Abstract
Cell division entails a sequence of processes whose specific demands for biosynthetic precursors and energy place dynamic requirements on metabolism. However, little is known about how metabolic fluxes are coordinated with the cell division cycle. Here, we examine budding yeast to show that over half of all measured metabolites change significantly through the cell division cycle. Cell cycle-dependent changes in central carbon metabolism are controlled by the cyclin-dependent kinase Cdk1, a major cell cycle regulator, and the metabolic regulator protein kinase A. At the G1/S transition, Cdk1 phosphorylates and activates the enzyme Nth1, which funnels the storage carbohydrate trehalose into central carbon metabolism. Trehalose utilization fuels anabolic processes required to reliably complete cell division. Thus, the cell cycle entrains carbon metabolism to fuel biosynthesis. Since the oscillation of Cdk-activity is a conserved feature of the eukaryotic cell cycle, we anticipate its frequent use in dynamically regulating metabolism for efficient proliferation.
Introduction
Across the kingdoms of life, cells coordinate metabolism, growth and division. This coordination increases the fitness of unicellular organisms living in changing environments, and allows multicellular organisms to shape and maintain their body plan. To coordinate metabolism, growth and division, cells have evolved extensive signaling networks that sense nutrient status. For example, receptors bind extracellular sugar to activate downstream molecules regulating growth and metabolism. In turn, metabolism gates the decision to divide. Cells deprived of essential nutrients will not pass the point of commitment to cell division, which is known as Start in yeast and the Restriction point in mammals (Broach, 2012; Johnson and Skotheim, 2013; Wang and Proud, 2009; Zaman et al., 2008).
In proliferative conditions, cells committed to division proceed through a coordinated sequence of processes, including DNA replication, mitosis and cytokinesis, that are collectively known as the cell cycle (Morgan, 2007). Each of these processes comes with specific and heterogeneous demands for biosynthetic precursors and energy, such as nucleotides for DNA synthesis (Buchakjian and Kornbluth, 2010; Vander Heiden et al., 2009). Thus, cell cycle progression necessarily places dynamic requirements on metabolism. Regulating metabolic fluxes to satisfy these periodic demands is likely essential to maximize fitness and survival. However, little is known regarding if and how metabolic fluxes are temporally coordinated with the cell cycle.
The need to accurately allocate resources during different phases of growth and division may be most acute for single cell organisms growing in nutrient-poor environments. Nutrient limitation has been used to control the growth rate of budding yeast in chemostat cultivations to probe the connection between metabolism and growth. Such studies have examined how the rate of growth affects cell physiology, protein composition, transcription, and metabolism (Brauer et al., 2008; Canelas et al., 2010; Castrillo et al., 2007; Gutteridge et al., 2010). Moreover, these studies link metabolism and growth rate to the activity of key signaling molecules including protein kinase A (PKA) and target of rapamycin (TOR) (Castrillo et al., 2007). In addition to the examination of these steady-state relationships, chemostat cultivations have also been used to examine a dynamic phenomenon known as ‘metabolic cycling’ (Burnetti et al., 2016; Klevecz et al., 2004; Kuenzi and Fiechter, 1969; Tu et al., 2005; Tu et al., 2007; Wittmann et al., 2005). Metabolically cycling populations of cells exhibit coordinated oscillations in metabolism and cell cycle phase, which suggests a link between these two processes. However, metabolic cycling is a complex phenomenon. Cell-to-cell communication of unknown origin synchronizes cell metabolism, drives periodic changes in the extracellular environment, and only partially synchronizes the cell cycle. In addition, the phase shift between cell and metabolic cycles varies in different conditions (Klevecz et al., 2004; Slavov and Botstein, 2011; Tu, 2010) and metabolic cycles have even been shown in the absence of cell cycle progression (Slavov et al., 2011). Thus, it remains unclear which changes in metabolism are driven by cell cycle progression and which might be intrinsic to a metabolic oscillator.
Here, to isolate the impact of cell cycle progression on cell metabolism, we examine dilute populations of cell cycle synchronized budding yeast. This allows us to exogenously control cell cycle progression and directly measure its effect on metabolism. We use dilute batch cultures to minimize the impact of cells on their environment, and thereby eliminate a potential feedback mechanism on cell growth and metabolism. To gain a global view of all changes in cellular metabolism, we employed untargeted metabolomics of cells growing on ethanol minimal medium. We found that more than half of the hundreds of detectable metabolites changed concentration significantly during the cell cycle, demonstrating that cell cycle progression per se drives large-scale changes in cell metabolism.
Metabolite variations through the cell cycle were not limited to biosynthetic pathways, but were also pronounced in central carbon metabolism. This included the cell cycle-dependent depletion of the storage carbohydrate trehalose. The storage carbohydrates trehalose and glycogen are highly abundant in slow growing cells and can serve as long-term carbon reserves during starvation (Francois and Parrou, 2001). Trehalose also acts as a molecular chaperone and stress protectant (Tapia and Koshland, 2014; Tapia et al., 2015). Moreover, several previous studies support a link between storage carbohydrate metabolism and the cell cycle. It was found that the length of G1 correlates with the amount of carbohydrates stored (Brauer et al., 2008; Paalman et al., 2003; Sillje et al., 1997), budding often correlates with decreases in storage carbohydrates (Kuenzi and Fiechter, 1969, 1972; Muller et al., 2003; Sillje et al., 1997), and the in vitro activities of several storage metabolism enzymes correlate with cell cycle phase (Vandoorn et al., 1988). Furthermore, trehalose utilization accelerates cell cycle entry when recovering from stationary phase (Shi et al., 2010). Taken together, these observations suggest coordination of carbohydrate storage metabolism and the cell cycle. In support of this hypothesis, cell cycle regulation of lipid storage utilization also accelerates entry into the cell cycle from stationary phase (Futcher, 2009; Kurat et al., 2009). However, no molecular mechanism linking storage carbohydrate metabolism and cell cycle control has yet been identified, and the function of storage carbohydrate oscillations remains unclear.
Here, we show that storage carbohydrate metabolism in budding yeast is directly regulated by the cyclin-dependent kinase (Cdk1), whose activity oscillates through the cell cycle to coordinate major cell cycle events including DNA replication and mitosis (Morgan, 2007). Thus, oscillation of Cdk1 activity can directly entrain fluxes in central carbon metabolism to meet the temporally specific biosynthetic demands of the cell cycle. Moreover, our work suggests that Cdk1-dependent storage carbohydrate oscillations allow cells to complete division in adverse nutrient conditions by providing an additional pool of biosynthetic precursors.
Results
Global changes in metabolism are coordinated with the cell cycle downstream of Start
Observing the global coordination of metabolism with cell cycle progression requires examining populations of cells synchronously progressing through the cell division cycle. However, many environmental perturbations used to synchronize the cell cycle, including centrifugation, changes in temperature, or the addition of a carbon source such as galactose to modify gene expression, immediately impact cell metabolism and thereby complicate analysis (Boer et al., 2010; Ewald et al., 2013; Strassburg et al., 2010). We therefore constructed a strain that allowed us to arrest cells in G1 and then synchronously release them into the cell cycle by adding to the media nanomolar amounts of estrogen, which is not a natural signaling molecule in yeast and does not induce any signaling or stress response (Figure S1C and (McIsaac et al., 2014; Ottoz et al., 2014)). Since passing the G1/S transition (Start) requires activity of Cdk1 in complex with one of the three G1 cyclins (CLN1/2/3) (Richardson et al., 1989), we deleted all three cyclins and placed CLN1 under a synthetic, hormone-inducible promoter (Ottoz et al., 2014). Upon removal of hormone these cells arrest in G1 as unbudded cells. Experiments were performed using ethanol minimal media in shake flasks, where cells grew with doubling times around five hours. In these slow-growth conditions, cells must de novo synthesize all biomolecules from their limited resources. We performed all experiments using exponentially growing cultures at low density to avoid nutrient depletion or cell density effects.
