Abstract
Many diseases that originate on mucosal membranes ensue from the action of polymicrobial communities of indigenous organisms working in concert to disrupt homeostatic mechanisms. Multi-level physical and chemical communication systems among constituent organisms underlies polymicrobial synergy and dictates the community’s pathogenic potential or nososymbiocity, that is, disease arising from living together with a susceptible host. Functional specialization of community participants, often originating from metabolic co-dependence, has given rise to several newly appreciated designations within the commensal-to-pathogen spectrum. Accessory pathogens, while inherently commensal in a particular microenvironment, nonetheless enhance the colonization or metabolic activity of pathogens. Keystone pathogens (bacterial drivers or alpha-bugs) exert their influence at low abundance by modulating both the composition and levels of community participants and by manipulating host responses. Pathobionts (or bacterial passengers) exploit disrupted host homeostasis to flourish and promote inflammatory disease. In this review we will discuss how commensal or pathogenic properties of organisms are not intrinsic features, and have to be considered within the context of both the microbial community in which they reside and the host immune status.
Keywords: polymicrobial synergy, keystone pathogen, accessory pathogen, pathobiont, nososymbiocity
The Boyz n the Hood, the Relevance of Dysbiotic Polymicrobial Communities
The fundamental tenets of classical medical microbiology, based on laboratory studies of organisms in isolation, have engendered significant successes against infectious diseases. A long-standing pillar of public health policy is vaccination against diphtheria and tetanus, diseases caused by colonization with a single exogenous pathogen. It is in some sense ironic therefore, that such advances may have hindered our understanding of many other human diseases which are not caused by a single organism, but rather by dysbiotic communities of indigenous organisms working in concert (Table 1). Appreciation of the polymicrobial nature of certain diseases, and the lack of a single, exogenous microbial target, is now well established, and necessitates specific considerations for diagnosis, treatment and prevention. Moreover, polymicrobial accumulations of bacteria attain quasi-organismal status due to functional specialization of constituent species and coordination of their behavior through sophisticated signaling systems. The concept of dysbiotic endogenous communities as etiological agents also challenges the long-held notion that bacteria can be divided into two categories: commensals and pathogens. After all, these are conceptual structures that microbiologists have imposed upon organisms, they do not necessarily represent intrinsic properties of the organisms themselves. It now appears more accurate to consider the health- or disease- associated properties of an organism as a spectrum from commensalism to pathogenicity encompassing many newly recognized categories such as accessory pathogens, pathobionts and keystone pathogens. Furthermore, when referring to the potential of an indigenous community to cause disease, ‘nososymbiocity’ (nosos [Greek for disease] arising from host-microbe symbiosis [literally ‘living together’ in Greek]) may be a more accurate term than ‘pathogenicity’, which implies the presence of causative pathogens without considering the contextual nature of the bacterial role in health vs. disease. Additionally, polymicrobial communities present unique challenges to the immune system. Whereas collections of commensal organisms may contribute to the programming and maintenance of homeostatic immunity, dysbiotic communities induce an immune response that is ineffective, uncontrolled and destructive. In this review, we describe more recently appreciated aspects of microbial pathogenic potential, and discuss how the relationships among specialized community participants establish an emergent overall behavior of microbial communities in the context of host recognition and response.
Table 1.
Examples of Interbacterial Interactions in Polymicrobial Community Diseases
| Disease | Interacting organismsa | Mechanism of interaction and outcome | Refs |
|---|---|---|---|
| Wound Infections | Staphylococcus aureus – Pseudomonas aeruginosa | In dual species biofilms expression of staphylococcal Panton-Valentine toxin and α-hemolysin is increased. P. aeruginosa responds to staphylococcal peptidoglycan by elevated production of quinolone signal (PQS) controlled virulence factors. | [71, 72] |
| Cystic fibrosis | P. aeruginosa - Burkholderia cenocepacia and Stenotrophomonas maltophilia | Diffusible signal factor (DSF) family molecules (cis-2-unsaturated fatty acids) produced by B. cenocepacia and S. maltophilia increase P. aeruginosa persistence and antibiotic resistance. | [73] |
| Otitis media | Haemophilus influenzae - Moraxella catarrhalis | AI-2 produced by H. influenzae increases mixed biofilm development and antibiotic resistance in M. catarrhalis. | [17] |
| Gastrointestinal infections | Bacteroides. thetaiotaomicron - EHEC | B. thetaiotaomicron produces fucosidases that generate fucose from host-derived glycans. Fucose increases EHEC virulence gene expression by activating FusKR. Short chain fatty acids and succinate produced by Bacteroides also increase virulence gene expression in EHEC. | [32, 34, 35] |
| Oropharyngeal candidiasis | Candida albicans – Streptococcus oralis and S. gordonii | Streptococcal AI-2, peptidoglycan and peroxide can increase Candida filamentation and dissemination. | [6, 74] |
| Localized aggressive periodontitis | Aggregatibacter actinomycetemcomitans – S. gordonii | A. actinomycetemcomitans preferentially utilizes lactate produced by S. gordonii, and peroxide generated by S. gordonii increases expression of virulence genes in A. actinomycetemcomitans. | [15, 28] |
| Chronic periodontitis | Porphyromonas gingivalis – S. gordonii | Co-adhesion initiates tyrosine (de) phosphorylation dependent signaling pathway in P. gingivalis that enhances dual species biofilm development. | [3, 4] |
Examples of organisms for which interspecies communication has been defined. These polymicrobial infections can involve several other species and additional synergistic interactions.
