Abstract
Trypanosomatid parasites are the causative agents of many neglected tropical diseases including the leishmaniases, Chagas disease, and human African trypanosomiasis. They exploit unusual vacuolar soluble pyrophosphatases (VSPs), absent in humans, for cell growth and virulence and as such, are drug targets. Here, we report the crystal structures of VSP1s from Trypanosoma cruzi and T. brucei, together with that of the T. cruzi protein bound to a bisphosphonate inhibitor. Both VSP1s form a hybrid structure containing an (N-terminal) EF-hand domain fused to a (C-terminal) pyrophosphatase domain. The two domains are connected via an extended loop of about 17 residues. Crystallographic analysis and size exclusion chromatography indicate that the VSP1s form tetramers containing head-to-tail dimers. Phosphate and diphosphate ligands bind in the PPase substrate-binding pocket and interact with several conserved residues, and a bisphosphonate inhibitor (BPH-1260) binds to the same site. Based on Cytoscape and other bioinformatics analyses it is apparent that similar folds will be found in most if not all trypanosomatid VSP1s, including those found in insects (Angomonas deanei, Strigomonas culicis), plant pathogens (Phytomonas spp.) and Leishmania spp. Overall, the results are of general interest since they open the way to structure-based drug design for many of the neglected tropical diseases.
Introduction
Trypanosomatid parasites are the causative agents of many neglected tropical diseases (1, 2) including the leishmaniases, Chagas disease and human African trypanosomiasis (sleeping sickness). The prevalence of sleeping sickness has been rapidly decreasing over the past 10 years with the introduction of drugs such as eflornithine, and visceral leishmaniasis is now being treated quite successfully with miltefosine; but for Chagas disease (caused by Trypanosoma cruzi), there are no truly effective treatments for the chronic stage of the disease so new drugs, and new drug leads, are of great interest(3).
In previous work, we reported that all of the trypanosomatid parasites possess large numbers of Ca/Zn/P-containing organelles, called acidocalcisomes(4). Using 31P nuclear magnetic resonance spectroscopy we found high levels of condensed inorganic phosphates (primarily di- and tripoly-phosphates) associated with the acidocalcisomes(5, 6) and in other work we discovered plant-like pyrophosphatases, as well as vacuolar soluble pyrophosphatases (VSPs). In the presence of divalent metal cations(7, 8) these enzymes hydrolyze pyrophosphate (a.k.a. diphosphate, PPi) and inorganic polyphosphates to orthophosphate (Pi). The role of one VSP, VSP1, in regulating pyrophosphate levels is considered critical in pH and osmoregulation in trypanosomes(8) and the trypanosomatid VSP1s are essential for parasite growth, virulence, and persistent infection, in animal models(7, 8). This led us to an interest in pyrophosphate analogs, bisphosphonates, as potential VSP1 inhibitors, and one bisphosphonate inhibitor active against T. brucei VSP1 (TbVSP1) gave a 40% survival in a mouse model of infection(9). There are, however, no structures reported for any trypanosomatid VSP1s which—based on bioinformatics—appear to be "Rosetta stone" sequences(10), proteins that contain two, functionally-related domains that are fused into one, larger protein. One domain is thought to be an EF-hand domain, the other is a phosphatase domain that can hydrolyze a diverse range of condensed phosphates—PPi through at least PolyP100—enzymes that are referred to here and elsewhere as "PPases".
Results and discussion
A bioinformatics view of PPases
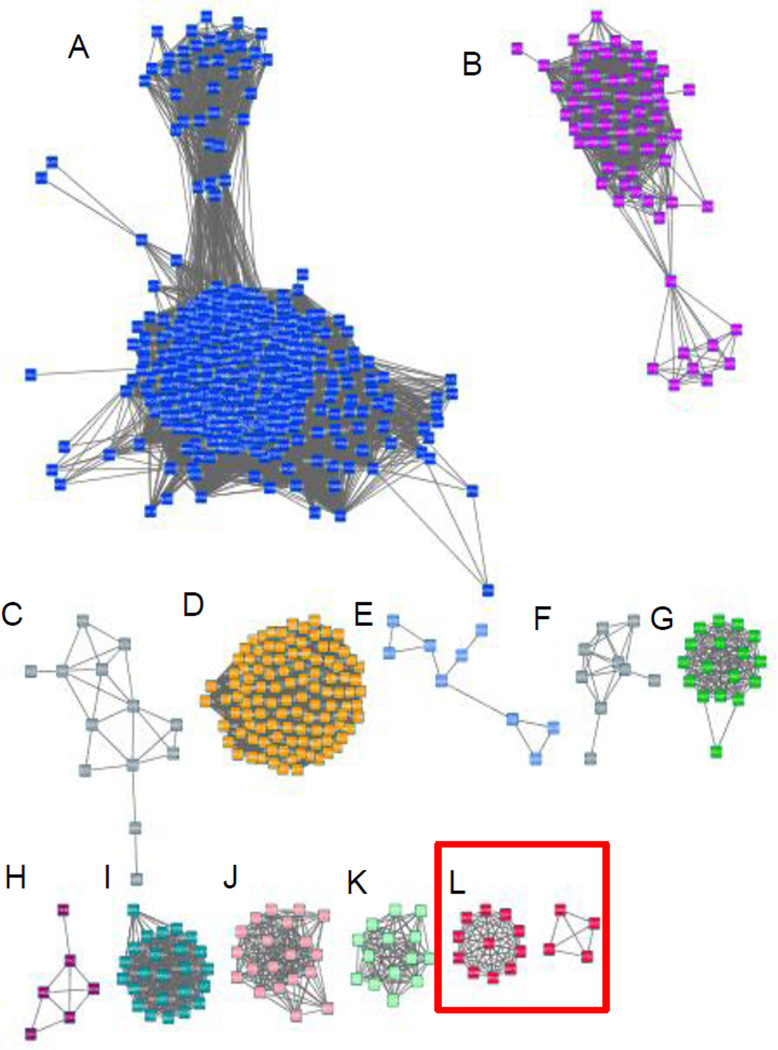
Soluble PPases, primarily diphosphatases, are divided into three families, depending on their sequences and structure: family I PPases occur in all kingdoms of life and prefer Mg2+ as their cofactor and they are usually dimeric or hexameric(11, 12); family II PPases are relatively rare, are found mainly in bacteria, and prefer Mn2+ as their cofactor(13); family III PPases belong to the haloalkane dehalogenases family and are found in some bacteria and are Ni2+ dependent(14). Trypanosomatid VSP1s are classified as family I PPases(7, 8, 15). Conventional family I PPases all have a single-domain structure, as first reported for the PPase from Saccharomyces cerevisiae (ScPPase; PDB ID code 1WGJ)(12). Although the trypanosomatid VSP1s have about 40% sequence identity to the other family I members, they are predicted to contain an additional EF-hand domain that precedes the catalytic domain(8). A Cytoscape(16) analysis of the family I pyrophosphatases using tools in the Enzyme Function Initiative(17) is shown in Figure 1 and comprises 6,620 sequences, based on the sequence of TcVSP1, constructed with an expectation-value (e-value) of 10−40. Major clusters are as shown in Figure 1. Cluster L contains a set of “fusion hybrids” or putative “Rosetta stone” proteins, 37 EF-hand/PPase domain proteins, shown by the red square. All of the organisms in cluster L are trypanosomatids; a full list is shown in Table S1. The list includes the major human pathogens T. cruzi, T. brucei, Leishmania spp., as well as trypanosomes found in insects (Angomonas deanei, Strigomonas culicis), and one that is an important plant pathogen (Phytomonas spp.). This widespread occurrence implies functional-relatedness between the two domains, and solving the structures of these fusion hybrids could be of interest in the context of future drug discovery since the proteins are absent in humans but essential for the trypanosomatid parasites.
