Abstract
Purpose
Vitamin D is well known for its effects on bone mineralisation but has also been attributed immunomodulatory properties. It positively influences human health, but in vivo data describing vitamin D effects on the human gut microbiome are missing. We aimed to investigate the effects of oral vitamin D3 supplementation on the human mucosa-associated and stool microbiome as well as CD8+ T cells in healthy volunteers.
Methods
This was an interventional, open-label, pilot study. Sixteen healthy volunteers (7 females, 9 males) were endoscopically examined to access a total of 7 sites. We sampled stomach, small bowel, colon, and stools before and after 8 weeks of vitamin D3 supplementation. Bacterial composition was assessed by pyrosequencing the 16S rRNA gene (V1–2), and CD8+ T cell counts were determined by flow cytometry.
Results
Vitamin D3 supplementation changed the gut microbiome in the upper GI tract (gastric corpus, antrum, and duodenum). We found a decreased relative abundance of Gammaproteobacteria including Pseudomonas spp. and Escherichia/Shigella spp. and increased bacterial richness. No major changes occurred in the terminal ileum, appendiceal orifice, ascending colon, and sigmoid colon or in stools, but the CD8+ T cell fraction was significantly increased in the terminal ileum.
Conclusion
Vitamin D3 modulates the gut microbiome of the upper GI tract which might explain its positive influence on gastrointestinal diseases, such as inflammatory bowel disease or bacterial infections. The local effects of vitamin D demonstrate pronounced regional differences in the response of the GI microbiome to external factors, which should be considered in future studies investigating the human microbiome.
Electronic supplementary material
The online version of this article (doi:10.1007/s00394-015-0966-2) contains supplementary material, which is available to authorized users.
Keywords: Vitamin D, Human gut microbiome, IBD, Inflammation, Gammaproteobacteria
Introduction
The human gut is home to about 1014 microorganisms, which are collectively referred to as the gut microbiome. These microorganisms work as biochemical factories, helping the host in nutrient acquisition, vitamin production, and toxin degradation. Bacterial enzymes are also involved in an array of metabolic pathways [1] and can even affect the central nervous system by converting amino acids, carbohydrates, and other ingested substances into bioactive compounds [2]. When it comes to energy acquisition, especially carbohydrate metabolism, the human genome is quite limited and a healthy gut microbiome is necessary to effectively digest the diverse carbohydrates [3].
The human gut microbiome is known to have a major impact on colonisation resistance against intestinal pathogens [4], on modulation of the intestinal immune system [5], and on host metabolism [6, 7]. At the current stage of knowledge, some factors, which influence the composition of the intestinal microbiome, have been identified, including diet [8–10], host genetics [11], environmental circumstances [12], and drugs, such as antibiotics or laxatives [13, 14]. Impairment of gut homoeostasis has been linked to many gastrointestinal diseases, such as inflammatory bowel diseases (IBD), intestinal infections, irritable bowel syndrome (IBS), and colon cancer [15–18] but also to extra-intestinal diseases, such as obesity, diabetes mellitus, autoimmune diseases, and neurological disorders [19]. For general health benefits but also for a number of diseases, supplementation with vitamin D has shown to be beneficial. Vitamin D is well known for its role on bone mineralisation but has also major effects on the immune and the cardiovascular system as well as on host defence against pathogenic microorganisms [15, 20–22]. In many parts of the world, about half of the population is assumed to have insufficient vitamin D levels (lower than 20 ng/mL serum) due to a lack of UV exposure and a diet-lacking vitamin D [23]. Sufficient levels of vitamin D have been associated with a lower risk of autoimmune diseases such as IBD, type 1 diabetes mellitus, and rheumatic diseases, but also neoplasia and infections such as tuberculosis and hepatitis C [21]. Elevating serum vitamin D levels to 42 ng/mL has been estimated to decrease disease rates in various cancers, cardiovascular diseases, diabetes mellitus, and infections by 10–50 % and the overall mortality rate by 18 % per year [24].
The hormonal activity of vitamin D has been attributed to the expression of the vitamin D receptor (VDR), which has its highest expression in CD8+ T cells compared with other immune cells [25], and to the vitamin D activating enzyme CYP27B1. VDR and CYP27B1 are found in several different cell types including kidneys, muscle, or prostate, but also in cells of the immune system [25, 26], supporting a prominent role of vitamin D in gut homoeostasis and immunity [25]. Given the beneficial effects of vitamin D, many people are using vitamin D supplementation as part of their regular diet, but effects of vitamin D on the human intestinal microbiome have not yet been studied. We hypothesised that some of the beneficial effects attributed to vitamin D might be mediated by the gastrointestinal microbiome. We recruited healthy human volunteers for an interventional study to assess whether vitamin D3 (vitD3) affects the human microbiome. Most microbiome studies are limited to easily accessible stool samples, but studies have indicated that there can be a pronounced difference between mucosal and stool microbiomes [13, 27]. Therefore, we sampled seven gastrointestinal regions in addition to stools to assess the mucosa-associated microbiome before and after 8 weeks of oral vitD3 supplementation.
Materials and methods
Study design, population, and medication
This study was designed as an interventional monocentric open pilot study. The inclusion criteria were: healthy volunteers, age between 18 and 40 years, body mass index (BMI) between 20 and 30 kg/m2, and non-smoker. The Ethics Committee at the Medical University of Graz, Austria, approved the study. The study was conducted from January 2012 to August 2012 at the Medical University of Graz according to the principles of Good Clinical Practice (ClinicalTrials.gov, NCT01538485) and ethical standards of the Declaration of Helsinki and all later amendments. All volunteers gave their written informed consent after being informed about the aim of the study, study procedure, and possible risks of participating in this study. Sixteen healthy volunteers, 9 men and 7 women, aged 25 ± 4 (mean ± SD) years participated in the study. All volunteers had a normal BMI (23 ± 3 kg/m2) with a mean weight of 69 ± 11 kg and height of 172 ± 8 cm.
After baseline assessment, each volunteer received a weekly dose of 980 IU/kg bodyweight of vitamin D3 (Oleovit D3, Fresenius Kabi, Graz, Austria) for 4 weeks representing a daily dose of 140 IU/kg bodyweight, but maximal 68,600 IU per week in total. For the remaining 4 weeks, each volunteer received a weekly vitD3 dose of 490 IU/kg bodyweight (daily dose of 70 IU/kg bodyweight), but maximal 34,300 IU per week in total.