LinkedIn: Community Infrastructure
The development of heterotypic bacterial communities is a highly orchestrated process dependent on mutual interspecies binding interactions and metabolic compatibility. The physical association (termed coadhesion or coaggregation) of microbial species provides the structural framework for a community [1]. Many of the pioneering studies of coadhesion, and indeed of tissue tropisms defined by substrate-specific adherence, involved the oral ecosystem where coadhesion drives the temporal and spatial development of the highly heterogeneous oral biofilms [2]. For example, oral colonization by the anaerobe Porphyromonas gingivalis is facilitated by coadhesion with the antecedent colonizer Streptococcus gordonii [3]. Coadhesion is mediated by two sets of adhesin-receptor pairs, the FimA and Mfa1 subunit fimbriae on P. gingivalis which respectively engage GAPDH and the major wall protein SspA/B on the streptococcal surface [3]. The binding interaction extends beyond passive attachment, and in P. gingivalis a tyrosine (de)phosphorylation-dependent signaling pathway is activated which induces a pattern of gene expression to prepare the organisms for a community environment [4]. Furthermore, both P. gingivalis and S. gordonii differentially regulate a significant proportion of their expressed proteomes, indicating extensive signaling between the organisms [5]. Cross-kingdom coadhesion between oral streptococci and Candida albicans is also well-documented. The functionally versatile streptococcal SspA/B protein mediates this attachment through binding to the glycosylphosphatidylinositol (GPI)-linked candida cell wall protein Als3 [6]. The Als3 protein is uniquely expressed on hyphae, and hence streptococci decorate the hyphal surfaces of C. albicans in mixed-species communities. Als3p also binds to multiple staphylococcal adhesins, and this partnership can enhance staphylococcal invasion into host tissues and consequently a disseminated staphylococcal infection [7].
The close physical association between coadhered organisms facilitates nutritional relationships. P. gingivalis produces isobutyric acid which stimulates growth of the oral spirochete Treponema denticola, while T. denticola produces succinic acid which enhances growth of P. gingivalis [8]. Contact with T. denticola also upregulates the expression of P. gingivalis adhesins and proteases [9]. Consequently, growth of P. gingivalis and T. denticola in a dual species biofilm produces a significantly larger biomass compared to the total of the individual monospecies biofilms, and the organisms are synergistically pathogenic in murine models of periodontal disease [8, 10]. Another advantage to the community lifestyle is the potential to benefit from the metabolic enzymes possessed by partner organisms. This process, known as syntrophy (literally meaning ‘eating together’), allows the sequential degradation of large heterogenous substrates. For example, a four-species consortium of oral streptococci can digest the salivary mucin MUC5B, whereas the same species individually are unable to degrade this complex oligomeric glycoprotein [11].
Physical associations among organisms are facilitated and augmented by chemical signaling [12] and, as recently recognized, electrical communication through pilus-based nanowires [13]. Chemical signaling can occur through diffusible signal factors (DSFs), quorum sensing with short (22–23 amino acid) peptides and with autoinducers (AIs), and other metabolites such as short chain fatty acids (SCFAs), lactate, indole and hydrogen peroxide [12]. Remarkably, some bacteria such as Escherichia coli and Aggregatibacter actinomycetemcomitans can also sense and respond to host catecholamine hormones (such as epinephrine and norepinephrine, as well as adrenaline and noradrenaline) [14]. Communication using these diffusible signals and its impact on microbial physiology and community development, has been extensively reviewed in recent articles [6, 12, 15, 16]. For example, AI-2 produced by Haemophilus influenzae and sensed by Moraxella catarrhalis leads to elevated dual-species biofilm mass containing higher numbers of viable bacteria, along with the induction of antibiotic resistance in M. catarrhalis [17].
A trend thus emerges of organisms favoring a lifestyle in a community with other compatible species, and indeed polymicrobial communities dominate the skin and mucosal surfaces of humans. That is not to imply that all interactions between microbes are symbiotic, as there are well-documented antagonistic interactions among various species, and consequently these organisms tend not to be found in close proximity in vivo [1, 16, 18]. Polymicrobial synergy and antagonism therefore shapes community composition and consequently nososymbiocity.
The Good, the Bad, and the Indifferent
An almost bewildering diversity and number of bacteria inhabit the human host, and yet in most people, most of the time, this is the normal healthy situation. In essence, host defense against non-self and bacterial expansion are balanced, and colonization is restricted to superficial layers of tissue. In some cases, such as the healthy gingiva, a low level of inflammation with controlled recruitment of neutrophils is maintained to constrain the bacterial challenge [19]. Moreover, the indigenous microbiota can play an active role in the maintenance of health and provide protection against exogenous pathogens. While the full range and mechanistic basis for such colonization resistance are not fully understood, both bacteria-bacteria and bacteria-host interactions are involved. Interbacterial interactions include the inhibitory action of toxic metabolites, bacteriocins, antibiotics, and type VI secretion systems, along with competition for space and nutrients [20]. Competition is more effective between organisms that occupy similar niches and have related metabolic requirements. As an illustration, strains of E. coli that inhabit the healthy gastrointestinal (GI) tract can outcompete the closely related murine organism Citrobacter rodentium, which serves as a model for enterohaemorrhagic (EHEC) and enteropathogenic (EPEC) E. coli [21]. Antagonism derives from competition for structurally similar carbohydrates, and consistent with this, Bacteroides species which have different nutritional requirements are unable to compete with C. rodentium [21]. Host innate and adaptive immune responses to indigenous communities can improve the protective functions of the mucosal barrier by enhancing tight junction formation and increasing production of antimicrobial peptides, cytokines/chemokines and secretory IgA antibodies which have been proposed to target preferentially commensals with pathobiont potential, at least in the context of colitis [20, 22]. Furthermore, it is now apparent that the indigenous microbial community is required for the normal development of a fully competent immune system (Box 1).
Box 1. Role of the Indigenous Microbiota in Host Immunity.
On the one hand constrained by the host immune system, on the other the indigenous microbiota regulates certain basic developmental features and functions of the immune system [75]. The use of germ-free mice, and mice with a knockout in genes controlling various bacterial recognition systems, has led to an appreciation that microbial colonization is required to engender a fully functional immune system. In the absence of the gut microbiota there is incomplete development of the gut-associated lymphoid tissues (GALT), including Peyer’s patches, crypt patches and isolated lymphoid follicles (ILFs). Deficiencies in the GALT, in which antigen is acquired and processed by antigen-presenting cells, will in turn impact lymphocyte functions. Indeed, the indigenous microbiota regulates the development of TH17 lymphocytes, a lineage of CD4+ TH cells that produce IL-17 and IL-22 and are important for gut and oral immune competence. In the mouse small intestine, colonization with a single organism, a segmented filamentous bacterium (SFB), related to Clostridia, is sufficient to produce TH17 cells [76]. The resident microbiota is also partially responsible for the development of another CD4+ TH subset, the FOXP3+ regulatory T cells (TRegs) [77]. As TRegs produce IL-10 they are important for the prevention of uncontrolled and sustained inflammatory responses. In addition to T cell maturation, microbial colonization is required for the generation of IgA-producing B-cells. Through a variety of mechanisms, resident organisms cause lamina propia dendritic cells and follicular dendritic cells to induce B cell differentiation [78]. Epithelial function is enhanced by bacterial colonization. MyD88-dependent bacterial signaling can improve repair of damaged epithelium and incite production of epithelial antimicrobial proteins such as RegIIIγ[79].
