Abstract
Objective
Autophagy dysfunction has been reported in osteoarthritis (OA) cartilage. The objective of this study was to investigate the role of microRNA-155 (miR-155), which is overexpressed in OA, in the regulation of autophagy in human chondrocytes.
Design
Rapamycin (50nM) and 2-deoxyglucose (2-DG) (5mM) were used to stimulate autophagy in primary human articular chondrocytes and in the T/C28a2 human chondrocyte cell line. Cells were transfected with LNA GapmeR or mimic specific for miR-155 and autophagy flux was assessed by LC3 western blotting and by Cyto-ID® dye quantification in autophagic vacuoles. Expression of predicted miR-155 targets in the autophagy pathway were analyzed by real-time PCR and western blotting.
Results
Autophagy flux induced by rapamycin and 2-DG was significantly increased by miR-155 LNA, and significantly decreased after miR-155 mimic transfection in T/C28a2 cells and in human primary chondrocytes. These effects of miR-155 on autophagy were related to suppression of gene and protein expression of key autophagy regulators including Ulk1, FoxO3, Atg14, Atg5, Atg3, Gabarapl1, and Map1lc3.
Conclusion
MiR-155 is an inhibitor of autophagy in chondrocytes and contributes to the pathogenesis of OA.
Keywords: Autophagy, microRNAs, cartilage, chondrocytes
Introduction
Osteoarthritis (OA) is the most common joint disease, characterized by articular cartilage degradation and changes in all other joint tissues 1, 2. Articular chondrocytes, the only cell type in adult cartilage, synthetize extracellular matrix (ECM) components and are responsible for cartilage homeostasis 3. Aging and OA are associated with profound changes in articular cartilage, including reduced cellularity and chondrocyte death as well as abnormal chondrocyte activation and differentiation. Such changes could be the result of either catabolic extracellular stimuli or cell-autonomous changes in homeostatic mechanisms.
Macroautophagy, hereafter termed autophagy, is a bulk self-degradation process via lysosomes and, as an adaptive response to stress conditions, its dysfunction has been associated with many age-related diseases 4. The autophagic pathway is controlled by multiple signalling pathways that converge on mammalian target of rapamycin (mTOR) and AMP-activated protein kinase (AMPK). The core autophagy initiation regulator is the Unc-51-like kinase (ULK) complex consisting of ULK, ATG13, FIP200 and ATG101. ULK1 phosphorylates Beclin1, an essential component of BECLIN1-VPS34-ATG14 complex for vesicle nucleation. Two ubiquitin-like conjugation systems are responsible for autophagosome elongation and maturation steps. Phosphatydilethanolamine (PE) is conjugated to the soluble form of microtubule-associated protein 1 light chain 3 (LC3-I) by ATG7 and ATG3 enzymes. The insoluble form, termed LC3-II, is stably linked to the autophagosomal membrane whose elongation is mediated by the ATG12/ATG5-ATG16 complex. Then, autophagosomes fuse with lysosomes, and the cargo is degraded and released in cytosol in order to be recycled 5.
Changes in autophagy that have been reported in aging and OA-affected cartilage include reduced numbers and size of autophagosomes and this is, in part, related to a reduced expression of the autophagy proteins ULK1, BECLIN1 and LC3 6 and mTOR overexpression 7. Several studies reported beneficial effects of autophagy in preventing chondrocyte death, OA-like changes in gene expression and cartilage degeneration 8–12. However, mechanisms underlying the autophagy failure in OA, specifically those related to the reduced expression of autophagy proteins, are still unclear.
MicroRNAs (miRs) are endogenous small-noncoding RNAs able to regulate gene expression by mediating mRNA degradation and/or blocking mRNA translation. They can modulate many cellular processes, including apoptosis, cell differentiation and metabolism and their deregulation has been associated with cancer and other pathologies, including OA 13–15. Several miRs have been identified as suppressors of factors involved in different stages of autophagy 16–19 while others have been shown to induce autophagy by inhibiting mTOR activity 20, 21.
We performed a genome-wide miRNA-sequencing study on human normal cartilage versus OA cartilage. MiR-155 was among the most significantly upregulated miRNA in OA. Here we investigated the role of miR-155 in regulation of autophagy in chondrocytes.
