Summary
A new paradigm has emerged in recent years characterizing transcription initiation as a bidirectional process, encompassing a larger proportion of the genome than previously thought. Past concepts of coding genes thinly scattered among a vast background of transcriptionally inert noncoding DNA have been abandoned. A richer picture has taken shape, integrating transcription of coding genes, enhancer RNAs, and various other noncoding transcriptional events. In this review we give an overview of recent studies detailing the mechanisms of RNA Pol II-based transcriptional initiation and discuss the ways in which transcriptional direction is established, as well as its functional implications.
Keywords: Transcriptional initiation, directionality, promoter, enhancer, eRNA, Nucleosome-Depleted Region
What is bidirectional transcription?
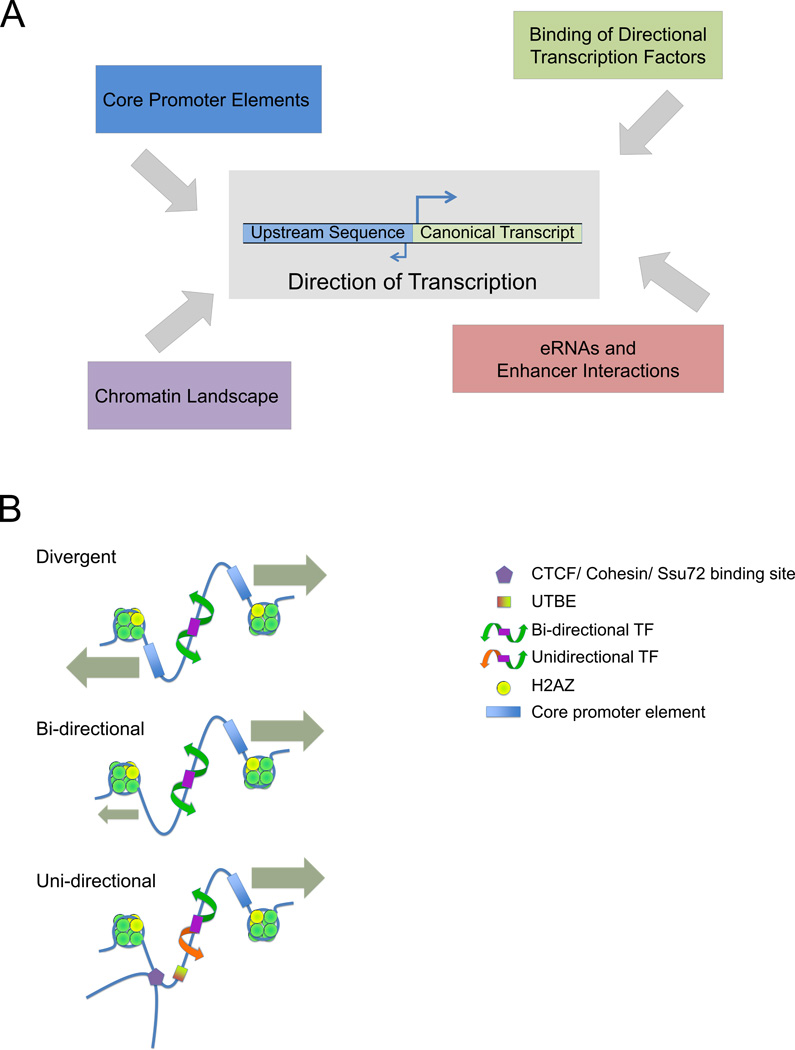
The determinants of transcriptional initiation are intricate and interwoven. What is clear from the high proportion of the human genome that is transcribed (estimated at 60%) in comparison to the small proportion that is coding (2%) [1, 2] is that transcriptional processes involve much more of the genome than was once thought. High-resolution analyses and detailed catalogs of transcription start sites (TSS) obtained using next-generation sequencing methods have shown that transcription initiation frequently occurs in both directions from a given promoter region [3, 4]. These studies have raised the question of whether transcription initiation is an inherently bidirectional or unidirectional process. In one model, biases in the direction of transcription arise as emergent properties from the complex regulatory restrictions placed upon inherently bidirectional promoter elements. In an alternative model, transcription at its core is unidirectional, with the appearance of bidirectionality arising due to the adjacent placement of individual unidirectional core promoters in opposite orientations. In the latter model, the similar needs of two separate gene promoters to coordinately regulate transcription factor (TF) recruitment might select for divergent transcript orientations. Transcription occurring in two directions from a single core promoter and divergent transcription originating from two distinct core promoters have not always been well distinguished in the literature. The conflation of these two categories has led to some ambiguity. Here we refer to transcription arising from a core promoter in opposite directions as bidirectional, whereas transcription of two outward facing transcripts from independent core promoters is termed divergent transcription. To some extent, the terminology that researchers in the field adopt depends on variability in the definitions and size estimates of what constitutes a promoter and how far divergent genes may lie from one another. In this review we discuss the evidence for each model to illustrate the current understanding of transcriptional initiation, and also consider the related issue of sense and antisense transcription at genes. Ultimately, we suggest a more nuanced view of promoters as non-directional, conducive regions of DNA prone to the occurrence of an open chromatin structure, the transcriptional potential of which is channeled either bidirectionally or unidirectionally in a context dependent manner. The regulatory constraints of the various layers of regulation then work additively to produce specific transcriptional states (Figure 1, Key Figure).
Figure 1. Key Figure. The determinants of directionality.
(A) The different factors that go into establishing directionality at a transcriptionally permissive site. (B) Contributors to divergent and unidirectional transcription located within an NDR. For outward-facing, head-to-head coding genes, most transcripts arise from two separate core promoter elements. Bidirectional transcription from a single core element may be characterized by unstable transcripts in the antisense direction in the case of coding genes, and in both directions in the case of enhancers. Finally, a subset of promoters show predominantly unidirectional transcription. As the majority of TFs have bidirectional activities, the number of truly unidirectional promoters may be relatively small. These categories may overlap and vary depending on differential conditions and tissue types. In each case, the presence or absence of transcription is subject to secondary regulation including TF expression and binding, CTCF and cohesin mediated looping, Ssu72 mediated 5’ to 3’ gene looping, and modifications to H2A.Z which may further promote or antagonize the progression of RNA Pol II.
