SUMMARY
Ultraviolet (UV)-induced DNA damage, a major risk factor for skin cancers, is primarily repaired by nucleotide excision repair (NER). UV-radiation Resistance Associated Gene (UVRAG) is a tumor suppressor involved in autophagy. It was initially isolated as a cDNA partially complementing UV sensitivity in Xeroderma Pigmentosum (XP), but this was not explored further. Here we show that UVRAG plays an integral role in UV-induced DNA damage repair. It localizes to photolesions and associates with DDB1 to promote the assembly and activity of the DDB2-DDB1-Cul4A–Roc1 (CRL4DDB2) ubiquitin-ligase complex, leading to efficient XPC recruitment and global-genomic NER. UVRAG depletion decreased substrate handover to XPC and conferred UV-damage hypersensitivity. We confirmed the importance of UVRAG for UV-damage tolerance using a Drosophila model. Furthermore, increased UV-signature mutations in melanoma correlate with reduced expression of UVRAG. Our results identify UVRAG as a regulator of CRL4DDB2-mediated NER and suggest that its expression levels may influence melanoma predisposition.
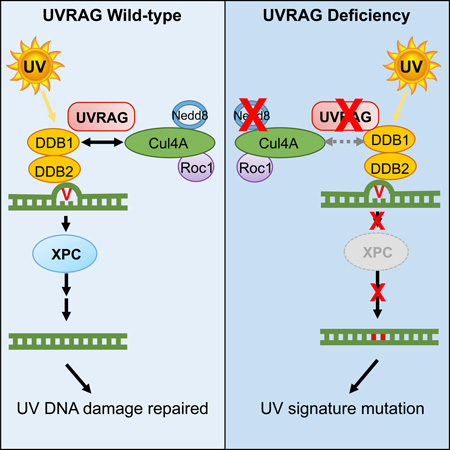
Graphical abstract
E-TOC
UV-induced DNA damage is a risk factor for skin cancers. Yang et al. demonstrate that UVRAG targets DDB1 and activates the DDB1-containing CRL4DDB2 ubiquitin-ligase complex, resulting in efficient photolesion repair. Reduced levels of UVRAG correlates with increased UV-signature loads in skin melanoma, which could potentially influence melanoma predisposition and progression.
INTRODUCTION
Exposure to ultraviolet (UV) is the main cause of skin cancer development (Garibyan and Fisher, 2010). DNA is the major target of UV-induced cellular damage. When left unrepaired, it leads to accumulation of “UV-signature” mutations, mainly C>T/G>A transitions at dipyrimidine sites, and induction of skin cancer (Hodis et al., 2012). Indeed, the most abundant somatic mutations present in melanomas, the most dangerous form of skin cancer, are UV-induced photodamages, as discovered in recent genome-wide association studies (Hodis et al., 2012; Pleasance et al., 2010), suggesting that cellular responses to UV-induced DNA damage may not function fully in this UV-related fatal disease. The most important mechanism that protects DNA against UV radiation is nucleotide excision repair (NER), which removes helix-distorting adducts on DNA (Marteijn et al., 2014). The importance of NER in melanoma is clearly demonstrated by the genetic disease Xeroderma Pigmentosum (XP), which is defective in NER and has a 1,000-fold greater risk of developing melanoma (Emmert and Kraemer, 2014; Spatz et al., 2010). Indeed, polymorphisms in NER-related genes have been shown to predict melanoma survival (Emmert and Kraemer, 2013; Li et al., 2006; Li et al., 2013; Wei et al., 2003).
There are two distinct sub-pathways of NER, global-genomic NER (GG-NER) and transcription-coupled NER (TC-NER), which differ in initial steps of damage recognition, but converge to use a common set of effectors for DNA incision, oligonucleotide removal, and nick ligation (Marteijn et al., 2014). Unlike TC-NER that selectively repairs DNA lesions on the actively transcribed genes, GG-NER scans the whole genome for damage via the DDB1-DDB2 (UV-damaged DNA binding proteins 1 and 2) heterodimers, designated UV-DDB, and via XPC (xeroderma pigmentosum group C), which repairs photolesions regardless of the transcriptional status (Kamileri et al., 2012). Upon UV-irradiation, UV-DDB recognizes and binds DNA lesions, then recruits the Cullin 4A (Cul4A)-Roc1 ubiquitin ligase (CRL4) complex (Marteijn et al., 2014; Sugasawa et al., 2005). This UV-DDB-Cul4A–Roc1 complex (referred to as CRL4DDB2) catalyzes the ubiquitination of histones and/or recruits chromatin remodelers at the sites of UV lesions, ensuing lesion handover from UV-DDB to XPC (Cleaver et al., 2009; Duan and Smerdon, 2010). Thus, CRL4DDB2 is essential in the initial detection of UV-damaged chromatin DNA. CRL4DDB2 activation is also regulated by the covalent attachment of Nedd8 to Cul4A, which is negatively influenced by CAND1 (Cullin-associated-and-neddylation-dissociated 1) (Bennett et al., 2010). Disruption of any component of the UV-DDB-Cul4-Roc1-XPC axis leads to a failure to repair UV-induced damage, resulting in genomic instability and increased cancer development (Scharer, 2013). Although the core NER reaction is well studied, the regulatory mechanisms that safeguard the integrity of an efficient NER and its high importance for the cumulative UV-like mutagenesis in skin cancer, particularly melanoma, is still elusive.
UV-radiation Resistance Associated Gene (UVRAG) was initially isolated in 1997 as a cDNA partially complementing UV sensitivity in Xeroderma Pigmentosum (XP) (Perelman et al., 1997), hence the name UVRAG. However, this initial important observation has not been further explored. UVRAG contains four major domains: a proline-rich (PR) domain, a lipid-binding C2 domain, a coiled-coil domain (CCD), and a C-terminal domain presumed to be unstructured (Liang et al., 2006). We have previously identified UVRAG as a multivalent trafficking adaptor involved in autophagic, endocytic, and secretory trafficking pathways (He et al., 2013; Liang et al., 2006; Liang et al., 2008). Our recent data further show that the C-terminal region of UVRAG is involved in centrosome stability and regulates DNA-PK (DNA-dependent protein kinase) (Zhao et al., 2012). Significantly, all of these activities of UVRAG are genetically separable and functionally independent, suggesting biological connection and coordinated regulation of the different processes under diverse environmental cues. Notably, none of these identified activities of UVRAG explain its link to UV-induced DNA damage.
Herein, we demonstrate that UVRAG interacts specifically with the UV-induced photolesion sensor DDB1 in vivo; this interaction allows UVRAG to be recruited to the damaged foci after UV exposure, promoting complex assembly, Cul4A neddylation, and ubiquitin ligase activity of the CRL4DDB2 complex in GG-NER. Inactivation of UVRAG inhibits efficient substrate handover from UV-DDB to XPC in the NER cascade, rendering cells ultrasensitive to UV-induced tissue and DNA damage in vitro and in vivo. Finally, reduced levels of UVRAG are associated with increased UV-signature loads in cutaneous but not UV-shielded melanoma and could potentially influence melanoma predisposition and disease progression.
RESULTS
Essential Role of UVRAG in Protecting Cells from UV Damage
To elucidate the molecular functions of UVRAG in UV-sensitivity and skin cancer, we set out to examine the role of UVRAG in UV-induced damage in melanoma cells by colony-forming assays. Knockdown of UVRAG in A375 human and B16 mouse melanoma cells drastically sensitized melanoma cells to UV-irradiation and UV-mimetic drugs (i.e., NFZ and 4-NQO), but not to other drugs (e.g., CPT and MMS) (Figure 1A–C, S1A–C, and S1H–M). UV sensitivity was abolished by re-expression of shRNA-resistant WT UVRAG. In contrast, re-expression of UVRAG (L286F) point-mutant, identified in two independent melanoma exome studies (Berger et al., 2012; Hodis et al., 2012), failed to confer UV resistance (Figure 1A–C, S1A–C and S1H–M). In accord, ectopic expression of UVRAGWT, but not UVRAGL286F, caused increased cell tolerance to UV-irradiation and to UV-mimetics (Figure S1D–G and N–Q), indicative of UVRAGL286F being a loss-of-function mutation in UV protection. These results indicate that UVRAG, as its name suggests, may play a role in protecting cells from UV damage.
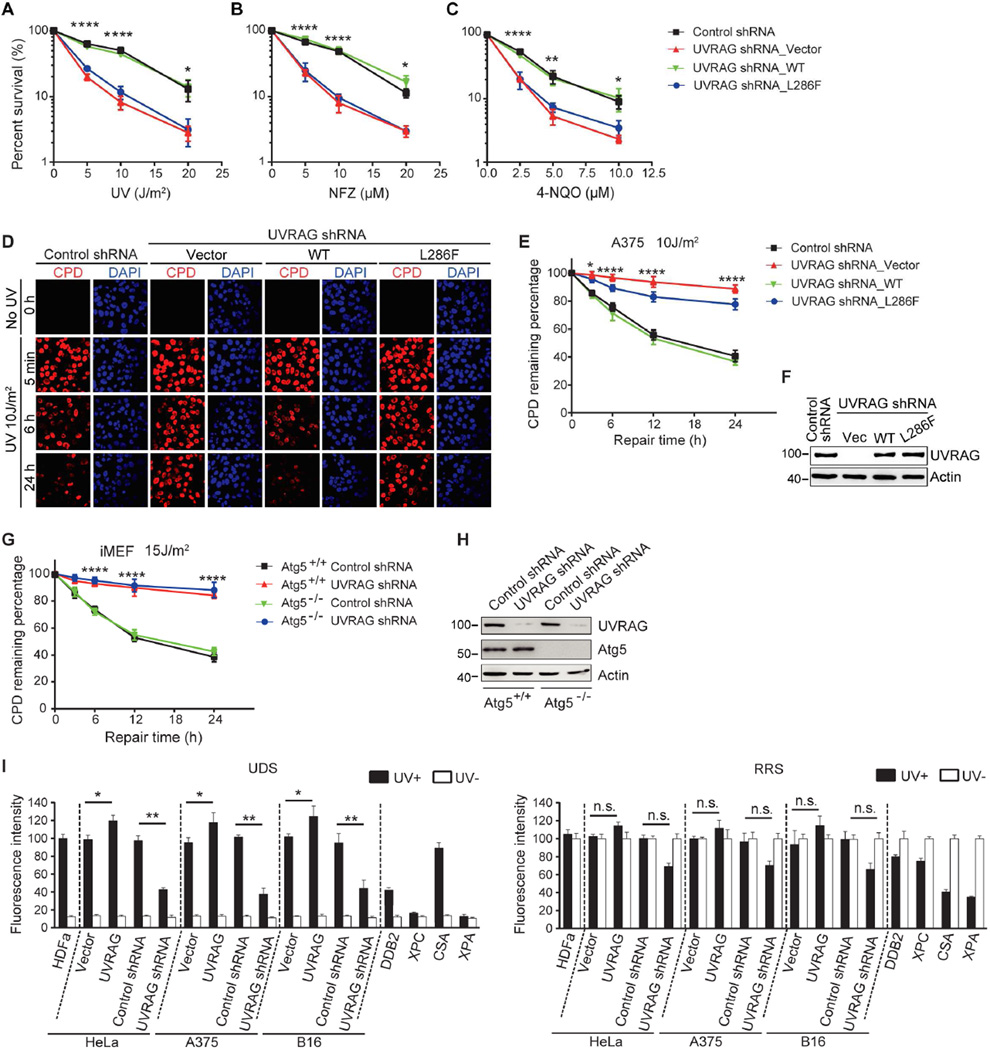
Figure 1. The Role of UVRAG In UV-irradiation Sensitivity and NER.
