Abstract
Allosteric HIV-1 integrase inhibitors (ALLINIs) have recently emerged as a promising class of antiretroviral agents and are currently in clinical trials. In infected cells, ALLINIs potently inhibit viral replication by impairing virus particle maturation but surprisingly exhibit a reduced EC50 for inhibiting HIV-1 integration in target cells. To better understand the reduced antiviral activity of ALLINIs during the early stage of HIV-1 replication, we investigated the competitive interplay between a potent representative ALLINI, BI/D, and LEDGF/p75 with HIV-1 integrase. While the principal binding sites of BI/D and LEDGF/p75 overlap at the integrase catalytic core domain dimer interface, we show that the inhibitor and the cellular cofactor induce markedly different multimerization patterns of full-length integrase. LEDGF/p75 stabilizes an integrase tetramer through the additional interactions with the integrase N-terminal domain, whereas BI/D induces protein–protein interactions in C-terminal segments that lead to aberrant, higher-order integrase multimerization. We demonstrate that LEDGF/p75 binds HIV-1 integrase with significantly higher affinity than BI/D and that the cellular protein is able to reverse the inhibitor induced aberrant, higher-order integrase multimerization in a dose-dependent manner in vitro. Consistent with these observations, alterations of the cellular levels of LEDGF/p75 markedly affected BI/D EC50 values during the early steps of HIV-1 replication. Furthermore, genome-wide sequencing of HIV-1 integration sites in infected cells demonstrate that LEDGF/p75-dependent integration site selection is adversely affected by BI/D treatment. Taken together, our studies elucidate structural and mechanistic details of the interplay between LEDGF/p75 and BI/D during the early stage of HIV-1 replication.
Graphical abstract
The virally encoded integrase (IN) is an important therapeutic target for treating HIV-1 infected patients as its function is essential for viral replication. A tetramer of IN assembles on the ends of viral DNA to form the stable synaptic complex and catalyzes a two-step covalent integration of the DNA product of reverse transcription into host chromosomes.1 The cellular chromatin associated protein LEDGF/p75 binds the HIV-1 IN tetramer and navigates the preintegration complex (PIC), which also contains additional viral and cellular proteins, to active genes during integration.2–6
HIV-1 IN is composed of three domains, including the N-terminal domain (NTD), which coordinates the architecturally important Zn2+ ion, the catalytic core domain (CCD) that binds two Mg2+ ions required for the catalytic activities, and the C-terminal domain, which exhibits a SH3-like fold (reviewed in refs 7 and 8). All three domains directly bind viral DNA and contribute to the functional tetramerization of IN (reviewed in refs 9–14). This tetramer is further stabilized through interactions with cellular cofactor LEDGF/p75.15,16 LEDGF/p75 tethers the HIV-1 intasome to chromatin through its bimodal binding. Interactions with HIV-1 IN are mediated by the C-terminal integrase binding domain (IBD) of LEDGF/ p75, which inserts a small loop in a V-shaped cavity located at the IN CCD-CCD dimer interface.17 The N-terminal domain of LEDGF/p75 comprising the PWWP domain, nuclear localization signal, AT hooks, and highly charged regions preferentially engages chromatin sites containing the H3K36me3 post-translational modification.18–20 This histone mark is found at active genes and has been shown to correlate positively with HIV-1 integration sites.21
HIV-1 IN catalytic function has been exploited as a drug target. Three inhibitors raltegravir, elvitegravir, and dolutegravir are in clinical use. More recent efforts have led to the identification of allosteric IN inhibitors (ALLINIs, also called LEDGINs, NCINIs, INLAIs, or MINIs) that bind in the clinically unexploited dimer interface of HIV-1 IN.22–27 Crystallographic studies have revealed significant overlap between ALLINI and LEDGF/IBD binding to the IN CCD-CCD dimer interface.22,24,26,28–31 For example, both the ALLINI carboxylic acid and LEDGF/IBD Asp-366 establish key hydrogen bonds with the backbone amides of IN residues Glu-170 and His-171. Accordingly, ALLINIs are able to inhibit the IN-LEDGF/p75 interaction in vitro (reviewed in refs 8 and 32). ALLINI interactions extend to both IN subunits, thus allowing the inhibitor to bridge between interacting subunits and induce additional protein–protein interactions that lead to higher-order, aberrant IN multimerization. The sizes of the IN multimers formed in the presence of ALLINIs significantly exceed that of the tetrameric form needed for catalysis.25,33,34 In infected cells, ALLINIs inhibit both early and late stages of HIV-1 replication with the most potent antiviral activity observed as a result of improper maturation of virus particles.26,28,35–37 During maturation, the antiviral activity of ALLINIs has been linked to the promotion of aberrant IN multimers, which in turn leads to the mislocalization of ribonucleoprotein complexes outside of the capsid core and results in eccentric, noninfectious virions. Of note, during the late stage of replication, LEDGF/p75 does not play any significant role as its depletion or overexpression does not affect ALLINI potencies in virus producer cells.28,34,38,39
In contrast to potent inhibition by ALLINIs during the late stage of replication, the compounds are significantly less effective during early steps of HIV-1 infection. Additionally, the depletion of endogenous LEDGF/p75 has been shown to enhance inhibitor activity in target cells.28,34,38,40 These experiments have provided initial clues that there could be competition between BI/D and LEDGF/p75 for interaction with IN during the early steps of HIV-1 replication. Here, we have used complementary biochemical/biophysical approaches coupled with virology experiments to investigate competition between LEDGF/p75, BI/D, and IN during the early stage of HIV-1 replication. Differential hydrogen/deuterium exchange-mass spectrometry (HDX MS) experiments show that LEDGF/p75 and BI/D induce markedly different multimerization patterns of full length IN. Furthermore, we have found that LEDGF/p75 can competitively reverse aberrant IN multimerization induced by BI/D in vitro. Consistent with these observations, we found a reverse correlation between increasing cellular levels of LEDGF/p75 and BI/D potency in target cells. Finally, genome-wide sequencing experiments have demonstrated that LEDGF/p75 dependent integration site selection is adversely affected by treatments of target cells with BI/D. Together these findings support the idea of the competitive interplay between LEDGF/p75 and BI/D during the early steps of HIV-1 infection.
RESULTS AND DISCUSSION
Available HIV-1 IN crystal structures17,28 are limited to BI/D or LEDGF/IBD binding to CCD–CCD dimers. These structures have been informative and have revealed both important similarities and differences between the inhibitor and the cellular protein interactions with the CCD–CCD dimer interface. However, these truncated structures are unable to explain the different multimerization patterns observed for full-length IN in the presence of BI/D or LEDGF/p75.15,34 LEDGF/p75 stabilizes a functional tetrameric form of IN, whereas BI/D promotes aberrant, inactive higher-order IN multimerization. Therefore, to investigate how the inhibitor and LEDGF/p75 induce differential multimerization of full-length IN, we employed HDX MS analysis.
