Summary
Deubiquitinating enzymes (DUBs) recognize and cleave linkage-specific polyubiquitin (polyUb) chains, but mechanisms underlying specificity remain elusive in many cases. The severe acute respiratory syndrome (SARS) coronavirus papain-like protease (PLpro) is a DUB that cleaves ISG15, a two-domain Ub-like protein, and Lys48-linked polyUb chains, releasing diUbLys48 products. To elucidate this specificity, we report the 2.85 Å crystal structure of SARS PLpro bound to a diUbLys48 activity-based probe. SARS PLpro binds diUbLys48 in an extended conformation via two contact sites, S1 and S2, which are proximal and distal to the active site, respectively. We show that specificity for polyUbLys48 chains is predicated on contacts in the S2 site and enhanced by an S1-S1′ preference for a Lys48 linkage across the active site. In contrast, ISG15 specificity is dominated by contacts in the S1 site. Determinants revealed for polyUbLys48 specificity should prove useful in understanding PLpro deubiquitinating activities in coronavirus infections.
Graphical Abstract
Highlights
-
•
A Lys48 linkage-specific diubiquitin activity-based probe selectively labels SARS PLpro
-
•
The structure of a diUbLys48∼SARS PLpro complex reveals an extended di-Ub conformation
-
•
S2-S1 and S1-S1′ interactions make SARS PLpro specific for K48-linked polyubiquitin
-
•
SARS PLpro recognizes Lys48-linked polyUb chains and ISG15 via distinct manners
Békés et al. present a high-resolution crystal structure of a SARS virus PLpro∼diUbLys48 complex that reveals an extended conformation of the Lys48-linked diUb unit and shows the biochemical basis for SARS PLpro’s preference for Lys48-linked polyUb chains.
Introduction
Viruses can dampen the host anti-viral response by hijacking the ubiquitin (Ub) system (Bhoj and Chen, 2009, Isaacson and Ploegh, 2009) by expressing factors such as viral deubiquitinating enzymes (DUBs) that antagonize Ub-dependent pro-inflammatory pathways (Bailey-Elkin et al., 2014b, Capodagli et al., 2011, Mielech et al., 2014). For instance, the severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) human coronaviruses encode viral polypeptide processing proteases that can also catalyze deubiquitinating and deISGylating activities. The relevant host substrates of these viral DUBs and their mechanisms of target selection remain poorly defined.
Ub-dependent signaling pathways are regulated by the type of Ub modification (mono-ubiquitin [monoUb] or polyubiquitin [polyUb]) or type of Ub chain linkage utilized (Pickart, 2001), as polyUb chains can be formed by conjugation to any of Ub’s seven lysine (Lys) residues or its N-terminal methionine (Komander and Rape, 2012). DUB-mediated cleavage of Ub chains or conjugates serves as a critical regulator or antagonist of Ub-driven signaling pathways (Reyes-Turcu et al., 2009).
While the molecular basis for monoUb specificity for many human and viral DUBs was revealed in structures of monoUb-bound DUB complexes, the molecular basis of Ub chain linkage specificity is understood only for a handful of DUBs (Keusekotten et al., 2013, Mevissen et al., 2013, Sato et al., 2015). In these cases, specificity is dictated by direct readout of a particular isopeptide-linkage via binding the primed (S1′) Ub and unprimed, or proximal, (S1) Ub across the DUB active site. In one case, Lys11-specificity of the OTUD2 catalytic core is achieved by recognizing S1′ Ub and S1 Ub across the DUB active site, an activity that is aided by another domain that contacts a third Ub in a distal S2 site (Mevissen et al., 2013). Lys48-linked Ub chains are highly abundant in cells (Kirkpatrick et al., 2006), and their conjugation to other proteins targets them for proteasomal degradation (Chau et al., 1989), but the molecular basis for DUB recognition of Lys48-linked Ub chains remains unclear.
Ub-based chemical probes have been widely used to profile DUB activities and to stabilize monoUb-bound DUB complexes for structure determination (Ekkebus et al., 2013, Hemelaar et al., 2004). Total chemical synthesis of di-ubiquitin (diUb) activity-based probes (Ub-ABPs), with Cys-reactive warheads at the isopeptide linkage of choice (Mulder et al., 2014) enabled profiling of DUB linkage-specificities across S1-S1′; however these ABPs are not suitable for characterizing DUBs that recognize Ub chain topology through alternative mechanisms.
We and others recently showed that a coronavirus DUB, SARS papain-like protease (PLpro), but not MERS PLpro, preferentially recognizes and releases diUbLys48 units during cleavage of polyUb chains by an alternative mechanism that relies on SARS PLpro recognition of diUb via at least two binding sites in S2-S1, rather than S1-S1′ (Békés et al., 2015, Ratia et al., 2014). The structure of SARS PLpro bound to monoUb revealed surfaces required for S1 recognition and plausible explanations for Ub chain specificity and potential surfaces important for S2 recognition (Ratia et al., 2014), but the molecular basis for SARS PLpro Lys48-Ub chain specificity remains unknown. We report the crystal structure of SARS PLpro bound to a diUbLys48-ABP. The structure reveals SARS PLpro DUB recognition of an extended Lys48-linked diUb chain via distinct S1Ub and S2Ub binding sites, and biochemical studies show that S2Ub binding is most important for polyUb processing. The molecular basis for diUbLys48 recognition by a Lys48-specific DUB has remained unclear, and in this case, SARS PLpro Ub chain specificity is dominated by indirect readout of a unique diUb chain conformation at a site distal from the active site.
Results
A DiUbLys48 Activity-Based Probe Preferentially Labels SARS PLpro
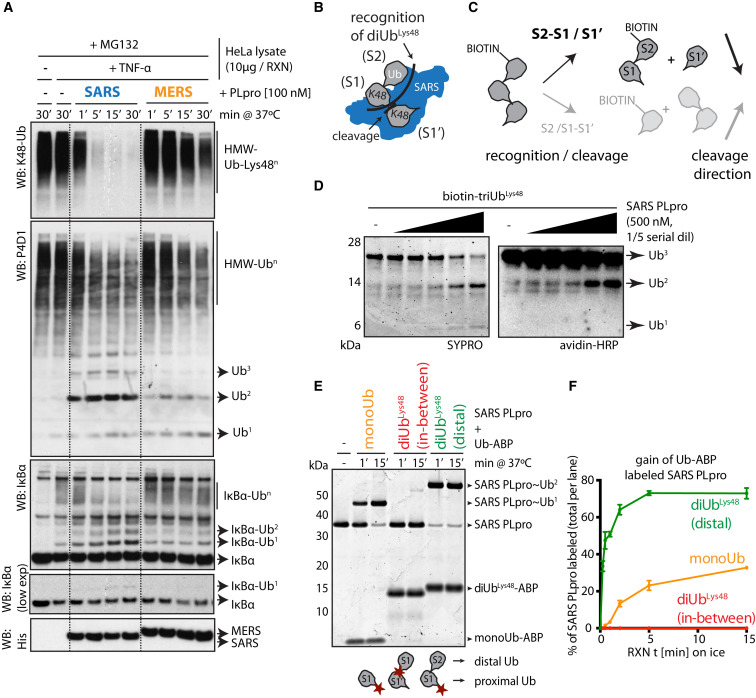
SARS PLpro and MERS PLpro are efficient deubiquitinating enzymes, on par with other human DUBs (Báez-Santos et al., 2014, Békés et al., 2015, Ratia et al., 2014); however, SARS PLpro rapidly removes Lys48-linked Ub chains from conjugated substrates, including polyubiquitinated IκBα stabilized by tumor necrosis factor α (TNF-α)/MG132 treatment (Figure 1 A), and is unique in its recognition and release of diUbLys48 units from chains of three or more Ubs (Békés et al., 2015). This unique activity is striking when compared to the related MERS PLpro, since they are structurally similar and share 52% amino acid sequence homology (Báez-Santos et al., 2014). While SARS PLpro efficiently cleaves higher-molecular-weight (HMW) polyUbLys48 conjugates, it exhibits poor activity in cleavage assays using free diUb chains or mono- or di-ubiquitinated substrates, such as IκBα (Figure 1A). These data supported a model whereby SARS PLpro uses distal Ub binding site (S2) to recognize diUbLys48 across S2-S1 (Figure 1B), rather than across S1-S1′, as is typical for most DUBs.
Figure 1.
Distal diUbLys48 ABP Labels SARS PLpro
(A) Ub-conjugate cleavage in TNF-α-treated HeLa cell lysates by SARS and MERS PLpro. Dotted lines added for clarity.
(B and C) Schematics of (B) Ub chain recognition by SARS PLpro and (C) recognition and cleavage of biotin-tagged triUbLys48.
(D) Cleavage of biotin-triUbLys48 by SARS PLpro. Cleavage intermediates detected by avidin-HRP reveal biotin on the diUb product.
(E) Qualitative labeling of SARS PLpro by Ub-ABPs (cartoons at bottom with red stars indicate warhead positions).
(F) Quantitative labeling of SARS PLpro by Ub-ABPs indicating percent of SARS PLpro labeled as derived from gels in Figure S2C. Error bars represent ±SEM.
See also Figures S1 and S2.
To provide evidence for this model, a singly N-terminal biotin-tagged triUbLys48 chain (Figure 1C) was generated (Figures S1A–S1G) and cleaved using SARS PLpro. Analysis of cleavage intermediates shows that the N-terminal biotin-label is retained on the diUb product (Figure 1D, right), suggesting that tri-ubiquitin (triUb) recognition requires binding via S2-S1, in a distal-to-proximal direction (Figure 1C, top schematics). In contrast, cleavage intermediates produced by other USP-family DUBs contain mixtures of mono- and diUb products bearing the biotin tag (Figure S2A). USP21CD and USP2CD show little preference, while MERS PLpro displays a slight preference.