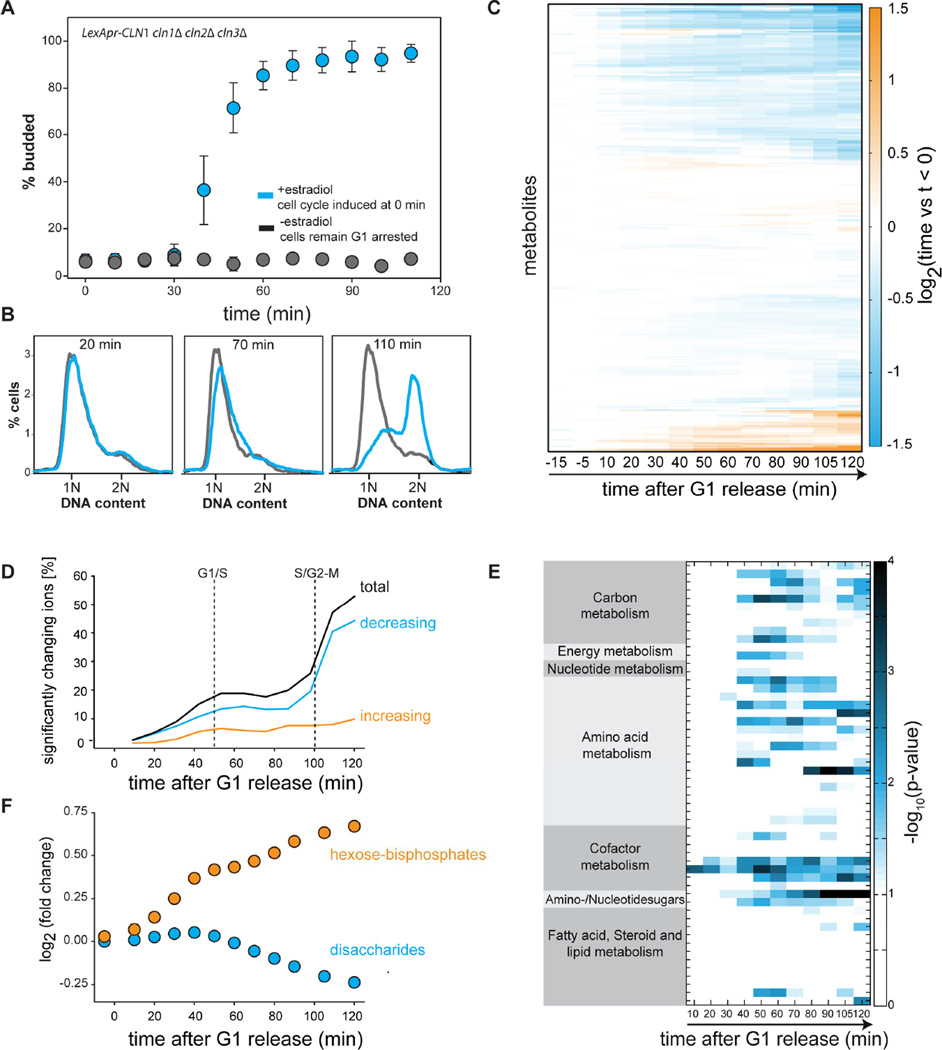
To investigate how metabolism is affected by the cell cycle, we used our strain to drive synchronous progression through the G1/S transition (Figure 1A–B) and performed untargeted metabolomics using flow-injection mass spectrometry. Approximately half of the hundreds of metabolites that were detected changed their concentration significantly (p ≤ 0.01, FDR corrected 2 sample t-test) by more than 15% in the course of the cell cycle (Figure 1C–D and Table S4). Since both arrested and cycling cells accumulate biomass at a similar rate (Figure S1A), our results strongly suggest that the observed metabolite dynamics are primarily associated with the cell cycle. Major changes in metabolite concentration occur around the G1/S and mitotic entry transitions (Figure 1D). To identify the pathways that were most affected by cell cycle progression we used pathway enrichment analysis. This identified synthesis pathways for precursors of amino acids, cell wall components, and lipids (Figure 1E and S1D). An increase in cell wall synthesis and lipid metabolism is most likely due to the rapid increase in the rate of surface expansion during bud growth in the S/G2/M phases of the cell cycle. Surprisingly, we saw only minor changes in nucleotide synthesis pathways despite their periodic utilization through the cell cycle. This is most likely due to extensive feedback regulation that ensures homeostasis in nucleotides and their precursor concentrations despite changes in fluxes through the pathway during DNA synthesis (Hofer et al., 2012; Mathews, 2014).
Figure 1. Global changes in metabolism are coordinated with cell cycle progression.
(A) A strain expressing a single G1 cyclin controlled by a synthetic hormone-inducible promoter was arrested in G1 and synchronously released into the cell cycle at t=0 minutes (blue). Control cells were mock treated and remained arrested (grey). The budding index denotes the fractions of cells in S/G2/M phases of the cell cycle. Error bars indicate standard deviations from four biological replicates. (B) DNA content of cells released from a G1 arrest (blue) or control cells (grey). (C) Heatmap of metabolite changes relative to the average of 3 time points prior to release from G1 arrest. (D) Fraction of metabolites that change significantly following synchronous release from G1 arrest. Significant is defined relative to t<0 and requires a change in concentration greater than 15%, and to have p < 0.01 for a 2-sample t-test that was false discovery rate corrected. Dotted lines denote the G1/S (50% budding) and S/G2-M (50% 2N DNA) transitions. (E) Metabolic pathway enrichment analysis identifies pathways whose metabolites change significantly through the cell cycle. (F) Following release from G1, trehalose concentrations decrease while sugar-phosphates in glycolysis, such as fructose-bis-phoshate, increase. See Figure S1.
In addition to many biosynthetic pathways, we observed extensive cell cycle-dependent changes in central carbon metabolism (glycolysis, tricarboxylic acid (TCA) cycle, pentose-phosphate pathway). This is consistent with the hypothesis that fluxes through central carbon metabolism are adjusted to provide precursors for biosynthetic pathways through the cell cycle. Strikingly, these changes in central carbon metabolism coincided with a significant decrease in trehalose, a disaccharide that serves as an important carbohydrate reserve for the cell. Indeed, 20% of the cell’s dry weight can be composed of trehalose and glycogen, the other storage carbohydrate (Francois and Parrou, 2001). Since storage metabolism is directly connected to glycolysis, the observed decrease in trehalose likely leads to the increase of hexose-phosphates and downstream changes in central carbon metabolism (Figure 1F). These results are consistent with several previous studies that have suggested a link between oscillations in storage carbohydrate concentrations and the cell cycle (Kuenzi and Fiechter, 1969; Muller et al., 2003; Sillje et al., 1999; Sillje et al., 1997). However, no molecular mechanism linking storage carbohydrate metabolism and cell cycle control has yet been identified, and the role of storage carbohydrates for cell cycle progression remains unclear.
The G1/S transition drives storage carbohydrate utilization
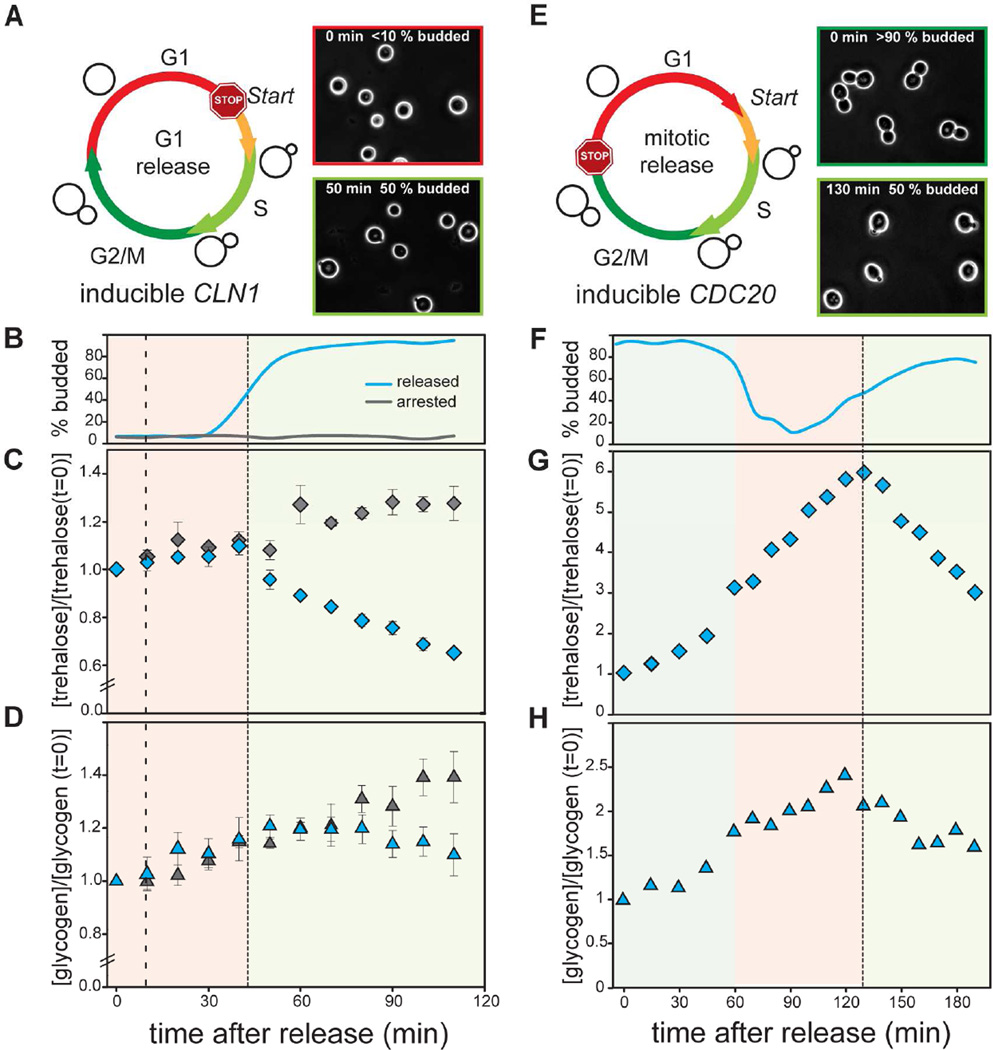
To confirm that storage carbohydrate metabolism is directly linked to the cell cycle and downstream of Start, we used a biochemical approach to examine storage carbohydrate levels in cell cycle synchronized populations. Cells were synchronized in G1 and released into the cell cycle while trehalose and glycogen were measured every 10 minutes using enzymatic assays (Figure 2A–D). Both trehalose and glycogen concentrations began to decrease just as cells started budding, which is approximately 34 minutes after Start in these conditions (Figure S1B). In contrast, control cells, in which CLN1 was not induced, continued to accumulate trehalose and glycogen (Figure 2C–D). This is consistent with previous data showing that trehalose utilization correlated with budding when G1 cells, isolated using size fractionation, were released into the cell cycle (Sillje et al., 1997). This confirmed that cell cycle progression per se, and not time or growth, is responsible for the observed storage utilization.
Figure 2. Storage carbohydrates oscillate during the cell division cycle.
(A) Cells were arrested in G1. ϩ0% of all cells were in G1 at the time of release. Cells synchronously passed the G1/S transition approximately 50 minutes after release (50% budded denoted by dashed line). (B) Budding index, (C) Normalized trehalose concentration, and (D) normalized glycogen concentration for cells either released from (blue) or maintained in (grey) a G1 arrest. Error bars indicate standard error of the mean from four biological replicates. (E) Cells expressing a hormone inducible CDC20 allele were arrested in mitosis (>90% budded) and then synchronously released into the cell cycle at t=0 minutes. Dashed line indicates where 50% of the cells reached the G1/S transition and budded (t=130 minutes). (F) Budding index, (G) trehalose and (H) glycogen concentrations measured after release from mitotic arrest. All concentrations were normalized to 1 at t=0 minutes. See also Figure S2 and Table S1.
