SUMMARY
Innate lymphoid cells (ILCs) serve as sentinels in mucosal tissues, sensing release of soluble inflammatory mediators, rapidly communicating danger via cytokine secretion, and functioning as guardians of tissue homeostasis. Although ILCs have been studied extensively in model organisms, little is known about these “first responders” in humans, especially their lineage and functional kinships to cytokine-secreting T helper (Th) cell counterparts. Here, we report gene regulatory circuitries for four human ILC–Th counterparts derived from mucosal environments, revealing that each ILC subset diverges as a distinct lineage from Th and circulating natural killer cells, but shares circuitry devoted to functional polarization with their Th counterparts. Super-enhancers demarcate cohorts of cell identity genes in each lineage, uncovering new modes of regulation for signature cytokines, new molecules that likely impart important functions to ILCs, and potential mechanisms for autoimmune disease SNP associations within ILC–Th subsets.
INTRODUCTION
Innate lymphoid cells (ILCs) are a heterogeneous population of lymphocytes that lack antigen-specific receptors. ILCs respond to soluble mediators released into the tissue microenvironment when homeostasis is perturbed by pathogens or allergens (Artis and Spits, 2015; Diefenbach et al., 2014; Eberl et al., 2015; Rankin et al., 2013). Upon receiving danger signals, ILCs produce cytokines and chemokines that provide a frontline defense against infections, contribute to tissue repair, and regulate adaptive immunity. Based on cytokine production, ILCs are divided into three populations: (i) ILC1s, which produce IFN-γ; (ii) ILC2s, which produce type 2 cytokines; and (iii) ILC3s, which produce IL-22 and/or IL-17.
ILCs distribution is widespread in tissues but are of low abundance in blood (Gasteiger et al., 2015). ILC1s share many properties with conventional natural killer (cNK) cells, including IFN-γ production, but have distinct cytolytic capacities and, unlike cNKs, do not recirculate in blood. ILC categories functionally mirror those for T helper subsets, with ILC1s resembling Th1 cells, ILC2s resembling Th2s, and ILC3s resembling Th17s. (Wang et al., 2015). The similarities indicate that functional modules converging on cytokine production evolved to enable innate and adaptive arms of the immune response with shared core programs under the control of distinct activation pathways and temporal kinetics, providing the functional flexibility required to face nearly any pathogen.
In addition to signature cytokines, ILC lineages have been defined in mouse by transcriptome analyses and ontogenic relationships (Klose et al., 2014; Rankin et al., 2015; Robinette et al., 2015). ILC1 and cNK cells share many transcripts, with a major distinction being expression of EOMES in cNKs (Daussy et al., 2014; Klose et al., 2014). Lineage tracing in mice has clarified developmental relationships among ILC subsets. Similar to T cells, all ILC subsets and cNKs originate from common lymphoid progenitors (CLP), which give rise to common innate lymphoid progenitors (CILP) with restricted potential to generate ILCs and cNKs (Yu et al., 2014). CILPs differentiate into progenitors with more restricted potential, such as the NK progenitor (NKP), the common helper lymphoid progenitor (CHILP), and the innate lymphoid cell progenitor (iLCP), which together give rise to all ILCs (Constantinides et al., 2014; Klose et al., 2014; Xu et al., 2015).
Specification of ILC lineages from precursors depends on distinct transcription factors (TFs), some of which also mediate polarization of Th cells. RORγT and the aryl hydrocarbon receptor (AHR) coordinate both ILC3 and Th17 differentiation (Diefenbach et al., 2014; Quintana, 2013; Stockinger et al., 2011; Wang et al., 2015). TBX21 (TBET) is necessary for development of Th1, ILC1, and cNK cells, while EOMES is uniquely required for cNK differentiation (Diefenbach et al., 2014; Eberl et al., 2015). GATA3 specifies ILC2 and Th2 lineages, but is also required early in ILC lineage specification and later for ILC3 homeostasis (De Obaldia and Bhandoola, 2015; Diefenbach et al., 2014; Tindemans et al., 2014). Additional TFs, such as ID2, act at the CHILP stage, driving ILC development by antagonizing T lineage specifying functions of E2A family TFs (Klose et al., 2014).
Compared with mouse, human ILC subsets are under-characterized. Human ILC3s are primarily defined by their ability to produce IL-22 in response to IL-23 (Cella et al., 2010). Human ILC3s also produce GM-CSF and IL-26, as well as two other soluble mediators, LIF and BAFF, which are not substantially expressed by Th17 cells or mouse ILC3s (Cella et al., 2010). Although developmental data are limited, tonsillar ILC3s express RORC plus AHR, and appear to derive from a CD34+c-Kit+RORC+ hematopoietic progenitor (Montaldo et al., 2014). However, assignment of human ILC3s to a distinct lineage remains controversial due to reports that they are intermediates in a linear differentiation pathway for cNKs (Hughes et al., 2014). ILC3s also exhibit some functional plasticity under certain conditions in vitro that stimulate IFN-γ production (Cella et al., 2010).
Human ILC1s include a major subset in mucosal epithelium (intraepithelial ILC1s, iILC1s), which produce IFN-γ in response to IL-15 and IL-12 (Fuchs et al., 2013). Unlike cNKs, iILC1s are unresponsive to IL-18, have limited cytolytic capacity, and express markers of intraepithelial residency, such as CD103, CD160, CD49a, and CD101. Like Th1s, most tonsillar iILC1s express T-BET and low amounts of EOMES. Fate mapping in mouse has shown that iILC1s are a distinct lineage from cNKs (Constantinides et al., 2015; Constantinides et al., 2014). Although a second ILC1 subset has been reported in human (Bernink et al., 2013), these IFN-γ producing cells are found at very low frequencies in mucosal-associated lymphoid tissues. Similar to mouse, human ILC2s express GATA3 and produce IL-5 and IL-13 (Mjosberg et al., 2011), but these cells are found mostly in inflamed tissues from subjects with allergies.
Despite functional and ontogenic similarities, differences exist between ILC and Th counterparts, especially their dependence on pathogen-derived antigens. Th priming requires signals from TCR and co-stimulatory receptors, which drive expansion, differentiation, and cytokine expression. ILC effector responses depend solely on cytokine microenvironments, endowing them with more rapid response profiles. Distinctions in cytokine expression also exist between human ILC–Th counterparts – Th17s produce IL-17 and -22, while ILC3s secrete only the latter.
A comprehensive analysis of gene expression and regulatory pathways for human ILCTh subsets is lacking. In this regard, epigenetic landscapes, which reflect patterns of regulatory element activity, provide rich information about lineage and functional relationships (Gosselin et al., 2014; Kanno et al., 2012; Koues et al., 2015; Mercer et al., 2011; Mukasa et al., 2010). We now report regulatory circuitries for gene expression programs in human ILC3, iILC1, Th17, and Th1 cells from tonsil, a mucosal-associated lymphoid tissue. Based on their regulatory logic and transcriptomes, we find that ILC3s indeed constitute a unique lineage in humans. ILCs and Th cells employ a combination of overlapping and divergent enhancers to express shared genes, while activating signature expression programs that mediate unique functions of innate and adaptive immunity. Our epigenome-centric approach identified a collection of ILC super-enhancers that regulate the expression of cell identity genes and functions, including the IL-22/26 locus in ILC3s.