Sample collection and storage
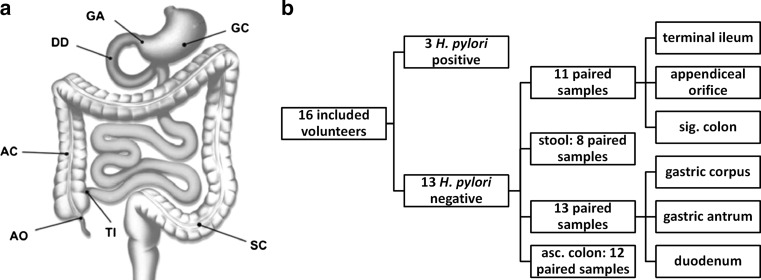
Before vitD3 supplementation was started, all volunteers underwent a gastroduodenoscopy followed by a colonoscopy on the next day. Stools were collected before bowel preparation for gastroduodenoscopy and immediately stored at −80 °C until DNA isolation. After 8 weeks of vitD3 supplementation, both endoscopic procedures and stool sampling were repeated. As a laxative, all volunteers received 2 litres of MOVIPREP® (Norgine, Marburg, Germany), a polyethylene glycol-based electrolyte solution for bowel preparation. During gastroduodenoscopy, biopsies were taken from the gastric corpus (GC), the gastric antrum (GA), and the descending part of the duodenum (DD) (Fig. 1a). During colonoscopy, biopsies were taken from the terminal ileum (TI), the appendiceal orifice region (AO), the ascending colon (AC) at 10 cm distal to the ileocecal valve, and the sigmoid colon (SC) at 30 cm proximal to the anal canal (Fig. 1a). Two biopsies were taken from each region, immediately frozen and stored at −80 °C until DNA isolation, and two biopsies from each region were immediately placed on ice in complete RPMI media containing 10 % foetal calf serum (cRPMI-10), glutamine, and penicillin/streptomycin (Life Technologies, Darmstadt, Germany) for flow cytometry. Flow cytometry analysis was started within 1 h after the biopsy.
Fig. 1.
Sampling scheme. a Biopsied body regions (drawing generously provided by Tauschmann et al. [46]). b Paired samples passed quality filtering. A subgroup of 3 volunteers was analysed separately due to a H. pylori infection in their stomach. GC gastric corpus, GA gastric antrum, DD duodenum, TI terminal ileum, AO appendiceal orifice, AC ascending colon, SC sigmoid colon
Isolation of lamina propria mononuclear cells and flow cytometry
Gastrointestinal biopsies were immediately transferred to a dithiothreitol/EDTA solution (Sigma, Hamburg, Germany, and Life Technologies, Darmstadt, Germany) and incubated for 15 min at 37 °C. The samples were finely sliced and digested in a collagenase A solution (Roche, Mannheim, Germany) for 1 h at 37 °C. The resulting cell suspension was passed through a 70-µm cell strainer (BD Biosciences, San Jose, CA, USA) and collected in a cRPMI-10 filled tube (Life Technologies, Darmstadt, Germany). After centrifugation, the pellet was resuspended in PBS (Life Technologies, Darmstadt, Germany) and cell viability was confirmed by staining an aliquot with 0.4 % trypan blue (Sigma, Hamburg, Germany) for microscopy.
For the quantification of CD3+CD4−CD8+ T cells (CD8+ T cells) in lamina propria by flow cytometry, the following monoclonal antibodies were used: anti-CD3 PerCP-Cy5.5 (eBiosciences, USA) and anti-CD4 PE and anti-CD8 V500 (BD Biosciences, San Diego, CA, USA). Positive signals were defined by the use of isotype controls and the fluorochrome minus one (FMO) method.
Vitamin D and safety parameter measurements
The vitamin D levels (calcidiol, 25(OH)D) were measured by using a commercially available enzyme-linked immunosorbent assay (ELISA, IDS, Bolden, UK) with an intra- and inter-assay coefficient of variation of 5.6 and 6.4 %, respectively.
Safety parameters including serum calcium levels, urine calcium, urine calcium/creatinine ratio, complete blood cell count, serum phosphorus, serum albumin, and PTH were assessed at baseline and every 4 weeks thereafter by standard laboratory methods. C-reactive protein was measured by using a Tina quant CRP immunoturbidimetric assay (Roche COBAS INTEGRA, Mannheim, Germany).
DNA isolation, 16S rRNA gene PCR amplification, and FLX sequencing
Isolation of total genomic DNA, PCR, and FLX sequencing were performed as recently described in Kump et al. [28]. Briefly, a MagnaLyser Instrument (Roche Diagnostics, Mannheim, Germany) and a MagNA Pure LC 2.0 Instrument (Roche Diagnostics) with the MagNA Pure LC DNA Isolation Kit III (bacteria, fungi) were used for automated DNA isolation according to the manufacturer’s instructions. For library preparation, 454 one-way read strategy (Lib-L kit, Primer A, Primer B; Roche 454 Life Science, Branford, CT, USA) amplifying the 16S rRNA hyper variable regions V1–2 with the target specific primers F27-AGAGTTTGATCCTGGCTCAG and R357-CTGCTGCCTYCCGTA was used. FLX fusion primers with FLX adaptor, key, and MID sequences were used for amplification (supplementary table S1). For each sample, a PCR was carried out with 50 ng total genomic DNA in a 25-µL reaction volume, performed in triplicate. Amplicons were purified according to Kump et al. [28] and pooled and sequenced using the GS FLX Titanium Sequencing Kit XLR70 (Roche 454 Life Science, Branford, CT, USA) according to manufacturer’s instructions.
Sequencing data analysis
Raw sequences were preprocessed and filtered using MOTHUR version 1.31.2 according to the standard Schloss 454 SOP (see http://www.mothur.org/wiki/454_SOP) [29]. In more detail, reads were denoised using PyroNoise [30], chimera filtered with UCHIME [31], pyrosequencing errors were reduced with pre.cluster [32], and non-bacterial sequences were also excluded. High-quality reads were aligned to the SILVA database [33, 34], and taxonomy was assigned by MOTHUR’s implementation of the ribosomal database project (RDP) classifier [35] followed by binning into phylotypes based on taxonomy using a minimum probability cut-off of 80 %. As we were using short reads, we only classified the reads down to the genus level. The shared file, MOTHUR’s OTU-table format was then converted into biom format and passed on to QIIME’s v.1.7 core_diversity.py command using non-phylogenetic parameters [36]. The GC was rarefied to 1178 sequences/sample, GA to 2125, DD to 2441, TI to 1647, AO to 2215, AC to 2014, SC to 2188, and stools to 1788 sequences/sample.