To show that a storage switch at the G1/S transition is not due to our synchronization method, we also synchronized cells in mitosis using the same estrogen-inducible promoter to control the expression of CDC20, a regulatory subunit of the anaphase promoting complex (LexApr-CDC20) (Figure 2E). Cells were released into the cell cycle from a mitotic arrest and we measured trehalose and glycogen levels (Figure 2F). Surprisingly, Cdc20-arrested cells also accumulated trehalose (Figure S2A–C). However, just as in our previous experiment, storage carbohydrate utilization began at the G1/S transition of the next cell cycle (Figure 2G–H).
Taken together, these results confirm that the metabolic switch from storage synthesis to storage consumption is linked to the cell cycle. While this is consistent with previous observations in fed-batch and chemostat cultures (Kuenzi and Fiechter, 1969, 1972; Muller et al., 2003; Paalman et al., 2003), we now show that this is governed by the cell cycle control network and is downstream of the Start transition at the G1/S boundary. This result contradicts previous hypotheses that the storage switch is upstream of Start and drives entry into the cell cycle (Futcher, 2006; Sillje et al., 1997).
The trehalase Nth1 is phosphorylated by Cdk1 and PKA1
We next sought to identify the molecular mechanism through which storage metabolism is linked to the cell cycle. The trehalose utilization switch coincides with the G1/S transition, downstream of Start. Since Start coincides with a rapid, positive feedback-driven increase in Cdk1 activity that is sustained through the rest of the cell cycle (Doncic et al., 2011; Skotheim et al., 2008), we hypothesized that Cdk1 was directly responsible for triggering storage utilization. Supporting this hypothesis, two Cdk1 target screens, where Cdk1 was inhibited and the resulting changes in protein phosphorylation were determined, identified enzymes responsible for trehalose and glycogen utilization (Holt et al., 2009; Ubersax et al., 2003).
Since changes in trehalose concentrations were more pronounced than in glycogen concentrations (Figure 2), we decided to focus on the regulation of trehalose metabolism. We performed all mutant analyses in strains unable to utilize glycogen (gph1Δ) so that all storage carbohydrate utilization proceeds through the trehalose pathway. In addition, we removed the putative trehalase Nth2 (nth2Δ) so that the trehalase Nth1 is solely responsible for trehalose utilization. We note that yeast also have an additional trehalase Ath1, that is necessary for consuming extracellular trehalose, but is not implicated in cytosolic trehalose catabolism, and was not identified in Cdk1 target screens (Table S2).
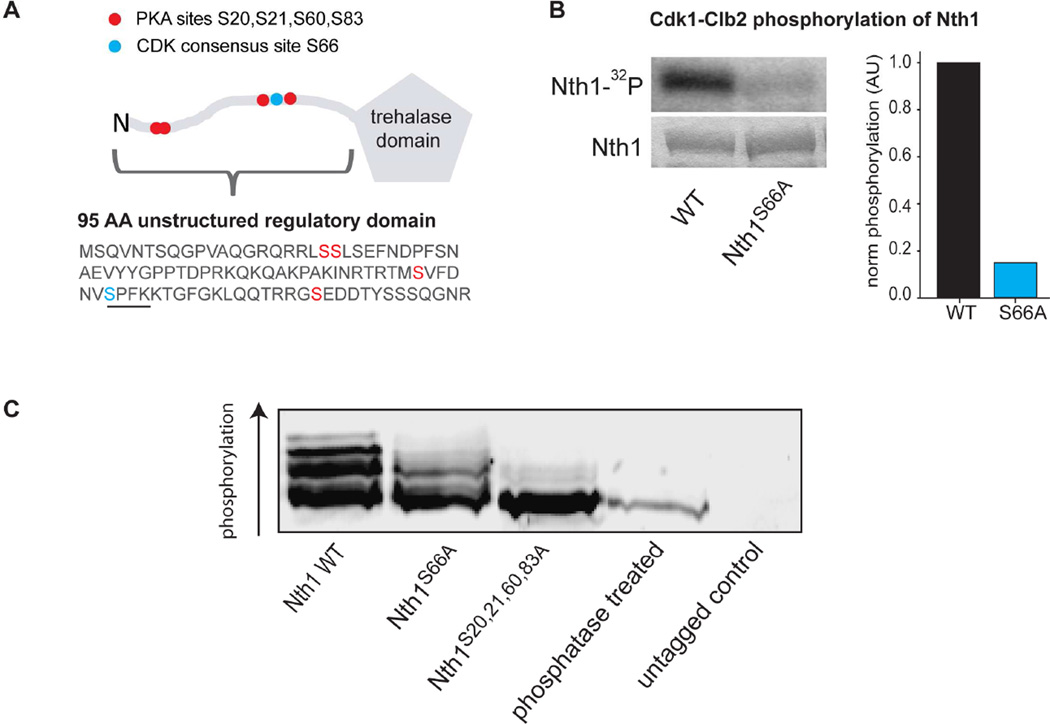
To test whether Cdk1 phosphorylates the trehalase Nth1 directly, we performed an in vitro phosphorylation assay. We purified Nth1 and obtained Cdk1 in complex with the B-type cyclin Clb2 from yeast (Koivomagi et al., 2011), whose activity begins to increase at budding and continues until mitosis (Grandin and Reed, 1993). Indeed, Cdk1-Clb2 directly and specifically phosphorylates Nth1 at the consensus site S66 (Figure 3A–B). To determine if B-type cyclin activity is required for Nth1 activity in vivo, we overexpressed stabilized Sic1 to inhibit B-type cyclin-Cdk1 complexes. This results in cells arrested post-Start with high G1 cyclin activity (Haase and Reed, 1999). In these cell trehalose is not utilized (Figure S2D–F).
Figure 3. Cdk1 phosphorylates the trehalase Nth1.
(A) Schematic and partial sequence of Nth1, which has an unstructured N-terminal regulatory region containing known PKA phosphorylation sites (red) and a Cdk1 consensus phosphorylation site (blue). The Cdk1 consensus motif is underlined. (B) Autoradiogram (top), coomassie gel (bottom), and quantification (right) for an in vitro phosphorylation assay using purified Cdk1-Clb2-Cks1 complex, purified Nth1 (wild type and S66A) and radiolabeled ATP. (C) Western blot of indicated Nth1-3xFlag proteins after separation on a PhosTag-SDS gel.
To determine if Nth1 is phosphorylated on S66 in vivo, we grew cells expressing FLAG-tagged Nth1 protein (Nth1WT) or a protein where the S66 Cdk1 site was substituted with alanine (Nth1ΔCDK). We grew cells asynchronously and examined the Nth1 phosphorylation pattern using PhosTag-SDS-PAGE-Western analysis (Figure 3C) which enhances the separation of phosphoisoforms (Kinoshita et al., 2006). Nth1WT existed in several phosphorylated species and one unphosphorylated band, indicating that Nth1 is phosphorylated on multiple sites in our conditions (Figure 3C and S3D). The most highly phosphorylated form of Nth1 was not found in cells expressing Nth1ΔCDK, confirming that S66 is phosphorylated in vivo.
The metabolic regulator protein kinase A (PKA) has been implicated in trehalose metabolism and been shown to phosphorylate Nth1 on at least four sites (Figure 3A) (Panni et al., 2008; Schepers et al., 2012; Shi et al., 2010; Veisova et al., 2012), which do not include S66. Consistently, Nth1ΔCDK still forms several different phospho-isoforms showing that additional sites are phosphorylated in vivo. To determine whether the PKA sites were phosphorylated in vivo during growth on ethanol, we constructed mutants where the wild type NTH1 was replaced by an NTH1ΔPKA allele in which the PKA sites (S20, S21, S60, and S83) were substituted with alanine. Mutating the PKA sites eliminated Nth1 phosphorylation almost completely, confirming the identity of the PKA sites in vivo (Figure 3C). Interestingly, the Cdk1 site was only very weakly phosphorylated on Nth1ΔPKA. This suggests that PKA phosphorylation may be required for Cdk1 to phosphorylate Nth1 efficiently in vivo.
Trehalose utilization during the cell cycle is controlled by Cdk1 phosphorylation
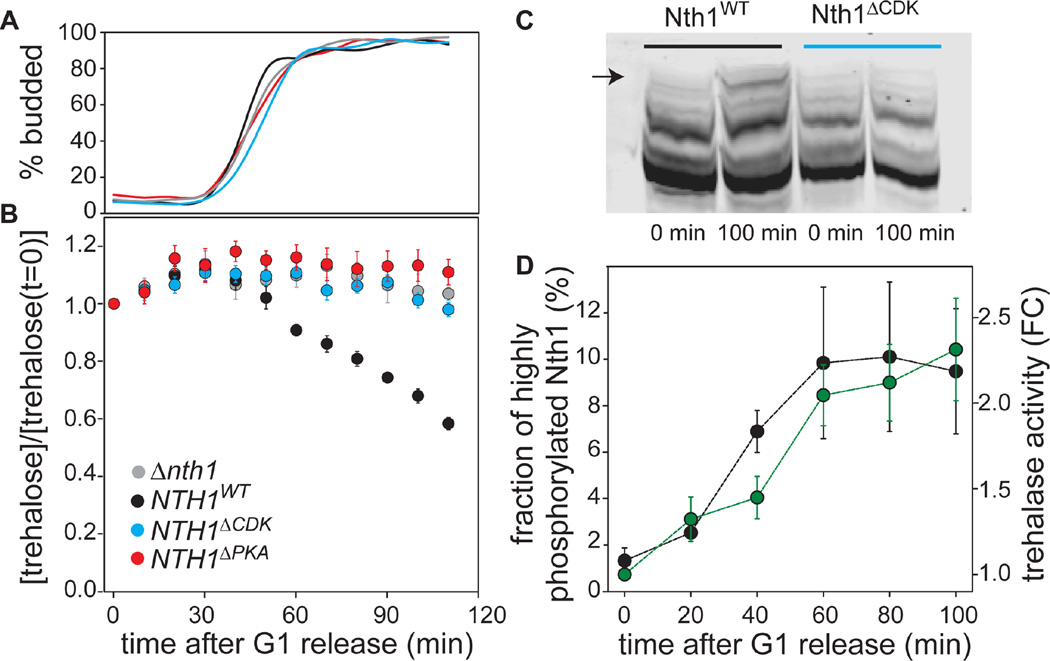
To test if phosphorylation of Nth1 is necessary for the storage carbohydrate switch at G1/S, we synchronized NTH1ΔCDK or NTH1ΔPKA cells and examined trehalose content following release into the cell cycle. Unlike wild type, neither NTH1ΔCDK nor NTH1ΔPKA cells were able to utilize trehalose following the G1/S transition (Figure 4A–B). Both these phospho-site mutations phenocopied nth1Δ even though wild type levels of protein were expressed (Figure 4B and S3A). NTH1 S66E mutant cells behaved just like the alanine mutations indicating that glutamic acid substitution does not mimic phosphorylation at S66 (Figure S3C and Table S1). Similarly, NTH1 5E mutant cells, where all Cdk1 and PKA sites were mutated to glutamate, did not show any activity (data not shown). Finally, the concentration of wild type Nth1 protein was found to be constant during the cell cycle, confirming that enzyme activity is regulated by phosphorylation rather than abundance (Figure S3B).