(A–C) UV sensitivity of A375 cells upon UVRAG inhibition. A375 cells expressing control shRNA or UVRAG-specific shRNA were transduced with empty retroviral vector (UVRAG shRNA_Vector), with retroviral vector expressing WT human UVRAG (UVRAG shRNA_WT), or with retroviral vector expressing UVRAG L286F point mutant (UVRAG shRNA_L286F). Cells were exposed to the indicated doses of UV-C (A), UV mimetic agents nitrofurazone (NFZ; B) and 4-nitroquinoline-1-oxide (4-NQO; C), followed by colony survival assay. Data are mean ± SD from three independent experiments. *p < 0.05; **p < 0.01; ****p < 0.0001 (UVRAG shRNA_Vector vs. Control shRNA).
(D–F) UVRAG is required for UV-induced CPD repair. A375 cells expressing control shRNA or UVRAG-specific shRNA were transduced with empty retroviral vector (UVRAG shRNA_Vector), WT human UVRAG (UVRAG shRNA_WT), or the L286F mutant (UVRAG shRNA_L286F). Cells were UV-C treated and recovered for a period of time as indicated. UV-induced DNA damage was visualized using CPD counterstaining. Representative images are shown in (D). Quantification of the percentage of remaining CPD per cell relative to that of 0 hr after UV-C in each sample is plotted (E). UVRAG expression was assessed by immunoblotting and compared to actin levels (F). Data shown represent mean ± SD; n = 200 cells, data pooled from three independent experiments. Scale bar, 20 µm. *p < 0.05; ****p < 0.0001 (UVRAG shRNA_Vector vs. Control shRNA).
(G and H) UVRAG knockdown delayed UV-induced CPD repair in Atg5-knockout iMEFs. The Atg5+/+ and Atg5−/− immortalized MEF cells were transfected with control shRNA or UVRAG-specific shRNA and then subjected to UV-C. The percent distribution of CPD foci before UV, and 5 min, 6 hr, and 24 hr post-UV was determined (G). Protein levels of UVRAG and Atg5 are shown (H). Scale bar, 20 µm. Data are mean ± SD from three independent experiments. ****p < 0.0001 (Atg5−/− UVRAG shRNA vs. Atg5−/− Control shRNA).
(I) Effect of UVRAG on UDS and RRS activity. A375, B16, and HeLa cells expressing an empty vector, Flag-UVRAG, control shRNA, or UVRAG-specific shRNA were UV-C-irradiated and subjected to UDS (left) and RRS (right) assays. UDS and RRS activities in normal HDFα cells, DDB2-deficient, XPC-deficient, CSA-deficient, and XPA-deficient cells served as controls. Filled bars, UV-irradiated; open bars, no UV. Typical UDS and RRS images are provided in Figure S3E. Data shown represent mean ± SD from three independent experiments. *p < 0.05; **p < 0.01; n.s., not significant.
See also Figure S1–S3 for additional information.
UVRAG Is Required for UV-induced Photolesion Repair Independently of Autophagy
Chromatin-associated cyclobutane pyrimidine dimers (CPDs) are a sensitive and representative marker of UV-induced DNA damage (Marteijn et al., 2014). We found that UV-irradiation induced comparable levels of CPDs in control and in UVRAG-depleted melanoma cells (5 min time-point in Figure 1D–F and S2C and 2D). However, more CPDs retained 24 hr post-UV in UVRAG-depleted cells, which were rescued by introducing UVRAGWT but not UVRAGL286F (Figure 1D–F and S2C and 2D). Consistently, ectopic expression of UVRAGWT, but not UVRAGL286F, enhanced the clearance rate of UV-induced damage (Figure S2A and 2B, S2E and 2F). These data support a role for UVRAG in photodamage repair.
To explore whether the UV damage-protecting role of UVRAG might be related to autophagy, we examined the effect of UVRAG on CPD levels in autophagy-deficient Atg5-knockout immortalized MEFs (iMEF) (Kuma et al., 2004). UVRAG depletion induced a marked increase in CPD-foci in these cells, regardless of the autophagy status (Figure 1G and 1H). Analogous results were obtained when cells were treated with Bafilomycin A1 (Baf-A1) to block autophagosome degradation (Figure S3A and 3B). These data indicate that UVRAG-mediated suppression of UV-induced photolesions does not necessarily require functional autophagy machinery. While these observations do not exclude a role for autophagy in UV-induced damage, such event would be downstream of a critical UVRAG-dependent step in maintaining genetic stability upon UV exposure. We next determined whether UVRAGL286F, defective in UV protection as shown above, also impairs the ability of UVRAG to promote autophagy by measuring subcellular distribution of the autophagy marker GFP-LC3 and levels of the autophagosome-associated LC3 (LC3-II). UVRAGL286F enhanced rapamycin-induced autophagy to a similar extent as UVRAGWT, as evidenced by increased GFP-LC3 puncta per cell, increased LC3-II conversion, and increased response to the late-stage autophagy inhibitor Baf-A1 (Figure S3C and 3D). These results indicate that UVRAG directly regulates photolesion repair through a mechanism independent of UVRAG-mediated autophagy.
UVRAG Interacts with the CRL4DDB2 E3-Ligase Complex In GG-NER
UV-induced DNA damage is predominantly repaired by NER (Garibyan and Fisher, 2010). We used two different assays, i.e., unscheduled DNA synthesis (UDS) and recovery of RNA synthesis (RRS), to specifically measure two major NER pathways, GG-NER and TC-NER, respectively (Hasegawa et al., 2010). GG-NER-deficient (DDB2 and XPC) cells, TC-NER-deficient (CSA) cells, and GG- and TC-NER-deficient XPA cells were included as controls. UVRAG knockdown resulted in a significant reduction in UDS, as occurred in DDB2- and XPC-deficient cells (Figure 1I and Figure S3E). No significant effect was observed on TC-NER rate upon UVRAG knockdown (Figure 1I and Figure S3E). In accord, ectopic expression of UVRAG specifically promoted the rate of UDS, but not that of RRS, after UV treatment (Figure 1I). While these observations do not exclude a role for UVRAG in TC-NER, they support a major role for UVRAG in GG-NER. Unlike UVRAG, knockdown of Beclin1, an interactor of UVRAG in autophagy (Liang et al., 2006), did not affect GG-NER (Figure S3F), again suggesting that UVRAG has an autophagy-independent function to regulate NER.
To elucidate the mechanism by which UVRAG functions in GG-NER, we immunoaffinity-purified Flag-UVRAG before and after UV exposure of A375 cells and analyzed UVRAG-interacting proteins by mass spectrometry (Figure S4A and 4B). DDB1 was identified with high confidence as a prominent candidate. DDB1 forms a heterodimer with DDB2 and serves as a substrate adaptor of Cul4A–Roc1 ubiquitin-ligase (CRL4) complex in GG-NER (Angers et al., 2006; He et al., 2006). An interaction between recombinant UVRAG and DDB1 was detected in vitro, supporting their direct interaction (Figure S4C). UVRAG was co-immunoprecipitated with DDB1 both endogenously and exogenously (Figure 2A and S4D). The formation of the UVRAG-DDB1 complex in response to UV irradiation was further evaluated by the single-molecule pull-down (SiMPull) assay (Jain et al., 2011). Cells co-expressing Flag-DDB1 and UVRAG-GFP were UV-irradiated; the lysates were applied to single-molecule imaging chambers coated with anti-Flag or control antibodies (Figure S4E). UVRAG-GFP fluorescence spots, which marked individual immobilized DDB1-UVRAG complexes, were drastically increased in the anti-Flag coated chambers after UV as compared to the untreated and the control channels (Figure S4F and 4G), indicative of an increased UVRAG-DDB1 complex formation upon UV-irradiation. In fact, both endogenous and exogenous UVRAG can be co-immunoprecipitated with Cul4A, Roc1, and DDB2, and to a greater extent after UV-irradiation (Figure 2A, and S4H and 4I), suggesting that UVRAG associates with CRL4DDB2 in NER. Consistently, no interaction was detected between Beclin1 and DDB2, suggesting that Beclin1 is not involved in the UVRAG-CRL4DDB2 complex formation (Figure S4J).