Assaying the transcriptome
High resolution methods to define the transcriptome have revealed that transcription initiates not only in the expected location downstream of promoters, but also within promoter regions upstream of coding sequences and bidirectionally at active enhancers. Both these sources of noncoding transcription generally produce short unstable RNAs that are rapidly degraded [5, 6]. Transcription has also been observed to originate within transcript bodies [7], and from the 3' ends of genes in antisense orientation [8](Figure 2). Numerous techniques have been used to detect nascent transcripts (Table 1). Unstable transcripts can be identified when RNA degradation pathways are inhibited, causing the persistence of unstable RNAs [9, 10]. These experiments have been used to interrogate the genomic sites of transcriptional initiation and classify them broadly into 3 types based on their bidirectional potential: stable/stable, stable/unstable, and unstable/unstable. These categories reflect the functional directionality of a promoter but don't specify whether initiation actually occurs in both directions. For an in-depth account of the various noncoding transcripts that have been described and the techniques that have been used to identify them, see the review by Steinmetz et al [11].
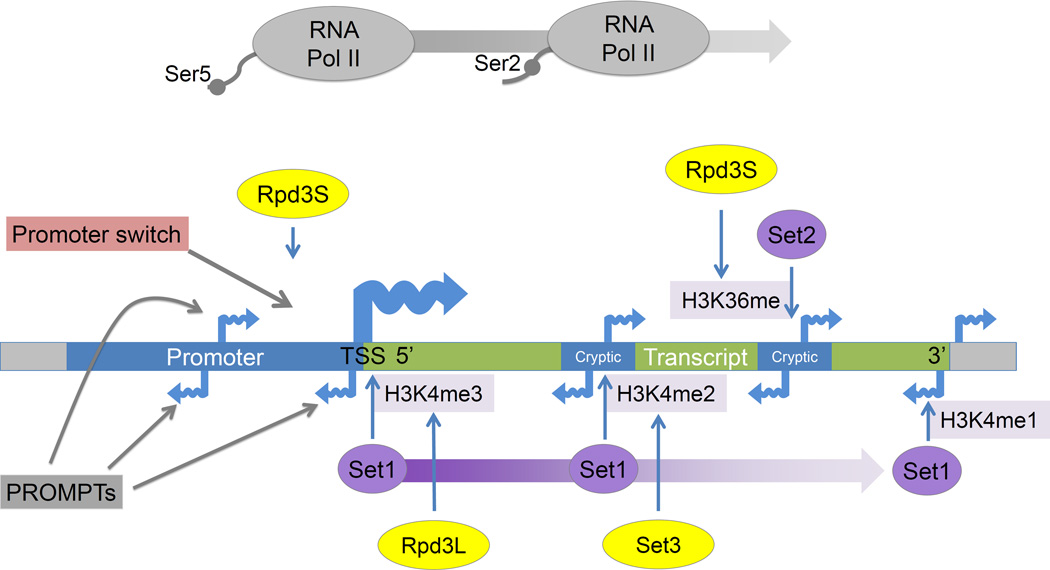
Figure 2. Regulation by covalent chromatin modifications and noncoding transcription.
The interplay between histone methyltransferase (purple) and histone deacetylase (yellow) activities influences noncoding transcription. During transcription RNA Pol II is phosphorylated on its C terminal domain (CTD) first at serine 5 (Ser5P) near the 5' end of genes and then at serine 2 (Ser2P), allowing RNA Pol II to enter into the elongation phase of transcription, and establishing correct chromatin structure across transcribed regions [89]. Set1 interacts with RNA Pol II Ser5P and creates a gradient of H3K4 methylation starting with tri-methylation at the 5’ end and ending with mono-methylation at the 3’ end. These methyl marks are then targeted by two separate histone deacetylases. Rpd3L (the larger complex) deacetylates histones with H3K4me3 marks at the 5’ ends of transcripts [90]. Set3 deacetylates histones with H3K4me2 marks within gene bodies and represses 5’ proximal cryptic transcripts [91]. Set2 mediated H3K36 methylation takes place within gene bodies and towards the 3’ regions of the genes. This H3K36 modification is then targeted by Rpd3S (small complex) which removes the acetylation that co-occurs with transcriptional elongation, thereby suppressing cryptic transcripts originating within gene bodies near the 3’ end [92]. These transcripts are thought to arise because of chromatin remodeling concomitant with transcription, leading to increased accessibility of intragenic regions to TFs and Pol II [93]. Rpd3S has also been implicated in the repression of antisense transcripts, thereby promoting sense strand directed transcription [60].
Table 1.
Genomic techniques for assaying transcriptional initiation.