RESULTS
Transcriptome Profiling of Human ILC and Th Cells
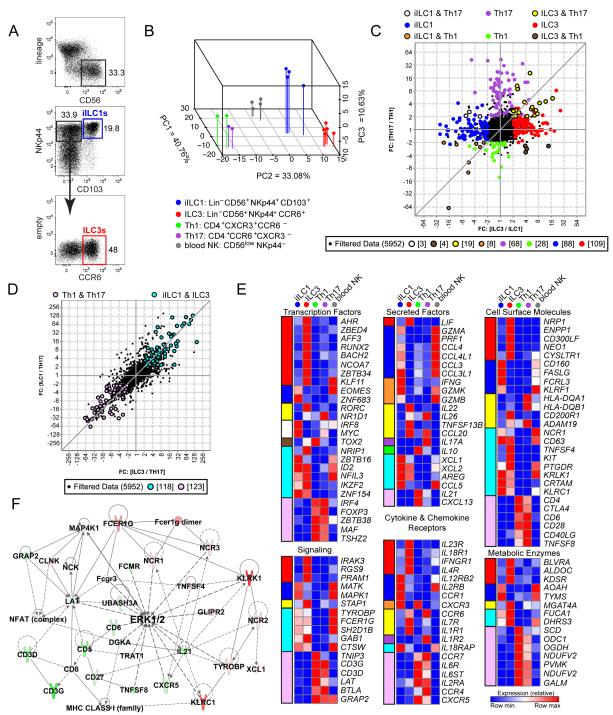
To interrogate unique and overlapping pathways in ILC and Th cells from a mucosal environment, we isolated lymphocytes from pediatric tonsils. This source provides a sufficient number of purified ILCs for transcriptome and low-input epigenome analyses. We divided lineage-negative cells that expressed CD56 and NKp44 into CD103+iILC1s and CCR6+ILC3s (Figure 1A). The latter subset produces IL-22 in response to IL-23, confirming its functional categorization (Cella et al., 2010). CD4-enriched tonsillar lymphocytes were divided into CXCR3+Th1 and CCR6+Th17 cells, as described (Annunziato et al., 2007; Zielinski et al., 2012). ILC2 and Th2 subsets were at sparingly low abundance in tonsils and could not be analyzed. As a comparator for ILCs, we isolated the prototypic innate lymphoid subset, cNK cells (CD56lowNKp44−), from human blood.
Figure 1. Transcriptome analysis of primary human tonsillar ILCs and Th cells.
(A) Strategy for iILC1 and ILC3 sorting.
(B) Supervised PCA of genes with most variable expression among iILC1, ILC3, Th1, Th17, and blood cNK cell replicates (top 15%).
(C) Plot comparing the fold change of ILC3 and iILC1 to the fold change of Th17 and Th1 depicts transcripts differentially expressed among ILCs and Th subsets. Colored circles highlight transcripts expressed at least two fold higher in a single tonsillar subset compared with the other three subsets. Colored circles with black borders denote transcripts that are shared by both an ILC and Th subset with expression >2 fold higher than cells of their respective lineage.
(D) Transcripts shared by ILC subsets that were increased by two fold compared to Th subsets and vice versa.
(E) Heatmaps highlighting the average expression of selected transcripts that were differentially expressed in each cell type as shown in panels C and D. Color coding at the left of each heatmap designates the categories of cell type-restricted expression patterns, retaining colors shown in panels C and D.
(F) The highest scoring IPA network generated from transcripts differentially expressed in ILCs versus Th cells, as in panel D. Transcripts represented in red were upregulated in ILCs versus Th cells at least two-fold in microarray data, and those in green were similarly downregulated. Darker colors indicate greater fold changes among upregulated or downregulated transcripts.
See also Figure S1.
To explore relationships between cell types, we performed supervised principal components analysis (PCA) on expression array data, focusing on the top 15% most differentially expressed genes. A reproducible separation was observed between ILC3, ILC1, cNK, and Th cells (Figure 1B). Consistent with prior studies (Cosmi et al., 2008; Ramesh et al., 2014), gene expression in Th1 and Th17 replicates from different tonsils was highly related, with a single module of genes providing a primary distinction (PC3). PCA clearly segregated innate cells from their Th functional counterparts; i.e., iILC1s were distinct from Th1s and ILC3s were distinct from Th17s. Thus, ILC and T lineage commitment dominated functional relationships in several transcriptional modules. PCA also revealed that blood cNKs segregated substantially from their functional counterpart, iILC1s. When comparisons were broadened to entire transcriptomes, unsupervised hierarchical clustering of array data separated all tonsillar subsets from blood cNKs (Figure S1A). These analyses suggested that the mucosal microenvironment, in addition to lineage specification, had an influence on expression programs in lymphoid cells. Indeed, differentially expressed genes in iILC1s, when compared with blood cNKs, generated a signature that was present in all four tonsillar subsets (Figures S1B and C). As such, we restricted most comparisons to cells derived from tonsils, using blood cNKs as external comparators.
Although PCA underscored distinctions in transcriptional modules between innate and adaptive counterparts, ILC and Th subsets share overlapping developmental pathways and functions. Therefore, we asked which genes were uniquely overexpressed by each subset and which were shared by multiple subsets. Comparisons of fold change expression profiles revealed important features of lineage specification and functional polarization. First, “unique” transcripts were most abundant in ILC3s (~100), followed by iILC1s, Th17s, and Th1s (Figure 1C). Second, the number of “unique” transcripts was far greater than that shared between any ILC and any Th subset. Third, among the latter transcript category, ILC3+Th17 was dominant (19 transcripts). These data suggested that, despite similar functionality, ILC and Th counterparts have fundamentally distinct transcriptional programs. Indeed, ILCs shared more differentially expressed genes with each other (118) than with their Th counterparts (27) and vice versa (123) (Figures 1C and D). These data suggest that ILC and T lineage factors distinguish the identity of cytokine-producing lymphocytes to a greater extent than factors governing their functional polarization.
In this regard, Figure 1E highlights a set of genes whose expression distinguished innate and adaptive subsets, spotlighting six biological categories. Importantly, genes encoding lineage determinants, receptors, and effector molecules aligned with the predicted functionality of each subset. For example, expression of RORC, CCR6, and IL26 was shared between ILC3s and Th17s, whereas CXCR3 and IFNG were shared between iILC1s and Th1s. Although TBX21, a “master” TF for Th1/ILC1 lineages, was expressed two-fold higher in iILC1 than ILC3 cells, the probe set had low absolute values and fell below cut-off criteria. In addition, T cell markers, such as CD4 and CD28, were expressed by Th1 and Th17 cells, while many NK cell markers, such as KLRK1, KLRC1, NCR1, and CRTAM, were expressed by iILC1s and ILC3s.
Several features of this analysis should be emphasized. First, consistent with patterns of “unique” genes, ILC3s expressed a substantial number of “unique” TFs. These included not only known ILC3 factors (RORC, AHR), but also several whose roles in ILC3 biology were less clear (e.g., RUNX2, BACH2, and ZBTB34). Second, consistent with their primary activation modes (cytokines for ILCs, antigen for T cells), ILCs expressed more cytokine receptors at substantially higher levels than Th cells, with a notable exception. Both Th subsets specifically expressed IL6R and IL6ST, which form the IL6 receptor, suggesting that this proinflammatory cytokine may regulate Th but not ILC responses. Third, when compared with Th subsets, ILCs expressed a distinct set of metabolic genes, particularly those encoding enzymes with anti-microbial functions (e.g., AOAH, BLVRA, and FUCA1).
We next performed Ingenuity Pathway Analysis (IPA) on genes with greater than twofold differences in ILC3s vs Th17s, iILC1s vs Th1s, or ILC3s vs iILC1s (Figures S1D-F). These analyses indicated that Th-enriched genes connected to pathways for TCR signaling, while ILC-enriched genes coordinated pathways for cytokine signaling or attenuated TCR signaling (e.g., cyclosporin A in the IPA database). Pathway distinctions were more apparent when input transcripts were restricted to those differentially expressed by both ILC subsets compared to both Th subsets, generating a network composed almost entirely of signaling molecules (Figure 1F). Differentially expressed transcripts created an innate activation module for ILCs centered on FCER1G (FcRγ) and TYROBP (DAP12), while the Th module centered on CD3 components. Despite distinct receptor origins, both pathways converged on the MAP kinase, ERK.
Distinct pathways also emerged when individual ILC-Th subsets were compared. Pathways augmented in ILC3s versus Th17s were connected to ETS family TFs (designated SPI1 by the IPA database), which are known to regulate cNK cell development (Ramirez et al., 2012) (see Fig. 3), or the IL1β cytokine (Figure S1D). The known potential of human ILC3s to convert into ILC1s also became evident. Namely, IL15- and T-BET-regulated pathways emerged when ILC3s were compared with Th cells, albeit these pathways had a lower z-score than ILC3–iILC1 comparisons (Figures S1D-F). Together, transcriptome arrays confirmed and revealed modules of gene expression that coordinate lineage specification, signal transduction, and functional polarization of human ILC/Th subsets in a mucosal microenvironment.