Figure 1.
Sequence similarity network based on the TcVSP1 sequence and constructed with an expectation-value (e-value) of 10−40; 6,620 sequences were used to produce the network. Each node represents sequences with at least 30% similarity. Major clusters are: A, Arthropoda+Ascomycota+Basidiomycota; B, Chlorophyta+Streptophyta; C, Proteobacteria; D, Streptophyta; E, Actinobacteria (Mycobacterium spp.); F, Bacteroidetes; G, Bacillariophyta+Eustigmatophyceae+Phaeophyceae; H, Apicomplexa (Plasmodium spp.); I, Ascomycota; J, Cyanobacteria; K, Euryarchaeota; L, Trypanosomatidae (T. brucei, T. cruzi).
VSP1 structure determination
We first expressed, purified and crystallized VSP1 from T. brucei, TbVSP1. Full details are given in the Methods Section. We obtained rod-shaped TbVSP1 crystals that diffracted to 2.5 Å and belonged to the space group C2, with unit-cell parameters a=199.92 Å, b=70.14 Å, c=141.76 Å, β=106.38°. However, molecular replacement (MR) approaches to solve the TbVSP1 structure failed. We thus next expressed, purified, and crystallized TcVSP1, this time using Se-methionine in the growth medium. We obtained crystals that diffracted to 2.62 Å (Table 1 and Table S2) and obtained the structure of this protein (PDB ID code 5CUV) using the MAD method. The structure so obtained was then used as a search model to solve the TbVSP1 structure using the MR method. The electron densities for all (20) ligands observed in the structures to be discussed below (citrate, D-malate, phosphate, diphosphate, a bisphosphonate) are shown in Figure S1 (contoured at 1σ and 2σ) together with their local ligand density fit (LLDF) values.
Table 1.
Summary of data processing and refinement statistics
| Se-Met TcVSP | TbVSP | TcVSP-BPH1260 | ΔN1-TcVSP-PPi | |
|---|---|---|---|---|
| PDB ID | 5CUV | 5CUY | 5CUU | 5CUX |
| Data collection | ||||
| Space group | C2221 | C21 | C2221 | P3221 |
| Wavelength (Å) | 0.97896 | 0.97622 | 1.00000 | 1.00000 |
| Resolution (Å) | 25-2.62 (2.71-2.62) | 25-2.5 (2.59-2.5) | 25-2.96 (3.07-2.96) | 25-2.8 (2.9-2.8) |
| Unit-cell | ||||
| a/b/c (Å) | 100.5/103.0/157.4 | 199.9/70.1/141.8 | 100.8/102.6/158.8 | 122.9/122.9/127.7 |
| α/β/γ (°) | 90/90/90 | 90/106.38/90 | 90/90/90 | 90/90/120 |
| No. of unique reflections | 24891 (2467) | 64763 (6191) | 17519 (1718) | 27934 (2741) |
| Redundancy | 11.5 (11.3) | 3.3 (3.1) | 4.4 (4.5) | 7.3 (7.5) |
| Completeness (%) | 99.9 (100.0) | 98.7 (94.7) | 99.9 (100) | 99.9 (100) |
| Average I/σ(I) | 26.68(5.9) | 15.39 (2.8) | 28.5 (3.56) | 42.5 (4.9) |
| Rmergea (%) | 8.6 (44.4) | 9 (38.3) | 6.2 (50.0) | 5.7 (49.5) |
| Refinement | ||||
| No. of reflections | 23085 (2136) | 61721 (5068) | 16551 (1390) | 26928 (2351) |
| Rwork(95% of data) | 0.178 (0.262) | 0.194 (0.327) | 0.194 (0.308) | 0.198 (0.284) |
| Rfree (5% of data) | 0.235 (0.311) | 0.252 (0.353) | 0.274 (0.363) | 0.233 (0.329) |
| R.m.s.d. bonds (Å) | 0.0196 | 0.0159 | 0.0157 | 0.0178 |
| R.m.s.d. angles (°) | 2.003 | 1.835 | 1.808 | 2.208 |
| Dihedral angles (%) | ||||
| Most favored | 91.4 | 93.3 | 90.6 | 93.8 |
| Allowed | 6.1 | 5.3 | 7.0 | 4.7 |
| Disallowed | 2.5 | 1.4 | 2.4 | 1.6 |
| No. of non-H atoms/average B (Å2) | ||||
| Protein | 6221/42.8 | 12632/50.3 | 6187/78.4 | 4254/58.9 |
| Water | 346/43.5 | 686/49.6 | 121/59.8 | 118/59.0 |
| Ligand | 9/58.3 | 52/103.7 | 67/118.0 | 27/102.7 |
| Ion | 4/47.4 | 4/52.0 | 2/87.5 | 35/115.8 |
Values in parentheses are for the highest resolution shell.
Rmerge = ∑hkl∑i|Ii(hkl)-<I(hkl)>| / ∑hkl∑iIi(hkl).
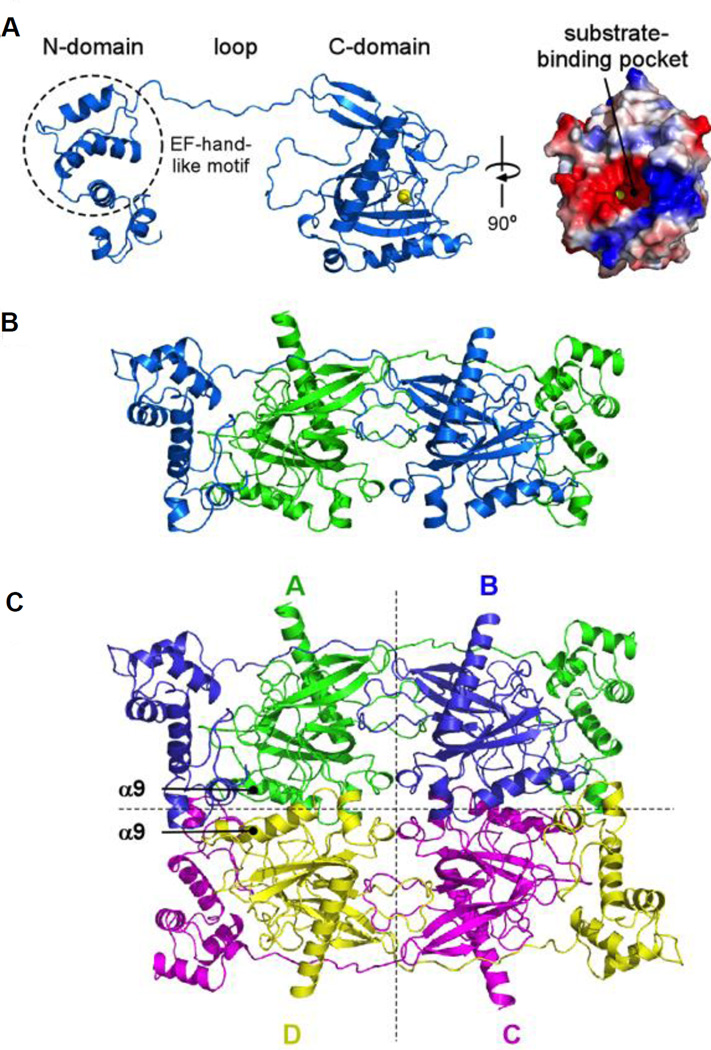
We show in Figure 2A the structure of native TcVSP1 in the absence of any added substrate or inhibitor ligands. The structure clearly shows the presence of the EF-hand N-terminal domain, and the C-terminal PPase domain, the substrate binding pocket (Figure 2A, to the right) being relatively solvent-exposed. The two chains in the TcVSP1 dimer pack in a “head-to-tail” fashion with the EF-hand domain in one chain interacting with the PPase domain in the second molecule in the dimer, Figure 2B. As can also be seen in Figures 2A and 2B, there is a long (~17 residue, ~32 Å) polypeptide chain linker connecting the two domains whose centers of mass are separated—in a single chain—by ~67 Å. The orthorhombic TcVSP1 crystal thus contains two polypeptide chains in an asymmetric unit, and the two dimers are related by crystallographic symmetry to constitute a tetramer, as shown in Figure 2C. The TbVSP1 crystal also contains four polypeptide chains, a tetramer that has the same arrangement as that found in TcVSP1.