Methods
Cell culture of human chondrocytes
Normal human cartilage was harvested during autopsy from the femoral condyles and the tibial plateaus from 4 adult donors (age < 50) without history of joint diseases and with macroscopically normal cartilage surfaces. The collection of human tissue was under the approval by the Scripps Human Subjects Committee. Chondrocytes were isolated as described previously 22 and cultured in DMEM supplemented with 10% calf serum with antibiotics. First or second-passage cells were used in this study.
Immortalized human primary chondrocytes, T/C-28a2 23, were obtained from Dr. Mary Goldring and maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Life Technologies, NY, USA) supplemented with 10% Fetal Bovine Serum (FBS) (Life Technologies), with antibiotics at 37°C in the presence of 5% CO2.
Cell transfection and experimental design
Primary chondrocytes and T/C28a2 cells were seeded in 24-well plates at a density of 5 × 104 cells/well and in 96-well plates at a density of 1 × 104 cells/well without antibiotics. The next day, LNA™ GapmeR control (LNA nc) and LNA miR-155 (40nM) (Exiqon) and Mission® microRNA mimic control (mimic nc) and mimic miR-155 (10nM) (Sigma Aldrich) were transfected into cells using Lipofectamine RNAiMAX (Invitrogen) for 24 hours.
To assess autophagic flux, time course experiments were conducted in cells stimulated with rapamycin (RAPA, 50nM), 2-deoxy-glucose (2-DG, 5mM) or vehicle in the presence or absence of chloroquine (CQ, 25μM). Maximal induction of autophagy was achieved at different time points depending on the cell type and treatment employed; subsequently, these specific time points were selected for miR-155 functional experiments as indicated for each experiment. Briefly, primary human chondrocytes were treated with RAPA for 12 hours or maintained for 24 hours under basal conditions. T/C28a2 cells were treated with RAPA or 2-DG for 4 and 2 hours, respectively. Then, RNA or protein samples were collected for qPCR and western blot analysis, or cells were subjected to Cyto-ID dye detection as described below. Doses of the various agents were selected to be non-toxic based on prior studies 24–26. In addition, at completion of each experiment cell morphology was monitored and there was no apparent cell shrinkage, rounding or detachment from the culture plates.
Bioinformatics prediction of miR-155 targets
TargetScan and miRWalk databases were used to predict potential targets of miR-155 directly and indirectly involved in the autophagic pathway.
RNA isolation, cDNA synthesis and real-time PCR (qPCR)
Samples were collected in Qiazol (Qiagen) at room temperature and after the addition of chloroform, centrifuged at 12,000g at 4°C for 15 minutes. RNA was extracted by using miRneasy minikit (Qiagen) to separate small RNAs from total RNA. Two different fractions were used to assess miR-155 levels and gene expression of potential miR-155 targets, respectively. MicroRNA reverse transcription was conducted with TaqMan MicroRNA RT kit (Life Technologies) and qPCR was performed with TaqMan Universal Mastermix (Life Technologies) following kit instructions. Specific primers for miR-155 and RNU48 (internal control) were purchased from Life Technologies. RNA reverse transcription and qPCR were performed with TaqMan Reverse Transcription kit (Life Technologies) and LightCycler 480 Probes Master (Roche), respectively. Pre-designed primers for GAPDH (internal control), FOXO3, RICTOR, ATG14, ATG3, GABARAPL1, MAP1LC33, ULK1 and ATG5 were obtained from Life Technologies.
Protein isolation and Western Blotting
Cells were lysed in RIPA buffer, sonicated at 4°C a nd centrifuged at 12,000g for 15 minutes. Proteins were separated on 4%–20% SDS-polyacrylamide gels and transferred to nitrocellulose membranes (Amersham), blocked in Odyssey blocking buffer 1X (LI-COR Bioscience) for 60 minutes at room temperature and incubated overnight with primary antibodies diluted in Odyssey Blocking Buffer 1:1 with phosphate-buffered saline-PBS and 0.1% Tween 20. Antibodies for RICTOR (1:1000), ULK1 (1:1000), FOXO3 (1:1000), ATG14 (1:1000), P-AKT (Ser473) (1:2000), AKT (1:1000), ATG5 (1:1000) ATG3 (1:1000), P-S6 (S235/S236) (1:2000), S6 (1:1000), LC3A/B (1:1000) were purchased from Cell Signaling. Antibody for Gabarapl1 (1:500) was purchased from Fisher Scientific and that for β-actin (1:5000) from Abcam. After three washes with Tris-buffered saline–0.1% Tween (TBST), membranes were incubated with IR Dye labelled secondary antibody (goat anti-Mouse 1:10000 and goat anti-Rabbit 1:5000, LI-COR Biosciences) diluted in Odyssey Blocking Buffer 1:1 with PBS, 0.1% Tween 20 and 0.01% SDS. Subsequently, the membranes were washed 3 times with TBST, kept in water and visualized by Odyssey Scanner (LI-COR Biosciences). Densitometry was performed using LI-COR Odyssey Software Version 4.0 (LI-COR Biosciences).