Figure 4. Cdk1 and PKA1 phosphorylation are required for Nth1 activation.
(A) Budding index, and (B) normalized trehalose concentration for cells synchronously released from a G1 arrest. Error bars denote the standard error of the mean for three biological replicates with two technical replicates each. All trehalose concentrations were normalized to the concentration at t=0 minutes. Grey denotes nth1Δnth2Δgph1Δ, black, blue and red denote wild type NTH1, NTH1ΔPKA, and NTH1ΔCDK alleles, respectively, in an nth2Δgph1Δ background. (C) Phostag-Western blot showing Nth1 and Nth1ΔCDK phospho-isoforms before (0 min) and after (100 min) the G1/S transition. (D) Black dots denote the fraction of Nth1 that is highly phosphorylated based on quantification of Phostag-Western blots of Nth1 after G1 release (quantified band indicated by arrow in C). Error bars denote standard deviations from three technical replicates. Green dots denote trehalase activity measured from cell extracts (fold change relative to before release). Error bars denote standard deviations from two biological and two technical replicates.
Further supporting the role of Cdk1 in trehalose utilization, we found that S66 phosphorylation is required to trigger the full utilization of trehalose during exit from stationary phase (Figure S4), a transition previously associated only with PKA regulation (Francois and Parrou, 2001; Schepers et al., 2012). We note that when stationary phase cells are exposed to an abrupt increase in glucose, trehalose utilization begins almost immediately, significantly before budding (Figure S4C), and budding is slightly delayed in nth1Δ, NTH1ΔCDK, and NTH1ΔPKA cells (Figure S4B), consistent with previous reports (Shi et al., 2010; Sillje et al., 1999). Thus it is likely that the high PKA activity associated with glucose addition (Broach, 2012; Conrad et al., 2014) is sufficient for partial activation of Nth1. However, the full activation of Nth1 still requires the Cdk1 phosphorylation site (Figure S4C). Thus, utilization of trehalose depends on phosphorylation of Nth1 on both Cdk1 and PKA sites.
Having shown that both Cdk1 and PKA are required for storage utilization, we next sought to identify which of these kinases is responsible for the timing of Nth1 activation at the G1/S transition. While it is well established that Cdk1 activity oscillates during the cell cycle (Morgan, 2007), there are also reports of cell-cycle dependent spikes in cyclic AMP, which could periodically activate PKA (Muller et al., 2003; Smith et al., 1990). To investigate the temporal dynamics of Nth1 phosphorylation, we released NTH1-3xFLAG cells from G1. The phosphorylation pattern of wild-type Nth1, but not of Nth1ΔCDK, changed during the cell cycle (Figure 4C, S4D). We quantified the fraction of highly phosphorylated wild type Nth1 following synchronous release from G1. The highly phosphorylated form, which most likely corresponds to the active form of the enzyme, increases approximately tenfold as cells enter the division cycle at G1/S (Figure 4D, S4D). At G1/S we also observed an increase of trehalase activity in cell extracts (Figure 4D). Thus, the G1/S transition triggers both increased Nth1 phosphorylation on S66 and trehalase activity.
Taken together, our data show that Nth1 is an integration point of PKA and Cdk1 signaling. While both PKA and Cdk1 phosphorylation are required for Nth1 activity, our data suggest that phosphorylation by PKA is constitutive in our constant growth conditions, while phosphorylation by Cdk1 oscillates with the cell cycle and is responsible for activating the trehalase activity downstream of Start.
Mutants deficient in carbohydrate storage change the cell cycle dynamics of central carbon metabolism
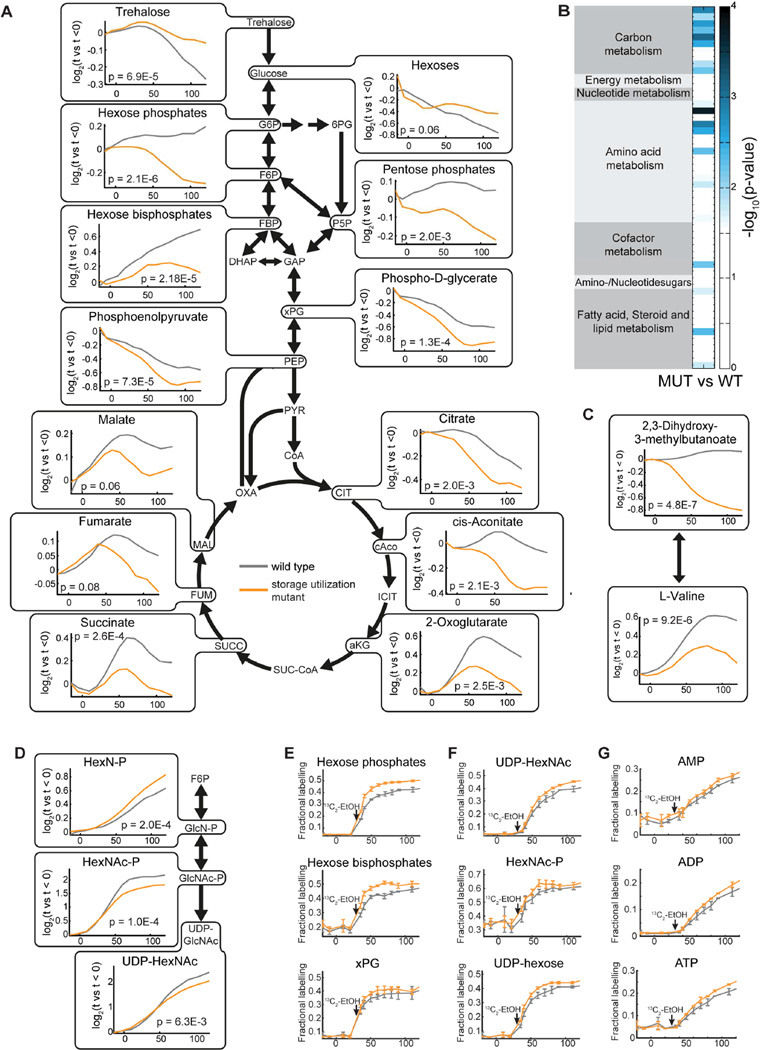
After determining the molecular mechanism triggering the storage carbohydrate switch, we next sought to investigate its function. Since storage carbohydrates are funneled into central carbon metabolism in a cell cycle-dependent manner, we hypothesized that there should be global cell cycle-dependent changes in the metabolome of cells unable to utilize storage carbohydrates. We therefore analyzed the metabolite dynamics in synchronously released nth1Δ nth2Δ gph1Δ cells in which trehalose and glycogen utilization is blocked. Compared to wild type cells, storage utilization mutant cells exhibited significant changes in the dynamics of central carbon metabolites and amino acid metabolism during cell cycle progression (p ≤ 0.01, repeated measure ANOVA). Most detected metabolites in central carbon metabolism were significantly lower in storage mutants than in wild type cells throughout the cell cycle, confirming that storage utilization funnels glucose into central carbon metabolism and most likely fuels glycolytic flux (Figure 5A–B). Moreover, we observed significant changes in the biosynthesis of valine, leucine, isoleucine, glycine, serine, and aminoglycans (Figure 5C–D, S5).
Figure 5. Cell cycle-dependent changes in metabolites in wild type and in carbohydrate storage utilization mutant cells.
(A) Changes in central carbon metabolism through the cell cycle in wild type and storage mutant cells. (B) Metabolic pathway enrichment analysis, comparing wild type and storage mutant cells through the cell cycle, identifies pathways with significant changes in metabolites. (C) Changes in valine biosynthesis, and (D) amino sugar metabolism through the cell cycle in wild type and storage mutant cells. Temporal profiles represent mean values of 2 individual biological and 2 technical replicates. The corresponding p-values are from a repeated measure corrected ANOVA comparing wild type and nth1Δnth2Δgph1Δ storage utilization mutants. (E–G) Fractional labeling of metabolites in (E) glycolysis (F) cell wall synthesis, and (G) nucleotides following addition of 13C ethanol to the media at 30 minutes after release. The addition of 13C ethanol resulted in 50 % of the ethanol in the media being labeled (see also Figure S6).
Next, we sought to investigate the relative contribution of trehalose utilization and ethanol uptake to central carbon metabolism and biosynthetic pathways. However, metabolite level dynamics are not sufficient to determine fluxes and origins of metabolites. Moreover, the highly dynamic nature of metabolism during the cell cycle prevents the determination of nutrient uptake rates or intracellular fluxes, which are not at steady state. We therefore performed a 13C carbon tracing experiment to examine the contribution of ethanol to the dynamic metabolite pools. Briefly, we traced labeled ethanol that was added to the media when cells start to bud (30 minutes after release), and metabolic differences become visible (Figure 5A–D). After 13C labeled ethanol addition, 50% of the ethanol in the media was isotopically labeled (Figure S6A). We then used untargeted and targeted metabolomics to measure kinetics of incorporation of 13C into metabolites (Table S5–6). Metabolites in glycolysis (Figure 5E, S6B) and cell wall synthesis (Figure 5F, S6E), and nucleotides (Figure 5G, S6E) had a higher fractional labeling in nth1Δ nth2Δ gph1Δ cells than in wild type cells, which indicates that the relative contribution of ethanol is increased in mutant cells during S/G2/M. This confirms that upon blockage of trehalose and glycogen utilization, intracellular fluxes are rearranged such that biosynthetic requirements are increasingly fueled by ethanol.