Figure 3. Enriched TF motifs in ILC versus Th regulomes.
(A) Fold change p-values for enrichment of TF motifs comparing enhancers that are differentially active in ILC versus Th cells.
(B) Heatmap showing the log p-values for enrichment of TF motifs in enhancers that exhibit augmented H3K27ac in one cell type versus the other three.
(C) Relative expression of selected members for TF families identified in panel B.
(D) Heat maps (left) showing the binding of BATF or IRF4 in GM12878 B cells (http://genome.ucsc.edu/ENCODE/) at enhancers (+ 2 kb) identified as ILC3- or Th17-enriched. The right bar graphs quantify numbers of enhancers that bind BATF and/or IRF4 in the indicated cell types.
(E) UCSC Genome Browser view of human IL22 (top) as described in Figure 2C. The cell type specificity of each boxed enhancer is shown at the top. The middle tracks denote sites of TF binding in GM12878 cells. The bottom tracks show data for BATF and IRF4 in mouse Th17 cells (Li et al., 2012) at conserved enhancer regions.
See also Figure S3.
ILC–Th Regulomes
Gene expression programs in ILC and Th subsets likely reflect distinct patterns of enhancer activation that, in turn, correspond to differences in TF profiles. Even genes that are co-expressed in ILC and Th subsets may be regulated by different sets of enhancers responding to distinct environmental cues (e.g., cytokines in ILCs and TCR ligation in Th cells). To probe the regulatory logic that specifies ILC/Th lineages and their functions, we performed regulome analysis using methods for ultra-low input chromatin profiling. We first identified regions of open chromatin, which demarcated active and poised regulatory elements, using an Assay for Transposase-Accessible Chromatin followed by high throughput sequencing (ATAC-seq) (Buenrostro et al., 2013). For Th1 cells, we used published DNase-seq data to identify accessible chromatin (GSM1024749). The activation status of each element was assessed using Chromatin Immunoprecipitation coupled with sequencing (ChIP-seq) for two histone modifications enriched at active promoters (H3K4me3) or enhancers (H3K27ac). To exclude architectural elements, most of which were ATAC+, known CTCF binding regions were removed from our master list of regulatory regions (Koues et al., 2015).
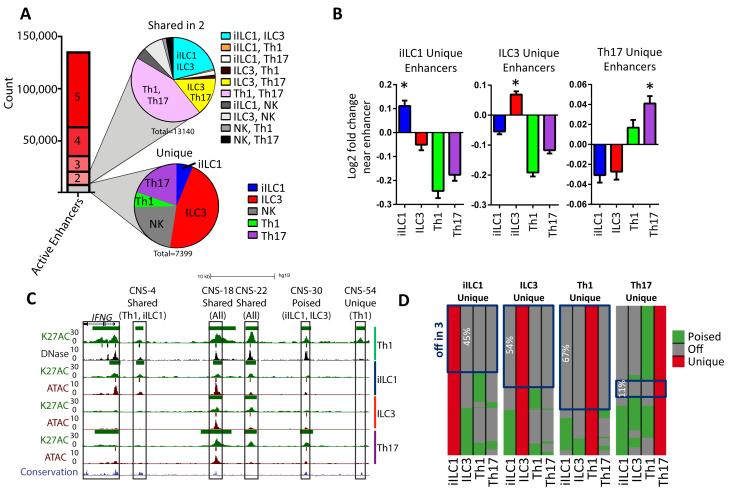
We focused epigenome analyses on enhancers, which generally are dominant regulatory elements for determining the expression status of target genes. Enhancers were defined as accessible chromatin regions that were >2 kb from known transcriptional start sites (TSSs) and were considered active if they coincided with H3K27ac peaks. These analyses revealed ~140,000 enhancers that were active in at least one of the five cell types profiled (Figure 2A), namely tonsillar iILC1s, ILC3s, Th1s, Th17s, and blood cNKs. We also defined the regulatory circuitry of each cell type by linking enhancers to their putative target gene(s) (Table S1) using pattern recognition algorithms to assign concordant changes in chromatin and expression for genes within 250 kb of each enhancer (Koues et al., 2015).
Figure 2. Unique and shared enhancers in human ILC and Th subsets.
(A) The stacked bar graph shows numbers of active enhancers that were either unique (gray) or shared (red shades) by the indicated number of cell types. The pie graphs depict cell type distributions for unique (bottom) or shared (top) active enhancers.
(B) Fold change expression of genes (+ SD) located most proximally to enhancers uniquely active in the indicated cell type. For each panel, expression in the indicated cell type was compared to average expression in the other three subsets. *Denotes statistical significance (p<0.01) using a one-way anova.
(C) UCSC Genome Browser views of the human IFNG locus, showing tracks of DNAse-, ATAC- and H3K27ac ChIP-seq data for the indicated cell types, displayed as reads per million (RPM) values. For human Th1 cells, published DNAse-seq data are shown (GSE29692). A track for mammalian sequence conservation is provided at the bottom. Colored bars above tracks represent statistically significant called peaks. Known IFNG regulatory regions (CNSs) are boxed and categorized by cell type restriction in their activities.
(D) Activity status (poised versus off) in other cell types for enhancers that were designated as uniquely active in a given subset. Boxes highlight the subset of enhancers that are off in other cell types.
Only ~5% of identified enhancers were uniquely active in a single cell type. The majority of cell type-specific enhancers were found in ILC3s, further underscoring their distinctive identity at both gene expression and regulome levels (Figure 2A). Expression of genes located most proximally to cell type-specific enhancers was augmented when compared with average expression in the other cell types (Figures 2B and S2A). Some enhancers were active in two cell types (~10%). Consistent with gene expression patterns, the majority of these enhancers were shared by Th1/Th17, followed by iILC1/ILC3 and ILC3/Th17 cells (Figure 2A). Thus, a larger proportion of the regulome used by each subset was independent of its functional polarization. Instead, enhancer activation patterns were dominated by factors that coordinated divergence between Th and ILC lineages.
We next assessed whether enhancers active in only one subset were either off (DNase/ATAC−H3K27ac−) or poised (DNase/ATAC+H3K27ac−) in other cell types. Figures 2C and S2B highlight examples of different enhancer categories for the IFNG and ZEB2 loci, respectively. As expected, the average expression of genes associated with poised enhancers was significantly lower than those near active enhancers (Figure S2C). A subset of enhancers uniquely active in a single cell type were poised in at least one other cell type (Figure 2D), especially for Th17s. This pattern suggests that each lineage retains some remnants of regulomes from an earlier common precursor, and/or they retain some degree of functional plasticity. Together, enhancer profiling defined the gene regulatory circuitry for ILC-Th cells, revealing a complex blend of elements that tracked cleanly with lineage specification or remained poised in a manner that reflected functional or developmental kinships.
Distinct TFs Dominate ILC and Th Regulomes
TF expression is a critical determinant of enhancer activation during lineage specification or cellular activation. Many lineage decisions or biological responses are dominated by a single TF or related family. To understand more completely the TFs coordinating ILC and Th regulomes, we analyzed enhancer categories for enrichment of sequence motifs using HOMER. When analyses focused on innate versus adaptive lineages, enhancers with augmented H3K27ac levels in Th cells were enriched for motifs bound by BATF/AP1, IRF, NFAT, and EGR (Figure 3A), TFs activated by TCR signaling (Glasmacher et al., 2012; Li et al., 2012). In contrast, enhancers hyperactive in both ILC subsets were enriched for RUNX and ETS motifs. Consistent with this finding, RUNX3 is essential for ILC development in mouse (Ebihara et al., 2015). These new data also predict that an ETS family member is required for development, homeostasis, or function of ILC subsets.