Cyto-ID dye detection
Cyto-ID assay (Enzo) measures autophagic vacuoles and monitors autophagic flux in live cells using a dye that selectively labels autophagic vacuoles. The dye has been optimized through the identification of titratable functional moieties that allows minimal staining of lysosomes while exhibiting bright fluorescence upon incorporation into pre-autophagosomes, autophagosomes, and autolysosomes (autophagolysosomes) 27. Fluorescence was measured at 360 Ex/460 Em (Hoechst 33342 nuclear stain) and 480 Ex/530 Em (Cyto-ID green autophagy detection reagent) with a plate reader (TECAN Safire II from Tecan Systems). Cyto-ID intensity was first normalized to Hoechst intensity and then to the control sample in order to evaluate the fold changes.
Statistical analysis
The data are reported as mean ± standard deviation (SD) of the indicated number of independent experiments. Some experiments were conducted with primary chondrocytes samples from four different donors, each performed either in duplicate or in triplicate. All datasets were assessed for normal population distribution using the Kolmogorov-Smirnov test, and homogeneity of variance using Bartlett’s test. Analysis of variance (ANOVA) was used to compare group means from datasets that met all the assumptions of this test, followed by planned pairwise comparisons using the Bonferroni procedure for these selected pairs of groups. The overall ANOVA tests were performed at the alpha = 0.05 level of significance, and the alpha levels of the subsequent individual comparisons were adjusted so as to preserve the familywise error rate at the 0.05 level. Not-normally distributed populations were compared using the nonparametric Kruskal-Wallis test with Dunn’s method. Statistical analysis was conducted using Prism (GraphPad Software version 5.0). P < 0.05 was considered statistically significant.
Results
MiR-155 modulates autophagy in T/C28a2 cells
In a Next Generation Sequencing study, miR-155 was found to be one of the most highly upregulated miRs in human OA knee cartilage compared to normal cartilage 28. Searching miR target databases revealed that miR-155 seed-complementary sequences are present in several autophagy-related genes and identified GABARAPL1, ATG3, ATG5 and FOXO3 as putative targets. In light of this, and previous findings of dysfunctional autophagy in OA 29, we analyzed the potential role of miR-155 in chondrocyte autophagy.
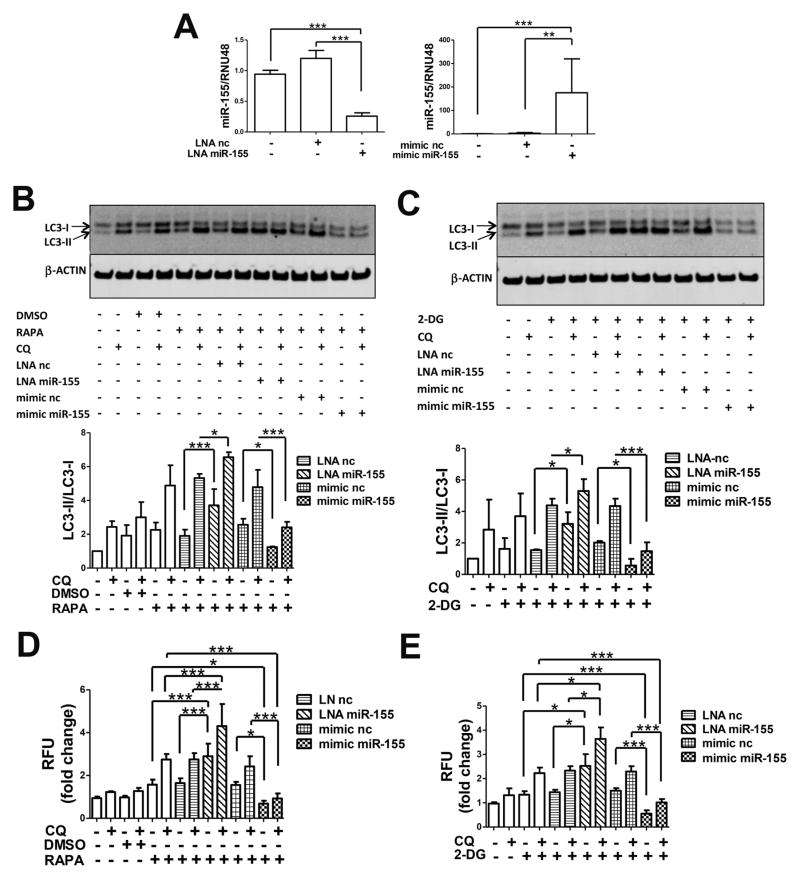