Figure 2. Overall structure of TcVSP1.
The monomeric (A), dimeric (B), and tetrameric (C) configurations of TcVSP1 are shown in cartoon models.
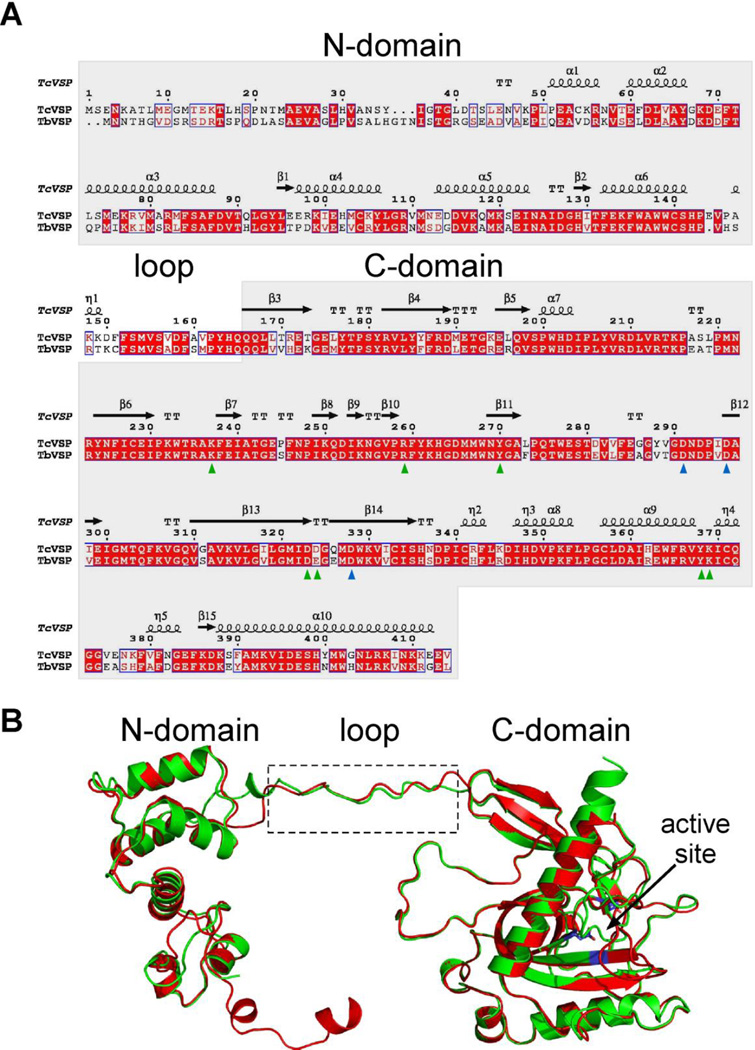
The subunit arrangements of TcVSP1 and TbVSP1 found in the tetramers are very different to those found in the hexameric E. coli PPase (EcPPase) and dimeric ScPPase. The crystal structures indicate that the trypanosomatid VSP1s form tetramers via two sets of interactions: A–B (C–D) and A–D (B–C), Figure 2C. The buried surface areas of the A–B and A–D interfaces are 3917.1 and 910.7 Å2 in TcVSP, and 3953 and 931.2 Å2 in TbVSP. The A–B pair involves interactions between the bottom of two C-domains, while the A–D dimer is formed through C-domains containing strand α9 and several loops, Figure 2C. Several residues in the loop (Figure 2A) extend into the neighboring monomer and appear to be important in A–B dimer formation, as discussed in more detail below. Sequence and structure alignments of the TbVSP1 and TcVSP1 proteins were performed using the Clustal W(18) and Espript 3(19) programs and are shown in Figures 3A and 3B. The protein sequence identity between the two VSPs is 69.8%, and the structures superimpose on each other very well with a C • r.m.s.d. (root mean square deviation) of 0.623 Å, Figure 3B. The Mg-coordinated residues are shown in Figure 3A by blue triangles, the Pi/PPi-interacting residues are shown in green. Since the Mg-coordinating Asp residues are strictly conserved (Mg2+ or other divalent cations are essential for hydrolysis in family I PPases), these three Asp residues are shown in Figure 3B to indicate the active site location.
Figure 3. Sequence and structure alignment of TcVSP1 and TbVSP1.
(A) Sequence alignment performed by using the Clustal W18 and Espript 319 programs. Blue triangles, divalent cation-interacting residues; green triangles, PPi-interacting residues. (B) Structure superimposition of TcVSP1 (green) and TbVSP1 (red) Blue, divalent cation-binding Asps.
So, compared with all other family I pyrophosphatases, as shown above, the trypanosomatid VSP1s possess an EF-hand N-terminal domain, together with an extended "linker" sequence of about 17 residues (Figure 2A). This linker sequence in the dimer spans the C-terminus domain of the other monomer to form a head-to-tail dimer, Figure 2B.
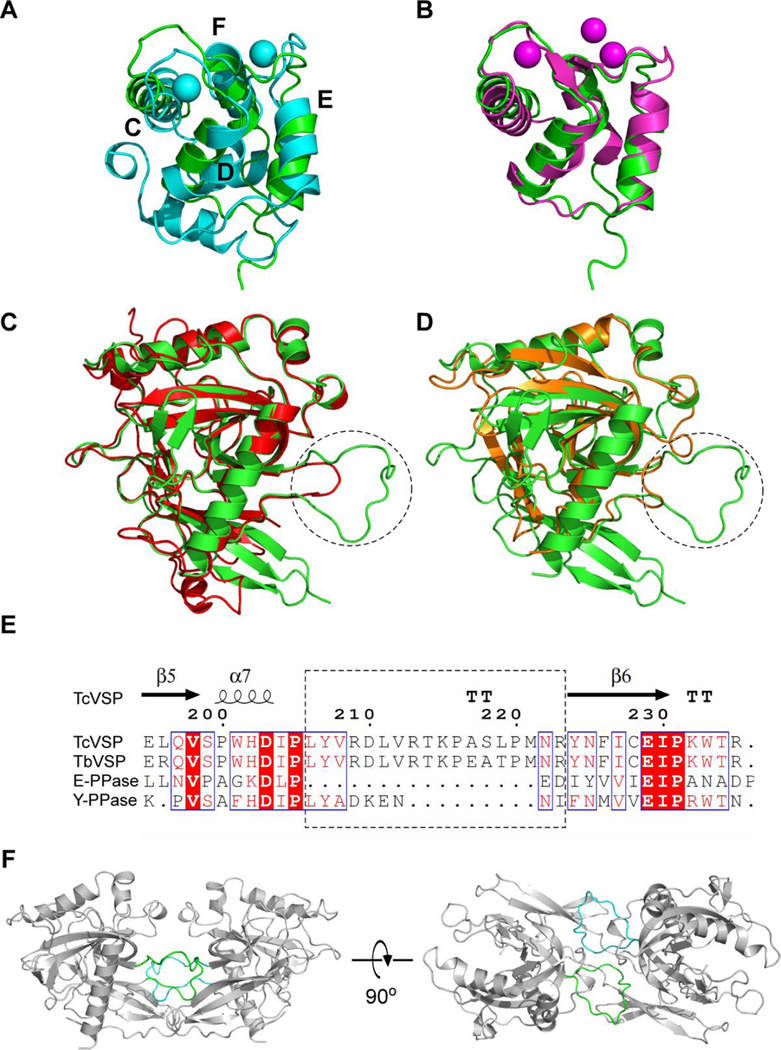
The N-terminal EF-hand domain is very similar to that found in the classic EF-hand domain protein parvalbumin (ex rat, PDB ID code 1RWY) where there is a 2.78 Å root mean square deviation (r.m.s.d.) for Cα atoms between TcVSP1 and parvalbumin, Figure 4A. The term “EF”-hand domain originates from helices E and F in parvalbumin which as can be seen in Figure 4A(20), contains two helix-loop-helix motifs. These EF-hand domains typically bind to Ca2+ (21). The structure is very similar to that found in human calmodulin, Figure 4B.