The importance of colonization resistance is evident from cases where the indigenous microbiota is disrupted or suppressed, thereby allowing colonization by exogenous pathogens and outgrowth of indigenous pathobionts. For instance, the administration of broad-spectrum antibiotics in hospitalized patients can lead to substantial increases in the abundance of pathobionts with potential for systemic dissemination and induction of septic shock [20]. Exogenous pathogens are equipped with tools to overcome colonization resistance, such as more efficient utilization of nutrients or iron acquisition [20]. While these are not always considered classical virulence factors, they are, nonetheless, essential for the early stages of the disease process. For example, EHEC strains which possess the eut operon encoding the enzymes necessary for utilization of ethanolamine as a nitrogen source in the ruminant reservoir may have a competitive advantage in microenvironments rich in indigenous E. coli strains that lack, or poorly express, this operon [23].
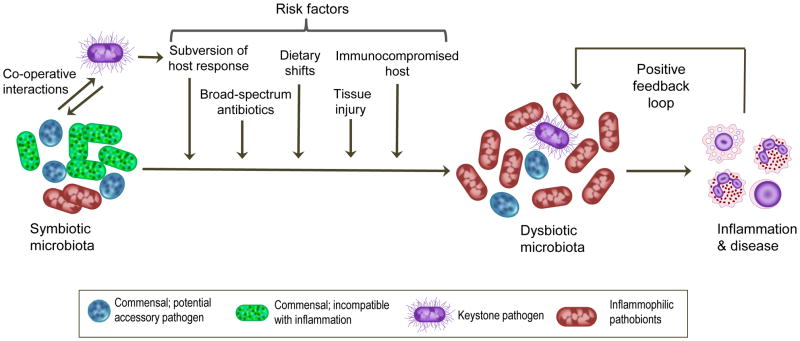
Notwithstanding the benefits of the indigenous microbiota, an increase in nososymbiocity is the basis of a number of diseases (Table 1). A variety of factors can underlie the transition of a microbial community to a dysbiotic state (see below and Figure 1, Key Figure). For instance, alterations in host status affecting immune competence or nutrient availability could modulate polymicrobial community composition, the metatranscriptional landscape, or translocation to previously sterile sites. As a community develops, changes to the microenvironment such as availability of oxygen or pH can provide physiological support for the outgrowth or over-representation of dysbiotic organisms [3, 16]. Given the complexity of microbial communities, along with metabolic partitioning within the community, a metastable state is likely to prevail. Thus, once a dysbiotic condition has been induced, disease potential could persist for an extended time. Below we review in greater detail and with specific examples a triad of protagonists in a dysbiotic community, accessory pathogens, keystone pathogens, and pathobionts.
Figure 1. Factors That Promote Dysbiosis.
Mucosal inflammatory disease is induced under certain conditions by a polymicrobial community, in which different members have distinct and synergistic roles that promote destructive inflammation. Keystone pathogens — which are aided by accessory pathogens in terms of nutritional and/or colonization support — initially subvert host immunity leading to the emergence of a dysbiotic microbiota, in which commensal-turned pathobionts overactivate the inflammatory response and cause tissue destruction. Inflammation can exacerbate dysbiosis through several mechanisms ([56–60]; see text for details), hence inflammation and dysbiosis positively reinforce each other. Additional risk factors that shift the balance towards dysbiosis and the emergence of inflammophilic pathobionts include (but are not limited to) frequent use of antibiotics, high-fat diet, tissue injury, and immune deficiencies. These factors could promote dysbiosis by acting individually or in combination.
Lending a Helping Hand: Accessory Pathogens
As a concept associated with pathogenicity, virulence can be meaningfully defined only in the context of a susceptible host. Accordingly, whether an organism can behave as a ‘pathogen’ or ‘commensal’ may not necessarily be predicted from its capacity, or lack thereof, to express classical ‘virulence factors’, such as cytotoxins, capsular polysaccharide, or invasins. Therefore, with diseases caused by dysbiotic microbial communities, virulence may refer to any microbial trait that can elevate the pathogenicity of the community. In other words, a microbial function that by itself may not contribute to disease, may do so in an interactive multi-species community, characterized by functional specialization of constituent organisms. This notion has recently shifted our perception away from a commensal-pathogen duality to a more nuanced, context-dependent view of microbial pathogenic potential [24].
Accessory pathogens are a subset of organisms that, while generally perceived as commensal, under certain conditions can act synergistically to support or enhance the virulence of disease-associated organisms (Figure 1). Traditionally an archetypal oral commensal, S. gordonii is now appreciated as an accessory pathogen par excellence. Not only does the organism provide an attachment substratum for colonization by the keystone periodontal pathogen P. gingivalis, but dual species communities of S. gordonii and P. gingivalis are more pathogenic in animal models compared to either species alone [25, 26]. Remarkably, S. gordonii also promotes the pathogenicity of A. actinomycetemcomitans, another important periodontitis-associated organism, by cross-feeding with L-lactate, a metabolic by-product of S. gordonii [27]. To enable cross-feeding, A. actinomycetemcomitans employs a detoxification and dispersion (or ‘fight and flight’) response to reduce exposure to inhibitory levels of streptococcal H2O2 while keeping at an optimal ‘safe’ distance from S. gordonii [28]. Such ‘fight and flight’ interactions can lead to defined spatial arrangements of the various species within communities, thereby optimizing the function of the community as a whole [28].