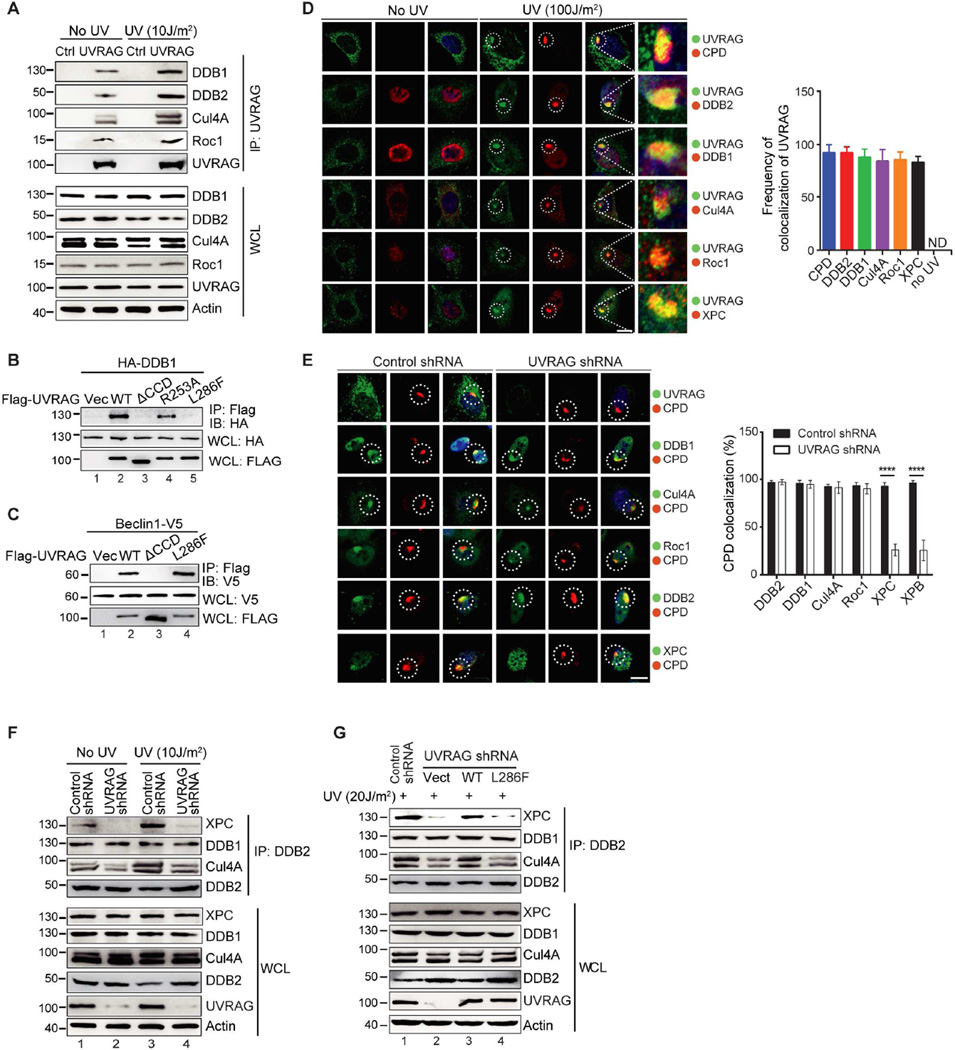
Figure 2. UVRAG Forms A Complex with CRL4DDB2 E3 Ligase and Is Required for Efficient Recruitment of XPC to Photolesions.
(A) Interaction between endogenous UVRAG and the CRL4DDB2 E3-ligase proteins under basal condition and UV-C treatment. Whole cell lysates (WCL) of 293T cells were used for IP with control serum (Ctrl) or anti-UVRAG antibody, followed by immunoblotting (IB) with the indicated antibodies. The bottom panel shows endogenous protein expression with actin as a loading control.
(B) UVRAG L286F mutant is defective in DDB1 binding. 293T cells were transfected with HA-DDB1 and Flag-UVRAG WT or its mutants. WCL were immunoprecipitated with anti-Flag followed by IB with anti-HA.
(C) Interaction of UVRAG ΔCCD and L286F mutants with Beclin1. 293T cells were transfected with Beclin1-V5 and Flag-UVRAG WT or its mutants. WCL were immunoprecipitated with anti-Flag, followed by IB with anti-V5 antibody.
(D) Recruitment of UVRAG to UV-induced CPD sites and its colocalization with the UV-DDB-Cul4A–Roc1 complex proteins. HeLa cells were micropore UV-C (100 J/m2) irradiated (pore size: 5 µm), immunostained with anti-UVRAG (green), anti-CPD (red, 1st row), anti-DDB2 (red, 2nd row), anti-DDB1 (red, 3rd row), anti-Cul4A (red, 4th row), anti-Roc1 (red, 5th row), or anti-XPC (red, 6th row), followed by confocal microscopy. The insets highlight co-localization of UVRAG with the GG-NER proteins at CPD sites. Right panel shows quantification of the frequency of UVRAG colocalizing with UV-induced CPD or GG-NER proteins. Data represent mean ± SD from three independent experiments (n = 200). See also Figure S5A for multi-cell images. Scale bar, 20 µm.
(E) UVRAG Knockdown inhibited XPC recruitment to the UV-induced CPD sites. HeLa cells stably expressing control shRNA or UVRAG shRNA were micropore UV-C (100 J/m2) irradiated (pore size: 5 µm), and immunostained with anti-CPD (red) and anti-UVRAG (1st row), anti-DDB1 (2nd row), anti-Cul4A (3rd row), anti-Roc1 (4th row), anti-DDB2 (5th row), or anti-XPC (6th row). Right panel shows quantification of the colocalization of the indicated GG-NER proteins with CPD foci. Data shown represent mean ± SD; n = 200 cells obtained by gathering data from three independent experiments. ****p < 0.0001. See also Figure S5H for multi-cell images. Scale bar, 20 µm.
(F) UVRAG knockdown inhibited endogenous DDB2 interaction with XPC. HeLa cells expressing control shRNA or UVRAG shRNA were treated with or without UV. WCL were then used for IP with anti-DDB2 and IB for XPC and the CRL4DDB2 complex subunits.
(G) Effect of UVRAG on the complex assembly of DDB2 with XPC and with the CRL4DDB2 ubiquitin-ligase proteins. A375 cells stably expressing UVRAG-specific shRNA complemented with an empty vector, or with WT UVRAG or the L286F mutant were UV-C treated. WCL, 30 min post-UV irradiation, were used for IP with anti-DDB2 followed by IB with the indicated antibodies.
See also Figure S4–S5 for additional information.
Using several UVRAG and DDB1 deletion mutants, we identified residues 230–305 in the coiled-coil domain of UVRAG and the β-propeller C domain (BPC) of DDB1 as mediators of their interaction (Figure S4K–O). Notably, we have previously shown that UVRAGΔCCD, albeit defective in DDB1 interaction, preserved the activity in double-strand break (DSB) repair that engages the C-terminal residues 584–699 (Zhao et al., 2012), suggesting that UVRAG association with DDB1 is structurally and functionally distinguishable from its DSB repair activities. Importantly, the L286F mutation in the CCD was deficient in DDB1 binding, whereas alanine substitution of another conserved residues (R253) impaired, but did not completely eliminate, DDB1-binding (Figure 2B). This result is in agreement with the fact that the DDB1-binding defective L286F is impaired in repair of UV-induced DNA damage (Figure 1D and 1E, S2A and 2B). Given that UVRAG CCD is also engaged in binding Beclin1 (Liang et al., 2006), we examined whether the Beclin1 binding activity of UVRAG is affected by the L286F mutation. Both UVRAGWT and UVRAGL286F, but not UVRAGΔCCD, co-immunoprecipitated with Beclin1 (Figure 2C). This was consistent with previous observations demonstrating that UVRAGL286F, albeit defective in UV protection, remains competent for autophagy.
UVRAG Is Required for Efficient Recruitment of NER Factors to UV-induced Damage Sites
Despite overall cytoplasmic distribution of UVRAG under normal conditions, we observed specific recruitment of WT, but not the DDB1-binding defective L286F and ΔCCD mutants of UVRAG, to CPD foci, where it co-localized with DDB2, DDB1, Cul4A, Roc1, and XPC after UV-irradiation (Figure 2D and S5A–D). Knockdown of DDB1 or deficiency of DDB2 inhibited UVRAG translocation to UV-damaged sites (Figure S5E–G). But in TCR-related CSA-knockout cells, UVRAG still accumulated at CPD, again indicative of a specific role of UVRAG in GG-NER (Figure S5E and 5F). Depletion of either XPC or the downstream XPA did not change the association of UVRAG with UV-damaged sites, suggesting that UVRAG translocates to CPD sites prior to XPC (Figure S5E and 5F). Conversely, knockdown of UVRAG significantly reduced UV-induced XPC recruitment, as well as the downstream NER effector XPB, to the damaged sites, though not appreciably affecting that of DDB2, DDB1, Cul4A, and Roc1 (Figure 2E and S5H). Indeed, DDB1 and Cul4A were able to be co-immunoprecipitated with DDB2 in UVRAG-depleted cells, whereas XPC interaction with DDB2 was reduced, even after UV-irradiation (Figure 2F). Endogenous DDB2-XPC interaction in UVRAG- depleted cells could be restored by re-expression of UVRAGWT, but not UVRAGL286F (Figure 2G). Therefore, UVRAG is required for effective association of XPC with DDB2 after UV-irradiation in vivo. Our results conform to a model where UVRAG translocates to UV-induced DNA-lesions through DDB1, and UVRAG interaction with the DDB1-containing CRL4DDB2 complex then helps recruit XPC.