Figure 4.
EF-hand and PPase domains in TcVSP1 compared with similar structures in other proteins. TcVSP1 (PDB ID code 5UCV, residues 72–150) superimposed on (A) rat parvalbumin (PDB ID code 1RWY) and (B) human calmodulin (PDB ID code 3UCW). Ca2+ and Mg2+ in 1RWY and 3UCW structures are shown as spheres. The presence of 4 helices in 1RWY make up the two EF-hand motifs. (C) TcVSP1 superimposed on EcPPase (PDB ID code 4UM4). (D) TcVSP1 superimposed on ScPPase (PDB ID code 2IHP). (E) Clustal W18 and Espript 319 alignments of TcVSP1, TbVSP1, EcPPase, and ScPPase. The dashed circles in (C), (D) indicate the α7-β6 loops, shown in the dashed box in (E). (F) A–B dimeric configuration that highlights the α7-β6 loops in chain A (green) and B (cyan). These loops are absent in the E. coli and yeast proteins and contribute to A–B dimer formation.
The C-terminal domain is the catalytic PPase domain and adopts a similar structure to that is seen in other family I PPases. For example, the Cα r.m.s.d. values between the PPase domains are 1.18 Å between E. coli PPase (EcPPase, PDB ID code 4UM4) and TcVSP1, and 0.622 Å between the ScPPase (PDB ID code 2IHP) and TcVSP1, Figures 4C and 4D, respectively. The loop regions in the hexameric EcPPase and dimeric ScPPases are much shorter, Figure 4E, suggesting these proteins lack the interactions required to form the dimers that are found in the VSP1s. Such A–B dimer contacts are shown in Figure 4F.
Protein oligomerization
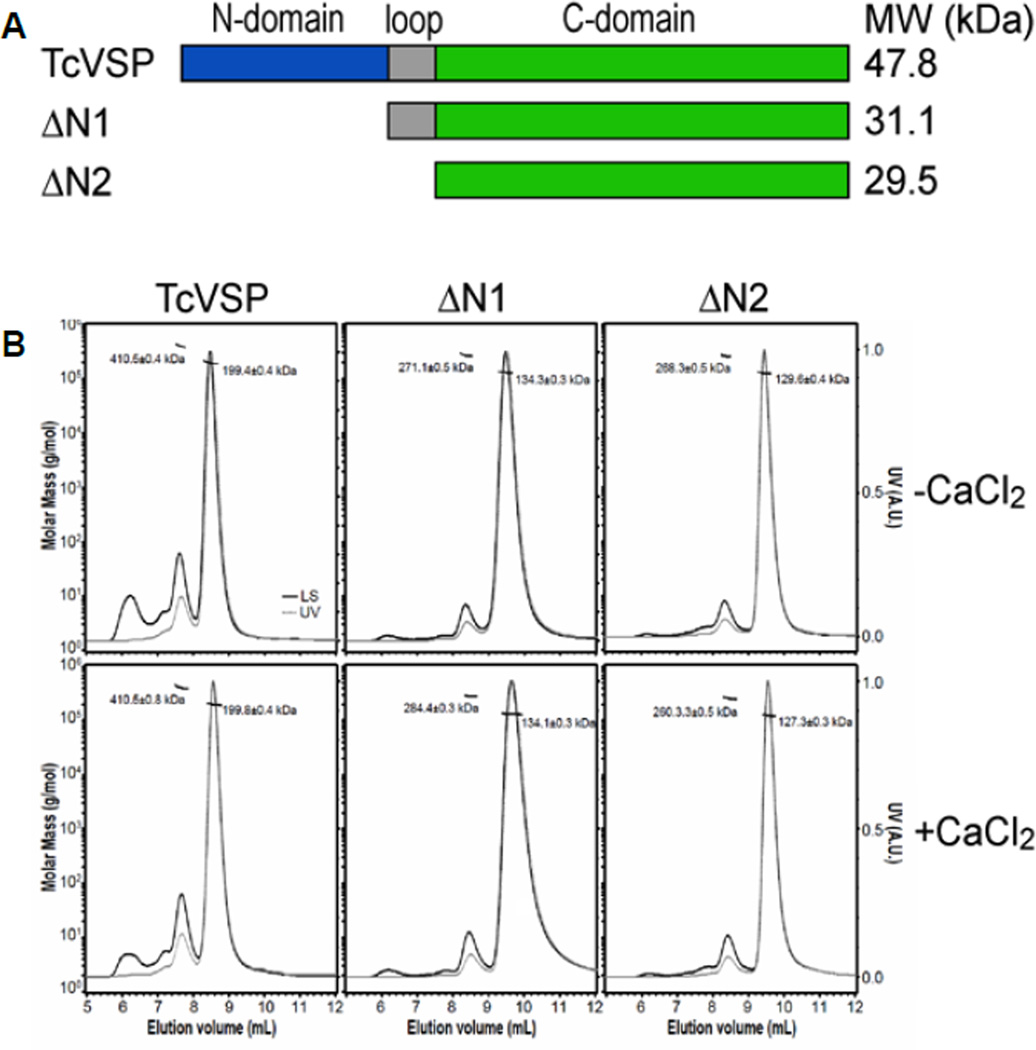
To examine the oligomerization state in solution of the VSP1 proteins described previously (8), we carried out size-exclusion chromatography coupled with multi-angle light scattering (SEC-MALS), in addition to analytical ultracentrifugation (AUC). The SEC-MALS results, Figure 5B, show a major peak for TcVSP1 that corresponds to a molecular weight of 199.4 ± 0.4 kDa, indicating a tetramer. A small fraction (~11%) corresponds to a molecular weight of 410.5 ± 0.4 kDa, indicating the presence of an octamer. TbVSP1 elutes primarily as a tetramer with a molecular weight of 208.5 ± 0.4 kDa. However, there was some aggregation as evidenced by the presence of several high molecular weight elution peaks. The SEC-MALS results were validated by the results of analytical ultracentrifugation (AUC) experiments, Figures S2A and S2B. So, SEC-MALS, AUC as well as the x-ray results all point to a tetrameric arrangement for both proteins.
Figure 5. SEC-MALS analysis of full-length and N-terminal truncated TcVSP1s.
