Abstract
As tumours evolve, the daughter cells of the initiating cell often become molecularly heterogeneous and develop different functional properties and therapeutic vulnerabilities. In glioblastoma (GBM), a lethal form of brain cancer, the heterogeneous expression of the epidermal growth factor receptor (EGFR) poses a substantial challenge for the effective use of EGFR-targeted therapies. Understanding the mechanisms that cause EGFR heterogeneity in GBM should provide better insights into how they, and possibly other amplified receptor tyrosine kinases, affect cellular signalling, metabolism and drug resistance.
Intratumoural heterogeneity is a characteristic of most cancers; consequently, a tumour is likely to harbour a small population of cells that are resistant to most available treatments1. The mechanisms that contribute to tumour heterogeneity (FIG. 1) are now starting to be understood. Glioblastoma (GBM) is a highly heterogeneous tumour2 and was one of the first cancers to be profiled through The Cancer Genome Atlas (TCGA) project3 (supervised by the National Institute of Health (NIH)), making it one of the most genomically well-characterized types of cancer. Mutations in genes that occur at frequencies ≥5% above baseline levels are also likely to have been identified in genome wide profiling studies4 and so further contribute to the rich data set that can be used to understand GBM heterogeneity.
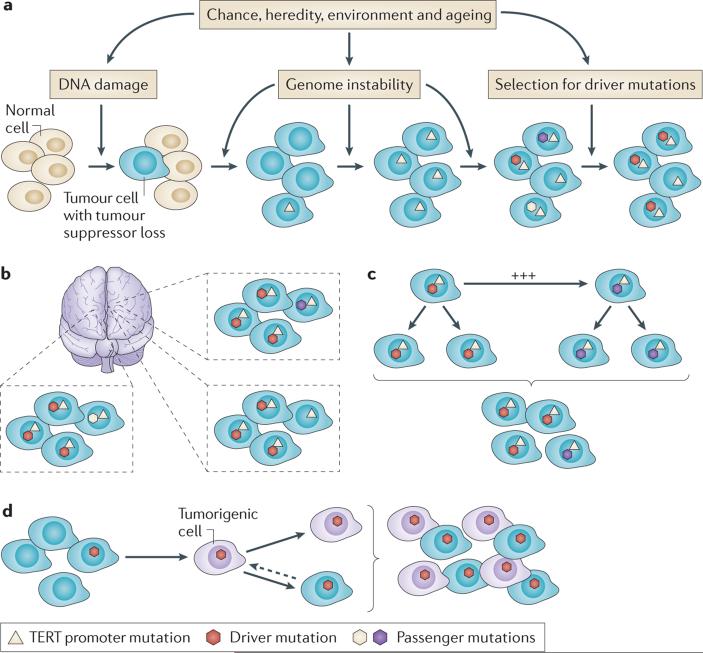
Figure 1. Multiple mechanisms regulate the development, progression and maintenance of tumour heterogeneity.
a | Clonal selection proposes that genome instability, usually as a consequence of the loss of a key tumour suppressor protein, such as telomerase reverse transcriptase (TERT), creates an environment in which new mutations are generated and mutations that increase the fitness for which tumour cells are selected. b | Regional differences in the microenvironment, due to differences in tissue architecture and nutrient and oxygen levels, leads to different selection pressures. c | The effects of clonal cooperation balance those of clonal selection to maintain heterogeneity. A subpopulation of tumour cells with lower fitness can be maintained if it increases the growth of another subpopulation through non-cell autonomous signalling. d | Differences in cell differentiation hierarchies, that is the cancer stem cell hypothesis, predict that a subpopulation of tumorigenic cells self-renews and gives rise to more-differentiated, functional and molecularly distinct non-tumorigenic progeny.
Receptor tyrosine kinases (RTKs) are crucial regulators of the growth factor signalling that controls cellular proliferation, metabolism and survival in response to environmental cues5,6. Therefore, it is not surprising that genetic alterations of RTKs, including those in the epidermal growth factor receptor (EGFR), have major roles in the development and progression of GBM7. RTKs have a remarkably heterogeneous cellular pattern within individual tumours8,9. In this Opinion article, we focus on EGFR amplification and mutations in GBM (see REF. 10 for a detailed and up-to-date summary of integrated GBM genomics). We also highlight the causes and consequences of intratumoural EGFR heterogeneity in GBM, focus on the general mechanisms that drive this feature, link these mechanisms with specific molecular events and examine their impact on signal transduction, cellular metabolism and resistance to EGFR-targeted therapy.
Tumour heterogeneity
The term tumour heterogeneity can be used to describe multiple forms of tumour variability, including intertumoural mutational pattern variation, intratumoural histological variation and intratumoural mutational polyclonality. In GBM, intertumoural mutational patterns are relatively stereotypical and are less heterogeneous than the range of mutations observed in many other types of cancer4,11. By contrast, individual GBM tumours display striking histological variation12. The full extent of intratumoural mutational polyclonality in GBM tumours is not yet known but, at present, the amount of intratumoural mutational heterogeneity seems to be similar to the amount that is observed in most cancer types.
In cancer, spontaneous somatic mutations, combined with sequential selection for aggressive subclones (that is, cells that can survive and/or proliferate in a compromised microenvironment), drive the growth of single cancer cells into complex, heterogeneous tumour masses13. In this paradigm, which is known as clonal evolution, new mutations are produced with increasing frequency as the tumour progresses, which makes late-stage cancer progressively more difficult to treat. Much recent data, including next-generation sequencing data, support the clonal evolution model as a major underpinning of tumour progression, heterogeneity and drug resistance14–21 (FIG. 1a). However, these same data show that clonal evolution in human solid tumours is not a linear process in which only one clone dominates as the tumour evolves.
Heterogeneity is also shaped by the local microenvironment. First, direct cell–cell interactions between inflammatory, stromal, endothelial22 and tumour cells, as well as autocrine and paracrine responses to secreted factors23, all impart selection pressures within the tumour microenvironment. Second, physical features, including neuro-anatomical structures and the proximity of the tumour to blood vessels and/or the leptomeninges, can also affect nutrient and oxygen levels in tumour cells. Therefore, regional heterogeneity can develop as tumour cells experience distinct selection pressures in different parts of the tumour (FIG. 1b). Third, cooperation between tumour subclones may also maintain intratumoural mutational heterogeneity24–26 (FIG. 1c). Last, cancer therapy also shapes tumour heterogeneity by either expanding or collapsing tumour cell sub-populations depending on the treatment (discussed below).