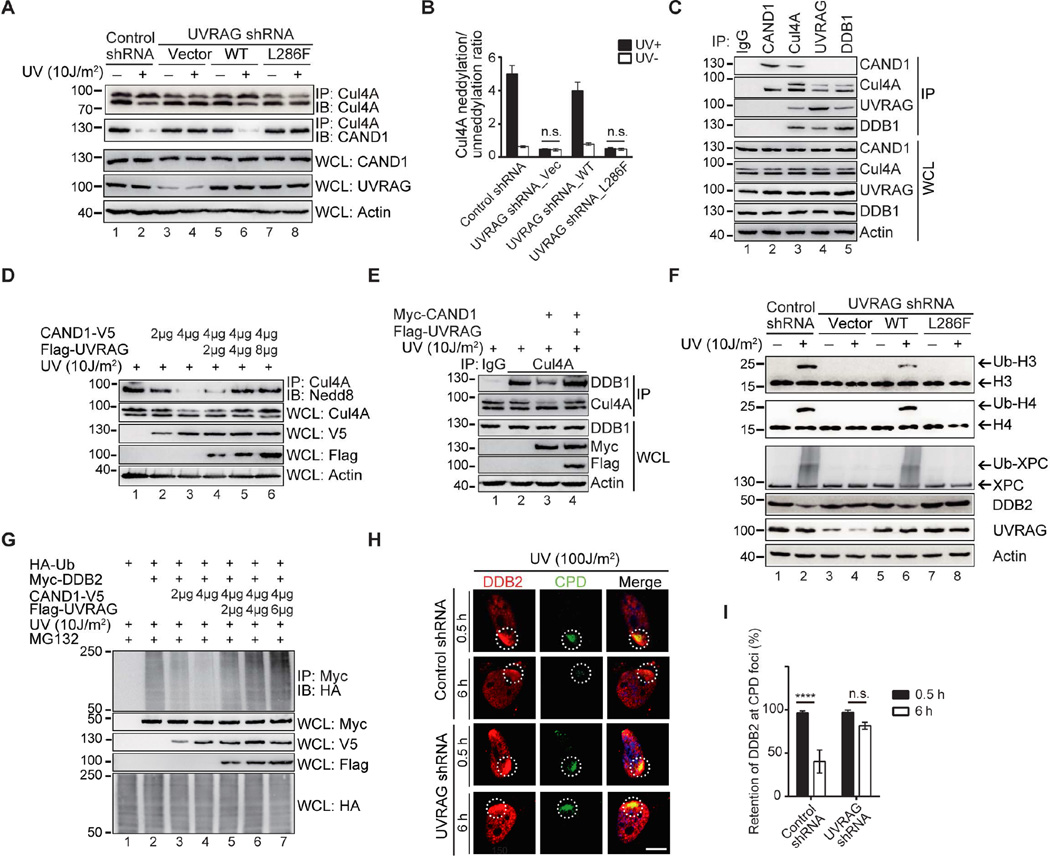
UVRAG Promotes the Assembly and the Cul4A Neddylation of CRL4DDB2 E3-Ligase Complex
DDB1 functions in NER as part of a complex with Cul4A–Roc1 ubiquitin ligase (Angers et al., 2006). Furthermore, NEDD8 modification of Cul4A represents a critical mechanism to activate the Cul4A–based E3-ligase by inducing structural reorganization to promote the processivity of ubiquitin transfer (Bennett et al., 2010). Since UVRAG promotes DDB1-dependent NER, we evaluated the effects of UVRAG on assembly and activity of the CRL4DDB2 complex. As shown in Figure 3A, expression of UVRAG, but not of UVRAGL286F, resulted in increased reciprocal co-immunoprecipitation of DDB1 with Cul4A without affecting their overall levels in cells, while accompanied by increased neddylation of Cul4A. In contrast, less Cul4A co-immunoprecipitated with DDB1 in UVRAG depleted cells, concomitant with a clear reduction in Cul4A neddylation (Figure 3B). Re-introducing UVRAGWT but not UVRAGL286F reverted the assembly and the neddylation of DDB1-Cul4A complex (Figure 3B). In accord, we also observed that UVRAG expression accelerated the NEDD8 modification of Cul4A after UV-irradiation in cells expressing HA-Cul4A (Figure 3C). In contrast, removal of UVRAG or expression of UVRAGL286F in UVRAG-depleted cells caused a lack of UV-induced Cul4A neddylation (Figure 3D, Figure 4A, and 4B). We next tested whether the change in Cul4A neddylation is specific to UVRAG per se. Using in vitro neddylation assay with purified recombinant proteins of UVRAG, DDB1, and Cul4A–Roc1, we observed a dose-dependent increase in Cul4A neddylation by UVRAG in the presence of DDB1 as expected (Figure 3E). Notably, in UV-irradiated cells in vivo, the positive effect of UVRAG on UV-induced Cul4A neddylation was abrogated by depletion of DDB2 (Figure S6A), suggesting that DDB2 acts upstream of UVRAG during the UV damage-induced Cul4A modification. These results indicate that UVRAG, through its interaction with DDB1, is necessary to promote the assembly and the neddylation of the CRL4DDB2 E3-ligase, which is needed for GG-NER.
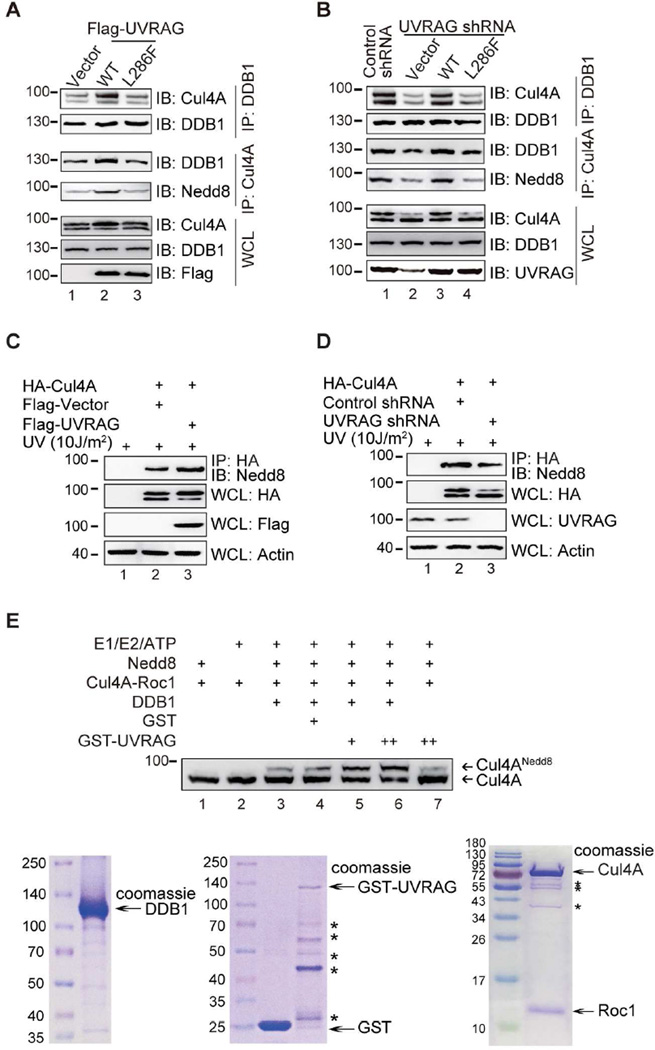
Figure 3. UVRAG Promotes the Assembly and Cul4A Neddylation of CRL4DDB2 E3 Ligase In GG-NER.
(A) Ectopic expression of UVRAG facilitates the assembly of the DDB1-Cul4A complex. A375 cells stably expressing WT Flag-UVRAG or the L286F mutant were co-immunoprecipitated with anti-DDB1 or anti-Cul4A followed by immunoblotting with indicated antibodies. Western blot analyzed of the amount of endogenous Cul4A and DDB1.
(B) UVRAG deficiency impaired the complex assembly of DDB1 and Cul4A. A375 cells were stably expressing control shRNA or expressing UVRAG shRNA complemented with empty vector, WT UVRAG, or the L286F mutant. WCL were immunoprecipitated with anti-DDB1 or anti-Cul4A followed by IB with indicated antibodies.
(C) UVRAG promotes Cul4A neddylation. 293T cells transfected with HA-Cul4A along with Flag-vector or Flag-UVRAG were treated with UV. WCL were immunoprecipitated with anti-HA followed by IB with anti-Nedd8 antibody. Actin serves as a loading control.
(D) Deficiency of UVRAG inhibits Cul4A neddylation. The 293T cells expressing control shRNA or UVRAG shRNA were transfected with HA-Cul4A followed by UV treatment. WCL were immunoprecipitated with anti-HA followed by IB with anti-Nedd8 antibody.
(E) UVRAG promotes Cul4A neddylation in vitro. Purified recombinant GST-UVRAG, DDB1, and Cul4A–Roc1 proteins (Coomassie gels at the bottom) were mixed with the neddylation components (indicated on the top of the figure). The reaction mixtures were detected for Cul4A and its Nedd8 modification. *, degradation products of proteins.
Figure 4. UVRAG Antagonizes CAND1 and Promotes the Ubiquitin-mediated Proteolysis of CRL4DDB2 E3 Ligase In GG-NER in vivo.
(A) Effect of UVRAG on UV-induced Cul4A neddylation and Cul4A–CAND1 interaction. A375 cell lines stably expressing control shRNA, or expressing UVRAG-specific shRNA complemented with empty vector, WT UVRAG, or with its DDB1-binding defective L286F mutant were UV-C treated. WCL were immunoprecipitated with anti-Cul4A followed by IB with anti-Cul4A or anti-CAND1 antibody.
(B) Densitometric quantification of the neddylated Cul4A/un-neddylated Cul4A ratio under the indicated condition in (A). Data shown represent mean ± SD from three independent experiments. n.s., not significant.
(C) UVRAG and CAND1 bind to Cul4A in a mutually exclusive manner. WCL prepared from A375 cells were immunoprecipitated with control IgG, anti-CAND1, anti-Cul4A, anti-UVRAG, or anti-DDB1, followed by IB with the indicated antibodies.
(D) UVRAG antagonizes the inhibitory effect of CAND1 on Cul4A neddylation. A375 cells transiently transfected with different amounts of Flag-UVRAG and/or CAND1 were UV-C treated. WCL were immunoprecipitated anti-Cul4A antibody followed by IB with anti-Nedd8 antibody.
(E) UVRAG antagonizes the inhibitory effect of CAND1 on the DDB1-Cul4A interaction. A375 cells transiently transfected with Flag-UVRAG and/or myc-CAND1 were UV-C treated. WCL were immunoprecipitated with control or anti-Cul4A antibody followed by IB with anti-DDB1 and Cul4A antibodies.
(F) Effect of UVRAG on UV-induced histones and XPC ubiquitination, and DDB2 degradation. A375 cell lines stably expressing control shRNA, or expressing UVRAG-specific shRNA complemented with empty vector, WT UVRAG, or with its DDB1-binding defective L286F mutant were treated with UV-C irradiation. WCL were immunoblotted for histone H3/H4 ubiquitination, XPC ubiquitination, and DDB2 degradation.
