Abstract
The aberrant activity of Ras homologous (Rho) family small GTPases (20 human members) has been implicated in cancer and other human diseases. However, in contrast to the direct mutational activation of Ras found in cancer and developmental disorders, Rho GTPases are activated most commonly by indirect mechanisms in disease. One prevalent mechanism involves aberrant Rho activation via the deregulated expression and/or activity of Rho family guanine nucleotide exchange factors (RhoGEFs). RhoGEFs promote formation of the active GTP-bound state of Rho GTPases. The largest family of RhoGEFs is comprised of the Dbl family RhoGEFs with 70 human members. The multitude of RhoGEFs that activate a single Rho GTPase reflect the very specific role of each RhoGEF in controlling distinct signaling mechanisms involved in Rho activation. In this review, we summarize the role of Dbl RhoGEFs in development and disease, with a focus on Ect2, Tiam1, Vav and P-Rex1/2.
Keywords: Rac1, RhoA, Cdc42, guanine nucleotide exchange factors, cancer, mouse models
Introduction
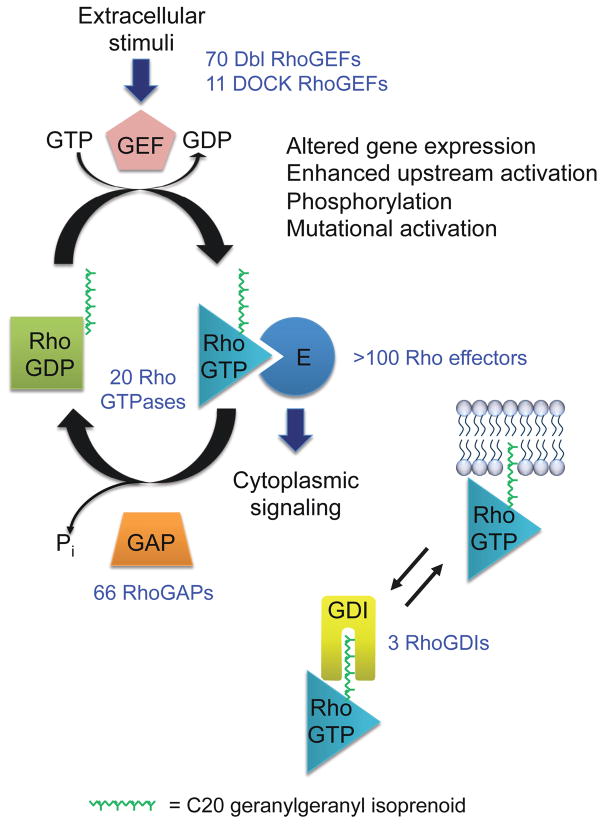
Ras homologous (Rho) family proteins (20 human members) comprise a major branch of the Ras superfamily of small GTPases,1 with RhoA, Rac1 and Cdc42 the most extensively studied and characterized.2 Rho GTPases specifically regulate actin organization, cell motility, polarity, growth, survival and gene transcription.3 Rho GTPases are binary switches that cycle between an active GTP-bound and an inactive GDP-bound state (Figure 1).4 Rho guanine nucleotide exchange factors (RhoGEFs) accelerate the intrinsic exchange activity of Rho GTPases to stimulate formation of Rho-GTP.5 Rho GTPase activating proteins (RhoGAPs) stimulate the intrinsic GTP hydrolysis activity of Rho GTPases, resulting in the inactive GDP-bound state. Rho guanine nucleotide-dissociation inhibitors (GDIs) comprise a third class of regulatory proteins.6 RhoGDIs bind Rho GTPases in a GDP-bound state and also extract and sequester Rho GTPases from the cell membrane where Rho GTPases can be activated. Once activated, the GTP-bound GTPase then associates with effector targets that number over 100.7–9 In this review, we focus on the classical Dbl family RhoGEFs and their role in development and disease. In particular, we focus on the RhoGEFs with the best validated roles in cancer (Vav1/2/3, Ect2, Tiam1/2, P-Rex1/2) and their analyses in cell culture, mouse models and human cancers.
Figure 1. Rho GTPase regulation.
The human Rho GTPase family is comprised of 20 members. The majority cycle between GDP-bound inactive and GTP-bound active states. Unlike RhoGEFs and RhoGAPs that possess shared domains and sequence identity, allowing precise determination of the number encoded in the human genome, the majority of Rho GTPase effectors lack a common well-defined recognition domain/motif. The information here applies mainly to RhoA, Rac1, Cdc42 and related isoforms. Not all Rho GTPases are regulated by GEFs, GAPs and/or GDIs, not all are posttranslationally modified by a geranylgeranyl isoprenoid lipid, and some do not have known membrane targeting elements.
The discovery of RhoGEFs: diverse in numbers and structure
The first RhoGEF was identified initially as an oncogene in mammalian cells (Table 1 and Figure 2). Using the same NIH 3T3 mouse fibroblast focus formation assay that led to the discovery of mutant Ras in human cancer,10 analysis of genomic DNA isolated from a human diffuse B-cell lymphoma resulted in the discovery of the DBL oncogene that encoded an N-terminally truncated and activated protein.11 Dbl was also detected earlier as an oncogene that caused the tumorigenic growth of NIH 3T3 cells after transfection with genomic DNA from the MCF-7 human breast carcinoma cell line (designated mcf2)12, and only later was it determined to be identical to Dbl.13 Dbl was subsequently shown to catalyze the GTP/GDP exchange activity of Cdc42.14 Additional NIH 3T3 focus formation and related biological assays identified Dbl-related proteins, in particular Vav, Tiam1 and Ect2 (Table 1 and Figure 2). That RhoGEFs were discovered initially as oncoproteins provided the first suggestion that Rho GTPases may also have a function in oncogenesis.
Table 1.
Discovery of Dbl RhoGEFs as oncogenes
| Gene | Description | References |
|---|---|---|
| Dbl/ Mcf2 | Identified as an oncogene that caused NIH 3T3 nude mouse tumorigenic growth after transfection of genomic DNA from the MCF-7 human breast carcinoma cell line (designated mcf2); oncogene in an NIH 3T3 focus formation assay using genomic DNA from a primary human diffuse B-cell leukemia; identified in an NIH 3T3 focus formation screen using genomic DNA from a human nodular poorly differentiated lymphoma | 11, 12, 83 |
| Vav | Identified as an oncogene that caused NIH 3T3 nude mouse tumorigenic growth after transfection by genomic DNA from a human esophageal carcinoma. Since this represented the sixth oncogene identified from this group, it was given the name of the sixth letter of the Hebrew alphabet. | 63 |
| Ect2 | Identified as an oncogene in an NIH 3T3 focus formation assay using a cDNA expression library from the Balb/MK mouse keratinocyte cell line; designated epithelial cell transforming cDNA 2 | 39, 176 |
| Tim | Identified as an oncogene in an NIH 3T3 focus formation assay using a cDNA expression library from a human mammary epithelial cell line; designated transforming immortalized mammary oncogene | 177 |
| Net1 | Identified as an oncogene in an NIH 3T3 focus formation assay using a cDNA expression library from a human neuroepithelioma cell line; designed neuroepithelial cell transforming gene 1 | 178 |
| Lbc | Identified as an oncogene that caused NIH 3T3 nude mouse tumorigenic growth after transfection by genomic DNA from an acute phase CML; designated lymphoid blast crisis oncogene | 179 |
| Lfc | Identified as a transforming gene in an NIH 3T3 focus formation assay in cDNA expression library from the 32D mouse hematopoietic cell line; designated Lbc’s first cousin | 180 |
| Tiam1 | Identified by retrovirus insertion for genes that induced T cell lymphoma in vitro; designated T cell lymphoma invasion and metastasis 1 | 51 |
| Ost/ Dbs | Identified as a transforming gene in an NIH 3T3 focus formation assay using a cDNA expression library generated from the rat osteosarcoma cell line ROS; identified as a transforming gene in an NIH 3T3 focus formation assay in cDNA expression library from the mouse hematopoietic cell line 32D; designated Dbl’s big sister | 181, 182 |
| Lsc/ Lip | Identified as a transforming gene in an NIH 3T3 focus formation assay using a cDNA expression library generated from the murine myeloid progenitor cell line B6SUtA1; designated Lbc’s second cousin; identified as a transforming gene in an NIH 3T3 focus formation assay in genomic DNA from a pleiomorphic liposarcoma | 177, 183 |
| Clg | Identified as a common-site lymphoma/leukemia guanine nucleotide exchange factor proviral integration site in mouse leukemias | 184 |
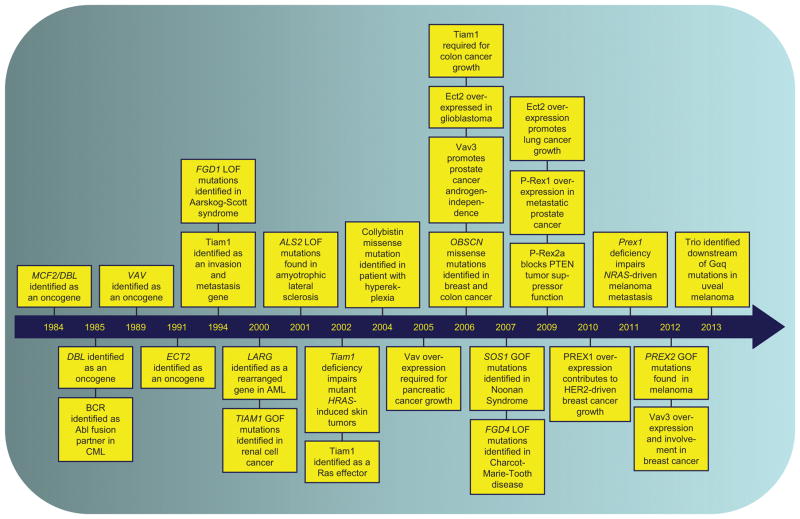
Figure 2. Key discoveries linking Dbl RhoGEFs to human disease.
We highlight representative discoveries in the study of RhoGEFs that link their aberrant function to human disease. Gain-of-function, GOF; loss-of-function, LOF.
Dbl and the Dbl-related proteins share a ~200 amino acid catalytic Dbl homology (DH; also called RhoGEF) domain and an immediately adjacent regulatory ~100 amino acid pleckstrin homology (PH) domain5 (Figure 3). Additional RhoGEFs with this tandem DH-PH domain structure were identified by genetic and biochemical approaches and by in silico database searches. There are 70 human Dbl family RhoGEFs, many of which have conserved orthologs found in all vertebrate species and in invertebrates, including Drosophila, C. elegans, S. cerevisiae and S. pombe. The nomenclature for many Dbl RhoGEFs is complicated by the different names used in independent discoveries, by different names for orthologs of different species, by the existence of different gene products due to alternative RNA splicing and by establishment of the ARHGEF gene family nomenclature by the HUGO Gene Nomenclature Committee (http://www.genenames.org/genefamilies/ARHGEF). We have compiled a summary of Dbl RhoGEF nomenclature, based on the names most commonly used in the literature together with the additional names used for each RhoGEF (Supplementary Table 1).
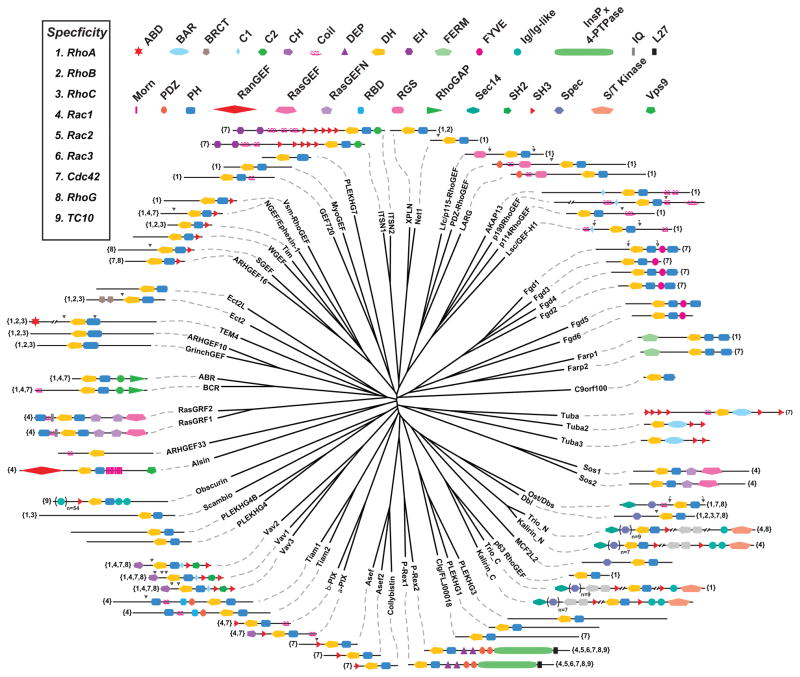
Figure 3. The human Dbl RhoGEF family.
The 72 DH domains found within the 70 members of human Dbl family RhoGEFs were aligned using ClustalX174 and a phylogenetic tree was subsequently drawn using the program FigTree (http://tree.bio.ed.ac.uk/software/figtree/). The protein domain architecture for each full-length Dbl protein is shown as annotated by SMART (http://smart.embl.de/) or described in the published literature. Black arrowheads indicate sites of genetic truncation known to activate RhoGEF function, while bracketed arrowheads designate biologically or catalytically active fragments of a GEF. Numbers within braces next to a Dbl protein indicate the reported Rho GTPase specificity of the DH domain. Please refer to Supplementary Table 1 for other names utilized for these RhoGEFs.
In addition to their common structural elements that define them as RhoGEFs, there are some distinctive features that mark individual RhoGEF proteins. Two RhoGEFs possess tandem sets of DH-PH domains (Trio and Kalirin) and four RhoGEFs (Tuba1, 2, 3 and ARHGEF33) lack an apparent PH domain (Figure 3).5 Dbl RhoGEFs also diverge significantly in the N- and C-terminal sequences flanking the DH/PH domains. These flanking sequences commonly contain a diversity of protein-protein or protein-lipid interaction domains and motifs whose functions are to regulate intrinsic RhoGEF catalytic activity, determine subcellular localization and/or facilitate complex formation with other proteins (Figure 3).