We next took advantage of linkage-specific diUb activity-based probes that place warheads at the isopeptide linkage (Figure 1E, “in-between”; Mulder et al., 2014) or proximal end (Figure 1E, “distal diUbLys48,” green, right cartoon; Flierman et al., 2016). The distal diUb-ABP bears an isosteric non-hydrolyzable triazole linker in lieu of the native isopeptide linkage (Figure S2B). In labeling assays with SARS PLpro (Figure 1E), the distal diUbLys48-ABP (green) reacted well, monoUb-ABP (orange) reacted slowly, and the in-between diUbLys48-ABP (red) reacted poorly, and quantification shows distal diUbLys48-ABP adduct forms most efficiently in comparison to other probes (Figures 1F and S2C). The observation that SARS PLpro formed adducts least efficiently with the in-between diUbLys48-ABP probe (compare to monoUb-ABP), suggests that it might bind the in-between diUbLys48-ABP via S2-S1, preventing it from binding and reacting via S1-S1′ interactions (Figure S2D). Importantly, the in-between diUbLys48-ABP efficiently labels other DUBs that do not exhibit diUb preferences (Figures S2E and S2F). With the ideal reagent in hand, we set out to determine the structural basis for diUbLys48 recognition by SARS PLpro.
Crystal Structure of SARS PLpro Bound to a DiUbLys48-ABP
SARS PLpro was cross-linked to the distal diUbLys48-ABP, purified and crystallized. Crystals diffracted to 2.85 Å, and a structure of SARS PLpro-diUbLys48-ABP (Figure 2 A) was determined by molecular replacement (Supplemental Experimental Procedures). Two SARS PLpro-diUbLys48-ABP complexes occupy the asymmetric unit. The model was refined to an Rwork/Rfree of 23.2/26.4 with good stereochemistry (Table 1 ). One of the two complexes exhibits continuous electron density, while the other is less ordered with some discontinuity. Electron density is evident for the propargyl warhead of diUbLys48-ABP and active site Cys112 of SARS PLpro in both complexes (Figure S3A), but the diUb Lys48 isopeptide-mimic triazole linkage is weaker in one complex (Figure S3B).
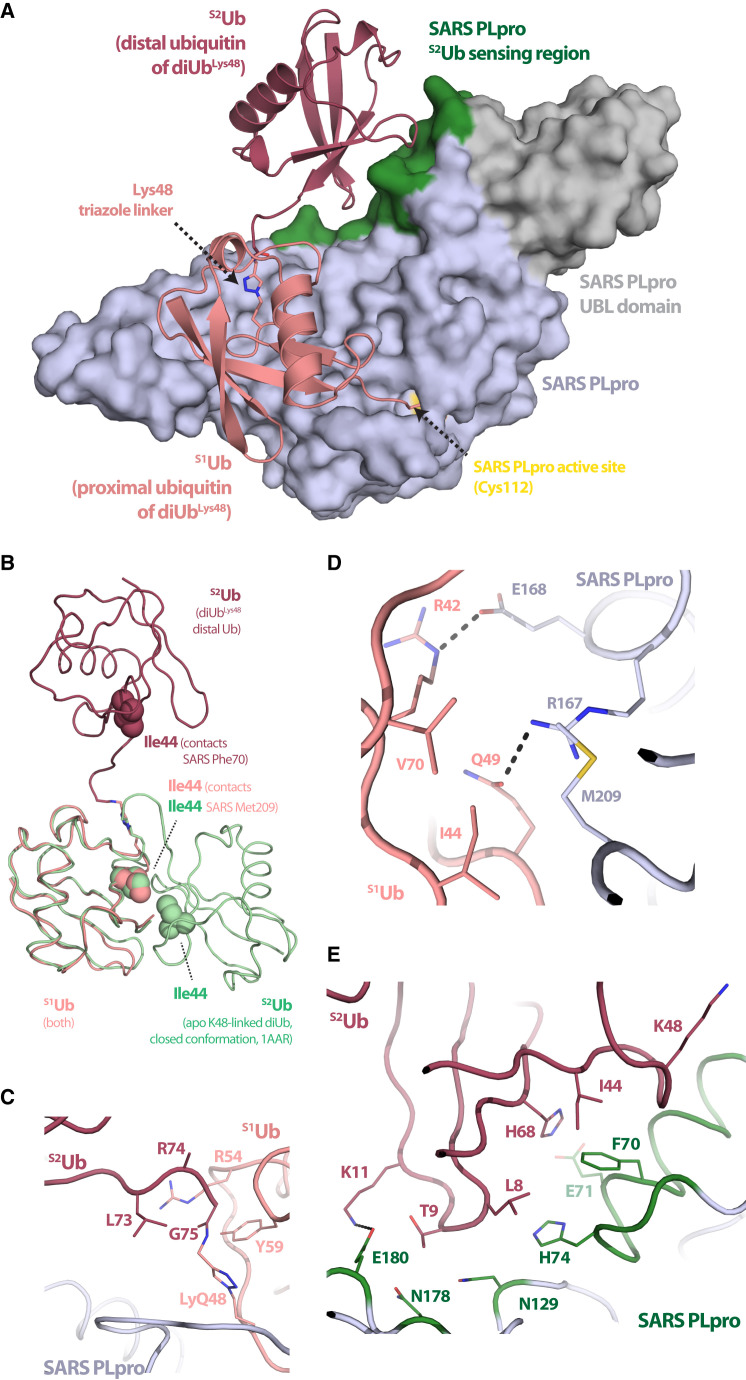
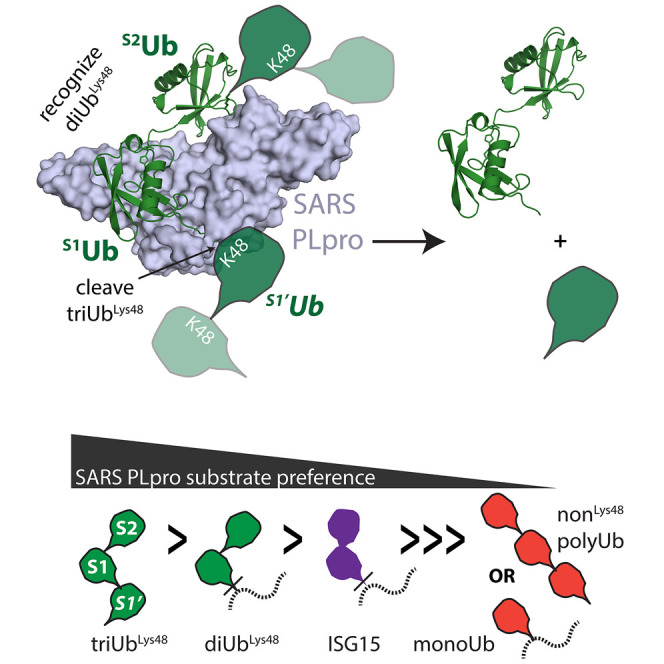
Figure 2.
Crystal Structure of SARS PLpro Bound to a diUbLys48-ABP
(A) Cartoon and surface representations of SARS PLpro covalently bound to diUbLys48-ABP. SARS PLpro shown in surface representation with the USP-family DUB domain colored blue-white, the N-terminal Ubl domain in gray, the S2Ub binding motif in green, and the active site cysteine (C112) in yellow. Cartoon representation of Lys48-linked diUb with proximal Ub (S1Ub) in salmon and distal Ub (S2Ub) in raspberry.
(B–E) Cartoon representation of interaction surfaces between SARS PLpro and diUbLys48; color as in (A). (B) Structure-based alignment of SARS-PLpro-diUbLys48 and a closed conformation of diUbLys48 (PDB 1AAR). Ub Ile44 (hydrophobic patch) that contact SARS PLpro shown as spheres. (C) View of the covalent triazole linkage between S1Ub-Lys48 and S2Ub-Gly75 indicating minimal contacts to SARS PLpro. (D) View of contacts between the SARS PLpro palm domain (M209 and Arg167/Asp168) and the S1Ub, highlighting both hydrophobic and polar interactions (S1Ub-Ile44 and -Gln49/Arg42), respectively. (E) View of contacts between the SARS PLpro S2Ub binding motif (Phe70, Glu71, and His74) and the S2Ub-Ile44 hydrophobic patch (S2Ub-Ile44, -Leu8, and -His68).
See also Figure S3.
Table 1.
Crystallographic Data and Refinement Statistics
| Data Collectiona | |
|---|---|
| Source | APS 24IDE |
| Wavelength (Å) | 0.9791 |
| Number of crystals | 1 |
| Space group | P21 |
| Cell dimensions | |
| a,b,c (Å) | 72.98, 68.24, 119.02 |
| α,β,γ (°) | 90.0, 103.21, 90 |
| Resolution (Å) | 50–2.85 (2.95–2.85) |
| Completeness (%) | 98.0 (100.0) |
| Total reflections | 86,261 (7,690) |
| Unique reflectionsa | 50,843 (5,048) |
| Wilson B-factor | 66.9 |
| Redundancy | 3.3 (3.0) |
| Rmerge (%) | 7.0 (53.5) |
| CC1/2 (%) | 99.7 (57.0) |
| CCa (%) | 99.9 (85.2) |
| < I >/σ(I) | 13.99 (1.97) |
| Refinementb | |
| Resolution (Å) | 50–2.85 (2.95–2.85) |
| Reflectionsc (work/free) | 50,864/2,590 |
| Rwork/Rfree (%) | 23.2 (36.5)/26.4 (40.3) |
| Number of atoms | 7366 |
| Protein | 7298 |
| Ligand | 13 |
| Water | 55 |
| Average B factors (Å2) | 79.8 |
| Protein | 79.9 |
| Ligand | 73.6 |
| Water | 58.1 |
| Rmsd | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.60 |
| MolProbityd | |
| Favored (%) | 93.3 (856) |
| Allowed (%) | 99.7 (853) |
| Outliers (%) | 0.3 (3) |
| Clash score | 100th percentile |
| MolProbity score | 100th percentile |
| PDB code | 5E6J |
Unique reflections for data collected and refinement include anomalous data.