We next focused on enhancers that were augmented in a single cell type versus the other three. A full accounting of enriched TF motifs for cell type-specific enhancers is provided in Table S2. “Master” TFs for differentiation into functionally distinct subsets were enriched in the predicted lineages. A motif recognized by RORγT was enriched in ILC3- and Th17-specific enhancers, whereas enhancers unique to iILC1 and Th1 cells were enriched for a T-box motif, to which T-BET binds (Figure 3B). In addition to distinctions described above for shared enhancers, motifs for PRDM1 were enriched in iILC1- and Th1-specific enhancers, perhaps indicating a role for PRDM1, or related TFs, in type-1 effector functions. Indeed, iILC1s expressed high levels of ZNF683 (Figure 1E), a PRDM1 homologue that is necessary and sufficient for IFN-γ production in human CD8+ effector-type cells (Vieira Braga et al., 2015). Th subsets were also enriched for HMG/TCF motifs, which are targets of TCF7, a TF essential for T cell development and function (Weber et al., 2011). Consistent with results from this unbiased analysis, members of TF families enriched in cell type-specific enhancers exhibited parallel expression patterns (Figures 3C and S3A). Of note was the reciprocal expression pattern for two AP1 family members, BATF and FOSL2, which function as a transcriptional activator or repressor, respectively (Ciofani et al., 2012). Accordingly, the AP1 motif was enriched in active Th enhancers, where the activating factor BATF was expressed, while the repressive FOSL2 predominated in ILCs.
To validate some predictions, we leveraged public ChIP-seq data for human BATF and IRF4, whose motifs were enriched in Th-specific enhancers. Although ChIP-seq data were derived from a transformed human B cell (GM12878), these TFs were bound at a higher percentage of Th17-specific compared with ILC3-specific enhancers (Figure 3D). Indeed, many Th17-specific enhancers bound both BATF and IRF4, likely reflecting the importance of composite sites for their stable association at some enhancers (Glasmacher et al., 2012; Li et al., 2012). We also examined BATF and IRF4 binding at elements that exhibited preferential activity in given cell types (Figures 3E and S3B). For example, enhancers positioned near the IL22 cytokine gene, expressed by ILC3s and Th17s, displayed a range of cell type specificity. However, two enhancers preferentially activated in Th17 cells were bound by BATF and IRF4, while an enhancer with higher activity in ILC3s was associated with SPI1 in the GM12878 B cells (Figure 3E). The two enhancers with augmented activity in Th17s were conserved in mouse and were bound by BATF and IRF4 in mouse Th17 cells. Thus, epigenome analyses identified a cohort of TFs that coordinate enhancer activation to regulate distinct gene expression programs in innate and adaptive lymphoid effectors from human mucosal tissue.
Human ILC and Th Super-enhancers
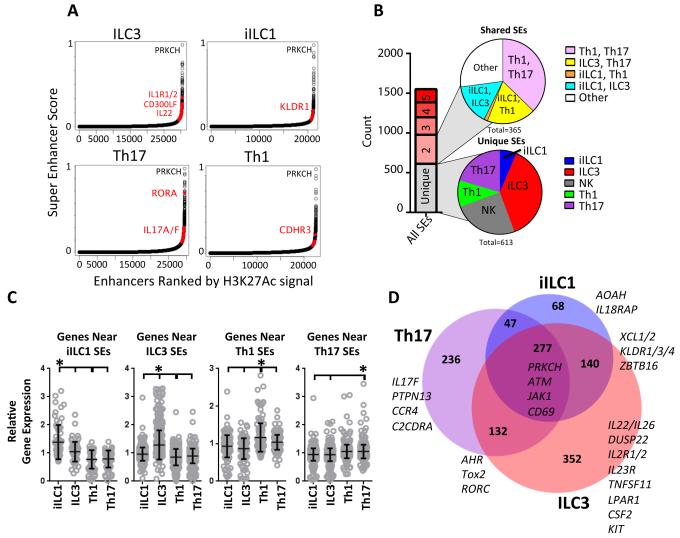
Super-enhancers (SEs) encompass a class of regulatory elements devoted to maintaining high expression levels for genes associated with cell identity (Whyte et al., 2013). SEs are distinguished by their extreme loads and broad distributions of H3K27ac, as well as their dramatic enrichment for lineage-specifying TFs. To further define genes that contribute to unique identities of ILC and Th cells in human, we used published algorithms and standard cut-off criteria to generate lists of SE regions from H3K27ac data (Table S3) (Hnisz et al., 2013). Ranked plots of H3K27ac+ regions, which represent both super- and conventional-enhancers (CE) are shown in Figure 4A, with cell type-specific SEs highlighted in red.
Figure 4. Super-enhancer distribution in human ILC and Th subsets.
(A) Rank order of increasing H3K27ac enrichment at enhancer loci for each ILC–Th subset. Red dots denote SEs that are uniquely called in each cell type and selected genes associated with these SEs.
(B) SE distributions. Stacked bar shows active enhancers that are unique (gray) or shared by the indicated number of cell types (red shades). Pie graphs depict distributions for unique SEs (bottom) or those shared by two cell types (top).
(C) Relative expression levels of genes within 30 kb of cell type-specific SEs. Statistical significance (paired T-test): *p≤ 0.05.
(D) Venn diagram showing unique and shared categories of SEs with a selected set of associated genes highlighted.
See also Figure S4.
Consistent with their role in cell identity, many SEs (>30%, Figure 4B) were cell type-specific, which is in contrast to our analyses of CEs (~5% unique, Figure 2A). However, like our findings with CEs, ILC3s had the most unique SEs, further confirming their striking distinction from the other cell types, while iILC1s and Th1s had the fewest unique SEs. As an example, an ILC3-specific SE encompassed the KIT receptor locus (Figure S4A), which was selectively expressed in this cell type. One of the few iILC1-specific SEs spanned AOAH (Figure S4B), which encodes an enzyme known to detoxify bacterial lipopolysaccharide (Munford and Hunter, 1992), revealing an anti-microbial function for this innate mucosal subset. A cNK-specific SE was associated with the perforin (PRF1) gene. This SE was absent in iILC1s (Figure S4C), consistent with the cytolytic capacities of these two type-1 innate subsets.
SEs shared in two cell types were most evident in Th1–Th17, a finding that also emerged from CE analysis. Two Th-shared SEs spanned the TNFSF8 (Figure S4D) and TNFRSF8 loci, reflecting the Th-specific expression of both CD30 and its ligand, respectively, which form an autoregulatory loop in activated T cells. A more general analysis revealed that genes encompassed by or flanking (+ 20 kb) a cell type-specific SE exhibited significantly elevated expression in that cell lineage compared with the others (Figure 4C). A comparison of SEs in Th17s, ILC3s, and iILC1s identified 277 gene loci with important functions in both innate and lymphoid lineages, including CD69, which mediates tissue residency (Figure 4D) (Shiow et al., 2006). 132 genes were encompassed by SEs active in ILC3s and Th17s, including several TFs (AHR, RORC) that play a role in differentiation of IL22 producing cells, and TOX2, whose role in these functional subsets was unclear. SE-associated genes shared by iILC1s and ILC3s encoded ZBTB16, a TF important for ILC lineage commitment (Ishizuka et al., 2016), as well as the XCL chemokines (Figure S4E), whose roles in ILC biology were unknown. Similar comparisons of Th- versus ILC-restricted SEs illuminated additional features of regulatory circuits that contrast Th and ILC identities (Figure S4F). We cannot exclude the possibility that some regions were called SEs in one cell type, but barely missed the SE cut-off in one or more other cell types, which can be discerned from focused inspections of the primary data. Notwithstanding, our identification of SEs in human lymphoid cells highlights unique and shared epigenetic landscapes that endow ILCs with aspects of cell identity distinguishing them from their Th and NK counterparts.
SEs Associated with Signature Cytokines and Surface Markers
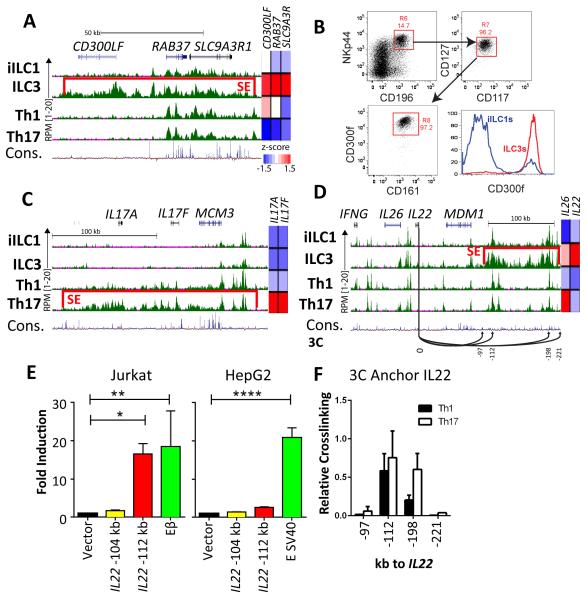
In addition to expected loci, SE analysis uncovered relationships that may provide new insights or tools for studying ILC biology. One example was the CD300LF locus, which encodes a phospholipid-responsive inhibitory receptor (Choi et al., 2011), and was spanned by an ILC3-specific SE (Figure 5A). Accordingly, CD300LF mRNA was expressed at a significantly higher level by ILC3s and the protein was detected on their cell surface (Figures 5A and B), further defining the distinct phenotype of this innate subset.