| Technique | Method | Results | References |
|---|---|---|---|
| RNA Pol II ChIP Seq |
Chromatin is fragmented, Pol II is immuno- precipitated, and the interacting DNA is sequenced. |
Identifies DNA that is bound by RNA Pol II in a non-strand specific fashion at ~200 bp resolution. |
[81, 82] |
| cap analysis of gene expression (CAGE)/ SMORE-seq/ TIF-seq |
RNA is treated with 5' cap- specific enzyme tobacco acid pyrophosphatase (TAP) and subjected to cDNA sequencing. |
Identify 5' ends from stable capped RNAs at single nucleotide resolution. |
[18, 83, 84] |
| global run-on sequencing (GRO-seq) and GRO-cap |
Run on assay which restarts RNA Pol II in vitro in the presence of a labeled nucleotide (BrUTP) in order to purify nascent RNA. |
Identify nascent RNAs and post initiation pause sites at ~50 bp resolution. Since transcription is restarted in vitro, it can detect unstable transcripts which would normally be rapidly degraded in vivo. |
[9, 85, 86] |
| precision nuclear run-on and sequencing (PRO-seq) |
Run on assay which restarts RNA Pol II in vitro using biotin-labeled ribonucleoside triphosphate analogs. By supplying only 1 of the 4 nucleotides at a time, run-on transcription is limited and resolution is improved over GRO-seq. |
Identifies nascent RNAs and post-initiation pause sites at <50 bp resolution. Since in GRO-seq type assays transcription is restarted in- vitro, it can detect unstable transcripts which would normally be rapidly degraded in vivo. |
[87] |
| native elongating transcript sequencing (NET-seq) |
RNA Pol II associated RNA is purified, and the associated RNA is sequenced. |
Identifies nascent RNA at single nucleotide resolution. When combined with CTD phosphorylation specific immunoprecipitation, different populations of RNA can be identified based on the modification status of their transcribing RNA Pol II. |
[60, 73, 88] |
| RNA-seq combined with inhibition of RNA degradation pathways |
Different components of various RNA degradation pathways are inhibited (such as exosome components) to enable the isolation of unstable RNAs. |
Identifies RNAs regardless of stability. Resolution is variable depending on the RNA seq method employed. |
[6] |
eRNAs
In mammalian cells, transcriptional activity at enhancers is widespread and dynamically regulated, generally producing unstable transcripts in both directions when actively functioning as an enhancer [12]. It should be noted though that the majority of putative enhancers identified by chromatin profiling have not been experimentally validated as being functional. A small subset of enhancers produce stable long noncoding RNAs (lncRNAs) as one of their transcript pairs. During changing conditions or cell states, enhancer RNA (eRNA) production is the most rapid and salient transcriptional response, preceding even the transcription of TFs in response to the change [13]. The question of whether the majority of eRNAs are functional remains open. Post transcriptional knockdown of a handful of eRNAs has revealed cases where they are necessary for enhancing transcription at interacting genes [14] and for promoter-enhancer loop formation [15]. However, there are also many instances where knockdown of these eRNAs does not inhibit the function of the enhancer [16]. On average, however, eRNA transcription is a good predictor of enhancer activity [12]. For further discussion of the potential functions of eRNAs see Li et al [17].
PROMPTs
Within promoter regions, noncoding RNAs termed PROMPTs (promoter upstream transcripts, Figure 2) have been detected after depletion of components of the exosome, an RNA degradation complex [4]. Similar transcripts have been noted by other groups and have been termed bidirectional noncoding RNAs (BNCs) [18], cryptic unstable transcripts (CUTs) [3] or stable unannotated transcripts (SUTs) [3, 11]. In mammalian cells, PROMPTs have been observed to be transcribed in both the sense and antisense directions [10, 19]. These transcripts are generally transcribed by RNA polymerase II (RNA Pol II) but can originate upstream from Pol I and Pol III transcribed genes also [4]. Antisense PROMPT transcription has been reported to be correlated [10] and anti-correlated [4] with downstream coding genes. Skewing in initiation direction may reflect trade-offs where the presence of activated open chromatin generally recruits more of the transcription machinery, but also where a transcript's abundant expression may monopolize the pool of available RNA Pol II. In contrast to stable mRNAs, most PROMPTs and eRNAs are depleted for 5' splice sites [5] and enriched for polyadenylation sites [6], features which target them for early transcriptional termination and degradation. While the majority of PROMPTs are rapidly degraded, some stable noncoding transcripts produced from promoter regions have been shown to be functional [20, 21]. Some promoter transcripts are reproducibly observed in specific tissues, cell lineages, and cancers, while others are ubiquitous [22].
The similarity between PROMPTs and eRNAs supports the characterization of promoters as a specific type of a general class of origin of transcription, one in which a stable transcript with coding potential is produced. It has been suggested that promoters and enhancers should be viewed as a unified category of transcriptional initiation sites that are differentially regulated [10, 19, 23]. Generally, eRNA transcription occurs bidirectionally, with both directions producing roughly equivalent levels of RNA [12]. In contrast, while it is likely that most promoters produce PROMPTs, or antisense transcripts, transcription is generally skewed towards the sense direction [22]. While depletion of exosome components leads to increases for both eRNAs and PROMPTs, eRNA increases are significantly higher [12]. In some cases, intragenic enhancers can produce multi-exonic enhancer RNAs (meRNAs) which are spliced and polyadenylated just like coding genes, but are unlikely to have coding potential [24]. Interestingly, not only are enhancers being recognized as resembling promoters, but promoters have been characterized with enhancer functions. Like enhancers, promoters often interact with other promoters, and in these cases, can have enhancer-like effects on their interacting partners [24, 25]. In this context, both elements should be regarded as sites of transcriptional initiation that are differentially characterized by the types of transcripts they produce.
Core promoter elements work synergistically to establish transcriptional directionality
Core promoter elements are vital components in determining whether transcriptional initiation occurs and the direction in which it occurs. The most widely recognized of these sequences, the TATA box, has often been regarded as a directional element, in part due to a strong bias in its appearance at sites of asymmetric, directional transcription [18]. In humans, TATA box like features occur at about 29% of unidirectional promoters compared with only about 9% of bidirectional promoters [26]. Some experiments suggest that the TATA sequence orientation matters. For example, inversion of the TATA box in the yeast S. cerevisiae HIS4 promoter in vivo causes a failure of HIS4 transcription [27]. However, examples of inverted TATA elements can be found in natural genomic contexts [28]. In vitro, isolated TATA boxes support bidirectional transcription [28] (Figure 1B, Key Figure). In yeast, the TATA box is able to promote transcription in both orientations but the orientation does affect the level of transcriptional output [29]. Inversion of asymmetric TATA boxes in reporter plasmids transfected into human cells still produced transcriptional activation of the downstream genes they regulated [30]. These experiments indicate that the position of the TATA box in relation to other promoter elements is more important for determining directionality than the TATA box orientation itself.