Statistics calculated with Phenix; highest shell in parentheses.
Reflections includes Bijvoet pairs.
Calculated with the program MolProbity.
The diUbLys48-ABP-bound SARS PLpro structure reveals the basis for SARS PLpro catalytic domain recognition of proximal (S1Ub) and distal (S2Ub) Ub molecules within the context of a diUbLys48 unit. The SARS PLpro catalytic domain includes an N-terminal Ub-like (Ubl) domain that is dispensable for SARS PLpro activity (Békés et al., 2015, Mielech et al., 2014), followed by classical palm and finger DUB domains, as described for SARS PLpro (Ratia et al., 2006) and other USP-family member DUBs (Reyes-Turcu et al., 2009). The DUB catalytic module superposes well between diUbLys48-ABP-bound SARS PLpro and structures of apo (PDB: 2FE8) or monoUb-bound SARS PLpro (PDB: 4MM3; Figures S3C and S3D), with root-mean-square deviation (rmsd) values of 0.56 Å and 0.44 Å over 255 amino acids (Ser61-Ile315), respectively.
The orientation of diUbLys48 bound to SARS PLpro is different from prior structures of Lys48-linked Ub chains, whether bound or unbound; this is most apparent when our structure is compared to the “closed” conformation of diUbLys48 (PDB: 1AAR; Cook et al., 1992; Figure 2B). It appears that SARS PLpro stabilizes Lys48-linked Ub chains in an extended conformation, akin to conformations of Lys63- or Met1-linked diUb (Komander et al., 2009), yet distinct from those as well. Although SARS PLpro contacts S2Ub and S1Ub, it makes few contacts to the interface between S2Ub and S1Ub or the isopeptide analog (Figure 2C).
The position of S1Ub within SARS PLpro-diUbLys48-ABP is similar to the monoUb-SARS PLpro structure (Ratia et al., 2014), including contacts to the S1Ub C terminus; the S1Ub-Ile44 patch via Met209 of SARS, and polar contacts to S1Ub-Gln49 and S1Ub-Arg42 by the SARS PLpro palm domain via Arg167 and Asp168, respectively (Figure 2D). The related viral DUB, MERS PLpro, recognizes S1Ub in a similar manner, yet specific contacts to the S1Ub-Ile44 patch are not identical (Bailey-Elkin et al., 2014a). Additionally, SARS PLpro cradles S1Ub with its fingers domain, with S1Ub interaction surfaces comprising the largest buried interaction surface area (∼890 Å2). In comparison to apo SARS, both structures with monoUb and diUb bound reveal similar displacements of the BL2-loop (Figure S3E) that accommodates the Ub C-terminal tail in the active site (Ratia et al., 2014). When compared to the monoUb-bound SARS PLpro complex, a small conformational change is observed in S1Ub with respect to displacement of a loop between Ub amino acids 51–57 that is next to Lys48 and the triazole linkage (Figures S3F and S3G). To query if displacement could be due to the triazole linkage, our diUb structure was compared to Lys48-linked diUb (PDB 1AAR) revealing that amino acids 51–57 adopt a similar conformation to that observed in Lys48-linked diUb (Figure S3F), despite dissimilar S2Ub conformations (Figures 2B and S3G).
Recognition of S2Ub involves contacts centered on a hydrophobic interface between the S2Ub-Ile44 patch and a SARS PLpro α helix between the palm domain and N-terminal Ub-like (Ubl) domain that spans amino acids 62–74 (Figure 2E). The buried surface area in the S2Ub-SARS interface is smaller than the S1Ub-SARS interface (∼540 Å2 and ∼890 Å2, respectively) but includes contacts to S2Ub-Ile44, -His68, and -Leu8 by SARS PLpro residues Phe70, Glu71, and His74. S2Ub-Lys48 is exposed on the surface (Figure 2E), suggesting that additional Lys48-linked Ub molecules could be accommodated in the context of a polyUb chain. Other contacts to the S2Ub core include SARS PLpro residues Asn129, Asn178, and Glu180 from the palm domain, with the latter contacting S2Ub-Lys11 (Figure 2E).
Differential Contributions of S2Ub and S1Ub Binding Sites for PolyUbLys48 Cleavage
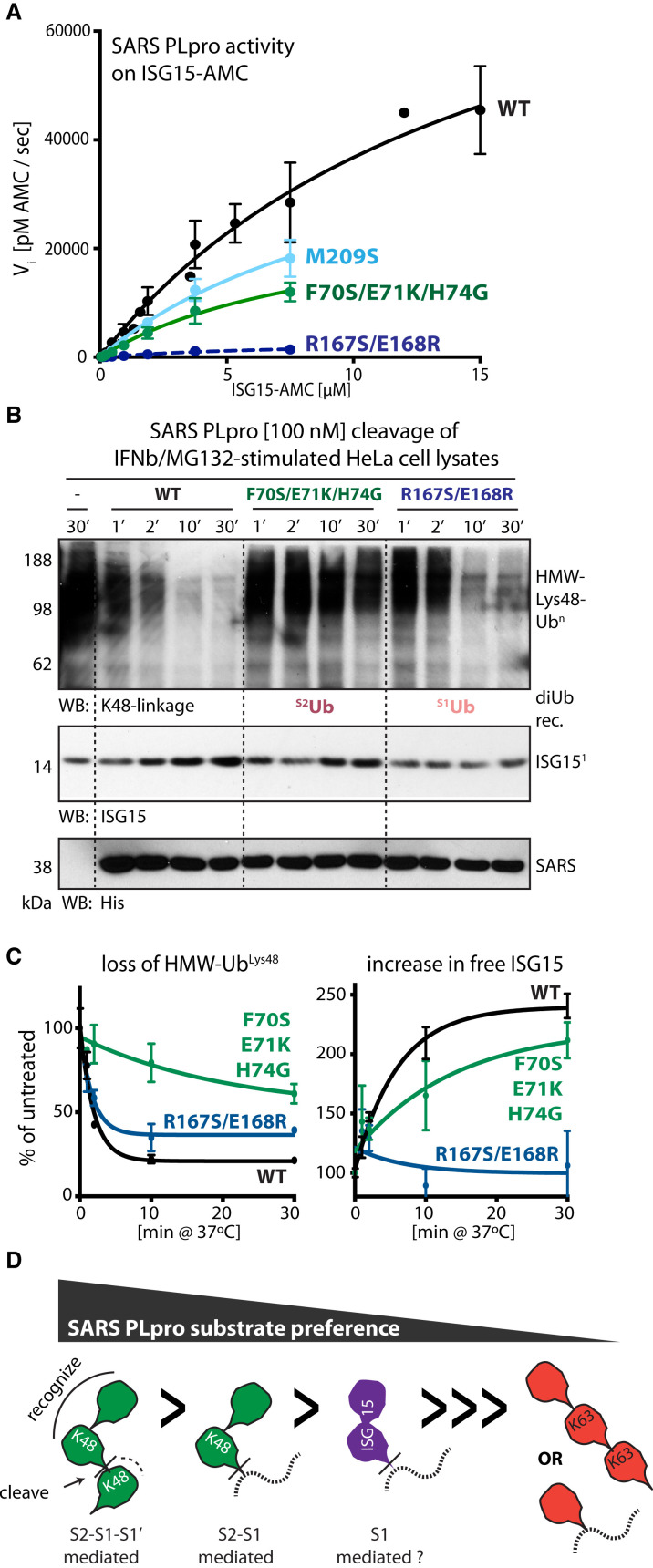
We next queried if proximal and distal Ub recognition are important for Ub chain processing by generating SARS PLpro mutants (Figure S4A) and assaying their activity on polyUb chains. Non-conservative substitutions of a cluster of residues in SARS PLpro that are in proximity to the S2Ub-Ile44 hydrophobic patch (e.g., SARS PLpro F70S/E71K/H74G) or individual substitutions F70S and H74G greatly reduce Ub chain cleaving activity by SARS PLpro, as assayed on pentaUbLys48 (Figure 3 A, top, green; Figure S4B) and tetraUbLys48 (Figure S4C). In contrast, SARS PLpro N178A/E180K and E180K substitutions, residues that contact S2Ub-Lys11, have a less pronounced effect on pentaUbLys48 (Figure 3A, top, green; Figure S4B) or tetraUbLys48 (Figure S4C). These data suggest the importance of distal S2Ub contacts, as the S1Ub binding surface remains intact in these mutants. Further supporting a dominant role for S2Ub interactions is the observation that mutation of residues surrounding the S1Ub-Ile44 patch have a modest effect compared to S2Ub-Ile44-disrupting mutations for pentaUbLys48 (Figure 3A, bottom, blue; Figure S4B) and tetraUbLys48 (Figure S4C) especially the M209S, R167S, R167S/E168R mutants. The E168R mutant, and a helix-swap mutant that replaces SARS residues with those in MERS PLpro (R167S/E168R+helix), have somewhat diminished activities. And while the N178A/E180K (S2Ub-contacting via Lys11) and R167S/E168R+helix mutants (S1Ub-contacting via Gln49/Arg42) display diminished polyUb cleaving ability, their defects are less than that observed for the F70S/E71K/H74G (S2Ub-contacting via Ile44) mutant. The catalytic mutant (C112A) has no activity.