Non-genetic mechanisms also have an important role in tumour heterogeneity. The cancer stem cell model suggests that a hierarchical organization of tumours exists in which cancer cells give rise to both tumori genic and non-tumorigenic progeny — a process that generates phenotypic cellular heterogeneity27–29. Based on mechanisms that are poorly understood22,27,28, the relative proportion of tumorigenic versus non-tumorigenic cells can vary between tumour types and between individual tumours of the same type (FIG. 1d). The factors that make a cell tumorigenic or non-tumorigenic remain unclear, but epigenetic mechanisms that influence differentiation states23, may activate oncogenic pathways that can lead to cellular dedifferentiation30,31 and result in the genetic alteration of a bona fide stem cell27 might all have a role. At present, the role of cellular dedifferentiation in gliomagenesis remains unclear32. Some genetic mouse models indicate that neural stem cell and/or progenitor cell populations, such as the subventricular zone of the brain, are more likely to undergo malignant transformation in response to oncogenic perturbations than differentiated cells, such as the astrocytes and neurons of the cerebral cortex or striatum33–35. By contrast, other experimental mouse models indicate that mature and highly differentiated cells can also undergo malignant transformation in response to oncogenic perturbations31,36,37. Neural stem cells, astrocytes and even differentiated neurons were capable of generating malignant gliomas in response to lentiviral transduction of oncogenes, and showed high expression of stem and progenitor cell markers, regardless of the cell of origin31. Future studies will be needed to further determine the role of dedifferentiation in gliomagenesis.
RTK amplification and mutation in GBM
The genomic ‘portrait’ of GBM is rich and can be viewed from multiple perspectives, each yielding decidedly different views as to the essential molecular features and potential drug targets. In this Opinion article, we consider that the genetic alteration of growth factor receptor signalling pathways is an essential component of most adult GBMs. The Pan Cancer project of TCGA, the goals of which include the identification of actionable driver mutations through the comparative analysis of 3,000 tumours across 12 cancer types, similarly concluded that genetically altered RTKs, and their downstream effectors, are the most abundant targetable driver mutations in GBM38. This is also consistent with mouse models of GBM, in which the reconstitution of the genetically altered components of growth factor signalling pathways, in the same molecular context as in clinical samples, recreates histologically identical GBMs36,37. Thus, independent, converging and compelling lines of evidence indicate that the RTK lesions detected in GBM, and their associated biochemical alterations, are crucial driver events in GBM development.
Importantly, not all GBMs are driven by RTK alterations. Identifying potential mutations in shared nodes of convergence that are downstream of RTKs in adult GBMs — such as isocitrate dehydrogenase 1 (IDH1) mutations, tumour protein p53 (TP53) mutations in low-grade gliomas and epigenomic mutations in paediatric GBMs — may provide crucial new insights into the fundamental molecular underpinnings of GBM pathogenesis.
Heterogeneity of RTK mutations in primary GBM
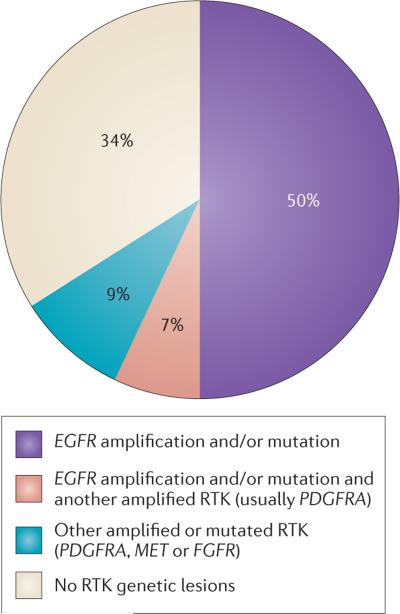
To date, integrated exome sequencing and DNA copy number analysis of 251 GBMs has been carried out through TCGA, providing a bird's eye view of the diversity of RTK genetic alteration7. This survey focused on primary GBMs — de novo GBMs that did not arise from lower grade gliomas. This form of GBM is, unfortunately, the most common clinical presentation of the disease, representing up to 95% of cases in some epidemiological studies39. RTK amplifications and/or mutations had occurred in 66% of the primary GBM samples that were tested by TCGA. Amplifications and/or mutations in EGFR were the only RTK lesions observed in 50% of all de novo primary GBMs. EGFR genetic alterations also coexist with other RTK lesions, but this occurs in only a small proportion (7%) of tumours. Therefore, EGFR genetic alterations — including mutations, rearrangements, alternative splicing and focal amplifications — are the dominant RTK lesions in GBM, occurring in 57% of tumours7 and, overall, are the most common oncogene alteration in these tumours. Amplification of platelet derived growth factor receptor alpha polypeptide (PDGFRA) occurs in 13% of GBMs, and nearly half of these tumours also contain amplifications and/or mutations in EGFR7 (FIG. 2). MET amplifications and fibroblast growth factor receptor (FGFR) mutations, including fusion genes40,41, occur in approximately 2% of the GBMs that have been analysed to date (FIG. 2).
Figure 2. Receptor tyrosine kinase genetic alterations in primary glioblastoma.
Re-analysis of The Cancer Genome Atlas (TCGA) data demonstrates that epidermal growth factor receptor (EGFR) amplifications and/or mutations are the dominant receptor tyrosine kinase (RTK) lesions in adult primary glioblastomas7. FGFR, fibroblast growth factor receptor; PDGFRA, platelet derived growth factor receptor alpha polypeptide.
RTK alterations usually coexist with mutations that activate other core regulatory pathways, including downstream components of growth factor receptor signalling pathways7. A frequency distribution of the coexistence of mutations and copy number alterations stratified by the RTK genotype demonstrates the frequent co-occurrence of mutations in PI3K and deletion of PTEN, as well as the co-occurrence of mutations and/or deletion of cyclin-dependent kinase inhibitor 2A (CDKN2A; encoding both INK4A and ARF) with all of the detectable RTK alterations7. This is consistent with the required cooperation of multiple core pathways for tumour formation in genetically engineered mouse models of GBM3,7,8.
Thus, the genomic portrait suggests a surprisingly straightforward picture: EGFR is the dominant, but not the exclusive, RTK lesion in primary GBMs; EGFR alterations are usually the sole RTK lesions; and RTK lesions usually occur in the context of other PI3K-pathway activating alterations and in the presence of CDKN2A loss and inactivation.