Figure 5. Novel SE-regulated loci.
(A) UCSC Genome Browser view of RPM-normalized H3K27ac data for the CD300LFSE with relative expression for CD300LF in each cell type shown at the right. Red bar denotes designated SE.
(B) Flow cytometric analysis for CD300f protein (encoded by the CD300LF gene) on tonsillar ILC3s and iILC1s. A small CD300f+ population within iILC1s may represent converting ILC3s.
(C) UCSC Genome Browser view of RPM-normalized H3K27ac data for the IL17A/F-SE with relative expression for IL17A/F in each cell type shown at the right.
(D) UCSC Genome Browser view of RPM-normalized H3K27ac data for the IL22/26-SE with relative expression for IL22/26 in each cell type shown at the right. Arrows represent interactions tested by 3C assays.
(E) Luciferase reporter assays for enhancer activity in two regions from the IL22/26-SE. See panel D for locations of tested elements. Control vectors were the Tcrb enhancer (Eβ) for Jurkat T cells and the SV40 enhancer for HepG2 cells. Experiments were performed in triplicate. Shown are mean values (+ SD) for fold change versus a vector containing only the minimal SV40 promoter, normalized by Renilla luciferase. Statistical significance (one-way anova): *p< 0.05, **p< 0.01, ***p< 0.001.
(F) 3C assays in Th1 and Th17 cells from human blood. The assays test relative crosslinking efficiency of the IL22 promoter region (labeled “0” in D) and selected enhancer or control regions within or flanking the SE, as designated in panel D.
We also identified an SE in Th17 cells that spanned its signature IL17A/IL17F cytokine locus, as well as the neighboring MCM3 gene, which is expressed in all cell types (Figure 5C). In contrast to mice, which express Il17 in both Th17s and a subset of ILC3s, humans express these cytokines only in Th17s (Figure 1E). Only a subset of enhancers active in Th17s were in regions of high sequence conservation, perhaps reflecting evolutionary revisions to the regulatory landscape of this locus that endow Th17-specific expression in humans. Indeed, the vast majority of active IL17A/F enhancers were completely off in ILC3s, with the exception of several that remained poised, suggesting a minimal potential for activation-induced IL-17 expression by agonists. Future studies should allow the identification of CEs within the SE that mediate divergent IL17 expression patterns in mouse and human.
In human, the signature ILC3 cytokine locus spans IL22 and IL26, which lie between the ubiquitously expressed MDM1 gene and the Th1/iILC1-restricted cytokine gene, IFNG (Figure 5D). Regulation of the latter gene has been studied extensively in both mouse and human (Ansel et al., 2003; Balasubramani et al., 2010; Collins et al., 2012); however, much less is known about the cis-elements controlling IL22/26. Strikingly, we identified an ILC3-specific SE distal to MDM1, which precisely paralleled expression of IL22 (Figure 1E). In contrast to the composite SE element, many individual CEs in this region, including some within the SE, were shared with other cell types. The most related H3K27ac landscape was in Th17s, potentially explaining their expression of IL26 (Figure 1E). We predicted that the SE and its composite CEs were critical elements regulating cell type-restricted expression of IL22/26. Indeed, a CE with the most intense deposition of H3K27ac (SE-112), which was conserved in mammals, exhibited robust enhancer activity in a human T cell, but not in a hepatoma cell line (Figure 5E). A second region (SE-104), which was less conserved and had lower levels of H3K27ac, was inert in both cell types. We also measured cross-linking efficiencies of the IL22 promoter with SE elements using chromosome conformation capture (3C), which provided a readout for association between the promoter and selected distal regions. As shown in Figure 5F, the IL22 promoter associated efficiently with two conserved CEs, including SE-112, compared with two regions outside of the SE, which lacked active enhancers. One element, termed IL22-198, exhibited significantly higher cross-linking in cultured Th17 versus Th1 cells, reflecting higher IL22 expression in the former. However, this region did not function as an enhancer in Jurkat T cells, perhaps due to the lack of Th17 polarization in this transformed line. Thus, a novel regulatory region, including an ILC3-specific SE, controlled the expression of IL22 and IL26 in both innate and adaptive lymphoid cells inhabiting human mucosae.
ILC Super-enhancers Are Enriched for Autoimmune-associated SNPs
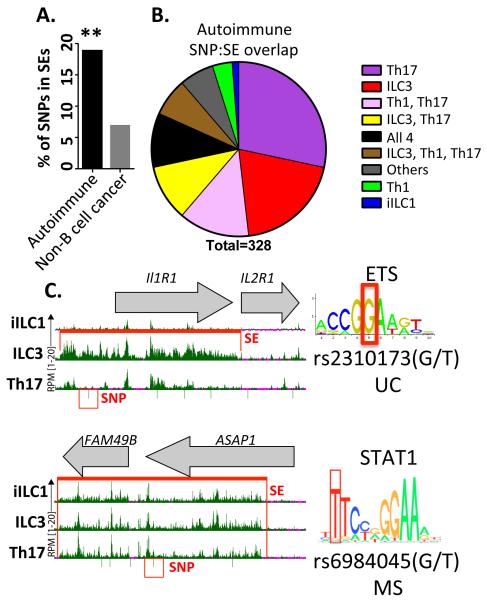
Most disease-associated differences in human genomes co-localize with regulatory elements rather than coding exons (Maurano et al., 2012). These elements include SEs, which control the cohort of genes for cell identity and their signature functions. For ILC and Th cells, obvious signature functions include their secretion of pro-inflammatory mediators (e.g., IL-17 and IFN-γ), dysregulation of which leads to a spectrum of autoimmune conditions. We investigated whether SEs for lymphoid cells were enriched for SNPs linked to autoimmunity by GWAS. As shown in Figure 6A, ~20% of reported autoimmune SNPs were encompassed by an SE identified in any ILC–Th subset. In contrast, a significantly lower proportion of SNPs for non-B cell cancers were found in this SE collection (~5%).
Figure 6. Autoimmune-associated SNPs found in ILC–Th super-enhancers.
(A) Proportion of total SNPs associated with autoimmunity or non-B cell cancers found in SEs from the four ILC-Th subsets. Statistical significance (permutation analysis): **p≤ 0.01.
(B) Number of autoimmune SNPs in called SEs divided into categories based on cell type distributions.
(C) UCSC Genome Browser view of two loci associated with an ILC3-specific (IL1R1/IL2R1, top) or an ILC-Th SE region (ASAP1/FAM49B, bottom) with the autoimmune disease SNPs highlighted. Right panels show predicted TF motifs potentially disrupted by the autoimmune SNP (UC, ulcerative colitis; MS, multiple sclerosis).
When categorized by cell type distributions, Th17- and ILC3-specific SEs had the most autoimmune SNPs, followed by shared SEs found in either Th1–Th17 or ILC3–Th17 cells (Figure 6B). We next compared the gene content of SNP-associated SEs for four disorders: Crohn disease (CD), ulcerative colitis (UC), rheumatoid arthritis (RA) and type 1 diabetes (T1D). SNPs associated with CD and UC were primarily restricted to ILC3 and Th17 SEs (Table 1). A subset of these disease-associated SNPs was localized to SEs regulating genes implicated in autoimmunity, including IL23R (ILC3-specific) and STAT3 (ILC3+Th17). Our analyses also identified loci harboring genes with ILC-specific SEs that were linked to autoimmunity by their functions in other cell types. One example, ATG16L1, had been linked to autoimmunity via its expression in Paneth and dendritic cells (Stappenbeck et al., 2011). In contrast to UC and CD, SNPs identified for RA and T1D were enriched in Th1–Th17 SEs. Many genes linked to these SNP-containing SEs have been implicated in RA or T1D, including IL2RA/B, CTLA4, and CSF2 (Marson et al., 2015).