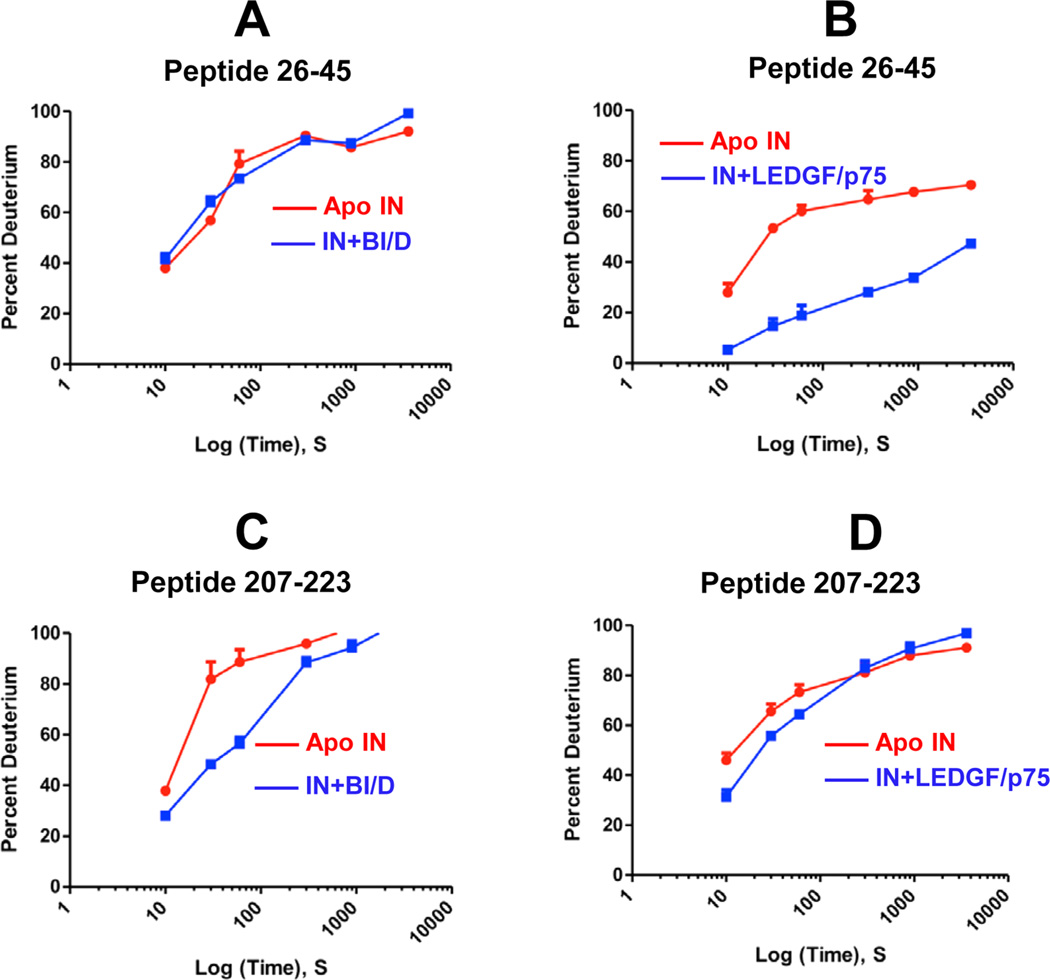
Representative HDX results are depicted in Figure 1, and the HDX signature comparison for full-length IN in the presence of BI/D or LEDGF/p75 is given in Figure 2. Comprehensive differential HDX perturbation maps for all detected peptides are shown in Figures S1 and S2. These results reveal both similarities and differences between IN-BI/D and IN-LEDGF/ p75 complexes. As expected based on previous crystal structures,17,28 both BI/D and LEDGF/p75 induced strong protections or decreased exchange in the IN CCD (residues 154–202). However, HDX experiments have also revealed additional regions in full length IN that are protected upon BI/ D or LEDGF/p75 binding, which differ markedly between the two complexes. For example, the IN NTD peptides spanning residues 24–45 were strongly protected by LEDGF/p75 but not BI/D (compare A and B in Figure 1, also see Figure 2). Conversely, BI/D induced strong protections in the segment connecting the CCD with the CTD (residues 203–223), whereas the corresponding peptides were not detectably affected by LEDGF/p75 binding (compare C and D in Figure 1; also see Figure 2). The C-terminal peptides encompassing residues 242–256 additionally exhibited significant protections in the presence of BI/D but not LEDGF/p75. IN peptides 226–241 and 257–288 could be resolved in the presence of BI/D but not LEDGF/p75 and were significantly protected by the inhibitor.
Figure 1.
Deuterium uptake plots for representative peptides of HIV-1 IN in the presence of BI/D or LEDGF/p75. The plots show levels of D2O uptake for indicated time points for IN peptides. The error bars represent the SD for three independent experiments.
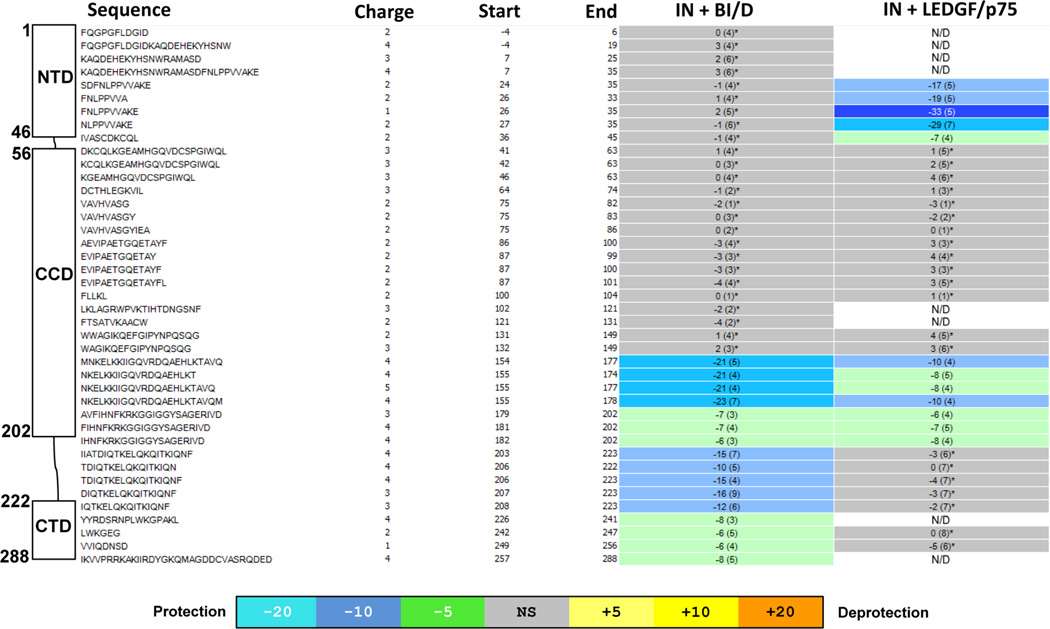
Figure 2.
HDX signature comparison for full-length IN in the presence of BI/D or LEDGF/p75. Amino acid sequences, start and end amino acid numbers, and charge states for each detected peptide are shown. The schematic presentation IN domains with respective peptides are given on the left. The peptides that did not show statistically significant difference when apo-IN is compared with IN-BI/D or IN-LEDGF/p75 complexes based on the software HDX Workbench45 are colored gray. The regions that revealed statistically significant reduction in deuterium uptake are colored green or blue according to the heat map coloring scheme used by the software HDX Workbench. N/D: these peptides were not detecteds.
In HDX experiments, the slower D2O uptake correlates with stronger hydrogen bonds for amides in that region and suggests that the select protein region has been stabilized by either direct binding of the cognate ligand or allosteric protein conformational changes induced by ligand binding. Based on the available crystal structures,17,28 it is logical to conclude that the protections seen in the IN CCD are due to direct binding of BI/D or LEDGF/p75. Of note, BI/D binding effects extend to additional protein segments, which include the segments connecting the CCD with the CTD and the CTD. We interpret these protections as BI/D induced protein conformational changes and/or protein–protein interactions that lead to higher-order multimerization of IN. Interestingly, when BI/D HDX results are compared to previous results obtained with the ALLINI-2 compound,33 similarities and differences are seen. As expected, similar CCD segments showed protection surrounding the inhibitor binding site. Additionally, similar protections were seen in the C-terminus of IN, demonstrating the importance of this region in aberrant multimerization of IN. However, equally important are the key differences seen for each inhibitor. Unlike for ALLINI-2, BI/D induced strong protections in the segment connecting the CCD with the CTD (residues 203–223). This highlights the fact that different ALLINIs are capable of promoting unique higher order, aberrant IN multimers.