(A) The top panel shows the truncation positions. (B) SEC-MALS results obtained in the absence and presence of 0.1 mM CaCl2. The horizontal thin line segments represent the calculated molar masses and their corresponding molecular weights (left ordinate axis). The dashed grey line and the solid black line correspond to the normalized UV absorbance at 280 nm (right ordinate axis) and the intensity of light scattering, respectively.
The N-terminal domain
We next consider the structures of the N-terminal domains in more detail. A search of known protein structures using the DALI program(22) shows that the N-terminal domains of TcVSP1 and TbVSP1 are most similar to the N-terminal domain of human calmodulin (PDB ID code 3UCW). This is shown in Figure 4B for TcVSP1 where there is a 1.57 Å Cα r.m.s.d over 71 residues with calmodulin (in the presence of Mg2+). Calmodulin forms a dimer and binds to metal ions. In previous work we hypothesized that TbVSP1 exploits an EF-hand like motif in the N-terminal domain to coordinate Ca2+ to form larger complexes with molecular weights of up to ~600 kDa(8). However, here we were unable to model any metal ion into the N-terminal domains in TcVSP1 or TbVSP1 even though the crystals were grown in the presence of high concentrations (10 mM) of Ca2+. Moreover, the results of the SEC-MALS and AUC experiments showed that the oligomerization status of both TcVSP1 and TbVSP1 were not significantly altered by the presence of Ca2+ (Figures 5B and S2). Interestingly, a protein sequence alignment (Figure S3A) indicates that the VSP1 EF-hand like motifs lack several Asp or Glu residues that are used to coordinate Ca2+ in the homologous calmodulin EF-hand motif(23, 24), as shown in the x-ray structures in Figures S3B. It thus appears that the N-terminal domain in the VSP1s will have very limited ability to bind Ca2+, making its role rather elusive. However, one possibility is that it might be involved in tetramer formation.
We thus next determined the structure of a TcVSP1 that lacked the N-terminal domain (ΔN1-TcVSP1-PPi; PDB ID code 5CUX). The protein crystallized in the same tetrameric form as that of the full-length protein and the buried surface areas of the A–B and A–D interfaces were 2481.6 and 834.1 Å2, respectively, strongly suggesting that it is the C-terminal domain rather than the N-terminal domain that drives tetramer formation, consistent with a role of the α7-β6 loop in initial dimer formation. Both ΔN1-TcVSP1 and ΔN2-TcVSP1 (the N-domain and loop deleted TcVSP1) variants displayed comparable activity to that of the full-length proteins, Table S3, as reported previously(8). And, as shown in the SEC-MALS results in Figure 5B and the AUC results in Figure S2C and D, both proteins form tetramers in solution. So there is no evidence for the N-terminus being involved in oligomerization (with or without Ca2+). Another possibility is that the N-terminus might be involved in acidocalcisome-targeting, but in earlier work(8) using a green fluorescent protein ΔN1 construct we found no evidence for acidocalcisome targeting, so further work (beyond the scope of the current article) is required in order to clarify the role of the N-terminal domain in the VSP1 protein.
Ligand-bound complex structures
We next investigated how ligands bind to the PPase active site. In previous work it has been shown that four divalent metal ions are required for family I PPase catalysis. In our crystallization buffers, Mg2+ was present (at 10 mM) but only one cation could be modeled into the substrate-binding pocket of both full-length VSP1 apo-structures. The Mg2+ observed is bound to the so-called M1 site where it is coordinated by three strictly conserved Asp residues, Figure S4. The absence of the other metal ions might be due to their relatively low concentrations in the mother liquor.
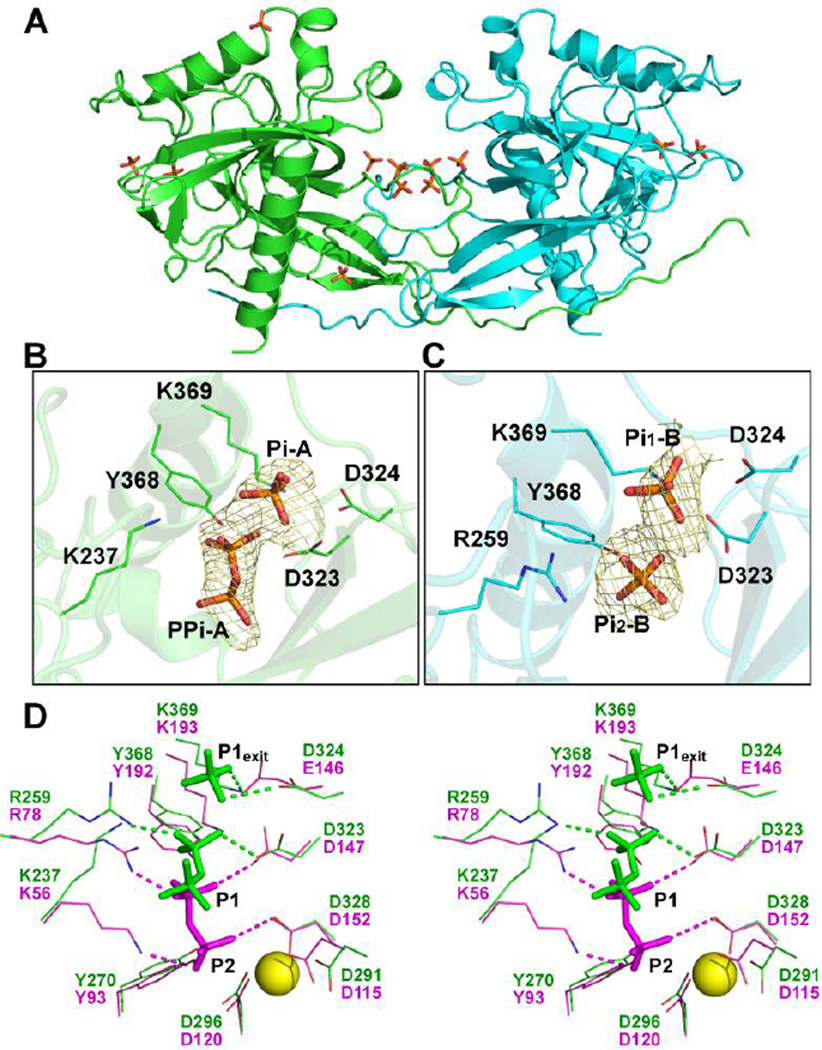
Several Pi and PPi were then modeled into the ΔN1-TcVSP1-PPi structure, Figure 6A. In chain A, one Pi (Pi-A) and one PPi (PPi-A) were found in the substrate-binding pocket, Figure 6B. One phosphate group of PPi-A (PA) is located near the P1 (leaving group) binding site, with O1A, O2A, and O3A within hydrogen bonding distances from the side-chains of residues Y368 (O1A), K237 (O2A), and K369 (O3A), Figure 6B. Pi-A is located in a position that corresponds to P1exit in the ScPPase complex structure(25) and is the expected Pi product. In chain B, two Pi are found, in the positions corresponding to Pi-A and the PA group of PPi-A, Figure 6C. The location of PPi-A is slightly displaced from that found in the ScPPase, and the P2 (substrate) site is empty, Figure 6C. The P2 site has previously been proposed to contain four divalent metal ions(12). Since no metal ions were observed in the ΔN1 structure, it is likely that PPi binding affinity for the P2 site is reduced. The side-chains of K237 and R259 in chain A, which would bind to P2 and P1, deviate from their reported positions in yeast PPase and turn outwards, Figure 6D. These two residues are involved in product release and thus appear to be of high mobility.