(G) UVRAG antagonizes CAND1-mediated inhibition of DDB2 ubiquitination. 293T cells were transfected with Myc-DDB2 and HA-ubiquitin (Ub), along with different amount of CAND1-V5 and/or Flag-UVRAG. At 48 hr post-transfection, cells were UV-irradiated and treated with MG132. WCL were used for IP with anti-Myc followed by IB with anti-HA.
(H, I) UVRAG deficiency impedes UV-induced DDB2 degradation and CPD repair. A375 cells stably expressing control shRNA or UVRAG shRNA were UV-C treated for 30 min and let recovered for a period of time as indicated. DDB2 levels and UV-induced CPD damage were determined by immunostaining with anti-DDB2 and anti-CPD, respectively (H). The retention of DDB2 at CPD foci is quantified (I). Scale bar, 10 µm. ****p < 0.0001; n.s., not significant.
See also Figure S6 for additional information.
CAND1 has previously been suggested to sequester unneddylated Cullins and compete with the substrate adaptors for binding to the Cullin-RING complexes, rendering them in an inactive form (Bennett et al., 2010; Hu et al., 2004; Liu et al., 2002). We thus asked whether UVRAG might also antagonize the inhibitory effect of CAND1 on Cul4A sequestration. Despite their apparent interaction, we detected less Cul4A co-immunoprecipitated with CAND1 upon ectopically expressed UVRAGWT, but not UVRAGL286F, in a dose-dependent manner (Figure S6B and 6C). Consistently, depletion of UVRAG prevented dissociation of Cul4A from CAND1 upon UV-irradiation, which was reverted by re-expression of UVRAGWT but not UVRAGL286F (Figure 4A). Importantly, no CAND1 was detected in the UVRAG immunocomplex, and conversely no UVRAG and no DDB1 was detected in the CAND1 immunocomplex, whereas Cul4A was readily detected in both CAND1 and UVRAG-DDB1 complexes, suggesting mutually exclusive binding of CAND1 and UVRAG to Cul4A (Figure 4C). Additionally, CAND1 selectively associated with unneddylated Cul4A as seen previously (He et al., 2006; Hu et al., 2004), whereas UVRAG and DDB1 could associate with Cul4A regardless of Cul4A neddylation status (Figure 4C). In agreement with the previous report (He et al., 2006), CAND1 expression resulted in decreased levels of Cul4A neddylation and a concomitant reduction in Cul4A bound to DDB1 (Figure 4D and 4E). By contrast, co-expression of UVRAG reverted the CAND1-mediated inhibition of Cul4A neddylation as well as Cul4A–DDB1 association, while UVRAGL286F failed to do so (Figure 4D and 4E and Figure S6D). Together, these results consistently indicate that UVRAG is essential for the dynamic complex assembly and activation of the CRL4DDB2 complex in NER.
UVRAG Interaction with CRL4DDB2 Is Required for Ubiquitin-mediated Proteolysis in NER
The CRL4DDB2 complex is required to remodel chromatin at UV lesions by catalyzing the ubiquitination of histones and NER factors, a prerequisite for XPC CPD site recruitment in GG-NER (Duan and Smerdon, 2010). We next examined the effects of UVRAG on ubiquitin-mediated proteolysis by the CRL4DDB2 complex in NER. We detected the slower migrating forms of histones H3/H4 after UV-irradiation (Figure 4F), corresponding to ubiquitinated histones (Figure S6E), which were barely detectable in UVRAG-knockdown cells and in UVRAGL286F-complemented cells, but not in UVRAGWT-complemented cells (Figure 4F). As previously reported (Sugasawa et al., 2005), UV-irradiation triggered strong XPC ubiquitination, but was also abrogated in UVRAG-depleted cells (Figure 4F). Re-expression of UVRAGWT, but not UVRAGL286F, restored XPC ubiquitination after UV-irradiation (Figure 4F), which was shown to enhance its DNA-binding affinity (Sugasawa, 2006). Moreover, we noted that DDB2 levels rapidly decreased after UV-irradiation. However, depletion of UVRAG or re-expression of UVRAGL286F prevented DDB2 degradation (Figure 4F). Consistent with the previous finding that DDB2 is polyubiquitinated by the CRL4DDB2 E3-ligase and ultimately degraded to allow spatial access of XPC to DNA lesion (El-Mahdy et al., 2006), we observed that UV-induced reduction of DDB2 levels was suppressed by the proteasome inhibitor MG132 in UV-irradiated cells co-expressing Myc-DDB2 and HA-ubiquitin (Figure S6F). Likewise, overexpression of UVRAG promoted the amount of ubiquitin immunoprecipitated with DDB2 in UV-irradiated cells, whereas depletion of UVRAG decreased the rapid turnover of DDB2 and stabilized DDB2 after UV irradiation (Figure S6F and 6G and Figure 4F). In agreement with the CAND1 results above, expression of UVRAG alleviated the inhibitory effect of CAND1 on Cul4A–mediated DDB2 ubiquitination upon UV irradiation (Figure 4G). Finally, in a complementary approach, we analyzed dynamic association of DDB2 with photolesions. Knockdown of UVRAG produced a significant delay of removal of DDB2 from CPD foci and a concomitant reduction in CPD repair (Figure 4H and 4I). These results indicate that UVRAG interaction with DDB1 promotes substrate ubiquitination mediated by the CRL4DDB2 E3-ligase after UV-irradiation, which destabilizes damage-containing nucleosomes to displace substrates from DNA lesions, facilitating recruitment of downstream effectors to initiate NER. In line with this, inactivation of UVRAG had minimal effect on the repair of UV-damaged naked DNA in vitro, as did DDB2 deficiency (Figure S6H), suggesting that like UV-DDB (Rapic Otrin et al., 1998), UVRAG is important for the repair of photolesions within a chromatin context by a mechanism that involves the CRL4DDB2 E3-ligase activation.
In accord, ectopic expression of UVRAG reduced UV-induced cell death in melanoma cells, as shown by increased rates of clonogenic survival, but not in cells depleted of DDB1 and Cul4A (Figure S6I–L). Analogous results were obtained in cells treated with the neddylation inhibitor MLN4924 (Figure S6M and 6N). Thus, UVRAG protects cells from UV-mediated cell death in a DDB1- and Cul4A–dependent manner.
UVRAG Inhibits UV-induced Damage in vivo
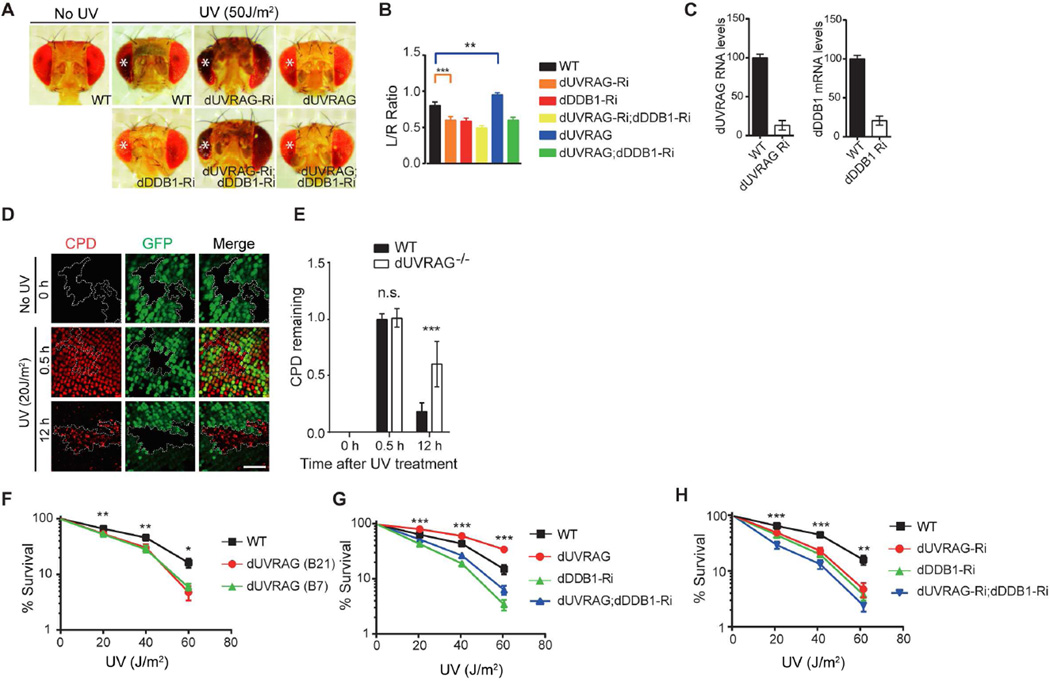
UVRAG is highly conserved, with closely related orthologs present in fly (designated as dUVRAG) and mammals (Lee et al., 2011; Lorincz et al., 2014; Takats et al., 2014). To test UVRAG-mediated UV protection in vivo, we used developing Drosophila pupal retina model to evaluate UV-induced damage by comparing the tissue loss in the irradiated eye vs. the untreated one in the same animal (Kelsey et al., 2012). Depletion of dUVRAG by RNAi exclusively in the retina resulted in a significant loss of ommatidial structures compared to control RNAi, as seen by inhibition of DDB1 (dDDB1), suggesting both genes essential in UV-protection (Figure 5A–C). Combined interference with both dUVRAG and dDDB1 showed similar effects, leading to an eye of reduced size. Ectopic expression of dUVRAG prevented UV-induced loss of ommatidia, yet this effect was largely suppressed when dDDB1 was silenced (Figure 5A–C), further supporting a DDB1-dependent function of UVRAG in UV protection.
Figure 5. UVRAG-mediated UV DNA Damage Repair in Drosophila.
(A) Effect of UVRAG on UV-induced tissue loss in Drosophila retina. Shown are representative examples of each genotype following UV irradiation (254 nm, 50 J/m2) of the left eye marked by asterisks (*). Knockdown of dUVRAG by the eye driver GMR-Gal4 enhanced the UV-induced tissue loss (GMR>dUVRAGRNAi), whereas overexpression of dUVRAG (GMR>dUVRAG) reduces UV-triggered tissue loss in the retina. The protective effect of UVRAG was lost when dDDB1 was knockdown by RNAi (GMR>dUVRAG; dDDB1RNAi).