Interestingly, we could not detect tangible changes in the labeling kinetics of most amino acids. This suggests that the differences in cell cycle dynamics of concentrations in these pathways are not directly explained by fluxes from trehalose, but are due to more complex regulatory and compensatory mechanisms.
Trehalose utilization partially decouples cell cycle progression from nutrient availability
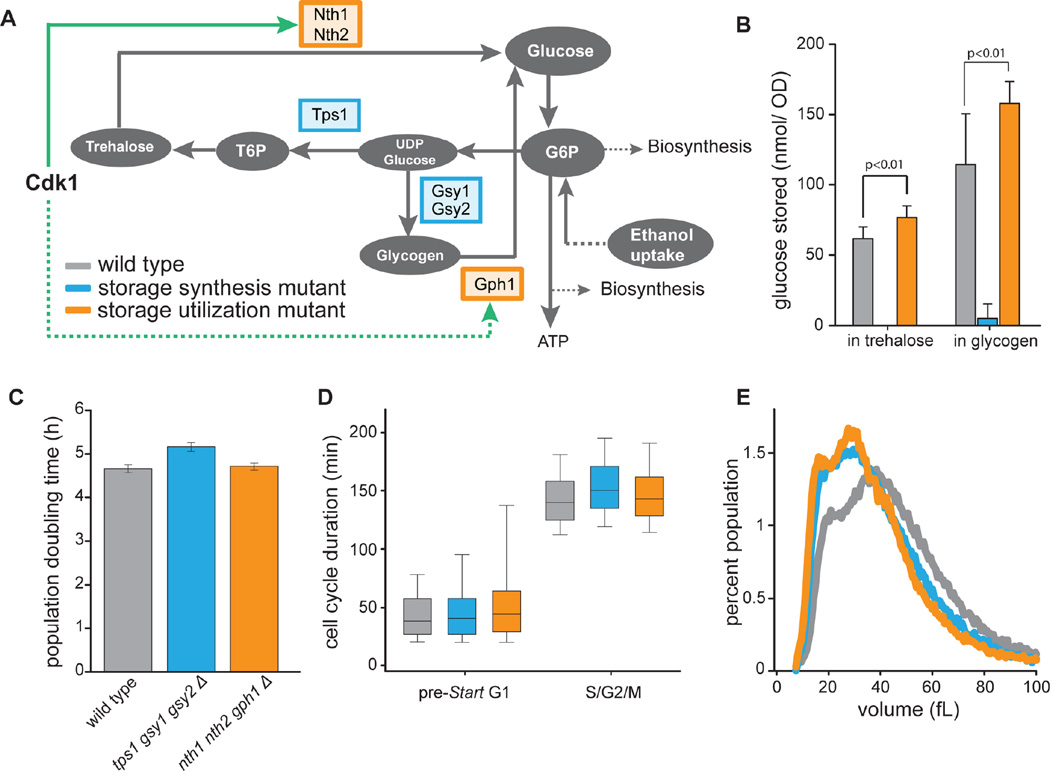
To understand the impact of altered metabolism on cell physiology in storage utilization mutants, we determined their growth rate in batch, and the duration of cell cycle phases in single cells during asynchronous exponential growth on ethanol. Growth rates as well as the relative duration of cell cycle phases of storage utilization mutants were very similar to wild type cells, consistent with the hypothesis that mutants are able to mostly compensate for the inability to turn over storage carbohydrates in these growth conditions (Figure 6). We observed a slightly wider distribution of G1 duration in storage utilization mutants. Since we did not find this phenotype in a storage synthesis mutant, we attribute this mostly to technical variation. Interestingly, we further observed that nth1Δ nth2Δ gph1Δ cells have a median size that is 10% smaller than wild type cells growing asynchronously on ethanol (Figure 6D). To further corroborate these results we also determined sizes of a mutant that cannot synthesize trehalose and glycogen (tps1Δ gsy1Δ gsy2Δ). This mutant was also smaller, indicating that storage turnover, and not the concentration of storage carbohydrates was responsible for this size phenotype (Figure 6A–D). This size phenotype suggested that even during exponential growth, when the contribution of trehalose and glycogen utilization to biosynthesis is rather small compared to ethanol uptake, the absence of storage utilization during S/G2/M cannot be completely compensated for.
Figure 6. Cell cycle kinetics for storage carbohydrate utilization and synthesis mutants.
(A) Schematic of carbohydrate storage metabolic pathways and their regulation by Cdk1. We examined mutants incapable of either synthesizing (tps1Δgsy1Δgsy2Δ; blue) or utilizing (nth1Δnth2Δgph1Δ; orange) storage carbohydrates. (B) Trehalose and glycogen content of asynchronous cultures. Error bars indicate standard deviations from 3 biological replicate experiments with 3 technical replicates each. (C) Population doubling times for wild type and mutants deficient in storage carbohydrate turnover. Error bars indicate standard error of the mean for 9 biological replicates. (D) Distribution of cell cycle phase durations for wild type (grey), storage synthesis mutants (blue) and storage utilization mutants (orange) measured in single cells. Using nuclear Whi5-GFP as a cell cycle marker, we determined the distribution of durations of pre-Start G1 (nuclear Whi5) and S/G2/M (cytoplasmic Whi5) for first generation mother cells. For each strain, at least 300 cells were analyzed. Bar and whiskers plot denotes 5th, 25th, median, 75th, and 95th percentiles. (E) Size distribution of cells growing asynchronously on ethanol. The distributions represent the mean of 3 biological replicates where ~50,000 cells were analyzed in a Coulter Counter.
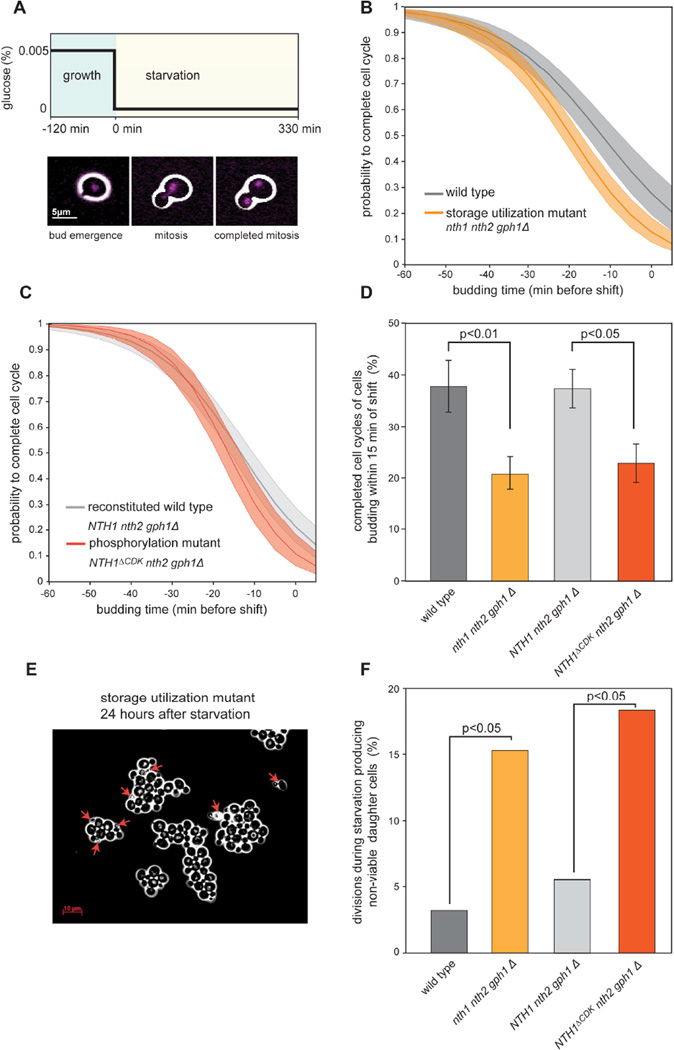
When nutrient supply decreases, the relative contribution of storage carbohydrates to biosynthesis necessarily increases. We therefore predicted that storage utilization mutants would be sensitive to nutrient downshifts specifically in the S/G2/M phases of the cell cycle. To test this hypothesis, we grew cells in a microfluidic imaging platform and subjected them to a sudden drop in carbon supply, which resulted in growth arrest (Figure 7A, S7, and Movie 1). Ten minutes after the carbon removal, neither wild type nor storage utilization mutant cells initiated budding (Figure S7), which is consistent with nutrients being sensed by the Start commitment point in late G1 (Turner et al., 2012). We examined whether cells that were in S/G2/M (budded) at the time of the shift were able to complete mitosis within the monitored 5.5 hours after the shift (separation of nuclei, Figure 7A and Movie 1). The longer a cell had been in S/G2/M before the shift, the higher its probability of completing mitosis (Figure 7B). Consistent with the hypothesis that storage utilization promotes completion of the cell cycle, utilization mutant cells had a significantly lower probability of completing mitosis than wild type cells (Figure 7B, D). Cells that did not complete mitosis generally arrested as budded, mono-nucleated cells. Whether these cells successfully completed S-phase and entered a controlled G2 arrest remains to be investigated. We also observed very few instances where cells attempted and failed at anaphase (partially separated nuclei) and these cells typically died shortly after.
Figure 7. Storage carbohydrate utilization partially decouples cell cycle progression from nutrient availability.