Reflective of the contextual-dependence of microbial influence, both on other organisms and on the host, P. gingivalis can function as an accessory pathogen in lower airway infection. P. gingivalis enhances the pathogenicity of Pseudomonas aeruginosa in obstructive pulmonary disease exacerbations [29, 30]. Further, P. gingivalis promotes the ability of P. aeruginosa to invade respiratory epithelial cells and modulates its apoptosis-inducing capacity, potentially enhancing the ability to establish infection [29, 30]. Moreover, AI-2 produced by oropharyngeal bacteria modulates gene expression of P. aeruginosa and appears to contribute to lung pathology [31]. Indeed, P. aeruginosa, while unable to produce AI-2, can sense and respond to the signal. Co-culture of P. aeruginosa with AI-2–producing strains of staphylococci and streptococci enhances virulence gene expression and increases tissue damage caused by P. aeruginosa in a rat lung model [31]. Staphylococci and streptococci are often present in the lungs of cystic fibrosis (CF) patients where they may thus act as accessory pathogens and contribute to a polymicrobial etiology of CF.
In the gut, Bacteroides thetaiotaomicron produces fucosidases that use host-derived glycans to generate fucose, which activates the fucose sensor FusKR of EHEC. FusKR activation, in turn, leads to the expression of genes that modulate EHEC pathogenicity and metabolism [32]. Additionally, certain short-chain fatty acids (e.g., acetate and butyrate) produced by intestinal Bacteroides species can increase virulence gene expression in other organisms, such as the SPI-1 invasion genes in Salmonella enterica serovar Typhimurium [33] and the locus of enterocyte effacement (LEE) genes that are required for cell adherence of EHEC and induction of attaching and effacing lesions [34]. Succinate, a common metabolite of gastrointestinal Bacteroides also promotes nososymbiocity. EHEC virulence gene expression is enhanced by succinate through the transcription factor Cra, which is functionally sensitive to succinate concentrations [35]. However, non-pathogenic E. coli can also possess Cra, and the extent to which succinate-mediated interactions are important for disease in the presence of multiple microbial interactions remains to be determined. Indeed, it is important to emphasize that the above examples are components of complex metabolic interactions among bacteria, and that metabolites are likely utilized by other community members which may be critical for disease development. Interactions between typical and accessory pathogens represent only a part of the entire picture of microbial interactions in disease development. For example, the influence of keystone pathogens in microbiota transition may be the result of multiple interactions among community constituents.
Given the widespread distribution and numerical abundance of oral streptococci and gastrointestinal Bacteroides, it is unlikely that these organisms evolved as accessory pathogens; rather their role is more likely unwitting, rather than willing, participants. Hence, while exogenous pathogens employ strategies to overcome colonization resistance, endogenous bacteria with pathogenic potential exploit the inherent metabolic and/or colonization properties of their microbial neighbors to increase the nososymbiocity of the community.
Master Manipulators: Keystone Pathogens
In the ecological literature, the term ‘keystone’ was introduced to characterize species whose influence on their communities is disproportionately large relative to their abundance, thus forming the ‘keystone’ of the community’s structure [36]. Accordingly, the keystone-pathogen hypothesis holds that certain low-abundance microbes can promote the formation of and stabilize dysbiotic and disease-provoking communities [37] (Figure 1). In remodeling a normally symbiotic microbiota into a dysbiotic one, keystone pathogens can cause both quantitative (increased biomass) and qualitative alterations to the microbiota. The latter involve changes in microbial composition, the metatranscriptome and the metaproteome. Keystone pathogen-induced alterations to the microbiota may be mediated through effects on the host (subversion of the immune response) or – more directly – on the microbial community via interspecies interactions with other member species [19, 38–42].
Consistent with the keystone-pathogen concept, a recent study on the intestinal innate immune response of gnotobiotic zebrafish showed that combinations of distinct bacterial species induce immune responses that do not reflect the numerically dominant species [43]. Rather, different species exert different ‘per capita’ immunostimulatory effects, suggesting that knowledge of specific properties of individual species may provide predictive insights into the function of multi-species communities [43]. Moreover, the fact that minor members can exert dominant effects challenges the Occam’s razor interpretation (which is inherent in compositional analyses) that the relative abundances of different taxa or species can have predictive value for pathology. Using a time-series algorithm called LIMITS (Learning Interactions from MIcrobial Time Series) to infer ecological interaction networks in the gut microbiome, a recent study identified keystone species with disproportionate influence on the gut microbiome structure despite their under-representation according to metagenomics abundance data [44]. Interestingly, one of two such species identified is Bacteroides fragilis for which there is experimental evidence that it may act as a keystone species (below). The other putative keystone species identified is Bacteroides stercosis [44], which has also been associated with increased risk of colon cancer [45].