The available crystal structures of LEDGF/IBD with two domain (NTD-CCD) fragments of HIV-2 and maedi-visna virus INs41,42 help to interpret the HDX results for the full-length IN-LEDGF/p75 complex. These crystal structures have shown that LEDGF/IBD in addition to binding the IN CCD, also engages the NTD. For example, HIV-2 IN residues Glu6, Glu10, and Gln35 interact with LEDGF/IBD Lys401, Arg405, and Val435, respectively.41 While the peptides from the first N-terminal 23 amino acid segment of HIV-1 IN could not be resolved in our HDX experiments, the strong protections seen in the HIV-1 IN segment 24–36 are likely to reflect LEDGF/ p75 binding to the HIV-1 IN NTD. HDX experiments have additionally allowed us to map the interacting interfaces in full length LEDGF/p75 with the protections being observed exclusively in the C-terminal IBD (residues 351–377 and 399–410, Figure S3). Taken together, our HDX experiments have revealed expected similarities as well as marked differences between BI/D and LEDGF/p75 interactions with full-length HIV-1 IN.
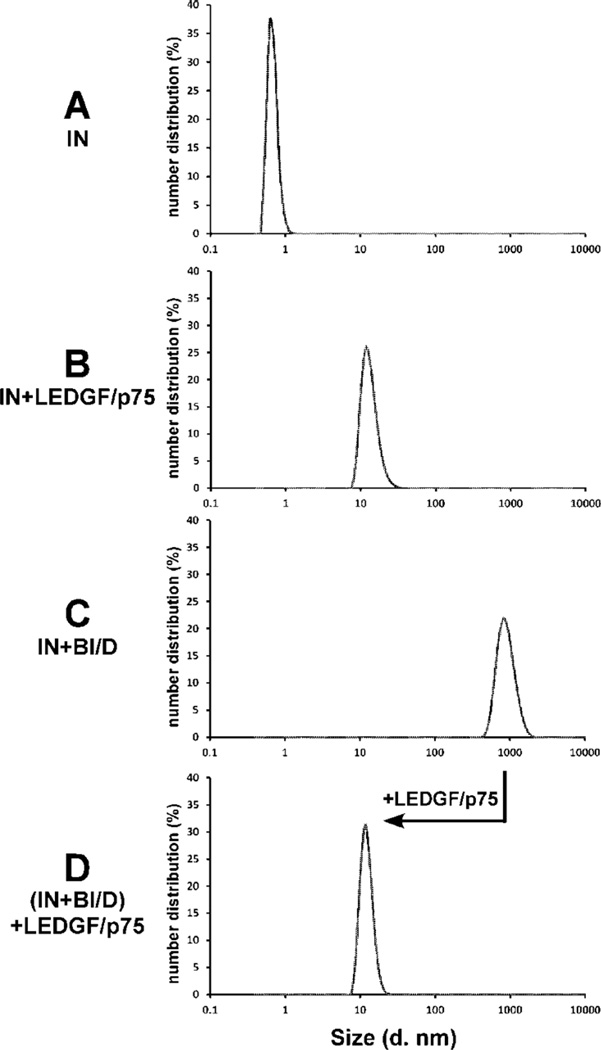
Next, we performed complementary dynamic light scattering (DLS) experiments to further compare BI/D and LEDGF/p75 induced IN multimerization (Figure 3). The DLS experiments allow for detection and estimation of sizes of large particles formed in solution.43–45 The DLS signals for BI/D, IN, or LEDGF/p75 alone were indistinguishable from the buffer alone sample with a single peak corresponding to a diameter size of ~1 nm, indicating the small sizes of the inhibitor and apo-proteins (for clarity only the DLS signal for IN alone is shown in Figure 3A). The IN-LEDGF/p75 complex exhibited a peak with a diameter of ~11 nm (Figure 3B), which is consistent with the estimated size of an IN tetramer46 bound to two molecules of LEDGF/p75. In contrast, as expected,34 the addition of BI/D to IN resulted in much larger particles with an estimated diameter size of ~825 nm (Figure 3C). Surprisingly, the addition of LEDGF/p75 to the BI/D-induced aberrant IN multimers markedly reduced the particle sizes, and the resulting DLS signal was similar to that observed for the IN-LEDGF/p75 complex without added inhibitor (compare D with B in Figure 3). These results indicate that LEDGF/p75 can effectively reverse the BI/D-induced aberrant IN multimerization in vitro.
Figure 3.
DLS analysis of effects of BI/D and LEDGF/p75 on multimerization of full-length HIV-1 IN. (A) IN alone. (B) LEDGF/ p75 was added to IN and incubated for 30 min. (C) BI/D was added to IN and incubated for 30 min. (D) BI/D was added to IN and incubated for 30 min as in C, and then LEDGF/p75 was added to the mixture and incubated for an additional 30 min.
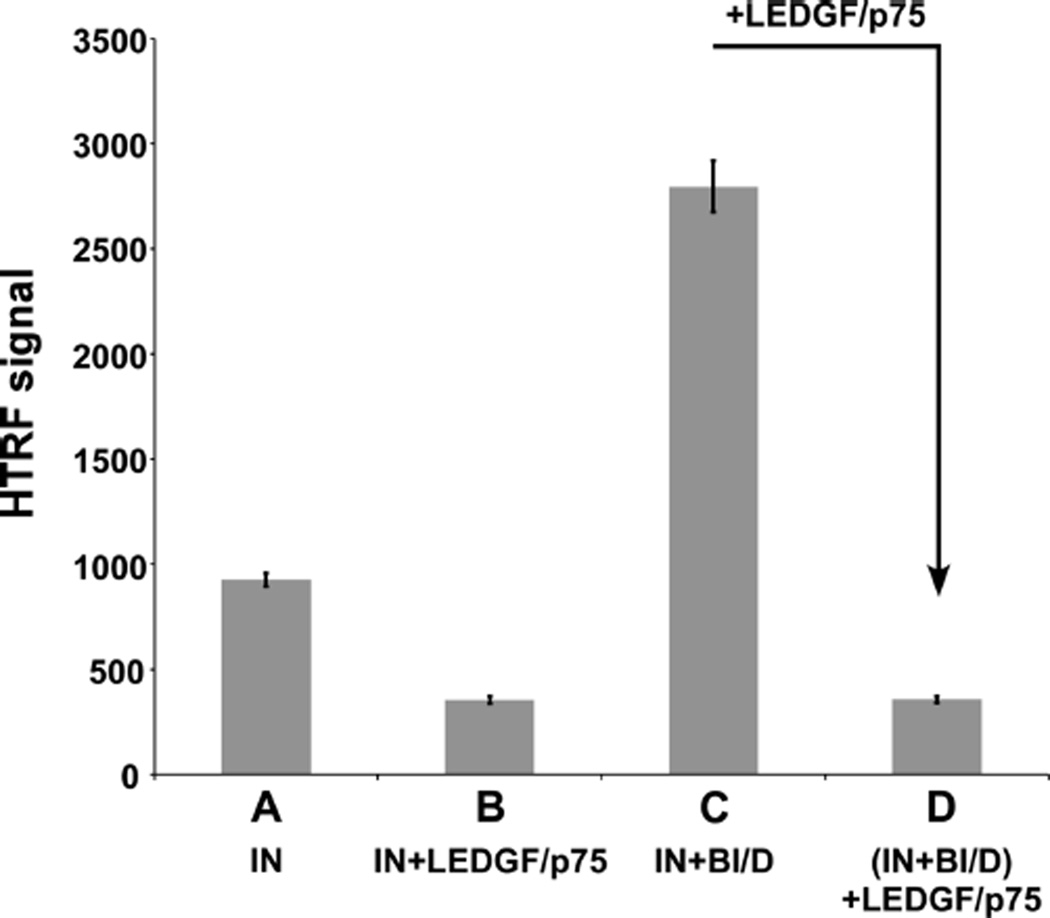
We have also used a homogeneous time-resolved fluorescence (HTRF)-based assay31 to examine how BI/D and LEDGF/p75 affect IN subunit–subunit interactions (Figure 4). The IN alone sample had a higher HTRF signal than the IN-LEDGF/p75 complex (compare A with B in Figure 4). Free IN exists as a dynamic mixture of monomers, dimers, and tetramers as well as higher order oligomers which are not functionally active, all of which collectively contribute to the HTRF signal. LEDGF/p75 binding stabilizes IN tetramers and thus reduces the overall extent of dynamic exchange between IN subunits, which likely accounts for the observed reduction in HTRF signal. In contrast, the addition of BI/D to IN markedly enhanced the HTRF signal due to aberrant, higher-order protein multimerization (Figure 4C). However, the subsequent addition of LEDGF/p75 to BI/D-induced aberrant IN multimers reduced the HTRF signal (compare C and D in Figure 4) to similar levels seen with the IN-LEDGF/p75 complex (compare D with B in Figure 4). Importantly, similar observations were made with other representative ALLINIs including both quinoline-based (ALLINI-2 and LEDGIN-6) and pyridine-based (KF116) compounds (Figure S4), indicating that the competitive interplay with LEDGF/p75 is a generic mechanism of ALLINI-induced IN multimerization.
