Summary
Telomerase is the ribonucleoprotein enzyme that replenishes telomeric DNA and maintains genome integrity. Minimally, telomerase activity requires a templating RNA and a catalytic protein. Additional proteins are required for activity on telomeres in vivo. Here we report that the Pop1, Pop6, and Pop7 proteins, known components of RNase P and RNase MRP, bind to yeast telomerase RNA and are essential constituents of the telomerase holoenzyme. Pop1/Pop6/Pop7 binding is specific and involves an RNA domain that is highly similar to a protein-binding domain in the RNAs of RNase P/MRP. The results also show that Pop1/Pop6/Pop7 function to maintain the essential components Est1 and Est2 on the RNA in vivo. Consistently, addition of Pop1 allows for telomerase activity reconstitution with wild type telomerase RNA in vitro. Thus, the same chaperoning module has allowed the evolution of functionally and, remarkably, structurally distinct RNPs, telomerase and RNases P/MRP, from unrelated progenitor RNAs.
Introduction
The ends of eukaryotic chromosomes are capped by telomeres, which ensure that these physiological ends are not perceived as DNA double-strand breaks (Palm and de Lange, 2008). The underlying telomeric DNA is composed of particular short direct repeats to which a complex set of proteins bind. In order to maintain the functionality of telomeres and ensure genome stability, a certain minimal tract of those telomeric repeats must be present at each individual telomere (Hockemeyer and Collins, 2015; Sarek et al., 2015). However, the conventional replication machinery alone is unable to completely duplicate the DNA at physical ends of chromosomes, an effect dubbed the end-replication problem that affects all organisms with linear chromosomes (Watson, 1972). In virtually every system tested, this problem is solved by the presence of a specialized catalytic ribonucleoprotein (RNP) complex called telomerase (Greider and Blackburn, 1985; Egan and Collins, 2012). In humans, telomerase activity is subject to strict developmental control and loss of this regulation is strongly associated with cancer etiology (Schmidt and Cech, 2015). A minimal telomerase that shows activity in vitro contains a catalytic protein subunit related to reverse transcriptases and a constitutively associated RNA moiety that provides the template for telomeric repeat synthesis (Greider and Blackburn, 1987). However, in all organisms, additional subunits are essential for its function on telomeres in vivo (Egan and Collins, 2012; Schmidt and Cech, 2015). Intriguingly, whereas the basic function of telomerase at telomeres via a reverse transcriptase-like mechanism is highly conserved in all organisms, the composition of the species-specific RNPs varies significantly. While the telomerase RNAs from various organisms do harbor conserved structural elements that are associated with catalytic protein binding (Qi et al., 2013), they differ significantly in their overall sizes, predicted secondary structures as well as in how they are transcribed, matured, and how their 3′-ends are stabilized (Egan and Collins, 2012; Podlevsky and Chen, 2012). The reasons and evolutionary origin of this variability are unknown, and in order to understand the biology of it, a knowledge of the complete composition of a number of different telomerase RNPs is required.
In budding yeast, the core RNA moiety of telomerase is Tlc1, a 1158 nt lncRNA for which a general secondary structure prediction has been compiled (Dandjinou et al., 2004; Zappulla and Cech, 2004). The catalytic protein Est2 associates in a central domain that includes the templating sequence, a characteristic pseudo-knot, and a template limiting stem (Singer and Gottschling, 1994; Livengood et al., 2002; Seto et al., 2003; Chappell and Lundblad, 2004). Other known subunits that associate with sub-elements of the RNA include Est1 (Lundblad and Szostak, 1989; Seto et al., 2002), the RNA stabilizing Sm7-complex (Seto et al., 1999), and the yeast Ku complex (Stellwagen et al., 2003). The Est3 subunit is also essential for telomerase-mediated telomere lengthening in vivo (Lendvay et al., 1996), but it is not clear whether this protein interacts directly with the RNA. The relatively large size of the Tlc1 RNA appeared to be an obstacle for an in vitro reconstitution of telomerase as only a shortened and simplified RNA yielded robust and reproducible activity (Zappulla et al., 2005). It has been proposed that the yeast telomerase RNP differs from other large RNPs in that the long RNA subunit serves as a flexible scaffold and that the overall architecture would not be determined by the RNA structure, as is the case for example in RNases P/MRP or ribosomes (Zappulla and Cech, 2006). This idea is supported by the fact that the Tlc1 RNA allows permutations of large parts of the RNA and certain predicted long stems can be stiffened (Zappulla and Cech, 2004; Lebo and Zappulla, 2012). These possibilities were complicated by recent phylogenetic and mutational analyses of fungal telomerase RNAs that suggested the presence of additional essential elements (Gunisova et al., 2009; Lubin et al., 2012; Laterreur et al., 2013). Moreover, their function as well as the complete composition of the native holoenzyme remained elusive.
The results shown here demonstrate that the protein Pop1 and the Pop6/Pop7 protein heterodimer associate with high affinity to a sub-domain on the yeast telomerase RNA, previously called TeSS (Telomerase Stimulating Structure). We now show that this domain corresponds to a protein-binding platform known as the P3 domain in the RNAs of eukaryotic, and the Tlc1 P3 domain is functionally interchangeable with the P3 domains of RNase P/MRP in vivo. In addition, our data show that purified and active budding yeast telomerase RNPs expressed at native levels contain the Pop1 and the Pop6/Pop7 proteins. Moreover, their presence is required for stabilizing the Est1 and Est2 proteins on the RNP and, accordingly, for telomerase activity in vivo. Finally, adding recombinant Pop1 to an in vitro telomerase reconstitution system results in readily detectable activity even with the wild-type full-length telomerase RNA. Therefore, our results show that, similar to eukaryotic RNase P/MRP RNPs, the yeast telomerase RNP is built upon and depends on a P3-like domain and proteins associated with it. Thus, an identical central organizing module can forge the architecture of completely different RNPs that hitherto had no structural similarity.
Results
The Pop1/Pop6/Pop7 proteins associate with yeast telomerase RNA
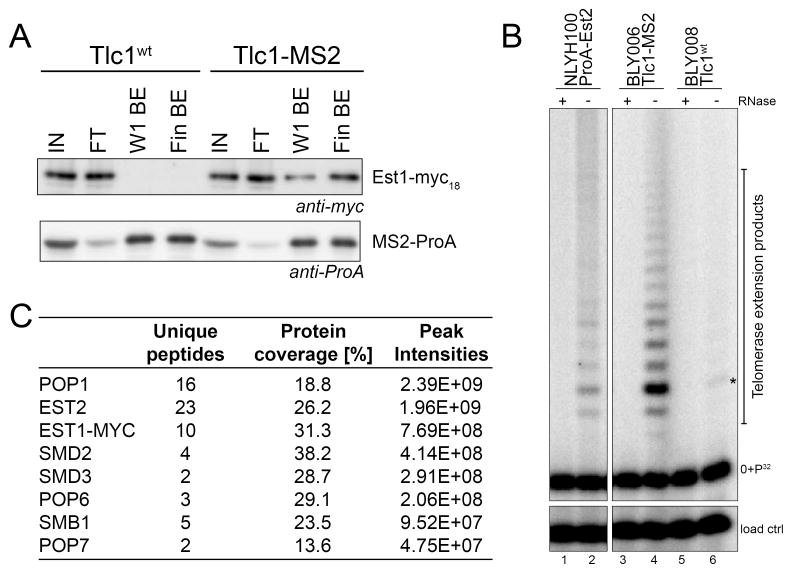
Given the large size of the Tlc1 RNA and the fact that the complete protein composition of the native yeast telomerase RNP is unknown, we reasoned that additional subunits could be associated with this RNA and affect telomerase function. In order to determine telomerase RNP composition, we used a fully functional tagged Tlc1 RNA that contains 10 MS2 stem-loop elements inserted near its 3′-end and that is expressed at endogenous levels (Gallardo et al., 2011). The same cells also expressed an MS2-ProA fusion protein that was used to enrich for telomerase RNPs on IgG beads. As quality control for these preparations, we asked that the known telomerase associated protein Est1, which carried a Myc tag in these experiments, was co-enriched and that the final preparations contained high telomerase activity (Figure 1A, B).