Table 1. Selected autoimmune-associated SNPs in SEs.
Shown are reference IDs for SE-SNPs categorized by autoimmune disease association. SNPs are classified by cell type SE designations, closest gene to the SNP, whether the gene is linked by eQTL (*), and predicted disruption of TF motifs or coding potential. Details are provided in Supplemental Experimental Procedures.
| SNP | Cell Type | Gene | Motif† |
|---|---|---|---|
| Crohn's | |||
| rs10210302 | iILC1 | ATG16L1 * | HLX1‡ |
| rs17221417 | ILC3,Th1,Th17 | NOD2 * | NRSF‡ |
| rs744166 | ILC3,Th17 | STAT3 * | PLZF‡ |
| rs2301436 | Th17 | FGFR1OP * | GFI1‡ |
| rs6601764 | Th17 | KLF6 | ISL2‡ |
| rs1992660 | Th17 | PTGER4 * | PAX‡ |
| Crohn's + UC | |||
| rs4613763 | ILC3,Th17 | PTGER4 | ZNF143‡ |
| rs9292777 | Th17 | PTGER4 * | . |
| rs11209026 | ILC3 | IL23R * | missense |
| Ulcerative Colitis | |||
| rs2310173 | ILC3 | IL1R1 * | ETS‡ |
| Crohn's + RA + T1D | |||
| rs2476601 | Th17 | PTPN22 * | ETS‡ |
| T1D + UC | |||
| rs6897932 | iILC1,ILC3,Th17 | IL7R | missense |
| rs3024493 | Th1,Th17 | IL10 | TCF4‡ |
| Rheumatoid Arthritis | |||
| rs3761847 | iILC1,ILC3,Th1,Th17 | TRAF1 * | E2F‡ |
| rs657075 | ILC3 | CSF2 | YY1‡ |
| rs3093023 | ILC3,Th17 | CCR6 * | POU2F2‡ |
| rs10821944 | Th1,Th17 | ARID5B | FOX‡ |
| rs4937362 | Th1,Th17 | ETS1 | CTCF‡ |
| rs881375 | Th1,Th17 | TRAF1 * | p53‡ |
| T1D | |||
| rs7528684 | iILC1 | FCRL3 * | NF-κB‡ |
| rs11755527 | ILC3 | BACH2 | AP-1‡ |
| rs2281808 | Th1,Th17 | SIRPG * | FXR‡ |
| RA+T1D | |||
| rs743777 | iILC1,ILC3,Th1,Th17 | IL2RB * | ATF‡ |
| rs3184504 | ILC3 | SH2B3 * | missense |
| rs231735 | Th1,Th17 | CTLA4 * | PU.1‡ |
| rs61839660 | Th17 | IL2RA | Mef2‡ |
SNP linked to gene expression by eQTL study
Bold predicted functional mutation
predicted motif disruption
One mechanism by which SNPs contribute to pathogenesis is alteration of TF sites in key enhancers. Indeed, ~80% of autoimmune SNPs in the ILC–Th SEs were predicted to disrupt motifs for known TFs, two examples of which are provided in Figure 6C. The first highlights a UC-associated SNP found in an ILC3-specific SE spanning IL1R1, in which the polymorphic allele was predicted to disrupt an ETS site, a motif enriched in ILC-specific enhancers (Figure 3A). The second example was in an ILC+Th SE that spanned the ASAP1 gene for an ARF-GAP signaling mediator. The MS-associated SNP was predicted to disrupt a STAT1 motif, a TF expressed in most cells (Figure 3C). We conclude that SEs spanning ILC–Th identity genes are enriched for autoimmune SNPs, perhaps reflecting the “TF motif breaking” consequences of nucleotide diversity in humans.
DISCUSSION
The importance of ILCs in mammalian immunity has emerged from in vitro studies in human and functional analyses in mice. However, many outstanding questions remain, especially with regard to ontogeny, functional relationships, ILC plasticity, and species distinctions. By integrating transcriptome and epigenome platforms, we shed light on important facets of lineage and functional relationships between innate and adaptive Th counterparts derived from a human mucosal tissue. These analyses defined human ILC–Th gene expression programs and corresponding gene regulatory schematics, revealing important aspects of their developmental and functional relationships.
Key findings to emerge included: (i) human ILC3s exhibited the most distinctive signatures of gene expression and active regulomes compared with other ILC–Th cells, (ii) human iILC1s diverged as a unique lineage from cNK cells, (iii) Th and ILC regulomes defined gene circuitry nodes that associated with lineage bifurcation (shared in Th1–17 versus shared in ILC1–3), which were often distinct from circuitry nodes shared for functional polarization (Th17–ILC3 versus Th1-iILC1), (iv) coordinately regulated enhancers identified TF families that governed Th versus ILC regulomes, (v) SE patterns defined cell identity genes and regulatory regions for known functional mediators, and (vi) SEs spanning ILC–Th identity genes were enriched for autoimmune SNPs, perhaps revealing critical regulatory elements for differential gene expression in these diseases.
Lineage relationships between human ILC and cNK cells have been a subject of intense debate. Reports have provided evidence that tonsillar ILC3s develop from a lineage-restricted progenitor (Montaldo et al., 2014), or that ILC3s are an intermediate in the terminal differentiation of cNK cells from CD34+CD56+ progenitors (Hughes et al., 2014). Transcriptome, regulome, and SE profiles demonstrated that ILC3s were the most divergent of the innate and adaptive cell types examined, including when compared with cNK or Th17, their closest functional counterparts. However, a direct comparison with Th22 cells (Duhen et al., 2009), which produce only IL-22 but are not abundant in tonsils, will further enlighten ILC3 developmental and functional relationships.
Ontogenic connections between ILC1 and cNKs also have remained unclear. Lineage tracing in mice suggested that ILC1s, including iILC1s studied here, developed from an ID2+ZBTB16+ precursor shared with other ILCs but not cNKs (Constantinides et al., 2015; Constantinides et al., 2014). In contrast, independent data supported a model in which ILC1s were tissue resident cNK cells (Sojka et al., 2014). While our study cannot resolve this issue completely, expression programs and regulomes of iILC1s diverged significantly from blood cNKs. Such divergence in gene circuitry renders it unlikely that iLC1s and cNKs are the same cell type. Notwithstanding, microenvironmental factors, layered on top of lineage specification, may contribute to this divergent regulatory logic. In depth study of this issue will require extensive phenotypic characterization of the multiple IFN-γ producing cells that inhabit human mucosae.
Regulome analyses indicated that the identity of each lymphoid subset was sculpted from distinct and shared gene circuitry modules that imparted developmental kinship (Th versus ILC), or signature functionalities (Th17–ILC3 versus ILC1–Th1–cNK). The adaptive modules included circuitry for genes involved in TCR signaling and switches for these circuits were a cohort of enhancers targeted by TFs linked to TCR signaling, such as BATF and IRF4 (Glasmacher et al., 2012; Li et al., 2012). Innate modules were enriched for circuitry governed by signals from innate immune receptors, including those associated with FCER1G and TYROBP adaptors. Like TCR signaling, these receptor driven pathways converged on MAP kinases (e.g., ERK), but connected to distinct TFs. Enhancers linked to circuits in the innate module were enriched for RUNX–ETS composite sites rather than AP1–IRF composite elements found in Th enhancers. Of note, RUNX3 is essential for ILC1 and ILC3 development in mouse (Ebihara et al., 2015), but a requirement for ETS TFs in the ILC lineage remains unknown.