Figure 4.
HTRF-based assays of IN multimerization in the presence of LEDGF/p75 and BI/D. The HTRF signal observed due to IN multimerization was monitored. (A) IN alone. (B) IN+LEDGF/p75 incubated for 30 min. (C) IN+BI/D incubated for 30 min. (D) IN +BI/D was incubated for 30 min as in C, and then LEDGF/p75 was added to the mixture and incubated for an additional 30 min. The error bars represent the SD for three independent experiments.
We next extended the application of the HTRF-based assays to monitor dose-dependent effects of LEDGF/p75 on BI/D-induced aberrant IN multimerization. Table 1 shows that increasing concentrations of LEDGF/p75 significantly (~10 to 58-fold) reduced the EC50 values for BI/D induced aberrant IN multimerization. Of note, these effects were observed with the addition of low nanomolar concentrations of LEDGF/p75. For example, the addition of 2 nM LEDGF/p75 shifted the BI/D EC50 value from ~120 nM in the absence of the cellular protein to 1390 nM in its presence, suggesting that significantly tighter IN-LEDGF/p75 interactions are able to compete effectively with BI/D.
Table 1.
Effects of LEDGF/p75 on BI/D EC50 Values for Inducing Aberrant IN Multimerization
| [LEDGF] nM | BI/D EC50, µMa |
|---|---|
| 0 | 0.11 ± 0.0088 |
| 2 | 1.39 ± 0.18 |
| 10 | 2.62 ± 0.30 |
| 20 | 3.22 ± 0.33 |
| 50 | 4.29 ± 0.32 |
| 100 | 5.22 ± 0.27 |
| 200 | 7.02 ± 0.89 |
Average values from three idependent experiments are shown
We have previously34 determined a Kd of ~123 nM for BI/D binding to the HIV-1 IN CCD, which closely correlates with the BI/D EC50 value of ~110 nM for aberrant multimerization of full length IN in the absence of LEDGF/p75 (Table 1). Here, we have determined a Kd of 3.32 ± 0.31 nM for LEDGF/ p75 binding to full length HIV-1 IN using our HTRF-based assay (Figure S5). Taken together, our results show that LEDGF/p75 binds IN with significantly higher affinity than BI/ D and is thus able to outcompete inhibitor-induced aberrant IN multimerization in vitro.
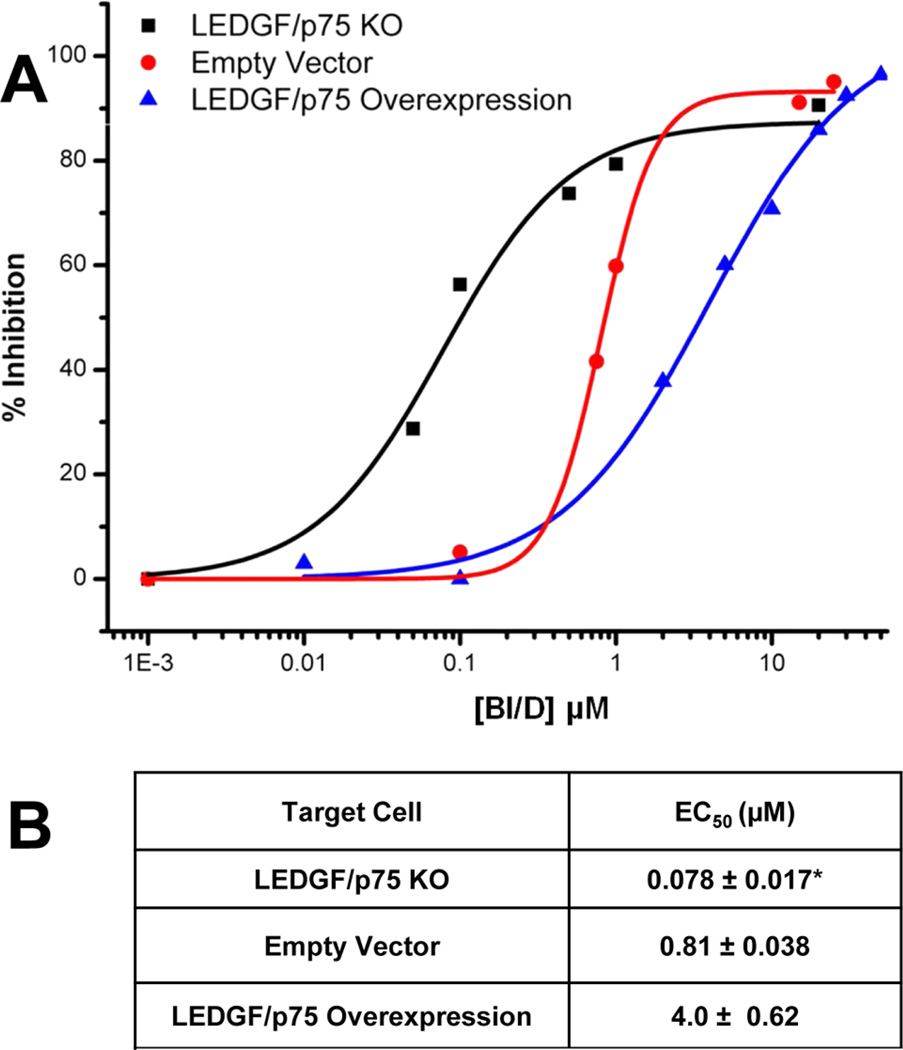
We next investigated the interplay between LEDGF/p75 and BI/D during the early steps of HIV-1 infection. Previous studies28,34 have shown that depletion of endogenous LEDGF/ p75 by either knockdown or knockout significantly increases the potency of BI/D. Here, we have extended these experiments by monitoring the effects of ectopic overexpression of LEDGF/p75 on BI/D potency during the early steps of HIV-1 infection. The results in Figure 5 show that increasing the cellular levels of LEDGF/p75 significantly reduces the potency of BI/D in infected cells. Immunoblotting of endogenous LEDGF/p75 was estimated to be around 2–3 µM in the nucleus of HEK293T cells, whereas ectopically expressed HA-LEDGF/p75 was around 9–30 µM (Figure S6). These results mirror closely the in vitro data in Table 1, indicating a negative correlation between the LEDGF/p75 cellular levels and the BI/D potency.
