Abstract
Phthalates are frequently used in personal care products and plasticizers and phthalate exposure is ubiquitous in the US population. Exposure to phthalates during critical periods in utero has been associated with a variety of adverse health outcomes but the biological mechanisms linking these exposures with disease are not well characterized. In this study, we examined the relationship of in utero phthalate exposure with repetitive element DNA methylation, an epigenetic marker of genome instability, in children from the longitudinal birth cohort CHAMACOS. Methylation of Alu and long interspersed nucleotide elements (LINE-1) was determined using pyrosequencing of bisulfite-treated DNA isolated from whole blood samples collected from newborns and 9 year old children (n=355). Concentrations of eleven phthalate metabolites were measured in urine collected from pregnant mothers at 13 and 26 weeks gestation. We found a consistent inverse association between prenatal concentrations of monoethyl phthalate, the most frequently detected urinary metabolite, with cord blood methylation of Alu repeats (β(95%CI):−0.14(−0.28,0.00) and −0.16(−0.31,−0.02)) for early and late pregnancy, respectively, and a similar but weaker association with LINE-1 methylation. Additionally, increases in urinary concentrations of di-(2-ethylhexyl) phthalate metabolites during late pregnancy were associated with lower levels of methylation of Alu repeats in 9 year old blood (significant p-values ranged from 0.003 to 0.03). Our findings suggest that prenatal exposure to some phthalates may influence differences in repetitive element methylation, highlighting epigenetics as a plausible biological mechanism through which phthalates may affect health.
Keywords: DNA methylation, phthalate, in utero exposure, Alu repeats, LINE-1 repeats
1. Introduction
Phthalates, diesters of phthalic acid, are used in a variety of common household products leading to near ubiquitous exposure in the U.S. population (Silva et al., 2004; Woodruff et al., 2011). For instance, many scented personal care items contain phthalates as additives; phthalates are also present in plastic products ranging from medical tubing and food packaging to vinyl flooring and children’s toys (CDC, 2009). Phthalates leach easily into the environment; humans are exposed to phthalates mainly through ingestion, inhalation, and dermal contact (Gaspar et al., 2014). Di-(2-ethylhexyl) phthalate (DEHP), a plasticizer, is one of the most widely used phthalates (Heudorf et al., 2007). The phthalate metabolite with the highest urinary concentrations in humans, monoethyl phthalate (MEP), is a metabolite of diethyl phthalate (DEP), a fragrance solvent found in personal care products like shampoos and cosmetics (Silva et al., 2004).
Phthalate exposure in humans has been associated with a number of adverse health outcomes, including asthma, inflammation, poorer birth outcomes, child growth, and sperm quality (Cai et al., 2015; Ferguson et al., 2015; Smarr et al., 2015; Teitelbaum et al., 2012; Whyatt et al., 2014). Several studies have identified adverse effects particularly in response to phthalate exposure during pregnancy (Kim et al., 2011; Valvi et al., 2015a; Whyatt et al., 2014), suggesting that the in utero period may be a critical window of vulnerability.
Epigenetic changes have emerged as a potential biologic mechanism through which in utero exposures can lead to adverse health outcomes later in life. Epigenetic marks, including DNA methylation, non-coding RNA, and histone modifications, influence changes in gene expression without changes in DNA sequence. In particular, DNA methylation of repetitive elements, Alu, and long interspersed nucleotide elements (LINE-1) have been inversely associated with exposure to a variety of chemicals including lipophilic persistent organic pollutants, perfluoroalkyl acids, and particulate matter (Bellavia et al., 2013; Huen et al., 2014; Watkins et al., 2014b). Lower levels of methylation of retrotransposable elements like LINE-1 and Alu have been linked to genomic instability that can influence disease etiology (Ayarpadikannan and Kim, 2014; Su et al., 2012).
Although animal studies have demonstrated consistent effects of phthalates on both global and site-specific methylation (Kang and Lee, 2005; Kostka et al., 2010; Martinez-Arguelles and Papadopoulos, 2015; Pogribny et al., 2008; Wu et al., 2010), data in human populations are more limited. Prenatal urinary phthalate metabolite concentrations have been associated with methylation of imprinted genes H19 and IGF2 (LaRocca et al., 2014) and LINE-1 repetitive elements (Zhao et al., 2015) in placental tissue. Thus far, no studies have examined the association of maternal phthalate exposure during pregnancy with methylation levels in children. The purpose of the present study is to determine the association of in utero phthalate exposure with methylation of Alu and LINE-1 repetitive elements in fetal and child blood collected from participants of the Center for Health Assessment of Mothers and Children of Salinas (CHAMACOS).
2. Materials and Methods
2.1 Study subjects
CHAMACOS is a longitudinal birth cohort study of pregnant Mexican-American women and their children living in the agricultural region of Salinas Valley, California. At the time of enrollment (1999–2000), pregnant women participating in this study were at least 18 years of age, less than 20 weeks gestation, Spanish or English speaking, eligible for low-income health insurance, receiving prenatal care at one of the participating community clinics, and planning to deliver at the local public hospital. We enrolled 601 pregnant women and 526 delivered live, singleton newborns (Eskenazi et al., 2003). DNA methylation was measured in children who had blood samples available for analysis. We analyzed DNA methylation in a total of 355 children (177 girls and 178 boys) at birth and/or age 9 years. Among the 355 children, 134 had methylation data at both time points while 110 children had only cord blood measurements and 111 had methylation measurements only at age 9. Children included in the study did not differ from all children in the cohort by other demographic and exposure variables (e.g. poverty level, marriage status, type of work during pregnancy, alcohol and smoking intake during pregnancy, prenatal exposure to dichlorodiphenyl trichloroethane (DDT), dichlorodiphenyldichloroethylene (DDE), and polybrominated diphenyl ethers (PBDEs)).
Study protocols were approved by the University of California, Berkeley and the Centers for Disease Control and Prevention (CDC) Committees for Protection of Human Subjects. Written informed consent was obtained from all mothers and assent was provided by the children at the 9-year assessment.
2.2 Interview
Trained bilingual, bicultural staff members interviewed CHAMACOS women twice during pregnancy (~13 weeks and 26 weeks gestation) and collected information on sociodemographics, mother’s reproductive and medical history, lifestyle exposures during pregnancy, occupational and residential history, exposures to pesticides and other environmental chemicals, and housing quality.
2.3 Phthalate metabolite measurements
Two urine samples were collected from mothers at the time of interview, aliquoted, barcoded and stored at −80°C at the University of California, Berkeley until samples were shipped on dry ice to CDC for measurement of phthalate metabolites. Prenatal phthalate metabolites concentrations were measured in maternal urinary samples at both time points (13 and 26 weeks gestation) for 334 mothers, at 13 weeks gestation only for 16 mothers and at 26 weeks gestation only for 5 mothers.
