cells defective in either the oncogenic KRAS allele or in both HIF-1α and HIF-2α, we recently reported that HIF-1α and HIF-2α work together to regulate metabolic genes signature overlapping with that of oncogenic KRAS5.
We have performed a global analysis of gene expression regulated by oncogenic KRAS, HIF-1α, HIF-2α, and both HIF-1α and HIF-2α together. These cell lines were applied in multiplex high-throughput screens with (i) an siRNA library targeting the druggable genome (7,784 targets) and (ii) small molecule libraries to identify “hits” that show toxicity only in cells that express the oncogenic KRAS or HIF transcription factors. Using Ingenuity Pathway Analysis (IPA), we analyzed how canonical cancer pathways are affected. We found druggable targets, canonical pathways targeted by small molecules, including natural products which may inhibit cancer cells with KRAS mutation and HIF activation. One prioritized marine natural product was validated in vitro and then exposed to a genetic zebrafish model system, giving an in vivo dimension to our screening platform.
RESULTS
Comparative Gene Expression Profiling of Isogenic HIF and KRAS Knockout Cells
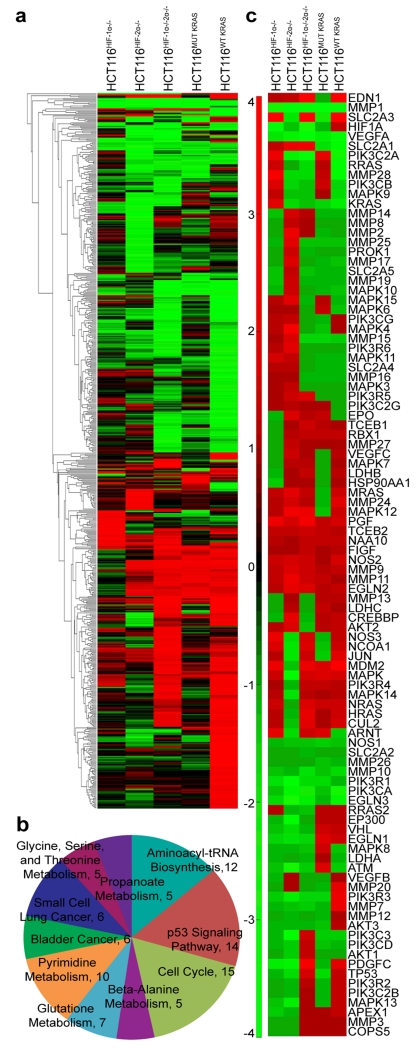
To determine whether HIF-1α and HIF-2α target genes are also downstream targets of oncogenic KRAS, we performed global gene expression analyses on Parental HCT116, HCT116HIF-1α−/−, HCT116HIF-2α−/−, HCT116HIF-1α-/-HIF-2α−/−, HCT116MUT KRAS, and HCT116WT KRAS cells. The parental HCT116 cell line contains an oncogenic KRAS allele and a wildtype KRAS allele. HCT116MUT KRAS has oncogenic KRAS gene, and the wild-type KRAS gene knocked out; whereas HCT116WT KRAS has wild-type KRAS gene, and with oncogenic KRAS knocked out4,5. Using a cut-off of >3.0-fold difference in gene expression between parental HCT116 versus the knockout cell lines, we found that global gene expression affected by oncogenic KRAS showed significant overlap with genes affected by both HIF-1α and HIF-2α (parental HCT116 versus HCT116WT KRAS in comparison to parental HCT116 versus HCT116HIF-1α-/-HIF-2α−/−) (Figure 1a). We present the credentials of the isogenic lines in Supplementary Figure S1, where the decrease in VEGFA and LDHA levels are confirmed. The shared genes fell into the following top 10 KEGG pathways6,7: cell cycle, p53 signaling pathway, aminoacyl-tRNA biosynthesis, pyrimidine metabolism, glutathione metabolism, small cell lung cancer, bladder cancer, glycine-serine-threonine metabolism, propanoate metabolism, and beta-alanine metabolism (Figure 1b).
Figure 1.
HCT116HIF-1α-/-HIF-2α−/− and HCT116WT KRAS cells demonstrate similar transcript profiles. (a) Global gene expression affected by oncogenic KRAS showed significant overlap with genes affected by both HIF-1α and HIF-2α (parental HCT116 versus HCT116WT KRAS in comparison to parental HCT116 versus HCT116HIF-1α-/-HIF-2α−/−). (b) GO analysis revealed top molecular functions associated with HCT116HIF-1α-/-HIF-2α−/−. (c) Similar to effects on global gene expression, gene perturbations specific to the HIF pathway are more similarly affected in the HCT116HIF-1α-/-HIF-2α−/− and HCT116WT KRAS cells; compared to HCT116HIF-1α−/−, HCT116HIF-2α−/−, or HCT116MUT KRAS cells.
We next examined whether the gene set involved in the canonical HIF pathway is regulated by oncogenic KRAS. Using IPA, we filtered for gene perturbations affecting the canonical HIF pathway. Genes in the HIF pathway involve growth factor receptors, signaling protein cascades including the Ras/Raf/MAPK and PI3K/AKT pathways, the HIF transcription factors and regulatory proteins (i.e., VHL, proteasome), and downstream target genes (glucose transporters, glycolysis enzymes, and angiogenic factors). Activation of growth factor receptors and signaling proteins would lead to the induction of HIF-1α and HIF-2α, and the consequent transactivation of target genes (Figure 1c and Supplementary Table S1). As shown in Supplementary Figures S1a and S1b, downstream HIF target genes (i.e., GLUT, MMPs, NOS, VEGF, EPO, LDH) are similarly downregulated (colored green) in both HCT116HIF-1α-/-HIF-2α−/− and HCT116WT KRAS cells. Our observation is consistent with published data that oncogenic KRAS positively regulates HIF transcriptional activity through the induction of HIF expression and activation5,8. Furthermore, we found that signaling genes upstream of HIF (MAPK and PI3K/AKT) are similarly downregulated (colored green) in both HCT116HIF-1α-/-HIF-2α−/− and HCT116WT KRAS cells; suggesting feedback regulation of these genes by HIF.
Genomic siRNA Screens for Gene Targets in Oncogenic KRAS and HIF Pathways
Since oncogenic KRAS and HIF-1α/HIF-2α gene signatures share significant overlap, we hypothesize that druggable gene targets can be identified, which inhibit oncogenic KRAS and HIF pathways. We performed an unbiased global search by using an arrayed siRNA library that targets 7,784 “druggable” genes with multiple coverage (4 siRNAs per gene) in 384-well format.