This diversity in flanking non-RhoGEF sequences distinguishes the regulation and role of RhoGEFs that otherwise activate the same set of Rho GTPases. For example, extracellular stimuli that first activate RhoGEFs downstream of G-protein coupled receptors (GPCRs) and subsequently cause activation of G protein beta-gamma subunits (P-Rex), of Ras (Tiam1), or of Src family tyrosine kinases (Vav) can all converge on downstream Rac1 activation through distinct RhoGEFs (Figure 4). These flanking sequences may also regulate subcellular localization and activation of spatially distinct cellular pools of Rho GTPases, leading to their utilization of distinct effectors. Finally, these flanking sequences may facilitate scaffolding functions for RhoGEFs that can further influence the effectors activated. For example, Tiam1 possesses a PH-CC-Ex globular domain15 that acts as a membrane targeting and protein-protein interaction domain.16–19 This scaffolding function can then influence the effector utilization of the activated Rho GTPase.20,21
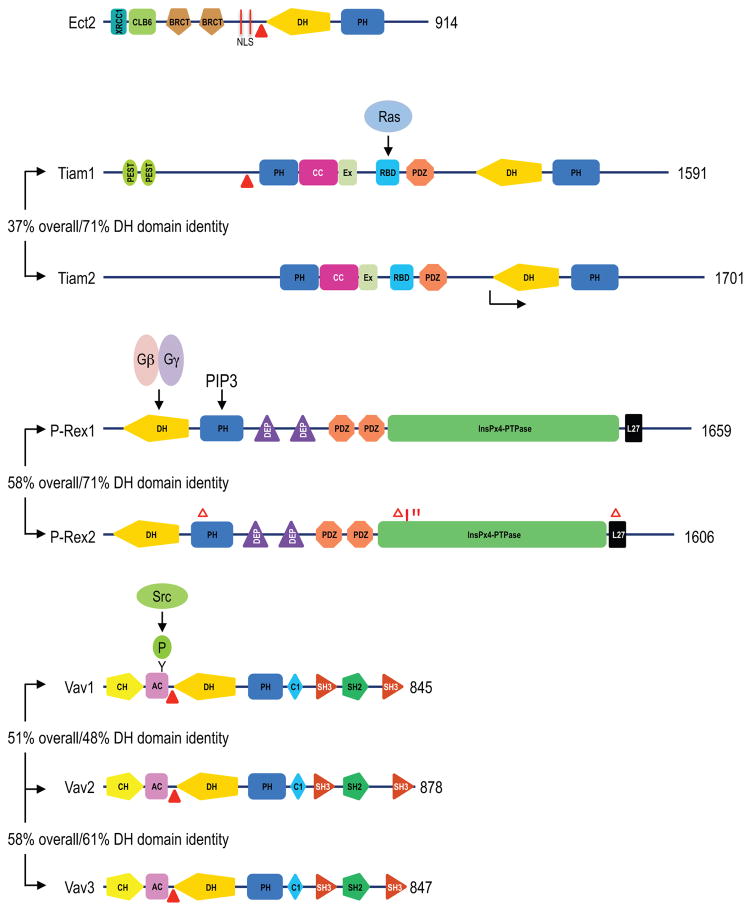
Figure 4. Mechanisms of RhoGEF activation.
Shown here are the “canonical” isoforms as identified in UniProt (http://www.uniprot.org/). Domain structures were determined, in part, by SMART (http://smart.embl-heidelberg.de/). Additional domains not identified in SMART were added based on previous sequence analyses of Ect238, Tiam116, P-Rex57 and Vav proteins.175 The number at the right end of each protein indicates the number of amino acids in the full length protein. Solid triangles indicate the position of N-terminal truncations that result in the formation of constitutively activated and transforming variants of Ect2, Tiam1, and Vav1-3. The arrow indicates the site of initiation of the shorter Tiam2S splice variant. The % values refer to amino acid sequence identity for the full length protein (overall) or the isolated DH domain, when compared to the first isoform shown. Gain-of-function P-Rex2a truncation (Δ) or missense (*) transforming mutations.99 Abbreviations: Ac, acidic domain; CC, coiled-coil; CH, calponin homology; Clb6 region, homology to yeast Clb6 B-type cyclin; Ex, extra region; NLS, nuclear localization sequence; PIP3, phosphatidylinositol (3,4,5)-triphosphate; XRCC1, homology to human repair protein XRCC1.
Humans also possess a second structurally and mechanistically distinct22 class of RhoGEFs, the dedicator of cytokinesis or DOCK family (11 human members) that act as GEFs for Rac and/or Cdc42, but not RhoA.23–25 DOCK RhoGEFs lack primary sequence homology with the DH domain and instead are characterized by a conserved Dock-homology region-2 (DHR-2; also called DOCKER, CZH2) that serves as the RhoGEF catalytic domain. DHR-2 domains exhibit no primary sequence homology to DH domains. Although plant species possess Rho-like GTPases (Rho of plants; Rop)26, and are regulated by DOCK RhoGEFs, RhoGAPs and RhoGDIs homologous to mammalian RhoGAPs27, they lack Dbl RhoGEFs. Instead, RopGEFs possess a structurally distinct plant-specific Rop nucleotide exchanger (PRONE) catalytic domain.28,29 Finally, although there are no Rho orthologs in prokaryotes, pathogenic bacteria possess effector proteins that can regulate their mammalian host Rho GTPases in part by mimicking the function of mammalian RhoGEFs.30,31
RhoGEFs and development
The combined number of human Dbl and DOCK RhoGEFs greatly exceeds the number of Rho GTPases, suggesting apparent redundancy in RhoGEF function. This is particularly striking for RhoA, where at least 28 Dbl RhoGEFs can activate this single GTPase (Figure 3). With six Rho GTPases constitutively GTP-bound and active, and not believed to be regulated by RhoGEFs (Rnd1-3, RhoH/TTF, RhoBTB1/2)32, the apparent redundancy in RhoGEFs is even more striking.
One approach to evaluate this apparent functional redundancy has been the generation of mice deficient in one or more RhoGEFs, which additionally addresses their involvement in development. The role of at least 26 Dbl family RhoGEFs in mouse development has been evaluated (Table 2), with only four (Sos1, Trio, AKAP13 and Ect2) found to be essential.33–36 However, these proteins all possess other functions independent of their RhoGEF catalytic activities, and embryonic lethality may not be due specifically to loss of the RhoGEF function. For example, when only the AKAP13 RhoGEF domain was disrupted, no lethality was seen, indicating that AKAP13 RhoGEF activity is not required for mouse development.37 AKAP13 is also a scaffolding protein that associates with the regulatory subunit of protein kinase A to spatially regulate its substrate utilization. AKAP13 additionally interacts with protein kinase C and D isoforms and with heterotrimeric Gα subunits; whether any of these interactions are specifically required during development remains to be determined. Sos1 is also a RasGEF activated downstream of receptor tyrosine kinases. Trio contains a serine/threonine kinase domain in addition to its two distinct DH-PH RhoGEF domains. Although Ect2 has no other known catalytic function aside from its RhoGEF activity, there are Ect2 non-RhoGEF sequences shown to be important for regulation of cytokinesis in vitro.38 Below we provide a summary of the developmental roles of RhoGEFs that have additional roles in cancer.
Table 2.
Roles of Dbl RhoGEFs in mouse development and tissue function
| Dbl Protein | Other Functionsa | Mammalian Phenotype Associated with Deficiency | References |
|---|---|---|---|
| Abr | RhoGAP | No abnormal phenotype detected Together with Bcr deficiency caused partial neonatal lethality, but viable mice have cerebellar and vestibular defects |
185, 186 |
| AKAP13 | Protein kinase A binding | Embryonic lethal at E10.5–11; cardiac developmental defect | 35, 187 |
| Alsin | RanGEF, Rab5GEF | Viable and fertile, no obvious developmental abnormalities | 188–192 |
| Asef1 | APCb binding | Viable and fertile, normal morphology and lifespan; impaired tumor angiogenesis and adenoma formation in Apcmin/+ mice | 193, 194 |
| Asef2 | APCb binding | Viable and fertile, normal morphology and lifespan; impaired intestinal adenoma formation in Apcmin/+ mice and reduced further when combined with Asef1 deficiency | 194 |
| Bcr | RhoGAP | Viable and fertile, but mice display abnormal neutrophil physiology | 195 |
| Collybistin | Viable and fertile, but increased levels of anxiety; abnormal spatial learning and synaptic plasticity | 196 | |
| Dbl | Abnormal dendrite morphology | 197 | |
| Ect2 | Embryonic lethal, perimplantation defect | 36 | |
| LARG | GAP for Gα12/13 | Reduced birthrate, but adult mice are viable with no defects in anatomy, survival or behavior; resistant to salt-induced hypertension When combined with p115-RhoGEF deficiency, causes embryonic lethality during midgestation, with multiple vascular defects |
198, 199 |
| p115-RhoGEF | GAP for Gα12/13 | Viable, normal life-span; altered marginal zone B cell and other hematopoietic cell functions; defect in activated neutrophil polarization, migration, and adhesion, peripheral leukocytosis with splenomegaly and extramedullary hematopoiesis; impaired neuronal innervation and motor dysfunction | 200–204 |
| Kalirin | 2nd DH-PH tandem domain; serine/threonine kinasec | Kalirin-7-specific knockout are viable and fertile, but display defects in long-term potentiation (LTP), behavioral alterations; DH1 knockout deficient in the three major isoforms (7,9 and 12) viable and fertile, with cortical morphology alterations, reduced hippocampus; LTP and memory defects | 205, 206 |
| Obscurind | Two serine/threonine kinase domains | Viable and fertile, no obvious physiological or pathological abnormalities; generally appear healthy but display mild myopathy and abnormal sarcoplasmic reticulum morphology | 207 |
| α–PIX | Viable, no gross changes in brain morphology; reduced numbers of mature lymphocytes and defective immune responses; abnormal spatial learning and dendrite morphology | 208, 209 | |
| P-Rex1 | Viable and healthy, slightly smaller size, with mild neutrophilia and a small liver Impaired melanoblast migration from the neural crest during embryogenesis |
60, 61 | |
| P-Rex2 | Viable and fertile, abnormal Purkinje cell dendrite morphology and impaired coordination; viable and fertile when combined with Prex1 −/−, but coordination is further impaired | 62 | |
| RasGRF1 | RasGAP | Viable and healthy; reduced long term potentiation, abnormal spatial learning, decreased body weight, impaired glucose tolerance and abnormal DNA methylation | 210–215 |
| RasGRF2 | RasGAP | Viable and fertile, but abnormal T cell physiology | 216, 217 |
| Sos1 | RasGEF | Embryonic lethal between E15.5 and birth, abnormal extraembryonic tissue morphology | 33, 218 |
| Sos2 | RasGEF | No abnormal phenotype detected | 219 |
| Tiam1 | Viable, but impaired oncogenesis; defect in embryonic brain development and decreased brain size | 54, 220 | |
| ARHGEF5 | Viable; dendritic cell migration defect | 221 | |
| Trio | Serine/threonine kinase | Embryonic lethality in late gestation, abnormal development of skeletal muscle and neural tissue | 34, 222 |
| Vav1 | Partial defect in T-cell development and function, normal B-lymphocyte development and function | 223–225 | |
| Vav2 | Normal T- and B-cell development and function, but B-cell defect in combination with Vav1 deficiency | 74, 75 | |
| Vav3 | Normal, but increased hematopoietic cell defects when combined with loss of Vav1 and Vav2 | 76 |
Based on sequence homology; not all have been validated to be catalytically active;
Adenomatous polyposis coli;
Kalirin-12 isoform, absent in Kalirin-7 isoform;
Knockout retains expression of the splice variant that contains only the kinase domain.
Ect2
Ect2 (Epithelial Cell Transforming Sequence 2) was originally discovered as an oncogene that transformed NIH 3T3 cells.39 However, the Ect2 protein found to transform these fibroblasts was a truncated version of the full-length protein, formed during DNA manipulation in vitro (Figure 4). The N-terminus of Ect2 contains two regions that are homologous to XCCR1 and Clb6 domains that have functions in the DNA damage response and cell cycle regulation, respectively. However, while required for Ect2 support of cytokinesis,38 no specific functions have been ascribed to these sequences. The N-terminus also contains two tandem BRCA1 C-terminal (BRCT) domains that have auto-inhibitory40,41 and phosphoprotein binding functions42,43 and two nuclear localization sequences (NLSs).44 The central catalytic portion of Ect2 consists of the tandem DH-PH domains. Finally, the C-terminal sequence of full-length Ect2 has no known domains or motifs but is required for the transforming activity of N-terminally truncated Ect2, and was recently implicated in modulating the stability of full length Ect2 protein.41,45,46 BRCT domains are not found in any other RhoGEFs and instead are found primarily in proteins involved in the DNA damage response network associated with cell cycle checkpoint functions.47,48 Although analyses of full length recombinant Ect2 protein indicate that it is selective for RhoA and its related isoforms (RhoB and RhoC) in vitro, cell-based studies suggest that Ect2 can also regulate Rac and Cdc42 activity.45
Recently we showed that Ect2-deficient mice are not viable.36 Whereas heterozygous Ect2+/− mice displayed normal development and lifespan, no Ect2−/− embryos were found at birth or as early as embryonic day 8.5. Our subsequent characterization of the defect in vitro demonstrated that isolated homozygous Ect2−/− blastocysts displayed abnormal outgrowth at day E3.5, indicating that Ect2 is required for peri-implantation development. The requirement for Ect2 at such an early stage of development suggests that it may play a key role in nearly all cells. Unlike a majority of Dbl RhoGEFs, there are no Ect2-related isoforms, which perhaps is a basis for its essential requirement in development.
The best-characterized normal function of Ect2 is its role in cytokinesis.40 Ect2 has been shown to regulate RhoA during cytokinesis49, the final step in both meiosis and mitosis. Therefore, it is likely that Ect2 is required for development because of its key role in normal cell division. Consistent with this possibility, a deficiency in MgcRacGAP (RacGAP1), a RhoGAP that facilitates Ect2 localization to the central spindle during cytokinesis, also produced a similar embryonic lethality phenotype.50 Thus, despite the existence of at least 27 other RhoGEFs that can activate RhoA (Figure 3), an Ect2 deficiency cannot be compensated for by other RhoGEFs to promote cytokinesis. However, since the Ect2 sequences flanking the DH-PH module can serve scaffolding functions and facilitate protein interactions, it is possible that non-GEF functions contribute to the critical role of Ect2 in development. A definitive resolution of this issue will require analyses of mice that harbor a RhoGEF-dead Ect2 allele.
Tiam1
Tiam1 (T-cell Lymphoma Invasion and Metastasis 1) was found by retroviral insertional mutagenesis of T lymphoma cells and then screened for invasiveness.51 Tiam1 contains a PH domain followed by a coiled-coil motif and extra region (Ex) that together comprise a protein- and membrane-binding domain,15 a Ras-binding domain (RBD),52 a PSD-95/DlgA/ZO-1 (PDZ) domain, then the tandem DH-PH domains (Figure 4). Although the exact mechanism(s) of Tiam1 activation is not clear, it is apparent that it can be regulated through intramolecular inhibition (coiled-coil motif with the DH domain), protein-protein interactions (Ras binding), and cellular localization (phospholipids with the PH domain) (Figure 4). Tiam1 is a Rac-selective GEF and was also shown to function as an effector of Ras.52 Although Rac1 deficiency causes severe embryonic lethality,53 Tiam1 is non-essential for mouse development.54 Thus, other GEFs that activate Rac can compensate for Tiam1 loss-of-function, for example the related Tiam2 protein (37% overall identity, 71% DH domain identity).55,56
P-Rex1/2
P-Rex1 (PtdIns(3,4,5)P3-dependent Rac Exchanger) was discovered by purifying proteins from the cytosol of neutrophils and assaying for proteins that activate Rac1 in the presence of phosphatidylinositol (3,4,5)-triphosphate (PtdIns(3,4,5)P3/PIP3).57 Two different groups of researchers identified P-Rex2 by searching for proteins with sequence identity to P-Rex1.58,59 P-Rex1 and P-Rex2 are very similar proteins (58% identity) that share amino-terminal DH-PH domains, followed by two Dishevelled/Egl-10/Pleckstrin (DEP) and two PDZ domains, and then a C-terminal sequence that displays homology to inositol polyphosphate-4-phosphatase (IP4P) (Figure 4). P-Rex1 and likely P-Rex2 are both regulated through GPCR activation and Gβγ activation and phosphoinositol 3-kinase (PI3K)-stimulated formation of PIP3. P-Rex1 function is attributed primarily to Rac activation, although P-Rex1 can also activate Cdc42 and other Rho family GTPases in biochemical assays.57
P-Rex1-deficient mice are healthy and viable but weigh less, with significantly smaller livers, than P-Rex1+/+ mice60. It was also shown that P-Rex1−/− mice on a pure C57BL6 background displayed a “white belly”, white feet and tail, a phenotype with 100% penetrance.61 This phenotype was caused by a defect in melanoblast migration. The belly, feet, and tail are the most distal points of melanoblast migration from the neural crest during mouse embryogenesis, leading to the anatomical pattern of this pigmentation defect.