If TATA elements have the potential to stimulate bidirectional transcription, what then accounts for the significant enrichment of TATA boxes at sites of unidirectional transcription? In yeast, TATA-containing genes tend to exhibit either high or low expression levels, are enriched for genes up-regulated in response to environmental stress, and are depleted for housekeeping genes [31]. Additionally, TATA boxes have been associated with tissue specific promoters [32, 33]. If TATA-regulated genes are less likely to be constitutively expressed, it is possible that divergent transcription occurs from these promoters only under specific conditions. The promoters of genes localized near telomeres are also enriched for TATA boxes [31]. The expression levels of these genes may need to be more dynamically regulated, as telomere-adjacent gene regions show less evolutionary conservation compared to centrally located regions [34]. These results support a model where the regulatory requirements of TATA containing genes are more variable and demanding, selecting for their independent regulation from potential upstream transcripts.
Core promoters lacking TATA boxes also impart directional preferences to the transcription originating from them. Unidirectional transcription (as measured by luciferase reporter assays) from the insulin-degrading enzyme (IDE) gene promoter is achieved through the presence of an upstream transcription blocking element (UTBE) [35, 36](Figure 1B, Key Figure). The downstream core promoter in isolation can promote transcription in both the sense and antisense directions. However, in the presence of a UTBE, antisense transcription is abrogated [36]. When promoters cloned into luciferase reporters in both orientations and transfected into 4 different cell lines were assayed, some showed strong directional preferences that varied based on cell line, and deleting one TSS often increased activity from the oppositely oriented one [26]. This indicates that the directionality associated with some promoter sequences is not solely due to intrinsic sequence factors, but relies on interactions with trans-acting cell specific factors.
Some core promoter elements are more likely to be associated with regions of bidirectional transcription. In particular, bidirectional promoters are more likely to have a higher GC content and to fall within CpG islands. In humans, about 90% of bidirectional promoters are found within CpG islands compared to only about 45% of unidirectional promoters [37]. Other elements slightly more common in unidirectional promoters include the downstream core promoter element (DPE) and the initiator (Inr), while the CCAAT box is almost twice as likely to occur in bidirectional promoters [26]. Characterization of the enhancer-associated unstable/unstable TSSs reveals that they display a lower CpG frequency than other bidirectional transcripts. However, they have core promoter elements (including TATA boxes and Inrs), predominantly bind the same TFs, and exhibit a canonical TSS structure with a nucleosome-depleted region (NDR) bordered by two well-positioned nucleosomes [9] suggesting a similar mode of transcriptional regulation as promoters.
For higher eukaryotes, bidirectional transcription seems to be most closely associated with the production of noncoding RNA in at least one direction. There is some debate concerning whether human promoters are inherently unidirectional or bidirectional [38, 39]. To some extent, the answer to this question rests on which cell types are being examined, the thresholds used to call antisense transcription and the exact methods of detection. Independent studies support the idea that bidirectional transcription is most often a result of separate core promoter elements flanking a NDR. For divergent transcripts with well defined core promoter elements, it may be that despite the potential for bidirectional activity at individual core elements, the majority of these transcripts face some selective pressure to independently regulate expression, favoring the maintenance of individual promoters which retain greater potential for independent regulation.
At sites of bidirectional transcription in yeast, two pre-initiation complexes (PICs) generally flank the NDR in an inverted orientation [40]. Similarly, in mice, bidirectional transcription involves the formation of 2 distinct PICs, more TF binding, a larger, more distinctly defined NDR, and on average higher gene expression [41]. When only one core promoter is present however, an explanation is needed for the asymmetric binding of RNA Pol II on opposite strands, which is needed to allow for bidirectional transcription (Figure 1B). Non-consensus binding of PIC components to the NDR has been suggested to explain binding of general TFs to promoter regions without any discernable recognition sequences [42]. Such binding was promoted by the TF Reb1 in yeast and inhibited by CTCF in human cells [43]. NDRs with two PICs are further characterized by a greater occupancy of the −1 nucleosome [43], suggesting that the chromatin structure at the NDR promotes opening of a transcription bubble and is conducive to bidirectional transcription, with PICs forming in both directions at the edges of the flanking nucleosomes. Secondary regulation may then generate a predominant direction of transcription either through pre- or post- initiation regulation. The fact that promoters can be unidirectional in some tissue types and bidirectional in others [22] supports the notion that core promoter sequences allow for bidirectional transcription but that this capacity is then regulated by secondary mechanisms such as cell-type specific TFs that promote either one or both transcripts in the pair in response to the different needs of the cell.
Transcription factors can modulate the directionality of transcription initiation sites
The binding motifs for a number of TFs, including NF-Y, Nrf-1, YY1, GABP, MYC, E2F1, and E2F4 are overrepresented in bidirectional promoters [44]. These factors may act as determinants of transcriptional direction, not just passively associate with it. For example, the introduction of a GABP binding site into unidirectional promoters caused the appearance of bidirectional transcription in 67% of tested promoters [45]. In addition to coding gene promoters, TFs are also associated with other origins of transcription. Sites producing unstable/unstable transcript pairs in B-cell derived lines showed histone modifications typical of enhancers (high levels of H3K4me1) and were preferentially bound by the immune specific transcription factor PU.1 [9]. Sites producing two stable divergently arranged coding transcripts showed enrichment for GABP (GA binding protein) localization [9]. Finally, sites producing stable transcripts only in one direction were associated with CTCF binding, which likely involves modulating chromatin structure [9, 46].
The notion of pioneer transcription factors raises the prospect of this special group of TFs being implicated in modulating directionality. Pioneer transcription factors possess the ability to open up local chromatin conformation upon binding their motifs, facilitating the binding of additional settler TFs in a cooperative and hierarchical fashion [47, 48]. While the majority of pioneer TFs open chromatin on both sides of their motifs, several (including Creb/ATF, Klf/Sp, NFYA, and Zfp161) open chromatin in a directional manner [49]. These directional motifs represent a plausible mechanism by which transcription from inherently bidirectional core promoter elements is converted to unidirectional activation in genomic contexts (Figure 1B, Key Figure).
