SUMMARY
Histone H2B monoubiquitination (H2Bub1) is centrally involved in gene regulation. The deubiquitination module (DUBm) of the SAGA complex is a major regulator of global H2Bub1 levels, and components of this DUBm are linked to both neurodegenerative diseases and cancer. Unexpectedly, we find that ablation of USP22, the enzymatic center of the DUBm, leads to a reduction, rather than an increase, in global H2bub1 levels. In contrast, depletion of non-enzymatic components, ATXN7L3 or ENY2, results in increased H2Bub1. These observations led us to discover two new H2Bub1 DUBs, USP27X and USP51, which function independently of SAGA and which compete with USP22 for ATXN7L3 and ENY2 for activity. Like USP22, USP51 and USP27X are required for normal cell proliferation, and their depletion suppresses tumor growth. Our results reveal that ATXN7L3 and ENY2 orchestrate activities of multiple deubiquitinating enzymes and that imbalances in these activities likely potentiate human diseases including cancer.
Keywords: H2B ubiquitination, USP22, USP27X, USP51, ENY2, ATXN7L3, SAGA
INTRODUCTION
Aberrations in levels of histone ubiquitination are associated with serious human afflictions, including developmental abnormalities, neurodegenerative diseases, and cancers (Weake and Workman, 2008). Monoubiquitination of H2B (H2Bub1) is conserved in all eukaryotes and is associated with gene activity (Minsky et al., 2008; Smith and Shilatifard, 2010). Cycles of H2B ubiquitination and deubiquitination at the 5’ ends of genes are important for transcription initiation (Henry et al., 2003). H2Bub1 is quite dynamic, as it disappears only minutes after transcription is blocked, whereas other modifications are more stable (Bonnet et al., 2014; Fuchs et al., 2014).
The deubiquitination module (DUBm) in SAGA is an important regulator of H2Bub1 levels in yeast (at K123), fly, and mammalian cells (at K120) (Henry et al., 2003; Lang et al., 2011; Mohan et al., 2014; Weake et al., 2008). The SAGA DUB module is highly conserved both in subunit composition and structural organization (Kohler et al., 2010; Lee et al., 2011; Samara et al., 2010; Weake et al., 2008; Zhang et al., 2008b; Zhao et al., 2008), The deubiquitinating enzyme USP22 (Ubp8 in yeast) closely associates with two adapter proteins, ATXN7L3 and ENY2 (Sgf11 and Sus1 in yeast, respectively) (Lan et al., 2015; Zhang et al., 2008b; Zhao et al., 2008), and a fourth protein, ATXN7 (Sgf73), anchors the DUB module to the larger SAGA complex. Proper structural organization of the entire module is crucial for DUB activity (Kohler et al., 2008; Kohler et al., 2010; Lan et al., 2015; Lang et al., 2011; Samara et al., 2010).
USP22 was originally described as part of an 11 gene ‘death from cancer signature’ that defines tumors with a cancer stem cell phenotype including aggressive growth, metastasis, and resistance to therapy (Glinsky, 2006). Several reports indicate that USP22 overexpression is linked to unfavorable outcomes in multiple types of cancers (He et al., 2015; Liang et al., 2014; Ning et al., 2012; Zhang et al., 2011). However, the mechanisms underlying USP22 overexpression in different malignancies and molecular links between histone deubiquitination and aggressive tumor growth are still not clear.
Here we report that in mammalian cells, depletion of USP22 leads to a surprising reduction, rather than the expected increase, in global H2Bub1 levels. Moreover, we discovered that ATXN7L3 and ENY2 cooperate with two other deubiquitinating enzymes, USP27X and USP51, to regulate global levels of H2B K120ub1. These new ubiquitin specific proteases (USPs) form catalytically active DUB modules that are independent of the SAGA complex. Furthermore, USP27X and USP51 activities are regulated by ATXN7L3 and ENY2 in the same manner as USP22, and the three deubiquitinating enzymes compete for these adapter proteins within cells. Importantly, ablation of USP27X or USP51 impacts cell proliferation and tumor growth, as does ablation of USP22. Our results demonstrate that ATXN7L3 and ENY2 act as master regulators of multiple H2B K120ub1 DUBs.
RESULTS
Loss of ATXN7L3 or ENY2, but Not USP22, Increases H2Bub1 Levels
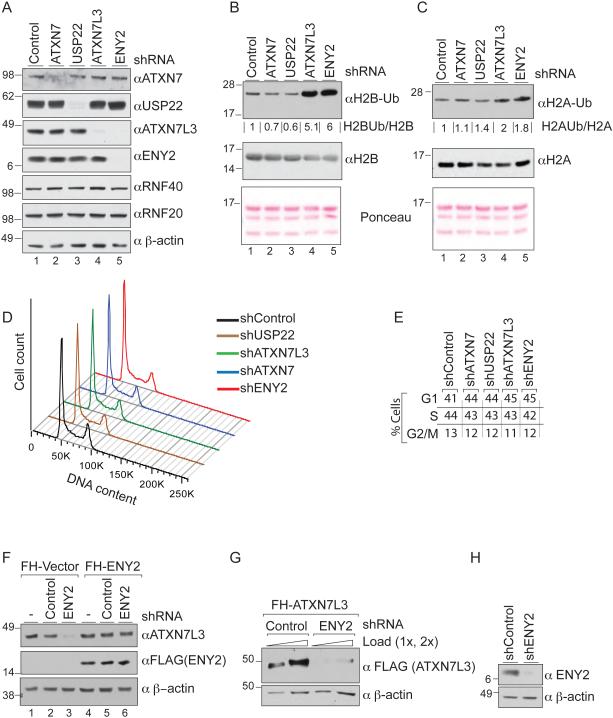
To investigate how different components of the mammalian SAGA DUBm regulate USP22 activity in vivo, we generated a series of 293T cell lines where each of the DUBm members, including USP22, ATXN7L3, ENY2, and the linker protein ATXN7, were depleted individually by shRNA. Immunoblot analyses of whole cell lysates (WCL) confirmed high efficiency of silencing for each of these factors (Figure 1A). We then monitored levels of H2B monoubiquitination at K120 (hereafter H2Bub1) by immunoblot as a readout of DUBm activity (Bonnet et al., 2014; Ingvarsdottir et al., 2005; Kohler et al., 2006; Lang et al., 2011; Mohan et al., 2014; Weake et al., 2008) (Figure 1B). As expected, depletion of ATXN7L3 or ENY2 led to a major increase (~5-6 fold) in H2Bub1 levels (Figure 1B, top panel; compare lane 1 to lanes 4 and 5). H2Aub1 levels were also slightly increased in ATXN7L3 or ENY2 depleted cells, as previously reported (Lang et al., 2011)(Figure 1C top panel). Surprisingly, ablation of USP22 or ATXN7 reproducibly led to a mild reduction of global H2Bub1 levels (Figure 1B, top panel compare lane 1 to lanes 2 and 3), indicating that loss of the adapter proteins has a greater effect on H2B deubiquitination than does loss of the catalytic subunit. These effects are not due to variation in expression of the H2B E3 ligases RNF20 and RNF40, as immunoblots show no detectable changes in levels of these proteins (Figure 1A). Additionally, the different levels of H2Bub1 in USP22 and ATXN7L3 or ENY2 knockdown (KD) cells are not due to changes in cell cycle distribution, since global H2Bub1 levels showed very little fluctuation through the cell cycle, except for a short period during M-phase (Figure S1A and B). Depletion of each SAGA DUBm component led to only a slight increase in G1 phase cells (Figure 1D and E). We confirmed our results in the breast cancer cell line MDA-MB-231, and again found greater effects on H2Bub1 levels upon loss of ATXN7L3 and ENY2 than upon loss of USP22 (Figure S1C).