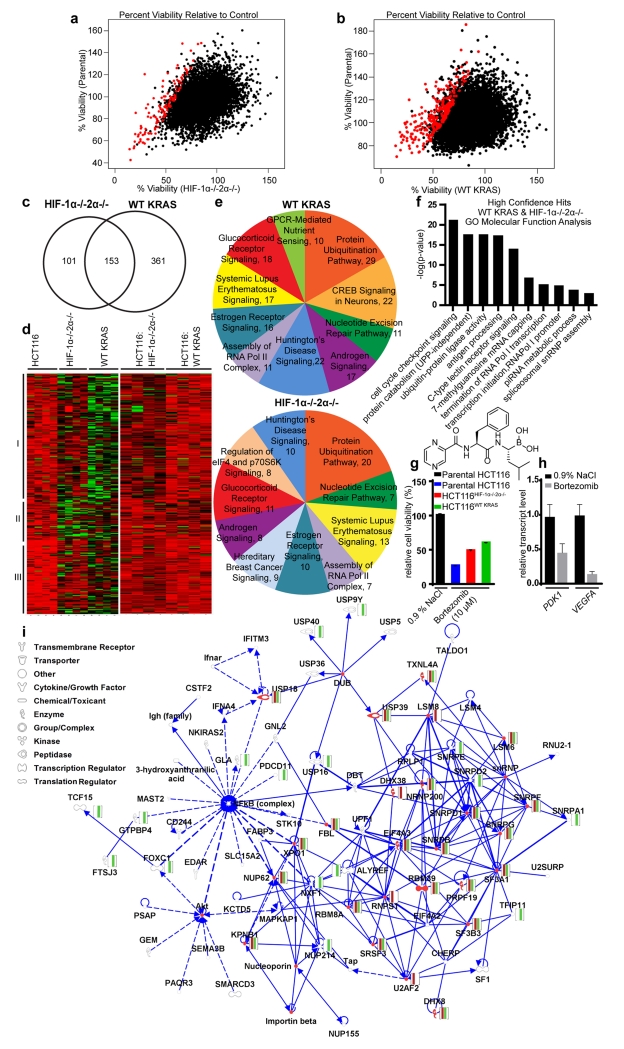
The isogenic lines were seeded at densities which yielded equivalent cellular confluency at the time of detection. We subjected the cells to the siRNA library (20 nM) and assessed differential cytotoxicity 96 h later (48 h to allow knockdown and additional 48 h to allow a downstream effect). If an siRNA inhibited oncogenic KRAS or HIF-1α/HIF-2α pathway, it should inhibit the parental cells and not the knockout cells. A correlation in siRNA-induced cytotoxicity was seen between parental HCT116 and HCT116HIF-1α−/−HIF-2α−/− (Figure 2a), and between parental HCT116 and HCT116WT KRAS (Figure 2b) cells. Hits were identified as those with a viability ratio of the parental HCT116 to one of the knockout cells (parental:ko) <0.6, p < 0.05 across replicates, and a minimum of two siRNAs causing differential toxicity (Supplementary Table S2). High-confidence hits (176 genes, highlighted in red) were identified as those with three or four siRNAs causing differential cytotoxicity (Figures 2a and 2b). Using Venn diagram (Figure 2c) and heatmap analyses (Figure 2d), all hits were grouped into those that affect the oncogenic KRAS pathway only, the HIF-1α/HIF-2α pathway only, or both oncogenic KRAS and HIF-1α/HIF-2α pathways. A total of 153 hits with statistically significant differential toxicity were shared between isogenic knockout lines (Figure 2c).
Figure 2.
Identification of novel gene targets that are involved in both oncogenic KRAS and HIF Pathways. Parental HCT116, HCT116HIF-1α-/-HIF-2α−/−, and HCT116WT KRAS cells were subjected to an siRNA library 7,784 “druggable” genes with multiple coverage (4 siRNAs per gene) and then assessed for differential cytotoxicity. (a,b) High-confidence hits (shown in red) were those where the viability ratio of the parental cells compared to one of the knockout cell lines (wt:ko) was <0.6 with p < 0.05 across replicates. Using Venn diagram (c) and heatmap analyses (d), hits can be grouped into those that affect oncogenic KRAS pathway only (I), HIF pathway only (II), or both oncogenic pathways (III). (e) IPA of top canonical pathways reveals significant overlap between the biofunctions of top hits. (f) GO analysis of top molecular functions. Effects of proteasome inhibitor bortezomib (10 μM) on (g) differential cell viability at and (h) HIF target gene expression (at 1.58 μM (IC90). (i) IPA of top networks of genes with differential toxicity in both HCT116HIF-1α-/-HIF-2α−/− (red), and HCT116WT KRAS (green) cells. Overlap between HIF/KRAS pathway and the inflammatory and PI3K/Akt pathways was highlighted, as the NFkB complex and Akt are among the top nodes of interacting hits within the network.
The top canonical pathways, biological processes, and networks of all hits were generated through the use of QIAGEN’s Ingenuity® iReport. A significant amount of overlap was seen between the top canonical pathways of siRNAs inhibiting KRAS and HIF-1α/HIF-2α pathways. The top ten canonical pathways included: protein ubiquitination, nucleotide excision repair (NER) (Supplementary Figure S3), systemic lupus erythematosus signaling, Huntington’s disease signaling, assembly of RNA polymerase II complex, estrogen receptor signaling, androgen signaling, and glucocorticoid receptor signaling pathways were all shared amongst hits (Figure 2e and Supplementary Table S3a, S3b). While most of the pathways contain proteins which have been shown to have interactions with either HIF-1α, HIF-2α, or KRAS, some of these pathways have not been strongly linked to HIF and KRAS pathways. The assembly of RNA polymerase II complex contains no proteins that have been shown to have direct interactions with HIF-1α, HIF-2α, or KRAS. Only one interaction (HIF1α with RAD23B)9 has been previously reported in the nucleotide excision repair pathway. Within the protein ubiquitination pathway, only two interactions with HIF1α (STUB1, PSMA4)10,11, two interactions with HIF2 α (PSMB1, STUB1)10,12 and one interaction with KRAS (PSMA30)13 have been reported. With 31 focus molecules per data set, ‘RNA Post-Transcriptional Modification, Cellular Assembly and Organization’ was the most significant network, with oncogenic KRAS and HIF networks also containing ‘Infectious Disease’ and ‘Cell Cycle’ functionalities, respectively (Supplementary Table S4). Further discussion and description of IPA outcomes including common upstream regulators (Supplementary Figure S4) can be found in the Supplemental Results and Supplemental Materials and Methods sections. A Gene Ontology (GO) analysis revealed that the RNA modification processes are largely associated with RNA splicing via the spliceosome and transesterification reactions15. Interestingly, when evaluating high confidence hits, major biological processes involved antigen and multi-organismal (symbiotic and viral) processes. High-confidence hits were evaluated to determine top biological processes and top molecular functions shared between active siRNAs. GO analysis of high confidence hits revealed that functions centered around cell cycle and growth signaling were among the top functions with KRAS/HIF-associated toxicity (Figure 2f). Since we identified the protein ubiquitination pathway, we then evaluated a proteasome inhibitor, bortezomib, in our isogenic screening system where it exhibited differential effects on cell viability (Figure 2g). Bortezomib also reduced levels of HIF-driven transcripts, including VEGFA (Figure 2h), consistent with previous reports that it is a HIF-1 inhibitor,14 while validating our screen.