Eleven phthalate metabolites were quantified including three low molecular weight (LMW) metabolites (MEP, mono-n-butyl phthalate (MBP), mono-isobutyl phthalate (MiBP)), four high molecular weight (HMW) metabolites of DEHP (mono(2-ethylhexyl) phthalate (MEHP), mono(2-ethyl-5-hydroxyhexyl) phthalate (MEHHP), mono(2-ethyl-5-oxohexyl) phthalate (MEOHP), mono(2-ethyl-5-carboxypentyl) phthalate (MECPP)), and four additional HMW metabolites of other parent phthalates (monobenzyl phthalate (MBzP), mono(3-carboxypropyl) phthalate (MCPP), monocarboxyoctyl phthalate (MCOP), monocarboxynonyl phthalate (MCNP)). Measurements were performed using solid phase extraction coupled with isotope dilution high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry (Silva et al., 2007). Quality control procedures included the use of laboratory and field blanks, calibration standards, and controls with high and low concentrations.
Concentrations below the limit of detection (LOD) with no corresponding instrumental signal were imputed from a log-normal distribution using the “fill-in” method described in Lubin, et al. (2004). Summary measurements (e.g. ΣLMW, ΣHMW, and ΣDEHP) were created as described elsewhere (Zota et al., 2014). Briefly, summary measures were calculated by first dividing concentrations of each metabolite by its molecular weight to generate molar sums, second adding the molar sums for each metabolite in the group, and third multiplying by the average molecular weight of the metabolites in that group to yield measurements expressed in μg/L.
Urinary dilution was accounted for by using either creatinine or specific gravity. Specific gravity was measured with a refractometer (National Instrument Company Inc., Baltimore, MD) while urinary creatinine was determined using a commercially available diagnostic enzyme method (Vitros CREA slides; ortho Clinical Diagnostics, Raritan, NJ). Phthalate metabolite concentrations (μg/L) were divided by creatinine levels (g/L) to yield creatinine adjusted phthalate metabolite concentrations expressed in μg/g creatinine. Specific gravity adjusted concentrations (μg/L) were produced using the formula: Pc=P[(1.024−1)]/(SG-1) where Pc is the specific gravity adjusted concentration, P is the phthalate concentration (μg/L), 1.024 is the median specific gravity of all samples, and SG is the specific gravity for the specific sample (Meeker et al., 2009). Phthalate metabolite concentrations adjusted for urinary dilution were used for descriptive analyses and simple correlation calculations. Previous studies have shown that the use of creatinine or specific gravity adjusted phthalate metabolite concentrations in regression models can introduce bias (Barr et al., 2005). Therefore, in regression models, we used unadjusted phthalate metabolite concentrations and we included maternal creatinine levels as a covariate in the model to adjust for urinary dilution. We chose to report results based on creatinine adjustment for ease of comparison to National Health and Nutrition Examination Survey (NHANES) data (Silva et al., 2004) and because a subset of women (n=77) were missing SG data. However, creatinine levels are known to change over the course of pregnancy (Adibi et al., 2008) and can be influenced by factors like muscle mass and age (Sauve et al., 2015), potentially influencing urinary adjustment of phthalate metabolite concentrations. Thus, we also report a sensitivity analysis accounting for SG instead of creatinine.
2.4 DNA methylation analyses
Blood specimens from CHAMACOS children were collected from umbilical cords (representing fetal blood) shortly after delivery and by venipuncture when children were 9 years old (mean=9.3 years, SD=0.3). Whole blood was collected in BD vacutainers (Becton, Dickinson and Company, Franklin Lakes, NJ) containing no anticoagulant. Following centrifugation of vacutainers, separated components were divided into serum and clot aliquots that were stored at −80°C. DNA was isolated from clots using a QIAamp Blood DNA Maxi kit (Qiagen, Inc., Santa Clarita, CA) as described previously (Holland et al., 2006). EpiTect Bisulfite Conversion Kits (Qiagen, Germantown, MD) were used for bisulfite conversion of 500 ng of DNA that was subsequently eluted into 20 μL Elution buffer. We used a non-CpG cytosine residue as an internal control to confirm bisulfite DNA conversion efficiency (99%). Alu and LINE-1 methylation levels were determined using a Pyromark Q96MD System (Qiagen) for pyrosequencing of PCR-amplified and bisulfite-treated DNA samples (Royo et al., 2007; Yang et al., 2004). The previously published method for Alu as described by Yang et al. (2004) reported only 3 CpG sites, however pyrosequencing read lengths have improved and now allow for determination of an additional CpG site. Here, we utilized methylation levels at all 4 CpG sites for subsequent analyses. Pyro Q-CpG Software (Qiagen) was used to calculate repetitive element methylation (%5-mC). All samples were run in triplicate for each time-point/subject. In order to minimize technical variability, we used several quality assurance procedures such as inclusion of internal standards, repeats, and positive and negative controls. To ensure low batch variability, all sample plates were run on the same day. Furthermore, all plates contained randomized encoded samples from different age groups to avoid experimental bias. The coefficients of variation (CV) for triplicates measures were 5 and 3 % for Alu and LINE-1, respectively. The intraplate CV’s were in a similar range.
2.5 Statistical analyses
We calculated Pearson’s correlation coefficient to examine the correlation of methylation between time points (birth versus age 9 years) and also the correlation between Alu and LINE-1 methylation. All urinary phthalate metabolite measurements were log10 transformed to approximate a normal distribution. To determine the relationship between different phthalate metabolite concentrations, we calculated Pearson’s correlation coefficients using creatinine-adjusted log10 transformed concentrations. Since phthalate metabolite concentrations can vary over time, an intraclass correlation coefficient (ICC) was calculated to determine the temporal variability of measured phthalate metabolite concentrations by subject. The ICC is defined as the ratio of intra-individual variance to total variance (sum of intra- and inter- individual variance). Correlations with absolute values ranging from 0.3 to 0.5 were considered moderate and those with absolute values ranging from 0.5 to 1.0 were considered strong.
We performed mixed effect regression models to determine the relationship of prenatal phthalate exposure with LINE-1 and Alu repeat methylation. Mixed effects models account for the correlated measures of methylation among the 4 adjacent CpG positions and for the triplicate measures at each position per individual, yielding a global estimate for the association of phthalate exposure with DNA methylation and have been utilized in previous studies of repeat element methylation (Byun et al., 2013; Huen et al., 2014). The following model was used:
where Yijk is the methylation level (Alu or LINE-1) for the i-th subject (i=1,..,.355) at the j-th CpG position (j=1,…,4), and the k-th replicate (k=1,…,3). X2,…,Xn and b2,…, bn, represent the covariates and their corresponding slopes for various models including variables like phthalate metabolite and creatinine levels. The sum of b0 and z0i represents the random intercept for the subject i and the sum of b1 and z1ij are the random slope for the i-th subject and the j-th CpG position. eijk is the residual error term.