P-Rex2 has two splice variants: a larger product that is very similar to P-Rex1 (P-Rex2a) and a smaller product (P-Rex2b) that does not contain the IP4P homology region. Whereas P-Rex1 is expressed in peripheral blood leukocytes, neither of the P-Rex2 variants is present there. Instead P-Rex2a is found predominantly in skeletal muscle, intestine, and brain, whereas P-Rex2b is found only in heart.59
P-Rex2-deficient mice are viable and fertile with normal appearance and weight. However, P-Rex2−/− mice displayed a mild defect with motor coordination that worsens with age.62 Furthermore, the P-Rex1−/− P-Rex2−/− double knockout mice exhibited defects in motor activity, posture, and gait consistent with cerebellar dysfunction that are stronger compared to the P-Rex1−/− or P-Rex2−/− single knockout mice.62
Vav1/2/3
Vav1 (named after the sixth letter in the Hebrew alphabet) was also discovered initially as an oncogene that caused growth transformation of NIH 3T3 cells. 63,64 Vav1-containing DNA was isolated from a human esophageal carcinoma, and the transforming variant also contained an N-terminal truncation of sequences upstream of the DH-PH domains. Two highly related isoforms, Vav2 and Vav3, share significant sequence identity (51 and 58% identity, respectively) and domain structure with Vav1 (Figure 4).65–68 Vav1/2/3 all contain the following domains: calponin homology (CH), an acidic (Ac) region, DH-PH, and a cysteine-rich zinc finger domain (C1) followed by a Src homology-2 and -3 (SH3-SH2-SH3) adaptor cassette69 (Figure 4). The active site of the DH domain is autoinhibited by a helix in the Ac region of Vav. Src and Syk kinases relieve the autoinhibition by phosphorylation of tyrosine 174 within the helix. Furthermore, the inhibitory conformation of the Ac region is stabilized through interactions of the CH domain with the Ac region and the DH-PH domains. Thus, maximal GEF activity of Vav is achieved in a multi-step process wherein the initial Src-dependent phosphorylation events that disrupt CH domain interactions are followed by phosphorylation on tyrosine 174.70,71 Although Vav function is attributed mainly to its ability to activate Rac, Vav proteins can also activate RhoA, Cdc42 and RhoG (Figure 4).
Vav2 and Vav3 are broadly expressed, whereas Vav1 is largely restricted to hematopoietic cells under normal conditions.72 Vav proteins are critical for the regulation of hematopoietic cell signaling by linking intracellular signaling to multi-subunit immune-recognition receptors (MIRRs).73 Vav1-deficient mice are viable but display a range of defects in the immune system, with the most severe defects found in T cell development and activation. By comparison, Vav2 deficiency did not perturb T cell development, and double knockout Vav1/2−/− mice exhibited similar T cell defects as seen in Vav1-deficient mice, indicating that Vav2 has no major function in T cell development in the absence of Vav1. In contrast, although Vav3-deficient mice also showed normal T cell development, combined deficiency of both Vav1/3 significantly exacerbated the T cell defect seen with Vav1 deficiency alone, indicating partially redundant roles for these two isoforms in T cell function. Finally, while combined Vav2/3 deficiency also showed no T cell abnormalities, the combined Vav1/2/3 deficiency (viable, no gross organ abnormalities) showed a severe impairment in T cell development, with no functional T cells, demonstrating that Vav2 can have a compensatory function in the absence of both Vav1 and Vav3.
Nonoverlapping B cell defects were observed in mice deficient in either Vav1 (B1 cell reduction, partial decrease in B cell proliferation upon B cell receptor (BCR) cross-linking) or Vav2 (reduced BCR responses). In contrast, combined Vav1/2 deficiency showed severe B cell maturation and response defects, demonstrating their nonredundant roles in B cell biology.74,75 Finally, mice deficient in all three Vav genes displayed additional B cell developmental defects absent in Vav2/3−/− double knockout mice.76 Thus, the Vav isoforms support both distinct and redundant functions in T and B cell development and activities, several of which cannot be compensated by other RhoGEFs.
RhoGEFs and cancer
The three Ras proteins (H-Ras, K-Ras4A/B, and N-Ras) are the founding members of the Ras superfamily of small GTPases1 and together they comprise the most frequently mutated oncoprotein family in human cancer (33%; http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/).10,77 However, with the exception of the recently described mutational activation of Rac1 in melanoma78,79, Rho family GTPases have not been reported to be mutated frequently in human cancer. Nevertheless, there is substantial experimental evidence that aberrant Rho GTPase function can contribute to cancer cell proliferation, invasion and metastasis.80 Instead, Rho GTPases are aberrantly activated more commonly by indirect mechanisms in cancer. These mechanisms include altered Rho localization mediated by GDIs; increased RhoGEF and/or decreased RhoGAP activity; altered gene expression of Rho GTPases, RhoGEFs, RhoGAPs, and RhoGDIs (Figure 1), and alternative splicing to generate constitutively active Rho GTPase isoforms (e.g., Rac1b).81,82
As mentioned above, although Dbl, Ect2 and Vav were identified originally as oncoproteins due to activation by N-terminal truncation, these altered RhoGEFs arose as an artifact of the DNA isolation/transfection procedure.83 Surprisingly, despite their potent transforming activities when assayed in NIH 3T3 mouse fibroblasts, such truncated, activated Dbl RhoGEFs have not been identified in human cancers. Instead, other mechanisms that lead to the deregulated expression and/or activation of full length RhoGEFs have been identified. Recent reviews have provided more comprehensive overviews of RhoGEF activation in cancer.4,84,85 Therefore, we have focused on the four Dbl RhoGEFs with the strongest evidence for their involvement in cancer growth (Supplementary Table 2) and have summarized representative studies for each below.
Ect2
Ect2 gene and/or protein overexpression has been described in glioblastoma,86–88 non-small cell lung cancer (NSCLC),89,90 esophageal,90 pancreatic,91 oral squamous cell,92 colorectal carcinomas,93 and other cancers (Supplementary Table 2).
The ECT2 gene is located in a region of chromosome 3q26 that is frequently altered in human tumors.94 Ect2 overexpression was associated with gene amplification in NSCLC, and this also correlated with an increase in the copy number of the gene encoding protein kinase C iota, PKCi (PRKCI).89 Ect2 knockdown was associated with decreased Rac1 activation, and constitutively activated Rac1 could compensate for the loss of Ect2, implicating this Rho GTPase in Ect2-dependent NSCLC anchorage-independent growth and invasion in vitro. Interestingly, the role of Ect2 in NSCLC was independent of Ect2 normal function in cytokinesis. Depletion of Ect2 expression in NSCLC did not result in any proliferation or multi-nucleation defects, phenotypes associated with Ect2 loss in normal cells.36
One mechanism that may account for a non-cytokinesis role for Ect2 in driving cancer growth under some conditions involves altered subcellular localization. Ect2 is one of two Dbl RhoGEFs (the other is Net1) with a nuclear-restricted localization in interphase cells. Fields and colleagues described regulation of NSCLC tumor growth and invasion through mislocalization of Ect2 to the cytoplasm, where it binds to a PKCi-Par6 complex to activate Rac1.89 They found that PKCi phosphorylated Ect2 at residue T328 adjacent to the Ect2 nuclear localization sequences (NLSs), and this was associated with Ect2 cytosolic mislocalization. Furthermore, a T328D phosphomimetic but not a T328A phosphodeficient Ect2 mutant could rescue loss of endogenous Ect2 to restore lung tumor cell growth and invasion in vitro. That cytoplasmic mislocalization is an important mechanism of Ect2 activation in cancer is consistent with the earlier observation that disruption of the Ect2 NLS motifs activated the transforming activity of full length Ect2 when evaluated in NIH 3T3 fibroblasts.44 Cytoplasmic Ect2 may then cause spatially inappropriate Rho GTPase activation to drive cancer growth.
Two studies showed that Ect2 expression correlated with poor prognosis in glioma patients and that Ect2 was important for glioma cell proliferation, migration, and invasion in vitro.87,88 However, acute depletion of Ect2 expression by siRNA in giloma tumor cell lines showed an increase in multinucleation, characteristic of a defect in cytokinesis. Therefore, the acute decrease in proliferation, migration and invasion may have been caused by the inability of cells to complete mitosis. In contrast, sustained shRNA depletion of Ect2 did not suppress proliferation in vitro, but orthotopic xenograft tumor growth was impaired and associated with increased mouse survival.95 Thus, tumor cells may adapt to acute Ect2 loss, such that Ect2-dependent long-term tumor growth is independent of the role of Ect2 in cytokinesis.
In summary, future studies to determine whether conditional loss of Ect2 ablates tumor growth in mouse models will provide further assessment of the driver function of Ect2 overexpression in cancer. Also, whether the mislocalization of Ect2 to the cytoplasm seen in NSCLC is the basis for a non-cytokinesis function that supports cancer cell growth will be important to evaluate in other human cancers. The potent oncogenic activities of the originally discovered N-terminally truncated Ect2, lacking the nuclear localization sequences, provides strong evidence that simple mislocalization may be an important driver of Ect2 activation in cancer. However, despite the implication of Ect2 as a driver in a variety of human cancers, the absence of any detection of N-terminally truncated Ect2 in human cancers argues that this cytoplasmic mislocalization mechanism may not hold true in the patient. In support of this possibility, in our own studies where we have established driver functions of full length Ect2 in ovarian and colorectal cancers, we have not found evidence for cytoplasmic mislocalization as the basis for the aberrant Ect2 function in these cancers (unpublished observations).
P-Rex
The first cancer-driving role of P-Rex1 was described in prostate cancer.96 P-Rex1 protein expression was elevated in metastatic but not primary prostate tumor cell lines and tumor tissue. RNAi silencing of P-Rex1 expression reduced Rac activity, and migration and invasion in vitro.
PREX1 mRNA overexpression has also been associated with breast cancer, in particular the estrogen receptor-positive luminal subtype and, to a lesser degree, HER2-positive tumors.97 Suppression of P-Rex1 expression was found to reduce HER2-stimulated Rac1 activation, motility, invasion, and tumorigenic growth.97,98
Increased P-Rex1 protein expression has also been seen in human melanoma tissue and cell lines relative to normal melanocytes,61 and RNAi suppression reduced Matrigel invasion of human melanoma cell lines. High P-Rex1 protein expression was associated with increased metastatic potential in immunocompromised mice. To assess a role for P-Rex1 in melanoma progression, P-Rex1-deficient mice were crossed to a mouse model of primary and metastatic melanoma (Tyr:NRasQ61K/INK4a−/−). Surprisingly, in contrast to the breast tumor studies, P-Rex1 deficiency did not reduce the incidence or latency of primary tumor formation. However, a significant reduction in metastasis was seen. While a mechanistic basis for these distinct roles for P-Rex1 in breast versus skin cancer progression has not been determined, one logical speculation is that this may reflect tissue type or genetic context differences in P-Rex1 function. More provocatively, it may reflect the differential activation of other Rho GTPases, aside from Rac1 (e.g., Cdc42), that then contribute to aberrant P-Rex1-dependent functions in breast versus melanoma growth and tumor progression.
Recently, in a whole-genome sequence analysis of melanoma, non-synonymous mutations in PREX2 were found with a 14% frequency in a cohort of 107 human melanomas.99 The mutations occurred throughout the gene, including interchromosomal translocations and nonsense mutations, with some encoding C-terminally truncated protein products due to premature termination codons. To assess the biological relevance of these mutations, P-Rex2 truncation or missense mutants were ectopically expressed in TERT-immortalized, mutant NRAS-expressing human melanocytes and shown to accelerate tumor formation in nude mice.
At least one P-Rex2 splice variant can promote cancer growth through a mechanism independent of its RhoGEF function.100 P-Rex2a can directly bind and inhibit the lipid phosphatase activity of the PTEN tumor suppressor, thereby causing activation of PI3K and Akt and subsequent stimulation of pro-growth and survival pathways. P-REX2A mRNA is preferentially overexpressed in PTEN-wildtype, PIK3CA-mutant breast tumors. Depleting P-Rex2a in PTEN-wildtype and PIK3CA-mutant MCF-7 breast cancer cells reduced phosphorylated AKT and cell growth in vitro.
The aberrant activation of both P-Rex1 and P-Rex2 in melanoma, together with the frequent mutational activation of their key substrate Rac1 in melanoma,78,79 provides perhaps the most compelling evidence for a critical driver function of a Rho small GTPase in cancer. Determining which effector(s) may then drive Rac1-dependent melanoma growth will be an important next step in these studies.
Tiam1
Tiam1 mRNA and/or protein overexpression has been described in many cancers that include melanoma,101 breast,102,103 colon,104,105 prostate,106 and renal cell carcinoma107 (Supplementary Table 2).
A role for Tiam1 in tumorigenesis has been evaluated in several mouse models of cancer. First, the consequence of Tiam1 deficiency was evaluated using the classical two-stage chemical carcinogenesis model for the initiation and promotion of cutaneous tumors.54 Tumor formation is induced using a single application of the mutagen 7,12-dimethylbenz[a]anthracene (DMBA), which is then followed by repeated applications of phorbol 12-myristate 13-acetate (PMA) tumor promoter. This treatment results in mutational activation of HRAS(Q61L) and the formation of benign papillomas that can progress to squamous cell carcinomas (SCC). Both the incidence of SCC formation and growth of SCC tumors were reduced in Tiam1−/− mice. Thus, TIam1 is an important contributor to H-Ras-induced SCC tumor growth. These results, together with the identification of Tiam1 as an effector of Ras52 and findings that Rac1 deficiency reduced mutant K-Ras-induced lung and pancreatic tumor formation56,108, all support Tiam1 as an important effector of Ras-driven cancer growth. Conversely, for those tumors that did arise, Tiam1 deficiency enhanced metastatic growth. Thus, while Tiam1 was required for tumor initiation and progression, it was antagonistic for malignant progression in these models.
The role of Tiam1 has also been evaluated in a mouse model of colon cancer, the Apc Min (multiple intestinal neoplasia) mice that harbor a truncated and loss-of-function mutant of the Apc tumor suppressor. Absence of Tiam1 resulted in significantly reduced formation and growth of polyps, but in enhanced invasion of malignant intestinal tumors.109
Finally, Tiam1 deficiency reduced mammary tumor initiation and reduced metastasis in transgenic mice whose tumors are driven by mouse mammary tumor virus (MMTV) promoter-regulated mutant ErbB2/Neu. However, it had no effect on the growth of mammary tumors in MMTV-Myc transgenic mice.110 Interestingly, although Tiam1 was originally discovered for its ability to promote T cell invasion in vitro51, in some tissues it has also been shown to inhibit progression to malignant growth.