Figure 1. USP22 loss does not lead to H2B deubiquitination defects in mammalian cells.
A) Immunoblots to demonstrate efficient silencing of the SAGA DUBm components in 293T cells. (B and C) Depletion of ATXN7L3 and ENY2, but not USP22 or ATXN7, leads to a robust increase of bulk H2Bub1 and a moderate increase of H2Aub1 levels; compare lanes 4 and 5 to 1, 2 and 3. (D) Depletion of each SAGA DUBm component leads to a mild increase in the proportion of G1-phase cells as indicated by DNA content. (E) Cell cycle distribution percentages. (F) Depletion of ENY2 severely impacts steady state levels of ATXN7L3 (compare lanes 2 and 3). This effect can be rescued by expression of exogenous ENY2 (compare lanes 3 and 6). (G) ENY2 is required for stability of exogenous ATXN7L3. (H) Efficiency of ENY2 silencing in panel G.
Depletion of ENY2 led to significant reduction of ATXN7L3 protein levels (Figure 1A, lanes 4 and 5), consistent with a previous report (Umlauf et al., 2013). ATXN7L3 levels were restored upon stable expression of exogenous FLAG-tagged ENY2 after depletion of endogenous ENY2 using shRNA specifically targeting the endogenous transcript (Figure 1F, compare lanes 3 and 6). Furthermore, ENY2 depletion reduced levels of exogenous ATXN7L3 (Figure 1G, H), indicating ENY2 affects ATXN7L3 stability post transcription. These results show that ATXN7L3 is tightly regulated by ENY2 interaction and suggest that no free ATXN7L3 exists in mammalian cells.
ATXN7L3 and ENY2 associate with USP27X and USP51
The strong effects of ATXN7L3 and ENY2 loss on H2B deubiquitination suggest these adaptors may facilitate the function of additional H2B DUBs. To test this possibility, we affinity purified ATXN7L3 interacting proteins from 293T cells that stably express low levels of FLAG- and V5-tagged ATXN7L3 (Figure 2A). As expected, the majority of the interacting proteins identified by mass spectrometry were components of SAGA (Figure 2B). In addition, several proteins not known to be part of SAGA were identified, including two additional USP proteins, USP27X and USP51.
Figure 2. ATXN7L3 interacts with USP27X and USP51.
(A) Schematic of tandem FLAG-V5 affinity purification. (B) FL-V5-ATXN7L3 associated proteins identified by mass spectrometry. (C) Silver stain and immunoblot analyses to confirm interactions of ATXN7L3 with USP27X and USP51. Bands corresponding to USP27X and USP51 are indicated by black lines. (D) Immunoprecipitations of nuclear extracts (NE) using an αATXN7L3 antibody or rabbit IgG followed by immunoblots for USP27X or USP51. Input represents 5% of the NE. (E) Schematic representation of the human USP27X locus. ATG indicates the predicted translation start site of human USP27X protein (A6NNY8) and CTG indicates the experimentally discovered start site. (F) Autoradiograph of 35S-labeled, in vitro translated hUSP27X. The arrow indicates expected size of the translated protein. (G) Protein domain organization in USP22, USP27X and USP51. The sequence identity between USP22 and USP27X and between USP22 and USP51 human proteins is indicated with the brackets on the right, and the percentage identity between the ZnF-UBP domains is indicated by brackets on the left. (H) Whole cell lysates (WCL) from the indicated cell lines were immunoblotted using USP27X and USP51 specific antibodies. β-actin; loading control. (I) USP27X and USP51 display predominantly nuclear localization. The KRRK NLS sequence in USP22 is 100% conserved in USP51 and is highly similar in USP27X (top panel). USP27X-FL-V5 or USP51-FL-V5 expressing cells stained with αV5 antibody (red) and αATXN7L3 antibody (green). Yellow indicates co-localization. DAPI was used for nuclear counterstaining (blue). Scale bar = 15 μm.
We generated polyclonal antibodies to USP27X and USP51 and after validating their specificity (Figures S3, S4), used these antibodies to confirm that both USP27X and USP51 co-purify with FLAG-HA-ATXN7L3 (Fig 2C). We further verified interactions between the endogenous proteins by immunoprecipitating ATXN7L3 from nuclear extracts (NE) and then immunoblotting for USP27X or USP51 (Figure 2D).
USP27X and USP51 Structure and Expression
Our USP51 specific antibody detects an ~80 kDa protein, matching the predicted size of USP51 (Uniprot: Q70EK9). However, the USP27X protein we detect is ~22 kDa larger than the predicted 50 kDa protein (Uniprot: A6NNY8) (Figure 2C), suggesting that USP27X may utilize an alternative translation start site upstream of the predicted ATG. Although no upstream, in frame ATG is present in USP27X, a ~72 kDa protein could be translated from an upstream CTG. Precedence exists for use of CTG as an initiation codon (Fritsch et al., 2012; Wegrzyn et al., 2008). To test this possibility, we cloned a region spanning the USP27X locus (Figure 2E) and performed in vitro transcription-translation reactions in the presence of 35S-labeled methionine. We observed a ~72 kDa 35S-labeled protein when the wild type locus was used as template, but did not detect any protein product when the predicted CTG start codon was changed to CTC (Figure 2F). We never detected a 50 kDa protein in vitro (by translation) or in vivo (by immunoblot).