Separate models were performed for each of the 11 phthalate metabolites and three summary measures (ΣLMW, ΣHMW, and ΣDEHP) during early and late pregnancy at each methylation time point, separately (birth and 9 years). Maternal creatinine levels were included as a covariate in the model to adjust for urinary dilution. We also used univariate models to identify other potential covariates associated with Alu or LINE-1 methylation (i.e. maternal age, maternal smoking during pregnancy). Maternal pre-pregnancy BMI was associated with some phthalate metabolite concentrations and Alu methylation in cord blood and was therefore included as a covariate in cord Alu methylation models. All other variables tested were not significantly associated with both phthalate metabolite concentrations and Alu or LINE-1 methylation and thus were not included in final models. Sensitivity analyses included: a) adjusting for specific gravity instead of creatinine to account for urinary dilution (specific gravity was included as a covariate in the regression model); b) accounting for cell heterogeneity using differential cell count in a subset of cord blood samples and Houseman estimation by minfi in 9 year olds (Yousefi et al., 2015); and c) analyzing phthalate exposure averaged over two prenatal time points (early and late pregnancy). Similar mixed models were also used to assess associations of child sex and age with methylation levels, irrespective of phthalate metabolite concentrations. All statistical analyses were carried out using STATA software, version 12.0 (College Station, TX). P-values less than 0.05 were considered significant and p-values less than 0.10 were reported as marginally significant.
3. Results
Table 1 describes characteristics of CHAMACOS children and their mothers. The majority of mothers were born in Mexico and more than half had lived in the United States for less than 5 years when their children were born. Most women were young, living within 200% of the poverty level, and very few of them smoked during pregnancy. More than half of the mothers were overweight (BMI: 25 – 29.9 kg/m2) or obese (BMI ≥30 kg/m2) prior to conception. Very few of the children were born preterm or with low birthweight. However, like their mothers, greater than 50% of the children were overweight or obese by age 9.
Table 1.
Demographics of CHAMACOS participants and their mothers, 1999–2000
| N | % | |
|---|---|---|
| Child sex | ||
| Boy | 178 | 50.1 |
| Girl | 177 | 49.9 |
| Child gestational age at birth | ||
| ≥ 37 weeks | 336 | 94.6 |
| 34-36 weeks | 19 | 5.4 |
| Child birthweight | ||
| Low birthweight (<2500g) | 344 | 3.1 |
| Normal birthweight (≥2500g) | 11 | 96.9 |
| Child obesity Status at 9 years | ||
| Normal (≤85th percentile) | 118 | 45.2 |
| overweight (>85th, < 95th percentile) | 39 | 14.9 |
| obese (≥95th percentile) | 104 | 39.8 |
| Maternal age at pregnancy | ||
| 18-24 | 157 | 44.4 |
| 25-29 | 111 | 31.4 |
| 30-34 | 55 | 15.5 |
| 35-45 | 31 | 8.8 |
| Number of years mother lived in US at pregnancy | ||
| Less than 1 year | 89 | 25.1 |
| 2 to 5 years | 98 | 27.6 |
| 6 to 10 years | 81 | 22.8 |
| 11 or more years | 49 | 13.8 |
| Entire Life | 38 | 10.7 |
| Maternal Prepregnancy BMI | ||
| Underweight (<18.5 kg/m2) | 3 | 0.9 |
| Normal (18.5 – 24.9 kg/m2) | 126 | 36.4 |
| overweight (25 – 29.9 kg/m2) | 137 | 39.6 |
| obese (≥30 kg/m2) | 80 | 23.1 |
| Mother Smoked During Pregnancy | ||
| No | 334 | 94.1 |
| Yes | 21 | 5.9 |
3.1 Phthalate Exposure
The distribution of creatinine-adjusted urinary phthalate metabolite concentrations measured in maternal samples collected at 13 and 26 weeks gestation are shown in Table 2. Phthalate metabolite concentrations in CHAMACOS mothers were comparable to those of pregnant women from the National Health and Nutrition Examination Survey (Woodruff et al., 2011). Detection frequencies ranged from 90% for MEHP to 99.7% for MEP, demonstrating a near ubiquitous exposure to phthalates in CHAMACOS participants. MEP was the metabolite with the highest concentrations at both time points. MEP and MBzP had the highest ICCs suggesting that exposures to their parent compounds DEP and benzylbutyl phthalate, respectively, were relatively stable between the two pregnancy time points but varied more broadly between the two time points for other phthalates. Most phthalate metabolites were at least moderately correlated with each other (Supplemental Table 1). Among LMW metabolites, MBP and MiBP were more strongly correlated with each other (r=0.53, p<0.005) than with MEP. DEHP metabolites were all highly correlated with each other (r’s from 0.72 to 0.95; p<0.005 for all six pairwise correlations). The non-DEHP HMW metabolites were moderately correlated with each other (r’s from 0.35 to 0.60) except for MCNP and MCOP, which were more strongly correlated (r=0.70, p<0.005).
Table 2.
Distribution of urinary concentrations of phthalate metabolites in CHAMACOS mothers during pregnancy (1999–2000)
| Phthalate metabolite (μg/g creatinine) | %>LODa | 13 weeks (n=350) | 26 weeks (n=339) | ICC (95%CI) | ||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| median(IQR) | min | max | median(IQR) | min | max | |||
| LMW | ||||||||
| MEP | 99.7 | 167.2(70.6-447.8) | 10.9 | 41791.0 | 166.1(65.6-396.3) | 0.6 | 9909.7 | 0.42(0.34,0.51) |
| MBP | 99.3 | 18.4(9.3-38.3) | 0 | 428.6 | 24.5(12-43.5) | 1.2 | 1282.2 | 0.02(0.00,0.92) |
| MiBP | 93.5 | 2.4(1.1-4.4) | 0 | 82.6 | 2.8(1.5-5.2) | 0 | 254.0 | 0.08(0.02,0.26) |
| ΣLMW | 220.2(107.0-533.9) | 14.2 | 45921.0 | 224.6(114.7-476.4) | 8.9 | 10885.0 | 0.42(0.33,0.51) | |
| DEHP | ||||||||
| MEHP | 89.6 | 3.1(1.4-6.7) | 0 | 272.3 | 3.7(1.9-6.9) | 0 | 104.4 | 0.06(0.01,0.30) |
| MEHHP | 99.4 | 12.8(6.6-24.6) | 0.7 | 846.7 | 15.9(8.7-28.6) | 0.2 | 865.7 | 0.07(0.01,0.28) |
| MEOHP | 98.3 | 8.7(4.7-15.9) | 0.1 | 630.2 | 12.6(6.9-22.2) | 0.1 | 651.6 | 0.06(0.01,0.28) |
| MECPP | 100.0 | 22.0(13.8-38) | 1.3 | 1135.6 | 26.9(17.4-48.5) | 5.2 | 1222.1 | 0.05(0.01,0.31) |
| ΣDEHP | 45.7(27.8-84.8) | 4.7 | 2798.3 | 59.5(35.3-104.6) | 7.9 | 2789.2 | 0.06(0.01,0.29) | |
| HMW | ||||||||
| MBzP | 98.3 | 6.4(3.3-12.4) | 0.3 | 133.0 | 7.6(4.4-13.9) | 0 | 168.6 | 0.32(0.23,0.42) |
| MCPP | 90.5 | 1.8(1.0-2.9) | 0 | 56.6 | 2.1(1.2-3.5) | 0 | 16.8 | 0.11(0.04,0.26) |
| MCOP | 96.4 | 2.8(1.7-4.4) | 0 | 162.3 | 3.2(1.9-5.1) | 0 | 33.2 | 0.01(0.00,1.00) |
| MCNP | 96.1 | 1.7(1.0-2.5) | 0.1 | 51.2 | 1.9(1.2-2.8) | 0 | 16.2 | 0.17(0.09,0.29) |
| ΣHMW | 66.0(37.4-107) | 8.4 | 2814.5 | 81.3(48.8-130.2) | 11.5 | 2807.6 | 0.06(0.01,0.29) | |
All measures were creatinine adjusted.