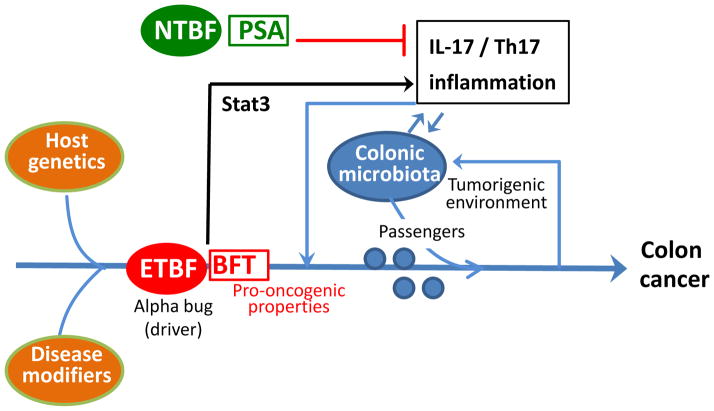
In colorectal cancer, B. fragilis, a low-abundance member of the colonic bacterial community (≤1–2%), has been proposed to be an ‘alpha-bug’ (a functional synonym for a keystone pathogen) which remodels the colonic microbiota and co-opts it in a collaborative manner to induce colon cancer [46]. In this regard, enterotoxigenic B. fragilis (ETBF), an organism associated with acute diarrheal disease and colorectal cancer in humans [47], induces formation of colon tumors in multiple intestinal neoplasia (Min) mice through a Stat3- and Th17-dependent inflammatory pathway [48] (Figure 2), which has been associated with poor prognosis in human colon cancer [49]. In contrast, nontoxigenic B. fragilis (NTBF) fails to activate the Stat3-Th17 pathway and cause colon cancer despite colonizing Min mice [48]. In fact, NTBF expresses polysaccharide A, a symbiosis factor that inhibits intestinal Th17 responses [50] (Figure 2). Antibiotic treatment of ETBF-colonized Min mice alters the carcinogenesis rate without interfering with ETBF colonization, suggesting that the interaction of ETBF with the colonic microbiota modulates the outcome of colon carcinogenesis [46]. The underlying mechanisms are uncertain, but it can be speculated that the ability of ETBF to induce Th17-mediated inflammation modifies the intraluminal environment in a manner that alters the colonic microbiota and its oncogenic potential. Alternatively or additionally, the environmental changes (inflammation and the growing tumor) may selectively suppress the growth of cancer-protective species while promoting the outgrowth of tumor-foraging opportunistic pathogens. The latter possibility has been proposed though a bacterial driver–passenger model for colorectal cancer, the driver aspect of which is related to the 'alpha-bug' hypothesis [51]. Specifically, ‘bacterial drivers’ deliver the ‘first hit’ (e.g., inflammation and DNA damage contributing to initiation of colorectal cancer) and promote the emergence of ‘bacterial passengers’, that is, opportunistic pathobionts that thrive in the tumorigenic environment and exacerbate pathology [51] (Figure 2).
Figure 2. Alpha-bugs and Colon Cancer.
Enterotoxigenic B. fragilis (ETBF) drives persistent Th17- and IL-17-dependent inflammation by activating signal transducer and activator of transcription 3 (STAT3) signaling in the colon, which leads to colonic hyperplasia and tumor formation in the multiple intestinal neoplasia (Min) mouse model. Min mice are susceptible to development of intestinal adenomas, hence ETBF-induced carcinogenesis requires a host with predisposing genetic traits or, in general, particular disease modifiers. ETBF also secretes a pro-oncogenic toxin (B. fragilis toxin; BFT). In contrast, non-enterotoxigenic B. fragilis (NTBF) produces polysaccharide A (PSA) which inhibits intestinal Th17 responses. According to the ‘alpha-bug’ hypothesis, alpha-bugs are not simply pro-oncogenic but can remodel the colonic bacterial community to a state that enhances mucosal inflammation and pro-oncogenic activity, resulting in colon cancer. Consistent with this, an alpha bug could be the ‘driver’ in the bacterial driver–passenger model for colorectal cancer, where the consequences of its actions (inflammatory tumorigenic environment) could lead to the emergence of opportunistic pathogens (’passengers’) that can exacerbate pathology leading to colon cancer.
In a mouse model of periodontitis, P. gingivalis fails to cause disease in germ-free mice despite colonizing this host [38]. In stark contrast, in conventional (specific-pathogen-free) mice, the same bacterium transforms the periodontal microbiota into a dysbiotic, periodontitis-causing community, as long as complement and Toll-like receptor signaling pathways remain intact [38, 39]. In this regard, the ability of P. gingivalis to induce subversive crosstalk between Toll-like and complement receptors on leukocytes leads to selective inhibition of antimicrobial responses and promotion of destructive inflammation, which generates nutritional substrates for the bacteria [24, 39, 52]. Thus, not only does the impairment of host immunity allow unchecked bacterial growth, but also the resulting inflammatory environment favors the development of a subset of species that can capitalize on the inflammatory spoils (tissue breakdown products such as collagen peptides).
Human ulcerative colitis is an inflammatory bowel disease that can be modeled in TRUC mice which are deficient in both the transcription factor T-bet and the adaptive immune response (T-bet−/− × Rag2−/−) [53]. The development of colitis in TRUC mice correlates strongly with the presence of Klebsiella pneumoniae and Proteus mirabilis. The two organisms together induce colitis in specific-pathogen-free wild-type mice but not in germ-free TRUC mice, suggesting that their colitogenicity requires the indigenous microbial community rather than a host with altered immune function [54]. Although K. pneumoniae and P. mirabilis can co-colonize germ-free mice at high levels, they constitute less than 1% of the total fecal microbiota of TRUC mice or of infected wild-type mice [54]. The nature of the interactions of K. pneumoniae and P. mirabilis with the gut microbiota is largely uncertain. Although the action of the two colitogenic organisms seems consistent with that of keystone pathogens, an alternative interpretation is that the indigenous microbiota may induce or prime immunological processes that facilitate destructive inflammatory responses by K. pneumoniae and P. mirabilis.
The observation that TRUC mothers could transmit colitis to cross-fostered wild-type pups [54] indicates that, at least under certain conditions, dysbiosis can be a cause, rather than a consequence, of inflammation. In a similar context, the colitogenic activity of an altered microbiota due to genetic deficiency in NLRP6 inflammasome could be transmitted to cross-fostered neonatal or co-housed adult wild-type mice [55]. In general, dysbiosis and inflammation can certainly reinforce each other regardless of which was the original cause. Intriguingly, not only can inflammation foster the growth of so-called ‘inflammophilic’ bacteria that thrive on inflammatory tissue breakdown products [56] but inflammation can also be sensed directly by bacteria to alter their virulence phenotype. For instance, interleukin-1β (IL- 1β) and tumor necrosis factor (TNF) can promote, respectively, the growth and virulence potential of certain pathogens that bind these cytokines through specific receptors [57, 58]. Similarly, P. aeruginosa can bind to interferon-γ through the outer-membrane porin OprF, thereby increasing the expression of quorum-sensing–dependent virulence determinants, PA-I lectin and pyocyanin, both of which can disrupt epithelial cell function [59]. Although these experiments were performed using high, non-physiological, concentrations of interferon-γ, the specificity of the interaction was validated by using seven other human cytokines at the same concentration. Moreover, acute gut inflammation leads to the generation of a respiratory electron acceptor (tetrathionate) for Salmonella, which thereby out-competes other microbes that rely on anaerobic fermentation [60]. It should be noted, however, that Salmonella is a ‘dominant’ rather than ‘keystone’ pathogen, since it causes gut infection and inflammation while becoming established as the dominant component of the microbiota [61].