Figure 1. Mass spectrometry of endogenously expressed telomerase RNPs identifies the Pop1/Pop6/Pop7 proteins as part of active telomerase complexes.
(A) Fractions from indicated steps during the purification as analyzed by western blotting. Blots were probed with anti Myc-antibodies (top) or anti-ProA antibodies (bottom). IN: input protein extract; FT: proteins in flow through; W1 BE: proteins retained on beads after the first wash; Fin BE: final proteins on the beads fraction that was used for mass spectrometry. The TLC1 alleles used are indicated on top. (B) Left: Telomerase activity assays from cells expressing a ProA-Est2 protein and enriched with IgG as positive control. Right: Final telomerase activity on the IgG beads using the extracts from the cells as in panel A. +/− RNase: sample treatment with RNase A. 0+P32: 32P end-labeled substrate oligo. Band labeled with a star is a background band migrating at the +3 position. (C) Combined results from three independent mass spectrometry determinations. Proteins identified are listed in order of the sum of peak-intensities (right column). The total number of unique peptides for each protein and the total protein coverage are indicated. Only proteins that were detected exclusively in the MS2-tagged Tlc1 fraction were considered and the top eight proteins are listed. For a more extensive list, see Figure S1A.
These preparations were then analyzed using mass spectrometry. The results revealed that the three proteins, Pop1, Pop6, and Pop7 were amongst the top prevalent proteins exclusive for preparations with MS2-tagged Tlc1 RNA (Figure 1C, Figure S1A). These proteins are common subunits of the yeast RNase P/MRP complexes, two related, highly conserved and essential RNPs that are involved in the processing of tRNA, rRNA and certain mRNAs (Esakova and Krasilnikov, 2010). Both RNPs contain a catalytic RNA subunit (Nme1 RNA for the RNase MRP and Rpr1 RNA for the RNase P) and ten and nine protein components, respectively. Eight of those proteins are the same for RNase P and RNase MRP and include the Pop1/Pop6/Pop7 trio identified here as well as in preparations derived with highly overexpressed telomerase (Lin et al., 2015).
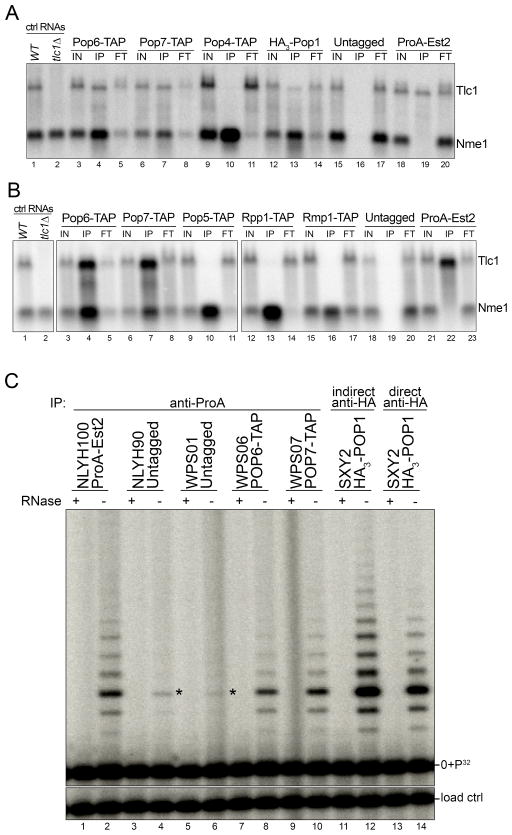
We used RNA co-immunoprecipitation with tagged components of the RNase MRP RNP in order to verify whether the detection of those proteins in the proteomic approach was specific. Indeed, precipitates obtained with extracts from cells in which either of the three proteins was tagged did contain Tlc1 RNA, as detected by northern blots (Figure 2A). However, none of four other components of the RNase MRP complex (Pop4, Pop5, Rmp1, Rpp1) brought down any Tlc1 RNA, even though the Nme1 RNA was readily detected, as expected (Figure 2A, 2B). Consistently, no peptide for any other component of the RNase P/ MRP RNPs were detected in any of the MS determinations (Figure S1A; see complete data sets at http://proteomecentral.proteomexchange.org). The efficiency of the immune-depletion of the Nme1 RNA from the extracts in these experiments was 59±15% when using Pop6-TAP and 53±5% for Pop7-TAP. Depletion of Tlc1 RNA in the same experiments was 46±7% for Pop6-TAP and 55±10% for Pop7-TAP (see methods for details). These very similar immunoprecipitation efficiencies combined with the fact that all RNase MRP RNPs do contain the Pop6/Pop7 proteins suggest that the Pop6/Pop7 proteins are constitutively associated with the telomerase RNA. As a second verification for the association of the Pop1, Pop6, and Pop7 proteins with telomerase, we assayed activity in immunoprecipitates generated with anti-ProA antibodies and extracts of strains expressing Pop6-TAP or Pop7-TAP, or with anti-HA antibodies in conjunction with HA3-Pop1 (Figure 2C). Importantly, telomerase activity could readily be detected in these precipitates, as it was detected in precipitates with ProA-Est2 as positive control, but no activity was detected when extracts from cells with TAP-tagged Pop4 were used (Figure S1B).
Figure 2. Binding of the Pop1/Pop6/Pop7 proteins to the Tlc1 RNA is specific.
(A) Northern analysis of immunoprecipitations using IgG covered beads with extracts from strains expressing the indicated TAP-tagged proteins. For the HA3-Pop1, anti-HA antibodies were used. The blots were hybridized with probes specific for the Tlc1 RNA and the Nme1 RNA at the same time. Ctrl RNAs on left: total RNAs from a wt strain or a strain that carried a tlc1Δ allele. IN: RNA from the input fraction IP: RNA from the immunoprecipitates; FT: RNA extracted from the unbound fraction. (B) Northern analysis as performed in panel (A). (C) Telomerase activity enrichment using a tagged Pop1, Pop6, or Pop7 protein. Top: antibodies used for immunoprecipitations with extracts derived from strains harboring the indicated tagged proteins. ProA-Est2 serves as positive control. Note that direct and indirect anti-HA refers to whether the anti-HA antibody was directly coupled to magnetic beads or whether antibody was first mixed with the slurry and then immunopurified with Protein A/G coupled magnetic beads. For the HA-tagged Pop1 protein, the latter technique appears more efficient in telomerase recuperation. Labeling as in Figure 1B. See also Figure S1B.
The telomerase RNA contains a functional P3-like domain
Proteins Pop6 and Pop7 associate with a specific substructure in the Nme1 and Rpr1 RNAs, a helix-loop-helix area called the P3 domain (Perederina et al., 2007; Perederina et al., 2010). This domain is a conserved essential feature of the protein-rich eukaryotic RNases P/MRP RNAs and it differentiates them from the bacterial and archaeal RNases P, which have a more simple protein composition (Lindahl et al., 2000; Ziehler et al., 2001; Piccinelli et al., 2005). The appearance of the P3 domains in eukaryotic RNases P/MRP was accompanied by the acquisition of three proteins that do not have homologues in bacterial and archaeal RNases P: Pop1, Pop6, and Pop7. Based on the results of structural, mutational, crosslinking and footprinting studies, it was proposed that the P3 RNA domain serves as a protein-binding hub in the eukaryotic RNases P/MRP (Ziehler et al., 2001; Perederina et al., 2007; Perederina et al., 2010; Hipp et al., 2012; Khanova et al., 2012; Fagerlund et al., 2015). Specifically, proteins Pop6 and Pop7 form a heterodimer that binds to the P3 domain and, together with the P3 RNA, form a platform that in turn binds the bigger Pop1 protein. This latter covers large parts of the RNase P/MRP holoenzyme structures and serves as a global RNP scaffold, interacting with other protein components as well as with several specific parts of RNA scattered across the structure (Fagerlund et al., 2015). Consistent with common function, the P3 domains of RNase P and RNase MRP are interchangeable (Lindahl et al., 2000).