In patients, increased Tiam1 protein expression was found in a subset of colon tumors104, and RNAi suppression reduced the growth of colon cancer cell lines in vitro and in vivo.109,111 Tiam1 overexpression was found to be inversely proportional to the degree of promoter methylation.105 In a small panel of breast cancer patient samples, Tiam1 protein expression levels were higher in grade III compared with grade II tumors102 and Tiam1 mRNA levels were significantly higher in tumour tissue from breast cancer patients who died from their disease compared with those who survived.112 However, in another study, Tiam1 protein expression levels decreased with the progression of breast carcinomas.103 The reasons for these opposing findings remain unclear but may be related to different patient populations whose diseases are driven by distinct mechanisms.
Much less is known regarding the involvement of Tiam2 in cancer. TIAM2 encodes two isoforms, one similar in structure to Tiam1 (designated long; Tiam2L), and another lacking N-terminal sequences upstream of the DH domain (designated short; Tiam2S).56 Interestingly, whereas mRNA for both isoforms could be detected in hepatocellular carcinoma (HCC), only the short isoform of Tiam2 protein was detected.113 Finally, although neither Tiam2S mRNA nor protein was detected in normal liver, HCC tumors and cell lines expressed elevated levels of Tiam2S protein, and further ectopic expression of Tiam2S enhanced the tumorigenic growth of an HCC cell line.
In summary, while altered Tiam1/2 expression has been observed in a variety of human cancers, the most compelling evidence for a driver function in cancer comes largely from model studies, where Tiam1 can either promote tumor progression or conversely antagonize invasion. The basis for these seemingly opposing roles for Tiam1 is unclear, especially since, as described above, it is the gain-of-function of another Rac-selective GEF (P-Rex1) that is essential for metastatic but not primary melanoma tumor growth.61 However, another possible explanation comes from the observation that Tiam1-Rac signaling restored E-cadherin-mediated adhesion in MDCK canine kidney epithelial cells, thereby reducing cell migration.114 Further, since remodeling of cell-cell junctions is critical for tumor cell invasion, Tiam1 activity may have opposing consequences in cells invading individually versus collectively. Whether these in vitro findings provide the explanation for the antagonistic role of Tiam1 in tumor cell invasion and metastasis remains to be determined.
Vav1
Although the Vav1 isoform originally identified by its ability to transform NIH 3T3 cells was activated by N-terminal truncation, and equivalent truncations of Vav2 and Vav3 also convert them to transforming proteins (Figure 4),67,115,116 no N-terminally truncated Vav proteins have been identified in cancer. Instead, Vav1 overexpression has been described in a spectrum of cancers, including neuroblastoma,117 lung,118 pancreatic,119 metastatic melanoma120 and B-cell chronic lymphocytic leukemia.121 Additionally, persistent phosphorylation has also been seen in some cancers, consistent with the observation that N-terminal phosphorylation can reversibly disrupt the N-terminal autoinhibitory function to activate Vav1.
Although Vav1 expression is normally restricted to hematopoietic cells, overexpression in several non-hematologic cancers has been described (Supplementary Table 2). For example, Vav1 overexpression was found in pancreatic tumor tissue and cell lines compared to normal pancreatic epithelial cells.119 Vav1 expression was inversely correlated with patient survival. RNAi silencing analyses found that Vav1 was necessary to support tumor growth in vivo and in vitro, and these pro-growth phenotypes were dependent on Vav1 RhoGEF activity. Vav1 support of tumor growth was associated with Rac1 activation of the PAK1 serine/threonine kinase, the NF-kappaB transcription factor, and the cyclin D1 regulator of G1 cell cycle progression.119 Similar expression of Vav1 has also been described in lung cancer and also shown to be required for tumor growth in vitro and in vivo.118
Vav1 is not the only family member to have a role in cancer: both Vav2 and Vav3 have been implicated. Vav2 is activated in metastatic melanoma through the chemokine receptor-ligand pair CXCR4-CXCL12 and promotes expression of metalloproteinases that lead to invasion.120 Recently, VAV3 gene transcription was found elevated primarily in estrogen and progesterone receptor-positive breast cancers.122 RNAi suppression analyses demonstrated that Vav3 was required for mouse mammary tumorigenesis and metastasis and that Vav2 showed a synergistic role with Vav3 in breast cancer growth and metastasis. Vav2/3 controlled a transcriptional program that involved upregulation of gene targets important for lung-specific metastasis. Interestingly, the abundance of Vav2/3 transcripts was modulated through both Rac1-dependent and -independent pathways.
The development of androgen resistance in prostate cancer cells limits the effectiveness of androgen-deprivation therapy. Vav3 was found to be overexpressed in prostate tumors and its overexpression was associated with cell line progression to androgen-independence both in vitro and in vivo.123–125 Vav3-driven androgen independence was RhoGEF-dependent, required nuclear localization,126 and was promoted in part through activation of PI3K-Akt signaling.127 Constitutively activated Rac1 alone could also drive androgen-independent growth in vitro and in vivo.125 High Vav3 protein expression was associated with disease progression and reduced survival, whereas RNAi depletion reduced the tumorigenic and metastatic growth of androgen-independent prostate tumor cells.128 Finally, suppression of Vav3 expression enhanced docetaxel-induced apoptosis in prostate cancer cells, further supporting a therapeutic value in blocking Vav3 function in prostate cancer.129
In summary, consistent with the mouse studies suggesting that the Vav isoforms exhibit both redundant and distinct functions in development, there appear to be Vav isoform-distinct functions in cancer. For example, Vav1 suppression alone was sufficient to impair growth of pancreatic cancer cells despite continued expression of Vav2, and Vav2 depletion did not alter growth.119 Thus, Vav2 cannot substitute for Vav1 in pancreatic cancer. Similarly, suppression of either Vav2 or Vav3 alone greatly reduced the metastatic growth of mouse mammary tumor cells, and concurrent ablation of both isoforms further abolished metastatic growth.122 The basis for why Vav2 and Vav3 may have distinct roles is not clear, since both activate RhoA, Rac1 and Cdc42. One possibility is that each activates spatially distinct subcellular pools of the Rho GTPases, leading to activation of distinct effector signaling networks. Another possibility is that the Vav isoforms activate an incompletely overlapping extended set of Rho GTPases. Analyses of RhoGEF specificity are often restricted to RhoA, Rac1 and Cdc42 but do not examine other family members. This tendency towards limited analysis of potential substrates remains an issue that needs to be addressed if the functions of specific RhoGEFs are to be fully understood.
RhoGEFs and other human diseases
FGD1, which encodes a Cdc42-specific RhoGEF, was originally identified as a mutated gene responsible for Aarskog-Scott Syndrome (ASS; also known as faciogenital dysplasia).130 ASS is an X-linked recessive developmental disorder characterized by short stature, by facial, skeletal, and urogenital anomalies, and by mild mental retardation. The ASS-associated FGD1 mutations cause loss-of-function (deletion, nonsense mutations causing expression of truncated proteins, missense mutations in DH-PH domains) and account for ~20% of individuals with ASS.130,131
Loss-of-function mutations in the related FGD4 gene are associated with one type (CMT4) of the Charcot-Marie-Tooth (CMT) disease group.132,133 Like FGD1, FGD4 encodes a Cdc42-specific GEF (Fgd4/Frabin). This heterogeneous disease group is characterized by damage to peripheral nerves that can result in loss of sensation and atrophy of muscles in the feet, legs, and hands. It is one of nine genes with causal association with CMT4, an autosomal recessive demyelinating form of CMT.
Mutations in OBSCN, the gene encoding the RhoGEF Obscurin, have been identified in melanoma and glioblastoma134 and may predispose towards these cancers and towards hereditary myopathies.135 OBSCN mutations have also been identified by exome sequencing of breast and colon cancers, supporting a possible driver function in these two cancers.136 Obscurin is a very large protein with GEF activity for both RhoA and TC10.
Alsin, a GEF for Rac and for the Rab5 small GTPase involved in vesicular transport, is encoded by the gene ALS2.137–139 Loss-of-function mutations in ALS2 have been found in a subset of individuals with amyotrophic lateral sclerosis (ALS),140–143 a disease characterized by progressive movement problems and muscle wasting caused by motor neuron death. Accordingly, Alsin expression is highest in motor neurons of the brain. ALS2 mutations have been associated with autosomal recessive juvenile onset ALS and the related conditions of infantile-onset ascending hereditary spastic paraplegia and of juvenile-onset primary lateral sclerosis. The exact contribution of Alsin, Rac1 and/or Rab5 to ALS diseases remains to be determined.
Germline gain-of-function mutations in SOS1 have been found in ~20% of individuals with Noonan syndrome.144,145 This syndrome is characterized by a range of distinctive facial characteristics, short stature, cardiac defects, bleeding disorders, skeletal malformations, and a diversity of other signs and symptoms. Sos1 has dual functions and can act as either a RacGEF or a RasGEF. However, the frequent findings in this disorder of gain-of-function mutations in RAS and in genes encoding its Raf effectors (BRAF and RAF1) argue that enhanced activity of Sos1 on Ras rather than on Rac contributes to the role of SOS1 mutations in Noonan syndrome.143,146,147
Loss-of-function mutations in Collybistin/ARHGEF9/hPEM2, a Cdc42-specific Dbl RhoGEF, have been identified in patients with X-linked mental retardation associated with epilepsy.148–152 Collybistin expression is neuronally restricted; Collybistin is found in neurons throughout the adult brain and spinal cord.
Additional Dbl RhoGEFs have also been associated with biology and disease.153 Altered expression and mouse model knockout studies implicate Kalirin, with an N- and C-terminal DH domain that activate Rac1 and RhoA respectively (Figure 3), in a number of neurological and cardiovascular diseases.154 These include Huntington’s Disease, Alzheimer’s, schizophrenia, depression, cocaine addiction and ischemic stroke.
Finally, many pathogenic bacteria employ a virulence strategy to subvert and hijack the function of Rho GTPases in their eukaryotic hosts. This strategy is mediated by a type III secretion system to inject virulence factors, or effectors, to facilitate cell invasion, intracellular survival, and modulation of the host immune responses.155 Among the effectors that target Rho GTPases are those that either mimic or antagonize the function of eukaryotic RhoGEFs.31 The first discovered was the Salmonella enterica serovar typhimurium protein SopE,156 with related RhoGEFs (SopE2, BopE2, CopE) later identified in other bacteria. Members of a second class of bacterial proteins that share a WxxxE sequence were first thought to act as straightforward Rho GTPase mimics,157 but were later found to also mimic RhoGEF activity. Examples include the Map protein from enteropathogenic E. coli, and IpgB1 and IpgB2 from Shigella flexneri, that mimic Cdc42, Rac1 and RhoA, respectively. These bacterial RhoGEFs lack sequence homology to the DH domain but nevertheless utilize a biochemical mechanism similar to that of Dbl family RhoGEFs.158 In contrast, another class of type III effectors antagonizes eukaryotic RhoGEF function.159 For example, it was determined that EspH binds directly to the DH-PH domain of multiple RhoGEFs, preventing their binding to Rho substrates. While the full impact of these bacterial Rho GTPase regulators remains to be determined, they provide yet another example of how aberrant RhoGEF function may contribute to human disease. Understanding the pressures that led to their convergent evolution may help to further define the roles of RhoGEFs in cancer and other diseases.
Perspectives
Given the essential involvement of Rho GTPases in virtually every aspect of normal cell physiology, it is not surprising that the aberrant activities of Rho GTPases are associated with cancer and other diseases. Since RhoGEFs are key mediators of Rho GTPase activation, it is also not surprising that aberrant RhoGEF activation is a major mechanism driving aberrant activation of their Rho GTPase targets. Finally, given the very divergent sequence and domain structures beyond their shared DH-PH domains, the diversity of mechanisms deregulating RhoGEF functions is also to be expected. Large challenges remain in unraveling the precise mechanisms and contributions of RhoGEFs to specific cancers and other diseases, but certainly improved understanding will help to promote therapeutic interference in their activities.
The possibility that RhoGEFs may be directly targeted is supported by the mechanism of the natural product brefeldin A, an ArfGEF inhibitor that binds to the interface between the Arf small GTPase and the catalytic domain of ArfGEF, freezing the complex in an abortive conformation that cannot complete nucleotide exchange.160–162 However, the significant conformational change seen in Arf upon association with its GEF and upon switching from the GDP- to GTP-bound states makes this interaction distinct from that of Rho GTPases. To date, there has been very limited success in developing pharmacologic inhibitors of RhoGEF function.4 Thus, whether potent and selective RhoGEF inhibitors can be developed remains to be determined. An alternative strategy for blocking RhoGEF activity has been the identification of small molecules that bind the GTPase and prevent GEF stimulation. Antagonists of GEF activation of Rho 163–165 and Ras 166–168 have been described. Whether such small molecule inhibitors of protein-protein interactions can achieve the potency and selectivity needed for clinical efficacy remains to be determined, but it is clear that we are at last on the way to finding out.
Finally, with the recent successful clinical development of numerous protein kinase inhibitors for cancer treatment (e.g., imatinib, erlotinib, vemurafenib), perhaps the most promising avenues for short term success in developing RhoGEF inhibitors may be blockade of the protein kinases that drive RhoGEF activation or that are activated by their Rho GTPase substrates. These may include inhibitors of the Src family protein tyrosine kinases169 that activate Vav family proteins; of the PKCi serine/threonine kinase170 responsible for Ect2 mislocalization to the cytoplasm; of PI3K171 to impair PIP3 production and activation of P-Rex; or of the PAK172 or ROCK173 serine/threonine kinases activated downstream of Rac1 or RhoA, respectively. Whether RhoGEFs themselves turn out to be druggable or not, their key roles in cancer warrant continued close attention to this large and interesting family of Rho GTPase regulators.