ICC - Intraclass Correlation Coefficient
LMW - low molecular weight
HMW - high molecular weight
LODs in μg/L are: MEP 0.6, MBP 0.4, MiBP 0.2, MEHP 0.5, MEHHP 0.2, MEOHP 0.2, MECPP 0.2, MBzP 0.3, MCPP 0.2, MCOP 0.2, MCNP 0.2
3.2 DNA Methylation
The distribution of Alu and LINE-1 methylation in cord and 9-year old blood collected from CHAMACOS boys and girls are shown in Table 3. Alu and LINE-1 methylation were slightly higher in boys compared to girls (Huen et al., 2014). This difference reached statistical significance for LINE-1 at both time points (p=0.02 and <0.0005 for cord and 9 year blood, respectively) and was marginally significant for Alu methylation in cord blood only (p=0.07). Mean Alu and LINE-1 methylation were slightly lower in 9-year old blood compared to cord blood but this trend was no longer significant after adjusting for cell composition. As was previously reported (Huen et al., 2014), Alu and LINE-1 methylation were not correlated with each other at either time point. Likewise, methylation in cord blood was not correlated with methylation in 9-year old blood for either repetitive element (Huen et al., 2014).
Table 3.
Alu and LINE-1 methylation in CHAMACOS boys and girls
| Mean(SD) %5mC
|
|||
|---|---|---|---|
| Boys | Girls | All | |
| Alu | |||
| Birtha | 25.38(0.61) | 25.22(0.7) | 25.3(0.66) |
| 9yrb | 25.27(0.81) | 25.24(0.6) | 25.26(0.71) |
| LINE-1 | |||
| Birth | 79.03(1.35) | 78.63(1.43) | 78.83(1.41) |
| 9yr | 78.88(1.27) | 78.03(1.26) | 78.42(1.33) |
Of the 244 cord blood samples, 122 were from boys and 122 were from girls.
Of the 245 9 year old blood samples, 114 were from boys and 131 were from girls.
3.3 Phthalates and DNA methylation
For exposure during early pregnancy, we observed a consistent trend of lower cord blood methylation (both Alu and LINE-1) with increases in phthalate metabolite concentrations (Table 4) but only a few of these associations reached statistical significance. The strongest association was between the LMW metabolite MEP and Alu methylation (β(95%CI):−0.14(−0.28,−0.00)) and a similar negative though non-significant trend was seen with LINE-1 methylation (β(95%CI):−0.23(−0.53,0.06)). The significant association of the LMW sum with Alu methylation is most likely driven by the relationship with MEP. The other two LMW metabolites, MBP and MiBP, were also negatively associated with LINE-1 methylation in cord blood. In contrast to what was observed for cord blood methylation, phthalate metabolite concentrations of MEP were positively associated with LINE-1 methylation in 9 year old blood (β(95%CI):0.59(0.24,0.94); Figure 1). We did not observe any other significant associations of phthalate metabolite concentrations during early pregnancy with Alu or LINE-1 methylation in 9-year old children.
Table 4.
Differences in repeat element methylation associated with a 10-fold increase in prenatal phthalate metabolite concentration during early pregnancy (13 weeks gestation, 1999–2000)
| Phthalate metabolite (μg/L) | Cord (n=239) | 9 yr (n=243) | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Alu | LINE-1 | Alu | LINE-1 | |||||
|
| ||||||||
| beta(95%CI) | p-val | beta(95%CI) | p-val | beta(95%CI) | p-val | beta(95%CI) | p-val | |
| LMW | ||||||||
| MEP | −0.14(−0.28,0.00) | 0.051 | −0.23(−0.53,0.06) | 0.125 | 0.03(−0.13,0.19) | 0.715 | 0.42(0.13,0.71) | 0.005 |
| MBP | −0.03(−0.21,0.16) | 0.781 | −0.47(−0.85, −0.08) | 0.017 | 0.05(−0.13,0.24) | 0.562 | −0.04(−0.38,0.30) | 0.818 |
| MiBP | 0.08(−0.08,0.24) | 0.321 | −0.34(−0.67, −0.01) | 0.044 | 0.00(−0.17,0.16) | 0.970 | 0.11(−0.19,0.42) | 0.459 |
| ΣLMW | −0.14(−0.29,0.01) | 0.067 | −0.28(−0.6,0.04) | 0.086 | 0.04(−0.14,0.21) | 0.678 | 0.43(0.11,0.75) | 0.009 |
| DEHP | ||||||||
| MEHP | −0.09(−0.24,0.06) | 0.237 | −0.13(−0.44,0.19) | 0.433 | −0.03(−0.18,0.12) | 0.672 | −0.03(−0.31,0.26) | 0.857 |
| MEHHP | −0.08(−0.27,0.11) | 0.400 | −0.22(−0.62,0.17) | 0.274 | −0.08(−0.29,0.12) | 0.428 | 0.13(−0.26,0.51) | 0.518 |
| MEOHP | −0.08(−0.26,0.11) | 0.411 | −0.11(−0.49,0.28) | 0.580 | −0.05(−0.24,0.14) | 0.620 | 0.04(−0.32,0.40) | 0.839 |
| MECPP | −0.14(−0.37,0.09) | 0.227 | −0.22(−0.7,0.25) | 0.361 | −0.1(−0.34,0.14) | 0.423 | 0.35(−0.10,0.80) | 0.128 |
| ΣDEHP | −0.11(−0.32,0.1) | 0.301 | −0.21(−0.65,0.23) | 0.346 | −0.09(−0.32,0.14) | 0.439 | 0.20(−0.23,0.63) | 0.368 |
| HMW | ||||||||
| MBzP | 0.09(−0.09,0.26) | 0.341 | −0.39(−0.76, −0.01) | 0.045 | −0.13(−0.33,0.06) | 0.177 | −0.27(−0.64,0.09) | 0.140 |
| MCPP | 0.08(−0.05,0.22) | 0.232 | −0.21(−0.5,0.08) | 0.157 | −0.14(−0.28,0.00) | 0.045 | 0.02(−0.24,0.27) | 0.892 |
| MCOP | −0.04(−0.27,0.18) | 0.708 | −0.23(−0.71,0.25) | 0.351 | −0.01(−0.22,0.20) | 0.936 | −0.34(−0.74,0.06) | 0.099 |
| MCNP | 0.06(−0.16,0.28) | 0.593 | −0.05(−0.51,0.42) | 0.850 | 0.01(−0.20,0.22) | 0.942 | −0.24(−0.64,0.16) | 0.239 |
| ΣHMW | −0.08(−0.31,0.14) | 0.462 | −0.31(−0.78,0.16) | 0.192 | −0.12(−0.36,0.12) | 0.325 | 0.05(−0.40,0.51) | 0.821 |
All models adjusted for creatinine as a covariate in the model. The cord Alu model also controlled for maternal pre pregnancy BMI.