Statistical analysis
To determine whether phylotypes (e.g. phyla, classes, order) differed significantly between the two time points, we used a paired t test since the study compared samples from the same patients. Subsequently, P values were corrected for multivariate testing using the Benjamini–Hochberg false discovery rate (FDR) procedure, as Bonferroni correction is too conservative for sequencing data. Corrected P values below 0.05 were considered statistically significant (*P < .05; **P < .01; ***P < .001). To determine whether the intervention had different effects in the biopsied regions, we calculated a Bray–Curtis distance matrix and used an analysis of similarity (ANOSIM) to test for significantly different clustering. ANOSIM was also used to test whether mucosal samples and stools differ significantly. Phylotype richness was compared before and after supplementation by using alpha rarefaction plots based on an observed species matrix and a nonparametric t test. The difference between CD8+ T cells were tested with a paired t test. All values are given as mean ± SD if not otherwise stated.
Results
Fifteen volunteers completed the whole study. One volunteer could not attend the second colonoscopy due to personal reasons but completed all other study procedures. Vitamin D levels (25(OH)D, calcidiol) increased significantly from 22.3 ± 13.1 ng/mL (before) to 55.2 ± 13.3 ng/mL after vitD3 supplementation (P < .0001). In all volunteers, all safety parameters, including serum and urine calcium and phosphate, as well as urine calcium/creatinine ratio, remained within a normal range and were not significantly altered (Table 1 and supplementary table S2). Throughout the study, we generated 1003,488 high-quality 16S amplicon reads in the range of 211–380 bp (average 264 bp) from mucosal and stool samples. Five microbiome samples were excluded from sequence data analysis as they were poorly covered (<900 sequences/sample). In a subgroup of 3 volunteers, a gastric Helicobacter pylori infection was found during routine clinical processing of biopsies and in the sequencing data. This subgroup was analysed separately as up to 90 % of the observed phylotypes in the GC were classified as Helicobacter spp. In total, we analysed 242 well-covered samples, resulting in 13 paired samples (before/after) for the upper GI tract (GC, GA, and DD), 12 paired samples for AC, 11 for TI, AO, and SC, and 8 paired stool samples (Fig. 1b).
Table 1.
Safety parameters
| Safety parameters | Before | After 8 weeks of vitD3 supplementation | P value |
|---|---|---|---|
| Calcidiol (ng/mL) ± SD | 22.3 ± 13.1 | 55.2 ± 13.3 | <0.0001 |
| Calcium (mmol/L) ± SD | 2.44 ± 0.09 | 2.43 ± 0.08 | 0.75 |
| Ca/creatinine ratio ± SD | 0.26 ± 0.18 | 0.20 ± 0.12 | 0.24 |
| PTH (pg/mL) ± SD | 39.5 ± 12.7 | 33.5 ± 14.7 | 0.14 |
We successfully increased vitamin D levels without significantly altering any of the measured safety parameters. Values are given as mean ± SD (n = 16). A paired t test was used to calculate P values
Phylogenetic landscape of the human GI tract
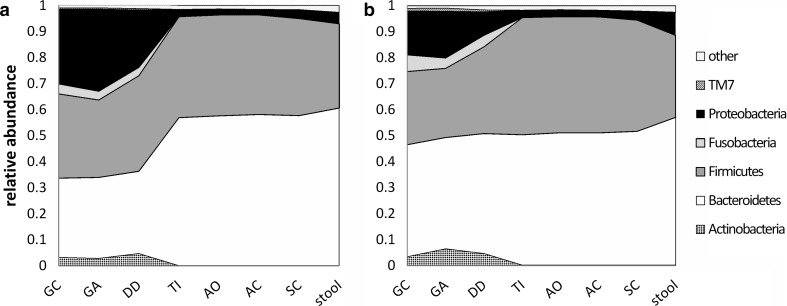
Bacteroidetes and Firmicutes were the dominating phyla which we found in the human gut microbiome (Table 2). We found significantly more Proteobacteria, Actinobacteria, TM7, and Fusobacteria in the upper (GC, GA, and DD) compared with the lower GI tract (TI, AO, AC, SC) and stools (P < .0001) (supplementary table S3–4). This probably reflects the oral microbial community which is considered the first input to the GI tract. The microbial communities of the TI and the AO region were more similar to the colon rather than the DD. The stool microbiome was more similar to the colon rather than the upper GI tract (Fig. 2a). A list of all found genera in the different regions of the GI tract is provided in supplementary table S5.
Table 2.
Dominating phyla
| Bacteroidetes | Firmicutes | |
|---|---|---|
| GC (%) | 36.30 ± 7.56 | 36.93 ± 9.34 |
| GA (%) | 37.46 ± 13.44 | 35.94 ± 12.92 |
| DD (%) | 32.06 ± 9.75 | 38.43 ± 14.48 |
| TI (%) | 58.44 ± 8.47 | 36.52 ± 8.40 |
| AO (%) | 57.99 ± 7.23 | 38.29 ± 8.07 |
| AC (%) | 57.86 ± 7.69 | 38.20 ± 7.57 |
| SC (%) | 58.16 ± 7.92 | 36.50 ± 8.26 |
| Stool (%) | 62.87 ± 9.66 | 31.43 ± 9.90 |
Values are given as mean ± SD
Fig. 2.
Mean relative abundance of phylogenetic landscape a before and b after vitD3 supplementation in 13 volunteers on the phylum level from the GC to the SC and from stool samples. We see a decline in Proteobacteria and an increase in Bacteroidetes in the upper GI tract (GC, GA, and DD). Lower regions (TI, AO, AC, and SC) and stools did not change on the phylum level. GC gastric corpus, GA gastric antrum, DD duodenum, TI terminal ileum, AO appendiceal orifice, AC ascending colon, SC sigmoid colon
Stools are often the only samples assessed in microbiome studies. In our study, the mucosal microbiome of the upper GI tract (GC, GA, DD) was significantly different from the stool microbiome (ANOSIM (genus level) P < .001). A list of the significantly different genera of the upper GI tract and stools is provided in supplementary table S6. The lower GI microbiome (TI, AO, AC, SC) was not significantly different from stools (ANOSIM (genus level) P = 0.105).