A global network was generated using IPA in which top networks with a minimum of 5 shared molecules were connected (Figure 2i). Top diseases and biofunctions within this network include RNA post-transcriptional modification, cellular assembly and organization, infectious disease, cell cycle, molecular transport, and RNA trafficking. Major nodes identified in these hits include the NFκB complex and Akt, suggesting that these pathways are involved with HIF/KRAS associated toxicity. It has been previously shown that NFκB is involved in the expression of HIF-1α16 and microtubule depolymerizational activation of HIF-1α occurs through a NFκB dependent mechanism17. Additionally, studies have demonstrated the role of HIF-1α in the binding of the NFκB complex to DNA18. Furthermore, KRAS has been shown to increase activation of the NFκB complex and enhances the protein DNA binding activity of NFκB complexes consisting of p50 and p65 subunits, further confirming this crosstalk19. The PI3K-Akt pathway, which promotes survival and growth, has been shown to associate with both HIF-1α and KRAS. Akt increases activation of HIF-1α protein20. Interference of HIF-1α was shown to decrease activation of Akt via PLGF protein21. Similarly, the presence of dominant negative Akt proteins in MC3T3-E1 cells was shown to decrease the activation of HIF-1α during ultrasound22. Several studies have also highlighted the involvement of KRAS in the activation and phosphorylation of Akt proteins, further supporting crosstalk between the HIF/KRAS and the PI3K/Akt pathways23-28. Additional top overlapping networks not connected to the top global network include those with functionality in RNA post-transcriptional modification, cell cycle, cellular assembly, organization, gene expression, DNA replication, recombination and repair, and cancer (Supplementary Table S5). The top diseases and biofunctions of active siRNAs were also assessed independently of shared network molecules to evaluate the global impact of the hits on a biological system based on activation z-score (Supplementary Table S5). Interestingly, many of our hits demonstrate some overlap with the functionalities of synthetic lethals targeting KRAS. Although significant amount of overlap is rarely seen between synthetic lethal screens, certain genes including POLR2B, PSMA2, POLR2G, PSMA, CDC6, E2F1, and PSMD14 have been previously reported29 (Supplementary Table S6).
Small Molecule Screen for Inhibitors of Oncogenic KRAS and HIF Pathways
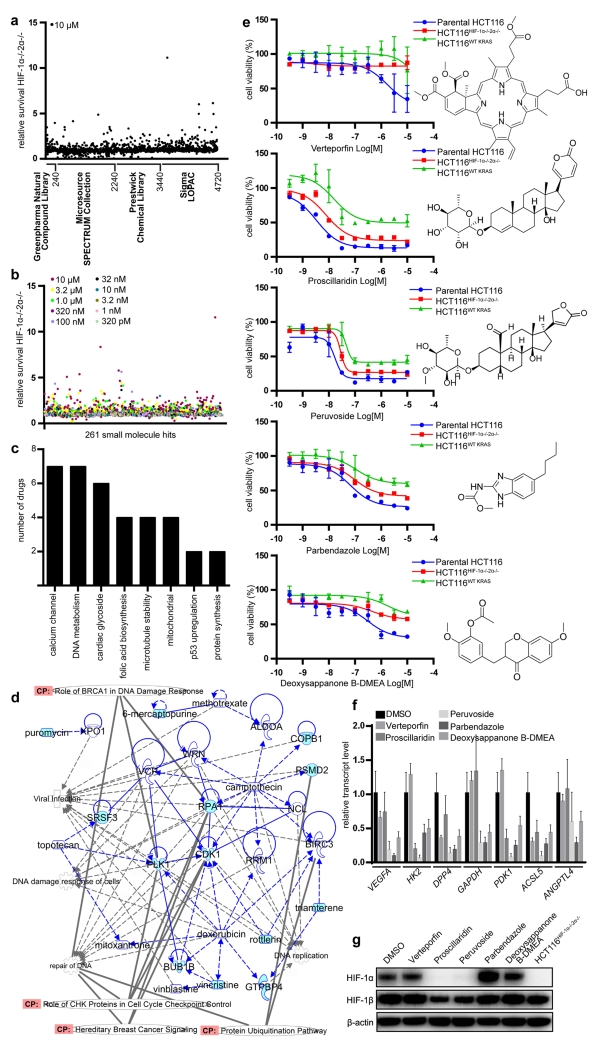
For proof-of-concept studies, we initially screened commercially available small molecule libraries composed of 4,720 natural products and other compounds (Figure 3a). Parental HCT116 or HCT116HIF-1α-/-HIF-2α-/- cells were treated with 10 μM of library compounds in 96-well plates and incubated under normoxic condition for 48 h, when cell viability was determined at equivalent cellular confluency. Putative “hits” were compounds where the viability ratio of the parental cells compared to the knockout cell lines was <0.6.
Figure 3.
Identification of small molecules that inhibit oncogenic KRAS and HIF pathways. (a) Commercially available libraries were screened for differential cytotoxicity in parental HCT116 versus HCT116WT KRAS and parental HCT116 versus HCT116HIF-1α-/-HIF-2α−/− cells with a viability ratio (wt:ko) < 0.6. (b) 261 small molecules which demonstrated differential toxicity or full toxicity at 10 μM (to account for false negatives) were further selected for a dose-response analysis. (c) Differentially active compounds exhibited overlap in their mechanisms of action. (d) Overlap between small molecule screen and siRNA library screen demonstrates similarities in functions and canonical pathways. Those genes and compounds not previously associated with HIF and KRAS are highlighted in blue. (e) Dose-response analysis for verteporfin, proscillaridin, peruvoside, parbendazole, and deoxysappanone B 7,3′-dimethyl ether acetate [deoxysappanone B-DMEA]. (f) HIF target gene expression in response to 16-h treatment of parental HCT116 cells with deoxysappanone B-DMEA (250 nM), proscillaridin (100 nM), peruvoside (100 nM), parbendazole (2 μM) and verteporfin (1 μM). (g) Proscillaridin (100 nM) and peruvoside (100 nM) abrogated HIF-1α protein levels completely, while partially suppressing HIF-1β. Verteporfin, parbendazole, and deoxysappanone B-DMEA had no effect on HIF-1α and HIF-1β protein levels.