The chromatin landscape and transcriptional directionality
By altering the chromatin landscape in the vicinity of the TSS, transcription in either direction can be promoted or prevented. In particular, the +1 nucleosome in the direction of transcription plays an important role by presenting a barrier to the progression of RNA Pol II. This barrier can be lowered by incorporation of the histone variant H2A.Z which has been proposed to destabilize the nucleosome and allow progression of RNA Pol II [50–52]. Covalent modifications on the N-terminal tail of H2A.Z could modulate chromatin accessibility downstream of the promoter. Mono-ubiquitination of H2A.Z has been shown to induce transcriptional repression [53], and its de-ubiquitination is necessary for the activation of androgen receptor mediated genes [54]. By contrast, acetylated H2A.Z is associated with increased levels of gene expression [55].
S. pombe cells lacking H2A.Z exhibit increased antisense transcription, implying a role for the histone in transcriptional repression [56]. However in S. cerevisiae, H2A.Z incorporated at the 3’ ends of gene bodies promoted overlapping antisense transcription [8]. It is not clear if these studies reveal a difference in the role of H2A.Z in these two species, or if they reflect different covalent modification states of H2A.Z promoting different functional outcomes. But they do suggest that altering H2A.Z incorporation at the +1 and −1 nucleosomes may be a mechanism for modulating the permissibility of nucleosomes bordering the NDR to polymerase progression, thereby promoting or inhibiting transcription in each direction from the TSS. Another histone variant incorporated into NDR proximal nucleosomes, H3.3, is also recruited to promoters and enhancers during transcriptional activation and causes destabilization of nucleosome structure [57]. At enhancers, asymmetric H2A.Z and H3.3 incorporation levels are associated with asymmetric Pol II enrichment [58].
Covalent modification of histones represents an integral component in the regulation of sense versus antisense transcription. Several chromatin remodelers and related factors have been shown to modulate antisense transcription originating from promoters. In S. cerevisiae, mutations in the histone chaperone complex Chromatin assembly factor 1 (Caf1) [59], the histone methyltransferase Set2 [60], the histone deacetylase Set3 [61], the histone deacetylase Rpd3S [60], and the chromatin remodeler Chd1 [62] all result in increased levels of divergent antisense transcripts. By modulating levels of antisense transcripts, these complexes help enforce directional transcription. Chd1 has also been shown to help overcome promoter-proximal stalling of RNA Pol II, facilitating productive elongation of sense transcripts [63]. In the opposing direction, deletion of Hda2 or the histone methyltransferase Set1 increases sense transcription by decreasing the presence of transcribed antisense RNA [64, 65].
Specific histone modifications characterize divergent human promoters, which are enriched for marks associated with transcriptional elongation (such as H3K4me2–3 and H3K27ac) in both the downstream and upstream directions, while unidirectional promoters lack this enrichment in the upstream direction [38]. Methylation marks across gene bodies recruit chromatin remodelers and histone deacetylases to modulate transcription initiating in those locations. The Set and Rpd3 histone methyltransferases and deacetylases exemplify the mechanisms by which the spread of co-transcriptional modifications can in turn regulate transcription (Figure 2). Processes leading to asymmetric chromatin enrichment patterns across transcriptional initiation sites can similarly be inhibitory or conducive to transcription in either direction.
A further level of chromatin-based regulation of transcriptional directionality involves chromatin loops. Polyadenylation complex factor Ssu72 facilitates the formation of gene loops between the 3’ and 5’ end of genes, and loss of these gene loops leads to increased levels of divergent transcription at yeast promoters [66]. In mammalian cells, CTCF and cohesin facilitate chromatin loop formation [67, 68], and display a biased association with unidirectional transcripts [46](Figure 2). In these genes, CTCF and the cohesin component Rad2 are often found a short distance (60–80 bp) upstream of the TSS and their enrichment level is anti-correlated with antisense transcription. These data emphasize the importance of loop formation in directing transcription. The many epigenetic factors involved in repressing antisense transcription support a view of promoters where an inherent predisposition towards bidirectional transcription must be actively controlled.
Chromosomal looping also occurs between enhancers and promoters, and this interaction is vital for the transcription promoting activities of enhancers. Surprisingly, enhancers and promoters not only share the ability to serve as sites of transcription initiation, but also share the ability to promote transcription at locations that they physically interact with. Just as enhancers increase the likelihood of transcription at interacting promoters, TSS that physically interact with enhancers through loops stimulate the production of eRNAs [69]. These analyses have also revealed that in some cases promoters can function as enhancers for other promoters [24, 25]. For a review of the determinants of promoter/enhancer interaction specificity see the review by van Arensbergen et al [70].
Functions of ncRNAs
Although there are many known examples of transcriptional regulation by ncRNA or antisense RNA, it has been difficult to ascribe functional relevance to the majority of PROMPTs and eRNAs. For example, deletion of exosome component RRP6 increases upstream antisense transcription and represses sense transcription of a certain subset of genes. Whether this is through titration of TFs and RNA Pol II away from the downstream gene, or effects of the transcribed PROMPT RNAs themselves remains to be determined [71]. The effects of overlapping transcription on coding gene transcription are likely to be highly context specific and depend on the direction of the overlapping transcription, the length of the coding gene, and the types of co-translational chromatin modifications. As an example, for many Set3 regulated genes, upstream originating overlapping transcription places Set1 dependent H3K4me2 over promoters and causes deacetylation by the Set3 complex, thereby repressing coding gene transcription. Loss of transcription from these overlapping transcripts or from internal antisense cryptic transcripts de-represses the coding genes [61] (Figure 2).