Figure 5.
Effects of varying cellular levels of LEDGF/p75 on BI/D potency in target cells. HEK293T cells were incubated with indicated concentrations of BI/D and then infected with VSV-G pseudotyped HIV-1 virions equivalent to 0.02 pg of p24 per cell. (A) Dose-dependent curves for BI/D activity in HEK293T LEDGF/p75 KO cells (black squares, data from ref 34), HEK293T cells transfected with either empty vector control (red circles) or with the plasmid that ectopically expresses HA-LEDGF/p75 (blue triangles). (B) Summary of the results depicted in A. *Data from ref 34.
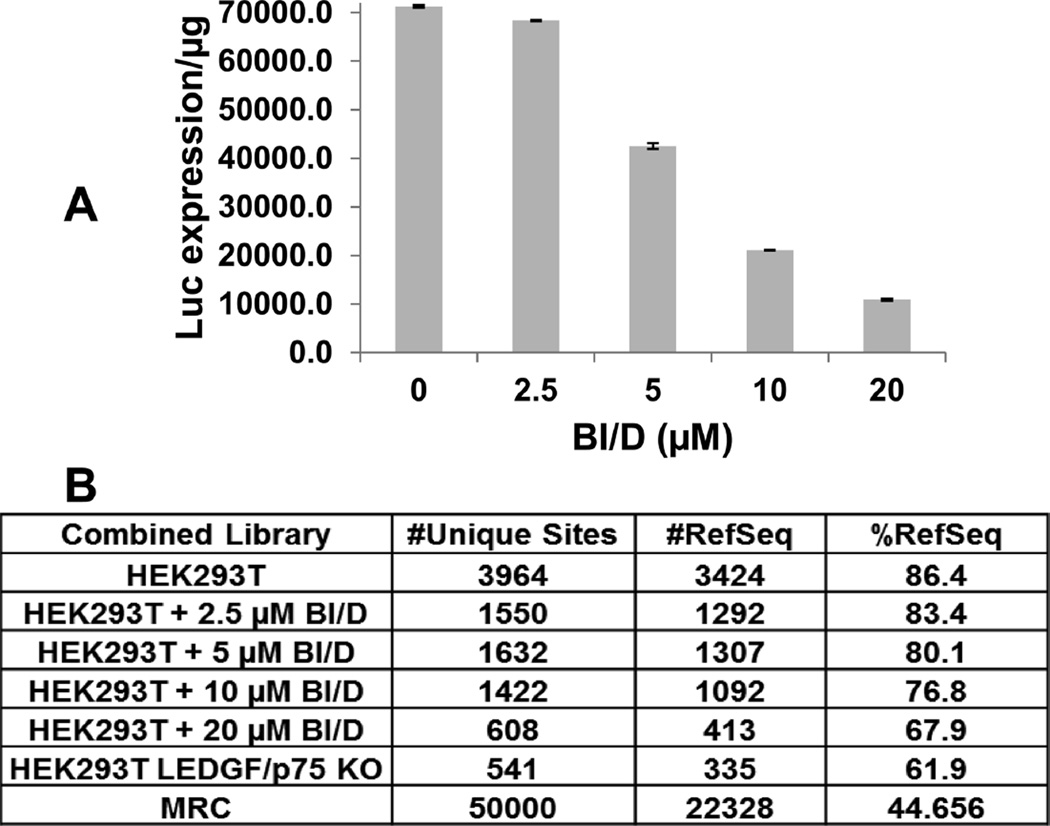
Our observations suggest that in order for the inhibitor to induce aberrant IN multimerization during the early steps of HIV-1 replication and thus impair integration in target cells, higher concentrations of BI/D are needed to outcompete the ability of LEDGF/p75 to bind and stabilize the functional tetramer of HIV-1 IN. Additionally, the role of LEDGF/p75 in navigating HIV-1 PICs to active genes during integration has been well established.2–6 Therefore, if sufficient amounts of inhibitor are added to target cells, the dependence on LEDGF/ p75 for integration site selection could be disrupted. To test this notion, we examined BI/D effects on the integration site selectivity of HIV-1. As expected,47 in the absence of the inhibitor, the majority of integration events mapped to reference sequences (RefSeq; 86.4%) in wild type HEK293T cells, whereas this value reduced to ~62% in LEDGF/p75 KO cells. The addition of BI/D inhibited HIV-1 expression in target cells and resulted in reduction of integration events into RefSeq genes in dose dependent manners (Figure 6). These results further support the mechanism for the competitive interplay between BI/D and LEDGF/p75 during the early steps of HIV-1 replication.
Figure 6.
Effects of BI/D on LEDGF/p75-dependent targeting of HIV-1 integration sites into chromatin. HEK293T cells were incubated with indicated concentrations of BI/D or diluent control DMSO and then infected with VSV-G pseudotyped HIV-1 virions equivalent to 10 pg of p24 per cell. Then, 48 h postinfection, 20% of the cells were harvested and used to measure luciferase activity per microgram of cellular protein (A). The remaining cells were cultured and harvested 7 days postinfection to isolate genomic DNA for Illumina sequencing. The number of unique HIV-1 integration sites and integration frequencies into reference sequences (RefSeq) are shown in B.
In summary, our data provide structural and mechanistic clues for the competitive interplay between BI/D and LEDGF/ p75 with IN, which underlies the BI/D EC50 values during the early steps of HIV-1 replication. While both BI/D and LEDGF/p75 bind to the same pocket at the IN CCD dimer interface,17,28 our HDX experiments have revealed marked differences in their respective interactions with full-length IN. The additional interactions of LEDGF/p75 with the IN NTD allows for stabilizing a tetrameric functional form of IN, whereas BI/D induces protein-protein interactions involving the IN C-terminal segments, which lead to aberrant, higher-order IN multimerization. Furthermore, we demonstrate that LEDGF/p75 binds HIV-1 IN with significantly higher affinity than BI/D and is able to competitively reverse BI/D induced aberrant IN multimerization in vitro. A previous study30 that used sedimentation velocity and turbidity assays showed that micromolar concentrations of ALLINI GSK1264 induced formation of insoluble aggregates for the preformed complex between quadramutated IN (C56S/F139D/F185H/C280S) and a truncated form of LEDGF326–530 containing the IBD. Here, we used wild type full length IN and LEDGF/p75 as well as BI/D concentration ranges that parallel its EC50 values in vitro and in cell culture. Our results show that full-length LEDGF/p75 can effectively outcompete aberrant IN multimerization induced by BI/D and thus adversely affect the inhibitor EC50 value during the early steps of infection.