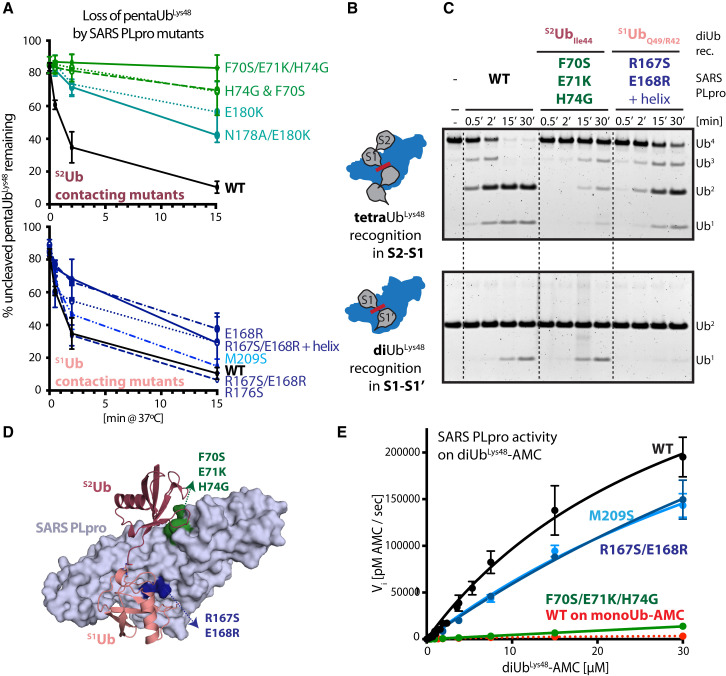
Figure 3.
Contribution of SARS PLpro S1Ub and S2Ub Sites to polyUbLys48 Cleavage
(A) Ub chain cleavage activities of SARS PLpro WT (black) and its mutants (S2Ub, top, green; S1Ub, bottom, blue) on pentaUbLys48. Representative gels used to derive graphs shown in Figure S3B. Error bars represent ±SEM.
(B) Schematics of tetraUbLys48 and diUbLys48 recognition by SARS PLpro.
(C) Gel-based cleavage assays of SARS PLpro WT and mutants on tetraUbLys48 (top) and diUbLys48 (bottom) indicating differential effects of diUb Lys48 and tetraUb Lys48 cleaving activities for S1Ub and S2Ub mutants of SARS PLpro. Additional mutants analyzed in Figure S4C.
(D) Cartoon and surface representation of SARS-PLpro∼diUbLys48 indicating the location of the S1Ub and S2Ub mutants.
(E) Michaelis-Menten kinetics of WT (black) and selected SARS PLpro mutants (M209S, hydrophobic S1 mutant, light blue; R167S/E168R, polar S1 mutant, dark blue; F70S/E71K/H74G, S2 mutant, green) on diUbLys48-AMC substrates and comparison to SARS PLpro WT on monoUb-AMC (in red). Extracted kinetic parameters (kcat and KM) are in Table 2.
See also Figure S4.
The relative contribution of S1Ub and S2Ub interactions within SARS PLpro was further probed by monitoring cleavage activity using tetraUbLys48 where cleavage depends on binding via S2-S1 (Figure 3B, top) and comparing this to diUbLys48 cleavage reactions that depend on binding via S1-S1′ (Figure 3B, bottom). It is worth noting that diUbLys48 cleavage by SARS PLpro requires 5-fold higher concentration of enzyme compared to tetraUbLys48 to observe activity. SARS PLpro wild-type (WT) activities were also compared to enzymes carrying mutations in the S1Ub and S2Ub binding sites using Lys48-linked diUbLys48 and tetraUbLys48 substrates (Figures 3C and 3D). As expected, the SARS PLpro S2Ub mutant (F70S/E71K/H74G) exhibits diminished activity against tetraUbLys48 (Figure 3C, top) yet retains WT-level activity on diUbLys48 (Figure 3C, bottom). Thus, S2Ub recognition is dispensable for diUbLys48 cleavage, consistent with an S1-S1′ binding mode being relevant for diUbLys48 cleavage. In contrast, the S1Ub mutant (R167S/E168R+helix) has diminished diUbLys48 cleavage activity yet retains the ability to cleave tetraUbLys48 chains with characteristic accumulation of diUb intermediates (Figure 3C). Single point mutants of the composite mutants (F70S for S2Ub, E168R for S1Ub) exhibit similar cleavage profiles (Figure S4C). These results suggest that mutations predicted to disrupt S1Ub recognition do not prevent cleavage of tetraUbLys48 when an intact S2Ub binding surface is present. As earlier, mutations within SARS PLpro predicted to disrupt contacts to S2Ub near Lys11 have a modest effect on tetraUbLys48 cleavage, and combining mutations designed to disrupt both S2Ub-Ile44 and -Lys11 patches are not additive (data not shown). Additionally, mutations designed to disrupt contacts to S2Ub, alone or in combination, do not display gain-of-function activity toward diUbLys48, suggesting that disrupting S2Ub interaction does not convert SARS PLpro into a DUB with stronger preference for S1-S1′ binding.
To quantify the contribution of S2Ub and S1Ub contacts with respect to diUbLys48 recognition by SARS PLpro, we turned to recently developed fluorogenic model diUbLys48-AMC substrates (containing a triazole-linker between Ub moieties), where the AMC fluorophore is conjugated to the proximal end of diUbLys48 (Flierman et al., 2016), enabling kinetic characterization of SARS PLpro by monitoring fluorescence during hydrolysis of the -AMC amide bond. Michaelis-Menten kinetic analysis of monoUb- and diUbLys48-AMC cleavage by SARS PLpro and its selected S2Ub and S1Ub mutants reveals that SARS PLpro cleaves the diUbLys48-AMC about ∼37-fold more efficiently than it cleaves monoUb-AMC (Figure 3E, black and red, respectively; Table 2 , compare apparent kcat/KM values of 3.3E+04 M−1s−1 for monoUb-AMC to 1.26E+06 M−1s−1 for diUbLys48-AMC). Moreover, kinetic comparison of SARS PLpro mutants reveals that loss of S2 interactions (F70S/E71K/H74G; Figure 3E, green; Table 2) results in an ∼33-fold loss of catalytic efficiency (kcat and KM could not be measured independently, and the S2 mutant could not be saturated by the diUbLys48-AMC substrate, indicative of a binding defect). Thus, the S2 mutant converts SARS PLpro into an S1-dependent, monoUb-based DUB. In contrast, mutation of either the hydrophobic interaction in the S1Ub site (M209S, Figure 3E, light blue; Table 2) or polar contacts to S1Ub (R167S/E168R, Figure 3E, dark blue; Table 2) results in a modest decrease in catalytic efficiency, an effect mainly driven by a 2- to 3-fold increase in KM without a corresponding loss in kcat (Table 2). Assaying additional point mutants at a single monoUb- or diUbLys48-AMC substrate concentration mirrors these trends (Figure S4D, top). Importantly, S2 mutants do not exhibit diminished monoUb-AMC cleavage rates (Figure S4D, bottom), consistent with gel-based experiments, as they maintained activity when processing diUbLys48 into monoUb.
Table 2.
Kinetic and Inhibition Parameters for SARS PLpro and Its Mutants on -AMC Substrates
| SARS PLpro Mutant | Kinetic Parameter | MonoUb-AMC | Triazole Linked DiUbLys48-AMC | Native DiUbLys48-AMC | ISG15-AMC |
|---|---|---|---|---|---|
| WT | Apparent kcat/KM [M−1s−1] | 3.33E+04 | 1.26E+06 | 1.01E+06a | 5.98E+05 |
| kcat [s−1] | 0.5042 ± 0.02839 | 42.02 ± 3.872 | n/a | 9.533 ± 1.218 | |
| KM [μM] | 15.12 ± 1.747 | 33.42 ± 4.869 | n/a | 15.94 ± 3.172 | |
| Fold kcat/KM over monoUb-AMC | 1.00 | 37.70 | 30.33a | 17.93 | |
| Michaelis-Menten curve fit (R2) | 0.9845 | 0.9668 | n/a | 0.9411 | |
| F70S E71K H74G (S2 mutant) | kcat/KM [M−1s−1] | — | 4.23E+04a | — | 2.94E+05 |
| kcat [s−1] | — | n/a | — | 2.748 ± 0.6693 | |
| KM [μM] | — | n/a | — | 9.359 ± 3.547 | |
| % kcat/KM of WT (per substrate) | — | 3.37 | — | 49.10 | |
| R167S E168R (S1 polar mutant) | kcat/KM [M−1s−1] | — | 6.50E+05 | — | 3.64E+04 |
| kcat [s−1] | — | 65.56 ± 22.38 | — | 0.318 ± 0.1184 | |
| KM [μM] | — | 100.8 ± 42.49 | — | 8.764 ± 5.614 | |
| % kcat/KM of WT (per substrate) | — | 51.63 | — | 6.08 | |
| M209S (S1 hydrophobic mutant) | kcat/KM [M−1s−1] | — | 7.06E+05 | — | 4.20E+05 |
| kcat [s−1] | — | 46.59 ± 10.29 | — | 4.774 ± 1.263 | |
| KM [μM] | — | 66.01 ± 19.61 | — | 11.88 ± 4.578 | |
| % kcat/KM of WT (per substrate) | — | 56.13 | — | 67.19 | |
| WT | Ki [μM] with monoUbb | NI | NI | NI | NI |
| Ki [μM] with diUbLys48 | 2.26 (0.9265) | 9.05 (0.8942) | 12.07 (0.8892) | 3.31 (0.8351) | |
| Ki [μM] with triUbLys48 | — | 10.57 (0.9509) | 10.11 (0.9066) | 4.08 (0.6709) | |
| Ki [μM] with ISG15 | NI | NI | NI | NI |
n/a, not applicable (kcat and KM cannot be independently calculated); NI, no detectable inhibition (IC50 > 100 μM or data do not converge).
Substrate not saturated, kcat/KM calculated from slope of linear graph.