All phthalate concentrations were log 10 transformed and low non detects were imputed
LMW - low molecular weight
HMW - high molecular weight
Figure 1.
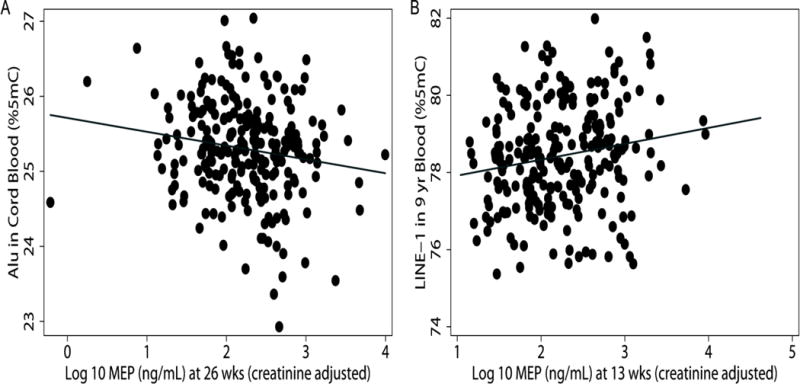
Scatter plot of prenatal MEP urinary concentration with (A) Alu methylation in cord blood and (B) LINE-1 methylation in 9 year old blood. For each ten-fold increase in MEP measured during late pregnancy, we observed 0.16 %5mC lower Alu methylation in cord blood (p=0.025). Each ten-fold increase of MEP during early pregnancy was associated with 0.42 %5mC high LINE-1 methylation in 9 year old blood (p=0.005).
During late pregnancy, the inverse association between MEP and LMW sum with Alu methylation in cord blood persisted (Figure 1) as did the weaker association with LINE-1 methylation in cord blood (Table 5). No other significant associations of phthalate metabolite concentrations during late pregnancy with repeat element methylation in cord blood were found. In blood from 9 year olds, we identified a significant inverse association between concentrations of the LMW metabolite MiBP during late pregnancy and Alu methylation (β(95%CI):−0.24(−0.41,−0.07)), but interestingly saw no association with MEP concentrations. We also found a consistent inverse relationship between DEHP metabolites and Alu methylation in 9-year old blood with a particularly strong association for MEHHP and also with MEOHP, which is formed from MEHHP. In contrast, LMW and DEHP metabolites were not significantly associated with LINE-1 methylation in 9-year old blood. However two HMW metabolites, MCOP and MCNP, were positively associated with LINE-1 methylation in 9-year old children.
Table 5.
Differences in repeat element methylation associated with a 10-fold increase in prenatal phthalate metabolite concentration during late pregnancy (26 weeks gestation, 1999–2000)
| Phthalate metabolite (μg/L) | Cord (n=239) | 9 yr (n=243) | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Alu | LINE-1 | Alu | LINE-1 | |||||
|
| ||||||||
| beta(95%CI) | p-val | beta(95%CI) | p-val | beta(95%CI) | p-val | beta(95%CI) | p-val | |
| LMW | ||||||||
| MEP | −0.16(−0.31, −0.02) | 0.025 | −0.20(−0.52,0.11) | 0.197 | −0.02(−0.18,0.13) | 0.761 | −0.09(−0.39,0.21) | 0.571 |
| MBP | −0.12(−0.32,0.07) | 0.225 | −0.06(−0.48,0.36) | 0.788 | −0.07(−0.29,0.14) | 0.490 | 0.07(−0.34,0.48) | 0.753 |
| MiBP | −0.14(−0.30,0.02) | 0.093 | −0.17(−0.52,0.17) | 0.330 | −0.24(−0.41, −0.07) | 0.006 | 0.21(−0.13,0.54) | 0.225 |
| ΣLMW | −0.19(−0.36, −0.02) | 0.025 | −0.25(−0.61,0.11) | 0.173 | −0.05(−0.23,0.13) | 0.613 | −0.06(−0.41,0.28) | 0.731 |
| DEHP | ||||||||
| MEHP | −0.01(−0.18,0.15) | 0.878 | −0.09(−0.45,0.27) | 0.616 | −0.18(−0.35, −0.02) | 0.032 | 0.18(−0.14,0.51) | 0.271 |
| MEHHP | −0.06(−0.26,0.13) | 0.525 | 0.00(−0.42,0.42) | 0.995 | −0.27(−0.47, −0.08) | 0.006 | 0.22(−0.16,0.61) | 0.252 |
| MEOHP | −0.05(−0.24,0.14) | 0.627 | 0.09(−0.32,0.5) | 0.662 | −0.30(−0.50, −0.11) | 0.003 | 0.27(−0.12,0.65) | 0.181 |
| MECPP | −0.02(−0.28,0.23) | 0.870 | 0.09(−0.45,0.63) | 0.753 | −0.2(−0.45,0.06) | 0.128 | 0.10(−0.4,0.59) | 0.701 |
| ΣDEHP | −0.04(−0.27,0.2) | 0.759 | 0.06(−0.44,0.56) | 0.807 | −0.26(−0.49, −0.02) | 0.033 | 0.18(−0.28,0.64) | 0.439 |
| HMW | ||||||||
| MBzP | −0.05(−0.24,0.14) | 0.598 | −0.22(−0.62,0.18) | 0.275 | −0.13(−0.32,0.06) | 0.168 | 0.07(−0.29,0.42) | 0.714 |
| MCPP | 0.00(−0.18,0.18) | 0.979 | −0.22(−0.6,0.15) | 0.244 | −0.06(−0.23,0.12) | 0.515 | 0.16(−0.18,0.5) | 0.367 |
| MCOP | −0.02(−0.20,0.17) | 0.845 | −0.20(−0.6,0.21) | 0.342 | −0.20(−0.39,0.00) | 0.053 | 0.41(0.03,0.79) | 0.037 |
| MCNP | −0.01(−0.22,0.19) | 0.897 | −0.38(−0.82,0.06) | 0.089 | 0.08(−0.17,0.34) | 0.516 | 0.62(0.14,1.1) | 0.012 |
| ΣHMW | −0.02(−0.27,0.24) | 0.896 | −0.02(−0.57,0.52) | 0.929 | −0.30(−0.55, −0.04) | 0.024 | 0.21(−0.3,0.71) | 0.429 |
All models adjusted for creatinine as a covariate in the model. The cord Alu model also controlled for maternal pre pregnancy BMI.