(B) Tissue loss was quantified by calculating the size ratio of UV-treated eye (left) to untreated eye (right). Data shown represent mean ± SD (n = 50) from three independent experiments. **p < 0.01; ***p < 0.001.
(C) RT-PCR analysis confirmed UVRAG and DDB1 knockdown in the wing discs of 3rd instar larvas of MS1096>dUVRAG-Ri and MS1096>dDDB1-Ri Drosophila, respectively.
(D) Impaired UV-induced DNA damage repair in dUVRAG-deficient cells of Drosophila eye imaginal discs. Larval eye discs containing WT dUVRAG (GFP-labeled) and dUVRAG null (UVRAGB21, black area marked by the dotted line) cells were stained with anti-CPD. No CPD staining was detected under normal conditions. Strong CPD induction was detected 30 min after UV in both dUVRAG and mutant cells, but persisted only in dUVRAG mutant cells. Scale bar, 50 µm.
(E) Quantification of CPD staining in eye imaginal discs. Data shown represent mean ± SD (n > 200) from three independent experiments. ***p < 0.001.
(F) Survival rates to UV-C irradiation (254 nm, 0–60 J/m2) of 3rd instar larvae of WT (control) or dUVRAG heterozygote mutants (UVRAGB21 and UVRAGB7). Data shown represent mean ± SD; n = 1000 larva for each genotype collected from three independent experiments. *p < 0.05, **p < 0.01.
(G) Survival rates to UV-C irradiation (254 nm, 0–60 J/m2) of 3rd instar larvae of the indicated genotypes. Data shown represent mean ± SD; n = 1000 larva for each genotype collected from three independent experiments. ***p < 0.001.
(H) Knockdown of dDDB1 by RNAi enhances the UV sensitivity of dUVRAGRNAi fly larvae as shown by their survival rates after UV-C. Data shown represent mean ± SD; n = 1000 larva for each genotype collected from three independent experiments. **p < 0.01, ***p < 0.001.
To validate that tissue damage reflects an impaired UV-induced damage repair, we assessed CPD levels in the retina of UVRAG mosaic eye clones. The non-labelled UVRAG-homozygous mutant cells were surrounded by the WT cells marked by GFP (Figure 5D). We found that shortly after UV-irradiation of eye imaginal discs, there were comparable levels of CPDs in WT and dUVRAG-depleted cells (Figure 5D and 5E). However, 12 hr post-irradiation, high levels of CPDs persisted in dUVRAG-depleted cells (Figure 5D and 5E). As seen with dDDB1 RNAi (dDDB1-Ri), the survival rate of dUVRAG mutant larva with allelic loss of UVRAG (UVRAGB7and UVRAGB21) or with dUVRAG RNAi (dUVRAG-Ri) was significantly reduced as compared to WT larva, while ectopic expression of dUVRAG conferred UV resistance in a largely DDB1-dependent manner (Figure 5F–5H). These data indicate that UVRAG plays a conserved role in UV-induced DNA damage repair in vivo.
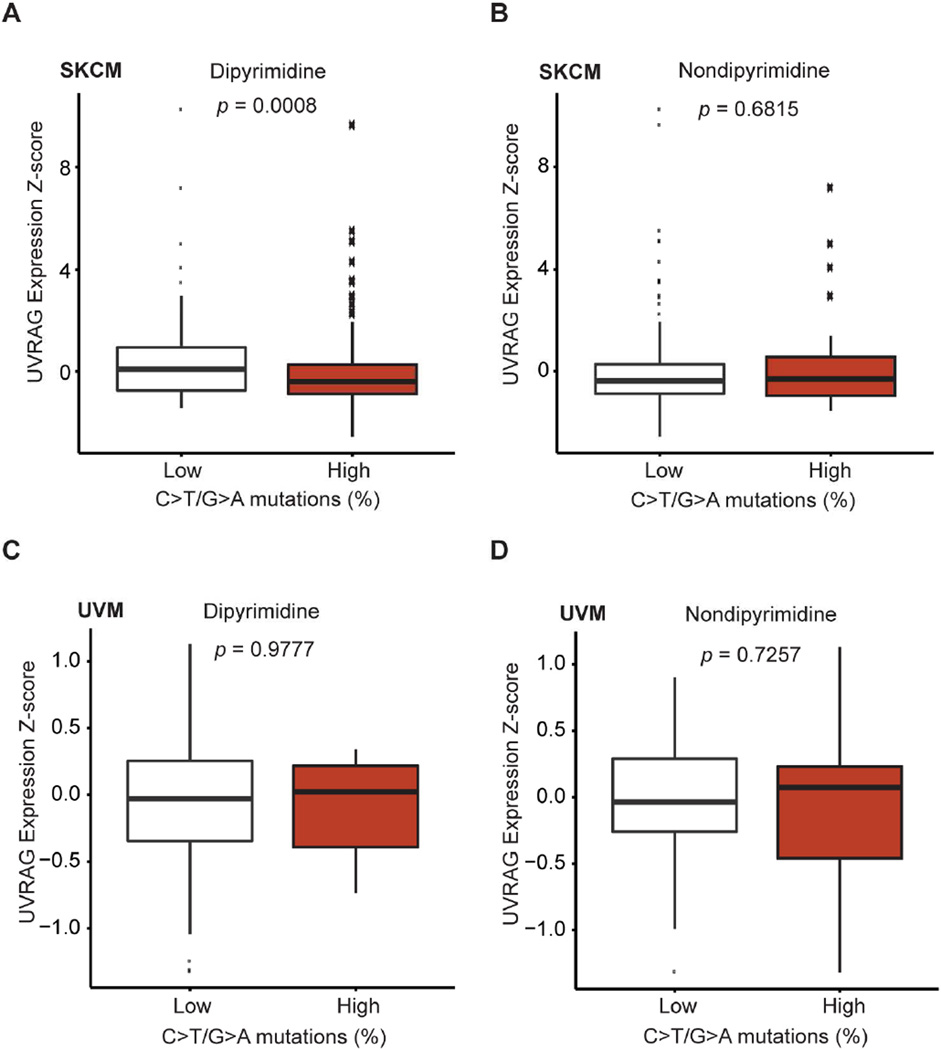
UVRAG Is Associated with Decreased UV-signature Loads in Melanoma
Recent landmark studies revealed an abundance of UV-induced DNA damage in melanoma genomes compared to most other types of tumors, thereby directly linking UV-irradiation to melanoma (Berger et al., 2012; Hodis et al., 2012; Pleasance et al., 2010). To explore the clinical relevance of UVRAG to UV-induced mutagenesis in melanoma, we analyzed all TCGA (The Cancer Genome Atlas) provisional melanoma patient datasets (Berger et al., 2012; Cerami et al., 2012; Gao et al., 2013; Hodis et al., 2012), and observed that higher UVRAG expression (Z-score) was associated with lower rates of UV-signature mutations, i.e., C>T/G>A transitions at dipyrimidine sites, in skin cutaneous melanoma (SKCM) (Figure 6A; p = 0.0008; two-sided, Wilcoxon rank-sum test), whereas no association detected for C>T/G>A transitions at non-dipyrimidine sites (Figure 6B; p = 0.6815; two-sided, Wilcoxon rank-sum test). Notably, the overall mutation rate at dipyrimidine sites was not significantly affected by UVRAG expression (Figure S7A; p = 0.2653; two-sided, Wilcoxon rank-sum test). Analogous assessment of “UV-signature” mutations in the UV-shielded uveal melanoma (UVM) revealed minimal association with UVRAG expression (Figure 6C and 6D), suggesting a specific role for UVRAG in melanoma UV-like mutagenesis. Indeed, a higher load of UV-specific mutagenesis was observed in UVRAGL286F melanoma (~94%) as compared with UVRAGWT ones (Figure S7B), consistent with its defective phenotype in NER. Univariate analysis revealed that SKCM patients with higher expression of UVRAG appeared to have increased overall survival compared to those with lower expression of UVRAG (Figure S7C; p = 0.0268). In fact, lower UVRAG expression appeared to be more frequent in SKCM cases with advanced tumor, node, and metastasis stage (Figure S7D). These data support an indispensable role of UVRAG in protecting against UV-induced mutagenesis in skin melanoma, which may further affect disease development and progression.
Figure 6. Negative Association of UVRAG Expression Z-score with UV-like Mutagenesis in Skin Melanoma.
(A and B) UVRAG expression Z-score in samples with high (> 50%) and low (≤ 50%) C>T/G>A transitions at dipyrimidines (A) and at non-dipyrimidines (B) for 340 available skin cutaneous melanoma (SKCM) TCGA primary tumors. Note the inverse relationship between UVRAG expression and C>T/G>A transition at dipyrimidines (A), but the absence of such a relationship at non-dipyrimidines (B). p, two-sided Wilcoxon rank-sum test.
(C and D) UVRAG expression Z-score in samples with high (> 50%) and low (≤ 50%) C>T/G>A transitions at dipyrimidines (C) and at non-dipyrimidines (D) for uveal melanoma (UVM) TCGA primary tumors (n = 80). p, two-sided Wilcoxon rank-sum test.
See also Figure S7 for additional information.