We selected 261 compounds for further testing, which included ones showing differential cytotoxicity and those that were toxic to both cell types at 10 μM concentration (the latter to incorporate potential false negatives which could demonstrate differential toxicity at lower concentrations) (Figure 3b). Several compounds showed up as hits from multiple libraries, serving as an internal validation including podophyllotoxin; niclosamide; anthothecol; camptothecin and its analogue, 10-hydroxycamptothecin; and methotrexate and its analogue, aminopterin. Using dose-response analysis, we identified 55 compounds (Supplementary Figure S5a-e and Supplementary Table S7) which showed differential cytotoxicity with dose titration to lower concentrations (Figure 3b). These compounds exhibited overlap in their mechanisms of action, including calcium channel regulation, DNA metabolism, cardiac glycoside functionality, folic acid biosynthesis, microtubule stability, p53 regulation, and protein synthesis regulation (Figure 3c). We generated a network in an effort to investigate overlap between the small molecule screen with the siRNA screen. Figure 3d demonstrates direct links between our screens, with functional (cell cycle checkpoints, DNA replication) and canonical pathway (protein ubiquitination pathway, BRCA1 DNA damage response, and hereditary breast cancer signaling) overlap also indicated. Interestingly, an increase incidence of colon cancer has been reported in BRCA1 mutation carriers, which is also part of hereditary breast cancer signaling30. Many molecules have shown previous associations with HIF, providing confidence to our screen. Others have not yet been previously associated with HIF and KRAS (highlighted in light blue), which warrant further investigation.
We further validated five compounds (verteporfin, proscillaridin, peruvoside, parbendazole, and deoxysappanone B 7,3′-dimethyl ether acetate) as they showed promising differential cytotoxicity for parental HCT116 cells over HCT116HIF-1α−/−HIF-2α−/− cells in terms of IC50 shift and/or efficacy and represented structural and biochemical diversity (Figure 3e). Verteporfin, parbendazole and deoxysappanone 7,3′-dimethyl ether acetate have not been previously identified as HIF inhibitors. All five compounds caused significant downregulation of HIF target genes (Figure 3f). Proscillaridin and peruvoside completely suppressed HIF-1α protein expression, producing “pharmacological HIF-1α knockouts” (Figure 3g), while others did not greatly affect protein levels, suggesting different mechanisms of HIF inhibition. We also measured differential cytotoxicity of parental HCT116, which has oncogenic KRAS mutation, versus the isogenic oncogenic KRAS knockout cell line (HCT116WT KRAS); and found that these five compounds are more effective in the presence of oncogenic KRAS (Figure 3e).
Characterization of Marine Natural Products as HIF Inhibitors in Vitro and in Vivo
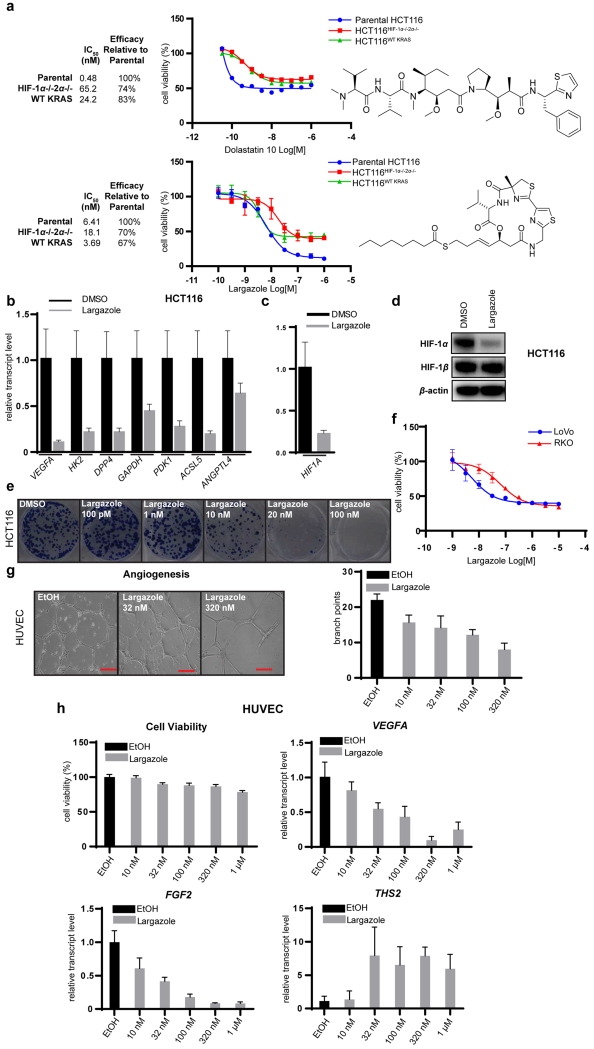
We next screened our library of marine natural products and identified dolastatin 1031 as an agent that preferentially acts on HIF-containing cells (Figure 4a). Dolastatin 10 demonstrated a strong decrease in potency in both HCT116HIF-1α-/-HIF-2α−/− and HCT116WT KRAS cells (IC50 shift from 0.48 to 65.2 and 24.2 μM, respectively), as well as a decrease in total efficacy (74% and 83%, respectively). We also found the class I HDAC inhibitor largazole32,33 to demonstrate significant differential cytotoxicity for parental HCT116 over HCT116HIF-1α-/-HIF-2α−/− and HCT116WT KRAS cells (Figure 4a). The potency of largazole is reduced by 3-fold in HCT116HIF-1α-/-HIF-2α−/− cells (IC50 shift from 6.41 to 18.1 μM), concomitant with reduced efficacy (70%). For cells lacking oncogenic KRAS the IC50s are comparable with parental cells; however the efficacy is reduced to 67%. The differential cytotoxicity of largazole for parental HCT116 over HCT116HIF-1α-/-HIF-2α−/− cells indicates that the antiproliferative activity of largazole is partially HIF-dependent and that this pathway represents a major mechanism of action. HIF target genes are potently downregulated by largazole (Figure 4b) in HIF-containing HCT116 cells. HIF-1α is also reduced at the transcriptional (Figure 4c) and protein level (Figure 4d). Largazole showed inhibitory activity in the HCT116 colony formation assay at 10 nM (Figure 4e) and reduced cytotoxicity against a HIF-independent cell line, RKO,34 compared with HCT116 and LoVo (Figure 4f). Largazole was also effective in an in vitro angiogenesis assay using human umbilical vein endothelial cells (HUVECs), decreasing the number of branch points of the tube network (Figure 4g) without affecting cell viability, while downregulating several proangiogenic factors (VEGFA, FGF2) and upregulating antiangiogenic factors (THS2) (Figure 4h).
Figure 4.