An important regulome stratum is the establishment of SEs that drive expression of genes for lineage identity and signature cell functions. Indeed, ILC–Th cells harbored a higher proportion of SEs unique to one cell type when compared with CEs, which were normally shared between two or more lineages. Cell type-specific SEs were linked to a number of “cell identity” genes whose functions in ILC biology were not recognized, including CD300L, AOAH, TNSF13B (BAFF), LIF, and TOX2. Notably, several of these genes were expressed in human, but not mouse ILC subsets (Robinette et al., 2015). Thus, our study highlights shared and divergent features of ILCs from different species and provides a rich resource for more in depth functional analyses. For example, functional dissection of an SE near the IL22–IL26–IFNG locus would be an important goal. If the SE or its composite elements were conserved in mouse, its spatial segregation from genes permits targeted deletion. Similarly, comparison of the SE spanning IL17A/F genes, which is only active in human Th17s, with syntenic regions in mice, where these genes are expressed in both ILC3 and Th17 cells, may reveal differences in the activities of individual enhancers that mediate innate versus adaptive expression patterns of the inflammatory cytokines IL17A/F. Many SEs shared in Th–ILC lineages likely rely on a combination of elements, which have shared or cell type-restricted activities. Dissection of such regulatory circuits will identify key enhancers that govern iILC1, ILC3, Th17, and Th1 cell identities.
EXPERIMENTAL PROCEDURES
Details for ATAC-, ChIP-seq, and informatics analysis are in Supplemental Experimental Procedures.
Cytometric and microarray analyses
Cells were sorted from 2-3 pooled donors for each replicate as described (Annunziato et al., 2007; Cella et al., 2010; Fuchs et al., 2013). RNA was isolated (RNeasy Plus Micro Kit, Qiagen), amplified, and hybridized to the Affymetrix Human Gene 1.0 ST arrays. RNA yields from each subset were comparable. Array data were analyzed as described (Robinette et al., 2015). Unsupervised clustering was performed in R using the hclust function. For CD300LF analyses, CD56–enriched tonsil cells were stained with a PercP-Cy5.5 lineage cocktail containing anti-CD3 (eBioscience), -CD19 (eBioscience), -CD34 (Biolegend) and -ILT3 (Biolegend). Cells were stained with the following combination of antibodies (Biolegend unless indicated): CD117-FITC; NKp44-PE (BD-Pharmingen); CD300f-eFluor660 (eBioscience); CD196-BV421; CD103-BV605; CD161-BV510; CD56-PE-Cy7 (BD-Pharmingen); CD127-biotin (eBioscience), followed by Streptavidin-APC-eFluor780. Data were acquired on an LSR-Fortessa (BD) and analyzed by FlowJo software (TreeStar).
ATAC- and ChIP-seq
ATAC sample preparation was carried out as described (Buenrostro et al., 2013). Libraries for 2-3 samples were pooled for PE50 sequencing (Illumina HiSeq2500). Inter-sample variability was minimal for data sets from a given cell type (r2>0.92). ATAC data for tonsillar Th17s recapitulated those published for their blood counterparts (>90% peaks; GSE29692). Ultra-low input ChIP-seq was carried out as described (Brind'Amour et al, 2015).
Luciferase
SE regions were PCR amplified and cloned into SV40 promoter driven PGL3 plasmid (Promega, France). Reporters were transfected into Jurkat cells by electroporation or HepG2 cells using Lipofectamine 2000 (InVitrogen).
Chromatin conformation capture
3C was performed on Bgl II digested chromatin from primary Th1 and 17 cells (107) as described (Majumder et al., 2015), and assayed using primer/probe combinations provided in Supplementary Experimental Procedures. Standard curves were prepared from Bgl II digested BACs spanning the IL22 locus and interactions between neighboring fragments within YWHAZ were used for normalization.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grants AI120606 (E.M.O., Colonna); AI115734, AI11852 (E.M.O.); AI095542, DE021255 (Colonna); CA188286 (J.E.P. and E.M.O.); CA176695 (Cella); TR000448 (WU-ICTS); and CA91842 (Siteman Cancer Center). We thank the Genome Technology Access Center for help with –omics analyses.
Footnotes
AUTHOR CONTRIBUTIONS
E.M.O. and M. Colonna conceptualized and supervised the project and designed experiments with all coauthors. Experiments and data analyses were performed by all authors. Specimens were processed by M. Cella, O.I.K. and S.I.P. The manuscript was written by E.M.O., M. Colonna, O.I.K., P.L.C., M. Cella, and M.L.R.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Annunziato F, Cosmi L, Santarlasci V, Maggi L, Liotta F, Mazzinghi B, Parente E, Fili L, Ferri S, Frosali F, et al. Phenotypic and functional features of human Th17 cells. J. Exp. Med. 2007;204:1849–1861. doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat. Immunol. 2003;4:616–623. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- Balasubramani A, Shibata Y, Crawford GE, Baldwin AS, Hatton RD, Weaver CT. Modular utilization of distal cis-regulatory elements controls Ifng gene expression in T cells activated by distinct stimuli. Immunity. 2010;33:35–47. doi: 10.1016/j.immuni.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernink JH, Peters CP, Munneke M, te Velde AA, Meijer SL, Weijer K, Hreggvidsdottir HS, Heinsbroek SE, Legrand N, Buskens CJ, et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat. Immunol. 2013;14:221–229. doi: 10.1038/ni.2534. [DOI] [PubMed] [Google Scholar]
- Brind’Amour J, Liu S, Hudson M, Chen C, Karimi MM, Lorincz MC. An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat. Comm. 2015;6:6033. doi: 10.1038/ncomms7033. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Meth. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M, Otero K, Colonna M. Expansion of human NK-22 cells with IL-7, IL-2, and IL-1beta reveals intrinsic functional plasticity. Proc. Natl. Acad. Sci., USA. 2010;107:10961–10966. doi: 10.1073/pnas.1005641107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SC, Simhadri VR, Tian L, Gil-Krzewska A, Krzewski K, Borrego F, Coligan JE. Cutting edge: mouse CD300f (CMRF-35-like molecule-1) recognizes outer membrane-exposed phosphatidylserine and can promote phagocytosis. J. Immunol. 2011;187:3483–3487. doi: 10.4049/jimmunol.1101549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins PL, Henderson MA, Aune TM. Diverse functions of distal regulatory elements at the IFNG locus. J. Immunol. 2012;188:1726–1733. doi: 10.4049/jimmunol.1102879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides MG, Gudjonson H, McDonald BD, Ishizuka IE, Verhoef PA, Dinner AR, Bendelac A. PLZF expression maps the early stages of ILC1 lineage development. Proc. Natl. Acad. Sci., USA. 2015;112:5123–5128. doi: 10.1073/pnas.1423244112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosmi L, De Palma R, Santarlasci V, Maggi L, Capone M, Frosali F, Rodolico G, Querci V, Abbate G, Angeli R, et al. Human interleukin 17-producing cells originate from a CD161+CD4+ T cell precursor. J. Exp. Med. 2008;205:1903–1916. doi: 10.1084/jem.20080397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daussy C, Faure F, Mayol K, Viel S, Gasteiger G, Charrier E, Bienvenu J, Henry T, Debien E, Hasan UA, et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J. Exp. Med. 2014;211:563–577. doi: 10.1084/jem.20131560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Obaldia ME, Bhandoola A. Transcriptional regulation of innate and adaptive lymphocyte lineages. Ann. Rev. Immnuol. 2015;33:607–642. doi: 10.1146/annurev-immunol-032414-112032. [DOI] [PubMed] [Google Scholar]
- Diefenbach A, Colonna M, Koyasu S. Development, differentiation, and diversity of innate lymphoid cells. Immunity. 2014;41:354–365. doi: 10.1016/j.immuni.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhen T, Geiger R, Jarrossay D, Lanzavecchia A, Sallusto F. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat. Immunol. 2009;10:857–863. doi: 10.1038/ni.1767. [DOI] [PubMed] [Google Scholar]