Focusing on RTK heterogeneity
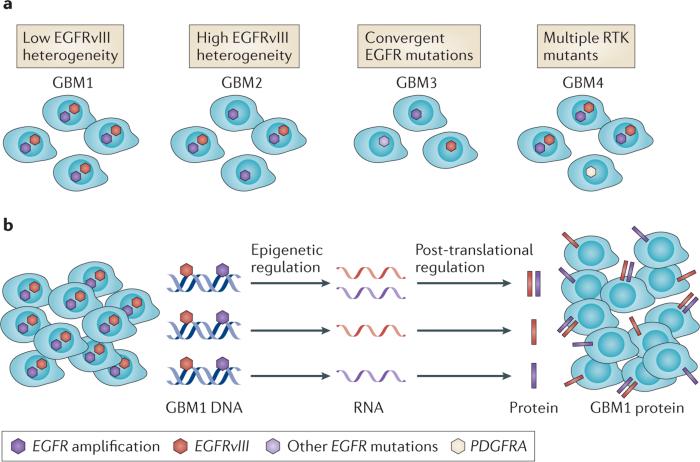
The RTK genetic alterations in primary GBM that are presented in FIG. 2 do not look heterogeneous; however, closer inspection reveals a substantial amount of RTK diversity (FIG. 3). Glioblastoma multiforme, the former name of GBM, was used to describe the striking cellular heterogeneity of the disease. Immunohistochemical staining of RTK alterations — such as EGFR variant III(EGFRvIII), which is the most common EGFR mutation in GBM and is characterized by the deletion of exons 2–7 that results in an in-frame deletion variant that has a truncated extracellular domain with ligand-independent constitutive activity12 (BOX 1) — shows similar cellular heterogeneity42–46. Consequently, DNA and RNA sequencing of bulk tumours provide only a limited insight into the RTK distribution among the many different cell subpopulations within the tumour. Therefore, the extent of RTK heterogeneity has, until recently, been poorly quantified. Single-cell DNA47 and RNA48 sequencing, and the bulk analysis of DNA and RNA that has been extracted from different regions of a tumour49,50, has begun to shed much needed light on the extent of intratumoural RTK heterogeneity in GBM.
Figure 3. Single-cell heterogeneity of receptor tyrosine kinase DNA, RNA and protein in glioblastoma.
a | Depicted are multiple varieties of intratumour receptor tyrosine kinase (RTK) DNA heterogeneity. b | Variable transcription of amplified and mutated RTKs adds an additional layer of complexity to glioblastoma (GBM). Recent single-cell sequencing studies indicate that individual GBM cells can contain multiple genetic RTK alterations43, but most GBM cells seem to express only one single-mutant RTK transcript44. At present, the underlying mechanisms are not understood. EGFR, epidermal growth factor receptor; EGFRvIII, epidermal growth factor receptor variant III; PDGFRA, platelet derived growth factor receptor alpha polypeptide.
Box 1. EGFR mutations in glioblastoma.
The epidermal growth factor receptor (EGFR) plays a crucial part in the biology of many different tumours; however, its specific genetic alterations vary depending on the type of tumour. For example, somatic mutations in the EGFR kinase domain — such as L858R in exon 21, in-frame deletions in exon 19 or insertions in exon 20 — are frequently found in non-small cell lung cancer, but are rarely found in glioblastoma (GBM). By contrast, a number of deletion mutations that involve the EGFR extracellular domain (ECD) are unique to GBM. These include the EGFR type I (amino-terminal deletion), type II (exons 14–15 deletion) and type III (exons 2–7 deletion) variants (EGFRvI, vII and vIII, respectively), of which EGFRvII and EGFRvIII have been confirmed to be constitutively active and oncogenic43,47,85. Low frequency cytoplasmic tail deletion mutants, such as those found in EGFRvIV (exons 25–27 deletion) and EGFRvV (exons 25–28 deletion), are also exclusive to GBM86 and are predicted to have attenuated ubiquitylation and degradation kinetics owing to the deletion of a CBL binding site at Y1045 (REF. 87). In addition, missense mutations that are located at the interface of the various domains of the EGFR ECD are another class of mutations identified in GBM and are found at ~14% frequency7. These mutations include R84K, A265V/D/T, P545L and G574V and are thought to favour an open, active EGFR confirmation88,89, which promotes constitutive receptor activity90.
RTK co-amplification
Fluorescent in situ hybridization using EGFR and PDGFRA DNA probes shows that some GBMs contain two different amplified RTKs51,52. In these rare tumours, most of the GBM cells contain either EGFR or PDGFRA amplifications, but not both, and usually the tumour cells containing PDGFRA amplifications are the minority population51. It is tempting to speculate that this pattern could arise from biclonal evolution, or that one of the RTK amplicons is progressively lost because it does not confer a sufficient growth advantage. However, the unequal segregation of amplified alleles, clonal cooperation and regional microenvironmental selection pressures51 might also contribute to this genetic heterogeneity. This example highlights the challenge of trying to infer clonal hierarchies from single molecular ‘snapshots’ of a tumour (FIG. 3a).
EGFR amplification with multiple mutations
A new single-cell sequencing approach has been used to identify unique, non-overlapping subclonal alterations in GBM47,53. In 71% of the samples that were studied, EGFR amplifications coexisted with at least one of a diverse range of EGFR variants, including structural alterations and/or missense extracellular domain mutations. In one particularly insightful example, up to 32 possible different clonal combinations, based on five distinct EGFR genomic lesions, were found47.
In depth single-nucleus sequencing analysis of two additional GBMs, integrated with bulk sequencing of the tumours, showed that the intratumoural heterogeneity of EGFR amplification and mutation arose through multiple routes47,53. In the first case, all of the tumour cells contained amplified wild-type EGFR and amplified EGFRvIII, but the levels of each varied dramatically among the tumour cells. EGFRvIII DNA can be located outside chromosomes in double minute DNA fragments2,7,54 at levels that can vary from cell to cell owing to their unequal segregation, which thus contributes to the intratumoural heterogeneity of EGFRvIII DNA levels (discussed below). In the second case, amplified wild-type EGFR and four additional amplified EGFR variants (BOX 1) were evident in the tumour, and these mutations were mutually exclusive, which suggests that the convergent evolution of independent EGFR mutants had occurred. These rare variants within the tumour were not detected by next-generation sequencing of the bulk tumour, raising the possibility that RTK intratumoural heterogeneity, at least with respect to EGFR (and perhaps other RTKs), might be even greater than is currently thought.
Transcription of amplified and mutated RTKs
The transcription of RTK mutants introduces an additional source of intratumoural heterogeneity (FIG. 3b). Single-cell RNA sequencing of GBM clinical samples showed that in one sample, most of the individual GBM cells that were expressing an EGFR transcript expressed only wild-type EGFR, EGFRvIII or EGFR with an exon 4 deletion (EGFR del4) RNA in a mutually exclusive manner48. A small number of GBM cells within a tumour seem to express both wild type EGFR and EGFRvIII proteins and, in these cells, wild type EGFR may phosphorylate EGFRvIII to activate downstream signalling44. Unfortunately, it is not yet technically feasible to carry out quantitative single-cell RNA and DNA sequencing on the same tumour cells from a clinical sample. However, this observation, although based on a small sample number and in need of repetition, raises the possibility that the heterogeneous expression of amplified and mutated RTK transcripts might be tightly regulated, thus contributing to the extensive intratumoural RTK heterogeneity of GBM (FIG. 3b).