To assess the phyla that were affected by vitD3 supplementation, we compared the relative phyla abundance before and after 8 weeks of vitD3 supplementation (Fig. 2a, b, supplementary table S3). Proteobacteria decreased significantly in the GC (P = .0348) and the GA (P = .0258), while Bacteroidetes increased significantly in the GC (P = .0013) and the DD (P = .0013). Other phyla like Actinobacteria, Firmicutes, and Fusobacteria were not significantly affected by vitD3 supplementation in any region of the GI tract.
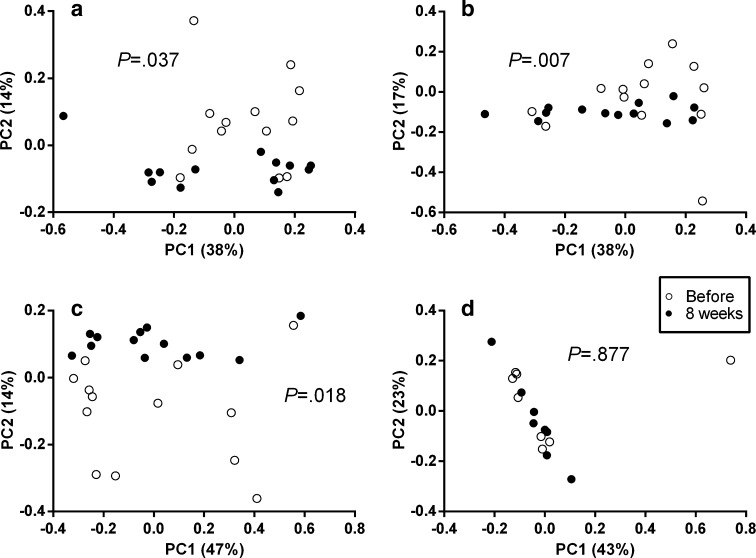
Beta-diversity measures represented by PCoA plots show how similar multiple samples are compared to each other. VitD3 supplementation changed the community structure in the upper GI tract (GC, GA, and DD samples) indicated by the significant separation of samples before and after vitD3 supplementation (Fig. 3a–c). No difference in community structure was observed in stools (Fig. 3d) or mucosal samples from the lower GI tract (TI, AO, AC, SC; see supplementary Fig. S7).
Fig. 3.
VitD3 supplementation changes bacterial community composition and community structure significantly in the upper GI tract in contrast to the lower GI tract. PCoA plots generated on a Bray–Curtis distance matrix on the genus level. a GC (n = 13), b GA (n = 13), c DD (n = 13), d stool (n = 8). a–c Distinct separation before and after vitD3 supplementation, whereas in stools, all samples cluster together (d). ANOSIM was used to test whether community composition is different between the groups. Samples taken before vitD3 supplementation are shown in white, and samples taken after supplementation are shown in black. GC gastric corpus, GA gastric antrum, DD duodenum, TI terminal ileum, AO appendiceal orifice, AC ascending colon, SC sigmoid colon
Reduction in Gammaproteobacteria
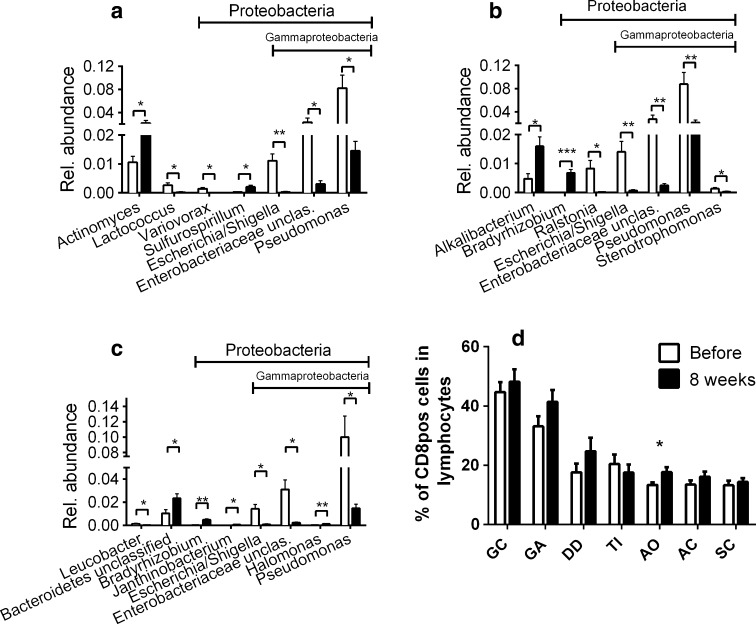
To assess whether all Proteobacteria were decreasing or whether just a particular phylotype (i.e. class, order, family or genera) was affected, we monitored the changes on all taxonomic levels in the upper GI tract (GC, GA, DD). Only the class of Gammaproteobacteria (GC, P = .045; GA, P = .034; DD P = .066), including Pseudomonas spp. (GC, P = .0280; GA, P = .0061; DD, P = .0316) and Escherichia/Shigella spp. (GC, P = .0022; GA, P = .0078; DD, P = .0190), showed a significant decline, while other Proteobacteria, like the Bradyrhizobium spp. (GA, P = .0003; DD, P = .0016) and Sulfurospirillum spp. (GC, P = .033), increased (Fig. 4a–c). Alphaproteobacteria increased significantly in relative abundance after 8 weeks of vitD3 supplementation in the GA (Fig. 4b, P = .0295). Besides the reduction in Gammaproteobacteria, Lactococcus spp. (P = .0434) and Variovorax spp. (P = .0164) decreased significantly. Actinomyces spp. (P = .0175) increased in the GC, while in the GA Alkalibacterium spp. (P = .0394) increased and Ralstonia spp. (P = .0342) decreased in relative abundance. The relative abundance of Leucobacter spp. (P = .0289) decreased and Janthinobacterium spp. (P = .0318) increased in the DD. Compared to the upper GI tract, observed changes in the lower GI tract were minor. In the TI, Roseburia spp. were significantly increased (P = .0421) and Peptostreptococcus spp. were decreased (P = .0423, supplementary Fig. S8), both representing the phylum Firmicutes. We also found a decline in an unclassified Clostridia genus in the AO region (P = .0438). Colonic mucosal samples showed no significant changes in any phylotype, whereas in stools, the class of Betaproteobacteria (P = .0234) decreased and Actinomyces spp. (P = .0073) increased in relative abundance (supplementary Fig. S8).