In diametric contrast to keystone pathogens, low-abundance symbionts with the ability to stabilize a microbiota in ways that suppress its nososymbiocity were referred to as ‘keystone stabilizers’ [62]. In this regard, B. thetaiotaomicron induces the antimicrobial peptide angiogenin, which kills opportunistic or pathogenic organisms but not B. thetaiotaomicron or other indigenous bacteria [63]. Moreover, B. thetaiotaomicron can suppress inflammation via PPARγ-dependent nuclear export of the p65 subunit of the NF-κB transcription factor [64]. However, as noted above, B. thetaiotaomicron can also upregulate virulence gene expression in EHEC [32], underscoring the contextual nature of classifying bacterial disease potential. In a gnotobiotic zebrafish model, low-abundance Shewanella (probiotic species used in aquaculture) acts as a keystone stabilizer by exerting a disproportionately large effect on intestinal neutrophil influx [43]. Specifically, Shewanella secretes an anti-inflammatory factor that overrides the pro-inflammatory effects of high-abundance species of the community [43]. Numerically minor constituents of microbial communities, therefore, can enhance or reduce nososymbiocity, leading to the notion that functional cataloging of community members is a more relevant prognostic measure than abundance determinations.
Cry Havoc and Let Slip the Pathobionts
Pathobionts are organisms that are generally benign within an indigenous community but become pathogenic when host-microbe homeostasis breaks down under certain conditions, such as antibiotic treatment, tissue damage, dietary shifts, and especially immune deficiencies [20, 65] (Figure 1). As indicated below with specific examples, these conditions can potentially promote the outgrowth of pathobionts and disrupt a symbiotic microbiota resulting in dysbiosis and inflammation. Additionally, the resulting environmental changes in affected tissues may induce the recruitment of circulating inflammatory cells that recognize as ‘danger’ indigenous bacteria that would otherwise be ignored by resident phagocytes. This can exacerbate inflammatory pathology, especially in immunocompromised hosts that cannot readily control indigenous bacteria.
Clostridium difficile can be found at low levels in the healthy human gut. However, its abundance is substantially increased and coincides with severe intestinal inflammation after treatment with broad-spectrum antibiotics that disrupt the indigenous microbiota [20]. Similarly, in a mouse model of C. difficile infection, antibiotic treatment promotes the ability of this organism to colonize the gut at high levels and induce inflammation. Antibiotic treatment also facilitates the outgrowth of vancomycin-resistant Enterococcus that can potentially disseminate and cause systemic organ infection [20].
Under normal conditions, the mouse host seems to be in balance with hemolysin hpmA-producing P. mirabilis in the gut. However, intestinal tissue damage causes CCR2-dependent recruitment of NLRP3 inflammasome-expressing blood monocytes, which respond to P. mirabilis with robust IL-1β release leading to exacerbation of inflammation [66]. In ligature-induced periodontitis in mice, tissue damage is associated with the expansion of γ-proteobacteria species (perhaps due to increased availability of nutrients from inflammatory tissue breakdown), which proactively aggravate destructive periodontal inflammation by activating the cytosolic receptor Nod1 [67].
A diet rich in milk-derived saturated fat (but not one rich in safflower oil-derived polyunsaturated fat) was shown to promote the generation of taurocholic acid, which in turn causes an increase in the abundance of Bilophila wadsworthia and development of colitis in IL-10-deficient mice [68].
Impaired IL-18 production in NLRP6 inflammasome-deficient mice is associated with an abnormal expansion of Prevotellaceae and the candidate division TM7, resulting in exacerbation of chemical colitis [55]. In leukocyte adhesion deficiency, the dysregulated host inflammatory response fosters a nutritionally favorable environment for bacterial growth and development of a compositionally unique microbiome of pathobionts, which can further exacerbate periodontal disease [69, 70]. Therefore, expansion of pathobionts, represents a potential tipping point in the development of nososymbiocity at mucosal surfaces. With the increase in metabolic activities of the pathobionts and spread into adjacent tissues, host-microbe homeostasis is unlikely to be readily restored without intervention to control a vigorous, tissue-damaging inflammatory response.
Concluding Remarks
Collectively, emerging evidence supports the concept that many inflammatory diseases may not be caused by individual bacteria perceived as ‘causative pathogens’, but rather by an entire microbial community under the influence of specific organisms and/or conditions (e.g., broad-spectrum antibiotic treatment, shifts in diet, or immune deficiencies) that can tip the balance from homeostasis to destructive inflammation (see Outstanding Questions). Evidence from mouse models of inflammatory disease suggests that even in hosts with normal immune systems, the immune response can potentially be subverted by keystone pathogens and subsequently be overactivated by pathobionts, thereby linking disrupted homeostasis to destructive inflammation. In this scenario, pathobionts act downstream of keystone pathogens, which in turn may act downstream of accessory pathogens that provide nutritional or colonization support. However, the ability of the pathobionts to cause disease is not necessarily reliant on the presence of keystone pathogens, which constitute one of several known mechanisms for homeostasis breakdown (Figure 1), suggesting that keystone pathogens are risk factors rather than causative agents of disease. As outlined above, the same bacteria may act as beneficial bacteria (keystone stabilizers) in one context and as accessory pathogens in another. Moreover, bacteria can act as accessory or keystone pathogens in different contexts. Therefore, the terms ‘accessory pathogens’, ‘keystone pathogens’, and ‘pathobionts’ refer to contextual properties of constituent members in a nososymbiotic community rather than to invariable intrinsic properties of specific bacterial species or strains thereof.
Outstanding Questions.
What is the full range and nature of the triggers that convert an indigenous microbial community to a dysbiotic state?
To what extent do interbacterial interactions defined in vitro or in animal models reflect the situation in human ecosystems?
Can community engineering suppress the influence of keystone pathogens, or enhance aspects of the immune response to prevent a dysbiotic switch?
Can detection of community alterations be used prospectively as a biomarker for disease development?
Can the pathogenic potential or nososymbiocity of indigenous polymicrobial communities be predicted by compositional or quantitative analyses irrespective of parallel analysis of the immune status of the host?