Other sources of overlapping transcription also modulate the likelihood of a gene being transcribed. In particular, a group of stochastically controlled "promoter switches" regulates the genes for the class I major histocompatibility complex (MHC) receptors in mouse and humans [72]. Within these promoters, antisense transcription upstream of the primary transcript is generally associated with a transcriptionally off state while sense-directed transcription originating upstream of the promoter represents an on state (Figure 2). Changes to the direction of transcription within the switch can thereby switch the activation state of the downstream gene. Similarly, antisense transcription originating from within gene bodies and converging on the promoter is a feature found within a group of low expression genes [73].
Some TFs have been shown to bind RNA as well as DNA [74, 75]. YY1 binds RNA at promoters and enhancers. When RNA transcription occurs, the nascent RNAs can bind YY1 and increase its occupancy at the NDR, creating positive transcriptional feedback. This RNA-mediated effect has led to the hypothesis that trapping of TFs (especially ones with RNA binding domains) at the TSS by RNA may be a general mechanism by which RNAs regulate transcriptional processes. This also provides a plausible function for upstream antisense transcription in recruiting TFs to the NDR so that they may promote downstream coding gene transcription [75]. In a different form of RNA based regulation, YY1 has also been shown to interact with a long intergenic noncoding RNA (lncRNA) transcribed from its own promoter region. When Linc-YY1 binds to YY1 it can de-repress YY1 target genes by causing the eviction of YY1 and PRC2 from gene promoters [20]. Two other well characterized lncRNAs that have been shown to bind PRC2 include COLDAIR in Arabidopsis which mediates repression of the flowering control gene FLC [76, 77] and HOTAIR in Drosophila which regulates the HOXD locus [78]. The individual transcriptional regulatory activities of a number of different lncRNAs have been previously discussed in a review [21].
Concluding remarks
The model that current knowledge in the field presents is one where a bidirectional core promoter element recruits the transcription machinery to an accessible NDR. Additional promoter elements, TF binding sites and chromatin features then specify the directionality of transcription originating from this location. Post-initiation factors can then modulate the elongation and stability of transcripts. These regulatory features may vary in response to changing environmental conditions and in metazoans, according to cell type. Finally, overlapping sense transcription and 3' end originating antisense transcription can spread chromatin signatures, which further encourage or repress transcription. The transcriptional output is likely to rely on a hierarchical ordering of these factors, the complex interactions of which are organism and context dependent (Figure 1A).
Several issues remain to be addressed (See Outstanding Questions Box). Given the propensity for bidirectional genes to show correlated expression, future experiments must determine the factors that promote concordant regulation of bidirectional transcripts. There is also a need to determine whether PROMPTs, especially sense strand PROMPTs (Figure 2), and promoter switches arise from core promoter element-like sequences. Further, the characterization of bidirectional promoters needs to distinguish between promoters from which transcription is initiated in both directions (such as a PROMPT-coding gene pair) and from which stable transcripts are produced in both directions (such as coding gene pairs) as these two categories are often conflated.
Outstanding Questions.
What factors promote concordant versus discordant regulation of bidirectional transcripts?
Is RNA-dependent regulation by transcription factors that have both RNA and DNA binding sites a common phenomenon?
How do eRNAs relate to other short ncRNAs such as miRNAs, and do they possess overlapping functions? If not how are they differentially recognized?
More work needs to be done to define how the directional specificities of specific core promoter elements and transcription factor binding sites are integrated to determine transcriptional direction. It remains to be determined which individual core promoter sequences are capable of initiating bidirectional transcription and whether they actually do so in their genomic contexts. For the majority of TFs, changing the orientation of their binding sites does not significantly change their influence on gene expression [79], and co-varying expression levels provides further evidence that TF binding increases the likelihood of transcription in both directions. Differential regulation of outward-facing transcripts in yeast is associated with the presence of insulator-like DNA binding factors Tbf1 and Mcm1 [80]. Two possibilities present themselves as a way of explaining the phenomenon of bidirectional transcription. In the first, antisense transcription is a by-product of forward transcription and an NDR, meaning that the cell may need to utilize mechanisms to prevent the accumulation of detrimental antisense transcripts. In the second, there is a function for antisense transcripts with a number of possible effects including the regulation of sense transcripts. While these functions do not need to be mutually exclusive, they could create a dichotomy between two different types of TSSs. If antisense transcription is a necessary outcome of forward transcription, a single core promoter might be the most commonly encountered situation. However, two independent promoters would allow for more precise regulation of the different transcripts. Ultimately, the regulation of transcriptional direction is accomplished by both pre and post initiation factors and integrates many factors such as promoting and repressing the initiation of transcription, regulating elongation and termination, and targeting unstable transcripts for rapid degradation.
Trends Box.
Most core promoters are bidirectionally competent with respect to initiation.
Promoters and enhancers share common features which are conducive to transcriptional initiation.
Transcriptional directionality is determined in an additive and hierarchical way.
Noncoding transcription can alter coding transcription levels by both spreading specific chromatin modifications and by producing functional RNAs.
Acknowledgments
Work in the Iyer lab has been supported in part by grants from the NIH (CA095548) and the Cancer Prevention and Research Institute of Texas (RP120194) to V.R.I.
Glossary
- +1 nucleosome
The first nucleosome that appears within the DNA encoding a transcript, appearing immediately downstream of a genes nucleosome depleted region.
- −1 nucleosome
The first nucleosome that appears upstream of a gene's nucleosome depleted region.
- Bi-directional transcription
Transcription emanating outward from a single core promoter region in opposite directions.
- Convergent transcription
Transcription originating from opposite strands of DNA with the direction of transcription converging.
- Core promoter
A DNA sequence, generally found within a nucleosome-depleted region, with the ability to independently initiate transcription.
- Core promoter element
A DNA sequence located within a core promoter region and regulates transcriptional output.
- Divergent transcription
Transcription emanating outward from two separate core promoter regions in opposing directions.
- Enhancer RNA (eRNA)
RNA that is usually bidirectionally transcribed from intergenic regions which act as enhancers.
- H2A.Z
A histone variant that can replace the canonical H2A histone within nucleosomes and is associated with sites of transcriptional initiation.