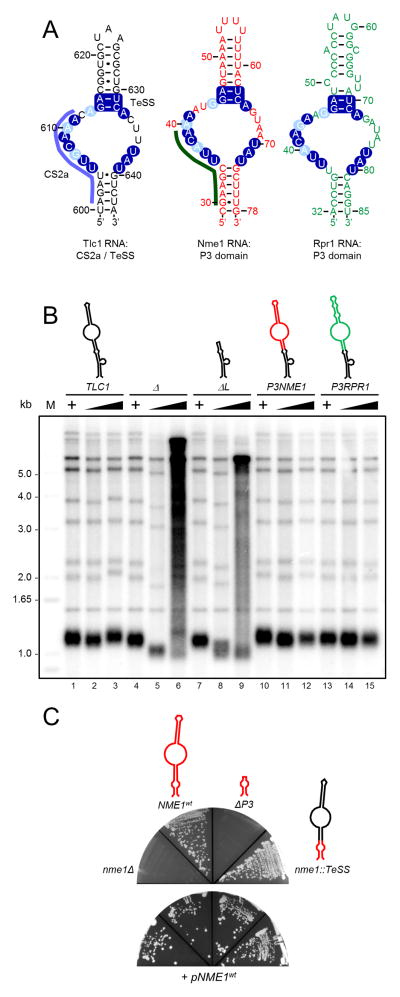
A comparison of the distal end of the stem IVc of the yeast telomerase RNA, a substructure that harbors a highly conserved sequence element CS2a and also includes TeSS (Gunisova et al., 2009; Laterreur et al., 2013), with the P3 domains in the Nme1 and Rpr1 RNAs shows striking similarities (Figure 3A and Figure S2A). These include a large internal loop flanked by two stems and the most critical nucleotides for function of the two P3 domains are also conserved in CS2a/TeSS (Figure S2A and Ziehler et al., 2001). The area with the most sequence conservation between the P3 elements and TeSS corresponds in fact to the conserved sequence element CS2a (Gunisova et al., 2009) and analogous similarities in this substructure can be found in a number of yeast species, including S. pombe and H. polymorpha (Figure S2B).
Figure 3. The CS2a/TeSS domain is a P3-like domain in Tlc1.
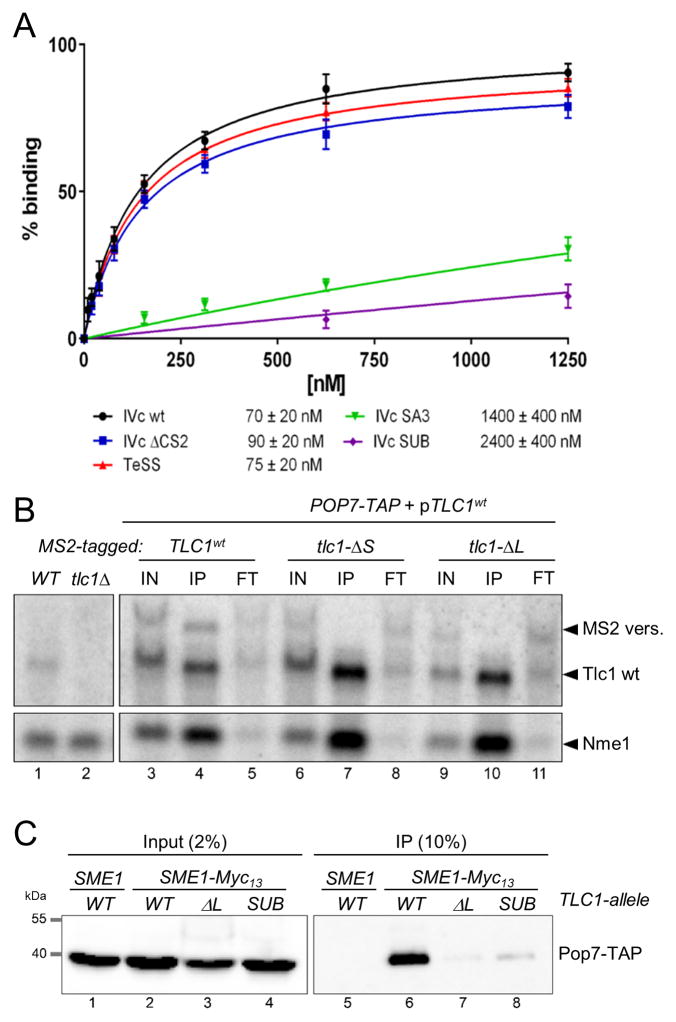
(A) Schematic representation of the TeSS structure at the distal end of stem IVc of Tlc1 (left), the P3 domain of Nme1 RNase MRP RNA (middle) and the P3 domain of the Rpr1 RNase P RNA (right). Dark blue shading: identical nucleotides, light blue: purines and pyrimidines are conserved. Purple blue line on left of Tlc1: CS2a (Conserved Sequence element 2a; Gunisova et al., 2009). Dark green line on Nme1 RNA: nucleotides protected by Pop6/Pop7 binding (Perederina et al., 2007). For more details, see Figure S2A). (B) Telomere length analyses in strains harboring the indicated TLC1 alleles. TLC1: wild type; Δ: tlc1Δ; ΔL: tlc1-ΔL allele that lacks the TeSS/P3 domain (see Figure S4); P3NME1: tlc1::P3NME1, the TeSS/P3 domain in the Tlc1 RNA was replaced with the one from the Nme1 RNA; P3RPR1; tlc1::P3RPR1, the TeSS/P3 domain in Tlc1 was replaced with the one from the Rpr1 RNA. +: Strain carried a wt TLC1 gene on a URA3 plasmid. Black wedges indicate outgrowth of strains after loss of the plasmid borne TLC1 gene; last lane reflects growth for 110 generations. Schematic structures on top are color coded as the nucleotides in panel (A). (C) Growth assays of cells that contain the indicated NME1 alleles: nme1Δ, complete deletion of NME1; ΔP3: nme1-ΔP3 allele that lacks the P3 domain; nme1::P3TLC1 the NME1 P3 domain was replaced with the TLC1 TeSS/P3 domain. Bottom plate: all strains contained a wt NME1 gene on a plasmid with the URA3 marker. Top plate: growth of cells after loss of the URA3, NME1 containing plasmid. Schematic structures and color coding as in panel A.
These striking similarities between the P3 domains in the RNAs of yeast RNases P/MRP and CS2a/TeSS in the telomerase RNA thus raised the possibility that CS2a/TeSS is in fact a P3-like domain. We thus asked whether the P3 domains in the Nme1 or the Rpr1 RNAs could replace the Tlc1 CS2a/TeSS (Figure 3B). In cells that lack Tlc1 altogether or that harbor a tlc1 allele in which the CS2a/TeSS is missing, telomeres shorten upon outgrowth, cells enter crisis and eventually, telomerase-independent mechanisms maintain telomeric DNA, as shown previously (Figure 3B, lanes 6 and 9 and (Singer and Gottschling, 1994; Gunisova et al., 2009)). Telomeres in cells carrying the hybrid Tlc1 RNAs with either the Nme1 P3 or the Rpr1 P3 in place of the CS2a/TeSS did not change in length (Figure 3B, lanes 12 and 15). These cells also grew like wt cells for extended periods, indicating that telomerase function with the hybrid RNAs is indistinguishable from that with a wt Tlc1 RNA. In addition, the CS2a/TeSS domain of the Tlc1 RNA can replace the P3 domain in the essential Nme1 RNA without effects on cell growth (Figure 3C).