Can microbial communities model more complex societies and facilitate testing of parameters that maintain community well-being?
Restoring immune function in immunocompromised patients and controlling environmental variables (e.g., antibiotics and diet) in patients regardless of immune status should contribute to immune homeostasis and health. Novel and potentially effective approaches to prevention and treatment of polymicrobial inflammatory diseases may be to interfere with the synergistic mechanisms that drive nososymbiocity, such as targeting relevant key interspecies interactions or the host signaling circuitry that is exploited for microbial subversion of the immune response.
Trends Box.
The emerging appreciation that certain inflammatory diseases are initiated by multi-species communities, wherein constituent organisms contribute to disease collectively rather than individually, challenges the notion that bacteria can be divided into two categories: commensals and pathogens.
A more nuanced understanding of microbial pathogenic potential views the health- or disease- associated properties of a given microbe as a spectrum that ranges from commensalism to pathogenicity, dependent upon the nature of its interactions with other community members and the host’s condition.
Within this spectrum, newly recognized categories include ‘accessory pathogens’, ‘keystone pathogens’, and pathobionts.
When referring to the potential of an indigenous community to cause disease, ‘nososymbiocity’ may be a more context-dependent and accurate term than ‘pathogenicity’.
Acknowledgments
The work of the authors is supported by NIH grants DE01111, DE012505, DE017921, DE016690, DE0130585 (RJL), DE015254, DE017138, DE021685, DE024716 and AI068730 (GH)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13:589–595. doi: 10.1016/j.tim.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Rosan B, Lamont RJ. Dental plaque formation. Microbes Infect. 2000;2:1599–1607. doi: 10.1016/s1286-4579(00)01316-2. [DOI] [PubMed] [Google Scholar]
- 3.Wright CJ, et al. Microbial interactions in building of communities. Mol Oral Microbiol. 2013;28:83–101. doi: 10.1111/omi.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright CJ, et al. Characterization of a bacterial tyrosine kinase in Porphyromonas gingivalis involved in polymicrobial synergy. Microbiologyopen. 2014;3:383–394. doi: 10.1002/mbo3.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuboniwa M, et al. Insights into the virulence of oral biofilms: discoveries from proteomics. Expert Rev Proteomics. 2012;9:311–323. doi: 10.1586/epr.12.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nobbs AH, Jenkinson HF. Interkingdom networking within the oral microbiome. Microbes Infect. 2015;17:484–492. doi: 10.1016/j.micinf.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlecht LM, et al. Systemic Staphylococcus aureus infection mediated by Candida albicans hyphal invasion of mucosal tissue. Microbiology. 2015;161:168–181. doi: 10.1099/mic.0.083485-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grenier D. Nutritional interactions between two suspected periodontopathogens, Treponema denticola and Porphyromonas gingivalis. Infect Immun. 1992;60:5298–5301. doi: 10.1128/iai.60.12.5298-5301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meuric V, et al. Treponema denticola improves adhesive capacities of Porphyromonas gingivalis. Mol Oral Microbiol. 2013;28:40–53. doi: 10.1111/omi.12004. [DOI] [PubMed] [Google Scholar]
- 10.Orth RK, et al. Synergistic virulence of Porphyromonas gingivalis and Treponema denticola in a murine periodontitis model. Mol Oral Microbiol. 2011;26:229–240. doi: 10.1111/j.2041-1014.2011.00612.x. [DOI] [PubMed] [Google Scholar]
- 11.Wickstrom C, et al. Proteolytic degradation of human salivary MUC5B by dental biofilms. Microbiology. 2009;155:2866–2872. doi: 10.1099/mic.0.030536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Short FL, et al. Polybacterial human disease: the ills of social networking. Trends Microbiol. 2014;22:508–516. doi: 10.1016/j.tim.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boesen T, Nielsen LP. Molecular dissection of bacterial nanowires. MBio. 2013;4:e00270–00213. doi: 10.1128/mBio.00270-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weigel WA, et al. Aggregatibacter actinomycetemcomitans QseBC is activated by catecholamines and iron and regulates genes encoding proteins associated with anaerobic respiration and metabolism. Mol Oral Microbiol. 2015;30:384–398. doi: 10.1111/omi.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murray JL, et al. Mechanisms of synergy in polymicrobial infections. J Microbiol. 2014;52:188–199. doi: 10.1007/s12275-014-4067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogt SL, et al. Chemical communication in the gut: Effects of microbiota-generated metabolites on gastrointestinal bacterial pathogens. Anaerobe. 2015;34:106–115. doi: 10.1016/j.anaerobe.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Armbruster CE, et al. Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. MBio. 2010;1:e00102–00110. doi: 10.1128/mBio.00102-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LeRoux M, et al. Bacterial danger sensing. J Mol Biol. 2015;427:3744–3753. doi: 10.1016/j.jmb.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darveau RP. The oral microbial consortium's interaction with the periodontal innate defense system. DNA Cell Biol. 2009;28:389–395. doi: 10.1089/dna.2009.0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamada N, et al. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamada N, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palm NW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertin Y, et al. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ Microbiol. 2011;13:365–377. doi: 10.1111/j.1462-2920.2010.02334.x. [DOI] [PubMed] [Google Scholar]
- 24.Lamont RJ, Hajishengallis G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med. 2015;21:172–183. doi: 10.1016/j.molmed.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuboniwa M, et al. Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Mol Microbiol. 2006;60:121–139. doi: 10.1111/j.1365-2958.2006.05099.x. [DOI] [PubMed] [Google Scholar]
- 26.Daep CA, et al. Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infect Immun. 2011;79:67–74. doi: 10.1128/IAI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramsey MM, et al. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 2011;7:e1002012. doi: 10.1371/journal.ppat.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stacy A, et al. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc Natl Acad Sci U S A. 2014;111:7819–7824. doi: 10.1073/pnas.1400586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan Y, et al. Oral bacteria modulate invasion and induction of apoptosis in HEp-2 cells by Pseudomonas aeruginosa. Microb Pathog. 2009;46:73–79. doi: 10.1016/j.micpath.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Li Q, et al. Porphyromonas gingivalis modulates Pseudomonas aeruginosa-induced apoptosis of respiratory epithelial cells through the STAT3 signaling pathway. Microbes Infect. 2014;16:17–27. doi: 10.1016/j.micinf.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 31.Duan K, et al. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol Microbiol. 2003;50:1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x. [DOI] [PubMed] [Google Scholar]