Comparison of the 636 amino acid (aa) sequence in this longer form of USP27X (Figure S2G) with the sequences of USP22 and USP51 revealed significant homology, including more than 82% identity between USP22 and USP27X and more than 70% identity between USP22 and USP51 based on ClustalW2 analyzes (Figure 2G). Further, Pfam analysis of protein domain structures (Finn et al., 2014) confirmed that in addition to the catalytic UCH domain, both USP27X and USP51 have an N-terminal Znf-UBP domain highly similar to that found in USP22, which is critical for USP22 interaction with ATXN7L3 and ENY2 and deubiquitination activity (Figure 2G and S2G) (Zhao et al., 2008).
To determine whether USP27X and USP51 are widely expressed, we performed immunoblots on lysates from three different cancer cell lines (neuroblastoma derived SH-SY5Y, colon cancer HCT116, and triple negative breast cancer MDA-MB-231), human ES cells (H1), and primary human astrocytes (Fig 2H). These USPs were clearly present in all lysates, albeit at varying levels, indicating that USP27X and USP51 are expressed in many tissue and cell types.
The nuclear localization signal (NLS) found in USP22 (Xiong et al., 2014) is conserved in USP51, and a highly similar sequence is found in USP27X (Figure 2I, top panel). We confirmed that V5-tagged USP51 and USP27X are largely nuclear, and co-staining confirms co-localization with endogenous ATXN7L3 (Figure 2I, bottom panel). Predominantly nuclear localization was further confirmed in live cells, using USP27X or USP51 fusions with GFP or mCherry respectively (Figure S5).
USP27X and USP51 Require ATXN7L3 and ENY2 for DUB Activity
The high degree of homology with USP22 and physical association with ATXN7L3 and ENY2 predict that USP27X and USP51 are also DUBs. To test this idea, we expressed wild type (WT) or mutant forms of USP22, USP27X and USP51 in which a conserved cysteine required for activity in USP22 is changed to serine in 293T cells (Figure 3A and B). Immunoblots revealed substantial reductions in global H2Bub1 levels in cells expressing WT USP22, USP27X or USP51 relative to cells transfected with an empty vector (Figure 3C, compare lane 1 to lanes 2, 3 and 4), indicating these proteins are indeed DUBs. Moreover, expression of catalytically dead proteins led to sizable increases in global levels of H2Bub1 (Figure 3C, compare lane 1 to lanes 5, 6 and 7), reflecting dominant negative effects of the mutant DUBs on H2B deubiquitination. Notably, co-expression of all three DUBs (either WT or mutant) did not further change H2Bub1 levels, compared to single DUB expression (Figure 3C, compare lanes 2-4 to lane 8 and lanes 5-7 to lane 9), suggesting that levels of the adapter proteins required for the activity of these DUBs may be limited in these cells.
Figure 3. USP27X and USP51 exhibit DUB activity in vivo and in vitro.
(A) Schematic of the catalytic domains of USP22, USP27X, and USP51. The asterisk indicates the catalytic cysteine (C185 for USP22, C285 for USP27X and C372 for USP51), which was changed to serine (C/S) to generate catalytically dead mutants. (B) Protein levels of expressed WT or mutant USP22, USP27X, and USP51 in 293T cells. (C) Expression of the WT USPs reduces global levels of H2Bub1 in 293T cells, whereas expression of catalytically dead USPs leads to H2Bub1 increase. (D) Experimental scheme for purification of USP27X- and USP51-containing DUB modules from insect cells (top), and colloidal coomassie staining of purified complexes (bottom). (E) ATXN7L3 and ENY2 are required for USP27X and USP51 catalytic activity. USP27X or USP51 alone or together with ATXN7L3 and ENY2 were purified from Sf21 cells for use in in vitro DUB reactions using Ubiquitin-AMC as a substrate. DUB activity was measured by the resultant fluorescence intensity. To test catalytic activity towards H2Bub1, nucleosomes were purified from 293T cells stably expressing FL-V5-H2B (F) and used in in vitro DUB reactions (G).
To further confirm that USP27X and USP51 are active DUBs, we expressed and purified recombinant forms of these proteins from insect cells (Figure 3D). In vitro DUB reactions using purified histones (Figure S3) or Ubiquitin-AMC (Figure 3E) as substrates indicated that neither USP27X nor USP51 were active in isolation. However, when co-expressed and co-purified with ATXN7L3 and ENY2 (Figure 3D), USP27X and USP51 form stable, stoichiometric complexes with these proteins (Figure 3D). Moreover, these recombinant DUB modules exhibited robust deubiquitination activity using Ubiquitin-AMC as a substrate, (Figure 3E), whereas no or background activity levels were detected when the DUB modules were reconstituted with catalytically dead versions of the USPs.
We next tested the activity of the USP27X or USP51 containing DUB modules towards nucleosomal H2Bub1. We purified nucleosomes expressing FLAG- and V5-tagged H2B (Figure 3F) as substrates, and again found that both USP27X and USP51 DUB modules showed strong activity towards H2Bub1 (Figure 3G, lanes 3 and 6), whereas the individual proteins or DUB modules reconstituted with mutant USPs had no detectable activity (Figure 3G, lanes 2 and 5 and lanes 4 and 7). Similar results were obtained using free histones as substrate (Figure S3). As was previously shown for the USP22 DUBm (Zhang et al., 2008a; Zhao et al., 2008), both USP27X and USP51 DUBm also deubiquitinated nucleosomal or free H2Aub1 (Figures 3G and S3A and B).
USP27X and USP51 Do Not Associate with SAGA
Since USP27X and USP51 are highly similar to USP22, and their activity is regulated by ATXN7L3 and ENY2, we reasoned that these DUBs might be part of alternative SAGA assemblies (Figure 4A). To explore this possibility, we purified the SAGA complex using FLAG- and HA-tagged GCN5 from 293T cells, and then probed the purified fractions using antibodies for known SAGA components, USP27X, or USP51. Immunoblots verified successful isolation of SAGA, as TRRAP, ATXN7, ATXN7L3 and USP22 were all present in the GCN5 pull downs (Figure 4B). However, neither USP27X nor USP51 were present, even though all three USPs are well expressed in these cells (Figure 4B, input lanes). We obtained very similar results upon isolation of SAGA through FLAG-tagged USP22 or FLAG-HA-tagged SPT3 from HeLaS3 cells (Figure S4A and B).
Figure 4. USP22, USP27X, and UPS51 compete for ATXN7L3 and ENY2.