Regional selection
Two recent studies indicate that RTK amplification, and the expression of amplified and mutated transcripts, can vary substantially in different parts of the same GBM. In the first study, a surgical intraoperative mapping scheme was used to collect spatially distinct regions from 11 samples of GBM, and they carried out copy number and transcriptome analyses of samples, including some samples from patients who were repeatedly biopsied over time, to assess intratumoural heterogeneity49. Different regions of the same tumour showed distinctive molecular signatures, and this was used to assemble a potential path of clonal evolution. In the second study50, RNA sequencing analysis of GBM biopsies that were taken with the aid of magnetic resonance imaging showed highly variable transcriptional patterns, including those of RTKs, in different regions of the tumour. These initial findings suggest that regions of the tumour could differ because of variations in the tumour microenvironment.
All of the studies discussed above begin to highlight the extent of regional intratumoural molecular heterogeneity in GBM. However, the full extent of regional heterogeneity of amplified and mutated RTKs remains unclear. Trying to reconstruct clonal hierarchies from molecular snapshots that are derived from either DNA copy number and DNA sequencing or RNA sequencing of bulk tumour samples remains a substantial challenge. Future studies that integrate single-cell RNA- or DNA-sequencing analysis from different regions of the same tumour, and that take into consideration protein expression levels, including how they change over time and in response to various treatments, will probably provide new insights into the regional intratumoural RTK heterogeneity and potentially into the different selection pressures that contribute to this.
Mechanisms of RTK heterogeneity
What are the specific molecular mechanisms by which the intratumoural heterogeneity of RTKs develops in GBM? Answering this question has been difficult because, unlike secondary GBMs that develop from lower grade gliomas and for which a sequence of mutations is beginning to emerge55, primary de novo GBMs are highly malignant on initial clinical presentation. Thus, directly observing the molecular sequence of events that drive GBM formation has not been possible in tumour samples from patients or in genetic mouse models.
One approach that has been adopted by some investigators uses mathematical modelling to infer the order of multistep mutational events in the genesis of primary GBMs. Based on the assumption that self-renewing cells can accumulate cancer-causing mutations through a stochastic process, dynamic mathematical models are constructed and the results are compared to the frequencies of mutations that are observed in TCGA data56. This approach has been recently used to infer that aneuploidy involving chromosomes 7 and 10 is an early event that is followed by CDKN2A or TP53 loss. Overexpression of PDGFA on chromosome 7 was implicated as a driving event in GBM formation because PDGFα overexpression is sufficient for GBM formation in a genetic mouse model57. The mathematical model also predicted that EGFR focal amplifications and mutations were late events. However, alternative mutational paths are likely to be relevant to GBM formation and progression.
Consistent with the clonal selection model (FIG. 1a), deletion and mutation of CDKN2A and/or TP53 could contribute to genome instability, thereby providing various cells with different mutations that will be subject to selective pressures. The capacity of cells to tolerate a high mutational load might occur as a result of mutations that alter cell-cycle regulation and/or senescence, enabling cells bearing damaged DNA to continue to proliferate (FIG. 4a). TCGA GBM data indicate that CDKN2A deletion or loss of expression was detected in 61% of the GBMs for which whole exome and copy number data were available. This loss was enriched in GBMs with EGFR amplifications, and/or mutations, and was found in 74% of tumours. This high frequency is consistent with genetic mouse models showing that combined EGFRvIII expression and CDKN2A loss are sufficient to transform mouse astro-cytes into GBMs36. Re-analysis of TCGA data7 shows that deletions and point muta-tions in TP53 are greatly enriched in GBMs with amplified EGFR and/or mutated EGFR that lack CDKN2A lesions (41%) compared with GBMs with EGFR amplifications and CDKN2A lesions (14%), suggesting an alternative source of genome instability that enables the development and clonal selection of EGFR amplification and mutation. Consistent with this, only 6% of the GBMs with EGFR amplifications and/or mutations contained both CDKN2A and TP53 genetic lesions7. The molecular mechanisms that underlie the aneuploidy of chromosome 7 — an event that is presumed to occur early in GBM development57 — the focal amplification, structural rearrangements and mutations of EGFR on chromosome 7, and whether these events are linked, remain unclear, but a number of molecular mechanisms are possible58.
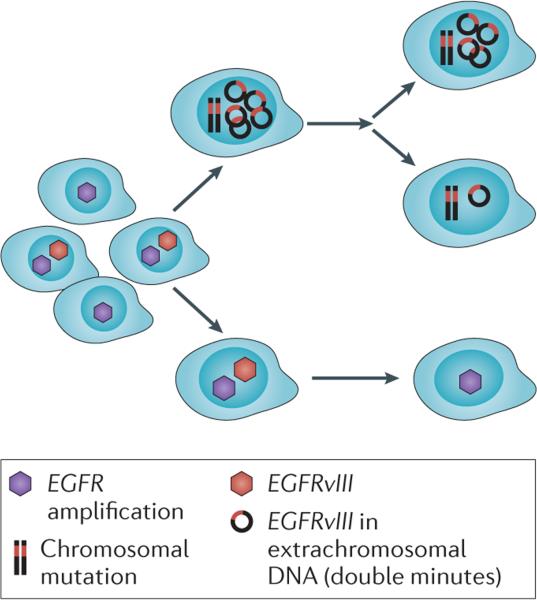
Figure 4. Interlacing mechanisms shape intratumoural receptor tyrosine kinase heterogeneity during glioblastoma progression.
Clonal selection of glioblastoma (GBM) cells expressing amplified receptor tyrosine kinases (RTKs), such as epidermal growth factor receptor (EGFR) and EGFR variant III (EGFRvIII), in the context of genome instability, is depicted. Heterogeneity is maintained through the unequal distribution of extrachromosomal EGFRvIII DNA in double minutes and through clonal cooperation between cells that express EGFRvIII and cells that do not.
Once EGFR amplifications, gene rearrangements and/or mutations have occurred, EGFR heterogeneity might be generated and maintained through a series of complimentary interlacing mechanisms. First, EGFRvIII strongly promotes tumour cell proliferation in a cell-autonomous manner59,60, therefore these cells are likely to have a proliferative advantage (FIG. 4). Second, the clonal cooperation between populations of GBM cells bearing different EGFR variants might maintain a heterogeneous state, and one study has shown that even a small number of GBM cells expressing EGFRvIII can contribute to the growth of wild-type EGFR GBMs through a novel interleukin 6 (IL-6)-dependent pathway26. Last, unequal segregation of EGFRvIII alleles on extrachromosomal DNA provides a compelling mechanism for introducing and maintaining EGFRvIII heterogeneity, especially because levels of EGFRvIII DNA could potentially be increased or decreased as an adaptive response to changes in the tumour environment2. Thus, RTK heterogeneity could be generated and maintained through clonal selection, clonal cooperation, regulation of extrachromosomal DNA and regional selection, resulting in a cellular composition that maximizes tumour growth (FIG. 4). This hypothesis is supported by data showing that EGFRvIII+ and EGFRvIII− cells that have been isolated and sorted from a single tumour each give rise to GBM neurospheres and tumours in vivo in which both cell types were present in the same ratio of EGFRvIII+/EGFRvIII− cells that was present in the parental tumour2.