The mechanism of action of BI/D elucidated here is different from its mode of action during the late stage of HIV-1 replication, where the inhibitor encounters minimal to no detectable competition from LEDGF/p75 and thus potently induces aberrant IN multimerization in virus particles. This latter mechanism has been exploited to develop improved pyridine-based multimerization selective IN inhibitors or MINIs.25 The rational strategy employed to design MINIs has prioritized enhancing the ability of the archetypal ALLINIs to better bridge between interacting IN subunits. Accordingly, MINIs potently inhibit the proper maturation of virus particles through modulating IN multimerization, while they are not effective inhibitors of IN-LEDGF/p75 interactions during early steps of HIV-1 replication.25 An alternative strategy for developing new classes of inhibitors could be through modifying current ALLINIs to improve their ability to bind tighter at IN sites which are also needed for interactions with LEDGF/p75. Such modifications could lead to new compounds with enhanced potency for inhibiting IN-LEDGF/p75 interactions and thus block the early steps of HIV-1 replication. The mechanistic findings reported here are expected to aid future efforts in the development of improved IN inhibitors.
MATERIALS AND METHODS
ALLINI BI/D and Recombinant Proteins
ALLINI BI/D, LEDGIN-6, ALLINI-2, and KF116 were synthesized as previously described.25,29,31,40 WT HIV-1 IN and LEDGF/p75 recombinant proteins with 6 × His or FLAG tags were expressed in E. coli cells and purified as described.31 Expression and purification of the complex between His-LEDGF/p75 and FLAG-HIV-1 IN was performed as follows. His-LEDGF/p75 was expressed in E. coli from pFT-1-LEDGF derived from pRSETB (Invitrogen).31 FLAG-HIV-1 IN was expressed in E. coli as previously reported.31 Cell pellets of both were mixed and lysed by sonication in buffer containing 500 mM NaCl, 50 mM HEPES at pH 7.5, 2 mM β-mercaptoethanol, 20 mM Imidazole, and 1 × Complete protease inhibitor (Roche). After centrifugation, the soluble lysate was filtered and purified using a 5 mL HisTrap (GE Healthcare) chromatography column. Bound protein was eluted using a linear gradient of 20 mM to 500 mM imidazole in the same buffer. Fractions containing both proteins were pooled and concentrated using a 100 kDa MWCO spin column. The proteins were then subjected to size exclusion chromatography using a HiLoad 16/60 Superdex 200 (GE Heathcare) in 500 mM NaCl, 50 mM HEPES at pH 7.5, and 2 mM β-mercaptoethanol. Fractions that contained the complex between His-LEDGF/p75 and FLAG-HIV-1 IN were pooled and concentrated using a 100 kDa MWCO spin column.
HDX
Solution-phase amide HDX experiments were carried out as described previously33 using a fully automated system, and sample handling was done using CTC HTS Twin PAL robots (LEAP Technologies), housed inside a 4 °C cabinet. In parallel reactions, 10 µM of preformed complex between 6 × His-LEDGF/p75 and FLAG-HIV-1 IN or 10 µM 6 × His-HIV-1 IN protein premixed with 1:5 molar excess of BI/D were subjected to HDX analysis. For the differential HDX experiments, 5 µL of 10 µM apo-IN and IN complexes with His-LEDGF/p75 or BI/D were mixed with 20 µL of D2O-containing HDX buffer (50 mM HEPES at pH 7.5, 1 M NaCl, 3 mM DTT) and incubated for a range of exchange times from 10 s to 1 h before quenching the deuterium exchange with an acidic quench solution (5 M Urea, 50 mM TCEP, and 1% v/v TFA at pH 2.4). Protease digestion was performed in-line with chromatography using an immobilized pepsin column. Mass spectra were acquired on an LTQ Orbitrap XL ETD mass spectrometer, and percent deuterium exchange values for peptide isotopic envelopes at each time point were calculated and processed using the HDX Workbench software.48
DLS Assays
For DLS experiments 500 nM BI/D or 3 µM LEDGF/p75 was added to 200 nM IN in the buffer containing 50 mM HEPES, at pH 7.4, 2 mM DTT, 2 mM MgCl2, and 1 M NaCl (buffer was filtered twice with 0.22 µm syringe driven filter from Millipore). DLS signals were recorded at RT (25 °C) using a Malvern Nano series zetasizer instrument, and particle size distribution was calculated.
HTRF-based Assays
Dose-dependent effects of BI/D and LEDGF/p75 additions on IN multimerization were monitored using reported HTRF-based assays.49 The HTRF format was also used to measure a Kd for IN-LEDGF/p75 binding. For this, increasing concentrations of FLAG-LEDGF/p75 were titrated in 20 nM His-IN solutions. The antibodies recognizing terminal FLAG and poly-His epitopes labeled with donor and acceptor fluorophores were used to monitor the FRET signal upon IN-LEDGF/p75 complex formation. The HTRF signal was recorded using a PerkinElmer Multimode EnSpire plate reader.
Cells, Viruses, and Antiviral Assays
HEK293T cells were transfected with the plasmid expressing HA-LEDGF/p7550 or its empty vector control. HIV-Luc virions were produced in HEK293T cells with pNL4–3.Luc.Env- and pCG-VSV-G using PolyJet DNA transfection reagent (SignaGen Laboratories). The target cells were preincubated with the indicated concentrations of BI/D or DMSO for 2 h, and then the cells were infected with HIV-1-Luc virions equivalent to 4 ng of HIV-1 p24 as determined by HIV-1 Gag p24 ELISA (ZeptoMetrix) following the manufacturer’s protocol. Two hours postinfection, the culture supernatant was removed, washed once with complete medium, and replaced with fresh complete medium with the inhibitor concentration maintained. The cells were cultured for 48 h, and the cell extracts were prepared using 1 × reporter lysis buffer (Promega). Luciferase activity was determined using a commercially available kit (Promega).
Estimation of Cellular Concentrations of LEDGF/p75
Cellular concentrations of endogenous LEDGF/p75 and ectopically expressed HA-LEDGF/p75 were estimated using whole cell HEK293T RIPA lysates. Cellular LEDGF/p75 and HA-LEDGF/p75 were detected by immunoblotting with LEDGF/p75 antibody (Abcam) and compared to a titration of recombinant purified protein. The average cellular volume of a HEK293T cell was calculated to be 1150 µm3 (assuming an average diameter of 13 µm and a spherical shape). Using E. coli as a basis (0.95 pg per cell and a 1.1 µm3 volume), the average weight of a HEK293T cell is 993 pg. Again using E. coli (90% water and 5.5% protein) and under conditions of perfect lysis and fractionation, these estimates yield 18 518 cells per microgram of RIPA extract. Taking the values of a volume of 1150 µm3, 18 518 cells per microgram of RIPA extract and the pmoles of LEDGF/p75 determined from the titration, one can derive an estimate of cellular concentration. Nuclear concentration was estimated based on HeLa cell percentage of nuclear volume of ~28% of the total volume. All numbers are taken from BioNumbers.51
Genome-wide Analysis of HIV-1 Integration Sites
Isolation of HIV-1 integration sites was performed using ligation-mediated PCR. Integration site sequencing, data analysis, and matched random control (MRC) generation were carried out as described.47,52
Supplementary Material
Acknowledgments
This work was supported in whole or in part by National Institutes of Health Grants AI110310 (to M.K. and J.R.F.), GM103368 (to M.K., P.R.G., and A.N.E.), AI052845 (to F.D.B.), and AI110270 (to J.J.K.).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.6b00167.