(A) Schematic representation of the SAGA complex. USP27X and USP51 are depicted as possible substitutes of USP22. (B) USP27X and USP51 are not part of SAGA complex. FLAG-HA-GCN5 associated proteins were analyzed by immunoblot for TRRAP, ATXN7, USP22, and ATXN7L3 (confirming successful SAGA purification), and USP27X and USP51. Asterisk indicates a nonspecific band. (C) FLAG- and V5-tagged USP27X and USP51 co-immunoprecipitate ATXN7L3 and ENY2 but not GCN5 or TAF10. (D) Loss of USP22 does not lead to USP27X or USP51 incorporation into SAGA as shown by immunoblots of SAGA purified using FLAG-GCN5. (E) Increased association of USP27X with ATXN7L3 in Usp22 KO cells. Whole cell lysates from WT or Usp22 KO mES cells were immunoprecipitated using an αATXN7L3 antibody or rabbit IgG, and purified fractions were immunoblotted for USP27X. Blots were quantified using ImageJ software. (F) USP22 and USP27X compete for ATXN7L3 binding. HA-USP22, HA-USP27X, and FLAG-ATXN7L3 were mixed in in vitro binding reactions with increasing amounts of HA-USP27X (0.1, 0.5, and 1 relative to USP22 amounts, based on colloidal blue stained gel) was added as a competition for USP22. USP22-ATXN7L3 or USP27X-ATXN7L3 complexes were precipitated with αFLAG (ATXN7L3) antibody. Amounts of recovered HA-USP22 and HA-USP27X were assessed by αHA immunoblot and quantified using ImageJ software (G) Simultaneous depletion of USP22, USP27X, and USP51 leads to increased global H2Bub1 in HCT116 cells. USP22 and USP51 were depleted in WT and USP27X KO cells (bottom panels), and the H2Bub1 signal was normalized to total H2B (top panels).
We also performed immunoprecipitations for FLAG-V5 tagged USP27X or USP51 from nuclear extracts and probed for SAGA components. As expected, ATXN7L3 and ENY2 associated with USP27X and USP51, but no other SAGA components, such as GCN5 or TAF10, were observed in the immunoprecipitations (Figure 4C, lanes 5 and 6). To determine whether USP22 blocks association of USP27X and USP51 with SAGA, we isolated GCN5-associated proteins after shRNA-mediated depletion of USP22 (Figure 4D, lanes 2 and 3 and 5 and 6). Again, no USP27X or USP51 was found associated with GCN5, indicating these DUBs are not part of SAGA.
USP22, USP27X, and USP51 Compete for Association with ATXN7L3 and ENY2
Since all three USPs require ATXN7L3 and ENY2 for activity, they might compete for these factors. To test this possibility, we immunoprecipitated endogenous ATXN7L3 from whole cell lysates of WT or Usp22 knockout (KO) mouse embryonic stem cells (mES cells) and assessed amounts of USP27X in the precipitated fractions (Figure 4E). We detected a substantial increase (~5 fold) in amounts of USP27X associated with ATXN7L3 in Usp22 KO cells relative to WT cells (Figure 4E, compare lanes 5 and 6). The differences in the amounts of USP27X associated with ATXN7L3 in WT and KO cells are not due to global changes in USP27X expression upon USP22 depletion (Figure 4E, compare lanes 1 and 2, long exposure). Similar results were obtained by comparing levels of ATXN7L3 co-immunoprecipitated with USP27X from control or USP22 stably depleted cells (Figure S4C). MudPIT analysis confirmed a ~5.5 fold increase of USP27X association with ATXN7L3 and a ~4 fold increase in USP51 association with ATXN7L3 upon depletion of USP22 (Figure S4D). Altogether, these results indicate that the three USPs compete for limiting amounts of ATXN7L3 within cells.
To further test if these DUBs compete for adapter binding, we separately expressed and purified all three USPs and ATXN7L3 from insect cells (Figure S4E) and performed competitive binding assays in vitro. As shown in Figure 4F, we found that association of USP22 and ATXN7L3 was greatly impaired upon addition of increasing amounts of USP27X (compare lane 7 to 8, 9 and10). Very similar results were obtained when USP51 was used as a competitor to USP22 (Figure S4F). These results demonstrate directly that the three USPs compete for adapter binding and likely explain the enhanced DUB activity observed upon USP22 or USP27X ablation (Figure 1B and S4G, top panels), as loss of one USP would free more ATXN7L3 and ENY2 for association with the others. Consistent with this idea, simultaneous depletion of USP22 and USP51 in USP27X KO HCT116 cells (Figure 4G) increased global H2Bub1 (Figure 4G, compare lanes 3 and 4) as was observed upon depletion of ATXN7L3 or ENY2 (Figure 1C).
USP27X and USP51 Associate with a Similar Spectrum of Proteins
To further assess if ATXN7L3 and ENY2 are stable components of USP27X and USP51 containing complexes, we performed gel filtration experiments. Immunoblots revealed that both USP27X and USP51 form tight complexes with ATXN7L3 and ENY2 that elute in fractions 13 to 15, corresponding to complexes of approximately 600-kDa, much smaller than the 2-MDa SAGA complex but larger than a tripartite, 150-kDa DUBm alone (Figure 5А and B).
Figure 5. USP27X and USP51 Associated Proteins.
(A) USP27X and (B) USP51 containing complexes were precipitated from 293T nuclear extracts and fractionated by size on a Superose 6 column. Eluted fractions were probed for ATXN7L3 and ENY2. (C) Profile of the FLAG and V5 purified USP27X and USP51 containing complexes after elution from αV5 resin. Asterisks indicate the bait proteins. (D) List of associated proteins identified by MudPIT. Presented are proteins identified in 3 independent experiments. Average cdNASF (Normalized Distributed Spectral Abundance Factor) is shown for each protein in the purification.
To identify proteins that associate with USP27X and USP51, we performed MudPIT analyses on FLAG- and V5-tagged USP27X or USP51 affinity purified fractions from 293T nuclear extracts (Figure 5C). As expected, ATXN7L3 and ENY2 were identified among the top interacting proteins for both USP27X and USP51 (Fig 5D). Consistent with our findings above, apart from ATXN7L3 and ENY2, DDB1 was the only protein in these purifications that was previously reported to associate with SAGA (Martinez et al., 2001). A protein related to ATXN7L3, ATXN7L3B, was also identified as a highly abundant protein in these purifications. ATXN7L3B is highly homologous to the N-terminal portion of ATXN7L3 that interacts physically with ENY2, but it lacks the C-terminal ZnF domain, which appears to be crucial for the histone deubiquitination function of ATXN7L3 (Lang et al., 2011). A number of proteins identified in one or both purifications, such as RBM39 (CAPERα) and TRIM28, function in transcriptional regulation and RNA processing (Dowhan et al., 2005; Friedman et al., 1996). Other proteins that co-purified with USP27X and USP51 with high reproducibility are involved in DNA repair and replication (Figure 5D).