Fig. 4.
Significantly changed phylotypes (genera) and changes in mucosal CD8+ T cell counts after vitD3 supplementation. a Gastric corpus, b gastric antrum, and c duodenum. d Percentage of CD8+ T cells of all lymphocytes isolated from mucosal biopsies (GC, P = .5168; GA, P = .1317; DD, P = .2058; TI, P = .5012; AO, P = .0315; AC, P = .2476; SC, P = .5773). Samples taken before vitD3 supplementation are shown in white, and samples taken after supplementation are shown in black. GC gastric corpus, GA gastric antrum, DD duodenum, TI terminal ileum, AO appendiceal orifice, AC ascending colon, SC sigmoid colon. P < .05 = *; P < .01 = **; P < .001 = ***; n = 13, bars = mean ± SEM. A paired t test was used to calculate P values, which were FDR corrected for multiple testing
Taken together, our data indicate that vitD3 supplementation significantly alters Gammaproteobacteria in the upper GI tract, whereas other GI regions seem to be less affected. CD8+ T cells may contribute to the decline in Gammaproteobacteria. Flow cytometry analysis of CD8+ T cell counts showed a significant increase in the AO (P = .0213) and a trend towards increased CD8+ T cells in the other regions after 8 weeks of vitD3 supplementation (Fig. 4d).
VitD3 increases richness in the gastric antrum and duodenum
Rarefaction analysis indicated sufficient capturing of the microbial diversity in all samples (Fig. 5 and supplementary Fig. S9). After vitD3 supplementation, we found a significantly increased phylotype richness in the GA (P = .004) and a trend towards an increased phylotype richness in the DD (P = .06) (Fig. 5). No change in phylotype richness was seen in GC, TI, AO, AC, SC or in stools (Fig. 5 and supplementary Fig. S9).
Fig. 5.
VitD3 supplementation increases phylotype richness in the gastric antrum and duodenum; alpha rarefaction curves of a the gastric corpus, b the gastric antrum, c the duodenum, and d stool. The number of observed species is plotted against the number of sequences per sample. We see an increase in richness in the gastric antrum (P = .004) and a trend in the duodenum (P = .06) but not in gastric corpus (P = .933) or in stool (P = .313). Rarefaction curves also indicate sufficient coverage
Helicobacter pylori reduction after 8 weeks of vitD3 supplementation
Microbiome sequence data and routine biopsy-processing indicated H. pylori infection in three volunteers. All three volunteers showed an overall decline in Helicobacter spp. after 8 weeks of vitD3 supplementation. We observed a trend towards a decline in Helicobacter spp. in the GA of all 3 volunteers, but changes in the GC and DD were inconsistent (supplementary Fig. S10). When we included the H. pylori infected subgroup into the overall microbiome data analysis, we found an additional increase in phylotype richness in the GC after 8 weeks of vitD3 supplementation (data not shown).
Discussion
The results of our study demonstrate the effect of an 8-week high-dose vitD3 supplementation on the human microbiome of the upper GI tract. These changes included a reduction in opportunistic pathogens and an increase in bacterial richness.
To the best of our knowledge, the effects of vitD3 on the human microbiome have not yet been studied. So far, the effects of a vitamin D supplementation on the gut microbiome have only been studied in murine colitis model. Vitamin D3 protected these mice from dextran sodium sulphate-induced colitis, which is a common model of IBD. The severity of colitis was significantly higher in VDR and CYP27B1 knockout mice [37]. Because the composition of the gut microbiome of mice is substantially different from the human microbiome [38], any interventional effects that are found in mouse studies cannot be translated directly to humans.
By taking a series of biopsies from the major regions of the human GI tract, we found an overall increase in phylotype richness together with a decrease in Proteobacteria mostly in the mucosa of the upper GI tract. In particular, the class of Gammaproteobacteria decreased significantly, including genera of typical opportunistic pathogens like Pseudomonas spp. and Escherichia/Shigella spp. Stools and mucosal regions of the lower GI tract showed only minor changes and support recent findings that mucosa-associated microbiomes differ significantly from stools [13] and that only biopsies offer the chance to analyse changes in the mucosa-associated microbiome in sufficient detail. Most human intestinal microbiome studies are based on data obtained from stools [8, 9, 16, 17, 39] or rectal/colonic mucosal samples [13, 18, 40]. These studies have investigated external influences, such as the effects of diet [8, 9, 41, 42], physical exercise [39], or drugs [14], and have identified associations with certain disease like IBD, IBS, or obesity [8, 18, 40]. But some interventional studies using therapeutics which affect the microbiome (e.g. prebiotics, omega-3 fatty acids) were not able to identify significant changes in stools [43, 44] since some effects might be limited to the upper GI tract. With our comprehensive sampling of the whole GI tract, we were able to show that the major effects of vitamin D on the human gastrointestinal microbiome were only present in the upper GI tract but not in mucosal samples of the colon, ileum, or stools. Future microbiome studies need to be aware that interventions such as vitamin D supplementation can have significantly different effects on different regions of the GI tract and that these effects cannot necessarily be detected in stools.
In general, the assessment of the microbial composition of samples from the gastrointestinal system is often affected by high inter-individual variation [45] and the methods used for sample processing (e.g. sample storage, DNA extraction, and primer choice). Previous studies have shown that freezing of stools prior to DNA extraction can lead to an increased ratio of Firmicutes to Bacteroidetes [46]. The proportion of Firmicutes reported in the literature varies considerably. Some studies report a microbial composition dominated by Firmicutes [27, 47, 48], whereas other studies report similar levels compared to the results in our study [45, 49, 50].
Our study also showed that the intestinal microbiome varies along the human GI tract and thus supports a previous study which found a similar bacterial distribution pattern in the ileum and colon of healthy humans [51]. The bacterial composition along the GI tract is determined by different niche factors (e.g. pH, oxygen, tension, nutrients), and microbiome data do not necessarily reflect the anatomical categorisation. In our study, the microbiome of the duodenum was more similar to the stomach rather than to the terminal ileum, which was more similar to colon samples. Different regions of the human GI tract vary in function, permeability, type, and number of immune cells [52, 53].