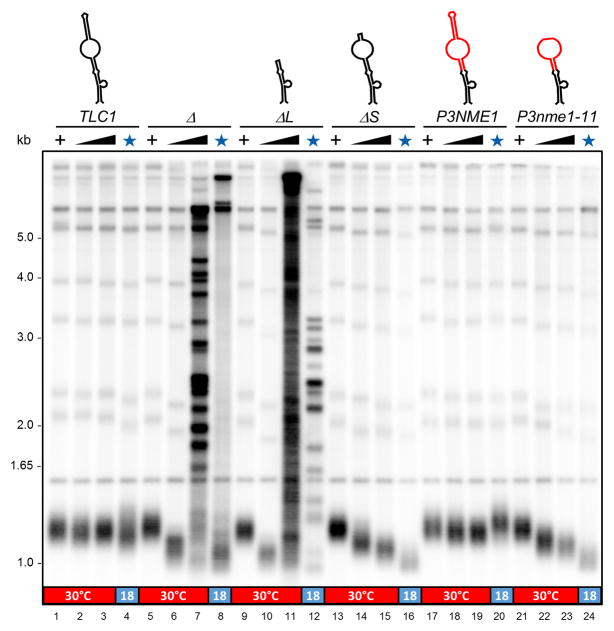
In order to demonstrate the functionality of a P3 domain on Tlc1 in vivo, we took advantage of a known conditional allele, nme1-11, that maps to the P3 domain in the Nme1 RNA (Shadel et al., 2000). The Nme1-11 RNA lacks the distal stem on P3 (nts. 45–64 in Figure 3A) and cells expressing the nme1-11 allele grow at 30°C, but do so very poorly at 17°C (Shadel et al., 2000). We thus constructed the nme1-11 allele into the hybrid Tlc1-P3NME1 RNA by deleting the corresponding nucleotides. If the function of the P3 domain of Nme1 in the telomerase RNP is affected by the mutation in the same way as in the RNase MRP, the tlc1::P3nme1-11 allele should cause a telomerase deficiency and a temperature-dependent shortening of telomeres. Cells harboring the Tlc1-P3Nme1wt RNA maintain normal length telomeres that are very similar as those in cells with a wt Tlc1 RNA, whether cells were grown at 30°C or 18°C for 110 generations (Figure 4, lanes 2–4 and 18–20). Cells that harbor the Tlc1-P3nme1-11 hybrid RNA and which are grown at 30°C manage to maintain telomeres, albeit at slightly shorter than wt lengths (Figure 4, lanes 22–23). These same cells grown at 18°C for 110 generations exhibit extremely short telomeres, significantly shorter than when grown at 30°C (Figure 4, compare lanes 23 with 24). We also constructed the equivalent of what the nme1-11 allele is in NME1 in the TeSS domain of TLC1 by deleting nucleotides 616–630 (Figure 3A), yielding the mutant allele called tlc1-ΔS (previously called SL-del; Gunisova et al., 2009; see predicted structure in Figure S4A). Remarkably, in cells expressing this Tlc1-ΔS RNA, a comparable temperature dependent decrease of telomere size is observed (Figure 4, compare lane 15 with 16). We conclude that the CS2a/TeSS domain in the Tlc1 RNA is a P3-like domain and functions analogously to the P3 domains in RNase P/MRP RNPs in vivo.
Figure 4. A cold sensitive allele of the NME1 P3 domain confers cold-sensitive telomere shortening.
Telomere length analyses in strains harboring the indicated TLC1 alleles. Lanes 1–4: TLC1; lanes 5–8: tlc1Δ; lane 9–12: tlc1-ΔL; lanes 13–16: tlc1-ΔL; lanes 17–20: tlc1-P3NME1; lanes 21–24: tlc1-P3nme1-11. Schematics of stem IVc structures and color coding as in Figure 3 (see also Figure S4A). +: Strain carried a wt TLC1 gene on a URA3 plasmid. Black triangles: growth of cells after loss of the wt TLC1 gene at 30°C for 30 and 110 generations. Lanes with the blue star: strains were grown at 18°C for 110 generations. The red/blue bar at bottom indicates the temperatures the respective strain was grown at.
The Pop6/Pop7 proteins bind directly to the Tlc1 P3 in vitro and in vivo
Previously, Pop6/Pop7 binding to the P3 domain of the Nme1 RNA was analyzed by in vitro reconstitution experiments, crosslinking, and structural analyses (Perederina et al., 2007; Perederina et al., 2010; Hipp et al., 2012; Khanova et al., 2012). Incubation of recombinant Pop6/Pop7 heterodimer with an RNA fragment mimicking the complete stem IVc of Tlc1 resulted in binding with an apparent Kd of about 70 nM (Figure 5A, Figures S3B, S3C), comparable to the P3 domain of Nme1 (about 150 nM, Perederina et al., 2007). Deleting portions of stem IVc of Tlc1 that have been described to be involved in Est1 association had no effect on Pop6/Pop7 binding (Figure 5A, mutants IVc ΔCS2 and TeSS, see Figure S3A for predicted structures). However, when the distal portion of stem IVc was mutated to interfere with the formation of the large internal loop (IVc SA3) or when the CS2a sequence was mutated (IVc SUB; previously called CS2a-sub, Gunisova et al., 2009), Pop6/Pop7 binding was dramatically reduced (Figure 5A, Figures S3B, S3C). These findings parallel the fact that when these latter two alterations were incorporated into the Tlc1 RNA in vivo, telomerase function at telomeres was completely lost (Gunisova et al., 2009; Laterreur et al., 2013) and telomerase activity recovered from these strains was dramatically reduced (Laterreur et al., 2013, and Figure S4B).
Figure 5. Binding characteristics of the Pop6/Pop7 heterodimer to the Tlc1 P3 domain in vitro and in vivo.
(A) Binding curves and apparent Kds of the Pop6/Pop7 heterodimer binding to indicated RNA oligonucleotides (see predicted structures, complete binding curve and gel assays in Figure S3). (B) Northern blot analysis of co-immunoprecipitated RNAs using IgG beads and extracts of strains that harbored TAP-tagged Pop7, a wt Tlc1 RNA and a 400 nt longer MS2-tagged Tlc1 RNA. The latter either contained a wt stem IVc, lanes 3–5; lacks the most distal stem-loop, MS2-tlc1-ΔS: lanes 6–8; or lacks the TeSS/P3 completely, MS2-tlc1-ΔL, lanes 9–11. IN: input (2.5%); IP: immunoprecipitates (10%); FT: flowthrough (2.5%). (C) Western blot of input (left) and immunoprecipitates (right) from strains harboring a Myc13-tagged Sme1 protein and Pop7-TAP. TCL1 alleles as indicated. See Figure S4A for predicted structures of the stem IVc in these mutated RNAs.
In order to determine the Pop6/Pop7 binding site in vivo, we engineered cells that expressed two versions of the Tlc1 RNA, one wt, and the other containing the MS2 stems near the 3′-end such that the two RNAs can be distinguished on northern blots by size. In these cells the Pop6 or Pop7 protein carried the TAP tag and we used extracts for RNA co-IPs (Figure 5B and see Figure S4A for the predicted RNA structures of all stem IVc alleles). When both Tlc1 RNAs had a wt stem IVc sequence, both RNAs were found in the precipitates of Pop7-TAP targeted immunoprecipitations (Figure 5B, lane 4), even though the MS2-version of the Tlc1 RNA was expressed at slightly lower levels (Figure 5B, lane 3). In addition and as expected, the Nme1 RNA was also immunoprecipitated. We then either completely (tlc1-ΔL) or partially (tlc1-ΔS) deleted the P3-like domain from stem IVc of the MS2-containing RNA (Figure S4A). Using these alleles in the same experiment as above showed that these versions of the MS2-tagged RNAs were not immunoprecipitated anymore, while the wt RNA remained in the precipitates just as the Nme1 RNA did (Figure 5B, lanes 7 and 10). Virtually identical results were obtained with Pop6-TAP as target protein (Figure S5A). As an independent confirmation of these association data, we also used co-IP experiments in a strain that contained a Myc-tagged Sme1 that is part of the Sm7-complex binding Tlc1 near its 3′-end (Seto et al., 1999). When we used anti-Myc antibodies for immunoprecipitations with extracts from a strain expressing this Sme1-Myc13 protein and a wt Tlc1 RNA, the Pop7-TAP protein co-immunoprecipitated (Figure 5C, lane 6). However, Pop7-TAP was virtually absent when the cells harbored the Tlc1-ΔL RNA (Figure 5C, lane 7) or the Tlc1-SUB RNA (Figure 5C, lane 8). Identical results were obtained when strains expressed Pop6-TAP (Figure S5B). The reduced detection of Pop7-TAP or Pop6-TAP was not due to a loss of the mutated Tlc1 RNAs from the strains, as very comparable amounts of Tlc1-ΔL or Tlc1-SUB RNAs can be detected in these strains (Figure S5C), as well as in previously reported strains (see Laterreur et al., 2013).