Ablation of USP22, USP27X, or USP51 Affects H2Bub1 Distribution and Regulation of Gene Expression
As H2Bub1 plays important roles in the regulation of gene expression, we investigated how depletion of the three DUBs impacts genome wide distribution of this epigenetic mark. To do this, we performed chromatin immunoprecipitation-sequencing (ChIP-seq) to compare H2Bub1 profiles in control MCF7 cells or cells depleted for USP22, USP27X, USP51 or ATXN7L3. As expected, depletion of each individual USP led to subtle decreases in global H2Bub1, whereas ATXN7L3 reduction led to a robust increase of global H2Bub1 levels (Figure 6A). Interestingly, ChIP-seq analyses revealed no significant changes in the distribution of H2Bub1 across the genome in ATXN7L3 depleted samples compared to control samples, despite the global increase of H2Bub1 in the depleted cells (Figure 6A). These results are in line with previously published data (Bonnet et al., 2014) describing ATXN7L3 as a global facilitator for H2B deubiquitination throughout the entire transcribed genome. Comparison of the profiles of H2Bub1 in the individual USP depleted cells revealed differential H2Bub1 enrichment over certain loci (examples in Figure 6C). Consistent with the global decreases in H2Bub1 observed by immunoblot, ChIP-Seq analyses uncovered more loci depleted of H2Bub1 than increased (Figure 6B, and tables S1-S3). Again, no significant redistribution of H2Bub1 enrichments was found in the DUB depleted cells relative to control cells, suggesting that these DUB modules may all contribute to maintenance of proper levels of H2Bub1 across the genome.
Figure 6. USP27X and USP51 are required for proper gene expression.
(A) Global levels of H2Bub1 upon depletion of USP22, USP27X, USP51, or ATXN7L3 in MCF7 cells, relative to total H2B. (B) H2Bub1 ChIP-seq analyses identified differential H2Bub1 enrichment at several loci in USP22, USP27X, or USP51 KD cells, but not in ATXN7L3 KD cells. MA plots indicate log2 fold change values (y-axis) against average log2 cpm (counts per million) values (x-axis). The green dots represent differentially enriched loci with FDR ≤ 0.05. The red lines mark log2 fold change at 1 and −1. (C) Genome browser tracks (IGV) showing H2Bub1 signal over HOXD10, HOXD12, HOXB3 and WNT5A loci upon the indicated depletions in MCF7 cells. Dashes (A, B and C) below the tracks indicate regions amplified in qPCR experiments in panel D. Scale on left (0-8) indicates the height of normalized H2Bub1 signal (see supplementary methods). (D) ChIP-qPCR of H2Bub1 levels at the indicated loci, upon depletion of USP22, USP27X, USP51, or ATXN7L3. Values (mean ± SD of three independent experiments) are expressed as a percentage of input. (E) Expression levels of WNT5A and the indicated HOX genes as measured by qRT-PCR, normalized to levels in shControl cells (set to 1). Error bars represent ± SD of three independent experiments. (F) Altered gene expression in USP22, USP27X, or USP51 depleted cells. MA plots indicate log2 fold change values (y-axis) against average log2 cpm (counts per million) values (x-axis). The red dots represent genes with FDR ≤ 0.05. The red lines mark log2 fold change at 1 and −1. (G) Venn diagram of significantly altered genes upon depletion of each USP.
Because H2Bub1 was previously shown to play an important role in regulation of HOX and WNT gene expression (Mohan et al., 2010; Zhu et al., 2005), and our ChIP-seq analysis revealed reduced H2Bub1 enrichment over some of these genes in USP depleted cells (Figure 6C), we sought to further examine how fluctuations in H2Bub1 enrichment at HOXB3, HOXD10, and WNT5A might alter their expression. qPCR experiments confirmed ~2-4 fold decreases in amounts of H2Bub1 at these loci in USP22 or USP27X depleted cells and a milder decrease in USP51 KD cells, whereas ATXN7L3 ablation led to ~2 fold increase (Figure 6D). These results suggest that the three DUBs can at least partially compensate for each other at these genes and that ablation of one USP likely allows increased activity of the others. Expression analyses (qRT-PCR) of HOXA9, 10, 11, HOXB3 and 4, or HOXD10, 11, 13, as well as WNT5A, revealed that all of these genes are down-regulated upon ablation of each USP or ATXN7L3 (Figure 6E), suggesting that dynamic H2B ubiquitination and deubiquitination are required for accurate expression of these loci.
To more fully identify genes or pathways misregulated upon depletion of USP22, USP27X, or USP51, we performed RNA-sequencing (RNA-seq) of RNA purified from shUSP22, shUSP27X, and shUSP51, or shControl treated MCF7 cells. The expression of ~8-10% of all expressed genes was significantly altered upon depletion of each individual USP (Figure 6F and tables S4, S5, and S6). These analyses revealed a substantial overlap of altered genes between the depletion of each of the three USPs, including 340 commonly affected genes (p-value 1.1e-130 by Fisher’s exact test) in USP27X and USP51 depleted cells, 283 (p-value 3.3e-34) commonly affected genes in USP22 and USP27X depleted cells, 249 commonly affected genes (p-value 1.1e-74) in USP22 and USP51 depleted cells, and 137 commonly altered genes in all three USP depletions (~10% of all altered genes) (Figure 6G). GO analyses revealed that axon guidance, ephrin receptor and Wnt/β-catenin signaling are among the top 10 most altered pathways in each USP-depleted cell line (Figure S6A). These results are in good agreement with the discovery that loss of function of Nonstop or Sgf11 (the fly orthologs of USP22 and ATXN7L3 respectively) cause photoreceptor axon-targeting defects in Drosophila (Weake et al., 2008). We also performed hierarchical clustering on differentially expressed genes in shUSP27X, shUSP51 or shUSP22 cells vs shControl cells, using log2 ratios, and then divided the genes into 10 groups, according to the dendogram (Figure S6B and Table S7), and performed a pathway analysis on each group. Genes associated with hepatic stellate cell activation, endocytic pathways, and actin based motility, as well as genes associated with DNA repair showed the most differential expression between USP22, USP27X, and USP51 depleted samples. Overall these data suggest that each USP may have some specific targets, but also that ablation of any one of the USPs influences pathways controlled by the other two.