A significantly higher number of CD8+ T cells in the upper GI (GC and GA) compared with lower GI sites [52] is part of the first defence line against foodborne pathogens. The significant decrease in Gammaproteobacteria that we found was limited to the mucosa of the upper GI tract and might have been driven by the increased CD8+ T cell fraction found after 8 weeks of vitD3 supplementation. CD8+ T cells have the highest expression level of VDR compared with other immune cells [25], which suggests that CD8+ T cell function is also regulated by vitamin D. High vitamin D levels have been associated with a decline in naïve CD8+ T cells and an increase in CD8+ effector T cells [54]. After vitD3 supplementation, we observed a trend towards elevated CD8+ T cell levels in all seven assessed GI regions, but we did not analyse the different CD8+ T cell subpopulations in this study. The stimulation of mucosal CD8+ T cells by vitD3 could contribute for the effects on Gammaproteobacteria together with other known effects of vitamin D, such as the induction of antimicrobial peptides (e.g. cathelicidin [55]) and the stimulation of certain cytokine production by mucosal dendritic cells [56]. In addition, murine CD8+ T cells [57] and human activated T cells [58] have been found to express 1α-hydroxylase (CYP27B1) which enables the production of calcitriol, the active hormonal state of vitD3. Calcitriol influences cytokine production in human CD8+ T cells [59] and is known to act as a stop signal in inflammatory processes [60, 61]. CD8+ T cells are also capable of directly destroying harmed or infected host cells, thus lowering the number of proinflammatory cells [62].
Reducing such an inflammatory environment by vitD3 could diminish the competitive advantage of opportunistic pathogens, such as Escherichia/Shigella spp. or Pseudomonas spp. which are evolutionary better adapted to inflammation and can outcompete commensal bacteria [63]. In return, a low inflammatory environment allows beneficial bacteria such as Bacteroidetes to outcompete opportunistic pathogens, resulting in increased phylotype richness which we found in this study.
An increase in bacterial richness could be particularly beneficial for gastrointestinal problems such non-alcoholic steatohepatitis or IBD which is characterised by decreased bacterial richness, lower vitamin D levels, and an overgrowth of pathogens [17, 50, 64–68]. Studies have reported beneficial effects of a vitamin D treatment in IBD patients [69] which could be to some extent due to the restoration of gut microbiome homoeostasis indicated by a reduction in opportunistic pathogens such as Enterobacteriaceae, which belong to the class of Gammaproteobacteria [17, 65–67, 70]. 16S rRNA amplicon sequencing as used in our study yields only relative abundances which cannot be used to differentiate between an increase in Bacteroidia and a decrease in Gammaproteobacteria. We assume that a decrease in Gammaproteobacteria is more likely because we found a decrease in several genera of Gammaproteobacteria, whereas no single genus of Bacteroidia was significantly altered after 8 weeks of vitD3 supplementation.
After vitD3 supplementation, we also found a decrease in overall abundance of Helicobacter spp. in the H. pylori-positive subgroup where approximately 90 % of all bacteria in the stomach were classified as Helicobacter spp. These are the first data to describe an effect of vitD3 on H. pylori infections and support the finding that CYP27B1 knockout mice which cannot produce calcitriol show a significantly higher relative abundance of Helicobacteriaceae compared to wild-type mice [37]. In these knockout mice, a calcitriol supplementation successfully decreased Helicobacteriaceae levels. The relevant role of vitD3 in H. pylori infections is also supported by the finding that H. pylori itself induces increased expression of the VDR [71].
VitD3 dosing in our study was above currently recommended dosing for vitamin D supplementation [72] and resulted in mean vitamin D levels of 55.2 ng/mL. The safety of such a high vitamin D supplementation has previously been reported [73]. We did not observed any adverse effects and no safety parameters, including serum and urine calcium and phosphate were significantly changed during 8 weeks of a high-dose vitD3 supplementation. At present, it is not advised to generally administer such high vitD3 doses for longer time periods in humans. Randomized intervention long-term trials are needed to evaluate the risks and benefits of such high vitD3 doses for human health, because this dose is not what the international guidelines state as recommended daily allowance. One limitation of our study is the relatively small sample size mainly because of the invasive procedures. Despite the relatively small study cohort and a high degree of inter-individual variation of the intestinal microbiome, we were able to observe a significant modulatory effect of vitD3 in the upper GI tract.
Conclusion
The marked reduction in Gammaproteobacteria, which include typical opportunistic pathogens and the increase in phylotype richness, supports the beneficial effect of a high-dose vitD3 supplementation on the human gut microbiome. This might in part explain the effects of a vitamin D-rich diet on IBD or bacterial infections and encourage studying the effects of vitamin D in these patients. The fact that the vitamin D effect on the microbiome was only evident in the upper GI tract should guide future microbiome studies in humans to also assess the upper GI tract in addition to stools and colonic mucosal samples.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (DOCX 17657 kb)
Acknowledgments
This work was supported by the Medical University of Graz, the European Association for the Study of Diabetes (EASD.org), the doctoral college of molecular inflammation (DK-MOLIN) and BioPersMed (COMET K-project 825329), which is funded by the Federal Ministry of Transport, Innovation and Technology (BMVIT) and the Federal Ministry of Economics and Labour/the Federal Ministry of Economy, Family and Youth (BMWA/BMWFJ) and the Styrian Business Promotion Agency (SFG). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank all volunteers who participated in the study and Ingeborg Klymiuk and Slave Trajanoski (Medical University of Graz) for their support in the laboratory and data analysis.
Compliance with Ethical Standards
Conflict of interest
None.
Contributor Information
Christoph Högenauer, Email: christoph.hoegenauer@medunigraz.at.