To investigate whether miR-155 affects the autophagic response in the T/C28a2 human immortalized cell line, we transfected cells with LNA nc and LNA miR-155, mimic nc and mimic miR-155 in order to downregulate and raise endogenous miR-155 levels, respectively, as validated by qPCR (Figure 1A). Autophagy is a highly dynamic process that involves the formation, maturation and degradation of autophagosomes 30. Autophagic flux can be monitored by measuring the conversion of LC3 from its soluble form (LC3-I) to the lipidated form that is bound to the autophagosome membrane (LC3-II) 31. We analyzed autophagy flux after treatment with the autophagy inducers RAPA or 2-DG. Both compounds are widely employed for autophagy stimulation in vitro and in vivo studies due to their ability to block the mTOR pathway or reduce intracellular ATP thus activating AMPK, respectively 32. Moreover, T/C28a2 cells were simultaneously treated with or without CQ, to block autophagy flux and allow accumulation of the autophagic protein LC3-II, given its fast protein turnover, thus affording a better observation by western blot analysis 32. As shown in Figure 1B and 1C, the autophagy flux, as evaluated by the LC3-II/LC3-I ratio, was enhanced by CQ alone, indicating basal autophagy activation under the culture conditions used. This was increased significantly in samples transfected with LNA miR-155 compared to those with LNA nc, and was decreased significantly in cells transfected with mimic miR-155, compared to mimic nc.
Figure 1.
MiR-155 modulates autophagy in T/C28a2 cells. (A) T/C28a2 cells were transfected for 24h with LNA miR-155 and LNA negative control (LNA nc) (40nM), mimic miR-155 and mimic negative control (mimic nc) (10nM). MiR-155 levels were determined by qPCR (n=4 independent experiments). (B) 24h after transfection cells were treated with DMSO or 50nM rapamycin (RAPA) and/or 25μM chloroquine (CQ) for 4h or (C) 5mM 2-deoxy-glucose (2-DG) and/or 25μM CQ for 2h and then harvested for LC3 and β-actin detection by western blotting. Representative images and relative quantification for LC3-II/LC3-I ratios are shown (n=5 independent experiments), (D) 24h after transfection cells were incubated for 18h with DMSO or 50nM RAPA and/or 25μM CQ or (E) 5mM 2-DG and/or 25μM CQ and stained with Cyto-ID (1:1000) and Hoechst 33342 (1:2000) (n=3 independent experiments). Values are expressed as mean ± SD, *P<0.05, **P<0.01,***P<0.001.
Consistent with these results on LC3 conversion, the downregulation of miR-155 by LNA miR-155 increased and mimic miR-155 transfection decreased the number of autophagic vacuoles as assessed in live cells by Cyto-ID dye incorporation, specifically targeting autophagosomes (Figures 1D and 1E).