Figure 6. Pi and PPi in the ΔN1-TcVSP1-PPi structure.
(A) The overall structure of ΔN1-TcVSP1-PPi (PDB ID code 5CUX). Two monomers are shown in green and cyan. Pi and PPi groups are shown as stick models. (B) Pi and PPi with electron densities contoured at 1.0 σ. Residues with side-chains within hydrogen bond distances of the phosphate groups are presented as line models. (C), As (B) but for the Pi groups in chain B. (D) Chain A of the ΔN1 structure (green; PDB ID code 5CUX) superimposed on ScPPase structure (magenta, PDB ID code 1E6A). The PPi, Pi, and phosphate group—interacting residues are shown. The hydrogen bonds are shown as dashed lines; Mg2+ was modeled from TcVSP1, and is shown as a yellow sphere.
An inhibitor-bound complex structure
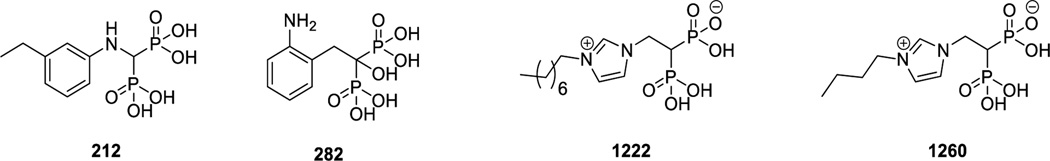
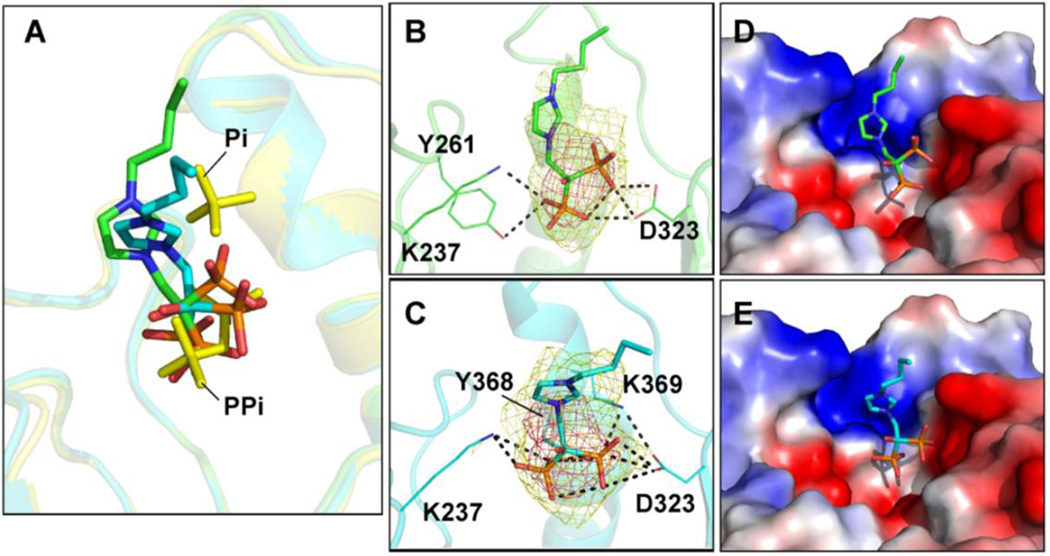
A long-term goal of our work is to develop trypanosomatid VSP1 inhibitors that are active in cells: drug leads. Here, we investigated the four VSP1 inhibitors shown in Figure 7. BPH-212 and BPH-282 are inhibitors that we reported previously(9); BPH-1222 and BPH-1260 are newer compounds(26) that were developed to inhibit farnesyl diphosphate synthase but which might also inhibit VSP1s since they both contain isosteres of diphosphate—the bisphosphonate group. We determined the activities of all four compounds against TcVSP1, the ΔN1 and ΔN2 deletion mutants, as well as TbVSP1. The activities of each protein using diphosphate or a ~100 residue polyphosphate (PolyP100) as substrate, in addition to the enzyme inhibition results, are shown in Table S3. None of the inhibitors had measurable activity against any of the enzymes when using diphosphate as substrate. However, the exopolyphosphatase activity was inhibited by BPH-212 (IC50 value ~5 µM), less so (~45 µM) by BPH-1260, which has a butyl side-chain, and not at all by BPH-1222, which has an octyl side-chain. So, the smaller the inhibitor, the better the inhibition. We attempted crystallization of both TcVSP1 and TbVSP1 with each bisphosphonate but only one compound, BPH-1260, yielded crystals that diffracted to <3.0 Å, only moderate resolution. Crystallographic data acquisition and processing results are shown in Table 1. The bisphosphonate group of BPH-1260 binds to the position corresponding to that of PPi-A in the ΔN1-TcVSP1-PPi structure, Figure 8A. The phosphonate groups make hydrogen bonds with the side-chains of Y261, K237, and D323 in both chains A and B, Figures 8B and 8C. Ligand electron densities are shown contoured at 1σ and 3σ. The ligand's n-butyl side-chains occupy a solvent-exposed polar surface pocket, as can be seen in the electrostatic surfaces shown in Figures 8D,E. Binding to this site will impose an energetic cost for longer side-chain containing species, such as BPH-1222, which being very amphipatic or detergent-like can form micelles in water. Binding is thus in all cases dominated by the Coulombic and hydrogen bond interactions of the bisphosphonate group with Y261, K237, and D323 while the decreased activity of the longer alkyl chain species is, we propose, due to the energetic penalty in moving the longer alkyl chains into a more polar environment.
Figure 7.
Structures of four VSP1 inhibitors.
Figure 8.
BPH-1260 in the TcVSP1 complex structure (A) Structure superimposition of TcVSP1-BPH-1260 (PDB ID code 5CUU; chain A, green; chain B, cyan) and ΔN1-TcVSP1-PPi (PDB ID code 5CUX; yellow). The ligands are presented as stick models. (B) BPH-1260 in chain A. (C) BPH-1260 in chain B.The side-chains of residues within hydrogen bond distances of BPH-1260 are shown as line models. Dashed lines indicate distance within 3.5 Å. Electron densities (2Fo −Fc) in (B), (C) contoured at 1σ, 3σ. (D) Electrostatic surface representation of BPH-1260 binding pocket in TcVSP1, chain A. Note that the n-butyl side-chain has few protein contacts and appears to extend into the solvent; longer chain-containing species are expected to be less potent inhibitors. (E) As (D) but for chain B.
Conclusions
The results we have obtained above are of interest for several reasons. First, we obtained the structures of vacuolar soluble pyrophosphatases from the trypanosomatid parasites T. cruzi (the causative agent of Chagas disease), and T. brucei (the causative agent of human African trypanosomiasis). Both structures are fusion hybrid proteins and show the presence of EF-hand domain structures separated by ~17 residues from the pyrophosphatase domain. Second, we carried out a bioinformatics survey of PPases finding that similar hybrid species are present in all trypanosomatids—varying from human pathogens, to trypanosomatids that colonize insect guts, to a plant pathogen. Third, we found that both TcVSP1 and TbVSP1 have two chains organized in a “head-to-tail” fashion in which the EF-hand domain in chain A is closely apposed to the PPase domain in chain B. These dimers then interact to form a tetramer, as observed by x-ray crystallography, SEC-MALS as well as AUC. Fourth, we found that removal of the EF-hand domain, or the EF-hand domain together with the linker, had no effect on PPase structure, or exopolyphosphatase activity. Fifth, we obtained the structures of Pi and PPi liganded proteins. Sixth, we obtained the structure of a bisphosphonate inhibitor complex finding that the bisphosphonate moiety bound into the active site occupied by PPi (in another family I PPase) and that larger inhibitors were less effective than smaller ones, due we propose to greater solvent exposure. The finding of these unusual “head-to-tail” dimeric structures as well as the solution of an inhibitor-bound structure will hopefully open the way to finding more potent and selective inhibitors/drug leads that target these unusual “Rosetta stone” proteins.