Thomas R. Pieber, Phone: +43 316 876 2131, Email: endo@medunigraz.at
References
- 1.Nicholson J, Holmes E, Kinross J. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1268. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 2.O’Shea EF, Cotter PD, Stanton C, et al. Production of bioactive substances by intestinal bacteria as a basis for explaining probiotic mechanisms: bacteriocins and conjugated linoleic acid. Int J Food Microbiol. 2012;152:189–205. doi: 10.1016/j.ijfoodmicro.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 3.Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawley TD, Walker AW. Intestinal colonization resistance. Immunology. 2013;138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gravitz L. Microbiome: the critters within. Nature. 2012;485:12–13. doi: 10.1038/485S12a. [DOI] [PubMed] [Google Scholar]
- 7.Tilg H, Moschen AR. Microbiota and diabetes: an evolving relationship. Gut. 2014;63:1513–1521. doi: 10.1136/gutjnl-2014-306928. [DOI] [PubMed] [Google Scholar]
- 8.Halmos EP, Christophersen CT, Bird AR, et al. Diets that differ in their FODMAP content alter the colonic luminal microenvironment. Gut. 2014 doi: 10.1136/gutjnl-2014-307264. [DOI] [PubMed] [Google Scholar]
- 9.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matijašić BB, Obermajer T, Lipoglavšek L, et al. Association of dietary type with fecal microbiota in vegetarians and omnivores in Slovenia. Eur J Nutr. 2014;53:1051–1064. doi: 10.1007/s00394-013-0607-6. [DOI] [PubMed] [Google Scholar]
- 11.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costello EK, Stagaman K, Dethlefsen L, et al. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorkiewicz G, Thallinger GG, Trajanoski S, et al. Alterations in the colonic microbiota in response to osmotic diarrhea. PLoS One. 2013;8:e55817. doi: 10.1371/journal.pone.0055817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2011;108:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ananthakrishnan AN, Cagan A, Gainer VS, et al. Higher plasma vitamin D is associated with reduced risk of Clostridium difficile infection in patients with inflammatory bowel diseases. Aliment Pharmacol Ther. 2014;39:1136–1142. doi: 10.1111/apt.12706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gagliani N, Hu B, Huber S, et al. The fire within: microbes inflame tumors. Cell. 2014;157:776–783. doi: 10.1016/j.cell.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 17.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 18.Gevers D, Kugathasan S, Denson L, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 20.Kovach MA, Ballinger MN, Newstead MW, et al. Cathelicidin-related antimicrobial peptide is required for effective lung mucosal immunity in Gram-negative bacterial pneumonia. J Immunol. 2012;189:304–311. doi: 10.4049/jimmunol.1103196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holick MF. Vitamin D: importance in the prevention of cancers, type 1 diabetes, heart disease, and osteoporosis. Am J Clin Nutr. 2004;79:362–371. doi: 10.1093/ajcn/79.3.362. [DOI] [PubMed] [Google Scholar]
- 22.Ananthakrishnan AN, Cagan A, Gainer VS, et al. Normalization of plasma 25-hydroxy vitamin D is associated with reduced risk of surgery in Crohn’s disease. Inflamm Bowel Dis. 2013;19:1921–1927. doi: 10.1097/MIB.0b013e3182902ad9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holick MF. The vitamin D deficiency pandemic and consequences for nonskeletal health: mechanisms of action. Mol Asp Med. 2008;29:361–368. doi: 10.1016/j.mam.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grant WB, Schuitemaker GE. Health benefits of higher serum 25-hydroxyvitamin D levels in The Netherlands. J Steroid Biochem Mol Biol. 2010;121:456–458. doi: 10.1016/j.jsbmb.2010.03.089. [DOI] [PubMed] [Google Scholar]
- 25.Veldman CM, Cantorna MT, DeLuca HF. Expression of 1,25-dihydroxyvitamin D(3) receptor in the immune system. Arch Biochem Biophys. 2000;374:334–338. doi: 10.1006/abbi.1999.1605. [DOI] [PubMed] [Google Scholar]
- 26.Battault S, Whiting SJ, Peltier SL, et al. Vitamin D metabolism, functions and needs: from science to health claims. Eur J Nutr. 2013;52:429–441. doi: 10.1007/s00394-012-0430-5. [DOI] [PubMed] [Google Scholar]
- 27.Stearns JC, Lynch MDJ, Senadheera DB, et al. Bacterial biogeography of the human digestive tract. Sci Rep. 2011;1:170. doi: 10.1038/srep00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kump PK, Gröchenig H-P, Lackner S, et al. Alteration of intestinal dysbiosis by fecal microbiota transplantation does not induce remission in patients with chronic active ulcerative colitis. Inflamm Bowel Dis. 2013;19:2155–2165. doi: 10.1097/MIB.0b013e31829ea325. [DOI] [PubMed] [Google Scholar]
- 29.Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quince C, Lanzén A, Curtis TP, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- 31.Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pruesse E, Quast C, Knittel K, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ooi J, Li Y, Rogers C, Cantorna M. Vitamin D regulates the gut microbiome and protects mice from dextran sodium sulfate-induced colitis. J Nutr. 2013;143:1679–1686. doi: 10.3945/jn.113.180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chung H, Pamp SJ, Hill JA, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clarke SF, Murphy EF, O’Sullivan O, et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut. 2014 doi: 10.1136/gutjnl-2013-306541. [DOI] [PubMed] [Google Scholar]
- 40.Carroll IM, Ringel-Kulka T, Keku TO, et al. Molecular analysis of the luminal- and mucosal-associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2011;301:G799–G807. doi: 10.1152/ajpgi.00154.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.White ANJ, Ng V, Spain CV, et al. Let the sun shine in: effects of ultraviolet radiation on invasive pneumococcal disease risk in Philadelphia, Pennsylvania. BMC Infect Dis. 2009;9:196. doi: 10.1186/1471-2334-9-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajkumar H, Mahmood N, Kumar M, et al. Effect of probiotic (VSL#3) and omega-3 on lipid profile, insulin sensitivity, inflammatory markers, and gut colonization in overweight adults: a randomized, controlled trial. Mediat Inflamm. 2014;2014:348959. doi: 10.1155/2014/348959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urwin HJ, Miles EA, Noakes PS, et al. Effect of salmon consumption during pregnancy on maternal and infant faecal microbiota, secretory IgA and calprotectin. Br J Nutr. 2014;111:773–784. doi: 10.1017/S0007114513003097. [DOI] [PubMed] [Google Scholar]