- 32.Pacheco AR, et al. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawhon SD, et al. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol Microbiol. 2002;46:1451–1464. doi: 10.1046/j.1365-2958.2002.03268.x. [DOI] [PubMed] [Google Scholar]
- 34.Nakanishi N, et al. Regulation of virulence by butyrate sensing in enterohaemorrhagic Escherichia coli. Microbiology. 2009;155:521–530. doi: 10.1099/mic.0.023499-0. [DOI] [PubMed] [Google Scholar]
- 35.Curtis MM, et al. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe. 2014;16:759–769. doi: 10.1016/j.chom.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Power ME, et al. Challenges in the quest for keystones. BioScience. 1996;46:609–620. [Google Scholar]
- 37.Hajishengallis G, et al. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hajishengallis G, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maekawa T, et al. Porphyromonas gingivalis manipulates complement and TLR signaling to uncouple bacterial clearance from inflammation and promote dysbiosis. Cell Host Microbe. 2014;15:768–778. doi: 10.1016/j.chom.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duran-Pinedo AE, et al. The periodontal pathogen Porphyromonas gingivalis Induces expression of transposases and cell death of Streptococcus mitis in a biofilm model. Infect Immun. 2014;82:3374–3382. doi: 10.1128/IAI.01976-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frias-Lopez J, Duran-Pinedo A. Effect of periodontal pathogens on the metatranscriptome of a healthy multispecies biofilm model. J Bacteriol. 2012;194:2082–2095. doi: 10.1128/JB.06328-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the Polymicrobial Synergy and Dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27:409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rolig AS, et al. Individual members of the microbiota disproportionately modulate host innate immune responses. Cell Host Microbe. 2015;18:613–620. doi: 10.1016/j.chom.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fisher CK, Mehta P. Identifying keystone species in the human gut microbiome from metagenomic timeseries using sparse linear regression. PLoS One. 2014;9:e102451. doi: 10.1371/journal.pone.0102451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore WE, Moore LH. Intestinal floras of populations that have a high risk of colon cancer. Appl Environ Microbiol. 1995;61:3202–3207. doi: 10.1128/aem.61.9.3202-3207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sears CL, Pardoll DM. Perspective: alpha-bugs, their microbial partners, and the link to colon cancer. J Infect Dis. 2011;203:306–311. doi: 10.1093/jinfdis/jiq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sears CL, et al. Bacteroides fragilis subverts mucosal biology: from symbiont to colon carcinogenesis. J Clin Invest. 2014;124:4166–4172. doi: 10.1172/JCI72334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu S, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tosolini M, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71:1263–1271. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
- 50.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tjalsma H, et al. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10:575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 52.Wang M, et al. Microbial hijacking of complement-toll-like receptor crosstalk. Sci Signal. 2010;3:ra11. doi: 10.1126/scisignal.2000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hajishengallis G. The inflammophilic character of the periodontitis-associated microbiota. Mol Oral Microbiol. 2014;29:248–257. doi: 10.1111/omi.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo G, et al. Tumor necrosis factor alpha binding to bacteria: evidence for a high-affinity receptor and alteration of bacterial virulence properties. Infect Immun. 1993;61:830–835. doi: 10.1128/iai.61.3.830-835.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Porat R, et al. Enhancement of growth of virulent strains of Escherichia coli by interleukin-1. Science. 1991;254:430–432. doi: 10.1126/science.1833820. [DOI] [PubMed] [Google Scholar]
- 59.Wu L, et al. Recognition of host immune activation by Pseudomonas aeruginosa. Science. 2005;309:774–777. doi: 10.1126/science.1112422. [DOI] [PubMed] [Google Scholar]
- 60.Winter SE, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stecher B, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stecher B, et al. 'Blooming' in the gut: how dysbiosis might contribute to pathogen evolution. Nat Rev Microbiol. 2013;11:277–284. doi: 10.1038/nrmicro2989. [DOI] [PubMed] [Google Scholar]
- 63.Hooper LV, et al. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4:269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 64.Kelly D, et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-γ and RelA. Nat Immunol. 2004;5:104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 65.Chow J, et al. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol. 2011;23:473–480. doi: 10.1016/j.coi.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seo SU, et al. Distinct commensals induce Interleukin-1beta via NLRP3 inflammasome in inflammatory monocytes to promote intestinal inflammation in response to injury. Immunity. 2015;42:744–755. doi: 10.1016/j.immuni.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiao Y, et al. Induction of bone loss by pathobiont-mediated nod1 signaling in the oral cavity. Cell Host Microbe. 2013;13:595–601. doi: 10.1016/j.chom.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Devkota S, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moutsopoulos NM, et al. Subgingival microbial communities in leukocyte adhesion deficiency and their relationship with local immunopathology. PLoS Pathog. 2015;11:e1004698. doi: 10.1371/journal.ppat.1004698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moutsopoulos NM, et al. Defective neutrophil recruitment in leukocyte adhesion deficiency type I disease causes local IL-17–driven inflammatory bone loss. Sci Transl Med. 2014;6:229ra240. doi: 10.1126/scitranslmed.3007696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pastar I, et al. Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS One. 2013;8:e56846. doi: 10.1371/journal.pone.0056846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Korgaonkar A, et al. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci U S A. 2013;110:1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Twomey KB, et al. Bacterial cis-2-unsaturated fatty acids found in the cystic fibrosis airway modulate virulence and persistence of Pseudomonas aeruginosa. ISME J. 2012;6:939–950. doi: 10.1038/ismej.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu H, et al. Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microbiol. 2014;16:214–231. doi: 10.1111/cmi.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kamada N, et al. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 76.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fritz JH, et al. Acquisition of a multifunctional IgA+ plasma cell phenotype in the gut. Nature. 2012;481:199–203. doi: 10.1038/nature10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hooper LV, et al. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]