(A) Experiment schematic: HTB2-mCherry histone labeled cells were grown on a microfluidics imaging platform in 50 mg/l glucose minimal medium and subjected to a sudden drop to 0 mg/l glucose. Images were taken every 5 minutes starting at 2 hours before and ending 5.5 hours after the media shift at t=0 minutes (see also Movie 1). (B–C) Probability for a cell to complete the cell cycle as a function of its cell cycle position at the time of the nutrient shift. The probability estimate and its 95% confidence interval (shaded area) were estimated using logistic regression of data from >400 cells. (B) Data from 3 biological replicate experiments for wild type (grey) and storage utilization mutant (orange) cells. (C) Data from 4 biological replicate experiments for reconstituted wild type NTH1 nth1 gph1 cells (light grey) and NTH1ΔCDK cells (red). (D) Fraction of the cells that entered the cell cycle (budded) within 15 minutes of nutrient removal that were able to complete mitosis. Error bars are estimated standard deviations from 100 bootstrap iterations; p-values calculated using a Fisher’s exact test. (E) Image of storage utilization mutant cells 24 hours after being exposed to nutrient removal as described in (A). Red arrows indicate dead cells. (F) Fraction of divisions that lead to non-viable progeny. Cells were imaged for 12 hours after nutrient depletion and daughter cells were monitored after division. p-values were calculated with a Fisher’s exact test.
To confirm that wild type cells respond better to a drop in the nutrient supply due to their control of carbohydrate storage metabolism by Cdk1, we repeated the starvation experiment with a strain expressing wild type NTH1 and another strain expressing the phosphorylation site mutant allele NTH1ΔCDK. Indeed, NTH1ΔCDK cells were less likely to complete division following nutrient deprivation than NTH1 wild type cells, confirming that activation of Nth1 by Cdk1 phosphorylation can explain the observed phenotype (Figure 7C). For cells budding within 15 minutes before the starvation (which are in the middle of S-phase), NTH1 wild type cells were twice as likely as NTH1ΔCDK mutant cells to successfully complete mitosis (Figure 7D).
Furthermore, we noticed that mutant cell colonies were highly enriched for dead daughter cells 24 hours after the experiment (Figure 7E and S7B). We thus repeated the starvation experiments and monitored cells for a longer period (12 hours after the shift). We observed that utilization mutant and phosphorylation mutant cells caught in S/G2/M were four times as likely as wild type strains to produce non-viable progeny, where daughter cells died within an hour following division (Figure 7F). Notably, cells that were in G1 at the time of carbon removal were not affected in their short-term viability since we did not detect any dead unbudded cells twelve hours after starvation.
These observations show that Cdk1-triggered storage utilization can provide an essential carbon pool to fuel DNA replication and mitosis and thereby partially decouple cell cycle progression from nutrient supply.
Discussion
Here, we showed that yeast metabolism is globally coordinated with cell cycle progression. By using synthetic promoters to express cell cycle regulators, we were able to synchronize cells and control cell cycle progression without perturbing their environment and their metabolism. We show that over 50% of the metabolome changes during cell cycle progression downstream of Start, the commitment point that triggers the G1/S transition. This causally links global changes in metabolism to cell cycle progression.
In addition to showing that cell cycle progression drives metabolic changes, we identified a specific mechanism through which the cell cycle regulatory network controls carbon metabolism. The main cell cycle kinase Cdk1 directly phosphorylates and activates the trehalase Nth1 to trigger the flux of storage carbohydrates into central carbon metabolism. While Nth1 activity also requires the nutrient signaling kinase PKA1, the timing of activation coincides with phosphorylation of the Cdk1-site at the G1/S transition. Moreover, Cdk1 activity is likely regulating other enzymes of the storage carbohydrate pathways. The enzyme responsible for glycogen utilization, Gph1, is likely activated at the G1/S transition and also contains a Cdk1 consensus site detected in the Cdk1 target screen (Ubersax et al., 2003) (Figure 2). We also note that trehalose concentrations do not increase during S/G2/M in mutant cells unable to utilize trehalose (Figure 4B). This implies that trehalose synthesis had been shut off at G1/S, which could be due to Cdk1 phosphorylating sites on the regulatory subunit of the trehalose synthase Tsl1 (Table S2).
The requirement of both PKA and Cdk1 phosphorylation for Nth1 activity demonstrates that carbon storage metabolism integrates information from multiple signaling pathways. Mechanistically, this signal integration occurs through the unstructured N-terminal tail of Nth1. Phosphorylation by PKA enables the binding of the 14-3-3 protein Bmh1 to allow trehalose access to the catalytic site (Macakova et al., 2013). Since the Cdk1 site on Nth1 is close to the two most important PKA sites, we speculate that S66 phosphorylation further enhances Bmh1 binding to increase trehalase activity. Since the importance of phospho-regulation of metabolic enzymes is becoming increasingly clear (Oliveira et al., 2012; Schulz et al., 2014), we anticipate that combinatorial regulation by cell cycle and nutrient sensing kinases may be a widespread mechanism coordinating metabolism with cell cycle progression.
We suggest that the purpose of storage oscillations is to ensure a sufficient supply of carbon precursors for cell cycle-dependent biosynthesis of macromolecules in nutrient-poor environments. In support of this hypothesis, we showed that the ability to use the storage carbohydrate pool in S/G2/M phases of the cell cycle increases a cell’s chance of completing division and producing viable progeny when exposed to a deteriorating nutrient supply. In response to acute starvation, wild type cells are better able to complete cell division than mutants deficient in storage carbohydrate utilization. However, some of these storage mutant cells were still able to complete mitosis after starvation. This suggests the presence of additional carbon stores (such as glutamate or lipids) that may be accessed under extreme circumstances.
Our analysis of yeast responding to an acute removal of extracellular carbon is likely related to more physiological conditions such as growth on a finite amount of nutrients. In this case, yeast proliferate rapidly up to the depletion of extracellular nutrients. Eventually, cells reach stationary phase, where nearly all cells are arrested in G1 (Werner-Washburne et al., 1993). Yet, how do cells support proliferation until complete depletion of the nutrient supply without more cells entering stationary phase in S, G2, or M phases of the cell cycle? We propose that the ability of cells to draw on carbohydrate stores to complete their division cycle allows them to continue proliferation up to the point of nutrient depletion without getting caught in an incomplete cell division cycle.
Our findings may also offer a new perspective on the extensively studied metabolic cycling phenomenon observed in chemostat cultivations, in which yeast cultures spontaneously synchronize their metabolism, cell cycle and transcriptional activity (Burnetti et al., 2016; Slavov and Botstein, 2011; Tu et al., 2005). While our data does not explain the complex population dynamics in metabolically synchronized chemostats, we suspect that Cdk1 regulation of carbon metabolism might be one of the links between the metabolic and cell division cycles in these conditions as well.
Most generally, we identified major cell-cycle dependent changes throughout various anabolic and catabolic pathways. We therefore propose that the cell cycle control network regulates metabolism to meet the specific requirements of eukaryotic cell division. Consistent with this view, it was recently shown that the anaphase promoting complex regulates the stability of metabolic enzymes in HeLa cells (Colombo et al., 2011), and that CDK regulates mitochondrial activity in yeast (Harbauer et al., 2014) and mammalian cells (Wang et al., 2014) in a cell-cycle dependent manner. Since the oscillation in CDK activity is a fundamental feature of eukaryotic cell division, we suspect that phosphorylation of specific enzymes by CDK generally contributes to how both cancerous and non-cancerous mammalian cells establish the distinct metabolism associated with growth and proliferation (Buchakjian and Kornbluth, 2010; Fiske and Vander Heiden, 2012; Lunt and Vander Heiden, 2011).
Experimental Procedures
Strains and Media
Strains were constructed using standard methods and are prototrophic in a Saccharomyces cerevisiae W303 background. For exogenous control of gene expression we used the recently reported LexA-estrogen-receptor-activation-domain (Lex-ER-AD) system (Ottoz et al., 2014). Minimal media with 1% ethanol or the specified amount of glucose was used throughout, except for Figure 2E–H and S2, which were performed on 1% ethanol synthetic complete medium. See Table S3 for strain details.
Cell cycle synchronization
Two sequential precultures were grown on media containing first 10 nM and then 5 nM β-estradiol (Sigma). LexApr-CDC20 strains were arrested for 6 hours, and LexApr-CLN1 strains were arrested for 15 hours in estradiol-free media. Cells were released by addition of 200 nM estradiol.
Metabolite measurements
Cells were harvested by filtration and metabolites were extracted in 70% ethanol as described in (Link et al., 2013). Metabolomics samples were analyzed by flow-injection analysis on an Agilent 6550 Q-TOF (Agilent, Santa Clara, CA) using the settings described in (Fuhrer et al., 2011). Ions were annotated to known metabolites using the KEGG sce database for S. cerevisiae (Kanehisa and Goto, 2000). If not stated otherwise, ion intensities were normalized by a univariate comparison between each individual measurement and the intensities of the 3 time points prior to release from G1 with foldchange = (log2(intensities(t)/intensities(t<0)) using a two-sample t-test, where p-values were corrected for false discovery rate as described in (Storey, 2002). Metabolic pathway enrichment analysis was performed using a hypergeometric test on significantly changing ions (p-value < 0.01, |log2(foldchange)| > 0.2) of this univariate comparison. To compare metabolite profiles of wild type and storage degradation mutants, we applied a repeated measure ANOVA and corrected the p-values as above for false discovery. For plotting, a 3-point moving average filter was applied on time profiles of metabolites. All metabolomics data analysis was performed using custom Matlab software (The Mathworks, Natick).
13C labeling experiments
30 minutes after release from G1 arrest, [U-13C] ethanol (Cambridge Isotope Laboratories, Andover, MA) was added to reach a ratio of 50% naturally labeled to 50% fully labeled ethanol. Metabolites were extracted as described above. For untargeted analysis, samples were measured and ions were annotated as above. Targeted metabolomics analysis was performed using ultra high-pressure chromatography-tandem mass spectrometry as in (Buescher et al., 2010; Ruhl et al., 2012).