Supplementary Material
Acknowledgments
We thank Adrienne D. Cox for careful and critical reading of our manuscript. We apologize to colleagues whose studies we did not cite due to space limitations. C.J.D. is supported by grants from the NIH, AACR/Pancreatic Cancer Action Network and the Lustgarten Pancreatic Cancer Foundation. D.R.C. was supported by a T32 training grant (CA71341) and an F31 predoctoral fellowship (CA159821).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 2.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 3.Hall A. Rho family GTPases. Biochem Soc Trans. 2012;40:1378–1382. doi: 10.1042/BST20120103. [DOI] [PubMed] [Google Scholar]
- 4.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Mata R, Boulter E, Burridge K. The ‘invisible hand’: regulation of RHO GTPases by RHOGDIs. Nat Rev Mol Cell Biol. 2011;12:493–504. doi: 10.1038/nrm3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bustelo XR, Sauzeau V, Berenjeno IM. GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. Bioessays. 2007;29:356–370. doi: 10.1002/bies.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bishop AL, Hall A. Rho GTPases and their effector proteins. Biochem J. 2000;348(Pt 2):241–255. [PMC free article] [PubMed] [Google Scholar]
- 9.Aspenstrom P. Effectors for the Rho GTPases. Curr Opin Cell Biol. 1999;11:95–102. doi: 10.1016/s0955-0674(99)80011-8. [DOI] [PubMed] [Google Scholar]
- 10.Cox AD, Der CJ. Ras history: The saga continues. Small GTPases. 2010;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eva A, Aaronson SA. Isolation of a new human oncogene from a diffuse B-cell lymphoma. Nature. 1985;316:273–275. doi: 10.1038/316273a0. [DOI] [PubMed] [Google Scholar]
- 12.Fasano O, Birnbaum D, Edlund L, Fogh J, Wigler M. New human transforming genes detected by a tumorigenicity assay. Mol Cell Biol. 1984;4:1695–1705. doi: 10.1128/mcb.4.9.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noguchi T, Galland F, Batoz M, Mattei MG, Birnbaum D. Activation of a mcf. 2 oncogene by deletion of amino-terminal coding sequences. Oncogene. 1988;3:709–715. [PubMed] [Google Scholar]
- 14.Hart MJ, Eva A, Evans T, Aaronson SA, Cerione RA. Catalysis of guanine nucleotide exchange on the CDC42Hs protein by the dbl oncogene product. Nature. 1991;354:311–314. doi: 10.1038/354311a0. [DOI] [PubMed] [Google Scholar]
- 15.Terawaki S, Kitano K, Mori T, Zhai Y, Higuchi Y, Itoh N, et al. The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module. EMBO J. 2010;29:236–250. doi: 10.1038/emboj.2009.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stam JC, Sander EE, Michiels F, van Leeuwen FN, Kain HE, van der Kammen RA, et al. Targeting of Tiam1 to the plasma membrane requires the cooperative function of the N-terminal pleckstrin homology domain and an adjacent protein interaction domain. J Biol Chem. 1997;272:28447–28454. doi: 10.1074/jbc.272.45.28447. [DOI] [PubMed] [Google Scholar]
- 17.Bourguignon LY, Zhu H, Shao L, Chen YW. CD44 interaction with tiam1 promotes Rac1 signaling and hyaluronic acid-mediated breast tumor cell migration. J Biol Chem. 2000;275:1829–1838. doi: 10.1074/jbc.275.3.1829. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka M, Ohashi R, Nakamura R, Shinmura K, Kamo T, Sakai R, et al. Tiam1 mediates neurite outgrowth induced by ephrin-B1 and EphA2. EMBO J. 2004;23:1075–1088. doi: 10.1038/sj.emboj.7600128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tolias KF, Bikoff JB, Kane CG, Tolias CS, Hu L, Greenberg ME. The Rac1 guanine nucleotide exchange factor Tiam1 mediates EphB receptor-dependent dendritic spine development. Proc Natl Acad Sci U S A. 2007;104:7265–7270. doi: 10.1073/pnas.0702044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchsbaum RJ, Connolly BA, Feig LA. Interaction of Rac exchange factors Tiam1 and Ras-GRF1 with a scaffold for the p38 mitogen-activated protein kinase cascade. Mol Cell Biol. 2002;22:4073–4085. doi: 10.1128/MCB.22.12.4073-4085.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchsbaum RJ, Connolly BA, Feig LA. Regulation of p70 S6 kinase by complex formation between the Rac guanine nucleotide exchange factor (Rac-GEF) Tiam1 and the scaffold spinophilin. J Biol Chem. 2003;278:18833–18841. doi: 10.1074/jbc.M207876200. [DOI] [PubMed] [Google Scholar]
- 22.Yang J, Zhang Z, Roe SM, Marshall CJ, Barford D. Activation of Rho GTPases by DOCK exchange factors is mediated by a nucleotide sensor. Science. 2009;325:1398–1402. doi: 10.1126/science.1174468. [DOI] [PubMed] [Google Scholar]
- 23.Cote JF, Vuori K. Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J cell Sci. 2002;115:4901–4913. doi: 10.1242/jcs.00219. [DOI] [PubMed] [Google Scholar]
- 24.Meller N, Merlot S, Guda C. CZH proteins: a new family of Rho-GEFs. J cell Sci. 2005;118:4937–4946. doi: 10.1242/jcs.02671. [DOI] [PubMed] [Google Scholar]
- 25.Pakes NK, Veltman DM, Williams RS. Zizimin and Dock guanine nucleotide exchange factors in cell function and disease. Small GTPases. 2013;4:22–27. doi: 10.4161/sgtp.22087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z, Watson JC. Molecular cloning and characterization of rho, a ras-related small GTP-binding protein from the garden pea. Proc Natl Acad Sci U S A. 1993;90:8732–8736. doi: 10.1073/pnas.90.18.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu G, Li H, Yang Z. Arabidopsis RopGAPs are a novel family of rho GTPase-activating proteins that require the Cdc42/Rac-interactive binding motif for rop-specific GTPase stimulation. Plant Physiol. 2000;124:1625–1636. doi: 10.1104/pp.124.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berken A, Thomas C, Wittinghofer A. A new family of RhoGEFs activates the Rop molecular switch in plants. Nature. 2005;436:1176–1180. doi: 10.1038/nature03883. [DOI] [PubMed] [Google Scholar]
- 29.Nagawa S, Xu T, Yang Z. RHO GTPase in plants: Conservation and invention of regulators and effectors. Small GTPases. 2010;1:78–88. doi: 10.4161/sgtp.1.2.14544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bulgin R, Raymond B, Garnett JA, Frankel G, Crepin VF, Berger CN, et al. Bacterial guanine nucleotide exchange factors SopE-like and WxxxE effectors. Infect Immun. 2010;78:1417–1425. doi: 10.1128/IAI.01250-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orchard RC, Alto NM. Mimicking GEFs: a common theme for bacterial pathogens. Cell Microbiol. 2012;14:10–18. doi: 10.1111/j.1462-5822.2011.01703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vega FM, Ridley AJ. SnapShot: Rho family GTPases. Cell. 2007;129:1430. doi: 10.1016/j.cell.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 33.Wang DZ, Hammond VE, Abud HE, Bertoncello I, McAvoy JW, Bowtell DD. Mutation in Sos1 dominantly enhances a weak allele of the EGFR, demonstrating a requirement for Sos1 in EGFR signaling and development. Genes Dev. 1997;11:309–320. doi: 10.1101/gad.11.3.309. [DOI] [PubMed] [Google Scholar]
- 34.O’Brien SP, Seipel K, Medley QG, Bronson R, Segal R, Streuli M. Skeletal muscle deformity and neuronal disorder in Trio exchange factor-deficient mouse embryos. Proc Natl Acad Sci U S A. 2000;97:12074–12078. doi: 10.1073/pnas.97.22.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayers CM, Wadell J, McLean K, Venere M, Malik M, Shibata T, et al. The Rho guanine nucleotide exchange factor AKAP13 (BRX) is essential for cardiac development in mice. J Biol Chem. 2010;285:12344–12354. doi: 10.1074/jbc.M110.106856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cook DR, Solski PA, Bultman SJ, Kauselmann G, Schoor M, Kuehn R, et al. The ect2 rho Guanine nucleotide exchange factor is essential for early mouse development and normal cell cytokinesis and migration. Genes Cancer. 2011;2:932–942. doi: 10.1177/1947601912437035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spindler MJ, Burmeister BT, Huang Y, Hsiao EC, Salomonis N, Scott MJ, et al. AKAP13 Rho-GEF and PKD-Binding Domain Deficient Mice Develop Normally but Have an Abnormal Response to beta-Adrenergic-Induced Cardiac Hypertrophy. PLoS One. 2013;8:e62705. doi: 10.1371/journal.pone.0062705. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Saito S, Tatsumoto T, Lorenzi MV, Chedid M, Kapoor V, Sakata H, et al. Rho exchange factor ECT2 is induced by growth factors and regulates cytokinesis through the N-terminal cell cycle regulator-related domains. J Cell Biochem. 2003;90:819–836. doi: 10.1002/jcb.10688. [DOI] [PubMed] [Google Scholar]
- 39.Miki T, Smith CL, Long JE, Eva A, Fleming TP. Oncogene ect2 is related to regulators of small GTP-binding proteins. Nature. 1993;362:462–465. doi: 10.1038/362462a0. [DOI] [PubMed] [Google Scholar]
- 40.Tatsumoto T, Xie X, Blumenthal R, Okamoto I, Miki T. Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J Cell Biol. 1999;147:921–928. doi: 10.1083/jcb.147.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim JE, Billadeau DD, Chen J. The Tandem BRCT Domains of Ect2 Are Required for Both Negative and Positive Regulation of Ect2 in Cytokinesis. J Biol Chem. 2004;280:5733–5739. doi: 10.1074/jbc.M409298200. [DOI] [PubMed] [Google Scholar]
- 42.Wolfe BA, Takaki T, Petronczki M, Glotzer M. Polo-like kinase 1 directs assembly of the HsCyk-4 RhoGAP/Ect2 RhoGEF complex to initiate cleavage furrow formation. PLoS Biol. 2009;7:e1000110. doi: 10.1371/journal.pbio.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burkard ME, Maciejowski J, Rodriguez-Bravo V, Repka M, Lowery DM, Clauser KR, et al. Plk1 self-organization and priming phosphorylation of HsCYK-4 at the spindle midzone regulate the onset of division in human cells. PLoS Biol. 2009;7:e1000111. doi: 10.1371/journal.pbio.1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saito S, Liu XF, Kamijo K, Raziuddin R, Tatsumoto T, Okamoto I, et al. Deregulation and mislocalization of the cytokinesis regulator ECT2 activate the Rho signaling pathways leading to malignant transformation. J Biol Chem. 2004;279:7169–7179. doi: 10.1074/jbc.M306725200. [DOI] [PubMed] [Google Scholar]
- 45.Solski PA, Wilder RS, Rossman KL, Sondek J, Cox AD, Campbell SL, et al. Requirement for C-terminal sequences in regulation of Ect2 guanine nucleotide exchange specificity and transformation. J Biol Chem. 2004;279:25226–25233. doi: 10.1074/jbc.M313792200. [DOI] [PubMed] [Google Scholar]
- 46.Liot C, Seguin L, Siret A, Crouin C, Schmidt S, Bertoglio J. APC(cdh1) mediates degradation of the oncogenic Rho-GEF Ect2 after mitosis. PLoS One. 2011;6:e23676. doi: 10.1371/journal.pone.0023676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leung CC, Glover JN. BRCT domains: easy as one, two, three. Cell Cycle. 2011;10:2461–2470. doi: 10.4161/cc.10.15.16312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gerloff DL, Woods NT, Farago AA, Monteiro AN. BRCT domains: A little more than kin, and less than kind. FEBS Lett. 2012;586:2711–2716. doi: 10.1016/j.febslet.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kimura K, Tsuji T, Takada Y, Miki T, Narumiya S. Accumulation of GTP-bound RhoA during cytokinesis and a critical role of ECT2 in this accumulation. J Biol Chem. 2000;275:17233–17236. doi: 10.1074/jbc.C000212200. [DOI] [PubMed] [Google Scholar]
- 50.Van de Putte T, Zwijsen A, Lonnoy O, Rybin V, Cozijnsen M, Francis A, et al. Mice with a homozygous gene trap vector insertion in mgcRacGAP die during pre-implantation development. Mech Dev. 2001;102:33–44. doi: 10.1016/s0925-4773(01)00279-9. [DOI] [PubMed] [Google Scholar]
- 51.Habets GG, Scholtes EH, Zuydgeest D, van der Kammen RA, Stam JC, Berns A, et al. Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell. 1994;77:537–549. doi: 10.1016/0092-8674(94)90216-x. [DOI] [PubMed] [Google Scholar]
- 52.Lambert JM, Lambert QT, Reuther GW, Malliri A, Siderovski DP, Sondek J, et al. Tiam1 mediates Ras activation of Rac by a PI(3)K-independent mechanism. Nat Cell Biol. 2002;4:621–625. doi: 10.1038/ncb833. [DOI] [PubMed] [Google Scholar]
- 53.Sugihara K, Nakatsuji N, Nakamura K, Nakao K, Hashimoto R, Otani H, et al. Rac1 is required for the formation of three germ layers during gastrulation. Oncogene. 1998;17:3427–3433. doi: 10.1038/sj.onc.1202595. [DOI] [PubMed] [Google Scholar]
- 54.Malliri A, van der Kammen RA, Clark K, van der Valk M, Michiels F, Collard JG. Mice deficient in the Rac activator Tiam1 are resistant to Ras-induced skin tumours. Nature. 2002;417:867–871. doi: 10.1038/nature00848. [DOI] [PubMed] [Google Scholar]
- 55.Hoshino M, Sone M, Fukata M, Kuroda S, Kaibuchi K, Nabeshima Y, et al. Identification of the stef gene that encodes a novel guanine nucleotide exchange factor specific for Rac1. J Biol Chem. 1999;274:17837–17844. doi: 10.1074/jbc.274.25.17837. [DOI] [PubMed] [Google Scholar]
- 56.Chiu CY, Leng S, Martin KA, Kim E, Gorman S, Duhl DM. Cloning and characterization of T-cell lymphoma invasion and metastasis 2 (TIAM2), a novel guanine nucleotide exchange factor related to TIAM1. Genomics. 1999;61:66–73. doi: 10.1006/geno.1999.5936. [DOI] [PubMed] [Google Scholar]
- 57.Welch HC, Coadwell WJ, Ellson CD, Ferguson GJ, Andrews SR, Erdjument-Bromage H, et al. P-Rex1, a PtdIns(3,4,5)P3- and Gbetagamma-regulated guanine-nucleotide exchange factor for Rac. Cell. 2002;108:809–821. doi: 10.1016/s0092-8674(02)00663-3. [DOI] [PubMed] [Google Scholar]
- 58.Donald S, Hill K, Lecureuil C, Barnouin R, Krugmann S, John Coadwell W, et al. P-Rex2, a new guanine-nucleotide exchange factor for Rac. FEBS Lett. 2004;572:172–176. doi: 10.1016/j.febslet.2004.06.096. [DOI] [PubMed] [Google Scholar]
- 59.Rosenfeldt H, Vazquez-Prado J, Gutkind JS. P-REX2, a novel PI-3-kinase sensitive Rac exchange factor. FEBS Lett. 2004;572:167–171. doi: 10.1016/j.febslet.2004.06.097. [DOI] [PubMed] [Google Scholar]