USP27X and USP51 Are Required for Tumor Growth in a Mouse Xenograft Model
The above studies also indicated that USP27X depletion impacts expression of genes involved in cell growth and cell cycle regulation (Figure S7), so we examined the proliferation of MCF7 breast cancer cells depleted for either USP27X or USP51. Equal numbers of cells expressing shRNAs that specifically target USP27X or USP51, but not USP22 (Figure 7A), or expressing control shRNA, were seeded and monitored for proliferation by cell counts 72 hours later. Depletion of either USP27X or USP51 significantly impacted MCF7 proliferation (Figure 7B). To further confirm the specificity of these effects, we performed rescue experiments using cell lines that stably express shRNA-immune constructs bearing either WT or catalytically dead (C285S) FLAG-tagged USP27X (Figure 7C). Expression of WT-USP27X on its own led to slightly increased proliferation, whereas expression of the C285S mutant led to slightly reduced proliferation. Importantly, expression of WT, but not mutant USP27X, prevented the proliferation defects caused by depletion of endogenous USP27X (Figure 7D). Together, these data clearly demonstrate that USP27X and USP51 are required for normal cellular proliferation.
Figure 7. USP27X and USP51 are required for xenograft tumor growth.
(A) Immunoblots to demonstrate silencing efficiency of USP27X and USP51 in MCF7 cells (B) Comparison of cell growth upon silencing of USP27X or USP51. 0.3×105 cells were seeded and cells were recounted after 72h. Plots show averages of 3 independent experiments. (C) Immunoblots comparing expression of WT and C285S mutant FLAG-V5-USP27X. (D) Endogenous USP27X was depleted in MCF7 cells expressing WT or C285S USP27X-V5-FL. Proliferation was measured by seeding of 0.3×105 cells each and then recounting after 48h. (E) USP27X silencing efficiency in MDA-MB-231-Luc cells. (F) USP27X is required for xenograft tumor growth. 1×105 MDA-MB-231-Luc cells stably expressing the indicated shRNA were injected into 8 week old NOD/SCID mice. The weights of shUSP27X expressing and control tumors are presented in (G). (H) Significant elevation of ATXN7L3 and ENY2 levels in breast cancer patients compared to healthy controls. Plot was generated based on log2 change in gene expression (708 cancer patients and 101 healthy individuals) per TCGA database. p-values in B, D, G and H are based on a Student’s t-test. (I) Model for USP22, USP27X, and USP51 function in cells. All 3 USPs require ATXN7L3 and ENY2 for activation, and in WT cells, these adapters are distributed between the different DUB complexes. Depletion of ATXN7L3 and ENY2 abolishes the activity of all 3 DUBs, leading to major increases in H2Bub1 levels. Depletion of individual USPs, however, leads to increased incorporation of ATXN7L3 and ENY2 into the remaining DUB complexes, increasing their activity.
Because USP22 is overexpressed in several highly aggressive cancers (Glinsky, 2006; Ji et al., 2015; Schrecengost et al., 2014), we next asked if depletion of USP27X impacts tumor formation and progression in a mouse xenograft model. USP27X depleted or control MB-MDA-231 cells were injected subcutaneously into immune-compromised mice (Figure 7E, F). Injection of control cells bearing non-targeting shRNA led to formation of tumors with an average mass of 600 mg by 30 days after injection, as expected. Injection of USP27X depleted cells, however, led to formation of significantly smaller tumors (Figure 7F, G). Very similar results were obtained using different shRNA vectors targeting USP27X (Figure S7), confirming reproducibility of these effects.
The reduced tumor burden upon depletion of USP27X prompted us to search the The Cancer Genome Atlas (TCGA) database to see if altered expression of USP27X or USP51 is associated with certain cancer phenotypes. We did not uncover any specific patterns for USP27X or USP51, but strikingly, increased expression of ENY2 or ATXN7L3 strongly correlates with the increased breast cancer incidence (Figure 7H). Taken together with previous findings by others that USP22 is overexpressed in many cancers and our findings that ATXN7L3 and ENY2 levels are normally limiting in cells, these results suggest that imbalances in the activities of USP22, USP27X and USP51 may directly impact cancer development and progression.
DISCUSSION
Global defects in H2Bub1 deubiquitination observed upon ATXN7L3 silencing previously led to the conclusion that SAGA is the major regulator of H2Bub1 in mammalian cells (Bonnet et al., 2014; Lang et al., 2011). Our results now reveal that in addition to USP22, ATXN7L3 and ENY2 activate two previously uncharacterized deubiquitinating enzymes, USP27X and USP51, which are not part of SAGA. Our work demonstrates that depletion of USP22, USP27X, or USP51 alone leads to a reduction in global H2Bub1 levels and that only simultaneous depletion of all three DUBs increases bulk levels of H2Bub1. Moreover, our work indicates that these three USPs compete for limiting amounts of ATXN7L3 and ENY2 in cells, and that imbalances in one USP affects activities of the others by changing availability of the adapter proteins.
Our ChIP-seq analyses revealed that ablation of each individual DUB had no apparent effect on the overall distribution of H2Bub1 across the genome, but rather caused differences in the enrichment of this mark over certain loci, which in turn correlated with changes in gene expression. H2Bub1 is reduced, rather than increased, at the majority of the altered loci. Substantial overlap in loci affected by USP22 and USP27X depletion (712 out of 1619 loci) suggests that these DUBs can at least partially compensate for, and likely compete with, each other in regulation of these genes. Although no data are available for the absolute amounts of USP27X and USP51 protein present in cells, a recent report indicates that USP22 is two times more abundant than ATXN7L3 in HeLa cells (Hein et al., 2015), further indicating that ATXN7L3 and ENY2 are likely limiting for H2Bub1 deubiquitination.
Although two H2B deubiquitinating enzymes also exist in yeast, Ubp8 and Ubp10, only Ubp8 is part of SAGA and associates with ATXN7L3 and ENY2 orthologs (Sgf11 and Sus1). Ablation of either Ubp8 or Ubp10 alone results in major increases in bulk levels of H2Bub1, in contrast to the slight decreases seen upon deletion of USP22, USP27X, or USP51 in mammalian cells. These results suggest that Ubp8 and Ubp10 likely target different pools of H2B for deubiquitination in yeast and that they do not compensate for one another. The same may also be true in flies, as deletion of the SAGA associated USP22 homolog Nonstop leads to a major increase in H2Bub1 levels (Weake et al., 2008). Why the USP22 family is expanded in mammalian cells is not yet clear.
MudPIT analyses revealed that USP27X and USP51 are incorporated into very similar complexes. The interacting partners of these DUBs suggest that they likely function in transcriptional regulation, RNA processing, and DNA replication and repair. Interestingly, one of the USP27X and USP51 associated proteins, C1QBP, was originally described as a mitochondrial protein (Dedio et al., 1998). Several recent studies, however, indicate C1QBP1 has important functions outside of the mitochondria in RNA splicing and in rRNA processing (Yoshikawa et al., 2011). Fibrillarin, a partner of C1QBP in rRNA processing, was also present in the USP27X and USP51 purifications. Therefore, these DUBs may be important for proper rRNA expression and processing.