DISCUSSION
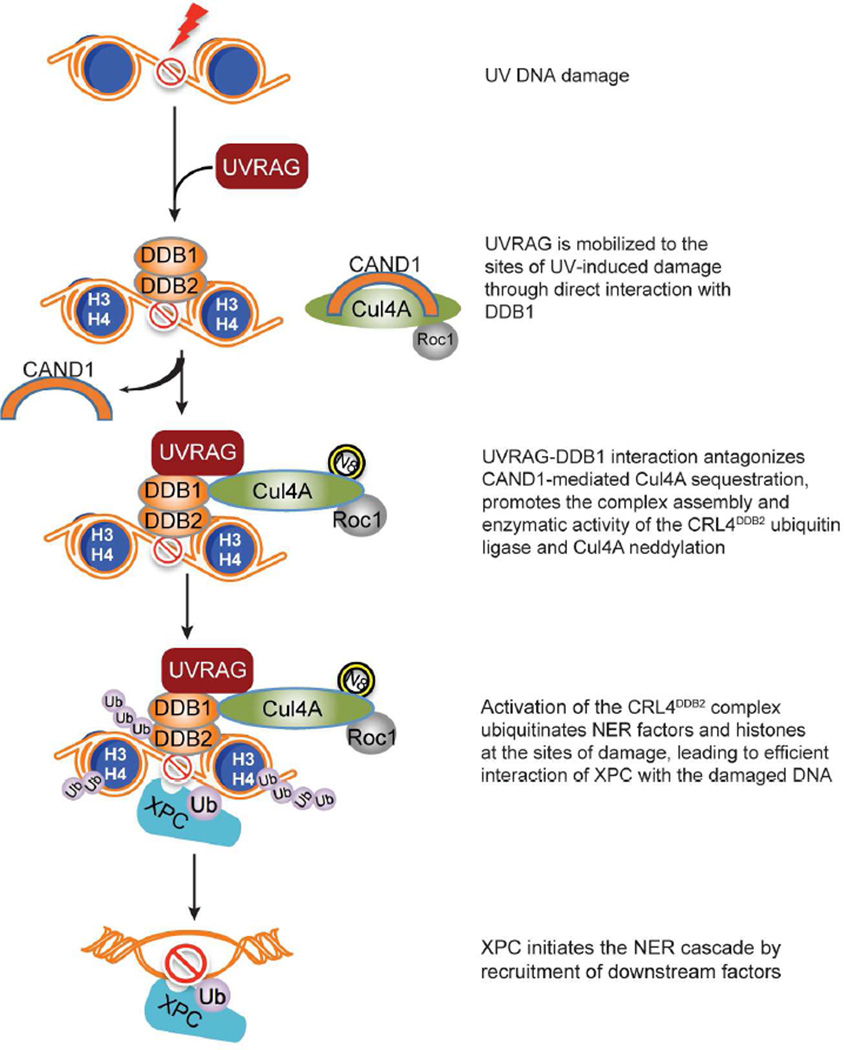
Herein, we demonstrated that UVRAG promotes UV-induced DNA damage repair by targeting DDB1. DDB1 interaction allows UVRAG to be recruited to the damaged foci after UV exposure, facilitating the assembly of the substrate-binding UV-DDB with the catalytic subunit Cul4A–Roc1 to form a functional CRL4DDB2 E3-complex. Consequently, activation of CRL4DDB2-driven ubiquitin-mediated proteolysis remodels the chromatin around the damaged sites, allowing the lesions accessible to downstream NER factors (Figure 7). Disruption of UVRAG-DDB1 interaction inhibited ubiquitination and association of XPC with UV-damaged CPD sites. Thus, UVRAG represents a key regulator in GG-NER. In addition, transgenic expression of UVRAG orthologue reduced levels of UV damage in Drosophila retina, indicating that the NER function of UVRAG occurs in vivo and not only in vitro. Furthermore, association of UVRAG with reduced UV-signature loads in skin melanoma underscores the importance of NER fidelity in antagonizing UV-associated genetic instability of skin cancer.
Figure 7. Model of UVRAG-mediated Regulation of the NER Pathway.
Upon UV-irradiation, UVRAG recruits to UV-induced DNA damage sites and associates with DDB1. The UVRAG-DDB1 interaction antagonizes CAND1 sequestration of Cul4A, enhances the modular complex assembly of the Cul4A–Roc1 ubiquitin ligase with the substrate receptor UV-DDB, resulting in Cul4A neddylation and enzyme activation. Consequently, ubiquitin-mediated proteolysis of histones H3/H4 and DDB2 leads to relaxation of the nucleosome around the photodamage, enabling access of XPC and downstream effectors to the damaged sites to initiate NER cascade and repair the damage. Hence, UVRAG inactivation and/or defective NER may play important roles in increased UV-mutagenesis in cutaneous melanoma.
Autophagic tumor suppressor UVRAG is a multitasking protein that can influence a plethora of homeostatic pathways, including membrane trafficking, centrosome integrity, and chromosomal stability (He et al., 2013; Liang et al., 2006; Liang et al., 2008; Zhao et al., 2012). Our results unraveled a nuclear function of UVRAG in ensuring UV-induced NER independent of its ability to regulate autophagy. UVRAG-mediated GG-NER is abrogated by point mutation (L286F) in UVRAG that blocks binding to DDB1 but remains competent for autophagy activation, suggesting that DDB1 interaction of UVRAG is specific and essential for NER. Moreover, autophagy loss or inhibition of autophagic flux could not forestall UV damage induced by UVRAG deficiency. Furthermore, it is the coiled-coil region that mediates the NER effect of UVRAG, which is separable from other interactions of UVRAG involved in the regulation of trafficking, centrosome, and DNA-PK (He et al., 2013; Zhao et al., 2012). These observations argue for an independent role of UVRAG-DDB1 interaction in the regulation of NER.
The regulatory effects of UVRAG are associated with activation of the DDB1-containing CRL4DDB2 ubiquitin-ligase complex by inducing the complex assembly and the Cul4A neddylation. Previous findings suggest that Cullin neddylation, and by extension CRL activity, is antagonized by the COP9 signalosome complex (CSN)-mediated deneddylation and CAND1-mediated Cul4A sequestration (Bennett et al., 2010). Although it remains possible that UVRAG may antagonize the effect of CSN to keep Cul4A in an active form, association of UVRAG with both neddylated and unneddylated Cul4A suggests that binding of UVRAG to the Cul4A complex is not rate-limiting for CSN-mediated deneddylation. In line with this, ectopic expression of UVRAG competes with CAND1 for Cul4A binding, whereas depletion of UVRAG results in increased Cul4A sequestration by CAND1, both in a DDB1-dependent manner in vivo. Thus, UVRAG activation of Cul4A may involve at least in part the release of Cul4A from CAND1 sequestration. Interestingly, our analysis of Cul4A neddylation in a cell-free system revealed that UVRAG could promote the neddylation reaction in the absence of CAND1, but not in the absence of DDB1, suggesting that it is the substrate adaptor, but not CAND1, that plays a more direct role in the neddylation process regulated by UVRAG. Our study is thus consistent with recent work highlighting a major role for substrate adaptor modules in the dynamic organization and activation of CRL complexes (Bennett et al., 2010).
Whereas Cul4A plays a positive role in UV-induced NER (Kamileri et al., 2012; Sugasawa, 2009), a previous study (Liu et al., 2009) reported that Cul4a deletion caused resistance against, rather than sensitivity to, UV-induced skin tumors in a mouse model. This paradox is unresolved but could be rationalized through the view that the Cul4A–based E3 ligase undergoes dynamic complex reorganization and cycles of neddylation in order to be fully functional in cells. It is possible that persistent inhibition of ubiquitin-mediated proteolysis due to genetic Cul4A inactivation can enhance stability of a subset of NER factors, thereby leading to prolonged NER signaling (El-Mahdy et al., 2006). It is also worth noting that Cul4A–based ubiquitin-ligase not only acts in NER, but functions widely by partnering with different factors under diverse environmental cues (Bennett et al., 2010). In our study, we focused on the role of UVRAG in DDB1 and Cul4A regulation in NER, a major defense mechanism against UV damage. Furthermore, treating melanoma cells with Cullin inhibitor sensitized cells to UV-induced damage. Thus, our findings demonstrate that at least in this context, UVRAG activated Cul4A–based E3 ligase functions to promote, rather than inhibit, DNA damage repair by NER.
NER fidelity constitutes an important melanoma risk biomarker and predicts melanoma survival (Emmert and Kraemer, 2013). Hence, the ability of UVRAG to promote NER is of importance with respect to melanoma and other UV-associated malignancies. Of note, UVRAG was initially discovered for its ability to complement UV-sensitivity of XP (Emmert and Kraemer, 2013; Perelman et al., 1997). In addition to other effects of UVRAG, the activating effects of UVRAG on DDB1-dependent NER may prevent environmental UV-induced DNA damage and thereof the development of melanoma. In fact, the NER-defective L286F mutation of UVRAG identified in melanoma patients was associated with a high mutation load dominated by UV-like mutagenesis (Berger et al., 2012; Hodis et al., 2012). Given that UVRAG expression is positively associated with reduced UV-signature in cutaneous melanoma, but not in UV-shielded melanoma, UVRAG may represent a predisposing factor for UV-associated genetic instability. Future studies on the role of UVRAG-DDB1 interactions in UV-induced genomic instability will advance our understanding of NER and its fundamental significance in skin cancers.
EXPERIMENTAL PROCEDURES
Unscheduled DNA synthesis (UDS) and Recovery of RNA synthesis (RRS) Assays
UDS detection was performed using a Click-iT DNA AlexaFluor Imaging kit (Life technologies), according to the manufacturer’s instructions. Briefly, after global irradiation (20 J/m2), cells on coverslips were incubated for 4h with 5 µM 5-ethynyl-2’-deoxyuridine (EdU), then washed with PBS, fixed and permeabilized before incubation for 30 min with the Click-iT reaction cocktail containing AlexaFluor Azide 488. After washing, the coverslips were mounted with mounting medium (Vectashield, Vector Labs, CA). Cell images were analyzed as for the RRS assay (see below). For each sample, at least 200 nuclei (non-S-phase) were analyzed per condition of three independent experiments. Of note, non S-phase cells can be easily differentiated from strong signals from scheduled DNA synthesis in S-phase cells.
For RRS, RNA detection was performed using a Click-iT RNA AlexaFluor Imaging kit (Life technologies), according to the manufacturer’s instructions. Briefly, cells were UV-C irradiated (10 J/m2) and incubated for 5 min (as a reference to show transcription is inhibited by UV irradiation) or for 4 hr at 37°C, followed by 2 hr incubation with 100 µM 5-ethynyl uridine (EU). Cells were then fixed and permeabilized in 4% formaldehyde and 0.5% Triton X-100 in PBS, and after washing with PBS, incubated for 30 min with the Click-iT reaction cocktail containing AlexaFluor Azide 488. Cells were then washed with PBST (0.05% Tween-20), and the coverslips were mounted with mounting medium (Vectashield, Vector Labs, CA). Images of the cells were obtained with a Nikon Eclipse C1 confocal microscope, and the average fluorescence intensity per nucleus was quantified by NIS-Elements software and normalized to the mock-treated cells. For each sample, at least 200 nuclei were analyzed from three independent experiments.