- 60.Welch HC, Condliffe AM, Milne LJ, Ferguson GJ, Hill K, Webb LM, et al. P-Rex1 regulates neutrophil function. Curr Biol. 2005;15:1867–1873. doi: 10.1016/j.cub.2005.09.050. [DOI] [PubMed] [Google Scholar]
- 61.Lindsay CR, Lawn S, Campbell AD, Faller WJ, Rambow F, Mort RL, et al. P-Rex1 is required for efficient melanoblast migration and melanoma metastasis. Nat Commun. 2011;2:555. doi: 10.1038/ncomms1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Donald S, Humby T, Fyfe I, Segonds-Pichon A, Walker SA, Andrews SR, et al. P-Rex2 regulates Purkinje cell dendrite morphology and motor coordination. Proc Natl Acad Sci U S A. 2008;105:4483–4488. doi: 10.1073/pnas.0712324105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Katzav S, Martin-Zanca D, Barbacid M. vav, a novel human oncogene derived from a locus ubiquitously expressed in hematopoietic cells. EMBO J. 1989;8:2283–2290. doi: 10.1002/j.1460-2075.1989.tb08354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bustelo XR, Ledbetter JA, Barbacid M. Product of vav proto-oncogene defines a new class of tyrosine protein kinase substrates. Nature. 1992;356:68–71. doi: 10.1038/356068a0. [DOI] [PubMed] [Google Scholar]
- 65.Henske EP, Short MP, Jozwiak S, Bovey CM, Ramlakhan S, Haines JL, et al. Identification of VAV2 on 9q34 and its exclusion as the tuberous sclerosis gene TSC1. Ann Hum Genet. 1995;59:25–37. doi: 10.1111/j.1469-1809.1995.tb01603.x. [DOI] [PubMed] [Google Scholar]
- 66.Schuebel KE, Bustelo XR, Nielsen DA, Song BJ, Barbacid M, Goldman D, et al. Isolation and characterization of murine vav2, a member of the vav family of proto-oncogenes. Oncogene. 1996;13:363–371. [PubMed] [Google Scholar]
- 67.Movilla NN, Bustelo XRX. Biological and regulatory properties of Vav-3, a new member of the Vav family of oncoproteins. Mol Cell Biol. 1999;19:7870–7885. doi: 10.1128/mcb.19.11.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trenkle T, McClelland M, Adlkofer K, Welsh J. Major transcript variants of VAV3, a new member of the VAV family of guanine nucleotide exchange factors. Gene. 2000;245:139–149. doi: 10.1016/s0378-1119(00)00026-3. [DOI] [PubMed] [Google Scholar]
- 69.Bustelo XR. Vav proteins, adaptors and cell signaling. Oncogene. 2001;20:6372–6381. doi: 10.1038/sj.onc.1204780. [DOI] [PubMed] [Google Scholar]
- 70.Aghazadeh B, Lowry WE, Huang XY, Rosen MK. Structural basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation. Cell. 2000;102:625–633. doi: 10.1016/s0092-8674(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 71.Yu B, Martins IR, Li P, Amarasinghe GK, Umetani J, Fernandez-Zapico ME, et al. Structural and energetic mechanisms of cooperative autoinhibition and activation of Vav1. Cell. 2010;140:246–256. doi: 10.1016/j.cell.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bustelo XR. Regulatory and signaling properties of the Vav family. Mol Cell Biol. 2000;20:1461–1477. doi: 10.1128/mcb.20.5.1461-1477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Turner M, Billadeau DD. VAV proteins as signal integrators for multi-subunit immune-recognition receptors. Nat Rev Immunol. 2002;2:476–486. doi: 10.1038/nri840. [DOI] [PubMed] [Google Scholar]
- 74.Tedford K, Nitschke L, Girkontaite I, Charlesworth A, Chan G, Sakk V, et al. Compensation between Vav-1 and Vav-2 in B cell development and antigen receptor signaling. Nat Immunol. 2001;2:548–555. doi: 10.1038/88756. [DOI] [PubMed] [Google Scholar]
- 75.Doody GM, Bell SE, Vigorito E, Clayton E, McAdam S, Tooze R, et al. Signal transduction through Vav-2 participates in humoral immune responses and B cell maturation. Nat Immunol. 2001;2:542–547. doi: 10.1038/88748. [DOI] [PubMed] [Google Scholar]
- 76.Fujikawa K, Miletic AV, Alt FW, Faccio R, Brown T, Hoog J, et al. Vav1/2/3-null Mice Define an Essential Role for Vav Family Proteins in Lymphocyte Development and Activation but a Differential Requirement in MAPK Signaling in T and B Cells. J Exp Med. 2003;198:1595–1608. doi: 10.1084/jem.20030874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sahai E, Marshall CJ. RHO–GTPase and cancer. Nat Rev Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 81.Jordan P, Brazao R, Boavida MG, Gespach C, Chastre E. Cloning of a novel human Rac1b splice variant with increased expression in colorectal tumors. Oncogene. 1999;18:6835–6839. doi: 10.1038/sj.onc.1203233. [DOI] [PubMed] [Google Scholar]
- 82.Schnelzer A, Prechtel D, Knaus U, Dehne K, Gerhard M, Graeff H, et al. Rac1 in human breast cancer: overexpression, mutation analysis, and characterization of a new isoform, Rac1b. Oncogene. 2000;19:3013–3020. doi: 10.1038/sj.onc.1203621. [DOI] [PubMed] [Google Scholar]
- 83.Eva A, Vecchio G, Diamond M, Tronick SR, Ron D, Cooper GM, et al. Independently activated dbl oncogenes exhibit similar yet distinct structural alterations. Oncogene. 1987;1:355–360. [PubMed] [Google Scholar]
- 84.Lazer G, Katzav S. Guanine nucleotide exchange factors for RhoGTPases: Good therapeutic targets for cancer therapy? Cell Signal. 2011;23:969–979. doi: 10.1016/j.cellsig.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 85.Barrio-Real L, Kazanietz MG. Rho GEFs and Cancer: Linking Gene Expression and Metastatic Dissemination. Sci Signal. 2012;5:pe43–pe43. doi: 10.1126/scisignal.2003543. [DOI] [PubMed] [Google Scholar]
- 86.Roversi G, Pfundt R, Moroni RF, Magnani I, van Reijmersdal S, Pollo B, et al. Identification of novel genomic markers related to progression to glioblastoma through genomic profiling of 25 primary glioma cell lines. Oncogene. 2006;25:1571–1583. doi: 10.1038/sj.onc.1209177. [DOI] [PubMed] [Google Scholar]
- 87.Sano M, Genkai N, Yajima N, Tsuchiya N, Homma J, Tanaka R, et al. Expression level of ECT2 proto-oncogene correlates with prognosis in glioma patients. Oncol Rep. 2006;16:1093–1098. [PubMed] [Google Scholar]
- 88.Salhia B, Tran NL, Chan A, Wolf A, Nakada M, Rutka F, et al. The guanine nucleotide exchange factors trio, Ect2, and Vav3 mediate the invasive behavior of glioblastoma. Am J Pathol. 2008;173:1828–1838. doi: 10.2353/ajpath.2008.080043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Justilien V, Fields AP. Ect2 links the PKCiota-Par6alpha complex to Rac1 activation and cellular transformation. Oncogene. 2009;28:3597–3607. doi: 10.1038/onc.2009.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hirata D, Yamabuki T, Miki D, Ito T, Tsuchiya E, Fujita M, et al. Involvement of epithelial cell transforming sequence-2 oncoantigen in lung and esophageal cancer progression. Clin Cancer Res. 2009;15:256–266. doi: 10.1158/1078-0432.CCR-08-1672. [DOI] [PubMed] [Google Scholar]
- 91.Zhang ML, Lu S, Zhou L, Zheng SS. Correlation between ECT2 gene expression and methylation change of ECT2 promoter region in pancreatic cancer. Hepatobiliary Pancreat Dis Int. 2008;7:533–538. [PubMed] [Google Scholar]
- 92.Iyoda M, Kasamatsu A, Ishigami T, Nakashima D, Endo-Sakamoto Y, Ogawara K, et al. Epithelial cell transforming sequence 2 in human oral cancer. PLoS One. 2010;5:e14082. doi: 10.1371/journal.pone.0014082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jung Y, Lee S, Choi HS, Kim SN, Lee E, Shin Y, et al. Clinical validation of colorectal cancer biomarkers identified from bioinformatics analysis of public expression data. Clin Cancer Res. 2011;17:700–709. doi: 10.1158/1078-0432.CCR-10-1300. [DOI] [PubMed] [Google Scholar]
- 94.Fields AP, Justilien V. The guanine nucleotide exchange factor (GEF) Ect2 is an oncogene in human cancer. Adv Enzyme Regul. 2010;50:190–200. doi: 10.1016/j.advenzreg.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weeks A, Okolowsky N, Golbourn B, Ivanchuk S, Smith C, Rutka JT. ECT2 and RASAL2 mediate mesenchymal-amoeboid transition in human astrocytoma cells. Am J Pathol. 2012;181:662–674. doi: 10.1016/j.ajpath.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 96.Qin J, Xie Y, Wang B, Hoshino M, Wolff DW, Zhao J, et al. Upregulation of PIP3-dependent Rac exchanger 1 (P-Rex1) promotes prostate cancer metastasis. Oncogene. 2009;28:1853–1863. doi: 10.1038/onc.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sosa MS, Lopez-Haber C, Yang C, Wang H, Lemmon MA, Busillo JM, et al. Identification of the Rac-GEF P-Rex1 as an essential mediator of ErbB signaling in breast cancer. Mol Cell. 2010;40:877–892. doi: 10.1016/j.molcel.2010.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Montero JC, Seoane S, Ocaña A, Pandiella A. P-Rex1 participates in Neuregulin-ErbB signal transduction and its expression correlates with patient outcome in breast cancer. Oncogene. 2011;30:1059–1071. doi: 10.1038/onc.2010.489. [DOI] [PubMed] [Google Scholar]
- 99.Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fine B, Hodakoski C, Koujak S, Su T, Saal LH, Maurer M, et al. Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science. 2009;325:1261–1265. doi: 10.1126/science.1173569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Uhlenbrock K, Eberth A, Herbrand U, Daryab N, Stege P, Meier F, et al. The RacGEF Tiam1 inhibits migration and invasion of metastatic melanoma via a novel adhesive mechanism. J cell Sci. 2004;117:4863–4871. doi: 10.1242/jcs.01367. [DOI] [PubMed] [Google Scholar]
- 102.Adam L. Tiam1 Overexpression Potentiates Heregulin-induced Lymphoid Enhancer Factor-1/beta -Catenin Nuclear Signaling in Breast Cancer Cells by Modulating the Intercellular Stability. J Biol Chem. 2001;276:28443–28450. doi: 10.1074/jbc.M009769200. [DOI] [PubMed] [Google Scholar]
- 103.Stebel A, Brachetti C, Kunkel M, Schmidt M, Fritz G. Progression of breast tumors is accompanied by a decrease in expression of the Rho guanine exchange factor Tiam1. Oncol Rep. 2009;21:217–222. [PubMed] [Google Scholar]
- 104.Minard ME, Ellis LM, Gallick GE. Tiam1 regulates cell adhesion, migration and apoptosis in colon tumor cells. Clin Exp Metastasis. 2006;23:301–313. doi: 10.1007/s10585-006-9040-z. [DOI] [PubMed] [Google Scholar]
- 105.Jin H, Li T, Ding Y, Deng Y, Zhang W, Yang H, et al. Methylation status of T-lymphoma invasion and metastasis 1 promoter and its overexpression in colorectal cancer. Hum Pathol. 2011;42:541–551. doi: 10.1016/j.humpath.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 106.Engers R, Mueller M, Walter A, Collard JG, Willers R, Gabbert HE. Prognostic relevance of Tiam1 protein expression in prostate carcinomas. Br J Cancer. 2006;95:1081–1086. doi: 10.1038/sj.bjc.6603385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Engers R. Rac Affects Invasion of Human Renal Cell Carcinomas by Up-regulating Tissue Inhibitor of Metalloproteinases (TIMP)-1 and TIMP-2 Expression. J Biol Chem. 2001;276:41889–41897. doi: 10.1074/jbc.M105049200. [DOI] [PubMed] [Google Scholar]
- 108.Heid I, Lubeseder-Martellato C, Sipos B, Mazur PK, Lesina M, Schmid RM, et al. Early requirement of Rac1 in a mouse model of pancreatic cancer. Gastroenterology. 2011;141:719–730. 730 e711–717. doi: 10.1053/j.gastro.2011.04.043. [DOI] [PubMed] [Google Scholar]
- 109.Malliri A, Rygiel TP, van der Kammen RA, Song JY, Engers R, Hurlstone AF, et al. The rac activator Tiam1 is a Wnt-responsive gene that modifies intestinal tumor development. J Biol Chem. 2006;281:543–548. doi: 10.1074/jbc.M507582200. [DOI] [PubMed] [Google Scholar]
- 110.Strumane K, Rygiel T, van der Valk M, Collard JG. Tiam1-deficiency impairs mammary tumor formation in MMTV-c-neu but not in MMTV-c-myc mice. J Cancer Res Clin Oncol. 2009;135:69–80. doi: 10.1007/s00432-008-0437-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu L, Zhang Q, Zhang Y, Wang S, Ding Y. Lentivirus-mediated silencing of Tiam1 gene influences multiple functions of a human colorectal cancer cell line. Neoplasia. 2006;8:917–924. doi: 10.1593/neo.06364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lane J, Martin TA, Mansel RE, Jiang WG. The expression and prognostic value of the guanine nucleotide exchange factors (GEFs) Trio, Vav1 and TIAM-1 in human breast cancer. Int Semin Surg Oncol. 2008;5:23. doi: 10.1186/1477-7800-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen JS, Su IJ, Leu YW, Young KC, Sun HS. Expression of T-cell lymphoma invasion and metastasis 2 (TIAM2) promotes proliferation and invasion of liver cancer. Int J Cancer. 2012;130:1302–1313. doi: 10.1002/ijc.26117. [DOI] [PubMed] [Google Scholar]
- 114.Hordijk PL, ten Klooster JP, van der Kammen RA, Michiels F, Oomen LC, Collard JG. Inhibition of invasion of epithelial cells by Tiam1-Rac signaling. Science. 1997;278:1464–1466. doi: 10.1126/science.278.5342.1464. [DOI] [PubMed] [Google Scholar]
- 115.Schuebel KE, Movilla N, Rosa JL, Bustelo XR. Phosphorylation-dependent and constitutive activation of Rho proteins by wild-type and oncogenic Vav-2. EMBO J. 1998;17:6608–6621. doi: 10.1093/emboj/17.22.6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Abe K, Rossman KL, Liu B, Ritola KD, Chiang D, Campbell SL, et al. Vav2 is an activator of Cdc42, Rac1, and RhoA. J Biol Chem. 2000;275:10141–10149. doi: 10.1074/jbc.275.14.10141. [DOI] [PubMed] [Google Scholar]
- 117.Hornstein I, Pikarsky E, Groysman M, Amir G, Peylan-Ramu N, Katzav S. The haematopoietic specific signal transducer Vav1 is expressed in a subset of human neuroblastomas. J Pathol. 2003;199:526–533. doi: 10.1002/path.1314. [DOI] [PubMed] [Google Scholar]
- 118.Lazer G, Idelchuk Y, Schapira V, Pikarsky E, Katzav S. The haematopoietic specific signal transducer Vav1 is aberrantly expressed in lung cancer and plays a role in tumourigenesis. J Pathol. 2009;219:25–34. doi: 10.1002/path.2579. [DOI] [PubMed] [Google Scholar]