Natural products derived from marine cyanobacteria as HIF inhibitors. (a) Dose-response curves using isogenic HCT116 cell lines for dolastatin 10 and largazole. (b-d) Largazole (20 nM) downregulated HIF target genes (b) and HIF-1α transcript (c) , and diminished HIF-1α protein (d) based on immunoblot analysis, while HIF-1β (d) was unaltered (16 h treatment). (e) Largazole inhibited HCT116 cell growth in colony formation assays. (f) Largazole demonstrated differential toxicity towards LoVo relative to the HIF-independent cell line, RKO. (g) Largazole inhibited angiogenesis in vitro in a dose-dependent manner, determined by matrigel assay using HUVECs (scale bar 200 μm). (h) Effect of largazole on HUVEC viability and transcripts of pro-angiogenic factors (VEGFA, FGF2) and anti-angiogenic factors (THS2).
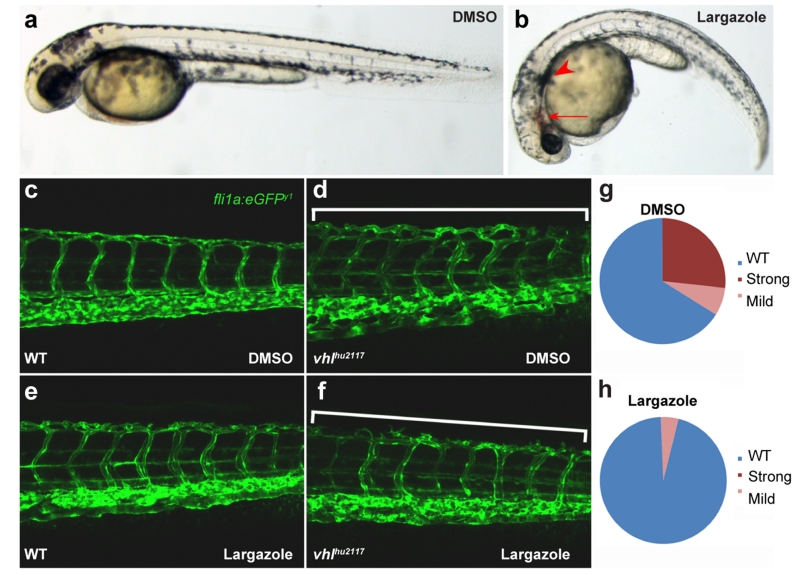
We next tested for the Hdac1 inhibitory function of largazole in vivo, through administration of 500 nM to zebrafish larvae from 19hpf. By 48hpf these larvae displayed phenotypes reminiscent of the zebrafish hdac1 mutant, colgate35,36, including blood pooling and pigment pattern defects (Figures 5a and 5b). We tested if largazole-mediated inhibition of Hdac1 would also have an antiangiogenic effect in zebrafish. To mimic pathogenic vascular defects, we exploited the excess vascularization seen in vhlhu2117 mutants. The Vhl protein is a negative regulator of Hif-1α, such that these mutants have excessive vascularization of the trunk and head, including a vascular plexus in the tail (Figures 5c and 5d), a process mediated by VEGFA37. Whilst application of 2 μM largazole from 36hpf until 132hpf had little effect on wild-type vascular patterning (Figure 5e), it significantly blocked the excessive vascular plexus in the tail of vhl mutants (Figures 5f-h).
Figure 5.
Hdac1 inhibitory function of largazole was evaluated in zebrafish angiogenic models. (a,b) Larvae treated with 500 nM largazole displayed blood pooling and pigment pattern defects, similar to that of hdac1 mutant, colgate phenotype. (c,d) To mimic pathogenic vascular defects, vhlhu2117 mutants exhibiting an increased vascularization were used. 2 μM largazole had little effect on basal vascularization of the wildtype embryo (e), while the vhlhu2117 vascularization was greatly minimized in the tail (f-h). The numbers of larvae showing the mutant phenotype after largazole treatment is significantly less than the expected 25% (p<0.01; Chi squared test; n=56 (DMSO), n=44 (Largazole)).
DISCUSSION
Our data demonstrated that isogenic knockouts of cancer genes can be used in multiplex screens to define targets and drugs affecting complex cancer pathways. Although isogenic knockouts have been used to find drugs affecting a single cancer pathway38,39, we show that multiple cancer pathways can be targeted at once by multiplex screens. As proof-of-concept, parental HCT116, HCT116HIF-1α-/-HIF-2α−/−, and HCT116WT KRAS cells were subjected to multiplex high-throughput screening (HTS) with an siRNA library targeting the druggable genome and with small molecule libraries to identify agents that show toxicity only in cells that express the oncogenic KRAS and HIF transcription factors.
From our siRNA screen, we identified eight canonical pathways to be associated with the oncogenic KRAS and HIF pathways: the protein ubiquitination, nucleotide excision repair, systemic lupus erythematosus signaling, Huntington’s disease signaling, assembly of RNA polymerase II complex, estrogen receptor signaling, androgen signaling, and glucocorticoid receptor signaling pathways. Our siRNA data is consistent with prior reports that HIF-α mediates transcription of target genes by directly associating with transcriptional cofactors and RNAPII40 and that proteasomal inhibitors inactivate HIF transcriptional activity41. Our data suggests that targeting these canonical pathways should be further developed for treating cancers with oncogenic KRAS mutation.
We have discovered small molecules that inhibit oncogenic KRAS and HIF pathways. Proscillaridin and peruvoside are cardiac glycosides and have previously been reported to exhibit cytotoxic activity in colorectal cancer cells42 and inhibit HIF-1α protein synthesis through inhibition of mitochondrial respiration43. We further showed that they also inhibit oncogenic KRAS pathway. We identified three novel oncogenic KRAS/HIF inhibitors: parbendazole, a deoxysappanone derivative, and verteporfin. Parbendazole is an inhibitor of microtubule assembly and antimicrotubule agents have been previously shown to inhibit HIF transcriptional activity by suppressing HIF-α translation and expression44. Our results suggest that parbendazole might inhibit HIF transcriptional activity independent of its antimicrotubule activity, as HIF protein expression was not affected in our cell-based system. Deoxysappanone is a component of green tea and has been shown to possess anti-diabetic properties by activating mitochondria45. Verteporfin is a benzoporphyrin used as a photosensitizer in photodynamic therapy to treat macular degeneration46. The mechanism of inhibition of HIF target gene expression by these compounds cannot be explained by effect on HIF protein expression. Further experiments are needed to elucidate how these agents regulate HIF transcriptional activity.