Trehalose and glycogen measurements
Cells were harvested by centrifugation and frozen in liquid nitrogen. Trehalose was extracted using boiling water. Trehalose concentrations were determined using a commercial kit (Megazyme). The glycogen extraction protocol was modified from (Muller et al., 2003). Briefly, cells were lysed in sodium carbonate and the glycogen digested overnight using amyloglucosidase. Glucose released from glycogen digestion was measured using a commercial kit (Megazyme). We normalized the amounts measured at each time point to the amount measured in the sample at t = 0 minutes. Absolute values (µmol/OD unit) are reported in Table S1.
Imaging and image analysis
Images were recorded on a Zeiss Axio-Observer Z1 with an automated stage. For the experiments described in Figure 7, cells were imaged on a commercial microfluidic system (CellAsic, EMD Millipore). For the single cell measurements described in Figures 6 and S1, cells were grown on 1.5 % agar patches containing 1% ethanol. Images were processed and cells segmented as described in (Doncic et al., 2013).
DNA content
Cells were stained with SYBR Green I and analyzed on a flow cytometer.
In vitro phosphorylation assay
N-terminally 6xHis-tagged recombinant Nth1 constructs were expressed in E. coli and purified by cobalt affinity chromatography. In vitro phosphorylation assays using purified Clb2/Cdk1/Cks1 complexes were performed as described in (Koivomagi et al., 2011). Figure 3B corresponds to an 8-minute reaction.
Phos-tag SDS-PAGE and Western analysis
Cells were lysed in urea buffer using a bead beater. The phosphorylated species of Nth1-3xFlag were resolved in 8% SDS-polyacrylamide gels supplemented with 100 µM Phos-tag reagent (FMS Laboratory, NARD institute, Japan). Nth1-3xFLAG was detected with ANTI-FLAG M2 antibody (Sigma) and Alexa Fluor 680 goat anti-mouse antibody (Life Technologies).
Trehalase activity assay
The trehalase assay was adapted from previously published protocols (Muller et al., 2003; Pernambuco et al., 1996; Uno et al., 1983; Veisova et al., 2012). Briefly, cells were lysed under native conditions. 20 µl extract were added to the assay buffer with trehalose to start the reaction. To stop the reaction, samples were boiled for 3 minutes. The glucose produced was measured using a commercial enzyme kit (Megazymes).
See also Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Cell cycle progression triggers global changes in metabolism
The cell cycle kinase Cdk1 activates the trehalase Nth1 at the G1/S transition
Active Nth1 releases trehalose to fuel biosynthesis during S/G2/M
Trehalose utilization enables cell cycle progression even during acute starvation
Acknowledgments
We thank Fabian Rudolf and Joerg Stelling for Lex-ER-AD constructs, Mart Loog for Clb2/Cdk1/Cks1 complexes, Mardo Koivomaegi for help with in vitro phosphorylation work, Andreas Doncic for help with image analysis, and Bruce Futcher for insightful discussions and inspiring this study with his essay (Futcher, 2006). This work was supported by the Swiss National Science Foundation (PBEZE3-134441 to JE), SystemsX.ch (IPhD grant to AK), a Burroughs Wellcome Fund Career Award at the Scientific Interface (JMS), the NIH (JS and JE, R21 DK095237) and the NSF (CAREER award to JS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: Conceptualization, J.C.E. and J.M.S.; Methodology, J.C.E., A.K., and N.Z.; Investigation, J.C.E. and A.K.; Writing – Original Draft, J.C.E. and J.M.S; Writing – Review & Editing, J.C.E., J.M.S., A.K., and N.Z.; Funding Acquisition, J.C.E., J.M.S., and N.Z.; Supervision, J.M.S. and N.Z.
References
- Boer VM, Crutchfield CA, Bradley PH, Botstein D, Rabinowitz JD. Growth-limiting intracellular metabolites in yeast growing under diverse nutrient limitations. Mol Biol Cell. 2010;21:198–211. doi: 10.1091/mbc.E09-07-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach JR. Nutritional control of growth and development in yeast. Genetics. 2012;192:73–105. doi: 10.1534/genetics.111.135731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchakjian MR, Kornbluth S. The engine driving the ship: metabolic steering of cell proliferation and death. Nat Rev Mol Cell Biol. 2010;11:715–727. doi: 10.1038/nrm2972. [DOI] [PubMed] [Google Scholar]
- Buescher JM, Moco S, Sauer U, Zamboni N. Ultrahigh performance liquid chromatography-tandem mass spectrometry method for fast and robust quantification of anionic and aromatic metabolites. Anal Chem. 2010;82:4403–4412. doi: 10.1021/ac100101d. [DOI] [PubMed] [Google Scholar]
- Burnetti AJ, Aydin M, Buchler NE. Cell cycle Start is coupled to entry into the yeast metabolic cycle across diverse strains and growth rates. Mol Biol Cell. 2016;27:64–74. doi: 10.1091/mbc.E15-07-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canelas AB, Harrison N, Fazio A, Zhang J, Pitkanen JP, van den Brink J, Bakker BM, Bogner L, Bouwman J, Castrillo JI, et al. Integrated multilaboratory systems biology reveals differences in protein metabolism between two reference yeast strains. Nat Commun. 2010;1:145. doi: 10.1038/ncomms1150. [DOI] [PubMed] [Google Scholar]
- Castrillo JI, Zeef LA, Hoyle DC, Zhang N, Hayes A, Gardner DCJ, Cornell MJ, Petty J, Hakes L, Wardleworth L, et al. Growth control of the eukaryote cell: A systems biology study in yeast. J Biol. 2007;6:4. doi: 10.1186/jbiol54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo SL, Palacios-Callender M, Frakich N, Carcamo S, Kovacs I, Tudzarova S, Moncada S. Molecular basis for the differential use of glucose and glutamine in cell proliferation as revealed by synchronized HeLa cells. P Natl Acad Sci USA. 2011;108:21069–21074. doi: 10.1073/pnas.1117500108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M, Schothorst J, Kankipati HN, Van Zeebroeck G, Rubio-Texeira M, Thevelein JM. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 2014;38:254–299. doi: 10.1111/1574-6976.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doncic A, Eser U, Atay O, Skotheim JM. An Algorithm to Automate Yeast Segmentation and Tracking. PLoS One. 2013;8 doi: 10.1371/journal.pone.0057970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doncic A, Falleur-Fettig M, Skotheim JM. Distinct interactions select and maintain a specific cell fate. Mol Cell. 2011;43:528–539. doi: 10.1016/j.molcel.2011.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald JC, Matta T, Zamboni N. The integrated response of primary metabolites to gene deletions and the environment. Mol Biosyst. 2013;9:440–446. doi: 10.1039/c2mb25423a. [DOI] [PubMed] [Google Scholar]
- Fiske BP, Vander Heiden MG. Seeing the Warburg effect in the developing retina. Nat Cell Biol. 2012;14:790–791. doi: 10.1038/ncb2554. [DOI] [PubMed] [Google Scholar]
- Francois J, Parrou JL. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 2001;25:125–145. doi: 10.1111/j.1574-6976.2001.tb00574.x. [DOI] [PubMed] [Google Scholar]
- Fuhrer T, Heer D, Begemann B, Zamboni N. High-Throughput, Accurate Mass Metabolome Profiling of Cellular Extracts by Flow Injection - Time-of-Flight Mass Spectrometry. Analytical Chemistry. 2011;83:7074–7080. doi: 10.1021/ac201267k. [DOI] [PubMed] [Google Scholar]
- Futcher B. Metabolic cycle, cell cycle, and the finishing kick to Start. Genome Biol. 2006;7:107. doi: 10.1186/gb-2006-7-4-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher B. TgI4 lipase: a big fat target for cell-cycle entry. Mol Cell. 2009;33:143–144. doi: 10.1016/j.molcel.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Grandin N, Reed SI. Differential function and expression of Saccharomyces cerevisiae B-type cyclins in mitosis and meiosis. Mol Cell Biol. 1993;13:2113–2125. doi: 10.1128/mcb.13.4.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteridge A, Pir P, Castrillo JI, Charles PD, Lilley KS, Oliver SG. Nutrient control of eukaryote cell growth: a systems biology study in yeast. BMC Biol. 2010;8:68. doi: 10.1186/1741-7007-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase SB, Reed SI. Evidence that a free-running oscillator drives G1 events in the budding yeast cell cycle. Nature. 1999;401:394–397. doi: 10.1038/43927. [DOI] [PubMed] [Google Scholar]
- Harbauer AB, Opalinska M, Gerbeth C, Herman JS, Rao S, Schonfisch B, Guiard B, Schmidt O, Pfanner N, Meisinger C. Mitochondria. Cell cycle-dependent regulation of mitochondrial preprotein translocase. Science. 2014;346:1109–1113. doi: 10.1126/science.1261253. [DOI] [PubMed] [Google Scholar]
- Hofer A, Crona M, Logan DT, Sjoberg BM. DNA building blocks: keeping control of manufacture. Crit Rev Biochem Mol Biol. 2012;47:50–63. doi: 10.3109/10409238.2011.630372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A, Skotheim JM. Start and the restriction point. Curr Opin Cell Biol. 2013;25:717–723. doi: 10.1016/j.ceb.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- Klevecz RR, Bolen J, Forrest G, Murray DB. A genomewide oscillation in transcription gates DNA replication and cell cycle. Proc Natl Acad Sci U S A. 2004;101:1200–1205. doi: 10.1073/pnas.0306490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivomagi M, Valk E, Venta R, Iofik A, Lepiku M, Morgan DO, Loog M. Dynamics of Cdk1 Substrate Specificity during the Cell Cycle. Molecular Cell. 2011;42:610–623. doi: 10.1016/j.molcel.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuenzi MT, Fiechter A. Changes in Carbohydrate Composition and Trehalase-Activity during Budding Cycle of Saccharomyces Cerevisiae. Arch Mikrobiol. 1969;64:396-&. doi: 10.1007/BF00417021. [DOI] [PubMed] [Google Scholar]