All phthalate concentrations were log 10 transformed and low non detects were imputed
LMW - low molecular weight
HMW - high molecular weight
3.4 Sensitivity analyses
Results remained unchanged when adjusting for specific gravity in comparison to models adjusting for creatinine. Likewise, models controlling for white blood cell proportions were very similar to crude models that did not account for blood cell counts, suggesting that cell heterogeneity was not a significant source of bias in these models. When we averaged phthalate metabolite concentrations over early and late pregnancy (pregnancy average model; Supplemental Table 2), the inverse relationship between MEP and Alu methylation in cord blood, which was observed in both models of early and late pregnancy phthalate exposure, remained significant. However, the strong inverse association of DEHP metabolites with 9-year Alu methylation identified in the late pregnancy analysis did not persist in the pregnancy average models, suggesting that late pregnancy may be a more sensitive period of exposure to DEHP.
4. Discussion
In this study, we examined the association of prenatal phthalate exposure during early and late pregnancy with repetitive element methylation of blood collected from CHAMACOS children at birth and age 9. We identified a consistent inverse association of MEP (at both pregnancy time points), the most frequently detected metabolite, with methylation of Alu repeats in cord blood. A similar but weaker association was also observed with LINE-1 methylation. Additionally, increases in urinary concentration of DEHP metabolites during late pregnancy were associated with lower levels of methylation of Alu repeats in 9-year old blood.
To our knowledge, only one other study has reported a relationship between phthalate exposure during pregnancy and repetitive element methylation. Zhao et al. (2015) reported an inverse association between exposure to DEHP during late pregnancy and LINE-1 methylation in placental samples. While we did not find the same association between urinary concentrations of DEHP metabolites with LINE-1 methylation in our study of cord and 9-year old blood samples, we did observe a significant association of the same metabolites with Alu methylation in 9-year old’s blood. Furthermore, both studies demonstrate inverse associations of several phthalate metabolites with methylation of repetitive elements during early development and childhood.
In this study, associations of prenatal phthalate metabolite concentrations observed with methylation of repeat elements varied between Alu and LINE-1 elements. This is consistent with other studies which have reported that the two measures have differential susceptibility to environmental exposures (Baccarelli et al., 2009; Pavanello et al., 2009; Tarantini et al., 2009; Wright et al., 2010). Additionally, they are not correlated with each other (Gao et al., 2012; Hou et al., 2010). Instead, LINE-1 and Alu methylation likely represent distinct measures of methylation in different parts of the methylome (Alexeeff et al., 2013; Price et al., 2012).
Several other environmental exposures have also been associated with lower levels of methylation of Alu and/or LINE-1 elements (Miao et al., 2014; Rusiecki et al., 2008; Wright et al., 2010). This inverse relationship between methylation of retrotransposable elements with environmental exposures may be biologically relevant. Among the 500,000 LINE-1 and 1.4 million Alu repeats, their methylation represents up to 50% of the global genomic methylation. Their high rates of methylation are likely responsible for the suppression of their transposition activity and thus lower levels of methylation of Alu and LINE-1 may lead to genomic instability (Xiao-Jie et al., 2015). Lower levels of methylation of LINE-1 and Alu elements has been associated with several cancers (Barchitta et al., 2014; Li et al., 2014; Salas et al., 2014) as well as some phthalate-related health outcomes such as obesity and sperm quality (Perng et al., 2013; Tian et al., 2014). However, it remains unclear how differences in DNA methylation of repetitive elements associated with prenatal phthalate exposures lead to health effects.
The most consistent association identified was between MEP urinary concentrations and Alu methylation in cord blood. MEP devolves from DEP, a compound commonly used in personal care products. Notably, Alu methylation in cord blood was significantly related to MEP concentrations at both pregnancy time points and also their average. This may be due to the relatively low temporal variability of MEP urinary concentrations, which, as seen in other studies (Valvi et al., 2015b), were moderately correlated between early and late pregnancy (ICC=0.42). Interestingly, this effect on methylation did not persist at age 9 years. Perhaps by late childhood post-natal exposures may be more influential on methylation since children’s phthalate exposures change as they get older (Sathyanarayana et al., 2008; Watkins et al., 2014a). Additionally, other studies have also reported associations of in utero DEP exposure with methylation. La Rocca and colleagues found that MEP concentrations during early pregnancy were associated with placental methylation of the maternally imprinted gene IGF2 (LaRocca et al., 2014).
The inverse association between urinary concentrations of DEHP metabolites and Alu methylation in 9-year olds blood was only observed during late pregnancy. The temporal variability of DEHP metabolite concentrations between the two pregnancy time points (ICCs from 0.05–0.07) was much higher in comparison to MEP, which has also been reported in other studies (Braun et al., 2012; Ferguson et al., 2014). These results suggest that late pregnancy may be a critical window of exposure for DEHP in relation to Alu methylation. In utero DEHP exposure has also been associated with differences in LINE-1 methylation in human placenta as well as adult rats but similar studies with Alu methylation have not been reported (Martinez-Arguelles and Papadopoulos, 2015; Zhao et al., 2015).
The primary strength of this study involves the prospective design allowing us to examine associations of exposures during two time points in pregnancy with differences in methylation in newborns and children. This study also has some limitations. First, methylation levels measured were made in blood, which can be biased by cell heterogeneity. However, in our study, results remained relatively unchanged when we accounted for white blood cell proportions in sensitivity analyses. Second, this study was limited to CHAMACOS participants, who were primarily Mexican-American children from low-income families. In the future, it will be important to confirm findings in other cohorts to demonstrate generalizability to other populations.
In summary, we identified several significant associations between exposure to both low and high molecular weight phthalates with methylation in CHAMACOS newborns and 9 year olds. These findings highlight a potential biological mechanism (DNA methylation) linking exposure to phthalates during pregnancy with later life health effects. Additional studies are needed to confirm findings and to further explore whether changes in methylation mediate effects of prenatal exposure on adverse health outcomes.