Functional impact of RTK heterogeneity
Until recently, it has been difficult to assess the effect of intratumoural RTK heterogeneity on cell signalling, metabolism and behaviour, including response to treatment. Recent technical advances61–63 have made it possible to quantify signalling differences and to assess variations in proliferative and apoptotic indices (through Ki-67 and terminal deoxy nucleotidyl transferase dUTP nick end labelling (TUNEL) assays, respectively) of distinct subpopulations of cells within a tumour and thus to begin to examine the functional consequences of RTK heterogeneity2. For example, EGFRvIII+ GBM cells are more proliferative and undergo less apoptotic cell death, both at baseline and in response to EGFR targeted therapies, when assessed in a patient derived xenograft (PDX) mouse model2. The EGFRvIII+ GBM cells in the tumour also took up more glucose, which is consistent with recent findings that the expression of EGFRvIII increases glycolytic metabolism64–68. If increased glycolysis is required to meet the increased demand for energy and carbon atoms for anabolic metabolism in GBM cells, then alterations in the levels of EGFRvIII expression might occur in response to environmental glucose levels. Clonal cooperation between EGFRvIII+ and EGFRvIII− GBM cells, as has been described26, might provide a mechanism by which GBMs maximize their growth in response to the nutrients that are available in the microenvironment. Future studies are needed to assess exactly how nutrient environments shape EGFR and other RTK heterogeneity in GBM, and how RTK heterogeneity in turn modulates tumour metabolism to maximize growth in response to the environment.
RTK heterogeneity and resistance
Radiation, cytotoxic chemotherapy and molecularly targeted treatments exert notable selection pressures on tumours, but how heterogeneity affects treatment response is poorly understood. In particular, the role of intratumoural RTK heterogeneity in cancer drug resistance has been difficult to determine, in part because tracking the fate of individual tumour cells in response to treatment presents a considerable technical and logistical challenge. Preclinical models suggest that intratumoural RTK heterogeneity might confer differential sensitivity to cytotoxic chemotherapies. For example, EGFRvIII promotes resistance to temozolomide and cisplatinum by, at least in part, activating mammalian target of rapamycin complex 2 (mTORC2) signalling69–71. Whether cytotoxic chemotherapy selects for resistant EGFRvIII+, mTORC2 activated tumour cells in patients with GBM who are undergoing standard treatments remains to be determined.
A number of mechanisms seem to have an important role in the development of resistance to RTK-targeted therapies, including insufficient drug exposure; second-site resistance-promoting mutations in the targeted RTK; mutations in downstream signalling effectors that keep the pathway activated; and bypass mechanisms through the amplification, mutation or upregulation of another RTK or its downstream effectors72.
Small molecule inhibitors of RTKs primarily bind to, and displace ATP from, a specialized domain of the receptor, thus blocking RTK activity and downstream function. Mutations that alter the binding of these drugs to the ATP pocket and/or increase the affinity of ATP versus the drug for this binding pocket prevent the tyrosine kinase inhibitors (TKIs) from blocking RTK activity73. Such mutations explain TKI resistance in other forms of cancer73; however, second-site EGFR mutations have not been detected in GBM.
Genetic disruption of the tumour suppressor gene PTEN and the phosphorylation of PTEN at Y240 limit suppression of the PI3K signalling pathway. Thus, cells with these mutations maintain PI3K signalling required for tumour growth74,75 and require higher doses of an EGFR TKI to turn off the crucial growth and survival signals76. PTEN deletion, mutation, or the phosphorylation of PTEN at Y240 show considerable intratumoural heterogeneity in GBM, and there is evidence of selection for tumour cells with Y240 phosphorylated PTEN in patients with GBM treated with the EGFR TKI erlotinib74.
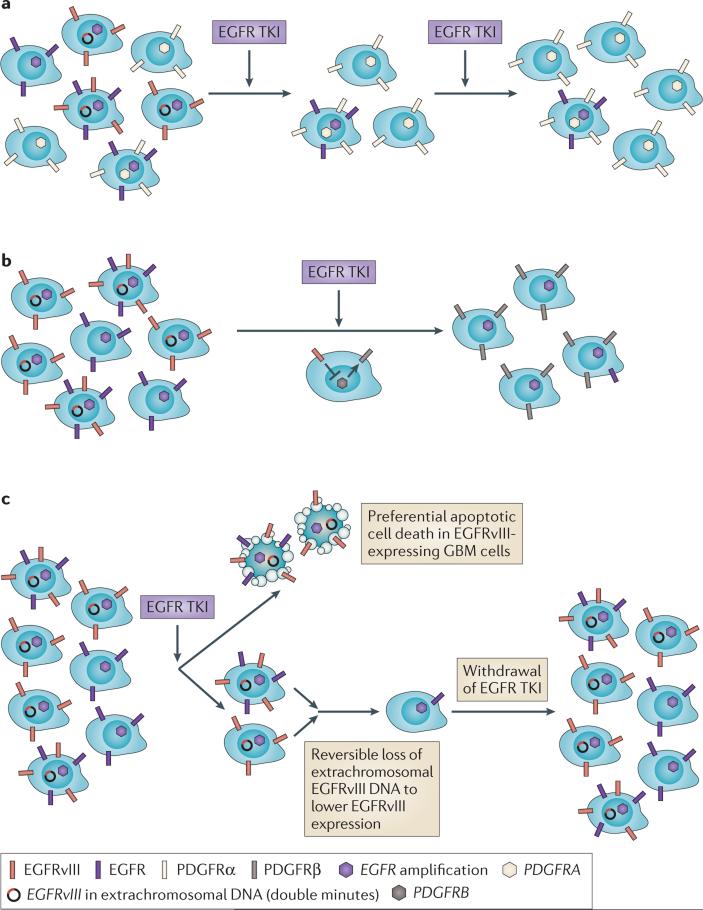
Resistance to TKIs can also occur through the amplification or mutation of other RTKs (FIG. 5a) or by feedback upregulation of a physiologically-regulated RTK pathway (FIG. 5b). The observation that some EGFR amplified GBMs also contain a population of PDGFRA amplified tumour cells that are selected for in response to EGFR TKIs, at least in experimental models51,52,77, provides evidence for one direct way in which RTK heterogeneity contributes to resistance. However, the incidence of RTK co-amplification may be underestimated if non-dominant amplified or mutated RTKs are present at a low level that is undetectable by next-generation sequencing of the bulk of a tumour.