Methods
Protein preparation
The genes encoding TcVSP1 and TbVSP1 were derived as described in our previous studies(7, 8). Genes were amplified by polymerase chain reaction (PCR) with the primers listed in Table S4 and then cloned into the pET46 Ek/LIC vector (Novagen, Madison, Wisconsin, USA). Recombinant plasmids were verified by sequencing and transformed into E. coli BL21(DE3). Recombinant E. coli cells were grown at 37 °C to an OD600 of 0.6. Protein expression was induced at 16 °C for 24 h by adding 1 mM IPTG. Cells were harvested by centrifugation at 5000×g at 4 °C for 20 min. Cell pellets were then resuspended in lysis buffer consisting of 25 mM Tris, pH 7.5, 150 mM NaCl, and 20 mM imidazole, then disrupted with a French press (GuangZhou JuNeng Biology and Technology Co. Ltd, Guangzhou, China). The supernatant was collected by centrifugation and purified by FPLC using Ni-NTA and DEAE columns (GE Healthcare, Uppsala, Sweden). The purified proteins were concentrated using a Centriprep (Millipore, Darmstadt, Germany) and purity was checked by SDS-PAGE. All purification procedures were performed at 4 °C.
Crystallization and data collection
Initial crystallization screening was performed manually using the sitting-drop diffusion method with 768 reservoir conditions (Hampton Research kits, Aliso Viejo, California, USA). The initial crystals were further optimized by adjusting reservoir compositions. Final crystallization conditions were as follows: (1) TcVSP1: 0.1 M DL-malic acid, pH 7.0, 10 % (w/v) PEG3350; (2) TbVSP1: 0.1 M potassium citrate tribasic, 16 % (w/v) PEG3350; (3) ΔN1-TcVSP1: 1 M magnesium sulfate, 0.1 M trisodium citrate dehydrate, pH 5.6. Prior to data collection, the crystals were mounted in a cryo-loop and flash-cooled in liquid nitrogen with cryo-protectants: (1) TcVSP1: 0.15 M DL-malic acid, pH 7.0, 15 % (w/v) PEG3350, 20 % (w/v) glycerol; (2) TbVSP1: 0.15 M potassium citrate tribasic, 20 % (w/v) PEG3350, 15 % (w/v) glycerol; (3) ΔN1-TcVSP1: 1.5 M magnesium sulfate, 0.15 M trisodium citrate pH 5.6, 15 % (w/v) glycerol. For the TcVSP1-BPH-1260 complex, TcVSP1 crystals were soaked with cryo-protectant containing 10 mM inhibitor for 3 hours at room temperature. X-ray diffraction data sets were collected at the National Synchrotron Radiation Research Center (NSRRC), Hsinchu, Taiwan. Diffraction images were processed by using the HKL-2000 program (27). Data collection statistics are shown in Table 1.
Structure determination and refinement
The multiple-wavelength anomalous diffraction (MAD) data sets of the selenium-containing TcVSP1 derivatives were collected at various wavelengths (Table S2). The initial TcVSP1 model was obtained by using SOLVE and RESOLVE(28) programs and further refined by using Coot(29) and CNS(30). The TbVSP1, TbVSP1-BPH-1260, and ΔN1-TcVSP1-PPi complex structures were solved by using molecular replacement (MR) using TcVSP1 as a search model, and refined as described above. Data collection and refinement statistics are summarized in Table 1. All protein structure figures were prepared by using the PyMOL program (http://pymol.sourceforge.net/).
Size exclusion chromatography-multi-angle light scattering (SEC-MALS)
Protein molecular weights were determined by static light scattering (SLS) using a Wyatt Dawn Heleos II multi-angle light scattering detector (Wyatt Technology) coupled to an AKTA Purifier UPC10 FPLC protein purification system and a WTC-030S5 size-exclusion column (Wyatt Technologies). 100 µL TcVSP1 (2.5 mg/mL), ΔN1-TcVSP1 (2.4 mg/mL), ΔN2-TcVSP1 (1.9 mg/mL), and TbVSP1 (4.7 mg/mL) were subjected to SEC-MALS analysis individually with a running buffer containing 25 mM Tris (pH 7.5), 150 mM NaCl and 0.02% NaN3 at a flow rate of 0.5 mL/min. 0.1 mM CaCl2 (final concentration) was added into the same running buffer to investigate the effects of Ca2+. BSA (2 mg/mL) was used for system calibration. The absolute molecular weights of the individual peaks observed in the size-exclusion chromatograms were determined by SLS in conjunction with their corresponding refractive indices using an online refractometer connected downstream from the SLS detector (Wyatt Optilab rEX). A standard value of the refractive index, dn/dc = 0.185 mL/g, was used for all proteins. The buffer viscosity, η= 1.0226 cP at 25 °C was calculated by using SEDNTERP(31). The reference refractive index, 1.3459 RIU, was obtained from the running buffer that passed through the reference cell.
Pyrophosphatase and polyphosphatase activities
VSP activities were assayed by measuring phosphate (Pi) release using the malachite green assay(32) with some modifications(7). For the pyrophosphatase (PPase) activity, reaction mixtures included 50 mM Tris-HCl (pH 8.5), 3 mM MgCl2, 100 µM PPi, 0.5 µg of purified VSP proteins (full-length or truncated) with or without a variable concentration of bisphosphonate of interest. For the exopolyphosphatase activity, reactions contained 50 mM MES (pH 6.5), 3 mM ZnCl2, 100 µM polyP100 (Kerafast), 0.5 µg of the VSP proteins with or without the bisphosphonate inhibitor. The reactions were performed in 96-well plates and started by the addition of the VSP enzyme. After incubation for 10 min at 30°C, the reactions were stopped by the addition of an equal volume of freshly prepared mixture of three parts of 0.045% malachite green and one part of 4.2% ammonium molybdate, which was filtered prior to use as described before(33). The absorbance (A) at 660 nm was read using a SpectraMax M2e plate reader (Molecular Devices, Sunnyvale, CA). For each substrate (PPi or polyP100), a blank value that corresponds to the same reaction conditions but without the enzyme was subtracted from the value obtained in the presence of the enzyme. After subtraction, the amount of Pi released was determined by comparison with a standard curve. The specific activity of VSP was defined as µmol Pi released min−1 per mg of protein (µmol min−1 × mg). The IC50 values for VSP inhibition were quantitated as described previously(9).
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China (31300615, 31470240, 31400678, and 31200053); by the United States Public Health Service (NIH grants CA158191, AI077538, and AI107663); by a Harriett A. Harlin Professorship, and by the University of Illinois Foundation/Oldfield Research Fund. Synchrotron data was collected at beamline BL-13C1 of the National Synchrotron Radiation Research Center, Taiwan supported by the National Science Council (NSC).
Footnotes
The authors declare no competing financial interest.
Additional details of ligand electron densities Figure S1); AUC sedimentation velocity analyses (Figure S2); EF-hand domain sequence and structure comparisons with calmodulin (Figure S3); Mg2+ coordination in PPases (Figure S4); complete list of all Trypanosomatids having the two-domain structure (Table S1); summary of Se-Met data collection statistics (Table S2); activities of VSP proteins and their inhibition by bisphosphonates (Table S3); primers used for protein production (Table S4). This material is available free of charge via the Internet at http://pubs.acs.org.