MiR-155 modulates autophagy in human primary chondrocytes
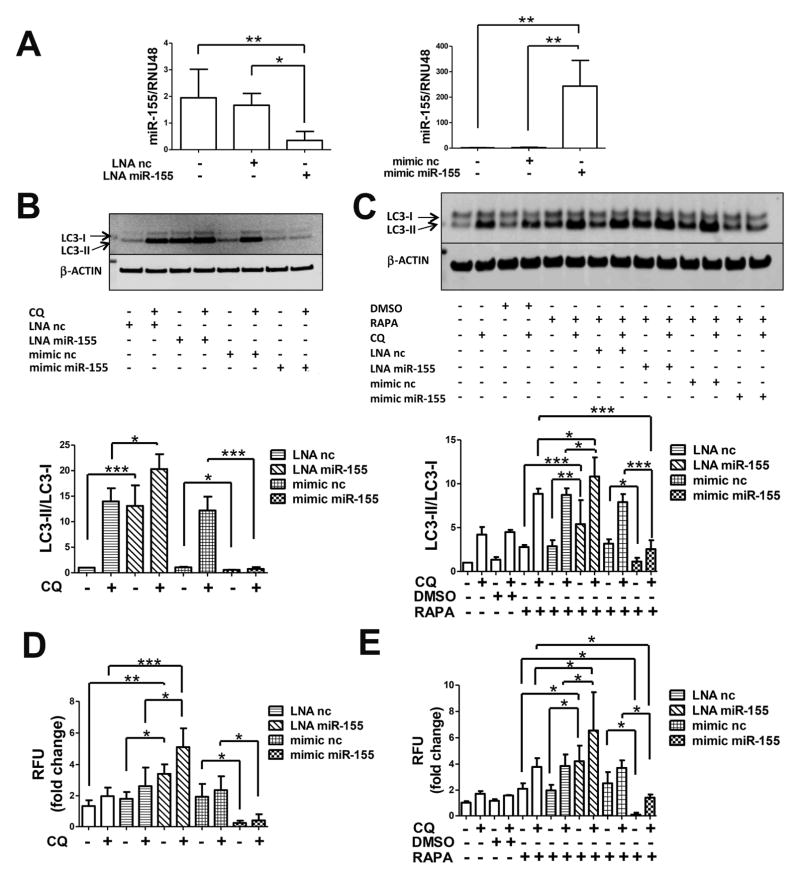
In order to confirm these observations, we evaluated the autophagic activity in primary chondrocytes transfected with LNA nc and LNA miR-155, mimic nc and mimic miR-155 to modulate cellular miR-155 levels (Figure 2A). A significant increase in basal autophagic flux was seen when cells were transfected with LNA miR-155 compared to the LNA nc while mimic miR-155 transfection resulted in a significant attenuation of LC3-I conversion (Figure 2B).
Figure 2.
MiR-155 modulates autophagy in human primary chondrocytes. (A) Cells were transfected for 24h with LNA miR-155 and LNA negative control (LNA nc), mimic miR-155 and mimic negative control (mimic nc). MiR-155 levels were determined by qPCR (n=4 independent experiments). (B) 24h after transfection cells were treated with or without 25μM chloroquine (CQ) for 12h or (C) DMSO or 50nM rapamycin (RAPA) and/or 25μM CQ for 12h and then harvested for LC3 and β-actin detection by western blotting. Representative images and relative quantification for LC3-II/LC3-I ratios are shown (n=4 independent experiments with separate chondrocyte preparations), (D) 24h after transfection cells were incubated for 18h with or without 25μM CQ or (E) DMSO or 50nM rapamycin (RAPA) and/or 25μM CQ and stained with Cyto-ID (1:1000) and Hoechst 33342 (1:2000) and intracellular fluorescence was measured (n=4 independent experiments with separate chondrocyte preparations). Values are expressed as mean ± SD, *P<0.05, **P<0.01,***P<0.001.
We also tested the effects of miR-155 modulation in RAPA-induced autophagy in chondrocytes after LNA and mimic transfection. Under this condition, autophagy flux was significantly increased by miR-155 downregulation and significantly reduced by mimic miR-155 transfection (Figure 2C). These results were confirmed by quantification of Cyto-ID dye intensity (Figures 2D and 2E).
MiR-155 regulates autophagy by suppressing MAP1LC3, GABARAPL1, ATG3, ATG5, ATG14, ULK1 and FOXO3
To characterize the mechanism underlying autophagy suppression by miR-155, we tested the bioinformatics-based prediction that miR-155 targets autophagy genes. We performed qPCR analysis on RNA from T/C28a2 cells and from human primary chondrocytes transfected with LNA nc, LNA miR-155, mimic nc and mimic miR-155.