Ki values were derived from IC50 values based on the equation Ki = IC50/(S/KM+1), assuming competitive inhibition, where S is the concentration of the substrate (based on Cer et al., 2009). Brackets show goodness of fit (R2) of IC50 values obtained from Prism’s log(inhibitor) versus normalized curve fit. Inhibition curves are shown in Figure S6A.
Structure-based alignment of the Lys48 linkage visible in the triazole-linked diUbLys48-ABP∼SARS-PLpro crystal structure to native Lys48 in free diUbLys48 (PDB: 1AAR) suggests that the triazole-linkage mimics the distance and geometry of a native isopeptide-bond (Figure S4E). To test if the triazole linkage is a good functional mimic of the native isopeptide-bond, we generated a native isopeptide-linked diUbLys48-AMC reagent (Figure S4F) and assayed initial cleavage rates for triazole-linked and native diUbLys48-AMC substrates (Figure S4G). Results indicate that the triazole linker is a faithful mimic of the isopeptide bond as initial cleavage rates are similar (despite native diUbLys48-AMC being contaminated with monoUb-AMC precursor; see Figure S4F).
SARS PLpro could not be saturated using the native substrate due to insufficient quantities, so inhibition studies were performed with triazole-linked or native isopeptide diUbLys48-AMC as substrates to calculate inhibition constants (Ki) using Lys48-linked Ub chains as inhibitors. Results in Table 2 (see also Figure S6A for inhibition curves) show that diUbLys48 and triUbLys48 inhibit triazole-linked or native diUbLys48-AMC cleavage with comparable Ki values (9 and 10 μM or 12 and 10 μM, respectively), values just ∼3-fold lower than the KM for diUbLys48-AMC as determined by Michaelis-Menten kinetics. Additionally, diUbLys48 inhibited monoUb-AMC hydrolysis with a Ki of ∼2.2 μM, a result consistent with kinetic assays where diUbLys48 recognition is preferred over monoUb. We were unable to observe inhibition of cleavage of diUbLys48-AMC substrates using monoUb or free ISG15 (Table 2).
Collectively, these results support the conclusion that distal S2Ub and proximal S1Ub binding surfaces are important for SARS PLpro activity, with S2Ub interactions being dominant, and that polyUb contacts to S2-S1 surfaces are preferred over interactions with S1-S1′ when processing Lys48-linked Ub chains.
S2-S1 Recognition by SARS PLpro Underlies Lys48 Ub Chain Linkage Specificity
DiUB can be linked via seven Ub lysine residues as well as its N terminus to alter their topology. Although dynamic, diUb conformations can be stabilized by interactions between Ub molecules, sometimes templated by their interacting partners (Ye et al., 2012). Linkage specificity for most DUBs characterized thus far is determined by contacts across the protease active site with diUb occupying S1-S1’ sites (Keusekotten et al., 2013, Mevissen et al., 2013, Sato et al., 2008, Sato et al., 2015). As shown previously, SARS PLpro is poor at cleaving diUb and does not strictly require a specific linkage across S1-S1′ (Békés et al., 2015). Given the extended conformation of diUbLys48 observed in complex with SARS PLpro and the paucity of contacts to the diUbLys48 linkage (Figure 2C), we hypothesized that SARS PLpro might indirectly sense linkage specificity by requiring that diUb occupies both S2Ub and S1Ub sites, a requirement that would place limits on the type of chain that could be accommodated because of differences in chain topology and distance between individual Ub molecules relative to the two Ub binding sites.
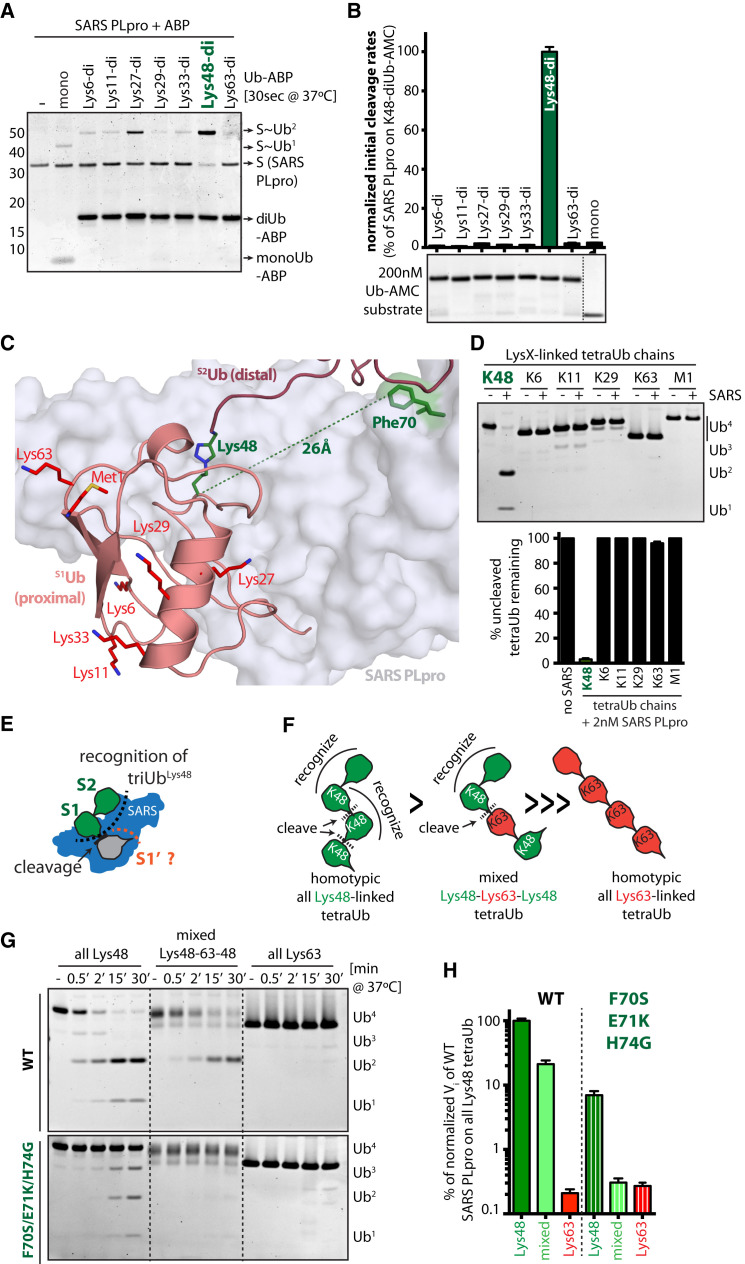
To assess S2-S1 linkage specificity of SARS PLpro, we tested a panel of linkage-specific distal diUb-ABPs to covalently label SARS PLpro. Although not as efficient as diUbLys48-ABP, diUbLys27-ABP exhibited better labeling efficiency compared other linkage-specific diUb-ABPs, which reacted at levels similar to monoUb-ABP (Figure 4 A and S5A). Since covalent activity-based probes are very reactive, we also tested linkage-specific diUb-AMC substrates by analyzing initial cleavage rates by SARS PLpro. These results suggest that SARS PLpro activity is highly restricted to diUbLys48-AMC cleavage (Figures 4B and S5B) as diUbLys48-AMC is cleaved ∼100-fold faster compared to other linkages. Each chain-forming residue in S1Ub is available for conjugation in our diUbLys48-SARS PLpro complex (Figure 4C), but the distance between S1Ub-Lys48 and the SARS-Phe70 S2Ub binding site is closest (26 Å) with each of the other sites requiring an additional 10 to 15 Å to span between the conjugated lysine and S2Ub binding site (Figure S5C). This raised the possibility that linkage-specificity across S2-S1 is enforced by restricting access to other topologies by requiring that SARS PLpro read the distance between diUb Ile44 hydrophobic patches via S2-S1 (Figure 2B), a requirement that is only satisfied by chains carrying Lys48 linkages. Accordingly, assaying homotypic linkage-specific tetra-ubiquitin (tetraUb) chains in endpoint cleavage assays also confirmed SARS PLpro to be Lys48-specific (Figure 4D).
Figure 4.
SARS PLpro Activity Is Restricted to Lys48-Linked Ub Chains with Specificity Dominated by S2-S1 Interactions
(A and B) S2-S1 linkage specificity of SARS PLpro probed by (A) SDS-PAGE analysis of cross-linking to linkage-specific diUb-ABPs (SYPRO-stained) and (B) release of AMC using diUb-AMC fluorogenic substrates with initial linear cleavage rates (Vi) plotted as percent of diUbLys48-AMC cleavage rate by WT SARS PLpro as derived from curves in Figure S4B.
(C) View of SARS-PLpro∼diUbLys48-ABP highlighting the location of S1Ub-Lys48 (dark green, sticks) and its proximity to Phe70 (green) in the SARS PLpro S2Ub binding site. Other chain-forming S1Ub residues (Lys-6, -11, -27, -29, -33, and -63 and Met1) shown as sticks (red).
(D) Linkage specificity of SARS PLpro assayed using homotypic tetraUb chains. Representative gel shown and bar graph indicating ±SEM from duplicate experiments.
(E) Schematics of triUbLys48 chain recognition by SARS PLpro via S2-S1 (green) and S1′ (gray).
(F) Cartoon of tetraUb chains, indicating SARS PLpro preferred sites of recognition (curved line) and cleavage (dotted line).
(G) Time-course for cleavage of tetraUb chains by SARS PLpro and its S2 mutant.
(H) Quantification of Vi as determined from duplicate experiments in Figure 4G. Error bars represent ±SEM.
See also Figure S5.