Accession codes
All structure coordinates (5CUV, 5CUY, 5CUU and 5CUX) are available in the PDB.
References
- 1.Bilbe G. Overcoming neglect of kinetoplastid diseases. Science. 2015;348:974–976. doi: 10.1126/science.aaa3683. [DOI] [PubMed] [Google Scholar]
- 2.Ekins S, Bunin BA. Computational Approaches and Collaborative Drug Discovery for Trypanosomal Diseases. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA; 2013. [Google Scholar]
- 3.Urbina JA, Docampo R. Specific chemotherapy of Chagas disease: controversies and advances. Trends in Parasitology. 2003;19:495–501. doi: 10.1016/j.pt.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Docampo R, de Souza W, Miranda K, Rohloff P, Moreno SN. Acidocalcisomes - conserved from bacteria to man. Nat. Rev. Microbiol. 2005;3:251–261. doi: 10.1038/nrmicro1097. [DOI] [PubMed] [Google Scholar]
- 5.Moreno B, Urbina JA, Oldfield E, Bailey BN, Rodrigues CO, Docampo R. 31P NMR spectroscopy of Trypanosoma brucei, Trypanosoma cruzi, and Leishmania major. Evidence for high levels of condensed inorganic phosphates. J. Biol. Chem. 2000;275:28356–28362. doi: 10.1074/jbc.M003893200. [DOI] [PubMed] [Google Scholar]
- 6.Urbina JA, Moreno B, Vierkotter S, Oldfield E, Payares G, Sanoja C, Bailey BN, Yan W, Scott DA, Moreno SN, Docampo R. Trypanosoma cruzi contains major pyrophosphate stores, and its growth in vitro and in vivo is blocked by pyrophosphate analogs. J. Biol. Chem. 1999;274:33609–33615. doi: 10.1074/jbc.274.47.33609. [DOI] [PubMed] [Google Scholar]
- 7.Galizzi M, Bustamante JM, Fang J, Miranda K, Soares Medeiros LC, Tarleton RL, Docampo R. Evidence for the role of vacuolar soluble pyrophosphatase and inorganic polyphosphate in Trypanosoma cruzi persistence. Mol. Microbiol. 2013;90:699–715. doi: 10.1111/mmi.12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lemercier G, Espiau B, Ruiz FA, Vieira M, Luo S, Baltz T, Docampo R, Bakalara N. A pyrophosphatase regulating polyphosphate metabolism in acidocalcisomes is essential for Trypanosoma brucei virulence in mice. J. Biol. Chem. 2004;279:3420–3425. doi: 10.1074/jbc.M309974200. [DOI] [PubMed] [Google Scholar]
- 9.Kotsikorou E, Song Y, Chan JM, Faelens S, Tovian Z, Broderick E, Bakalara N, Docampo R, Oldfield E. Bisphosphonate inhibition of the exopolyphosphatase activity of the Trypanosoma brucei soluble vacuolar pyrophosphatase. J. Med. Chem. 2005;48:6128–6139. doi: 10.1021/jm058220g. [DOI] [PubMed] [Google Scholar]
- 10.Date SV. The Rosetta stone method, Methods Mol. Biol. 2008;453:169–180. doi: 10.1007/978-1-60327-429-6_7. [DOI] [PubMed] [Google Scholar]
- 11.Harutyunyan EH, Oganessyan VY, Oganessyan NN, Avaeva SM, Nazarova TI, Vorobyeva NN, Kurilova SA, Huber R, Mather T. Crystal structure of holo inorganic pyrophosphatase from Escherichia coli at 1.9 A resolution. Mechanism of Hydrolysis. Biochemistry. 1997;36:7754–7760. doi: 10.1021/bi962637u. [DOI] [PubMed] [Google Scholar]
- 12.Heikinheimo P, Lehtonen J, Baykov A, Lahti R, Cooperman BS. The structural basis for pyrophosphatase catalysis. Structure. 1996;4:1491–1508. doi: 10.1016/s0969-2126(96)00155-4. [DOI] [PubMed] [Google Scholar]
- 13.Fabrichniy IP, Lehtio L, Salminen A, Zyryanov AB, Baykov AA, Lahti R, Goldman A. Structural studies of metal ions in family II pyrophosphatases: the requirement for a Janus ion. Biochemistry. 2004;43:14403–14411. doi: 10.1021/bi0484973. [DOI] [PubMed] [Google Scholar]
- 14.Lee HS, Cho Y, Kim YJ, Lho TO, Cha SS, Lee JH, Kang SG. A novel inorganic pyrophosphatase in Thermococcus onnurineus NA1. FEMS Microbiol. Lett. 2009;300:68–74. doi: 10.1111/j.1574-6968.2009.01766.x. [DOI] [PubMed] [Google Scholar]
- 15.Espiau B, Lemercier G, Ambit A, Bringaud F, Merlin G, Baltz T, Bakalara N. A soluble pyrophosphatase, a key enzyme for polyphosphate metabolism in Leishmania. J. Biol. Chem. 2006;281:1516–1523. doi: 10.1074/jbc.M506947200. [DOI] [PubMed] [Google Scholar]
- 16.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerlt JA, Allen KN, Almo SC, Armstrong RN, Babbitt PC, Cronan JE, Dunaway-Mariano D, Imker HJ, Jacobson MP, Minor W, Poulter CD, Raushel FM, Sali A, Shoichet BK, Sweedler JV. Enzyme Function Initiative. Biochemistry. 2011;50:9950–9962. doi: 10.1021/bi201312u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 19.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Cryst. Sect. D: Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 21.Gifford JL, Walsh MP, Vogel HJ. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 2007;405:199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- 22.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gagne SM, Li MX, Sykes BD. Mechanism of direct coupling between binding and induced structural change in regulatory calcium binding proteins. Biochemistry. 1997;36:4386–4392. doi: 10.1021/bi963076+. [DOI] [PubMed] [Google Scholar]
- 24.Senguen FT, Grabarek Z. X-ray structures of magnesium and manganese complexes with the N-terminal domain of calmodulin: insights into the mechanism and specificity of metal ion binding to an EF-hand. Biochemistry. 2012;51:6182–6194. doi: 10.1021/bi300698h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harutyunyan EH, Kuranova IP, Vainshtein BK, Hohne WE, Lamzin VS, Dauter Z, Teplyakov AV, Wilson KS. X-ray structure of yeast inorganic pyrophosphatase complexed with manganese and phosphate. Eur. J. Biochem. 1996;239:220–228. doi: 10.1111/j.1432-1033.1996.0220u.x. [DOI] [PubMed] [Google Scholar]
- 26.Xia Y, Liu Y-L, Xie Y, Zhu W, Guerra F, Shen S, Yeddula N, Fischer W, Low W, Zhou X, Zhang Y, Oldfield E, Verma I. A combination therapy for KRAS-driven lung adenocarcinomas using lipophilic bisphosphonates and rapamycin. Sci. Transl. Med. 2014;6 doi: 10.1126/scitranslmed.3010382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. Sect. D: Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 30.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Cryst. Sect. D: Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 31.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Cambridge: Royal Society of Chemistry; 1992. [Google Scholar]
- 32.Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA. An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 33.Shatton JB, Ward C, Williams A, Weinhouse S. A microcolorimetric assay of inorganic pyrophosphatase. Anal. Biochem. 1983;130:114–119. doi: 10.1016/0003-2697(83)90657-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.