Supplementary Material
Highlights.
Concentrations of 11 phthalate metabolites were measured in pregnant women.
Lower levels of methylation were found with higher in utero phthalate exposures.
DNA methylation may be a biologic mechanism linking phthalates and health effects.
Acknowledgments
We are grateful to the laboratory and field staff and participants of the CHAMACoS study for their contributions. We are thankful to Xiaoyun Ye, Manori Silva, Ella Samandar, Jim Preau, and Tao Jia, who performed the measurement of phthalate metabolite concentrations at CDC. This publication was made possible by grants from the National Institute of Environmental Health Science (NIEHS) [Po1 ES009605, Ro1 ES021369], from the US Environmental Protection Agency (EPA)[R82670901, and RD83451301]. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NIEHS, EPA, or CDC. Use of trade names is for identification only and does not imply endorsement by the CDC, the Public Health Service, or the US Department of Health and Human Services.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Potential conflicts of interest:
AB has served as a volunteer member of the Board for The organic Center, a non-profit organization that provides information for scientific research about organic food and farming.
Contributor Information
Karen Huen, Email: khuen@berkeley.edu.
Antonia M. Calafat, Email: aic7@cdc.gov.
Asa Bradman, Email: abradman@berkeley.edu.
Paul Yousefi, Email: yousefi@berkeley.edu.
Brenda Eskenazi, Email: eskenazi@berkeley.edu.
Nina Holland, Email: ninah@berkeley.edu.
References
- Adibi JJ, et al. Characterization of phthalate exposure among pregnant women assessed by repeat air and urine samples. Environ Health Perspect. 2008;116:467–73. doi: 10.1289/ehp.10749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeeff SE, et al. Association between blood pressure and DNA methylation of retrotransposons and pro-inflammatory genes. Int J Epidemiol. 2013;42:270–80. doi: 10.1093/ije/dys220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayarpadikannan S, Kim HS. The impact of transposable elements in genome evolution and genetic instability and their implications in various diseases. Genomics Inform. 2014;12:98–104. doi: 10.5808/GI.2014.12.3.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, et al. Rapid DNA Methylation Changes after Exposure to Traffic Particles. Am J Respir Crit Care Med. 2009 doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchitta M, et al. LINE-1 hypomethylation in blood and tissue samples as an epigenetic marker for cancer risk: a systematic review and meta-analysis. PLoS one. 2014;9:e109478. doi: 10.1371/journal.pone.0109478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr DB, et al. Urinary creatinine concentrations in the U.S. population: implications for urinary biologic monitoring measurements. Environ Health Perspect. 2005;113:192–200. doi: 10.1289/ehp.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellavia A, et al. DNA hypomethylation, ambient particulate matter, and increased blood pressure: findings from controlled human exposure experiments. J Am Heart Assoc. 2013;2:e000212. doi: 10.1161/JAHA.113.000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun JM, et al. Variability of urinary phthalate metabolite and bisphenol A concentrations before and during pregnancy. Environ Health Perspect. 2012;120:739–45. doi: 10.1289/ehp.1104139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun HM, et al. Evolutionary age of repetitive element subfamilies and sensitivity of DNA methylation to airborne pollutants. Part Fibre Toxicol. 2013;10:28. doi: 10.1186/1743-8977-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, et al. Human urinary/seminal phthalates or their metabolite levels and semen quality: A meta-analysis. Environ Res. 2015;142:486–94. doi: 10.1016/j.envres.2015.07.008. [DOI] [PubMed] [Google Scholar]
- N. C. f. E. H. Centers for Disease Control and Prevention, editor. CDC. Fourth National Report on Human Exposure to Environmental Chemicals. Atlanta, GA: 2009. [Google Scholar]
- Eskenazi B, et al. CHAMACoS, a longitudinal birth cohort study: lessons from the fields. J Childrens Healt. 2003;1:3–27. [Google Scholar]
- Ferguson KK, et al. Variability in urinary phthalate metabolite levels across pregnancy and sensitive windows of exposure for the risk of preterm birth. Environ Int. 2014;70:118–24. doi: 10.1016/j.envint.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KK, et al. Associations between Maternal Biomarkers of Phthalate Exposure and Inflammation Using Repeated Measurements across Pregnancy. PLoS one. 2015;10:e0135601. doi: 10.1371/journal.pone.0135601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, et al. Blood leukocyte Alu and LINE-1 methylation and gastric cancer risk in the Shanghai Women’s Health Study. Br J Cancer. 2012;106:585–91. doi: 10.1038/bjc.2011.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar FW, et al. Phthalate exposure and risk assessment in California child care facilities. Environ Sci Technol. 2014;48:7593–601. doi: 10.1021/es501189t. [DOI] [PubMed] [Google Scholar]
- Heudorf U, et al. Phthalates: toxicology and exposure. Int J Hyg Environ Health. 2007;210:623–34. doi: 10.1016/j.ijheh.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Holland N, et al. Paraoxonase polymorphisms, haplotypes, and enzyme activity in latino mothers and newborns. Environ Health Perspect. 2006;114:985–91. doi: 10.1289/ehp.8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, et al. Blood leukocyte DNA hypomethylation and gastric cancer risk in a high-risk Polish population. Int J Cancer. 2010;127:1866–74. doi: 10.1002/ijc.25190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen K, et al. Effects of age, sex, and persistent organic pollutants on DNA methylation in children. Environ Mol Mutagen. 2014;55:209–22. doi: 10.1002/em.21845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SC, Lee BM. DNA methylation of estrogen receptor alpha gene by phthalates. J Toxicol Environ Health A. 2005;68:1995–2003. doi: 10.1080/15287390491008913. [DOI] [PubMed] [Google Scholar]
- Kim Y, et al. Prenatal exposure to phthalates and infant development at 6 months: prospective Mothers and Children’s Environmental Health (MoCEH) study. Environ Health Perspect. 2011;119:1495–500. doi: 10.1289/ehp.1003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostka G, et al. Di-butyl phthalate-induced hypomethylation of the c-myc gene in rat liver. Toxicol Ind Health. 2010;26:407–16. doi: 10.1177/0748233710369124. [DOI] [PubMed] [Google Scholar]
- LaRocca J, et al. The impact of first trimester phthalate and phenol exposure on IGF2/H19 genomic imprinting and birth outcomes. Environ Res. 2014;133:396–406. doi: 10.1016/j.envres.2014.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, et al. The prognostic value of global DNA hypomethylation in cancer: a meta-analysis. PLoS one. 2014;9:e106290. doi: 10.1371/journal.pone.0106290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin JH, et al. Epidemiologic evaluation of measurement data in the presence of detection limits. Environ Health Perspect. 2004;112:1691–6. doi: 10.1289/ehp.7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Arguelles DB, Papadopoulos V. Identification of hot spots of DNA methylation in the adult male adrenal in response to in utero exposure to the ubiquitous endocrine disruptor plasticizer di-(2-ethylhexyl) phthalate. Endocrinology. 2015;156:124–33. doi: 10.1210/en.2014-1436. [DOI] [PubMed] [Google Scholar]