HDX results for IN binding to BI/D and LEDGF/p75, estimation of cellular levels of endogenous and ectopi-cally expressed LEDGF/p75 by immunoblotting, HTRF data for LEDGF/p75 binding to IN and its effects on ALLINI-induced aberrant protein multimerization (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Li M, Mizuuchi M, Burke TR, Jr, Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 3.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 4.Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. An essential role for LEDGF/p75 in HIV integration. Science. 2006;314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- 5.Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busschots K, Vercammen J, Emiliani S, Benarous R, Engelborghs Y, Christ F, Debyser Z. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J. Biol. Chem. 2005;280:17841–17847. doi: 10.1074/jbc.M411681200. [DOI] [PubMed] [Google Scholar]
- 7.Jaskolski M, Alexandratos JN, Bujacz G, Wlodawer A. Piecing together the structure of retroviral integrase, an important target in AIDS therapy. FEBS J. 2009;276:2926–2946. doi: 10.1111/j.1742-4658.2009.07009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng L, Larue RC, Slaughter A, Kessl JJ, Kvaratskhelia M. HIV-1 integrase multimerization as a therapeutic target. Curr. Top. Microbiol. Immunol. 2015;389:93–119. doi: 10.1007/82_2015_439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherepanov P. Integrase illuminated. EMBO Rep. 2010;11:328. doi: 10.1038/embor.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Krishnan L, Cherepanov P, Engelman A. Structural biology of retroviral DNA integration. Virology. 2011;411:194–205. doi: 10.1016/j.virol.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherepanov P, Maertens GN, Hare S. Structural insights into the retroviral DNA integration apparatus. Curr. Opin. Struct. Biol. 2011;21:249–256. doi: 10.1016/j.sbi.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Engelman A, Cherepanov P. The structural biology of HIV-1: mechanistic and therapeutic insights. Nat. Rev. Microbiol. 2012;10:279–290. doi: 10.1038/nrmicro2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krishnan L, Engelman A. Retroviral integrase proteins and HIV-1 DNA integration. J. Biol. Chem. 2012;287:40858–40866. doi: 10.1074/jbc.R112.397760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelman A, Cherepanov P. Retroviral Integrase Structure and DNA Recombination Mechanism. Microbiology spectrum. 2014;2:2. doi: 10.1128/microbiolspec.MDNA3-0024-2014. [DOI] [PubMed] [Google Scholar]
- 15.McKee CJ, Kessl JJ, Shkriabai N, Dar MJ, Engelman A, Kvaratskhelia M. Dynamic modulation of HIV-1 integrase structure and function by cellular lens epithelium-derived growth factor (LEDGF) protein. J. Biol. Chem. 2008;283:31802–31812. doi: 10.1074/jbc.M805843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kessl JJ, Li M, Ignatov M, Shkriabai N, Eidahl JO, Feng L, Musier-Forsyth K, Craigie R, Kvaratskhelia M. FRET analysis reveals distinct conformations of IN tetramers in the presence of viral DNA or LEDGF/p75. Nucleic Acids Res. 2011;39:9009–9022. doi: 10.1093/nar/gkr581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eidahl JO, Crowe BL, North JA, McKee CJ, Shkriabai N, Feng L, Plumb M, Graham RL, Gorelick RJ, Hess S, Poirier MG, Foster MP, Kvaratskhelia M. Structural basis for high-affinity binding of LEDGF PWWP to mononucleo-somes. Nucleic Acids Res. 2013;41:3924–3936. doi: 10.1093/nar/gkt074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 2012;8:e1002717. doi: 10.1371/journal.pgen.1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Nuland R, van Schaik FM, Simonis M, van Heesch S, Cuppen E, Boelens R, Timmers HM, van Ingen H. Nucleosomal DNA binding drives the recognition of H3K36-methylated nucleosomes by the PSIP1-PWWP domain. Epigenet. Chromatin. 2013;6:12. doi: 10.1186/1756-8935-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roth SL, Malani N, Bushman FD. Gammaretroviral integration into nucleosomal target DNA in vivo. Journal of virology. 2011;85:7393–7401. doi: 10.1128/JVI.00635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christ F, Voet A, Marchand A, Nicolet S, Desimmie BA, Marchand D, Bardiot D, Van der Veken NJ, Van Remoortel B, Strelkov SV, De Maeyer M, Chaltin P, Debyser Z. Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat. Chem. Biol. 2010;6:442–448. doi: 10.1038/nchembio.370. [DOI] [PubMed] [Google Scholar]
- 23.Fader LD, Malenfant E, Parisien M, Carson R, Bilodeau F, Landry S, Pesant M, Brochu C, Morin S, Chabot C, Halmos T, Bousquet Y, Bailey MD, Kawai SH, Coulombe R, LaPlante S, Jakalian A, Bhardwaj PK, Wernic D, Schroeder P, Amad M, Edwards P, Garneau M, Duan J, Cordingley M, Bethell R, Mason SW, Bos M, Bonneau P, Poupart MA, Faucher AM, Simoneau B, Fenwick C, Yoakim C, Tsantrizos Y. Discovery of BI 224436, a Noncatalytic Site Integrase Inhibitor (NCINI) of HIV-1. ACS Med. Chem. Lett. 2014;5:422–427. doi: 10.1021/ml500002n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsiang M, Jones GS, Niedziela-Majka A, Kan E, Lansdon EB, Huang W, Hung M, Samuel D, Novikov N, Xu Y, Mitchell M, Guo H, Babaoglu K, Liu X, Geleziunas R, Sakowicz R. New class of HIV-1 integrase (IN) inhibitors with a dual mode of action. J. Biol. Chem. 2012;287:21189–21203. doi: 10.1074/jbc.M112.347534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma A, Slaughter A, Jena N, Feng L, Kessl JJ, Fadel HJ, Malani N, Male F, Wu L, Poeschla E, Bushman FD, Fuchs JR, Kvaratskhelia M. A New Class of Multimerization Selective Inhibitors of HIV-1 Integrase. PLoS Pathog. 2014;10:e1004171. doi: 10.1371/journal.ppat.1004171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Rouzic E, Bonnard D, Chasset S, Bruneau JM, Chevreuil F, Le Strat F, Nguyen J, Beauvoir R, Amadori C, Brias J, Vomscheid S, Eiler S, Levy N, Delelis O, Deprez E, Saib A, Zamborlini A, Emiliani S, Ruff M, Ledoussal B, Moreau F, Benarous R. Dual inhibition of HIV-1 replication by integrase-LEDGF allosteric inhibitors is predominant at the post-integration stage. Retrovirology. 