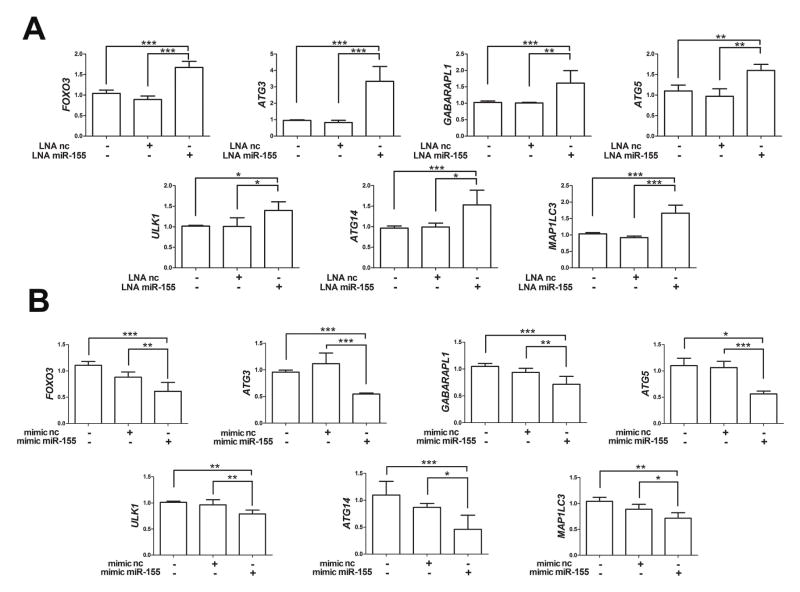
As shown in Figure 3A, LNA miR-155 but not LNA nc, led to a significant increase in the mRNA levels of GABARAPL1, ATG3, ATG5 and FOXO3. Conversely, mimic miR-155 but not mimic nc significantly reduced mRNA levels of GABARAPL1, ATG3, ATG5 and FOXO3 (Figures 3A and 3B). Furthermore, we analyzed autophagy-related genes that are not predicted targets of miR-155 and discovered that mRNA levels of ULK1, ATG14 and MAP1LC3 were modulated by miR-155 LNA and mimic (Figures 3A and 3B). Similar results were obtained in human primary chondrocytes (Supplemental Figure 1).
Figure 3.
MiR-155 regulates autophagy by suppressing gene expression of MAP1LC3, GABARAPL1, ATG3, ATG5, ULK1, ATG14, and FOXO3. T/C28a2 cells were transfected for 24h with LNA miR-155 and LNA negative control (LNA nc), mimic miR-155 and mimic negative control (mimic nc). qRT-PCR analysis of mRNA expression levels of direct and indirect targets of miR-155 in T/C28a2 cells following transfection with LNA (A) or (B) mimic (n=4 independent experiments). Values are expressed as mean ± SD, *P<0.05, **P<0.01, ***P<0.001.
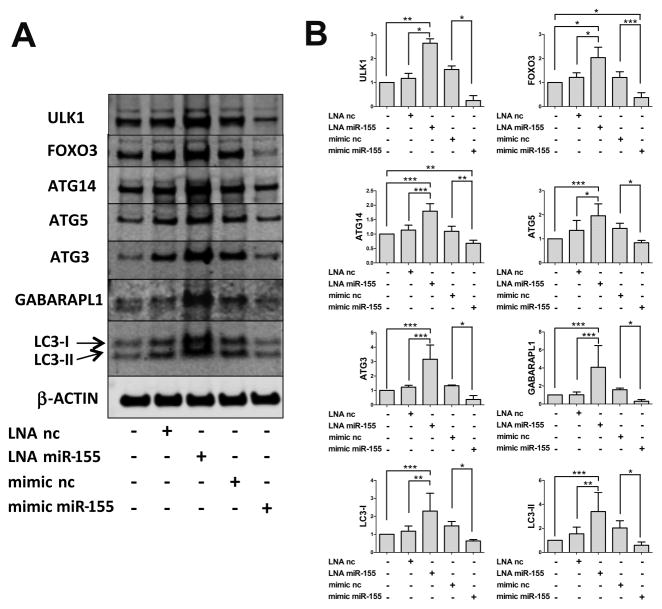
To confirm that the changes in mRNA levels of miR-155 targets corresponded to changes in protein expression, we performed western blotting on T/C28a2 cells transfected with LNA miR-155 or mimic miR-155. As shown in Figure 4, knockdown of endogenous miR-155 increased levels of GABARAPL1, ATG3, ATG5, FOXO3, ULK1, ATG14 and LC3 proteins and mimic miR-155 transfection strongly decreased these proteins.
Figure 4.