These data demonstrate that the Pop6/Pop7 heterodimer binds the telomerase RNA P3-like domain in vitro and in vivo. Furthermore, the Pop1 protein is also associated with active telomerase (Figure 2C) as it is with active RNase P/MRP RNPs. Collectively, this suggests that all three RNPs contain an analogous P3 module that consists of a highly related structure on the three RNAs plus the Pop1 and Pop6/Pop7 proteins that associate with this structure.
The Tlc1 P3 domain is required for stable Est1 and Est2 association with the Tlc1 RNA
We next wished to investigate the function of the Tlc1 P3 domain in the telomerase RNP. We surmised that if its function in telomerase was analogous to that in the RNase P/MRP RNPs, the P3 domain may be required for holoenzyme integrity. Therefore, we performed co-immunoprecipitation experiments with extracts from cells that contained an HA-tagged Sme1 protein that is part of the Sm7 complex binding the Tlc1 RNA near its 3′-end, various forms of the Tlc1 RNA, and in which both Est1 as well as Est2 proteins contained Myc tags. When we used anti-HA antibodies to pull down wt Tlc1 RNA, Est1 and Est2 could be readily detected in the precipitates, as expected (Figure 6A, lane 6; (Tucey and Lundblad, 2014)). However, when cells contained the Tlc1-ΔL or Tlc1-SUB RNAs, there was no more Est1 detectable in the precipitates and the amount of Est2 was significantly reduced (Figure 6A, lanes 7 and 8, respectively). As internal control for IP efficiency, the amounts of the three Tlc1 RNA versions in the precipitates were comparable (Figure 6A, lower northern panel). Therefore, the Tlc1 P3 domain is required for a stable association of Est2 and particularly Est1 on the Tlc1 RNA. Consistent with these results, immuno-precipitations performed with extracts from strains with a tagged Pop6 protein also yielded strong Est1 and Est2 co-enrichment, similar to what is observed with the tagged Sme1 protein (Figure S5D).
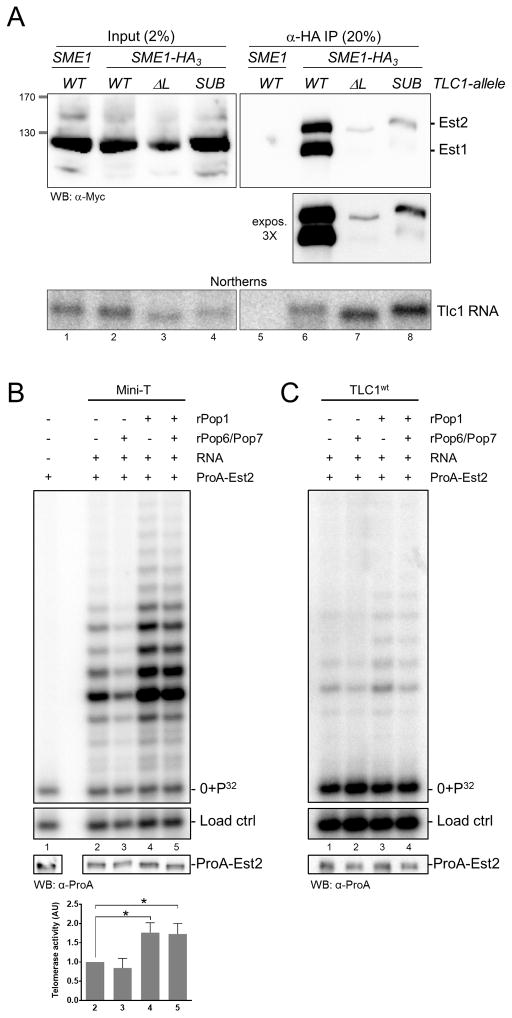
Figure 6. The Tlc1 TeSS/P3 domain is required for telomerase RNP integrity and function in vivo and in vitro.
(A) Western blots of input (left) and IPs (right) with extracts from strains harboring a HA3-tagged Sme1 protein and in which the Est1 as well as Est2 proteins carried a Myc12-tag. TLC1 alleles as indicated. Below, Northern blot of RNA extracted from equal amounts of the inputs (left) or IPs (right) as used for the Western, and which was hybridized to a TLC1-specific probe. (B) Telomerase activity assays with in vitro reconstituted RNPs using the Mini-T RNA (Zappulla et al., 2005). ProA-Est2 levels after the RRL reaction are indicated below. Recombinant rPop6/Pop7 heterodimer and/or rPop1 proteins were added as indicated. After the RRL assay, telomerase was enriched using IgG beads and activity was assayed as in Figure 1. Bar graph depicts quantified telomerase activities standardized to the one obtained without Pop protein addition (lane 2; * p < 0.05 as determined in an unpaired t-test with Welch’s correction). (C) Telomerase activity assays with in vitro reconstituted RNPs using a full-length wt Tlc1 RNA. Indications as in (B).
Recombinant Pop1 allows telomerase activity reconstitution in vitro
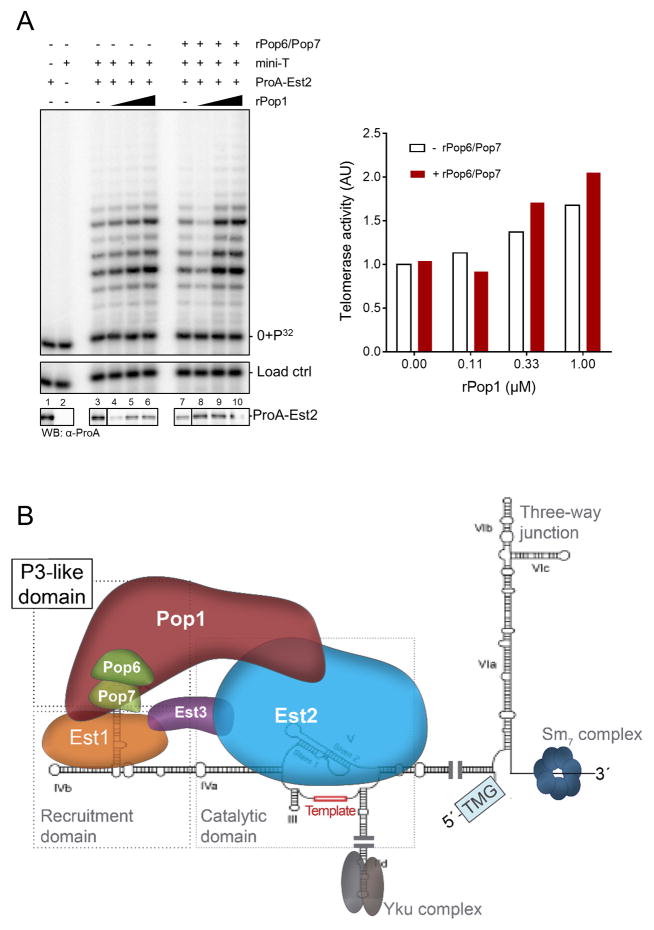
In vitro reconstitution of robust telomerase activity with a wt Tlc1 RNA has not been reported, but telomerase activity can be reconstituted using a shortened Tlc1 RNA called mini-T, combined with a transcription-translation system in rabbit reticulocyte lysates (RRL; Zappulla et al., 2005). If binding of the Pop1/Pop6/Pop7 proteins to the P3 domain is important for a stable association of Est2 on the RNA in vitro as well, their addition to the system should increase reconstituted RNP stability and detectable activity. We tested this possibility directly by setting up the in vitro reconstitution system using the mini-Tlc1 RNA as well as the wt Tlc1 RNA. Of note, the mini-Tlc1 RNA still includes the Tlc1 P3 domain studied here. When recombinant Pop1 or when all three Pop1/Pop6/Pop7 proteins were included in the reactions, telomerase activity with the mini-Tlc1 RNA increased by about 60% (Figure 6B, bottom panel). Moreover, in the presence of the Pop1 protein, telomerase activity can now be detected with wt Tlc1 RNA (Figure 6C). These increases in activity observed for the mini-Tlc1 or the wt Tlc1 RNA are not due to elevated levels of Est2 (Figure 6B, 6C bottom panels). Note that the addition of the Pop6/Pop7 proteins to the RRL mix (Figure 6B; lane 3) does not lead to a reproducible decrease in telomerase activity (see below, Figure 7A, lane 7). However, if Pop1 is added together with the Pop6/Pop7 proteins, the latter may increase the efficiency of Pop1 association with the P3 domain. We tested this possibility by setting up in vitro RRL-mediated telomerase reconstitution with mini-T RNA and varying concentrations of Pop1 in the presence or absence of Pop6/Pop7 (Figure 7A). While the stimulation of telomerase activity by Pop1 could be detected in both conditions, the stimulatory effect was about 20% stronger in the presence of Pop6/Pop7 in this assay (Figure 7A, right). These results therefore show that, as is the case for the in vivo activity, in vitro reconstitution of telomerase activity requires at least the Pop1 protein. Given that minimal telomerase activity is thought to depend on Est2 and Tlc1 RNA only, these data directly demonstrate the positive function of the Tlc1 P3 domain for reconstituting telomerase activity in vitro, a result that is entirely consistent with its function in vivo.