- 119.Fernandez-Zapico ME, Gonzalez-Paz NC, Weiss E, Savoy DN, Molina JR, Fonseca R, et al. Ectopic expression of VAV1 reveals an unexpected role in pancreatic cancer tumorigenesis. Cancer cell. 2005;7:39–49. doi: 10.1016/j.ccr.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 120.Bartolome RA, Molina-Ortiz I, Samaniego R, Sanchez-Mateos P, Bustelo XR, Teixido J. Activation of Vav/Rho GTPase signaling by CXCL12 controls membrane-type matrix metalloproteinase-dependent melanoma cell invasion. Cancer Res. 2006;66:248–258. doi: 10.1158/0008-5472.CAN-05-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Prieto-Sanchez RM, Hernandez JA, Garcia JL, Gutierrez NC, San Miguel J, Bustelo XR, et al. Overexpression of the VAV proto-oncogene product is associated with B-cell chronic lymphocytic leukaemia displaying loss on 13q. Br J Haematol. 2006;133:642–645. doi: 10.1111/j.1365-2141.2006.06094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Citterio C, Menacho-Marquez M, Garcia-Escudero R, Larive RM, Barreiro O, Sanchez-Madrid F, et al. The Rho Exchange Factors Vav2 and Vav3 Control a Lung Metastasis-Specific Transcriptional Program in Breast Cancer Cells. Sci Signal. 2012;5:ra71–ra71. doi: 10.1126/scisignal.2002962. [DOI] [PubMed] [Google Scholar]
- 123.Dong Z, Liu Y, Lu S, Wang A, Lee KS, Ridley A. Vav3 oncogene is overexpressed and regulates cell growth and androgen receptor activity in human prostate cancer. Mol Endocrinol. 2006;10:2315–2325. doi: 10.1210/me.2006-0048. [DOI] [PubMed] [Google Scholar]
- 124.Lyons LS, Burnstein KL. Vav3, a Rho GTPase guanine nucleotide exchange factor, increases during progression to androgen independence in prostate cancer cells and potentiates androgen receptor transcriptional activity. Mol Endocrinol. 2006;20:1061–1072. doi: 10.1210/me.2005-0346. [DOI] [PubMed] [Google Scholar]
- 125.Lyons LS, Rao S, Balkan W, Faysal J, Maiorino CA, Burnstein KL. Ligand-independent activation of androgen receptors by Rho GTPase signaling in prostate cancer. Mol Endocrinol. 2008;22:597–608. doi: 10.1210/me.2007-0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rao S, Lyons LS, Fahrenholtz CD, Wu F, Farooq A, Balkan W, et al. A novel nuclear role for the Vav3 nucleotide exchange factor in androgen receptor coactivation in prostate cancer. Oncogene. 2012;31:716–727. doi: 10.1038/onc.2011.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu Y, Mo JQ, Hu Q, Boivin G, Levin L, Lu S, et al. Targeted overexpression of vav3 oncogene in prostatic epithelium induces nonbacterial prostatitis and prostate cancer. Cancer Res. 2008;68:6396–6406. doi: 10.1158/0008-5472.CAN-08-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lin KY, Wang LH, Hseu YC, Fang CL, Yang HL, Kumar KJ, et al. Clinical significance of increased guanine nucleotide exchange factor Vav3 expression in human gastric cancer. Mol Cancer Res. 2012;10:750–759. doi: 10.1158/1541-7786.MCR-11-0598-T. [DOI] [PubMed] [Google Scholar]
- 129.Nomura T, Yamasaki M, Hirai K, Inoue T, Sato R, Matsuura K, et al. Targeting the Vav3 oncogene enhances docetaxel-induced apoptosis through the inhibition of androgen receptor phosphorylation in LNCaP prostate cancer cells under chronic hypoxia. Mol Cancer. 2013;12:12–27. doi: 10.1186/1476-4598-12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pasteris NG, Cadle A, Logie LJ, Porteous ME, Schwartz CE, Stevenson RE, et al. Isolation and characterization of the faciogenital dysplasia (Aarskog-Scott syndrome) gene: a putative Rho/Rac guanine nucleotide exchange factor. Cell. 1994;79:669–678. doi: 10.1016/0092-8674(94)90552-5. [DOI] [PubMed] [Google Scholar]
- 131.Genot E, Daubon T, Sorrentino V, Buccione R. FGD1 as a central regulator of extracellular matrix remodelling--lessons from faciogenital dysplasia. J cell Sci. 2012;125:3265–3270. doi: 10.1242/jcs.093419. [DOI] [PubMed] [Google Scholar]
- 132.Delague V, Jacquier A, Hamadouche T, Poitelon Y, Baudot C, Boccaccio I, et al. Mutations in FGD4 encoding the Rho GDP/GTP exchange factor FRABIN cause autosomal recessive Charcot-Marie-Tooth type 4H. Am J Hum Genet. 2007;81:1–16. doi: 10.1086/518428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Stendel C, Roos A, Deconinck T, Pereira J, Castagner F, Niemann A, et al. Peripheral nerve demyelination caused by a mutant Rho GTPase guanine nucleotide exchange factor, frabin/FGD4. Am J Hum Genet. 2007;81:158–164. doi: 10.1086/518770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Balakrishnan A, Bleeker FE, Lamba S, Rodolfo M, Daniotti M, Scarpa A, et al. Novel somatic and germline mutations in cancer candidate genes in glioblastoma, melanoma, and pancreatic carcinoma. Cancer Res. 2007;67:3545–3550. doi: 10.1158/0008-5472.CAN-07-0065. [DOI] [PubMed] [Google Scholar]
- 135.Perry NA, Ackermann MA, Shriver M, Hu LY, Kontrogianni-Konstantopoulos A. Obscurins: Unassuming giants enter the spotlight. IUBMB Life. 2013;1157 doi: 10.1002/iub.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 137.Otomo A, Hadano S, Okada T, Mizumura H, Kunita R, Nishijima H, et al. ALS2, a novel guanine nucleotide exchange factor for the small GTPase Rab5, is implicated in endosomal dynamics. Hum Mol Genet. 2003;12:1671–1687. doi: 10.1093/hmg/ddg184. [DOI] [PubMed] [Google Scholar]
- 138.Topp JD, Gray NW, Gerard RD, Horazdovsky BF. Alsin is a Rab5 and Rac1 guanine nucleotide exchange factor. J Biol Chem. 2004;279:24612–24623. doi: 10.1074/jbc.M313504200. [DOI] [PubMed] [Google Scholar]
- 139.Kanekura K, Hashimoto Y, Kita Y, Sasabe J, Aiso S, Nishimoto I, et al. A Rac1/phosphatidylinositol 3-kinase/Akt3 anti-apoptotic pathway, triggered by AlsinLF, the product of the ALS2 gene, antagonizes Cu/Zn-superoxide dismutase (SOD1) mutant-induced motoneuronal cell death. J Biol Chem. 2005;280:4532–4543. doi: 10.1074/jbc.M410508200. [DOI] [PubMed] [Google Scholar]
- 140.Devon RS, Helm JR, Rouleau GA, Leitner Y, Lerman-Sagie T, Lev D, et al. The first nonsense mutation in alsin results in a homogeneous phenotype of infantile-onset ascending spastic paralysis with bulbar involvement in two siblings. Clin Genet. 2003;64:210–215. doi: 10.1034/j.1399-0004.2003.00138.x. [DOI] [PubMed] [Google Scholar]
- 141.Eymard-Pierre E, Lesca G, Dollet S, Santorelli FM, di Capua M, Bertini E, et al. Infantile-onset ascending hereditary spastic paralysis is associated with mutations in the alsin gene. Am J Hum Genet. 2002;71:518–527. doi: 10.1086/342359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gray BN, Anderson JE, Burton MA, van Hazel G, Codde J, Morgan C, et al. Regression of liver metastases following treatment with yttrium-90 microspheres. Aust N Z J Surg. 1992;62:105–110. doi: 10.1111/j.1445-2197.1992.tb00006.x. [DOI] [PubMed] [Google Scholar]
- 143.Yang Y, Hentati A, Deng HX, Dabbagh O, Sasaki T, Hirano M, et al. The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet. 2001;29:160–165. doi: 10.1038/ng1001-160. [DOI] [PubMed] [Google Scholar]
- 144.Roberts AE, Araki T, Swanson KD, Montgomery KT, Schiripo TA, Joshi VA, et al. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat Genet. 2007;39:70–74. doi: 10.1038/ng1926. [DOI] [PubMed] [Google Scholar]
- 145.Tartaglia M, Pennacchio LA, Zhao C, Yadav KK, Fodale V, Sarkozy A, et al. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat Genet. 2007;39:75–79. doi: 10.1038/ng1939. [DOI] [PubMed] [Google Scholar]
- 146.Aoki Y, Matsubara Y. Ras/MAPK syndromes and childhood hemato-oncological diseases. Int J Hematol. 2013;97:30–36. doi: 10.1007/s12185-012-1239-y. [DOI] [PubMed] [Google Scholar]
- 147.Hadano S, Hand CK, Osuga H, Yanagisawa Y, Otomo A, Devon RS, et al. A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet. 2001;29:166–173. doi: 10.1038/ng1001-166. [DOI] [PubMed] [Google Scholar]
- 148.Harvey K, Duguid IC, Alldred MJ, Beatty SE, Ward H, Keep NH, et al. The GDP-GTP exchange factor collybistin: an essential determinant of neuronal gephyrin clustering. J Neurosci. 2004;24:5816–5826. doi: 10.1523/JNEUROSCI.1184-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Marco EJ, Abidi FE, Bristow J, Dean WB, Cotter P, Jeremy RJ, et al. ARHGEF9 disruption in a female patient is associated with X linked mental retardation and sensory hyperarousal. J Med Genet. 2008;45:100–105. doi: 10.1136/jmg.2007.052324. [DOI] [PubMed] [Google Scholar]
- 150.Kalscheuer VM, Musante L, Fang C, Hoffmann K, Fuchs C, Carta E, et al. A balanced chromosomal translocation disrupting ARHGEF9 is associated with epilepsy, anxiety, aggression, and mental retardation. Hum Mutat. 2009;30:61–68. doi: 10.1002/humu.20814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lesca G, Till M, Labalme A, Vallee D, Hugonenq C, Philip N, et al. De novo Xq11. 11 microdeletion including ARHGEF9 in a boy with mental retardation, epilepsy, macrosomia, and dysmorphic features. Am J Med Genet A. 2011;155A:1706–1711. doi: 10.1002/ajmg.a.34004. [DOI] [PubMed] [Google Scholar]
- 152.Shimojima K, Sugawara M, Shichiji M, Mukaida S, Takayama R, Imai K, et al. Loss-of-function mutation of collybistin is responsible for X-linked mental retardation associated with epilepsy. J Hum Genet. 2011;56:561–565. doi: 10.1038/jhg.2011.58. [DOI] [PubMed] [Google Scholar]
- 153.Miller MB, Yan Y, Eipper BA, Mains RE. Neuronal Rho GEFs in Synaptic Physiology and Behavior. Neuroscientit. 2013;19:255–273. doi: 10.1177/1073858413475486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mandela P, Ma XM. Kalirin, a key player in synapse formation, is implicated in human diseases. Neural Plast. 2012;2012:728161. doi: 10.1155/2012/728161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Galan JE. Common themes in the design and function of bacterial effectors. Cell Host Microbe. 2009;5:571–579. doi: 10.1016/j.chom.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hardt WD, Chen LM, Schuebel KE, Bustelo XR, Galan JES. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell. 1998;93:815–826. doi: 10.1016/s0092-8674(00)81442-7. [DOI] [PubMed] [Google Scholar]
- 157.Alto NM, Shao F, Lazar CS, Brost RL, Chua G, Mattoo S, et al. Identification of a bacterial type III effector family with G protein mimicry functions. Cell. 2006;124:133–145. doi: 10.1016/j.cell.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 158.Buchwald G, Friebel A, Galan JE, Hardt WD, Wittinghofer A, Scheffzek K. Structural basis for the reversible activation of a Rho protein by the bacterial toxin SopE. EMBO J. 2002;21:3286–3295. doi: 10.1093/emboj/cdf329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dong N, Liu L, Shao F. A bacterial effector targets host DH-PH domain RhoGEFs and antagonizes macrophage phagocytosis. EMBO J. 2010;29:1363–1376. doi: 10.1038/emboj.2010.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mossessova E, Corpina RA, Goldberg J. Crystal structure of ARF1*Sec7 complexed with Brefeldin A and its implications for the guanine nucleotide exchange mechanism. Mol Cell. 2003;12:1403–1411. doi: 10.1016/s1097-2765(03)00475-1. [DOI] [PubMed] [Google Scholar]
- 161.Robineau S, Chabre M, Antonny B. Binding site of brefeldin A at the interface between the small G protein ADP-ribosylation factor 1 (ARF1) and the nucleotide-exchange factor Sec7 domain. Proc Natl Acad Sci U S A. 2000;97:9913–9918. doi: 10.1073/pnas.170290597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Peyroche A, Antonny B, Robineau S, Acker J, Cherfils J, Jackson CL. Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol Cell. 1999;3:275–285. doi: 10.1016/s1097-2765(00)80455-4. [DOI] [PubMed] [Google Scholar]
- 163.Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci U S A. 2004;101:7618–7623. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Shang X, Marchioni F, Sipes N, Evelyn CR, Jerabek-Willemsen M, Duhr S, et al. Rational design of small molecule inhibitors targeting RhoA subfamily Rho GTPases. Chem Biol. 2012;19:699–710. doi: 10.1016/j.chembiol.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Shang X, Marchioni F, Evelyn CR, Sipes N, Zhou X, Seibel W, et al. Small-molecule inhibitors targeting G-protein-coupled Rho guanine nucleotide exchange factors. Proc Natl Acad Sci U S A. 2013;110:3155–3160. doi: 10.1073/pnas.1212324110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, et al. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc Natl Acad Sci U S A. 2012;109:5299–5304. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sun Q, Burke JP, Phan J, Burns MC, Olejniczak ET, Waterson AG, et al. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew Chem Int Ed Engl. 2012;51:6140–6143. doi: 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hocker HJ, Cho KJ, Chen CY, Rambahal N, Sagineedu SR, Shaari K, et al. Andrographolide derivatives inhibit guanine nucleotide exchange and abrogate oncogenic Ras function. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1300016110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Creedon H, Brunton VG. Src kinase inhibitors: promising cancer therapeutics? Crit Rev Oncog. 2012;17:145–159. doi: 10.1615/critrevoncog.v17.i2.20. [DOI] [PubMed] [Google Scholar]
- 170.Mochly-Rosen D, Das K, Grimes KV. Protein kinase C, an elusive therapeutic target? Nat Rev Drug Discov. 2012;11:937–957. doi: 10.1038/nrd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rodon J, Dienstmann R, Serra V, Tabernero J. Development of PI3K inhibitors: lessons learned from early clinical trials. Nat Rev Clin Oncol. 2013;10:143–153. doi: 10.1038/nrclinonc.2013.10. [DOI] [PubMed] [Google Scholar]
- 172.Coleman N, Kissil J. Recent advances in the development of p21-activated kinase inhibitors. Cell Logist. 2012;2:132–135. doi: 10.4161/cl.21667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rath N, Olson MF. Rho-associated kinases in tumorigenesis: re-considering ROCK inhibition for cancer therapy. EMBO Rep. 2012;13:900–908. doi: 10.1038/embor.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2. 0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 175.Bustelo XR. Regulation of Vav proteins by intramolecular events. Front Biosci. 2002;7:d24–30. doi: 10.2741/A766. [DOI] [PubMed] [Google Scholar]