When we screened a subset of our natural products that we isolated from marine cyanobacteria, we identified the microtubule-depolymerizing agent dolastatin 10,32 consistent with the differential cytotoxicity of the microtubule-stabilizing agent paclitaxel in our initial screen. We also identified the class I HDAC inhibitor largazole as a novel inhibitor of the oncogenic KRAS and HIF pathways.33,34 Largazole inhibited the expression of HIF-1α and HIF target genes (incl. VEGFA) and induced the antiangiogenic factor, THS2, which presumably contributes to its antiangiogenic and antitumor activity. The mechanism of largazole inhibition of HIF expression is at the transcriptional and protein level through modulation of gene expression and probably also decrease of HIF stability, suggested to be perturbed by HDAC inhibition. Isoforms HDAC1 and HDAC3 are reported to associate with HIF-1α and modulate the expression of HIF-1α target genes and thus regulate angiogenesis47,48, whereas isoform HDAC2 modulates transcriptional activity through its interaction with the tumor suppression gene p5349. It is likely that largazole also inhibits HIF transcriptional activity through its binding to HDACs; however, further experiments are needed.
Zebrafish provide a powerful model system that is extensively exploited for mechanistic studies and also shows promise for drug screening50. The relevance of the HIF inhibitory and antiangiogenic activity of largazole in vitro was demonstrated in vivo. Largazole mimicked the Hdac1 knockout phenotype in zebrafish consistent with in vitro enzymatic and cellular data that largazole is a HDAC inhibitor. Importantly, largazole was able to inhibit HIF-induced vascularization in a zebrafish mutant with activated HIF pathway, indicating that our cellular screens have predictive power for the identification of in vivo-active HIF inhibitors by sufficiently mimicking in vivo conditions. Our results further emphasize that zebrafish systems are well suited as an intermediate platform to predict in vivo bioavailability of HIF inhibitors. Largazole has shown in vivo activity in numerous studies with cancer and non-cancer applications33.
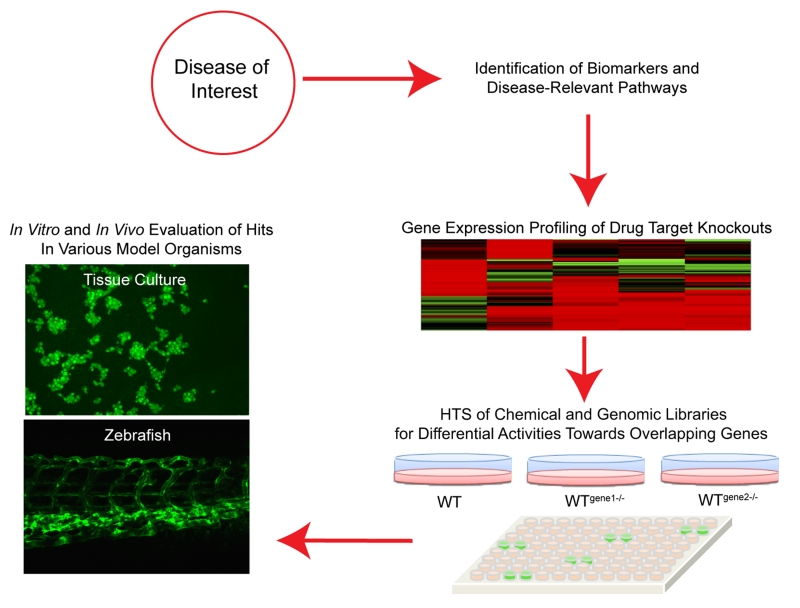
As a proof of concept, we demonstrated that multiplex screens of complex cancer pathways, oncogenic KRAS and HIF, can be used to find targets and drugs common to both pathways. Our high-throughput siRNA screen data shows that there are targets unique to each pathway (oncogenic KRAS or HIF). Drugs may be tailored to specific circumstances depending on the presence of certain pathway(s). Lastly, the general methodology presented here can be applied to other conditions and mutations through interrogating the corresponding pathways in vitro and in vivo (Figure 6). Coupling functional genomics with drug susceptibility or chemical-genetic interaction screens enables the identification of potential drug targets and small molecules as new tool compounds or starting points for drug discovery. Molecules prioritized in this manner can easily be validated in suitable zebrafish models due to the genetic tractability of the system and the predictive power of activity in higher organisms.
Figure 6.
Screening strategy for targeting a disease of interest using a multiplex platform.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the National Institutes of Health grant R01CA172310. We are grateful to the Agency for Science, Technology and Research (A*STAR) Singapore for funding through the Research Attachment Programme (M.S.B.) and to the University of Florida (UF) College of Pharmacy and UF Health (D. Guzick) for supporting the sabbatical visit at the IMCB (H.L.). J. J. Ma was partly funded by the SERI-IMCB Program on Retinal Angiogenic Disease (SIPRAD). We thank Y. Liu for technical assistance with many experiments.
Footnotes
ASSOCIATED CONTENT
Supplemental Materials and Methods, Supplemental Results, Supplementary Tables S1-S7, and Supplementary Figures S1-S5. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare the following competing financial interest(s): H. Luesch is co-founder of Oceanyx Pharmaceuticals, Inc., which is negotiating licenses for patents and patent applications related to largazole.