Figure 5. Impact of intratumoural receptor tyrosine kinase heterogeneity on the response to therapy in glioblastoma.
a | Epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI) resistance can be acquired through the co-amplification of EGFR with other amplified receptor tyrosine kinases (RTKS), such as platelet derived growth factor receptor alpha polypeptide (PDGFRA). b | Feedback activation of non-amplified, non-mutated RTKs are shown. For example, EGFRvIII transcriptionally represses PDGFRβ in tumour cells. EGFR TKIs derepress PDGFRβ, enabling tumour cells to switch their dependence to PDGFRβ for growth and survival. c | EGFRvIII-expressing tumour cells are notably more sensitive to EGFR TKI-mediated apoptotic cell death. EGFRvIII is eliminated from extrachromosomal DNA, lowering EGFRvIII expression and desensitizing tumour cells to the drug. Re-emergence of EGFRvIII on extrachromosomal DNA follows drug withdrawal. GBM, glioblastoma.
Feedback activation of PDGFRβ in response to EGFR TKIs is another mode of EGFR TKI resistance in GBM78 and is similar to some of the mechanisms that underlie the resistance of melanoma and colorectal cancer cells to the BRAF inhibitor vemurafinib77,79–82. In response to EGFR inhibition in cell lines, mouse models and patients with GBM, PDGFRβ is upregulated through transcriptional derepression78. The intratumoural heterogeneity of these resistance-promoting pathways remains to be determined.
Elimination of double minutes chromosomes containing EGFRvIII has recently been shown to be an unanticipated mechanism of GBM resistance to EGFR TKIs2 (FIG. 5c). Single-cell studies of a GBM PDX model demonstrated that EGFRvIII-expressing tumour cells were substantially more sensitive to erlotinib, and underwent apoptotic cell death in vivo upon exposure to the drug. However, a non-reversible down-regulation of EGFRvIII levels, which was mediated by the elimination of EGFRvIII-containing extrachromosomal DNA, was evident in tumours that were treated with erlotinib. A similar phenomenon was evident in patients who were treated with the EGFR TKI lapatinib. After treatment cessation, extrachromosomal EGFRvIII levels rose, resensitizing tumour cells to erlotinib — a finding with obvious therapeutic implications2. This result indicates that intratumoural RTK heterogeneity has a crucial role in EGFR TKI resistance in GBM.
Common themes
First and foremost, few clinical studies have been designed to enable the determination of whether RTK inhibitors, such as EGFR TKIs, inhibit their intended drug targets in the tumours of patients. Therefore, in the absence of detailed studies measuring intratumoural drug levels and target engagement, it is difficult to determine whether the failure of EGFR TKIs is simply a consequence of insufficient EGFR inhibition, or whether GBMs become dependent on alternative signalling networks for growth and survival. Quantitative analysis of intratumoural lapatinib levels and measurements of EGFR phosphorylation at Y1173 by multi-array immunoassays demonstrate that both intratumoural drug levels and the degree of EGFR inhibition in tumour tissues of patients with GBM are far below the amounts that are needed to cause tumour cell death83. In light of the fact that PTEN deficiency might cause tumour cells to require an even larger dose of an EGFR inhibitor to achieve the same level of pathway inhibition76, pharmacological failure might have a major role in the lack of durable efficacy that is observed in patients with GBM who have been treated with EGFR TKIs. Importantly, therapeutic resistance may still occur when the drug dose is limiting. If the intratumoural drug concentration is high enough to partially inhibit EGFR signalling, but is not high enough to cause extensive cell death, GBM cells adapt by eliminating extrachromosomal EGFRvIII DNA2 and/or derepressing PDGFRβ78.
Second, the preclinical data in GBM, and in other cancers, suggest that resistance to RTK targeted therapies could be partially alleviated by intermittent dosing schedules2. In a small study of patients with lung cancer who became resistant to gefitinib, a ‘drug holiday’ potently resensitized these patients to gefinitib and provided an extended period of disease control84. In addition, intermittent administration of TKIs might enable patients to tolerate far higher doses of a drug than the doses that are tolerated during continuous daily administration, possibly leading to increased intratumoural drug concentrations and better pathway inhibition in a larger fraction of tumour cells that are sensitive to the drug. This hypothesis should be tested in clinical trials.
Last, the presence of RTK heterogeneity strongly suggests the need for mechanism-based combination therapies. Consistent with Peter Nowell's prescient statement that genome instability combined with clonal selection leads to tumours bearing a population of cells that are resistant to any therapy the physician chooses13, Bozic and colleagues developed a mathematical model of the evolutionary dynamics of cancer. They demonstrated that, based on basal mutation rates, a minimum of dual therapy is required for disease control if no single mutation causes cross resistance to both of the drugs that are used1. Furthermore, for patients with a large disease burden, as is the case for many patients with GBM, three concurrent therapies are probably needed1. In line with Nowell's comments13, the earlier a tumour is treated then the less likely it is to be heterogeneous and to harbour resistance-promoting mutations. Importantly, in the mathematical model, effective combination therapy required concurrent, not sequential, administration of the drugs. In addition, the activity of other signalling pathways that modify signalling flux through the key effector pathway, and/or converge on common downstream nodes, are commonly altered in cancer and can profoundly alter the activity of EGFR mutations and therefore need to be considered when combination treatments are designed12.
Consider the current treatment for patients with GBM: following the initial surgical excision of as much of the tumour as possible, patients generally receive radiation therapy and temozolomide and are not given targeted therapies until after the tumours have failed to respond or relapsed, at which point heterogeneity is likely to be extensive. Nonetheless, at this stage of their treatment, patients are still assigned to experimental protocols that typically involve single targeted agents. Reconsideration of the timing of targeted combination treatment for patients with GBM, including at clinical presentation, is warranted.
The challenge for precision medicine
Cancer is possibly the most compelling scenario for precision medicine; it is a genetic disease in which current standards of care can be ineffective. Global efforts to sequence the cancer genome were motivated by the hope that treating patients, by matching the molecular composition of their tumour with a drug or combination of drugs that are optimized for their intended targets, would lead to better outcomes. Intratumoural heterogeneity, in particular the heterogeneity of amplified and mutated RTKs, presents a serious challenge. Single-cell RNA and DNA sequencing from geographically defined regions in GBM models, and from tumours of patients who have been treated with drugs alone and in combination, should provide important insights into how intratumoural RTK heterogeneity alters the response of patients to targeted therapies. Indeed, preclinical models have already yielded testable and scientifically sound hypotheses about how combinations of therapies might be used to overcome resistance that is mediated by heterogeneity. These include: first, treating tumours at the point of their lowest possible molecular diversity, such as right after initial surgical debulking. Second, testing mechanism-based combination therapies early and upfront. Third, ensuring that the drugs achieve the sufficient intratumoural levels that are required to block their intended drug targets and affect the growth and survival of tumour cells, which could be tested by direct analysis of tumour tissue or by non-invasive imaging surrogates. Last, reconsidering dosing schedules to include intermittent dosing, which might accommodate higher levels of drug infusion, possibly with lower toxic effects than are observed with daily drug dosing. The necessary elements with which to test the precision medicine hypothesis in the context of GBM are available in the form of a map of actionable driver mutations, the ability to stratify patients based on molecular phenotype and a supply of drugs that target some of the actionable driver mutations. There is also an interest among patients, oncologists and pharmaceutical companies in incorporating serial biopsies into the design of clinical trials to assess drug delivery and to identify the pharmacodynamic effects of drugs and the potential mechanisms of resistance against them. Incorporation of serial biopsies into clinical trials is costly and requires appropriate consent, but it may be important for developing better treatments for patients.