- Long Noncoding RNAs (lncRNAs)
Stable non coding RNAs that are produced either upstream of coding genes or from other intergenic initiation sites. Many have been shown to have structural or regulatory functions.
- Nucleosome-Depleted Region (NDR)
An area of the genome associated with the presence of a transcription start site which is depleted for nucleosomes in the middle and typically has strongly positioned nucleosomes on either side.
- Promoter
A regulatory region of DNA, occurring upstream of a transcript and consisting of a core promoter and core promoter elements, which is required for the proper regulation of gene expression.
- Promoter Upstream Transcripts (PROMPTs)
Noncoding transcripts originating from within a promoter region.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Z, et al. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Preker P, et al. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 2011;39:7179–7193. doi: 10.1093/nar/gkr370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Almada AE, et al. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–363. doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ntini E, et al. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat Struct Mol Biol. 2013;20:923–928. doi: 10.1038/nsmb.2640. [DOI] [PubMed] [Google Scholar]
- 7.Kaplan CD, et al. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 8.Gu M, et al. H2A.Z marks antisense promoters and has positive effects on antisense transcript levels in budding yeast. BMC Genomics. 2015;16:99. doi: 10.1186/s12864-015-1247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Core LJ, et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 2014;46:1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Preker P, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 11.Wei W, et al. Functional consequences of bidirectional promoters. Trends Genet. 2011;27:267–276. doi: 10.1016/j.tig.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arner E, et al. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015;347:1010–1014. doi: 10.1126/science.1259418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melo CA, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh CL, et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci U S A. 2014;111:7319–7324. doi: 10.1073/pnas.1324151111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hah N, et al. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, et al. Enhancer RNAs. Cell Cycle. 2014;13:3151–3152. doi: 10.4161/15384101.2014.962860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park D, et al. Simultaneous mapping of transcript ends at single-nucleotide resolution and identification of widespread promoter-associated non-coding RNA governed by TATA elements. Nucleic Acids Res. 2014;42:3736–3749. doi: 10.1093/nar/gkt1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seila AC, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou L, et al. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat Commun. 2015;6:10026. doi: 10.1038/ncomms10026. [DOI] [PubMed] [Google Scholar]
- 21.Albrecht AS, Orom UA. Bidirectional expression of long ncRNA/protein-coding gene pairs in cancer. Brief Funct Genomics. 2015 doi: 10.1093/bfgp/elv048. [DOI] [PubMed] [Google Scholar]
- 22.Balbin OA, et al. The landscape of antisense gene expression in human cancers. Genome Res. 2015;25:1068–1079. doi: 10.1101/gr.180596.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersson R, et al. A unified architecture of transcriptional regulatory elements. Trends Genet. 2015;31:426–433. doi: 10.1016/j.tig.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 24.Kowalczyk MS, et al. Intragenic enhancers act as alternative promoters. Mol Cell. 2012;45:447–458. doi: 10.1016/j.molcel.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 25.Leung D, et al. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature. 2015;518:350–354. doi: 10.1038/nature14217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trinklein ND, et al. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagawa F, Fink GR. The relationship between the "TATA" sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985;82:8557–8561. doi: 10.1073/pnas.82.24.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang W, et al. TATA elements direct bi-directional transcription by RNA polymerases II and III. Nucleic Acids Res. 1996;24:1158–1163. doi: 10.1093/nar/24.6.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lubliner S, et al. Core promoter sequence in yeast is a major determinant of expression level. Genome Res. 2015;25:1008–1017. doi: 10.1101/gr.188193.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu LC, et al. Upstream box/TATA box order is the major determinant of the direction of transcription. Nucleic Acids Res. 1991;19:6699–6704. doi: 10.1093/nar/19.24.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basehoar AD, et al. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 32.Engstrom PG, et al. Genomic regulatory blocks underlie extensive microsynteny conservation in insects. Genome Res. 2007;17:1898–1908. doi: 10.1101/gr.6669607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carninci P, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 34.Kellis M, et al. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 35.Zhang L, et al. An upstream promoter element blocks the reverse transcription of the mouse insulin-degrading enzyme gene. Biochem Biophys Res Commun. 2013;430:26–31. doi: 10.1016/j.bbrc.2012.11.052. [DOI] [PubMed] [Google Scholar]
- 36.Zhang L, et al. Transcriptional directionality of the human insulin-degrading enzyme promoter. Mol Cell Biochem. 2013;382:237–242. doi: 10.1007/s11010-013-1739-y. [DOI] [PubMed] [Google Scholar]
- 37.Yang MQ, Elnitski LL. Diversity of core promoter elements comprising human bidirectional promoters. BMC Genomics. 2008;9(Suppl 2):S3. doi: 10.1186/1471-2164-9-S2-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duttke SH, et al. Human promoters are intrinsically directional. Mol Cell. 2015;57:674–684. doi: 10.1016/j.molcel.2014.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andersson R, et al. Human Gene Promoters Are Intrinsically Bidirectional. Mol Cell. 2015;60:346–347. doi: 10.1016/j.molcel.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scruggs BS, et al. Bidirectional Transcription Arises from Two Distinct Hubs of Transcription Factor Binding and Active Chromatin. Mol Cell. 2015;58:1101–1112. doi: 10.1016/j.molcel.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Afek A, Lukatsky DB. Genome-wide organization of eukaryotic preinitiation complex is influenced by nonconsensus protein-DNA binding. Biophys J. 2013;104:1107–1115. doi: 10.1016/j.bpj.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Afek A, Lukatsky DB. Positive and negative design for nonconsensus protein-DNA binding affinity in the vicinity of functional binding sites. Biophys J. 2013;105:1653–1660. doi: 10.1016/j.bpj.2013.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin JM, et al. Transcription factor binding and modified histones in human bidirectional promoters. Genome Res. 2007;17:818–827. doi: 10.1101/gr.5623407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collins PJ, et al. The ets-related transcription factor GABP directs bidirectional transcription. PLoS Genet. 2007;3:e208. doi: 10.1371/journal.pgen.0030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bornelov S, et al. Different distribution of histone modifications in genes with unidirectional and bidirectional transcription and a role of CTCF and cohesin in directing transcription. BMC Genomics. 