SARS PLpro Lys48 Specificity Is Also Aided by S1-S1′ Interactions
Most linkage-specific DUBs characterized to date rely on S1-S1′ interactions to achieve linkage-specificity, although OTUD2 and OTUD3 have been shown to utilize both S2-S1 and S1-S1′ interactions to mediate Lys11- and Lys6/11-specificity, respectively (Mevissen et al., 2013). We noticed greater defects for diUbLys48-AMC cleavage for some SARS PLpro mutants (especially E180K, N178A/E180K, E168R, and R67S/E168R+helix) compared to gel-based assays using polyUbLys48 substrates. As diUbLys48-AMC requires only S2-S1 interactions for cleavage, we wondered if additional interactions outside S2-S1 might be responsible for the residual specificity and activity observed for cleavage of polyUbLys48 substrates.
To determine if a Lys48-linkage was also preferred across S1-S1′ (Figure 4E), we assayed SARS PLpro and its S2Ub-Ile44 mutant (F70S/E71K/H74G) for cleavage of homotypic Lys48- and Lys63-linked tetraUb chains and a mixed linkage tetraUb chain consisting of two Lys48-linked dimers linked by Lys63 (see schematics in Figure 4F). This latter mixed chain could be recognized by SARS PLpro in S2-S1, but its cleavage would require accommodation of a Lys63-linkage across S1-S1′. A time course reveals that SARS PLpro readily cleaves homotypic Lys48 tetraUb chains but is inactive on homotypic Lys63 tetraUb chains. Interestingly, the mixed chain is cleaved when the Lys63 linkage is presented across S1-S1′, but only when an intact S2 site is present (Figure 4G). Initial cleavage rates (Figure 4H) show that SARS PLpro is ∼5-fold slower in cleaving the mixed chain compared to the homotypic Lys48-linked chain, suggesting some specificity for a Lys48 linkage across S1-S1′. More importantly, the S2Ub-Ile44 mutant exhibits faster cleavage rates on homotypic Lys48 chains than on mixed or on homotypic Lys63-linked chains (Figures 4G and 4H). Taken together, these data suggest a measurable specificity for Lys48 across S1-S1′, even when diUbLys48 recognition via S2-S1 is compromised. Thus, Lys48-specificity of SARS PLpro is enforced by S2-S1 recognition but complemented by a preference for Lys48 linkages across S1-S1′, suggesting that SARS PLpro would be most active on polyUbLys48 chains. Consistent with the hypothesis of S2-S1-S1′ recognition of polyUbLys48 chains, di- and monoUb-conjugated IκBα accumulates during cleavage by SARS PLpro (Figure 1D, IκBα), while unmodified IκBα remains static (Figure 1D, lighter exposure IκBα). These data suggest that SARS PLpro acts efficiently on polyUbLys48 chains in a diUb-dependent manner yet generates mono- and diUb-conjugated substrate remnants that may no longer represent its preferred substrate.
In the absence of a crystal structure of a diUbLys48 unit occupying S1-S1′ sites of SARS PLpro, we analyzed our structure for loops predicted to be proximal to S1′ within SARS PLpro that differ in sequence from MERS PLpro, which displays little linkage-specificity across S1-S1′ (Békés et al., 2015). We identified two residues, W107 and A108, adjacent to the exit tunnel of the SARS PLpro active site and mutated them to residues observed in MERS PLpro (Figure S5D). Cleavage assays with this putative S1′ mutant reveals that W107L/A108S has diminished activity on triUbLys48, which requires interactions with S2-S1-S1′ (Figures S5E and S5F), but exhibits no significant loss of activity when cleaving diUbLys48-AMC, a substrate that is solely dependent on S2-S1 (Figure S5F). This mutant, however, loses activity on Ub-AMC and ISG15-AMC (whose recognition is primarily S1 dependent). Combined with the observation that a Lys48-linked isopeptide is preferred as a substrate, these data suggest that S1′-dependent recognition of Lys48-linkages by SARS PLpro is possible, although additional work will be required to explore this hypothesis.
Recognition of DiUbLys48 and ISG15 Requires Distinct Elements within SARS PLpro
ISG15 is a tandem ubiquitin-like (Ubl) molecule consisting of two Ubl folds linked by a flexible hinge (Narasimhan et al., 2005). ISG15 is implicated in anti-viral immunity, and it is a preferred substrate of SARS PLpro when compared to monoUb. Indeed, ISG15-AMC is cleaved ∼20-fold faster than Ub-AMC (Table 2) (Békés et al., 2015, Lindner et al., 2007, Ratia et al., 2014). DiUbLys48-AMC (apparent kcat/KM of 1.26E+06 M−1s−1) is only preferred by ∼2-fold compared to ISG15-AMC (apparent kcat/KM of 5.98E+05 M−1s−1), suggesting that the preferred substrate for SARS PLpro is Lys48-linked polyUb chains.
The activity of SARS PLpro on ISG15 has been shown to be dependent on the distal Ubl within ISG15 (Lindner et al., 2007). Because the S2 binding site is important for diUbLys48 cleavage as described here and as proposed previously (Ratia et al., 2014), we sought to directly compare mutations in S2 and S1 of SARS PLpro and their impact on ISG15 cleavage activity. SARS PLpro S1Ub and S2Ub mutants were used to cleave ISG15-AMC using Michaelis-Menten kinetics (Figure 5 A). Mutations have contrasting effects for ISG15-AMC cleavage (Figure 5A; Table 2). The S1 polar mutant (Figure 5A, dark blue) exhibits greater defects for ISG15-AMC cleavage with minimal effects diUbLys48-AMC cleavage. In contrast, the S2 mutant exhibits major defects for diUbLys48-AMC cleavage with less severe defects in ISG15-AMC cleavage. To confirm these differential effects, we utilized lysates prepared from IFNβ/MG132-treated cells that contained polyUbLys48 chains and ISG15-conjugated substrates and added recombinant SARS PLpro mutants (Figure 5B). Analyzing loss of HMW polyUbLys48-conjugates and the appearance of free ISG15, indicative of cleavage of ISGylated substrates, reveals contrasting effects for S2 and S1 mutants (Figure 5C). While the S2 mutant compromises polyUbLys48 chain, but not ISG15 cleavage, the S1 mutant has the opposite effect. Finally, we compared other SARS PLpro mutants for cleavage of diUbLys48-AMC and ISG15-AMC (Figure S6B). Overall, S2Ub site mutants (green) show minimal loss of ISG15-AMC cleavage activity, while S1Ub mutants (blue), particularly those containing the E168R mutation, have a pronounced loss-of-function effect. Thus, ISG15 recognition appears more dependent on interactions within S1 and perhaps an alternative S2, while diUbLys48 recognition is more dependent on contacts within S2. It is difficult to rationalize these effects in the absence of a structure of ISG15 bound to SARS PLpro, but it appears clear that ISG15 recognition differs in details when compared to diUbLys48 recognition (Figure S6C).
Figure 5.
Recognition of DiUbLys48 and ISG15 by SARS PLpro Appears Distinct
(A) Michaelis-Menten kinetics of WT (black) and selected SARS PLpro mutants (M209S, S1 mutant, light blue; R167S/E168R, S1 mutant, dark blue; F70S/E71K/H74G, S2 mutant, green) using ISG15-AMC. Extracted kinetic parameters (kcat and KM) in Table 2.
(B) Cleavage of HMW-UbLys48 (top, WB anti-K48) and ISG15-conjugates (bottom, WB ISG15) in lysates prepared from IFNβ/MG132-treated cells by SARS PLpro WT and S2 and S1 mutants.
(C) Quantification of loss of HMW-UbLys48 (left) and appearance of free ISG15 (right) from duplicate experiments shown in Figure 5B. Error bars represent ±SEM.
(D) Schematic representation of SARS PLpro substrate specificity. Dashed lines can indicate the -AMC substrate, a non-Lys48-linked Ub unit, or a protein substrate.
See also Figure S6.
Discussion
SARS PLpro appears unique among viral and human DUBs characterized thus far in its ability to recognize polyUb chains by reading units of a Lys48-linked diUb (Békés et al., 2015). Here, we reveal the structural basis for diUbLys48 recognition and specificity by SARS PLpro. Coupled with mutational, biochemical and kinetic data, our structure helps to explain the strict Lys48-linkage specificity exhibited by this viral DUB, which is primarily enforced by engaging the diUb module within S2-S1, and enhanced by a slight preference for Lys48-linked Ub across S1-S1′ (Figure 5D). To our knowledge, the diUbLys48-SARS PLpro structure represents the only available structure of a linkage-specific DUB bound to a Lys48-linked Ub chain, a result enabled by recently developed diUb activity-based probes (Flierman et al., 2016).
Instead of relying on diUb recognition across S1-S1′, as is common among other DUBs (Keusekotten et al., 2013, Mevissen et al., 2013, Sato et al., 2015) and endoproteases (Berger and Schechter, 1970), SARS PLpro recognizes diUb across S2-S1 binding surfaces that are tuned to recognize Lys48-linked Ub chains. This mode of Ub chain recognition has only been reported for the catalytic core of OTUD2, where an additional binding site mediates interaction with a Lys11-linked diUb (Mevissen et al., 2013). Through modeling, mutational analysis, and the crystal structure of a monoUb-bound SARS PLpro, the Mesecar group suggested that SARS PLpro is an S2-S1 mode DUB (Ratia et al., 2014) and that the distal Ub would be recognized by a hydrophobic surface in SARS PLpro, involving Phe70. Our current study illuminates the structural basis for this interaction. We provide biochemical evidence that the S2Ub interface has a dominant role in diUbLys48 recognition and polyUb chain cleaving activity, since mutating the distal S2Ub recognition surface is more detrimental than disrupting the proximal S1Ub recognition surface. We also provide evidence that SARS PLpro Lys48-specificity is complemented by a preference for Lys48-linked chains across S1-S1′.
The hydrophobic patches in Ub proteins are usually packed against each other in free UbLys48 chains (Fushman and Wilkinson, 2011). In the diUbLys48-SARS PLpro complex, diUbLys48 exhibits an extended conformation, with Ub hydrophobic patches separated by ∼30 Å and recognized by contacts provided by the S1Ub and S2Ub sites in the diUbLys48-SARS PLpro complex. As Ub chains are dynamic in solution (Ye et al., 2012), and because SARS PLpro is structurally similar in apo, monoUb, and diUbLys48 complexes, it is likely that SARS PLpro captures diUb units in this conformation.