2013;10:144. doi: 10.1186/1742-4690-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Bel N, van der Velden Y, Bonnard D, Le Rouzic E, Das AT, Benarous R, Berkhout B. The allosteric HIV-1 integrase inhibitor BI-D affects virion maturation but does not influence packaging of a functional RNA genome. PLoS One. 2014;9:e103552. doi: 10.1371/journal.pone.0103552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jurado KA, Wang H, Slaughter A, Feng L, Kessl JJ, Koh Y, Wang W, Ballandras-Colas A, Patel PA, Fuchs JR, Kvaratskhelia M, Engelman A. Allosteric integrase inhibitor potency is determined through the inhibition of HIV-1 particle maturation. Proc. Natl. Acad. Sci. U. S. A. 2013;110:8690–8695. doi: 10.1073/pnas.1300703110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng L, Sharma A, Slaughter A, Jena N, Koh Y, Shkriabai N, Larue RC, Patel PA, Mitsuya H, Kessl JJ, Engelman A, Fuchs JR, Kvaratskhelia M. The A128T resistance mutation reveals aberrant protein multimerization as the primary mechanism of action of allosteric HIV-1 integrase inhibitors. J. Biol. Chem. 2013;288:15813–15820. doi: 10.1074/jbc.M112.443390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta K, Brady T, Dyer BM, Malani N, Hwang Y, Male F, Nolte RT, Wang L, Velthuisen E, Jeffrey J, Van Duyne GD, Bushman FD. Allosteric Inhibition of Human Immunodeficiency Virus Integrase: LATE BLOCK DURING VIRAL REPLICATION AND ABNORMAL MULTIMERIZATION INVOLVING SPECIFIC PROTEIN DOMAINS. J. Biol. Chem. 2014;289:20477–20488. doi: 10.1074/jbc.M114.551119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kessl JJ, Jena N, Koh Y, Taskent-Sezgin H, Slaughter A, Feng L, de Silva S, Wu L, Le Grice SF, Engelman A, Fuchs JR, Kvaratskhelia M. A multimode, cooperative mechanism of action of allosteric HIV-1 integrase inhibitors. J. Biol. Chem. 2012;287:16801–16811. doi: 10.1074/jbc.M112.354373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jurado KA, Engelman A. Multimodal mechanism of action of allosteric HIV-1 integrase inhibitors. Expert Rev. Mol. Med. 2013;15:e14. doi: 10.1017/erm.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shkriabai N, Dharmarajan V, Slaughter A, Kessl JJ, Larue RC, Feng L, Fuchs JR, Griffin PR, Kvaratskhelia M. A critical role of the C-terminal segment for allosteric inhibitor-induced aberrant multimerization of HIV-1 integrase. J. Biol. Chem. 2014;289:26430–26440. doi: 10.1074/jbc.M114.589572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slaughter A, Jurado KA, Deng N, Feng L, Kessl JJ, Shkriabai N, Larue RC, Fadel HJ, Patel PA, Jena N, Fuchs JR, Poeschla E, Levy RM, Engelman A, Kvaratskhelia M. The mechanism of H171T resistance reveals the importance of N inverted question mark -protonated His171 for the binding of allosteric inhibitor BI-D to HIV-1 integrase. Retrovirology. 2014;11:100. doi: 10.1186/s12977-014-0100-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balakrishnan M, Yant SR, Tsai L, O’Sullivan C, Bam RA, Tsai A, Niedziela-Majka A, Stray KM, Sakowicz R, Cihlar T. Non-Catalytic Site HIV-1 Integrase Inhibitors Disrupt Core Maturation and Induce a Reverse Transcription Block in Target Cells. PLoS One. 2013;8:e74163. doi: 10.1371/journal.pone.0074163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desimmie BA, Schrijvers R, Demeulemeester J, Borrenberghs D, Weydert C, Thys W, Vets S, Van Remoortel B, Hofkens J, De Rijck J, Hendrix J, Bannert N, Gijsbers R, Christ F, Debyser Z. LEDGINs inhibit late stage HIV-1 replication by modulating integrase multimerization in the virions. Retrovirology. 2013;10:57. doi: 10.1186/1742-4690-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fontana J, Jurado KA, Cheng N, Ly NL, Fuchs JR, Gorelick RJ, Engelman AN, Steven AC. Distribution and Redistribution of HIV-1 Nucleocapsid Protein in Immature, Mature, and Integrase-Inhibited Virions: a Role for Integrase in Maturation. J. Virol. 2015;89:9765–9780. doi: 10.1128/JVI.01522-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fadel HJ, Morrison JH, Saenz DT, Fuchs JR, Kvaratskhelia M, Ekker SC, Poeschla EM. TALEN knockout of the PSIP1 gene in human cells: analyses of HIV-1 replication and allosteric integrase inhibitor mechanism. J. Virol. 2014;88:9704–9717. doi: 10.1128/JVI.01397-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Desimmie BA, Weydert C, Schrijvers R, Vets S, Demeulemeester J, Proost P, Paron I, De Rijck J, Mast J, Bannert N, Gijsbers R, Christ F, Debyser Z. HIV-1 IN/Pol recruits LEDGF/p75 into viral particles. Retrovirology. 2015;12:16. doi: 10.1186/s12977-014-0134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, Jurado KA, Wu X, Shun MC, Li X, Ferris AL, Smith SJ, Patel PA, Fuchs JR, Cherepanov P, Kvaratskhelia M, Hughes SH, Engelman A. HRP2 determines the efficiency and specificity of HIV-1 integration in LEDGF/p75 knockout cells but does not contribute to the antiviral activity of a potent LEDGF/p75-binding site integrase inhibitor. Nucleic Acids Res. 2012;40:11518–11530. doi: 10.1093/nar/gks913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hare S, Shun MC, Gupta SS, Valkov E, Engelman A, Cherepanov P. A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS Pathog. 2009;5:e1000259. doi: 10.1371/journal.ppat.1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hare S, Di Nunzio F, Labeja A, Wang J, Engelman A, Cherepanov P. Structural basis for functional tetramerization of lentiviral integrase. PLoS Pathog. 2009;5:e1000515. doi: 10.1371/journal.ppat.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berne BJ, Pecora R. Dynamic light scattering: with applications to chemistry, biology, and physics, Dover Publications, Mineola, NY. 2000 [Google Scholar]
- 44.Harding SE. Determination of diffusion coefficients of biological macromolecules by dynamic light scattering. Microscopy, Optical Spectroscopy, and Macroscopic Techniques. 1994;22:97–108. doi: 10.1385/0-89603-232-9:97. [DOI] [PubMed] [Google Scholar]
- 45.Murphy RM. Static and dynamic light scattering of biological macromolecules: what can we learn? Curr. Opin. Biotechnol. 1997;8:25–30. doi: 10.1016/s0958-1669(97)80153-x. [DOI] [PubMed] [Google Scholar]
- 46.Kotova S, Li M, Dimitriadis EK, Craigie R. Nucleoprotein intermediates in HIV-1 DNA integration visualized by atomic force microscopy. J. Mol. Biol. 2010;399:491–500. doi: 10.1016/j.jmb.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sowd GA, Serrao E, Wang H, Wang W, Fadel HJ, Poeschla EM, Engelman AN. A critical role for alternative polyadenylation factor CPSF6 in targeting HIV-1 integration to transcriptionally active chromatin. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E1054. doi: 10.1073/pnas.1524213113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pascal BD, Willis S, Lauer JL, Landgraf RR, West GM, Marciano D, Novick S, Goswami D, Chalmers MJ, Griffin PR. HDX workbench: software for the analysis of H/D exchange MS data. J. Am. Soc. Mass Spectrom. 2012;23:1512–1521. doi: 10.1007/s13361-012-0419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kessl JJ, Jena N, Koh Y, Taskent-Sezgin H, Slaughter A, Feng L, de Silva S, Wu L, Le Grice SF, Engelman A, Fuchs JR, Kvaratskhelia M. Multimode, cooperative mechanism of action of allosteric HIV-1 integrase inhibitors. J. Biol. Chem. 2012;287:16801–16811. doi: 10.1074/jbc.M112.354373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maertens G, Cherepanov P, Debyser Z, Engelborghs Y, Engelman A. Identification and characterization of a functional nuclear localization signal in the HIV-1 integrase interactor LEDGF/ p75. J. Biol. Chem. 2004;279:33421–33429. doi: 10.1074/jbc.M404700200. [DOI] [PubMed] [Google Scholar]
- 51.Milo R, Jorgensen P, Moran U, Weber G, Springer M. BioNumbers-the database of key numbers in molecular and cell biology. Nucleic Acids Res. 2010;38:D750–D753. doi: 10.1093/nar/gkp889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matreyek KA, Wang W, Serrao E, Singh PK, Levin HL, Engelman A. Host and viral determinants for MxB restriction of HIV-1 infection. Retrovirology. 2014;11:90. doi: 10.1186/s12977-014-0090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.