MiR-155 regulates autophagy by reducing protein levels of MAP1LC3, GABARAPL1, ATG3, ATG5, ULK1, ATG14, and FOXO3. T/C28a2 cells were transfected for 24h with LNA miR-155 and LNA negative control (LNA nc), mimic miR-155 and mimic negative control (mimic nc). Western blotting analysis of MAP1LC3, GABARAPL1, ATG3, ATG5, ULK1, ATG14, FOXO3 and β-ACTIN proteins. Representative images (A) and relative quantifications (B) are shown (n=4 independent experiments). Values are expressed as mean ± SD, *P<0.05, **P<0.01, ***P<0.001.
MiR-155 regulates mTOR activity
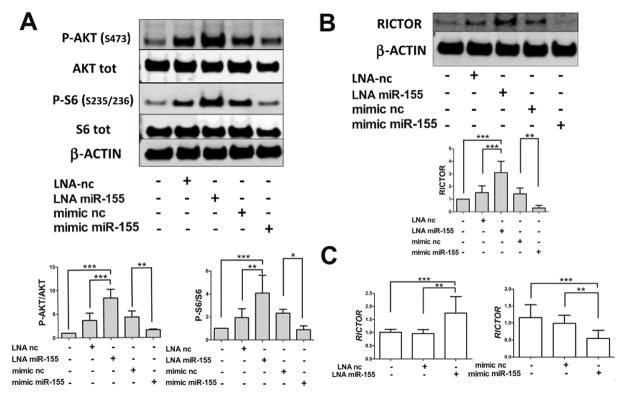
As mTOR is a critical negative regulator of autophagy 33–35, we examined whether it was involved in miR-155 effects. mTOR pathway activity was detected by analyzing the phosphorylation levels of ribosomal protein s6 (P-s6, S235/S236) and AKT (P-AKT, S473) in response to LNA miR-155 and mimic miR-155 transfection in T/C28a2 cells. Contrary to our expectations, P-S6 and P-AKT, in ratio to their total levels, were significantly increased following knockdown of miR-155 and significantly attenuated after mimic miR-155 transfection (Figure 5A). We next evaluated the expression of RICTOR, an essential mTOR complex-2 (mTORC2) component that can activate mTOR complex-1 (mTORC1) through phosphorylation of AKT (S473) 36, 37, which was identified as a miR-155 target in the bioinformatics analysis. As shown in Figures 5B and 5C, both RICTOR protein and mRNA levels were increased and decreased by LNA miR-155 and mimic miR-155 transfection, respectively.
Figure 5.
Effect of miR-155 on mTOR activity. T/C28a2 cells were transfected for 24h with LNA miR-155 and LNA negative control (LNA nc), mimic miR-155 and mimic negative control (mimic nc). (A) Western blotting analysis of P-AKT (Ser473), AKT total, P-S6 (Ser235/Ser236), S6 total and β-ACTIN levels by using specific antibodies. (B) Western blotting analysis of RICTOR and β-ACTIN. Representative images and relative quantifications are shown (n=4 independent experiments). (C) qRT-PCR analysis of mRNA expression levels of RICTOR are shown (n=4 independent experiments). Values are expressed as mean ± SD, *P<0.05, **P<0.01, ***P<0.001.
Discussion
Defective autophagy has recently emerged as a feature of articular cartilage in OA-affected joints in both human and in animal models. Studies in mouse models showed that in normal cartilage there is a constitutive or basal level of autophagy that is of higher magnitude as compared to liver in the same animal 38. With aging and OA development, the number of autophagosomes and the size of the autophagosomes decrease in cartilage. Conceptually, defective autophagy could be due to abnormal expression of autophagy proteins or abnormal signalling and regulation of autophagy activation. Several studies provided evidence for aging and OA-associated reduction in autophagy proteins, including ATG5, ULK1, BECLIN1 and LC3 6, 38. There is also evidence for abnormal autophagy signalling, most importantly, hyperactivation of mTOR, possibly due to reduced activation of AMPK 33, 39.
The present study was based on our discovery that miR-155 was one of the most upregulated miR in human OA cartilage 28 and aimed at determining the role of miR-155 in chondrocyte autophagy. MiR-155 appeared as a potential regulator of autophagy based on the presence of miR-155 seed-complementary sequences in the autophagy-related genes (ATG3, GABARAPL1, ATG5, ATG2B, LAMP2, FOXO3).