CPD Staining In Drosophila Eye Imaginal Disc
The third instar larvae were dissected and cultured in Shields and Sang M3 insect medium (Sigma S3652), which was supplemented with 10% FCS and 0.5% penicillin-streptomycin (Invitrogen). After PBS washing, imaginal discs were irradiated with UV bulb (20 J/m2) and recovered in Shields and Sang M3 insect medium for 30 min or 12 hr. All imaginal discs were fixed and stained with anti-CPD (TDM-2, Cosmo Bio) primary antibodies as described previously (Yan et al., 2010). The discs were then incubated with fluorescent-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) Confocal images were collected using a Nikon Eclipse C1 confocal microscope with 60X oil objectives. Images were processed using Adobe Photoshop. Approximate 200 dUVRAG fly mutant clones were randomly chosen for CPD-staining.
Supplementary Material
HIGHLIGHTS.
UVRAG is strictly required for global-genomic NER, independently of autophagy
UVRAG accumulates at photolesions and associates with DDB1
UVRAG expression inversely correlates with UV-like mutagenesis in melanoma
UVRAG-DDB1 interaction antagonizes CAND1 and activates CRL4DDB2 E3-ligase complex
Acknowledgments
We thank Ross Tomaino (Harvard Medical School, MA) for Mass Spectrometry Analysis. We also thank Dr. Jongkyeong Chung (Seoul National University, Korea) for providing the dUVRAG-mutant and overexpression Drosophila strains. We thank Dr. N. Mizushima for providing Atg5-knockout iMEF and Dr. J.U. Jung for providing SiMPull-related reagents. The results shown here are in whole or part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/. The authors wish to thank Dr. Martine Torres for her editorial assistance. This work was supported by the Margaret Early Trustee Foundation, American Cancer Society (RSG-11-121-01-CCG), and National Institutes of Health grants R01 CA140964 to C. Liang and AI073099 to J.U. Jung, and the GRL Program (K20815000001) from National Research Foundation of Korea to B.-H. Oh.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Y.Y. performed most experiments of this study and analyzed the data. S. H and F. L. conducted the bioinformatics analysis and S.H. performed the mass spectrometry analysis. Y.J. helped with colony survival assays. M.K. conducted protein purification. S.C. helps with SiMPull assay. Q.W., T.Z., S.P., D.O., and C. N. helped with data collection. B.F., J.J., B.O., G.A., and Z.Y. helped with data discussion. C.L designed and analyzed the experiments, and wrote the manuscript.
COMPETING FINANCIAL INTEREST STATEMENT
The authors declare no competing financial interests.
REFERENCES
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell. 2010;143:951–965. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, Ivanova E, Watson IR, Nickerson E, Ghosh P, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nature reviews. Genetics. 2009;10:756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- Duan MR, Smerdon MJ. UV damage in DNA promotes nucleosome unwrapping. J Biol Chem. 2010;285:26295–26303. doi: 10.1074/jbc.M110.140087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Mahdy MA, Zhu Q, Wang QE, Wani G, Praetorius-Ibba M, Wani AA. Cullin 4A–mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC. J Biol Chem. 2006;281:13404–13411. doi: 10.1074/jbc.M511834200. [DOI] [PubMed] [Google Scholar]
- Emmert S, Kraemer KH. Do not underestimate nucleotide excision repair: it predicts not only melanoma risk but also survival outcome. The Journal of investigative dermatology. 2013;133:1713–1717. doi: 10.1038/jid.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmert S, Kraemer KH. Do not underestimate nucleotide excision repair: it predicts not only melanoma risk but also survival outcome. J Invest Dermatol. 2014;133:1713–1717. doi: 10.1038/jid.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garibyan L, Fisher DE. How sunlight causes melanoma. Curr Oncol Rep. 2010;12:319–326. doi: 10.1007/s11912-010-0119-y. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Iwai S, Kuraoka I. A non-isotopic assay uses bromouridine and RNA synthesis to detect DNA damage responses. Mutation research. 2010;699:62–66. doi: 10.1016/j.mrgentox.2010.04.002. [DOI] [PubMed] [Google Scholar]
- He S, Ni D, Ma B, Lee JH, Zhang T, Ghozalli I, Pirooz SD, Zhao Z, Bharatham N, Li B, et al. PtdIns(3)P-bound UVRAG coordinates Golgi-ER retrograde and Atg9 transport by differential interactions with the ER tether and the beclin 1 complex. Nat Cell Biol. 2013;15:1206–1219. doi: 10.1038/ncb2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YJ, McCall CM, Hu J, Zeng Y, Xiong Y. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 2006;20:2949–2954. doi: 10.1101/gad.1483206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A–ROC1 ligase in response to DNA damage. Nat Cell Biol. 2004;6:1003–1009. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- Jain A, Liu R, Ramani B, Arauz E, Ishitsuka Y, Ragunathan K, Park J, Chen J, Xiang YK, Ha T. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473:484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamileri I, Karakasilioti I, Garinis GA. Nucleotide excision repair: new tricks with old bricks. Trends in genetics : TIG. 2012;28:566–573. doi: 10.1016/j.tig.2012.06.004. [DOI] [PubMed] [Google Scholar]
- Kelsey EM, Luo X, Bruckner K, Jasper H. Schnurri regulates hemocyte function to promote tissue recovery after DNA damage. J Cell Sci. 2012;125:1393–1400. doi: 10.1242/jcs.095323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Lee G, Liang C, Park G, Jang C, Jung JU, Chung J. UVRAG is required for organ rotation by regulating Notch endocytosis in Drosophila. Developmental biology. 2011;356:588–597. doi: 10.1016/j.ydbio.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Hu Z, Liu Z, Wang LE, Strom SS, Gershenwald JE, Lee JE, Ross MI, Mansfield PF, Cormier JN, et al. Polymorphisms in the DNA repair genes XPC, XPD, and XPG and risk of cutaneous melanoma: a case-control analysis. Cancer Epidemiol Biomarkers Prev. 2006;15:2526–2532. doi: 10.1158/1055-9965.EPI-06-0672. [DOI] [PubMed] [Google Scholar]
- Li C, Yin M, Wang LE, Amos CI, Zhu D, Lee JE, Gershenwald JE, Grimm EA, Wei Q. Polymorphisms of nucleotide excision repair genes predict melanoma survival. The Journal of investigative dermatology. 2013;133:1813–1821. doi: 10.1038/jid.2012.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Furukawa M, Matsumoto T, Xiong Y. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol Cell. 2002;10:1511–1518. doi: 10.1016/s1097-2765(02)00783-9. [DOI] [PubMed] [Google Scholar]
- Liu L, Lee S, Zhang J, Peters SB, Hannah J, Zhang Y, Yin Y, Koff A, Ma L, Zhou P. CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol Cell. 2009;34:451–460. doi: 10.1016/j.molcel.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz P, Lakatos Z, Maruzs T, Szatmari Z, Kis V, Sass M. Atg6/UVRAG/Vps34-containing lipid kinase complex is required for receptor downregulation through endolysosomal degradation and epithelial polarity during Drosophila wing development. BioMed research international. 2014;2014:851349. doi: 10.1155/2014/851349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15:465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- Perelman B, Dafni N, Naiman T, Eli D, Yaakov M, Feng TL, Sinha S, Weber G, Khodaei S, Sancar A, et al. Molecular cloning of a novel human gene encoding a 63-kDa protein and its sublocalization within the 11q13 locus. Genomics. 1997;41:397–405. doi: 10.1006/geno.1997.4623. [DOI] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapic Otrin V, Kuraoka I, Nardo T, McLenigan M, Eker AP, Stefanini M, Levine AS, Wood RD. Relationship of the xeroderma pigmentosum group E DNA repair defect to the chromatin and DNA binding proteins UV-DDB and replication protein A. Mol Cell Biol. 1998;18:3182–3190. doi: 10.1128/mcb.18.6.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5:a012609. doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatz A, Batist G, Eggermont AM. The biology behind prognostic factors of cutaneous melanoma. Curr Opin Oncol. 2010;22:163–168. doi: 10.1097/CCO.0b013e328337fe8f. [DOI] [PubMed] [Google Scholar]
- Sugasawa K. UV-induced ubiquitylation of XPC complex, the UV-DDB-ubiquitin ligase complex, and DNA repair. Journal of molecular histology. 2006;37:189–202. doi: 10.1007/s10735-006-9044-7. [DOI] [PubMed] [Google Scholar]
- Sugasawa K. The CUL4 enigma: culling DNA repair factors. Mol Cell. 2009;34:403–404. doi: 10.1016/j.molcel.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, Tanaka K, et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- Takats S, Pircs K, Nagy P, Varga A, Karpati M, Hegedus K, Kramer H, Kovacs AL, Sass M, Juhasz G. Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol Biol Cell. 2014;25:1338–1354. doi: 10.1091/mbc.E13-08-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Lee JE, Gershenwald JE, Ross MI, Mansfield PF, Strom SS, Wang LE, Guo Z, Qiao Y, Amos CI, et al. Repair of UV light-induced DNA damage and risk of cutaneous malignant melanoma. J Natl Cancer Inst. 2003;95:308–315. doi: 10.1093/jnci/95.4.308. [DOI] [PubMed] [Google Scholar]
- Yan D, Wu Y, Yang Y, Belenkaya TY, Tang X, Lin X. The cell-surface proteins Dally-like and Ihog differentially regulate Hedgehog signaling strength and range during development. Development. 2010;137:2033–2044. doi: 10.1242/dev.045740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Oh S, Li D, Ni D, Pirooz SD, Lee JH, Yang S, Lee JY, Ghozalli I, Costanzo V, et al. A dual role for UVRAG in maintaining chromosomal stability independent of autophagy. Developmental cell. 2012;22:1001–1016. doi: 10.1016/j.devcel.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.