Figure 7. Model for core structure of the active yeast telomerase RNP.
(A) Telomerase activity assays with in vitro reconstituted RNPs using the Mini-T RNA, increasing amounts of Pop1 with or without Pop6/Pop7 addition as indicated. Final Pop1 concentrations are 0 (lanes 3, 7); 0.11 μM (lanes 4, 8); 0.33 μM lanes 5, 9) and 1 μM lanes 6, 10). Quantification of relative telomerase activities is indicated on the right. No additional protein addition (lane 3) was set as 1; data are averages from two experiments. (B) Binding of the Pop6/Pop7 and Pop1 proteins are modeled on top of the newly identified TLC1 P3 domain of the yeast telomerase RNA. The presence of the Tlc1 P3 domain provides a protein binding platform that keeps the essential Est1 and Est2 proteins on the active RNP. Note that physical interactions between the Pop-proteins and Est1 or Est2 as well as the placement of Est3 in the model are hypothetical.
Discussion
Collectively, our results demonstrate that the yeast telomerase RNA contains an essential domain that is structurally and functionally equivalent to the P3 domains in the Rpr1 and Nme1 RNAs of yeast RNases P/MRP. In each of these divergent catalytic RNPs, the P3 RNA domain serves as binding platform for the Pop6/Pop7 and Pop1 proteins and in all three cases, the association of these proteins is required to stabilize the respective RNP. Consistently, all three P3 domains are fully functionally interchangeable (Figure 3 and Lindahl et al., 2000). All these components are absolutely essential for cell viability, given that they all are required for RNase P/MRP stability and enzymatic activity (Esakova and Krasilnikov, 2010).
In the case of the telomerase RNP, previous experiments had shown that complete removal of what we now call Tlc1 P3, as well as the substitution of sequences in the P3 internal loop (the tlc1-ΔL and the tlc1-SUB alleles) caused a complete loss of telomerase activity at telomeres in vivo (Gunisova et al., 2009). Moreover, the activity of immunopurified telomerase containing Tlc1 RNAs with these mutations was markedly reduced (Figure S4B). It is intriguing that the association site for the recruitment factor Est1 is in very close proximity of the P3 domain described here (Figure 7B; Seto et al., 2002). In this respect, our results also show that a loss of the P3 domain also causes a complete loss of Est1 and, to a large extent, the Est2 proteins from the RNA (Figure 6A), which explains the in vivo phenotype of these mutations. Several experiments indicate that it is the association of proteins on the Tlc1 P3 domain that stabilizes Est1 and Est2 proteins on the RNP and not the RNA structure alone. First, the presence or absence of the Est1 protein does not affect the level of telomerase activity as assayed in vitro, but mutations in the RNA that cause a loss of the P3 function cause a strong reduction in detectable activity (Figure S4B, Lingner et al., 1997; Laterreur et al., 2013). Second, the nme1-11 allele causes a cold sensitive phenotype because of a reduced binding of Pop1 to RNase MRP (Shadel et al., 2000). When this allele was engineered into the Tlc1-P3Nme1 allele, or when we used the corresponding deletion of the P3 region of the TLC1 gene itself, we observed a cold-sensitive shortening of telomeres, which strongly suggested that an analogous loss of Pop1 binding from the Tlc1 RNA causes a decrease in telomerase activity in vivo (Figure 4, Figure 5B). Third, the stimulation of the reconstituted telomerase activity with the mini-T RNA and the generation of detectable activity with wt Tlc1 RNA was dependent on the addition of the Pop1 protein and occurred in the absence of Est1 (Figures 6B, 6C). We therefore conclude that the Pop1-Pop6/Pop7 P3 module is an essential component of budding yeast telomerase and that it is required for stable Est1 and Est2 association to the active RNP.
The association of the Pop1 and Pop6/Pop7 proteins with telomerase initially was detected in partially purified active telomerase RNP preparations (Figure 1 and S1A). These assays also correctly identified known telomerase components, such as Est1, Est2, or Sm7-complex components (Smd2, Smb1, Smd3). However, we did not detect either of the Yku-complex proteins (Yku70 or Yku80) nor Est3 in our experiments. It has been estimated that there are only 10–30 telomerase RNPs per individual yeast cell (Mozdy and Cech, 2006) and given that we had to start with large cell cultures, it is possible that during RNP affinity purification, weakly associated proteins may have been reduced to levels below detection.
Previously, it was suggested that the role of Pop6/Pop7 in RNases P/MRP is limited to the stabilization of the binding of Pop1, which serves as the major scaffolding element. Of note, the binding of Pop1 to the Nme1 and Rpr1 RNAs does not require the presence of the Pop6/Pop7 heterodimer, but the presence of the latter strengthens Pop1 binding (Fagerlund et al. 2015). The in vitro reconstitution telomerase experiments with the mini-T RNA are in complete agreement with this proposed scenario (Figure 7A). It may thus be that the relatively high concentration of the components in the in vitro assays may partly alleviate the necessity of the additional stabilization of Pop1 binding by the Pop6/Pop7 heterodimer. In previous reconstitution experiments with the mini-T RNA, telomerase activity was limited to only one round of repeat addition (Zappulla et al., 2005; Figure 6B, lane 2). When Pop1 is added to these experiments, we now observe addition of at least two repeats (Figure 6B, lanes 4–5). It is therefore possible that the Tlc1 P3 domain is also important for a limited repeat addition processivity that has been suggested to occur on very short telomeres in vivo (Chang et al., 2007). However, we cannot exclude the possibility that these additional extension reactions are due to the increased stability of the complex.
A recent study on the composition of telomerase in relation to the cell cycle suggests that the association of the Est1 and Est2 proteins changes and that these proteins are only a very limited time together on the RNP (Tucey and Lundblad, 2014). While it is unknown how the proposed associations and dissociations of the proteins are regulated, given that the Tlc1 P3 domain affects the stability of both on the RNP, it could be the target of regulatory pathways in this respect. (Figure 6A, Figure 7B).