Single-agent targeted therapies, or even strategic combinations, are likely to fail if they are not dosed, sequenced and timed in a way that is designed to overcome the challenges that are imposed by intratumoural heterogeneity and tumour evolution. An understanding of the underpinning mechanisms will be needed to develop therapeutic strategies that are more likely to yield better outcomes for patients with GBM and other types of cancer.
Acknowledgements
This work is supported by grants from National Institute for Neurological Diseases and Stroke (NS73831 and NS080939-01), the National Cancer Institute (CA151819), The Ben and Catherine Ivy Foundation, the Defeat GBM Program of the National Brain Tumour Society and donations from the Ziering Family Foundation in memory of Sigi Ziering. W.K.C. is a fellow of the National Foundation for Cancer Research. The authors acknowledge the reading and comments by members of the Mischel laboratory.
Footnotes
Competing interests statement
The authors declare no competing interests.
Contributor Information
Frank B. Furnari, Ludwig Institute for Cancer Research and the Department of Pathology, University of California San Diego, La Jolla, California 92093, USA.
Timothy F. Cloughesy, Department of Neurology, David Geffen School of Medicine, University of California Los Angeles, California 90095, USA.
Webster K. Cavenee, Ludwig Institute for Cancer Research and the Department of Medicine, University of California San Diego, La Jolla, California 92093, USA.
Paul S. Mischel, Ludwig Institute for Cancer Research and the Department of Pathology, University of California San Diego, La Jolla, California 92093, USA.
References
- 1.Bozic I, et al. Evolutionary dynamics of cancer in response to targeted combination therapy. eLife. 2013;2:e00747. doi: 10.7554/eLife.00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nathanson DA, et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343:72–76. doi: 10.1126/science.1241328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gschwind A, Fischer OM, Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nature Rev. Cancer. 2004;4:361–370. doi: 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- 6.Lemmon MA, Schlessinger J, Ferguson KM. The EGFR family: not so prototypical receptor tyrosine kinases. Cold Spring Harb. Perspect. Biol. 2014;6:a020768. doi: 10.1101/cshperspect.a020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn GP, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26:756–784. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonavia R, Inda MM, Cavenee WK, Furnari FB. Heterogeneity maintenance in glioblastoma: a social network. Cancer Res. 2011;71:4055–4060. doi: 10.1158/0008-5472.CAN-11-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sturm D, et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nature Rev. Cancer. 2014;14:92–107. doi: 10.1038/nrc3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cloughesy TF, Cavenee WK, Mischel PS. Glioblastoma: from molecular pathology to targeted treatment. Annu. Rev. Pathol. 2014;9:1–25. doi: 10.1146/annurev-pathol-011110-130324. [DOI] [PubMed] [Google Scholar]
- 13.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 14.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welch JS, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding L, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shah SP, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 19.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sidransky D, et al. Clonal expansion of p53 mutant cells is associated with brain tumour progression. Nature. 1992;355:846–847. doi: 10.1038/355846a0. [DOI] [PubMed] [Google Scholar]
- 22.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Easwaran H, Tsai HC, Baylin SB. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol. Cell. 2014;54:716–727. doi: 10.1016/j.molcel.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marusyk A, et al. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature. 2014;514:54–58. doi: 10.1038/nature13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cleary AS, Leonard TL, Gestl SA, Gunther EJ. Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature. 2014;508:113–117. doi: 10.1038/nature13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inda MM, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24:1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 28.Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138:822–829. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 29.Jung P, et al. Isolation and in vitro expansion of human colonic stem cells. Nature Med. 2011;17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 30.Marumoto T, et al. Development of a novel mouse glioma model using lentiviral vectors. Nature Med. 2009;15:110–116. doi: 10.1038/nm.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedmann-Morvinski D, et al. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080–1084. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wee B, Charles N, Holland EC. Animal models to study cancer-initiating cells from glioblastoma. Front. Biosci. 2011;16:2243–2258. doi: 10.2741/3851. [DOI] [PubMed] [Google Scholar]
- 33.Zhu Y, et al. Inactivation of NF1 in CNS causes increased glial progenitor proliferation and optic glioma formation. Development. 2005;132:5577–5588. doi: 10.1242/dev.02162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson EL, et al. PDGFRα-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron. 2006;51:187–199. doi: 10.1016/j.neuron.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 35.Chen J, McKay RM, Parada LF. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149:36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bachoo RM, et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–277. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 37.Zhu H, et al. Oncogenic EGFR signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis. Proc. Natl Acad. Sci. USA. 2009;106:2712–2716. doi: 10.1073/pnas.0813314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ciriello G, et al. The molecular diversity of Luminal A breast tumors. Breast Cancer Res. Treat. 2013;141:409–420. doi: 10.1007/s10549-013-2699-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J. Neuropathol. Exp. Neurol. 2005;64:479–489. doi: 10.1093/jnen/64.6.479. [DOI] [PubMed] [Google Scholar]
- 40.Singh D, et al. Transforming fusions of [I]FGFR[/I] and [I]TACC[/I]genes in human glioblastoma. Science. 2012;337:1231–1235. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker BC, Engels M, Annala M, Zhang W. Emergence of FGFR family gene fusions as therapeutic targets in a wide spectrum of solid tumours. J. Pathol. 2014;232:4–15. doi: 10.1002/path.4297. [DOI] [PubMed] [Google Scholar]
- 42.Cloughesy TF, et al. Antitumor activity of rapamycin in a Phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med. 2008;5:e8. doi: 10.1371/journal.pmed.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishikawa R, et al. Immunohistochemical analysis of the mutant epidermal growth factor, deltaEGFR, in glioblastoma. Brain Tumor Pathol. 2004;21:53–56. doi: 10.1007/BF02484510. [DOI] [PubMed] [Google Scholar]
- 44.Fan QW, et al. EGFR phosphorylates tumor-derived EGFRvIII driving STAT3/5 and progression in glioblastoma. Cancer Cell. 2013;24:438–449. doi: 10.1016/j.ccr.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giacomini CP, et al. Breakpoint analysis of transcriptional and genomic profiles uncovers novel gene fusions spanning multiple human cancer types. PLoS Genet. 2013;9:e1003464. doi: 10.1371/journal.pgen.1003464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Del Vecchio CA, et al. [I]EGFRvIII[/I] gene rearrangement is an early event in glioblastoma tumorigenesis and expression defines a hierarchy modulated by epigenetic mechanisms. Oncogene. 2013;32:2670–2681. doi: 10.1038/onc.2012.280. [DOI] [PubMed] [Google Scholar]