- Eberl G, Colonna M, Di Santo JP, McKenzie AN. Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science. 2015;348:6566. doi: 10.1126/science.aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara T, Song C, Ryu SH, Plougastel-Douglas B, Yang L, Levanon D, Groner Y, Bern MD, Stappenbeck TS, Colonna M, et al. Runx3 specifies lineage commitment of innate lymphoid cells. Nat. Immunol. 2015;16:1124–1133. doi: 10.1038/ni.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD, Cella M, Colonna M. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-gamma-producing cells. Immunity. 2013;38:769–781. doi: 10.1016/j.immuni.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger G, Fan X, Dikiy S, Lee SY, Rudensky AY. Tissue residency of innate lymphoid cells in lymphoid and non-lymphoid organs. Science. 2015;350:981–985. doi: 10.1126/science.aac9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, Vander Lugt B, Khan AA, Ciofani M, Spooner CJ, Rutz S, et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science. 2012;338:975–980. doi: 10.1126/science.1228309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T, Briercheck EL, Freud AG, Trotta R, McClory S, Scoville SD, Keller K, Deng Y, Cole J, Harrison N, et al. The transcription Factor AHR prevents the differentiation of a stage 3 innate lymphoid cell subset to natural killer cells. Cell reports. 2014;8:150–162. doi: 10.1016/j.celrep.2014.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka IE, Chea S, Gudjonson H, Constantinides MG, Dinner AR, Bendelac A, Golub R. Single-cell analysis defines the divergence between the innate lymphoid cell lineage and lymphoid tissue-inducer cell lineage. Nat. Immunol. 2016;17:269–276. doi: 10.1038/ni.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Vahedi G, Hirahara K, Singleton K, O'Shea JJ. Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Ann. Rev. Immunol. 2012;30:707–731. doi: 10.1146/annurev-immunol-020711-075058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose CS, Flach M, Mohle L, Rogell L, Hoyler T, Ebert K, Fabiunke C, Pfeifer D, Sexl V, Fonseca-Pereira D, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157:340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- Koues OI, Kowalewski RA, Chang LW, Pyfrom SC, Schmidt JA, Luo H, Sandoval LE, Hughes TB, Bednarski JJ, Cashen AF, et al. Enhancer sequence variants and transcription-factor deregulation synergize to construct pathogenic regulatory circuits in B-cell lymphoma. Immunity. 2015;42:186–198. doi: 10.1016/j.immuni.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Spolski R, Liao W, Wang L, Murphy TL, Murphy KM, Leonard WJ. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature. 2012;490:543–546. doi: 10.1038/nature11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder K, Koues OI, Chan EA, Kyle KE, Horowitz JE, Yang-Iott K, Bassing CH, Taniuchi I, Krangel MS, Oltz EM. Lineage-specific compaction of Tcrb requires a chromatin barrier to protect the function of a long-range tethering element. J. Exp. Med. 2015;212:107–120. doi: 10.1084/jem.20141479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Housley WJ, Hafler DA. Genetic basis of autoimmunity. J. Clin. Invest. 2015;125:2234–2241. doi: 10.1172/JCI78086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer EM, Lin YC, Benner C, Jhunjhunwala S, Dutkowski J, Flores M, Sigvardsson M, Ideker T, Glass CK, Murre C. Multilineage priming of enhancer repertoires precedes commitment to the B and myeloid cell lineages in hematopoietic progenitors. Immunity. 2011;35:413–425. doi: 10.1016/j.immuni.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mjosberg JM, Trifari S, Crellin NK, Peters CP, van Drunen CM, Piet B, Fokkens WJ, Cupedo T, Spits H. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat. Immunol. 2011;12:1055–1062. doi: 10.1038/ni.2104. [DOI] [PubMed] [Google Scholar]
- Montaldo E, Teixeira-Alves LG, Glatzer T, Durek P, Stervbo U, Hamann W, Babic M, Paclik D, Stolzel K, Grone J, et al. Human RORgammat(+)CD34(+) cells are lineage-specified progenitors of group 3 RORgammat(+) innate lymphoid cells. Immunity. 2014;41:988–1000. doi: 10.1016/j.immuni.2014.11.010. [DOI] [PubMed] [Google Scholar]
- Mukasa R, Balasubramani A, Lee YK, Whitley SK, Weaver BT, Shibata Y, Crawford GE, Hatton RD, Weaver CT. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity. 2010;32:616–627. doi: 10.1016/j.immuni.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munford RS, Hunter JP. Acyloxyacyl hydrolase, a leukocyte enzyme that deacylates bacterial lipopolysaccharides, has phospholipase, lysophospholipase, diacylglycerollipase, and acyltransferase activities in vitro. J. Biol. Chem. 1992;267:10116–10121. [PubMed] [Google Scholar]
- Quintana FJ. Regulation of central nervous system autoimmunity by the aryl hydrocarbon receptor. Sem. Immunopath. 2013;35:627–635. doi: 10.1007/s00281-013-0397-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh R, Kozhaya L, McKevitt K, Djuretic IM, Carlson TJ, Quintero MA, McCauley JL, Abreu MT, Unutmaz D, Sundrud MS. Pro-inflammatory human Th17 cells selectively express P-glycoprotein and are refractory to glucocorticoids. J. Exp. Med. 2014;211:89–104. doi: 10.1084/jem.20130301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez K, Chandler KJ, Spaulding C, Zandi S, Sigvardsson M, Graves BJ, Kee BL. Gene deregulation and chronic activation in natural killer cells deficient in the transcription factor ETS1. Immunity. 2012;36:921–932. doi: 10.1016/j.immuni.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin L, Groom J, Mielke LA, Seillet C, Belz GT. Diversity, function, and transcriptional regulation of gut innate lymphocytes. Frontiers Immunol. 2013;4:22. doi: 10.3389/fimmu.2013.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin LC, Girard-Madoux MJ, Seillet C, Mielke LA, Kerdiles Y, Fenis A, Wieduwild E, Putoczki T, Mondot S, Lantz O, et al. Complementarity and redundancy of IL-22-producing innate lymphoid cells. Nat. Immunol. 2015;17:179–186. doi: 10.1038/ni.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, Gilfillan S, Colonna M. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 2015;16:306–317. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiow LR, Rosen DB, Brdickova N, Xu Y, An J, Lanier LL, Cyster JG, Matloubian M. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature. 2006;440:540–544. doi: 10.1038/nature04606. [DOI] [PubMed] [Google Scholar]
- Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, Zhong C, Chase JM, Rothman PB, Yu J, et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. Elife. 2014;3:e01659. doi: 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappenbeck TS, Rioux JD, Mizoguchi A, Saitoh T, Huett A, Darfeuille-Michaud A, Wileman T, Mizushima N, Carding S, Akira S, et al. Crohn disease: a current perspective on genetics, autophagy and immunity. Autophagy. 2011;7:355–374. doi: 10.4161/auto.7.4.13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger B, Hirota K, Duarte J, Veldhoen M. External influences on the immune system via activation of the aryl hydrocarbon receptor. Sem. Immunol. 2011;23:99–105. doi: 10.1016/j.smim.2011.01.008. [DOI] [PubMed] [Google Scholar]
- Tindemans I, Serafini N, Di Santo JP, Hendriks RW. GATA-3 function in innate and adaptive immunity. Immunity. 2014;41:191–206. doi: 10.1016/j.immuni.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Vieira Braga FA, Hertoghs KM, Kragten NA, Doody GM, Barnes NA, Remmerswaal EB, Hsiao CC, Moerland PD, Wouters D, Derks IA, et al. Blimp-1 homolog Hobit identifies effector-type lymphocytes in humans. Eur. J. Immunol. 2015;45:2945–2958. doi: 10.1002/eji.201545650. [DOI] [PubMed] [Google Scholar]
- Wang C, Collins M, Kuchroo VK. Effector T cell differentiation: are master regulators of effector T cells still the masters? Curr. Op. Immunol. 2015;37:6–10. doi: 10.1016/j.coi.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Weber BN, Chi AW, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, Bhandoola A. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Domingues RG, Fonseca-Pereira D, Ferreira M, Ribeiro H, Lopez-Lastra S, Motomura Y, Moreira-Santos L, Bihl F, Braud V, et al. NFIL3 Orchestrates the Emergence of Common Helper Innate Lymphoid Cell Precursors. Cell Rep. 2015;10:2043–2054. doi: 10.1016/j.celrep.2015.02.057. [DOI] [PubMed] [Google Scholar]
- Yu X, Wang Y, Deng M, Li Y, Ruhn KA, Zhang CC, Hooper LV. The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell precursor. Elife. 2014;3:e04406. doi: 10.7554/eLife.04406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski CE, Mele F, Aschenbrenner D, Jarrossay D, Ronchi F, Gattorno M, Monticelli S, Lanzavecchia A, Sallusto F. Pathogen-induced human TH17 cells produce IFN-gamma or IL-10 and are regulated by IL-1beta. Nature. 2012;484:514–518. doi: 10.1038/nature10957. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.