The structure of the bacterial RNase P is stabilized by a network of tertiary RNA-RNA interactions between auxiliary RNA elements (Reiter et al., 2010). During the evolutionary transition to the large protein-rich eukaryotic RNases P/MRP, most of these RNA elements have been lost and their role apparently was delegated to newly acquired protein components. It was suggested that the P3 RNA domain, a feature that is unique to the eukaryotic RNases P/MRP, provided an anchoring point for these proteins (Fagerlund et al., 2015 and references therein). It is quite remarkable that this module, complete with the associated proteins Pop6, Pop7, and Pop1, has found its way into the telomerase RNP, a totally unrelated and structurally distinct entity but in which the P3 domain plays a similar role. Curiously, the telomerase RNP permits that the stem IVc arm that comprises the P3 domain and the Est1 binding site be relocated to other locations on the RNA and it can even be supplied in trans (Zappulla and Cech, 2004; Lebo et al., 2015). This permissiveness to a permutation of certain parts of the RNP may be a consequence of the proposed modular assembly. It will be interesting to determine how the seemingly identical organizing P3 modules are accommodated in the overall structurally divergent RNPs and how wide-spread is this exchange of large functional modules between otherwise unrelated RNPs.
Experimental Procedures
Strains and Plasmids
Construction and genotypes of yeast strains and plasmid descriptions are provided in the Supplemental Experimental procedures and Supplemental Tables.
Native protein extracts and immunoprecipitations (IPs) for Mass Spectrometry
Native protein extracts for Mass Spectrometry were prepared as described in (Bajon et al., 2015). Detailed procedures can be found in the Supplemental Experimental procedures.
Mass Spectrometry
Samples from 3 independent mass spectrometry-IPs were subjected to trypsin digestion directly on beads and the resulting peptides were separated using a Dionex Ultimate 3000 nanoHPLC system as described in (Drissi et al., 2015). Raw data issued from the MS were processed, searched and quantified using the MaxQuant software package version 1.5.1.2 (Cox and Mann, 2008) employing the yeast W303_ALAV00000000 database (12/7/2012). The settings used for the MaxQuant analysis were as described (Drissi et al., 2015). Specific proteins were defined as those that were present in at least two mass spectrometry-IPs and identified exclusively in the TLC1-MS2 sample. The proteins were then sorted by sum of peak intensity. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Vizcaino et al., 2013) with the dataset identifier PXD003273.
Total protein extracts for co-immunoprecipitations, RNA-IP and northern blot analyses
Total protein extracts were prepared as described in ref Laterreur et al., 2013. Following immunoprecipitation of total protein extracts, RNA was extracted from the beads using a standard phenol-chloroform-isoamyl technique followed by ethanol precipitation in presence of salt and glycogen. Samples were then analyzed by northern blot using a TLC1-specific probe as well as a radiolabeled NME1-specific oligo. For calculating the fractions of the Nme1 and Tlc1 RNA in the immunoprecipitations (Figure 2A and B), the signals for the respective RNAs in the input (IN) and flowthrough (FT) fractions on Northern blots were quantified using a Typhoon FLA9000 apparatus and Quantity One software. IP signal was then calculated by using IN-FT/IN adjusted for loading and expressed as % of total. Values reported were derived from at least three independent experiments each for Pop6-TAP and Pop7-TAP.
Western-blots
After removal of TMG3 from IP beads, input and IP samples were mixed with 2x Laemmli loading buffer. Proteins were then analysed by Western blots using standard procedures. A more detailed description of the experiments can be found in the Supplemental Experimental Procedures.
Cold sensitivity and viability assays
Strains transformed with the various plasmids as indicated (see Supplemental table 2) were streaked on agar plates for extended growth at 18°C or 30°C for 5 and 2 days respectively. For the calculations of numbers of cell divisions, cells in an average size isolated colony were assumed to have divided 20 times. Genomic DNA from cells that have grown for the indicated number of generations were prepared and subjected to telomere length analysis. For the viability assays with hybrid Nme1 RNAs, transformed clones were grown at 30°C. For more details, see Supplemental Experimental procedures.
Telomere length analysis
Genomic DNA isolation was performed using standard phenol/chloroform extraction and southern blots were prepared as described previously (Laterreur et al., 2013). Data were acquired with a Typhoon FLA9000 (GE Healthcare).
In vitro telomerase reconstitution and assay conditions
Telomerase reconstitution assays were performed as reported (Zappulla et al., 2005) with minor modifications (see Supplemental Experimental procedures for details). When specified, RRL reactions were performed with the addition of 1 μM of recombinant proteins Pop6/Pop7 and/or Pop1 as indicated. Yeast telomerase activity assays were carried out as previously described (Friedman and Cech, 1999; Laterreur et al., 2013) with 30% of the IP-beads. In case of native cell extracts 10% of IP-beads were used. Relative activities were quantified by dividing the background corrected intensity of all extension products by the 0+P32 internal control using a Typhoon FLA9000 and Quantity One software. The activity in reactions without any additional proteins was set as 1. For Figure S4B, the obtained activity was also standardized over RNA abundance to obtain a relative telomerase activity (RTA; Laterreur et al., 2013). In this case, the wt RNA was set as 1.
Stem IVc RNAs and recombinant Pop1/Pop6/Pop7 proteins
RNA constructs IVc wt, IVc SUB, IVc SA3, IVc ΔCS2, and TeSS were produced by run-off transcription with T7 RNA polymerase using DNA templates based on synthetic oligonucleotides (Milligan et al., 1987). In vitro transcription and sequences of the template oligonucleotides are detailed in the Supplemental Experimental procedures. Pop 1 and Pop6/Pop7 proteins were expressed in E.coli and purified as previously described (Perederina et al., 2007; Fagerlund et al., 2015).
Gel mobility shift assays and estimation of dissociation constants
Prior to forming complexes with proteins, RNA was heated and refolded by slow-cooling to room temperature. After end-labeling of the refolded RNA, radiolabeled RNA was mixed with cold RNA. In order to estimate the binding constants, protein-RNA complexes were mixed at a 1:1 molar ratio; RNA concentrations varied from 10 nM to 10 μM. Radioactive RNA bands were quantified using PhosphorImager (Molecular Dynamics). For a detailed description of the assays, quantifications and dissociation constants determination, see Supplemental Experimental procedures.
Supplementary Material
Acknowledgments
We thank S. Benjira for help during project setup, D. Engelke, V. Lundblad, M. Schmitt, and D. Zappulla for valuable tools such as numerous yeast strains and plasmids, I. Berezin for help with protein purification, and members of the Wellinger lab for input. We also thank F.-M. Boisvert for use of his MS-facility. The authors declare that they have no competing financial interests. This work was supported by a grant from the Canadian Institutes of Health Research to RJW (CIHR 97874), the Canadian Research Chair in Telomere Biology and support by the CRCHUS to RJW. Support to ASK was provided by National Institutes of Health grant GM085149.