- Kuenzi MT, Fiechter A. Regulation of Carbohydrate Composition of Saccharomyces-Cerevisiae under Growth Limitation. Arch Mikrobiol. 1972;84:254-&. doi: 10.1007/BF00425203. [DOI] [PubMed] [Google Scholar]
- Kurat CF, Wolinski H, Petschnigg J, Kaluarachchi S, Andrews B, Natter K, Kohlwein SD. Cdk1/Cdc28-dependent activation of the major triacylglycerol lipase Tgl4 in yeast links lipolysis to cell-cycle progression. Mol Cell. 2009;33:53–63. doi: 10.1016/j.molcel.2008.12.019. [DOI] [PubMed] [Google Scholar]
- Link H, Kochanowski K, Sauer U. Systematic identification of allosteric protein-metabolite interactions that control enzyme activity in vivo. Nat Biotechnol. 2013;31:357–361. doi: 10.1038/nbt.2489. [DOI] [PubMed] [Google Scholar]
- Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- Macakova E, Kopecka M, Kukacka Z, Veisova D, Novak P, Man P, Obsil T, Obsilova V. Structural basis of the 14-3-3 protein-dependent activation of yeast neutral trehalase Nth1. Biochim Biophys Acta. 2013;1830:4491–4499. doi: 10.1016/j.bbagen.2013.05.025. [DOI] [PubMed] [Google Scholar]
- Mathews CK. Deoxyribonucleotides as genetic and metabolic regulators. Faseb J. 2014;28:3832–3840. doi: 10.1096/fj.14-251249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIsaac RS, Gibney PA, Chandran SS, Benjamin KR, Botstein D. Synthetic biology tools for programming gene expression without nutritional perturbations in Saccharomyces cerevisiae. Nucleic Acids Res. 2014;42:e48. doi: 10.1093/nar/gkt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO, editor. The Cell Cycle: Principles of Control. London: New Science Press; 2007. [Google Scholar]
- Muller D, Exler S, Aguilera-Vazquez L, Guerrero-Martin E, Reuss M. Cyclic AMP mediates the cell cycle dynamics of energy metabolism in Saccharomyces cerevisiae. Yeast. 2003;20:351–367. doi: 10.1002/yea.967. [DOI] [PubMed] [Google Scholar]
- Oliveira AP, Ludwig C, Picotti P, Kogadeeva M, Aebersold R, Sauer U. Regulation of yeast central metabolism by enzyme phosphorylation. Mol Syst Biol. 2012;8:623. doi: 10.1038/msb.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottoz DS, Rudolf F, Stelling J. Inducible, tightly regulated and growth condition-independent transcription factor in Saccharomyces cerevisiae. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paalman JW, Verwaal R, Slofstra SH, Verkleij AJ, Boonstra J, Verrips CT. Trehalose and glycogen accumulation is related to the duration of the G1 phase of Saccharomyces cerevisiae. FEMS Yeast Res. 2003;3:261–268. doi: 10.1111/j.1567-1364.2003.tb00168.x. [DOI] [PubMed] [Google Scholar]
- Panni S, Landgraf C, Volkmer-Engert R, Cesareni G, Castagnoli L. Role of 14-3-3 proteins in the regulation of neutral trehalase in the yeast Saccharomyces cerevisiae. FEMS Yeast Res. 2008;8:53–63. doi: 10.1111/j.1567-1364.2007.00312.x. [DOI] [PubMed] [Google Scholar]
- Pernambuco MB, Winderickx J, Crauwels M, Griffioen G, Mager WH, Thevelein JM. Glucose-triggered signalling in Saccharomyces cerevisiae: different requirements for sugar phosphorylation between cells grown on glucose and those grown on non-fermentable carbon sources. Microbiology. 1996;142(Pt 7):1775–1782. doi: 10.1099/13500872-142-7-1775. [DOI] [PubMed] [Google Scholar]
- Richardson HE, Wittenberg C, Cross F, Reed SI. An essential G1 function for cyclin-like proteins in yeast. Cell. 1989;59:1127–1133. doi: 10.1016/0092-8674(89)90768-x. [DOI] [PubMed] [Google Scholar]
- Ruhl M, Rupp B, Noh K, Wiechert W, Sauer U, Zamboni N. Collisional fragmentation of central carbon metabolites in LC-MS/MS increases precision of 13C metabolic flux analysis. Biotechnology and Bioengineering. 2012;109:763–771. doi: 10.1002/bit.24344. [DOI] [PubMed] [Google Scholar]
- Schepers W, Van Zeebroeck G, Pinkse M, Verhaert P, Thevelein JM. In vivo phosphorylation of Ser21 and Ser83 during nutrient-induced activation of the yeast protein kinase A (PKA) target trehalase. J Biol Chem. 2012;287:44130–44142. doi: 10.1074/jbc.M112.421503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz JC, Zampieri M, Wanka S, von Mering C, Sauer U. Large-scale functional analysis of the roles of phosphorylation in yeast metabolic pathways. Sci Signal. 2014;7:rs6. doi: 10.1126/scisignal.2005602. [DOI] [PubMed] [Google Scholar]
- Shi L, Sutter BM, Ye X, Tu BP. Trehalose is a key determinant of the quiescent metabolic state that fuels cell cycle progression upon return to growth. Mol Biol Cell. 2010;21:1982–1990. doi: 10.1091/mbc.E10-01-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillje HH, Paalman JW, ter Schure EG, Olsthoorn SQ, Verkleij AJ, Boonstra J, Verrips CT. Function of trehalose and glycogen in cell cycle progression and cell viability in Saccharomyces cerevisiae. J Bacteriol. 1999;181:396–400. doi: 10.1128/jb.181.2.396-400.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillje HH, ter Schure EG, Rommens AJ, Huls PG, Woldringh CL, Verkleij AJ, Boonstra J, Verrips CT. Effects of different carbon fluxes on G1 phase duration, cyclin expression, and reserve carbohydrate metabolism in Saccharomyces cerevisiae. J Bacteriol. 1997;179:6560–6565. doi: 10.1128/jb.179.21.6560-6565.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotheim JM, Di Talia S, Siggia ED, Cross FR. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature. 2008;454:291–296. doi: 10.1038/nature07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavov N, Botstein D. Coupling among growth rate response, metabolic cycle, and cell division cycle in yeast. Mol Biol Cell. 2011;22:1997–2009. doi: 10.1091/mbc.E11-02-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavov N, Macinskas J, Caudy A, Botstein D. Metabolic cycling without cell division cycling in respiring yeast. Proc Natl Acad Sci U S A. 2011;108:19090–19095. doi: 10.1073/pnas.1116998108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ME, Dickinson JR, Wheals AE. Intracellular and extracellular levels of cyclic AMP during the cell cycle of Saccharomyces cerevisiae. Yeast. 1990;6:53–60. doi: 10.1002/yea.320060106. [DOI] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. J Roy Stat Soc B. 2002;64:479–498. [Google Scholar]
- Strassburg K, Walther D, Takahashi H, Kanaya S, Kopka J. Dynamic transcriptional and metabolic responses in yeast adapting to temperature stress. OMICS. 2010;14:249–259. doi: 10.1089/omi.2009.0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia H, Koshland DE. Trehalose is a versatile and long-lived chaperone for desiccation tolerance. Curr Biol. 2014;24:2758–2766. doi: 10.1016/j.cub.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Tapia H, Young L, Fox D, Bertozzi CR, Koshland D. Increasing intracellular trehalose is sufficient to confer desiccation tolerance to Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2015;112:6122–6127. doi: 10.1073/pnas.1506415112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP. Methods in Enzymology. Elsevier Inc; 2010. Ultradian Metabolic Cycles in Yeast; pp. 857–865. [DOI] [PubMed] [Google Scholar]
- Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. Cyclic changes in metabolic state during the life of a yeast cell. Proc Natl Acad Sci U S A. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JJ, Ewald JC, Skotheim JM. Cell size control in yeast. Curr Biol. 2012;22:R350–R359. doi: 10.1016/j.cub.2012.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- Uno I, Matsumoto K, Adachi K, Ishikawa T. Genetic and biochemical evidence that trehalase is a substrate of cAMP-dependent protein kinase in yeast. J Biol Chem. 1983;258:10867–10872. [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandoorn J, Valkenburg JAC, Scholte ME, Oehlen LJWM, Vandriel R, Postma PW, Nanninga N, Vandam K. Changes in Activities of Several Enzymes Involved in Carbohydrate-Metabolism during the Cell-Cycle of Saccharomyces-Cerevisiae. Journal of Bacteriology. 1988;170:4808–4815. doi: 10.1128/jb.170.10.4808-4815.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veisova D, Macakova E, Rezabkova L, Sulc M, Vacha P, Sychrova H, Obsil T, Obsilova V. Role of individual phosphorylation sites for the 14-3-3-protein-dependent activation of yeast neutral trehalase Nth1. Biochem J. 2012;443:663–670. doi: 10.1042/BJ20111615. [DOI] [PubMed] [Google Scholar]
- Wang X, Proud CG. Nutrient control of TORC1, a cell-cycle regulator. Trends Cell Biol. 2009;19:260–267. doi: 10.1016/j.tcb.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Wang Z, Fan M, Candas D, Zhang TQ, Qin L, Eldridge A, Wachsmann-Hogiu S, Ahmed KM, Chromy BA, Nantajit D, et al. Cyclin B1/Cdk1 coordinates mitochondrial respiration for cell-cycle G2/M progression. Dev Cell. 2014;29:217–232. doi: 10.1016/j.devcel.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Washburne M, Braun E, Johnston GC, Singer RA. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol Rev. 1993;57:383–401. doi: 10.1128/mr.57.2.383-401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann C, Hans M, van Winden WA, Ras C, Heijnen JJ. Dynamics of intracellular metabolites of glycolysis and TCA cycle during cell-cycle-related oscillation in Saccharomyces cerevisiae. Biotechnol Bioeng. 2005;89:839–847. doi: 10.1002/bit.20408. [DOI] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.