While SARS PLpro exhibits an ∼5-fold preference for a Lys48-linkage in the S1-S1′ binding mode when an S2 site is also occupied, a lax requirement at S1′ is consistent with its function as an endopeptidase for viral pre-protein processing and perhaps cleavage of ISG15 substrates. The biological targets of SARS PLpro remain unclear, but the preference for polyUbLys48 chain cleavage into units of diUbLys48 suggests it is likely targeted to substrates that are modified by Lys48-linked polyUb chains.
The diUb-based recognition exhibited by SARS PLpro suggests that SARS PLpro could stabilize monoUb-modified substrate products (Békés et al., 2015), as they are not preferred substrates for SARS PLpro. Indeed, cleavage of Lys48-linked polyUb chains from IκBα by SARS PLpro led to an increase in di- and monoUb-conjugated forms of IκBα. Whether SARS PLpro cleaves other polyUbLys48-conjugated substrates to di- or monoUb-conjugated forms remains to be determined. Mono-ubiquitination at membranes and at the endoplasmic reticulum (ER) has been shown to regulate endocytosis and vesicle trafficking (Clague et al., 2012), which are also involved in coronavirus propagation. Given that SARS PLpro is ER localized, it is possible that SARS PLpro functions to stabilize mono-Ub “stubs” on ER substrates to provide an unknown advantage for the virus.
It remains unclear if the anti-inflammatory properties of SARS PLpro require all or a combination of its endopeptidase, DUB, or deISGylating activities. With mutations described herein, which bias activities in deubiquitination versus deISGylation, it may be possible to discern if both activities are important during SARS infection. With that said, it is worth noting that SARS viral titer levels peak at 16–20 hr in cell culture and in mouse model infection studies (Channappanavar et al., 2016, Totura and Baric, 2012), while interferon (IFN)-responsive genes, such as ISG15, are only induced later during infection (Channappanavar et al., 2016). As such, it is likely that the virus has already achieved its full replicative potential before SARS PLpro would encounter ISG15. Additionally, the function of substrate-conjugated ISG15 in anti-viral immunity in humans is now in question (Bogunovic et al., 2012), as a non-conjugatable form of ISG15 was shown to have similar activities as WT ISG15, likely via stabilization of USP18 (Zhang et al., 2015). Given the preference for polyUbLys48 and diUbLys48 over ISG15, it appears likely that polyUbLys48-conjugated substrates are the primary cellular targets of SARS PLpro. These observations suggest that SARS PLpro activities against ISG15 targets may not be as relevant for coronavirus infection as previously thought.
Although the identity of true SARS PLpro substrates remains to be determined, they could include host factors involved in anti-viral signaling, such as IκBα, or viral proteins targeted for degradation by host anti-viral E3 ligases. We propose a model where the most favored substrates for SARS PLpro would be Lys48-linked polyUb chains (S2-S1-S1′ dependent), followed by diUbLys48-conjugates (S2-S1 dependent), followed by ISG15-conjugates, with the least favored substrates being monoUb-conjugated substrates and other polyUb chains (only S1 dependent) (Figure 5D).
Experimental Procedures
Synthesis of the Singly Biotinylated TriUbLys48 Substrate
Biotinylated triUbLys48 was generated using procedures based on previously reported protocols (El Oualid et al., 2010) with modification described in Supplemental Experimental Procedures.
Cloning, Protein Expression, Purification, Crystallization, and Structure Determination of the SARS-PLpro∼DiUbLys48-ABP Complex
The generation of recombinant SARS PLpro was described elsewhere (Békés et al., 2015), with modifications as described in Supplemental Experimental Procedures. SARS PLpro (45 μM) was reacted with diUbLys48-PRG (45 μM) for 30 min at 37°C in 20 mM Tris (pH 8.0), 150 mM NaCl, and 5 mM DTT; purified by size-exclusion chromatography; concentrated to 11 mg/ml; and frozen in liquid nitrogen for storage (−80°C). Diffraction-quality crystals (∼50–100 μm) grew for 1 month at 12°C by hanging drop vapor diffusion against 0.1 M MES (2-(N-morpholino)-ethanesulfonic acid) (pH 5.5), 0.1 M lithium-acetate, and 12%–20% PEGs 4000/6000/8000. Single crystals were cryo-protected by addition of 20% ethylene glycol and flash cooled in liquid nitrogen. Diffraction data were collected from a single crystal and processed, and the structure was determined using methods reported in Supplemental Experimental Procedures.
Ub-ABP Labeling Assays
For qualitative assays, DUBs (1 μM) were incubated with excess activity-based probes (2–5 μM, monoUb-PRG, diUbLys48-VME [“in-between” diUb-ABP]; Mulder et al., 2014) or diUbLys48-PRG (“distal” diUb-ABP; Flierman et al., 2016) for indicated times at 37°C in 20 mM Tris (pH 8.0), 150 mM NaCl, and 5 mM DTT. Reactions were performed at least in duplicate. For linkage-specific distal diUb-ABPs, TAMRA-labeled probes were used in a 30-s labeling assay at 37°C. Reactions were quenched with loading sample buffer (4× LDS [Invitrogen], with 5 mM DTT), and analyzed by SDS-PAGE and SYPRO-staining. Gels were scanned to visualize the TAMRA-label (488 nM), imaged using Bio-Rad Gel-Doc, quantified by ImageJ software, and graphed with Prism. Error bars represent ±SEM.
Kinetic Assays with -AMC Substrates
To determine apparent kcat/KM for SARS PLpro and its mutants, monoUb-AMC, diUbLys48-AMC, and ISG15-AMC were prepared as 2-fold serial dilutions (starting at 30 μM: monoUb and diUbLys48 [triazole-linked]; at 15 μM: ISG15-AMC) in 20 mM Tris (pH 8.0), 150 mM NaCl, and 5 mM DTT. SARS PLpro was used at 10 nM (diUbLys48- and ISG15-AMC) or 50 nM (monoUb-AMC), and the final reaction volume was 10 μl. Substrates and DUBs were preincubated at 25°C for 1 min, and cleavage of UBL-AMCs was performed at 30°C using a Spectramax fluorescence plate reader running SoftMax Pro 5 (Molecular Devices) operated in kinetic mode in black, round-bottom 384-well plates (Corning, #3698). AMC fluorescence was monitored by excitation at 355 nm and emission at 460 nm over time for 5–10 min. Initial linear cleavage rates (Vi) were fitted by the Michaelis-Menten equation by Prism based on a free AMC standard curve. Experiments were performed at least in triplicate, and error bars indicate ±SEM. To compare triazole-linked or native diUbLys48-AMC, substrates were prepared as 2-fold serial dilutions of 3.75 μM (limited by the concentration of the native diUbLys48-AMC). Assays were performed in duplicate. To compare individual SARS PLpro mutants, monoUb-AMC, diUbLys48-AMC, and ISG15-AMC were used at 400 nM final concentrations, using 10 or 50 nM SARS PLpro. Data were plotted as percent of WT cleavage rates for each substrate (n = 3), and error bars indicate ±SEM. To compare linkage-specific diUb-AMC substrates using SARS PLpro, diUb-AMC substrates were used at 200 nM final concentrations, using 5 nM SARS PLpro. Data were plotted as percent of the diUbLys48-AMC cleavage rate (n = 3), and error bars indicate ±SEM.
Gel-Based Ub Chain Cleavage Assays
Ub chains (1 μM; 20 mM Tris [pH 8.0], 150 mM NaCl, and 5 mM DTT) were cleaved at 37°C for indicated times by SARS PLpro WT and its mutants at 10 nM for penta-, tetra-, and triUbLys48 or 50 nM of diUbLys48 cleavage. Reactions were quenched with loading sample buffer (4× LDS, Invitrogen) and analyzed by SDS-PAGE and SYPRO-staining. Gels imaged using a Bio-Rad Gel-Doc, quantified by ImageJ, cropped where indicated by heavy dashed lines, and graphed using Prism. Assays using SARS mutants were performed in batches; mutants were always compared to cleavage by WT. Loss of uncleaved substrate is expressed as a percent value of uncleaved substrate over the total Ub signal per lane. Initial cleavage rates were calculated from linear portions of curves showing loss of uncleaved substrate over time and expressed as percent of WT rates in arbitrary units. Reactions performed at least in duplicate. Error bars represent ±SEM.
DUB Assay in Lysates
Lysates from human interferon beta (IFN-β; 500 U/ml, 48 hr) or TNF-α (10 ng/ml, 10 min) and MG132 (10 μM, 40 min) stimulated HeLa cells (10 μg total lysate per reaction) were incubated with 100 or 50 nM DUBs, as indicated, in 20-μl reaction volumes with 25 mM DTT for indicated times. Reactions terminated by heating in SDS loading buffer, analyzed by SDS-PAGE and western blotting with indicated antibodies. Blots developed by horseradish peroxidase (HRP) chemiluminescence. Films were scanned, cropped where indicated by heavy dashed lines, quantified and graphed as described above. Light dashed lines in all figures included for clarity.
Author Contributions
M.B. initiated and performed the study with guidance from T.T.H. and C.D.L. G.J.v.d.H.v.N. synthesized diUbLysX-AMC substrates and biotin-triUbLys48; R.E. synthesized distal diUbLysX-ABP probes in H.O.’s laboratory. M.B. and C.D.L. determined the structure and wrote the manuscript.