Footnotes
Author Contributions:
Experiments were performed by BL, NL, AP, J.-F.N. and M.-L.D.; manuscript was written by RJW, ASK, BL and NL.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bajon E, Laterreur N, Wellinger RJ. A Single Templating RNA in Yeast Telomerase. Cell Rep. 2015;12:441–448. doi: 10.1016/j.celrep.2015.06.045. [DOI] [PubMed] [Google Scholar]
- Chang M, Arneric M, Lingner J. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 2007;21:2485–2494. doi: 10.1101/gad.1588807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell AS, Lundblad V. Structural Elements Required for Association of the Saccharomyces cerevisiae Telomerase RNA with the Est2 Reverse Transcriptase. Molecular and Cellular Biology. 2004;24:7720–7736. doi: 10.1128/MCB.24.17.7720-7736.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Dandjinou AT, Levesque N, Larose S, Lucier JF, Elela SA, Wellinger RJ. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr Biol. 2004;14:1148–1158. doi: 10.1016/j.cub.2004.05.054. [DOI] [PubMed] [Google Scholar]
- Drissi R, Dubois ML, Douziech M, Boisvert FM. Quantitative Proteomics Reveals Dynamic Interactions of the Minichromosome Maintenance Complex (MCM) in the Cellular Response to Etoposide Induced DNA Damage. Mol Cell Proteomics. 2015;14:2002–2013. doi: 10.1074/mcp.M115.048991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ED, Collins K. Biogenesis of telomerase ribonucleoproteins. RNA. 2012;18:1747–1759. doi: 10.1261/rna.034629.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esakova O, Krasilnikov AS. Of proteins and RNA: the RNase P/MRP family. RNA. 2010;16:1725–1747. doi: 10.1261/rna.2214510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerlund RD, Perederina A, Berezin I, Krasilnikov AS. Footprinting analysis of interactions between the largest eukaryotic RNase P/MRP protein Pop1 and RNase P/MRP RNA components. RNA. 2015;21:1591–1605. doi: 10.1261/rna.049007.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman KL, Cech TR. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes & Development. 1999;13:2863–2874. doi: 10.1101/gad.13.21.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo F, Laterreur N, Cusanelli E, Ouenzar F, Querido E, Wellinger RJ, Chartrand P. Live cell imaging of telomerase RNA dynamics reveals cell cycle-dependent clustering of telomerase at elongating telomeres. Molecular Cell. 2011;44:819–827. doi: 10.1016/j.molcel.2011.09.020. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- Gunisova S, Elboher E, Nosek J, Gorkovoy V, Brown Y, Lucier JF, Laterreur N, Wellinger RJ, Tzfati Y, Tomaska L. Identification and comparative analysis of telomerase RNAs from Candida species reveal conservation of functional elements. RNA. 2009;15:546–559. doi: 10.1261/rna.1194009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp K, Galani K, Batisse C, Prinz S, Bottcher B. Modular architecture of eukaryotic RNase P and RNase MRP revealed by electron microscopy. Nucleic Acids Res. 2012;40:3275–3288. doi: 10.1093/nar/gkr1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Collins K. Control of telomerase action at human telomeres. Nat Struct Mol Biol. 2015;22:848–852. doi: 10.1038/nsmb.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanova E, Esakova O, Perederina A, Berezin I, Krasilnikov AS. Structural organizations of yeast RNase P and RNase MRP holoenzymes as revealed by UV-crosslinking studies of RNA-protein interactions. RNA. 2012;18:720–728. doi: 10.1261/rna.030874.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laterreur N, Eschbach SH, Lafontaine DA, Wellinger RJ. A new telomerase RNA element that is critical for telomere elongation. Nucleic Acids Res. 2013;41:7713–7724. doi: 10.1093/nar/gkt514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebo KJ, Niederer RO, Zappulla DC. A second essential function of the Est1-binding arm of yeast telomerase RNA. RNA. 2015;21:862–876. doi: 10.1261/rna.049379.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebo KJ, Zappulla DC. Stiffened yeast telomerase RNA supports RNP function in vitro and in vivo. RNA. 2012;18:1666–1678. doi: 10.1261/rna.033555.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KW, McDonald KR, Guise AJ, Chan A, Cristea IM, Zakian VA. Proteomics of yeast telomerase identified Cdc48-Npl4-Ufd1 and Ufd4 as regulators of Est1 and telomere length. Nat Commun. 2015;6:8290. doi: 10.1038/ncomms9290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L, Fretz S, Epps N, Zengel JM. Functional equivalence of hairpins in the RNA subunits of RNase MRP and RNase P in Saccharomyces cerevisiae. RNA. 2000;6:653–658. doi: 10.1017/s1355838200992574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cech TR, Hughes TR, Lundblad V. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci U S A. 1997;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livengood AJ, Zaug AJ, Cech TR. Essential regions of Saccharomyces cerevisiae telomerase RNA: separate elements for Est1p and Est2p interaction. Mol Cell Biol. 2002;22:2366–2374. doi: 10.1128/MCB.22.7.2366-2374.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin JW, Tucey TM, Lundblad V. The interaction between the yeast telomerase RNA and the Est1 protein requires three structural elements. Rna. 2012;18:1597–1604. doi: 10.1261/rna.034447.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy AD, Cech TR. Low abundance of telomerase in yeast: implications for telomerase haploinsufficiency. RNA. 2006;12:1721–1737. doi: 10.1261/rna.134706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Perederina A, Esakova O, Koc H, Schmitt ME, Krasilnikov AS. Specific binding of a Pop6/Pop7 heterodimer to the P3 stem of the yeast RNase MRP and RNase P RNAs. RNA. 2007;13:1648–1655. doi: 10.1261/rna.654407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perederina A, Esakova O, Quan C, Khanova E, Krasilnikov AS. Eukaryotic ribonucleases P/MRP: the crystal structure of the P3 domain. EMBO J. 2010;29:761–769. doi: 10.1038/emboj.2009.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccinelli P, Rosenblad MA, Samuelsson T. Identification and analysis of ribonuclease P and MRP RNA in a broad range of eukaryotes. Nucleic Acids Res. 2005;33:4485–4495. doi: 10.1093/nar/gki756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlevsky JD, Chen JJ. It all comes together at the ends: Telomerase structure, function, and biogenesis. Mutation Research. 2012;730:3–11. doi: 10.1016/j.mrfmmm.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Li Y, Honda S, Hoffmann S, Marz M, Mosig A, Podlevsky JD, Stadler PF, Selker EU, Chen JJ. The common ancestral core of vertebrate and fungal telomerase RNAs. Nucleic Acids Res. 2013;41:450–462. doi: 10.1093/nar/gks980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragon A. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010;468:784–789. doi: 10.1038/nature09516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarek G, Marzec P, Margalef P, Boulton SJ. Molecular basis of telomere dysfunction in human genetic diseases. Nat Struct Mol Biol. 2015;22:867–874. doi: 10.1038/nsmb.3093. [DOI] [PubMed] [Google Scholar]
- Schmidt JC, Cech TR. Human telomerase: biogenesis, trafficking, recruitment, and activation. Genes Dev. 2015;29:1095–1105. doi: 10.1101/gad.263863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Livengood AJ, Tzfati Y, Blackburn EH, Cech TR. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes & Development. 2002;16:2800–2812. doi: 10.1101/gad.1029302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Umansky K, Tzfati Y, Zaug AJ, Blackburn EH, Cech TR. A template-proximal RNA paired element contributes to Saccharomyces cerevisiae telomerase activity. Rna. 2003;9:1323–1332. doi: 10.1261/rna.5570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle [published erratum appears in Nature 1999 Dec 23–30;402(6764):898] Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- Shadel GS, Buckenmeyer GA, Clayton DA, Schmitt ME. Mutational analysis of the RNA component of Saccharomyces cerevisiae RNase MRP reveals distinct nuclear phenotypes. Gene. 2000;245:175–184. doi: 10.1016/s0378-1119(00)00013-5. [DOI] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 2003;17:2384–2395. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucey TM, Lundblad V. Regulated assembly and disassembly of the yeast telomerase quaternary complex. Genes Dev. 2014;28:2077–2089. doi: 10.1101/gad.246256.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino JA, Cote RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD. Origin of concatemeric T7 DNA. Nat New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR. Yeast telomerase RNA: A flexible scaffold for protein subunits. Proceedings of the National Academy of Sciences. 2004;101:10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR. RNA as a flexible scaffold for proteins: yeast telomerase and beyond. Cold Spring Harbor Symposia on Quantitative Biology. 2006;71:217–224. doi: 10.1101/sqb.2006.71.011. [DOI] [PubMed] [Google Scholar]
- Zappulla DC, Goodrich K, Cech TR. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nature Structural & Molecular Biology. 2005;12:1072–1077. doi: 10.1038/nsmb1019. [DOI] [PubMed] [Google Scholar]
- Ziehler WA, Morris J, Scott FH, Millikin C, Engelke DR. An essential protein-binding domain of nuclear RNase P RNA. RNA. 2001;7:565–575. doi: 10.1017/s1355838201001996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.