2015;16:300. doi: 10.1186/s12864-015-1485-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soufi A, et al. Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sherwood RI, et al. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat Biotechnol. 2014;32:171–178. doi: 10.1038/nbt.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bonisch C, Hake SB. Histone H2A variants in nucleosomes and chromatin: more or less stable? Nucleic Acids Res. 2012;40:10719–10741. doi: 10.1093/nar/gks865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weber CM, et al. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol Cell. 2014;53:819–830. doi: 10.1016/j.molcel.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 53.Sarcinella E, et al. Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Mol Cell Biol. 2007;27:6457–6468. doi: 10.1128/MCB.00241-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Draker R, et al. USP10 deubiquitylates the histone variant H2A.Z and both are required for androgen receptor-mediated gene activation. Nucleic Acids Res. 2011;39:3529–3542. doi: 10.1093/nar/gkq1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu G, et al. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2013;12:180–192. doi: 10.1016/j.stem.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zofall M, et al. Histone H2A.Z cooperates with RNAi and heterochromatin factors to suppress antisense RNAs. Nature. 2009;461:419–422. doi: 10.1038/nature08321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen P, et al. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes Dev. 2013;27:2109–2124. doi: 10.1101/gad.222174.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Won KJ, et al. Proteogenomics analysis reveals specific genomic orientations of distal regulatory regions composed by non-canonical histone variants. Epigenetics Chromatin. 2015;8:13. doi: 10.1186/s13072-015-0005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marquardt S, et al. A chromatin-based mechanism for limiting divergent noncoding transcription. Cell. 2014;157:1712–1723. doi: 10.1016/j.cell.2014.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim T, et al. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell. 2012;150:1158–1169. doi: 10.1016/j.cell.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hennig BP, et al. Chd1 chromatin remodelers maintain nucleosome organization and repress cryptic transcription. EMBO Rep. 2012;13:997–1003. doi: 10.1038/embor.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Skene PJ, et al. The nucleosomal barrier to promoter escape by RNA polymerase II is overcome by the chromatin remodeler Chd1. Elife. 2014;3:e02042. doi: 10.7554/eLife.02042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Camblong J, et al. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 65.Camblong J, et al. Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev. 2009;23:1534–1545. doi: 10.1101/gad.522509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tan-Wong SM, et al. Gene loops enhance transcriptional directionality. Science. 2012;338:671–675. doi: 10.1126/science.1224350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hou C, et al. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proc Natl Acad Sci U S A. 2008;105:20398–20403. doi: 10.1073/pnas.0808506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tark-Dame M, et al. Depletion of the chromatin looping proteins CTCF and cohesin causes chromatin compaction: insight into chromatin folding by polymer modelling. PLoS Comput Biol. 2014;10:e1003877. doi: 10.1371/journal.pcbi.1003877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanyal A, et al. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Arensbergen J, et al. In search of the determinants of enhancer-promoter interaction specificity. Trends Cell Biol. 2014;24:695–702. doi: 10.1016/j.tcb.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Castelnuovo M, et al. Role of histone modifications and early termination in pervasive transcription and antisense-mediated gene silencing in yeast. Nucleic Acids Res. 2014;42:4348–4362. doi: 10.1093/nar/gku100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anderson SK. Probabilistic bidirectional promoter switches: noncoding RNA takes control. Mol Ther Nucleic Acids. 2014;3:e191. doi: 10.1038/mtna.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mayer A, et al. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell. 2015;161:541–554. doi: 10.1016/j.cell.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cassiday LA, Maher LJ., 3rd Having it both ways: transcription factors that bind DNA and RNA. Nucleic Acids Res. 2002;30:4118–4126. doi: 10.1093/nar/gkf512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sigova AA, et al. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015;350:978–981. doi: 10.1126/science.aad3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 77.Ietswaart R, et al. Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet. 2012;28:445–453. doi: 10.1016/j.tig.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 78.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sharon E, et al. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat Biotechnol. 2012;30:521–530. doi: 10.1038/nbt.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yan C, et al. Decoupling of divergent gene regulation by sequence-specific DNA binding factors. Nucleic Acids Res. 2015;43:7292–7305. doi: 10.1093/nar/gkv618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 82.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de Hoon M, Hayashizaki Y. Deep cap analysis gene expression (CAGE): genome-wide identification of promoters, quantification of their expression, and network inference. Biotechniques. 2008;44:627–628. 630–632. doi: 10.2144/000112802. [DOI] [PubMed] [Google Scholar]
- 84.Pelechano V, et al. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013;497:127–131. doi: 10.1038/nature12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Core LJ, et al. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kruesi WS, et al. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. Elife. 2013;2:e00808. doi: 10.7554/eLife.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kwak H, et al. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nojima T, et al. Mammalian NET-Seq Reveals Genome-wide Nascent Transcription Coupled to RNA Processing. Cell. 2015;161:526–540. doi: 10.1016/j.cell.2015.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Srivastava R, Ahn SH. Modifications of RNA polymerase II CTD: Connections to the histone code and cellular function. Biotechnol Adv. 2015;33:856–872. doi: 10.1016/j.biotechadv.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 90.Terzi N, et al. H3K4 trimethylation by Set1 promotes efficient termination by the Nrd1-Nab3-Sen1 pathway. Mol Cell Biol. 2011;31:3569–3583. doi: 10.1128/MCB.05590-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5' transcribed regions. Cell. 2009;137:259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Carrozza MJ, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 93.Lickwar CR, et al. The Set2/Rpd3S pathway suppresses cryptic transcription without regard to gene length or transcription frequency. PLoS One. 2009;4:e4886. doi: 10.1371/journal.pone.0004886. [DOI] [PMC free article] [PubMed] [Google Scholar]