Acknowledgments
The authors thank members of the T.T.H., C.D.L., and H.O. labs for reagents and discussions, particularly E. Wasmuth and L. Cappadocia for assistance in crystallography. NE-CAT beamlines are funded by the National Institutes of Health (NIH) National Institute of General Medical Sciences (NIGMS) (P41 GM103403) and the Advanced Photon Source is operated for the Department of Energy Office of Science by Argonne National Laboratory under contract DE-AC02-06CH11357. This work was supported in part by NIH/NIGMS F32GM100598 (M.B.), GM084244 and ES025166 (T.T.H.), GM065872 (C.D.L.), NIH National Cancer Institute P30 CA008748 (Sloan Kettering), NYU Laura & Isaac Perlmutter Cancer Center Support Grant’s Developmental Project Program P30 CA016087 (T.T.H.), NWO-VENI grant 722.014.002 (G.v.d.H.v.N), NOW-VICI grant 724.013.002 (H.O.), and ERC grant agreement number 281699 (H.O.). C.D.L. is an investigator of the Howard Hughes Medical Institute. The content is solely the responsibility of the authors and does not represent the official views of the NIH. M.B., T.T.H, C.D.L., R.E., and G.J.v.d.H.v.N. declare no competing financial interests. H.O. is part of the DUB Alliance that includes Cancer Research Technology and FORMA Therapeutics and is a founder and stakeholder of UbiQ, which holds intellectual property rights to technology for reagent generation.
Published: May 19, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and six figures and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2016.04.016.
Contributor Information
Huib Ovaa, Email: h.ovaa@nki.nl.
Tony T. Huang, Email: tony.huang@nyumc.org.
Christopher D. Lima, Email: limac@mskcc.org.
Accession Numbers
The accession number for the coordinates and structure factors reported in this paper is PDB: 5E6J.
Supplemental Information
References
- Báez-Santos Y.M., Mielech A.M., Deng X., Baker S., Mesecar A.D. Catalytic function and substrate specificity of the papain-like protease domain of nsp3 from the Middle East respiratory syndrome coronavirus. J. Virol. 2014;88:12511–12527. doi: 10.1128/JVI.01294-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Elkin B.A., Knaap R.C., Johnson G.G., Dalebout T.J., Ninaber D.K., van Kasteren P.B., Bredenbeek P.J., Snijder E.J., Kikkert M., Mark B.L. Crystal structure of the Middle East respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J. Biol. Chem. 2014;289:34667–34682. doi: 10.1074/jbc.M114.609644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Elkin B.A., van Kasteren P.B., Snijder E.J., Kikkert M., Mark B.L. Viral OTU deubiquitinases: a structural and functional comparison. PLoS Pathog. 2014;10:e1003894. doi: 10.1371/journal.ppat.1003894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Békés M., Rut W., Kasperkiewicz P., Mulder M.P., Ovaa H., Drag M., Lima C.D., Huang T.T. SARS hCoV papain-like protease is a unique Lys48 linkage-specific di-distributive deubiquitinating enzyme. Biochem. J. 2015;468:215–226. doi: 10.1042/BJ20141170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A., Schechter I. Mapping the active site of papain with the aid of peptide substrates and inhibitors. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1970;257:249–264. doi: 10.1098/rstb.1970.0024. [DOI] [PubMed] [Google Scholar]
- Bhoj V.G., Chen Z.J. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458:430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- Bogunovic D., Byun M., Durfee L.A., Abhyankar A., Sanal O., Mansouri D., Salem S., Radovanovic I., Grant A.V., Adimi P. Mycobacterial disease and impaired IFN-γ immunity in humans with inherited ISG15 deficiency. Science. 2012;337:1684–1688. doi: 10.1126/science.1224026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capodagli G.C., McKercher M.A., Baker E.A., Masters E.M., Brunzelle J.S., Pegan S.D. Structural analysis of a viral ovarian tumor domain protease from the Crimean-Congo hemorrhagic fever virus in complex with covalently bonded ubiquitin. J. Virol. 2011;85:3621–3630. doi: 10.1128/JVI.02496-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cer R.Z., Mudunuri U., Stephens R., Lebeda F.J. IC50-to-Ki: a web-based tool for converting IC50 to Ki values for inhibitors of enzyme activity and ligand binding. Nucleic Acids Res. 2009;37:W441–W445. doi: 10.1093/nar/gkp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau V., Tobias J.W., Bachmair A., Marriott D., Ecker D.J., Gonda D.K., Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- Clague M.J., Liu H., Urbé S. Governance of endocytic trafficking and signaling by reversible ubiquitylation. Dev. Cell. 2012;23:457–467. doi: 10.1016/j.devcel.2012.08.011. [DOI] [PubMed] [Google Scholar]
- Cook W.J., Jeffrey L.C., Carson M., Chen Z., Pickart C.M. Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2) J. Biol. Chem. 1992;267:16467–16471. doi: 10.2210/pdb1aar/pdb. [DOI] [PubMed] [Google Scholar]
- Ekkebus R., van Kasteren S.I., Kulathu Y., Scholten A., Berlin I., Geurink P.P., de Jong A., Goerdayal S., Neefjes J., Heck A.J. On terminal alkynes that can react with active-site cysteine nucleophiles in proteases. J. Am. Chem. Soc. 2013;135:2867–2870. doi: 10.1021/ja309802n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Oualid F., Merkx R., Ekkebus R., Hameed D.S., Smit J.J., de Jong A., Hilkmann H., Sixma T.K., Ovaa H. Chemical synthesis of ubiquitin, ubiquitin-based probes, and diubiquitin. Angew. Chem. Int. Ed. Engl. 2010;49:10149–10153. doi: 10.1002/anie.201005995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flierman D., van Noort G.J.v.d.H.v., Ekkebus R., Gerink P.P., Mevissen T.E.T., Hospenthal M.K., Komander D., Ovaa H. Non-hydrolyzable diubiquitin probes reveal linkage-specific reactivity of deubiquitylating enzymes mediated by s2 pockets. Cell Chem. Biol. 2016 doi: 10.1016/j.chembiol.2016.03.009. Published online April 6, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushman D., Wilkinson K.D. Structure and recognition of polyubiquitin chains of different lengths and linkage. F1000 Biol. Rep. 2011;3:26. doi: 10.3410/B3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemelaar J., Borodovsky A., Kessler B.M., Reverter D., Cook J., Kolli N., Gan-Erdene T., Wilkinson K.D., Gill G., Lima C.D. Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins. Mol. Cell. Biol. 2004;24:84–95. doi: 10.1128/MCB.24.1.84-95.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson M.K., Ploegh H.L. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe. 2009;5:559–570. doi: 10.1016/j.chom.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keusekotten K., Elliott P.R., Glockner L., Fiil B.K., Damgaard R.B., Kulathu Y., Wauer T., Hospenthal M.K., Gyrd-Hansen M., Krappmann D. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell. 2013;153:1312–1326. doi: 10.1016/j.cell.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick D.S., Hathaway N.A., Hanna J., Elsasser S., Rush J., Finley D., King R.W., Gygi S.P. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- Komander D., Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Komander D., Reyes-Turcu F., Licchesi J.D., Odenwaelder P., Wilkinson K.D., Barford D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner H.A., Lytvyn V., Qi H., Lachance P., Ziomek E., Ménard R. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Arch. Biochem. Biophys. 2007;466:8–14. doi: 10.1016/j.abb.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevissen T.E., Hospenthal M.K., Geurink P.P., Elliott P.R., Akutsu M., Arnaudo N., Ekkebus R., Kulathu Y., Wauer T., El Oualid F. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 2013;154:169–184. doi: 10.1016/j.cell.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielech A.M., Chen Y., Mesecar A.D., Baker S.C. Nidovirus papain-like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res. 2014;194:184–190. doi: 10.1016/j.virusres.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder M.P., El Oualid F., ter Beek J., Ovaa H. A native chemical ligation handle that enables the synthesis of advanced activity-based probes: diubiquitin as a case study. ChemBioChem. 2014;15:946–949. doi: 10.1002/cbic.201402012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan J., Wang M., Fu Z., Klein J.M., Haas A.L., Kim J.J. Crystal structure of the interferon-induced ubiquitin-like protein ISG15. J. Biol. Chem. 2005;280:27356–27365. doi: 10.1074/jbc.M502814200. [DOI] [PubMed] [Google Scholar]
- Pickart C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C., Mesecar A.D. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. USA. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratia K., Kilianski A., Baez-Santos Y.M., Baker S.C., Mesecar A. Structural Basis for the Ubiquitin-Linkage Specificity and deISGylating activity of SARS-CoV papain-like protease. PLoS Pathog. 2014;10:e1004113. doi: 10.1371/journal.ppat.1004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu F.E., Ventii K.H., Wilkinson K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Yoshikawa A., Yamagata A., Mimura H., Yamashita M., Ookata K., Nureki O., Iwai K., Komada M., Fukai S. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358–362. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- Sato Y., Goto E., Shibata Y., Kubota Y., Yamagata A., Goto-Ito S., Kubota K., Inoue J., Takekawa M., Tokunaga F., Fukai S. Structures of CYLD USP with Met1- or Lys63-linked diubiquitin reveal mechanisms for dual specificity. Nat. Struct. Mol. Biol. 2015;22:222–229. doi: 10.1038/nsmb.2970. [DOI] [PubMed] [Google Scholar]
- Totura A.L., Baric R.S. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2012;2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Blaser G., Horrocks M.H., Ruedas-Rama M.J., Ibrahim S., Zhukov A.A., Orte A., Klenerman D., Jackson S.E., Komander D. Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature. 2012;492:266–270. doi: 10.1038/nature11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Bogunovic D., Payelle-Brogard B., Francois-Newton V., Speer S.D., Yuan C., Volpi S., Li Z., Sanal O., Mansouri D. Human intracellular ISG15 prevents interferon-α/β over-amplification and auto-inflammation. Nature. 2015;517:89–93. doi: 10.1038/nature13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.