REFERENCES
- 1.Prior IA, Lewis PD, Mattos C. A Comprehensive survey of Ras mutations in Cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adjei AA. Blocking oncogenic Ras signaling for cancer therapy. J. Natl. Cancer Inst. 2001;93:1062–1074. doi: 10.1093/jnci/93.14.1062. [DOI] [PubMed] [Google Scholar]
- 3.Talks KL, Turley H, Gatter KC, Maxwell PH, Pugh CW, Ratcliffe PJ, Harris AL. The expression and distribution of the hypoxia-inducible factors HIF-1α and HIF-2α in normal human tissues, cancers, and tumor-associated macrophages. Am. J. Pathol. 2000;157:411–421. doi: 10.1016/s0002-9440(10)64554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agani F, Jiang BH. Oxygen-independent regulation of HIF-1: novel involvement of PI3K/AKT/mTOR pathway in cancer. Curr. Cancer Drug Targets. 2013;13:245–251. doi: 10.2174/1568009611313030003. [DOI] [PubMed] [Google Scholar]
- 5.Chun SY, Johnson C, Washburn JG, Cruz-Correa MR, Dang DT, Dang LH. Oncogenic KRAS modulates mitochondrial metabolism in human colon cancer cells by inducing HIF-1a and HIF-2a target genes. Mol. Cancer. 2010;9:293–303. doi: 10.1186/1476-4598-9-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kikuchi H, Pino MS, Zeng M, Shirasawa S, Chung DC. Oncogenic KRAS and BRAF differentially regulate hypoxia-inducible factor-1α and -2α in colon cancer. Cancer Res. 2009;69:8499–8506. doi: 10.1158/0008-5472.CAN-09-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bandau S, Knebel A, Gage ZO, Wood NT, Alexandru G. UBXN7 docks on neddylated cullin complexes using its UIM motif and causes HIF1α accumulation. BMC Biol. 2012;10:36. doi: 10.1186/1741-7007-10-36. Epub Apr 26, 2012. DOI: 10.1186/1741-7007-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo W, Zhong J, Chang R, Hu H, Pandey A, Semenza GL. Hsp70 and CHIP selectively mediate ubiquitination and degradation of hypoxia-inducible factor (HIF)-1α but not HIF-2α. J. Biol. Chem. 2010;285:3651–8663. doi: 10.1074/jbc.M109.068577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montagner M, Enzo E, Forcato M, Zanconato F, Parenti A, Rampazzo E, Basso G, Leo G, Rosato A, Bicciato S, Cordenonsi M, Piccolo S. SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature. 2012;487:380–384. doi: 10.1038/nature11207. [DOI] [PubMed] [Google Scholar]
- 12.Steunou AL, Ducoux-Petit M, Lazar I, Monsarrat B, Erard M, Muller C, Clottes E, Burlet-Schiltz O, Nieto L. Identification of the hypoxia-inducible factor 2α nuclear interactome in melanoma cells reveals master proteins involved in melanoma development. Mol. Cell Proteomics. 2013;12:736–748. doi: 10.1074/mcp.M112.020727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rolland T, Taşan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, Kamburov A, et al. A proteome-scale map of the human interactome network. Cell. 2014;159:1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Onnis B, Rapisarda A, Melillo B. Development of HIF-1 inhibitors for cancer therapy. J. Cell Mol. Med. 2009;13:2780–2786. doi: 10.1111/j.1582-4934.2009.00876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The Gene Ontology Consortium Gene Ontology Consortium: going forward. Nucl. Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belaiba RS, Bonello S, Zähringer C, Schmidt S, Hess J, Kietzmann T, Görlach A. Hypoxia up-regulates hypoxia-inducible factor-1α transcription by involving phosphatidylinositol 3-kinase and nuclear factor κB in pulmonary artery smooth muscle cells. Mol. Biol. Cell. 2007;18:4691–4697. doi: 10.1091/mbc.E07-04-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung YJ, Isaacs JS, Lee S, Trepel J, Neckers L. Microtubule disruption utilizes an NFκB-dependent pathway to stabilize HIF-1α protein. J. Biol. Chem. 2003;278:7445–7452. doi: 10.1074/jbc.M209804200. [DOI] [PubMed] [Google Scholar]
- 18.Rohwer N, Dame C, Haugstetter A, Wiedenmann B, Detjen K, Schmitt CA, Cramer T. Hypoxia-inducible factor 1α determines gastric cancer chemosensitivity via modulation of p53 and NF-κB. PLoS One. 2010;5:e12038. doi: 10.1371/journal.pone.0012038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizumoto Y, Kyo S, Kiyono T, Takakura M, Nakamura M, Maida Y, Moro N, Bono Y, Sakurai H, Inoue M. Activation of NF-κB is a novel target of KRAS-induced endometrial carcinogenesis. Clin. Cancer Res. 2011;17:1431–1450. doi: 10.1158/1078-0432.CCR-10-2291. [DOI] [PubMed] [Google Scholar]
- 20.Lee JW, Bae SH, Jeong JW, Kim SH, Kim KW. Hypoxia-inducible factor (HIF-1) α: its protein stability and biological functions. Exp. Mol. Med. 2004;36:1–12. doi: 10.1038/emm.2004.1. [DOI] [PubMed] [Google Scholar]
- 21.Okuyama H, Krishnamachary B, Zhou YF, Nagasawa H, Bosch-Marce M, Semenza GL. Expression of vascular endothelial growth factor receptor 1 in bone marrow-derived mesenchymal cells is dependent on hypoxia-inducible factor 1. J. Biol. Chem. 2006;281:15554–15563. doi: 10.1074/jbc.M602003200. [DOI] [PubMed] [Google Scholar]
- 22.Tang CH, Lu DY, Tan TW, Fu WM, Yang RS. Ultrasound induces hypoxia-inducible factor-1 activation and inducible nitric-oxide synthase expression through the integrin/integrin-linked kinase/Akt/mammalian target of rapamycin pathway in osteoblasts. J. Biol. Chem. 2007;282:25406–25415. doi: 10.1074/jbc.M701001200. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Lodish HF. Identification of K-ras as the major regulator for cytokine-dependent Akt activation in erythroid progenitors in vivo. Proc. Natl. Acad. Sci. U S A. 2005;102:14605–14610. doi: 10.1073/pnas.0507446102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliva JL, Zarich N, Martínez N, Jorge R, Castrillo A, Azañedo M, García-Vargas S, Gutiérrez-Eisman S, Juarranz A, Boscá L, Gutkind JS, Rojas JM. The P34G mutation reduces the transforming activity of K-Ras and N-Ras in NIH 3T3 cells but not of H-Ras. J. Biol. Chem. 2004;279:33480–33491. doi: 10.1074/jbc.M404058200. [DOI] [PubMed] [Google Scholar]
- 25.Chow HY, Jubb AM, Koch JN, Jaffer ZM, Stepanova D, Campbell DA, Duron SG, O’Farrell M, Cai KQ, Klein-Szanto AJ, Gutkind JS, Hoeflich KP, Chernoff J. p21-Activated kinase 1 is required for efficient tumor formation and progression in a Ras-mediated skin cancer model. Cancer Res. 2012;72:5966–5975. doi: 10.1158/0008-5472.CAN-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barceló C, Paco N, Morell M, Alvarez-Moya B, Bota-Rabassedas N, Jaumot M, Vilardell F, Capella G, Agell N. Phosphorylation at Ser-181 of oncogenic KRAS is required for tumor growth. Cancer Res. 2014;74:1190–1199. doi: 10.1158/0008-5472.CAN-13-1750. [DOI] [PubMed] [Google Scholar]
- 27.Young A, Lou D, McCormick F. Oncogenic and wild-type Ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer Discov. 2013;3:112–123. doi: 10.1158/2159-8290.CD-12-0231. [DOI] [PubMed] [Google Scholar]