- 45.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bahl MI, Bergström A, Licht TR. Freezing fecal samples prior to DNA extraction affects the Firmicutes to Bacteroidetes ratio determined by downstream quantitative PCR analysis. FEMS Microbiol Lett. 2012;329:193–197. doi: 10.1111/j.1574-6968.2012.02523.x. [DOI] [PubMed] [Google Scholar]
- 47.Hu J, Nomura Y, Bashir A, et al. Diversified microbiota of meconium is affected by maternal diabetes status. PLoS One. 2013;8:e78257. doi: 10.1371/journal.pone.0078257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Angelis M, Piccolo M, Vannini L, et al. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One. 2013;8:e76993. doi: 10.1371/journal.pone.0076993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davenport M, Poles J, Leung JM, et al. Metabolic alterations to the mucosal microbiota in inflammatory bowel disease. Inflamm Bowel Dis. 2014;20:723–731. doi: 10.1097/MIB.0000000000000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong VW-S, Tse C-H, Lam TT-Y, et al. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis—a longitudinal study. PLoS One. 2013;8:e62885. doi: 10.1371/journal.pone.0062885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Z, Geng J, Tang X, et al. Spatial heterogeneity and co-occurrence patterns of human mucosal-associated intestinal microbiota. ISME J. 2014;8:881–893. doi: 10.1038/ismej.2013.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tauschmann M, Prietl B, Treiber G, et al. Distribution of CD4(pos)-, CD8(pos)—and regulatory T cells in the upper and lower gastrointestinal tract in healthy young subjects. PLoS One. 2013;8:e80362. doi: 10.1371/journal.pone.0080362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davis GR, Santa Ana CA, Morawski SG, Fordtran JS. Permeability characteristics of human jejunum, ileum, proximal colon and distal colon: results of potential difference measurements and unidirectional fluxes. Gastroenterology. 1982;83:844–850. [PubMed] [Google Scholar]
- 54.Hwang YG, Hsu H, Lim FC, et al. Increased vitamin D is associated with decline of naïve, but accumulation of effector, CD8 T cells during early aging. Adv Aging Res. 2013;2:72–80. doi: 10.4236/aar.2013.22010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Borella E, Nesher G, Israeli E, Shoenfeld Y. Vitamin D: a new anti-infective agent? Ann N Y Acad Sci. 2014;1317:76–83. doi: 10.1111/nyas.12321. [DOI] [PubMed] [Google Scholar]
- 56.Bartels LE, Bendix M, Hvas CL, et al. Oral vitamin D3 supplementation reduces monocyte-derived dendritic cell maturation and cytokine production in Crohn’s disease patients. Inflammopharmacology. 2014;22:95–103. doi: 10.1007/s10787-013-0197-1. [DOI] [PubMed] [Google Scholar]
- 57.Ooi JH, McDaniel KL, Weaver V, Cantorna MT. Murine CD8+ T cells but not macrophages express the vitamin D 1α-hydroxylase. J Nutr Biochem. 2014;25:58–65. doi: 10.1016/j.jnutbio.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sigmundsdottir H, Pan J, Debes GF, et al. DCs metabolize sunlight-induced vitamin D3 to “program” T cell attraction to the epidermal chemokine CCL27. Nat Immunol. 2007;8:285–293. doi: 10.1038/ni1433. [DOI] [PubMed] [Google Scholar]
- 59.Willheim M, Thien R, Schrattbauer K, et al. Regulatory effects of 1alpha,25-dihydroxyvitamin D3 on the cytokine production of human peripheral blood lymphocytes. J Clin Endocrinol Metab. 1999;84:3739–3744. doi: 10.1210/jcem.84.10.6054. [DOI] [PubMed] [Google Scholar]
- 60.Kreutz M, Andreesen R, Krause SW, et al. 1,25-dihydroxyvitamin D3 production and vitamin D3 receptor expression are developmentally regulated during differentiation of human monocytes into macrophages. Blood. 1993;82:1300–1307. [PubMed] [Google Scholar]
- 61.Fritsche J, Mondal K, Ehrnsperger A, et al. Regulation of 25-hydroxyvitamin D3-1 alpha-hydroxylase and production of 1 alpha, 25-dihydroxyvitamin D3 by human dendritic cells. Blood. 2003;102:3314–3316. doi: 10.1182/blood-2002-11-3521. [DOI] [PubMed] [Google Scholar]
- 62.Murphy K. T cell-mediated cytotoxicity. In: Janeway CA, Travers JP, Walport M, Shlomchik MJ, editors. Janeway’s Immunobiology. 8. New York: Garland Science; 2011. p. 888. [Google Scholar]
- 63.Stecher B, Robbiani R, Walker AW, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pappa H, Gordon C, Saslowsky T. Vitamin D status in children and young adults with inflammatory bowel disease. Pediatrics. 2006;118:1950–1961. doi: 10.1542/peds.2006-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jellbauer S, Raffatellu M. An intestinal arsonist: pathobiont ignites IBD and flees the scene. Gut. 2014;63:1034–1035. doi: 10.1136/gutjnl-2013-305589. [DOI] [PubMed] [Google Scholar]
- 66.Mukhopadhya I, Hansen R, El-Omar EM, Hold GL. IBD-what role do Proteobacteria play? Nat Rev Gastroenterol Hepatol. 2012;9:219–230. doi: 10.1038/nrgastro.2012.14. [DOI] [PubMed] [Google Scholar]
- 67.Frank DN, Amand ALS, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jørgensen SP, Hvas CL, Agnholt J, et al. Active Crohn’s disease is associated with low vitamin D levels. J Crohns Colitis. 2013;7:e407–e413. doi: 10.1016/j.crohns.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 69.Jørgensen SP, Agnholt J, Glerup H, et al. Clinical trial: vitamin D3 treatment in Crohn’s disease—a randomized double-blind placebo-controlled study. Aliment Pharmacol Ther. 2010;32:377–383. doi: 10.1111/j.1365-2036.2010.04355.x. [DOI] [PubMed] [Google Scholar]
- 70.Willing BP, Dicksved J, Halfvarson J, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139(1844–1854):e1. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 71.Guo L, Chen W, Zhu H, et al. Helicobacter pylori induces increased expression of the vitamin D receptor in immune responses. Helicobacter. 2014;19:37–47. doi: 10.1111/hel.12102. [DOI] [PubMed] [Google Scholar]
- 72.Ross AC, Manson JE, Abrams SA, et al. The 2011 report on dietary reference intakes for calcium and vitamin D from the Institute of Medicine: what clinicians need to know. J Clin Endocrinol Metab. 2011;96:53–58. doi: 10.1210/jc.2010-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Prietl B, Treiber G, Mader JK, et al. High-dose cholecalciferol supplementation significantly increases peripheral CD4+ Tregs in healthy adults without negatively affecting the frequency of other immune cells. Eur J Nutr. 2013;53:751–759. doi: 10.1007/s00394-013-0579-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 (DOCX 17657 kb)