- 176.Miki T, Fleming TP, Bottaro DP, Rubin JS, Ron D, Aaronson SA. Expression cDNA cloning of the KGF receptor by creation of a transforming autocrine loop. Science. 1991;251:72–75. doi: 10.1126/science.1846048. [DOI] [PubMed] [Google Scholar]
- 177.Chan AM, McGovern ES, Catalano G, Fleming TP, Miki T. Expression cDNA cloning of a novel oncogene with sequence similarity to regulators of small GTP-binding proteins. Oncogene. 1994;9:1057–1063. [PubMed] [Google Scholar]
- 178.Chan AM, Takai S, Yamada K, Miki T. Isolation of a novel oncogene, NET1, from neuroepithelioma cells by expression cDNA cloning. Oncogene. 1996;12:1259–1266. [PubMed] [Google Scholar]
- 179.Toksoz D, Williams DA. Novel human oncogene lbc detected by transfection with distinct homology regions to signal transduction products. Oncogene. 1994;9:621–628. [PubMed] [Google Scholar]
- 180.Whitehead I, Kirk H, Tognon C, Trigo-Gonzalez G, Kay R. Expression cloning of lfc, a novel oncogene with structural similarities to guanine nucleotide exchange factors and to the regulatory region of protein kinase C. J Biol Chem. 1995;270:18388–18395. doi: 10.1074/jbc.270.31.18388. [DOI] [PubMed] [Google Scholar]
- 181.Horii Y, Beeler JF, Sakaguchi K, Tachibana M, Miki T. A novel oncogene, ost, encodes a guanine nucleotide exchange factor that potentially links Rho and Rac signaling pathways. EMBO J. 1994;13:4776–4786. doi: 10.1002/j.1460-2075.1994.tb06803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Whitehead I, Kirk H, Kay R. Retroviral transduction and oncogenic selection of a cDNA encoding Dbs, a homolog of the Dbl guanine nucleotide exchange factor. Oncogene. 1995;10:713–721. [PubMed] [Google Scholar]
- 183.Whitehead IP, Khosravi-Far R, Kirk H, Trigo-Gonzalez G, Der CJ, Kay R. Expression cloning of lsc, a novel oncogene with structural similarities to the Dbl family of guanine nucleotide exchange factors. J Biol Chem. 1996;271:18643–18650. doi: 10.1074/jbc.271.31.18643. [DOI] [PubMed] [Google Scholar]
- 184.Himmel KL, Bi F, Shen H, Jenkins NA, Copeland NG, Zheng Y, et al. Activation of clg, a novel dbl family guanine nucleotide exchange factor gene, by proviral insertion at evi24, a common integration site in B cell and myeloid leukemias. J Biol Chem. 2002;277:13463–13472. doi: 10.1074/jbc.M110981200. [DOI] [PubMed] [Google Scholar]
- 185.Kaartinen V, Gonzalez-Gomez I, Voncken JW, Haataja L, Faure E, Nagy A, et al. Abnormal function of astroglia lacking Abr and Bcr RacGAPs. Development. 2001;128:4217–4227. doi: 10.1242/dev.128.21.4217. [DOI] [PubMed] [Google Scholar]
- 186.Kaartinen V, Nagy A, Gonzalez-Gomez I, Groffen J, Heisterkamp N. Vestibular dysgenesis in mice lacking Abr and Bcr Cdc42/RacGAPs. Dev Dyn. 2002;223:517–525. doi: 10.1002/dvdy.10071. [DOI] [PubMed] [Google Scholar]
- 187.Kino T, Souvatzoglou E, Charmandari E, Ichijo T, Driggers P, Mayers C, et al. Rho family Guanine nucleotide exchange factor Brx couples extracellular signals to the glucocorticoid signaling system. J Biol Chem. 2006;281:9118–9126. doi: 10.1074/jbc.M509339200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Cai H, Lin X, Xie C, Laird FM, Lai C, Wen H, et al. Loss of ALS2 function is insufficient to trigger motor neuron degeneration in knock-out mice but predisposes neurons to oxidative stress. J Neurosci. 2005;25:7567–7574. doi: 10.1523/JNEUROSCI.1645-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hadano S, Benn SC, Kakuta S, Otomo A, Sudo K, Kunita R, et al. Mice deficient in the Rab5 guanine nucleotide exchange factor ALS2/alsin exhibit age-dependent neurological deficits and altered endosome trafficking. Hum Mol Genet. 2006;15:233–250. doi: 10.1093/hmg/ddi440. [DOI] [PubMed] [Google Scholar]
- 190.Devon RS, Orban PC, Gerrow K, Barbieri MA, Schwab C, Cao LP, et al. Als2-deficient mice exhibit disturbances in endosome trafficking associated with motor behavioral abnormalities. Proc Natl Acad Sci U S A. 2006;103:9595–9600. doi: 10.1073/pnas.0510197103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yamanaka K, Miller TM, McAlonis-Downes M, Chun SJ, Cleveland DW. Progressive spinal axonal degeneration and slowness in ALS2-deficient mice. Ann Neurol. 2006;60:95–104. doi: 10.1002/ana.20888. [DOI] [PubMed] [Google Scholar]
- 192.Deng HX, Zhai H, Fu R, Shi Y, Gorrie GH, Yang Y, et al. Distal axonopathy in an alsin-deficient mouse model. Hum Mol Genet. 2007;16:2911–2920. doi: 10.1093/hmg/ddm251. [DOI] [PubMed] [Google Scholar]
- 193.Kawasaki Y, Jigami T, Furukawa S, Sagara M, Echizen K, Shibata Y, et al. The adenomatous polyposis coli-associated guanine nucleotide exchange factor Asef is involved in angiogenesis. J Biol Chem. 2010;285:1199–1207. doi: 10.1074/jbc.M109.040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kawasaki Y, Tsuji S, Muroya K, Furukawa S, Shibata Y, Okuno M, et al. The adenomatous polyposis coli-associated exchange factors Asef and Asef2 are required for adenoma formation in Apc(Min/+)mice. EMBO Rep. 2009;10:1355–1362. doi: 10.1038/embor.2009.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Voncken JW, van Schaick H, Kaartinen V, Deemer K, Coates T, Landing B, et al. Increased neutrophil respiratory burst in bcr-null mutants. Cell. 1995;80:719–728. doi: 10.1016/0092-8674(95)90350-x. [DOI] [PubMed] [Google Scholar]
- 196.Papadopoulos T, Korte M, Eulenburg V, Kubota H, Retiounskaia M, Harvey RJ, et al. Impaired GABAergic transmission and altered hippocampal synaptic plasticity in collybistin-deficient mice. EMBO J. 2007;26:3888–3899. doi: 10.1038/sj.emboj.7601819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hirsch E, Pozzato M, Vercelli A, Barberis L, Azzolino O, Russo C, et al. Defective dendrite elongation but normal fertility in mice lacking the Rho-like GTPase activator Dbl. Mol Cell Biol. 2002;22:3140–3148. doi: 10.1128/MCB.22.9.3140-3148.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wirth A, Benyo Z, Lukasova M, Leutgeb B, Wettschureck N, Gorbey S, et al. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat Med. 2008;14:64–68. doi: 10.1038/nm1666. [DOI] [PubMed] [Google Scholar]
- 199.Mikelis CM, Palmby TR, Simaan M, Li W, Szabo R, Lyons R, et al. PDZ-RhoGEF and LARG Are Essential for Embryonic Development and Provide a Link between Thrombin and LPA Receptors and Rho Activation. J Biol Chem. 2013;288:12232–12243. doi: 10.1074/jbc.M112.428599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Girkontaite I, Missy K, Sakk V, Harenberg A, Tedford K, Potzel T, et al. Lsc is required for marginal zone B cells, regulation of lymphocyte motility and immune responses. Nat Immunol. 2001;2:855–862. doi: 10.1038/ni0901-855. [DOI] [PubMed] [Google Scholar]
- 201.Harenberg A, Girkontaite I, Giehl K, Fischer KD. The Lsc RhoGEF mediates signaling from thromboxane A2 to actin polymerization and apoptosis in thymocytes. Eur J Immunol. 2005;35:1977–1986. doi: 10.1002/eji.200425769. [DOI] [PubMed] [Google Scholar]
- 202.Rubtsov A, Strauch P, Digiacomo A, Hu J, Pelanda R, Torres RM. Lsc regulates marginal-zone B cell migration and adhesion and is required for the IgM T-dependent antibody response. Immunity. 2005;23:527–538. doi: 10.1016/j.immuni.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 203.Francis SA, Shen X, Young JB, Kaul P, Lerner DJ. Rho GEF Lsc is required for normal polarization, migration, and adhesion of formyl-peptide-stimulated neutrophils. Blood. 2006;107:1627–1635. doi: 10.1182/blood-2005-03-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zizer E, Beilke S, Bauerle T, Schilling K, Mohnle U, Adler G, et al. Loss of Lsc/p115 protein leads to neuronal hypoplasia in the esophagus and an achalasia-like phenotype in mice. Gastroenterology. 2010;139:1344–1354. doi: 10.1053/j.gastro.2010.06.041. [DOI] [PubMed] [Google Scholar]
- 205.Ma XM, Kiraly DD, Gaier ED, Wang Y, Kim EJ, Levine ES, et al. Kalirin-7 is required for synaptic structure and function. J Neurosci. 2008;28:12368–12382. doi: 10.1523/JNEUROSCI.4269-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Cahill ME, Xie Z, Day M, Photowala H, Barbolina MV, Miller CA, et al. Kalirin regulates cortical spine morphogenesis and disease-related behavioral phenotypes. Proc Natl Acad Sci U S A. 2009;106:13058–13063. doi: 10.1073/pnas.0904636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lange S, Ouyang K, Meyer G, Cui L, Cheng H, Lieber RL, et al. Obscurin determines the architecture of the longitudinal sarcoplasmic reticulum. J cell Sci. 2009;122:2640–2650. doi: 10.1242/jcs.046193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Missy K, Hu B, Schilling K, Harenberg A, Sakk V, Kuchenbecker K, et al. AlphaPIX Rho GTPase guanine nucleotide exchange factor regulates lymphocyte functions and antigen receptor signaling. Mol Cell Biol. 2008;28:3776–3789. doi: 10.1128/MCB.00507-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ramakers GJ, Wolfer D, Rosenberger G, Kuchenbecker K, Kreienkamp HJ, Prange-Kiel J, et al. Dysregulation of Rho GTPases in the alphaPix/Arhgef6 mouse model of X-linked intellectual disability is paralleled by impaired structural and synaptic plasticity and cognitive deficits. Hum Mol Genet. 2012;21:268–286. doi: 10.1093/hmg/ddr457. [DOI] [PubMed] [Google Scholar]
- 210.Brambilla R, Gnesutta N, Minichiello L, White G, Roylance AJ, Herron CE, et al. A role for the Ras signalling pathway in synaptic transmission and long-term memory. Nature. 1997;390:281–286. doi: 10.1038/36849. [DOI] [PubMed] [Google Scholar]
- 211.Itier JM, Tremp GL, Leonard JF, Multon MC, Ret G, Schweighoffer F, et al. Imprinted gene in postnatal growth role. Nature. 1998;393:125–126. doi: 10.1038/30120. [DOI] [PubMed] [Google Scholar]
- 212.Clapcott SJ, Peters J, Orban PC, Brambilla R, Graham CF. Two ENU-induced mutations in Rasgrf1 and early mouse growth retardation. Mamm Genome. 2003;14:495–505. doi: 10.1007/s00335-002-2258-4. [DOI] [PubMed] [Google Scholar]
- 213.Giese KP, Friedman E, Telliez JB, Fedorov NB, Wines M, Feig LA, et al. Hippocampus-dependent learning and memory is impaired in mice lacking the Ras-guanine-nucleotide releasing factor 1 (Ras-GRF1) Neuropharmacology. 2001;41:791–800. doi: 10.1016/s0028-3908(01)00096-x. [DOI] [PubMed] [Google Scholar]
- 214.Font de Mora J, Esteban LM, Burks DJ, Nunez A, Garces C, Garcia-Barrado MJ, et al. Ras-GRF1 signaling is required for normal beta-cell development and glucose homeostasis. EMBO J. 2003;22:3039–3049. doi: 10.1093/emboj/cdg280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Yoon B, Herman H, Hu B, Park YJ, Lindroth A, Bell A, et al. Rasgrf1 imprinting is regulated by a CTCF-dependent methylation-sensitive enhancer blocker. Mol Cell Biol. 2005;25:11184–11190. doi: 10.1128/MCB.25.24.11184-11190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Fernandez-Medarde A, Esteban LM, Nunez A, Porteros A, Tessarollo L, Santos E. Targeted disruption of Ras-Grf2 shows its dispensability for mouse growth and development. Mol Cell Biol. 2002;22:2498–2504. doi: 10.1128/MCB.22.8.2498-2504.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ruiz S, Santos E, Bustelo XR. RasGRF2, a guanosine nucleotide exchange factor for Ras GTPases, participates in T-cell signaling responses. Mol Cell Biol. 2007;27:8127–8142. doi: 10.1128/MCB.00912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Qian X, Esteban L, Vass WC, Upadhyaya C, Papageorge AG, Yienger K, et al. The Sos1 and Sos2 Ras-specific exchange factors: differences in placental expression and signaling properties. EMBO J. 2000;19:642–654. doi: 10.1093/emboj/19.4.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Esteban LM, Fernandez-Medarde A, Lopez E, Yienger K, Guerrero C, Ward JM, et al. Ras-guanine nucleotide exchange factor sos2 is dispensable for mouse growth and development. Mol Cell Biol. 2000;20:6410–6413. doi: 10.1128/mcb.20.17.6410-6413.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Yoo S, Kim Y, Lee H, Park S. A gene trap knockout of the Tiam-1 protein results in malformation of the early embryonic brain. Mol Cells. 2012;34:103–108. doi: 10.1007/s10059-012-0119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wang Z, Kumamoto Y, Wang P, Gan X, Lehmann D, Smrcka AV, et al. Regulation of immature dendritic cell migration by RhoA guanine nucleotide exchange factor Arhgef5. J Biol Chem. 2009;284:28599–28606. doi: 10.1074/jbc.M109.047282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Peng YJ, He WQ, Tang J, Tao T, Chen C, Gao YQ, et al. Trio is a key guanine nucleotide exchange factor coordinating regulation of the migration and morphogenesis of granule cells in the developing cerebellum. J Biol Chem. 2010;285:24834–24844. doi: 10.1074/jbc.M109.096537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zhang R, Alt FW, Davidson L, Orkin SH, Swat W. Defective signalling through the T- and B-cell antigen receptors in lymphoid cells lacking the vav proto-oncogene. Nature. 1995;374:470–473. doi: 10.1038/374470a0. [DOI] [PubMed] [Google Scholar]
- 224.Tarakhovsky A, Turner M, Schaal S, Mee PJ, Duddy LP, Rajewsky K, et al. Defective antigen receptor-mediated proliferation of B and T cells in the absence of Vav. Nature. 1995;374:467–470. doi: 10.1038/374467a0. [DOI] [PubMed] [Google Scholar]
- 225.Fischer KD, Zmuldzinas A, Gardner S, Barbacid M, Bernstein A, Guidos C. Defective T-cell receptor signalling and positive selection of Vav-deficient CD4+ CD8+ thymocytes. Nature. 1995;374:474–477. doi: 10.1038/374474a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.