- 28.Zeng M, Kikuchi H, Pino MS, Chung DC. Hypoxia activates the K-Ras proto-oncogene to stimulate angiogenesis and inhibit apoptosis in colon cancer cells. PLoS One. 2010;5:e10966. doi: 10.1371/journal.pone.0010966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steckel M, Molina-Arcas M, Weigelt B, Marani M, Warne PH, Kuznetsov H, Kelly G, Saunders B, Howell M, Downward J, Hancock DC. Determination of synthetic lethal interactions in KRAS oncogene-dependent cancer cells reveals novel therapeutic targeting strategies. Cell Res. 2012;8:1227–1245. doi: 10.1038/cr.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phelan CM, Iqbal J, Lynch HT, Lubinski J, Gronwald J, Moller P, Gharidian P, Foulkes WD, Armel S, Eisen A, Neuhausen SL, Senter L, Singer CF, Ainsworth P, Kim-Sing C, Tung N, Llacuachaqui M, Chornokur G, Ping S, Narod SA. Incidence of colorectal cancer in BRCA1 and BRCA2 mutation carriers: results from a follow-up study. Br. J. Cancer. 2014;110:530–534. doi: 10.1038/bjc.2013.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salvador-Reyes LA, Engene N, Paul VJ, Luesch H. Targeted natural products discovery from marine cyanobacteria using combined phylogenetic and mass spectrometric evaluation. J. Nat. Prod. 2015;78:1957–1962. doi: 10.1021/np500931q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taori K, Paul VJ, Luesch H. Structure and activity of largazole, a potent antiproliferative agent from the Floridian marine cyanobacterium Symploca sp. J. Am. Chem. Soc. 2008;130:1806–1807. doi: 10.1021/ja7110064. [DOI] [PubMed] [Google Scholar]
- 33.Hong J, Luesch H. Largazole: from discovery to broad-spectrum therapy. Nat Prod. Rep. 2012;29:449–456. doi: 10.1039/c2np00066k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dang DT, Chen F, Gardner LB, Cummins JM, Rago C, Bun F, Kantsevoy SV, Dang LH. Hypoxia-inducible factor-1α promotes nonhypoxia-mediated proliferation in colon cancer cells and xenografts. Cancer Res. 2006;66:1684–1693. doi: 10.1158/0008-5472.CAN-05-2887. [DOI] [PubMed] [Google Scholar]
- 35.Nambiar RM, Henion PD. Sequential antagonism of early and late Wnt-signaling by zebrafish colgate promotes dorsal and anterior fates. Dev. Biol. 2004;267:165–180. doi: 10.1016/j.ydbio.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 36.Nambiar RM, Ignatius MS, Henion PD. Zebrafish colgate/Hdac1 functions in the non-canonical Wnt pathway during axial extension and in Wnt-independent branchiomotor neuron migration. Mech. Dev. 2007;124:682–698. doi: 10.1016/j.mod.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Rooijen E, Voest EE, Logister I, Bussmann J, Korving J, van Eeden FJ, Giles RH, Schulte-Merker S. Von Hippel-Lindau tumor suppressor mutants faithfully model pathological hypoxia-driven angiogenesis and vascular retinopathies in zebrafish. Dis. Model Mech. 2010;3:343–353. doi: 10.1242/dmm.004036. [DOI] [PubMed] [Google Scholar]
- 38.Liu PQ, Tan S, Mendel MC, Murrills RJ, Bhat BM, Schlag B, Samuel R,, Matteo JJ, de la Rosa R, Howes K, Reik A, Case CC, Bex FJ, Young K, Gregory PD. Isogenic human cell lines for drug discovery: Regulation of target gene expression by engineered Zinc-Finger protein transcription factors. J. Biomol. Screen. 2005;10:304–313. doi: 10.1177/1087057104272663. [DOI] [PubMed] [Google Scholar]
- 39.Postel-Vinay S, Bajrami I, Friboulet L, Elliott R, Fontebasso Y, Dorvault N, Olaussen KA, André F, Soria J-C, Lord CJ, Ashworth A. A high-throughput screen identifies Parp1/2 inhibitors as a potential therapy for Ercc1-deficient non-small cell lung cancer. Oncogene. 2013;32:5377–5387. doi: 10.1038/onc.2013.311. [DOI] [PubMed] [Google Scholar]
- 40.Galbraith MD, Allen MA, Bensard CL, Wang X, Schwinn MK, Qin B, Long HW, Daniels DL, Hahn WC, Dowell RD, Espinosa JM. HIF1A employs CDK8-mediator to stimulate RNAPII elongation. Cell. 2013;153:1327–1339. doi: 10.1016/j.cell.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaluz S, Kaluzova M, Stanbridge EJ. Proteasomal inhibition attenuates transcriptional activity of hypoxia-inducible factor 1 (Hif-1) via specific effect on the Hif-1a C-terminal activation domain. Mol. Cell Biol. 2006;26:5895–5907. doi: 10.1128/MCB.00552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Felth J, Rickardson L, Rosén J, Wickström M, Fryknäs M, Lindsckog M, Bohlin L, Gullbo J. Cytotoxic effects of cardiac glycosides in colon cancer cells, alone and in combination with standard chemotherapeutic drugs. J. Nat. Prod. 2009;11:1969–1974. doi: 10.1021/np900210m. [DOI] [PubMed] [Google Scholar]
- 43.Zhang H, Qian DZ, Tan YS, Lee K, Gao P, Ren YR, Rey S, Hammers H, Chang D, Pili R, Dang CV, Liu JO, Semenza GL. Digoxin and other cardiac glycosides inhibit Hif-1a synthesis and block tumor growth. Proc. Natl. Acad. Sci U S A. 2008;105:19579–19586. doi: 10.1073/pnas.0809763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Escuin D, Kline ER, Giannakakou P. Both microtubule-stabilizing and microtubule-destabilizing drugs inhibit hypoxia-inducible factor-1α accumulation and activity by disrupting microtubule function. Cancer Res. 2005;65:9021–9028. doi: 10.1158/0008-5472.CAN-04-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li WL, Zheng HC, Bukuru J, De Kimpe N. Natural medicines used in the traditional Chinese medical system for therapy of diabetes mellitus. J. Ethnoparmacol. 2004;92:1–21. doi: 10.1016/j.jep.2003.12.031. [DOI] [PubMed] [Google Scholar]
- 46.Chen WS, Cao Z, Krishnan C, Panjawani N. Verteporfin without light stimulation inhibits yap activation in trabecular meshwork cells: Implications for glaucoma treatment. Biochem. Biophys. Res. Commun. 2015;466:221–225. doi: 10.1016/j.bbrc.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 47.Wu MZ, Tsai YP, Yang MH, Huang CH, Chang SY, Chang CC, Teng SC, Wu KJ. Interplay between Hdac3 and Wdr5 is essential for hypoxia-Induced epithelial-mesenchymal transition. Mol. Cell. 2011;5:811–822. doi: 10.1016/j.molcel.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 48.Geng H, Liu Q, Zue C, David LL, Beer TM, Thomas GV, Dai MS, Qian DZ. HIF1α protein stability is increased by acetylation at lysine 709*. J. Biol. Chem. 2012;287:35496–35505. doi: 10.1074/jbc.M112.400697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mizuno S, Yasuo M, Bogaard HJ, Kraskauskas D, Natarajan R, Voelkel NF. Inhibition of histone deacetylase causes emphysema. Am. J. Physiol. 2011;3:L402–413. doi: 10.1152/ajplung.00207.2010. [DOI] [PubMed] [Google Scholar]
- 50.MacRae CA, Peterson RT. Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 2015;14:721–731. doi: 10.1038/nrd4627. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.