- 47.Francis JM, et al. EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer Discov. 2014;4:956–971. doi: 10.1158/2159-8290.CD-13-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sottoriva A, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl Acad. Sci. USA. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gill BJ, et al. MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma. Proc. Natl Acad. Sci. USA. 2014;111:12550–12555. doi: 10.1073/pnas.1405839111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szerlip NJ, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc. Natl Acad. Sci. USA. 2012;109:3041–3046. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Snuderl M, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 53.Gini B, Mischel PS. Greater than the sum of its parts: single-nucleus sequencing identifies convergent evolution of independent EGFR mutants in GBM. Cancer Discov. 2014;4:876–878. doi: 10.1158/2159-8290.CD-14-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanborn JZ, et al. Double minute chromosomes in glioblastoma multiforme are revealed by precise reconstruction of oncogenic amplicons. Cancer Res. 2013;73:6036–6045. doi: 10.1158/0008-5472.CAN-13-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson BE, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng YK, et al. A mathematical methodology for determining the temporal order of pathway alterations arising during gliomagenesis. PLoS Comput. Biol. 2012;8:e1002337. doi: 10.1371/journal.pcbi.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ozawa T, et al. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell. 2014;26:288–300. doi: 10.1016/j.ccr.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crasta K, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishikawa R, et al. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc. Natl Acad. Sci. USA. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang HS, et al. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J. Biol. Chem. 1997;272:2927–2935. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- 61.Sun J, et al. A microfluidic platform for systems pathology: multiparameter single-cell signaling measurements of clinical brain tumor specimens. Cancer Res. 2010;70:6128–6138. doi: 10.1158/0008-5472.CAN-10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bendall SC, Nolan GP. From single cells to deep phenotypes in cancer. Nature Biotech. 2012;30:639–647. doi: 10.1038/nbt.2283. [DOI] [PubMed] [Google Scholar]
- 63.Shi Q, et al. Single-cell proteomic chip for profiling intracellular signaling pathways in single tumor cells. Proc. Natl Acad. Sci. USA. 2012;109:419–424. doi: 10.1073/pnas.1110865109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Masui K, et al. mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 2013;18:726–739. doi: 10.1016/j.cmet.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gini B, et al. The mTOR kinase inhibitors, CC214-1 and CC214-2, preferentially block the growth of EGFRvIII-activated glioblastomas. Clin. Cancer Res. 2013;19:5722–5732. doi: 10.1158/1078-0432.CCR-13-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Masui K, Cavenee WK, Mischel PS. mTORC2 in the center of cancer metabolic reprogramming. Trends Endocrinol. Metab. 2014;25:364–373. doi: 10.1016/j.tem.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang W, et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nature Cell Biol. 2012;14:1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang W, et al. EGFR-induced and PKCεmonoubiquitylation-dependent NF-κB activation upregulates PKM2 expression and promotes tumorigenesis. Mol. Cell. 2012;48:771–784. doi: 10.1016/j.molcel.2012.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nagane M, Levitzki A, Gazit A, Cavenee WK, Huang HJ. Drug resistance of human glioblastoma cells conferred by a tumor-specific mutant epidermal growth factor receptor through modulation of Bcl-XL and caspase-3-like proteases. Proc. Natl Acad. Sci. USA. 1998;95:5724–5729. doi: 10.1073/pnas.95.10.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tanaka K, et al. Oncogenic EGFR signaling activates an mTORC2–NF-κB pathway that promotes chemotherapy resistance. Cancer Discov. 2011;1:524–538. doi: 10.1158/2159-8290.CD-11-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Blaes J, et al. NDRG1 prognosticates the natural course of disease in WHO grade II glioma. J. Neurooncol. 2014;117:25–32. doi: 10.1007/s11060-013-1357-2. [DOI] [PubMed] [Google Scholar]
- 72.Groenendijk FH, et al. Sorafenib synergizes with metformin in NSCLC through AMPK pathway activation. Int. J. Cancer. 2014;136:1434–1444. doi: 10.1002/ijc.29113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ohashi K, Maruvka YE, Michor F, Pao W. Epidermal growth factor receptor tyrosine kinase inhibitor-resistant disease. J. Clin. Oncol. 2013;31:1070–1080. doi: 10.1200/JCO.2012.43.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fenton TR, et al. Resistance to EGF receptor inhibitors in glioblastoma mediated by phosphorylation of the PTEN tumor suppressor at tyrosine 240. Proc. Natl Acad. Sci. USA. 2012;109:14164–14169. doi: 10.1073/pnas.1211962109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mellinghoff IK, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N. Engl. J. Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 76.Vivanco I, et al. The phosphatase and tensin homolog regulates epidermal growth factor receptor (EGFR) inhibitor response by targeting EGFR for degradation. Proc. Natl Acad. Sci. USA. 2010;107:6459–6464. doi: 10.1073/pnas.0911188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stommel JM, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 78.Akhavan D, et al. De-repression of PDGFRβ transcription promotes acquired resistance to EGFR tyrosine kinase inhibitors in glioblastoma patients. Cancer Discov. 2013;3:534–547. doi: 10.1158/2159-8290.CD-12-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Corcoran RB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAFmutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Girotti MR, Marais R. Déjà vu: EGF receptors drive resistance to BRAF inhibitors. Cancer Discov. 2013;3:487–490. doi: 10.1158/2159-8290.CD-13-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Prahallad A, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 83.Vivanco I, et al. Differential sensitivity of glioma-versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov. 2012;2:458–471. doi: 10.1158/2159-8290.CD-11-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yano S, et al. Retreatment of lung adenocarcinoma patients with gefitinib who had experienced favorable results from their initial treatment with this selective epidermal growth factor receptor inhibitor: a report of three cases. Oncol. Res. 2005;15:107–111. [PubMed] [Google Scholar]
- 85.Humphrey PA, et al. Deletion-mutant epidermal growth factor receptor in human gliomas: effects of type II mutation on receptor function. Biochem. Biophys. Res. Commun. 1991;178:1413–1420. doi: 10.1016/0006-291x(91)91051-d. [DOI] [PubMed] [Google Scholar]
- 86.Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 2000;60:1383–1387. [PubMed] [Google Scholar]
- 87.Peschard P, et al. Structual basis for ubiquitin-mediated dimerization and activaiton of the ubiquitin protein ligase Cbl-b. Mol. Cell. 2007;27:474–485. doi: 10.1016/j.molcel.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 88.Ogiso H, et al. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- 89.Garrett TP, et al. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor α. Cell. 2002;11:763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- 90.Lee JC, et al. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med. 2006:e485. doi: 10.1371/journal.pmed.0030485. [DOI] [PMC free article] [PubMed] [Google Scholar]