- Meeker JD, et al. Urinary phthalate metabolites in relation to preterm birth in Mexico city. Environ Health Perspect. 2009;117:1587–92. doi: 10.1289/ehp.0800522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao M, et al. LINE-1 hypomethylation in spermatozoa is associated with Bisphenol A exposure. Andrology. 2014;2:138–44. doi: 10.1111/j.2047-2927.2013.00166.x. [DOI] [PubMed] [Google Scholar]
- Pavanello S, et al. Global and gene-specific promoter methylation changes are related to anti-B[a]PDE-DNA adduct levels and influence micronuclei levels in polycyclic aromatic hydrocarbon-exposed individuals. Int J Cancer. 2009;125:1692–7. doi: 10.1002/ijc.24492. [DOI] [PubMed] [Google Scholar]
- Perng W, et al. A prospective study of LINE-1DNA methylation and development of adiposity in school-age children. PLoS one. 2013;8:e62587. doi: 10.1371/journal.pone.0062587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogribny IP, et al. Mechanisms of peroxisome proliferator-induced DNA hypomethylation in rat liver. Mutat Res. 2008;644:17–23. doi: 10.1016/j.mrfmmm.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price EM, et al. Different measures of “genome-wide” DNA methylation exhibit unique properties in placental and somatic tissues. Epigenetics. 2012;7:652–63. doi: 10.4161/epi.20221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo JL, et al. Pyrosequencing protocol using a universal biotinylated primer for mutation detection and SNP genotyping. Nat Protoc. 2007;2:1734–9. doi: 10.1038/nprot.2007.244. [DOI] [PubMed] [Google Scholar]
- Rusiecki JA, et al. Global DNA hypomethylation is associated with high serum-persistent organic pollutants in Greenlandic Inuit. Environ Health Perspect. 2008;116:1547–52. doi: 10.1289/ehp.11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas LA, et al. LINE-1 methylation in granulocyte DNA and trihalomethane exposure is associated with bladder cancer risk. Epigenetics. 2014;9:1532–9. doi: 10.4161/15592294.2014.983377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyanarayana S, et al. Maternal and infant urinary phthalate metabolite concentrations: are they related? Environ Res. 2008;108:413–8. doi: 10.1016/j.envres.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve JF, et al. Creatinine and specific gravity normalization in biological monitoring of occupational exposures. J occup Environ Hyg. 2015;12:123–9. doi: 10.1080/15459624.2014.955179. [DOI] [PubMed] [Google Scholar]
- Silva MJ, et al. Urinary levels of seven phthalate metabolites in the U.S. population from the National Health and Nutrition Examination Survey (NHANES) 1999–2000. Environ Health Perspect. 2004;112:331–8. doi: 10.1289/ehp.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MJ, et al. Quantification of 22 phthalate metabolites in human urine. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;860:106–12. doi: 10.1016/j.jchromb.2007.10.023. [DOI] [PubMed] [Google Scholar]
- Smarr MM, et al. Parental urinary biomarkers of preconception exposure to bisphenol A and phthalates in relation to birth outcomes. Environ Health. 2015;14:73. doi: 10.1186/s12940-015-0060-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, et al. Genome-wide dynamic changes of DNA methylation of repetitive elements in human embryonic stem cells and fetal fibroblasts. Genomics. 2012;99:10–7. doi: 10.1016/j.ygeno.2011.10.004. [DOI] [PubMed] [Google Scholar]
- Tarantini L, et al. Effects of particulate matter on genomic DNA methylation content and iNoS promoter methylation. Environ Health Perspect. 2009;117:217–22. doi: 10.1289/ehp.11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitelbaum SL, et al. Associations between phthalate metabolite urinary concentrations and body size measures in New York City children. Environ Res. 2012;112:186–93. doi: 10.1016/j.envres.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, et al. Association of DNA methylation and mitochondrial DNA copy number with human semen quality. Biol Reprod. 2014;91:101. doi: 10.1095/biolreprod.114.122465. [DOI] [PubMed] [Google Scholar]
- Valvi D, et al. Prenatal Phthalate Exposure and Childhood Growth and Blood Pressure: Evidence from the Spanish INMA-Sabadell Birth Cohort Study. Environ Health Perspect. 2015a;123:1022–9. doi: 10.1289/ehp.1408887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvi D, et al. Variability and predictors of urinary phthalate metabolites in Spanish pregnant women. Int J Hyg Environ Health. 2015b;218:220–31. doi: 10.1016/j.ijheh.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Watkins DJ, et al. Variability and predictors of urinary concentrations of phthalate metabolites during early childhood. Environ Sci Technol. 2014a;48:8881–90. doi: 10.1021/es501744v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins DJ, et al. Associations between serum perfluoroalkyl acids and LINE-1 DNA methylation. Environ Int. 2014b;63:71–6. doi: 10.1016/j.envint.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyatt RM, et al. Asthma in inner-city children at 5–11 years of age and prenatal exposure to phthalates: the Columbia Center for Children’s Environmental Health Cohort. Environ Health Perspect. 2014;122:1141–6. doi: 10.1289/ehp.1307670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff TJ, et al. Environmental chemicals in pregnant women in the United States: NHANES 2003–2004. Environ Health Perspect. 2011;119:878–85. doi: 10.1289/ehp.1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright RO, et al. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ Health Perspect. 2010;118:790–5. doi: 10.1289/ehp.0901429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, et al. Dynamic epigenetic changes involved in testicular toxicity induced by di-2-(ethylhexyl) phthalate in mice. Basic Clin Pharmacol Toxicol. 2010;106:118–23. doi: 10.1111/j.1742-7843.2009.00483.x. [DOI] [PubMed] [Google Scholar]
- Xiao-Jie L, et al. LINE-1 in cancer: multifaceted functions and potential clinical implications. Genet Med. 2015 doi: 10.1038/gim.2015.119. [DOI] [PubMed] [Google Scholar]
- Yang AS, et al. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi P, et al. Estimation of blood cellular heterogeneity in newborns and children for epigenome-wide association studies. Environ Mol Mutagen. 2015 doi: 10.1002/em.21966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, et al. Prenatal phthalate exposure, infant growth, and global DNA methylation of human placenta. Environ Mol Mutagen. 2015;56:286–92. doi: 10.1002/em.21916. [DOI] [PubMed] [Google Scholar]
- Zota AR, et al. Temporal trends in phthalate exposures: findings from the National Health and Nutrition Examination Survey, 2001–2010. Environ Health Perspect. 2014;122:235–41. doi: 10.1289/ehp.1306681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