Currently, we do not know how USP27X or USP51 is targeted to chromatin. The zinc finger domain of ATXN7L3 may help direct these USPs to chromatin, as this domain mediates association of the SAGA DUBm with nucleosomes in vitro (Lang et al., 2011) and in yeast (Koehler et al., 2014; Morgan et al., 2016).
USP22 overexpression is associated with highly aggressive cancers, and our data indicate that USP27X also contributes to tumor growth. Our finding that depletion of each of the three USPs significantly alters the expression of many genes involved in adhesion and cell motility raises the possibility that USP27X and USP51 also play roles in controlling cell migration, as does USP22 (Nonstop) (Glinsky, 2006; Weake et al., 2008). Due to the lack of ChIP grade antibodies, we have not yet been able to define loci directly bound by USP27X, USP51, or USP22, so we cannot specify genes and pathways directly governed by these DUBs. USP27X and USP51 may also target non-histone substrates important for normal cell identity, growth, or behavior, as previously reported for USP22 (Atanassov and Dent, 2011; Atanassov et al., 2009; Lin et al., 2012). Interestingly, mutations in the USP27X locus were recently described as a primary cause of intellectual disabilities in humans (Hu et al., 2015). Additional work is needed to define the molecular basis of these defects.
Our data emphasize the importance of balance between the three ATXN7L3 and ENY2 associated DUBs (Figure 7I). Given that USP27X ablation limits growth of xenograft tumors and that ENY2 is highly overexpressed in many cancers, the consequences of USP22 overexpression must be evaluated in terms of possible effects on ENY2 and ATXN7L3 availability and downstream impacts on USP27X and USP51 activity.
EXPERIMENTAL PROCEDURES
In vitro Deubiquitination Assays
Nucleosomes, purified histones, or ubiquitin-AMC were used as substrates of USP27X or USP51 DUB modules isolated from Sf21 insect cells in in vitro assays as previously described in (Lan et al., 2015).
Gel filtration and Mass Spectrometry
FL-V5-USP27X and FL-V5-USP51 interacting proteins were affinity purified using the FLAG epitope from 293T whole cell lysates, and the eluted complexes were loaded onto a Superose 6 column and fractionated by size as described (Mohan et al., 2014).
Mass spectrometry and MudPIT analyses were done as in (Byrum et al., 2012; Washburn et al., 2001)
More detailed experimental procedures used in this work are presented in the supplemental information.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. L. Tora for the αATXN7L3 antibody, and Dr. D. Reinberg for the pINTO-N-FH expression vector. This work was largely supported by NIH grant R01 GM096472 to S.Y.R.D. B.A. acknowledges support from CSCDB at MDACC. A.J.T acknowledges the UAMS Proteomics Core Facility and NIH grants R01GM106024, R33CA173264, UL1TR000039, P20GM103429 and P20GM103625. R.M and J.L.W acknowledge support from NIH grant RO1GM 099945 and from the Stowers Institute for Medical Research. Sequencing was done by the Science Park NGS Core, supported by CPRIT Core Facility Support Grant RP120348. Animal services were supported by the MDACC Support grant from the NCI, CA16672. FACS was performed at the Science Park FACS core, partially supported by the Center for Environmental and Molecular Carcinogenesis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
B.A. Performed most of the experiments and analyzed the data. S.Y.R.D. aided in study design, data interpretation, and manuscript writing. R.M. and J.L.W. performed gel filtration experiments and assisted with analyses of proteomics data. X.J.L. performed in vitro DUB assays. E.M. and X.K helped with plasmid subcloning. W.L preformed ChIP experiments. Y.Z., L.F., S.D.B., S.G.M., A.J.T. and M.P.W. performed mass spectrometry, MudPIT, and proteomics data analyses. Y.L. and K.L. performed bioinformatic analyses. E.K. and L.W. provided Usp22 KO mES cells. X.K., T.D. and D.T. helped perform xenograft experiments.
REFERENCES
- Atanassov BS, Dent SY. USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator FBP1. EMBO Rep. 2011;12:924–930. doi: 10.1038/embor.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassov BS, Evrard YA, Multani AS, Zhang Z, Tora L, Devys D, Chang S, Dent SY. Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol. Cell. 2009;35:352–364. doi: 10.1016/j.molcel.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet J, Wang CY, Baptista T, Vincent SD, Hsiao WC, Stierle M, Kao CF, Tora L, Devys D. The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev. 2014;28:1999–2012. doi: 10.1101/gad.250225.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrum S, Smart SK, Larson S, Tackett AJ. Analysis of stable and transient protein-protein interactions. Methods in Mol. Biol. 2012;833:143–152. doi: 10.1007/978-1-61779-477-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedio J, Jahnen-Dechent W, Bachmann M, Muller-Esterl W. The multiligand-binding protein gC1qR, putative C1q receptor, is a mitochondrial protein. J Immunol. 1998;160:3534–3542. [PubMed] [Google Scholar]
- Dowhan DH, Hong EP, Auboeuf D, Dennis AP, Wilson MM, Berget SM, O'Malley BW. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERalpha and CAPERbeta. Mol. Cell. 2005;17:429–439. doi: 10.1016/j.molcel.2004.12.025. [DOI] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ., 3rd KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- Fritsch C, Herrmann A, Nothnagel M, Szafranski K, Huse K, Schumann F, Schreiber S, Platzer M, Krawczak M, Hampe J, et al. Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res. 2012;22:2208–2218. doi: 10.1101/gr.139568.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Hollander D, Voichek Y, Ast G, Oren M. Cotranscriptional histone H2B monoubiquitylation is tightly coupled with RNA polymerase II elongation rate. Genome Res. 2014;24:1572–1583. doi: 10.1101/gr.176487.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinsky GV. Genomic models of metastatic cancer: functional analysis of death-from-cancer signature genes reveals aneuploid, anoikis-resistant, metastasis-enabling phenotype with altered cell cycle control and activated Polycomb Group (PcG) protein chromatin silencing pathway. Cell Cycle. 2006;5:1208–1216. doi: 10.4161/cc.5.11.2796. [DOI] [PubMed] [Google Scholar]
- He Y, Jin YJ, Zhang YH, Meng HX, Zhao BS, Jiang Y, Zhu JW, Liang GY, Kong D, Jin XM. Ubiquitin-specific peptidase 22 overexpression may promote cancer progression and poor prognosis in human gastric carcinoma. Trnsl. Res. 2015;165:407–416. doi: 10.1016/j.trsl.2014.09.005. [DOI] [PubMed] [Google Scholar]
- Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, et al. A Human Interactome in Three Quantitative Dimensions Organized by Stoichiometries and Abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Haas SA, Chelly J, Van Esch H, Raynaud M, de Brouwer AP, Weinert S, Froyen G, Frints SG, Laumonnier F, et al. X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol. Psychiatry. 2015;21:133–48. doi: 10.1038/mp.2014.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsdottir K, Krogan NJ, Emre NC, Wyce A, Thompson NJ, Emili A, Hughes TR, Greenblatt JF, Berger SL. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol. Cell. Biol. 2005;25:1162–1172. doi: 10.1128/MCB.25.3.1162-1172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji M, Shi H, Xie Y, Zhao Z, Li S, Chang C, Cheng X, Li Y. Ubiquitin specific protease 22 promotes cell proliferation and tumor growth of epithelial ovarian cancer through synergy with transforming growth factor beta1. Oncol. Rep. 2015;33:133–140. doi: 10.3892/or.2014.3580. [DOI] [PubMed] [Google Scholar]
- Koehler C, Bonnet J, Stierle M, Romier C, Devys D, Kieffer B. DNA binding by Sgf11 protein affects histone H2B deubiquitination by Spt-Ada-Gcn5-acetyltransferase (SAGA) J. Biol. Chem. 2014;289:8989–8999. doi: 10.1074/jbc.M113.500868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Pascual-Garcia P, Llopis A, Zapater M, Posas F, Hurt E, Rodriguez-Navarro S. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol. Biol. Cell. 2006;17:4228–4236. doi: 10.1091/mbc.E06-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat. Cell. Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- Kohler A, Zimmerman E, Schneider M, Hurt E, Zheng N. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell. 2010;141:606–617. doi: 10.1016/j.cell.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan X, Koutelou E, Schibler AC, Chen YC, Grant PA, Dent SY. PolyQ expansions in ATXN7 affect solubility but not activity of the SAGA deubiquitinating module. Mol. Cell. Biol. 2015;35:1777–87. doi: 10.1128/MCB.01454-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G, Bonnet J, Umlauf D, Karmodiya K, Koffler J, Stierle M, Devys D, Tora L. The tightly controlled deubiquitination activity of the human SAGA complex differentially modifies distinct gene regulatory elements. Mol. Cell. Biol. 2011;31:3734–3744. doi: 10.1128/MCB.05231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Sardiu ME, Swanson SK, Gilmore JM, Torok M, Grant PA, Florens L, Workman JL, Washburn MP. Combinatorial depletion analysis to assemble the network architecture of the SAGA and ADA chromatin remodeling complexes. Mol. Syst. Biol. 2011;7:503. doi: 10.1038/msb.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang JX, Ning Z, Gao W, Ling J, Wang AM, Luo HF, Liang Y, Yan Q, Wang ZY. Ubiquitinspecific protease 22induced autophagy is correlated with poor prognosis of pancreatic cancer. Onco. Rep. 2014;32:2726–2734. doi: 10.3892/or.2014.3508. [DOI] [PubMed] [Google Scholar]
- Lin Z, Yang H, Kong Q, Li J, Lee SM, Gao B, Dong H, Wei J, Song J, Zhang DD, et al. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol. Cell. 2012;46:484–494. doi: 10.1016/j.molcel.2012.03.024. [DOI] [PubMed] [Google Scholar]
- Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat. Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, Zhang Y, Washburn MP, Florens L, Shilatifard A. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom) Genes Dev. 2010;24:574–589. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan RD, Dialynas G, Weake VM, Liu J, Martin-Brown S, Florens L, Washburn MP, Workman JL, Abmayr SM. Loss of Drosophila Ataxin-7, a SAGA subunit, reduces H2B ubiquitination and leads to neural and retinal degeneration. Genes Dev. 2014;28:259–272. doi: 10.1101/gad.225151.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MT, Haj-Yahya M, Ringel AE, Bandi P, Brik A, Wolberger C. Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science. 2016;351:725–728. doi: 10.1126/science.aac5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning J, Zhang J, Liu W, Lang Y, Xue Y, Xu S. Overexpression of ubiquitin-specific protease 22 predicts poor survival in patients with early-stage non-small cell lung cancer. Eur. J. Histochem. 2012;56:e46. doi: 10.4081/ejh.2012.e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, Wolberger C. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science. 2010;328:1025–1029. doi: 10.1126/science.1190049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrecengost RS, Dean JL, Goodwin JF, Schiewer MJ, Urban MW, Stanek TJ, Sussman RT, Hicks JL, Birbe RC, Draganova-Tacheva RA, et al. USP22 regulates oncogenic signaling pathways to drive lethal cancer progression. Cancer Res. 2014;74:272–286. doi: 10.1158/0008-5472.CAN-13-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell. 2010;40:689–701. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf D, Bonnet J, Waharte F, Fournier M, Stierle M, Fischer B, Brino L, Devys D, Tora L. The human TREX-2 complex is stably associated with the nuclear pore basket. J. Cell Sci. 2013;126:2656–2667. doi: 10.1242/jcs.118000. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Weake VM, Lee KK, Guelman S, Lin CH, Seidel C, Abmayr SM, Workman JL. SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. Embo J. 2008;27:394–405. doi: 10.1038/sj.emboj.7601966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol. Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Wegrzyn JL, Drudge TM, Valafar F, Hook V. Bioinformatic analyses of mammalian 5'-UTR sequence properties of mRNAs predicts alternative translation initiation sites. BMC Bioinformatics. 2008;9:232. doi: 10.1186/1471-2105-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Wang Y, Gong Z, Liu J, Li W. Identification of a functional nuclear localization signal within the human USP22 protein. Biochem. Biophys. Res. Commun. 2014;449:14–18. doi: 10.1016/j.bbrc.2014.04.133. [DOI] [PubMed] [Google Scholar]
- Yoshikawa H, Komatsu W, Hayano T, Miura Y, Homma K, Izumikawa K, Ishikawa H, Miyazawa N, Tachikawa H, Yamauchi Y, et al. Splicing factor 2-associated protein p32 participates in ribosome biogenesis by regulating the binding of Nop52 and fibrillarin to preribosome particles. Mol. Cell. Proteomics. 2011;10:M110 006148. doi: 10.1074/mcp.M110.006148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Pfeiffer HK, Thorne AW, McMahon SB. USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2A. Cell Cycle. 2008a;7:1522–1524. doi: 10.4161/cc.7.11.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell. 2008b;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yao L, Zhang X, Ji H, Wang L, Sun S, Pang D. Elevated expression of USP22 in correlation with poor prognosis in patients with invasive breast cancer. J. Cancer Res. Clin. Oncol. 2011;137:1245–1253. doi: 10.1007/s00432-011-0998-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol. Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.