Our data show that miR-155 suppresses autophagy flux in T/C28a2 cells and primary human chondrocytes as measured by LC3 conversion and by utilizing a Cyto-ID cationic amphiphilic tracer dye that specifically recognizes autophagosomes 27. Accordingly, the number of autophagosomes was significantly increased when miR-155 was blocked by LNA and it was reduced when cells were treated with miR-155 mimic. These autophagy modulating effects of miR-155 LNA and mimic were observed under basal culture conditions and under cell treatment with rapamycin in human chondrocytes and after treatment with rapamycin and 2-DG in T/C28a2 cells.
We explored mechanisms by which miR-155 suppressed autophagy and discovered that multiple factors involved in the autophagic cascade are miR-155 targets. By transfecting LNA and mimic specific for miR-155 in T/C28a2 and in human primary chondrocytes, we observed a negative correlation between miR-155 and its target mRNAs, including ATG3, GABARAPL1, ATG5, FOXO3 and other autophagy-related factors not predicted by miR target databases, including ULK1, MAP1LC3 and ATG14 (Figure 3 and Supplemental Figure 1). These data suggest that miR-155 is able to modulate the autophagy activity by regulating post-transcriptionally several components involved in different stages of autophagy, from the induction to elongation steps. Furthermore, consistent with changes in mRNA levels, miR-155 modulated protein levels of these factors. It is conceivable that miR-155 influences the expression of MAP1LC3, ATG14 and ULK1 by suppression of the transcriptional factor FOXO3 40, 41.
Moreover, by investigating potential involvement of mTOR as a critical negative regulator of autophagy 33–35 in miR-155-mediated autophagy suppression, we discovered that miR-155 inhibits mTOR activity rather than activating it, as demonstrated by detection of phosphorylation levels of ribosomal protein s6, as a downstream substrate of mTORC1, and of AKT, as an upstream regulator of mTORC1. This unexpected result can be explained by miR-155 effects on gene and protein expression of RICTOR, a critical component of mTORC2 36, which induces mTORC1 activation via AKT phosphorylation 37. We found that LNA and mimic miR-155 transfection increased and reduced protein and mRNA levels of RICTOR, respectively. This data indicates that the regulation of downstream autophagy-related factors by miR-155 is sufficient to affect the level of autophagy, independently of mTOR activity.
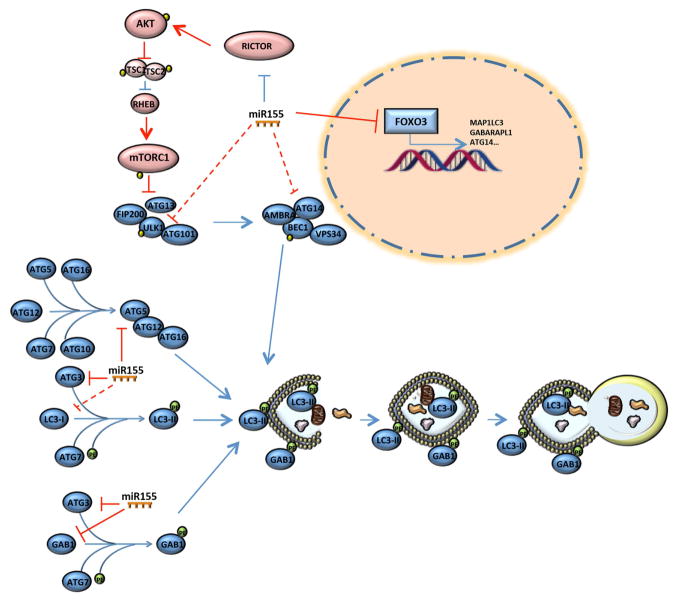
Taken together, these data identify miR-155 as a potent regulator of autophagy under physiological conditions. MiR-155 is able to modulate this process via multiple mechanisms as proposed in the model shown in Figure 6. In pathologic conditions when miR-155 levels are upregulated, as in OA cartilage, the autophagy flux is compromised. We propose that a critical factor might be an exact threshold level of miR-155 that controls the autophagic response to cellular metabolic requests.
Figure 6.
Proposed model of miR-155-mediated suppression of autophagy. MiR-155 targets the expression of several autophagy-related key factors: ULK1, involved in the initiation phase; ATG14, belonging to the BECLIN1 complex responsible of the phagophore nucleation; ATG5, ATG3, LC3 and GABARAPL1, crucial components in the elongation and maturation steps; FOXO3 suggested as critical transcription factor of autophagy-related genes, including MAP1LC3, ULK1, ATG14 and GABARAPL1. Even though miR-155-mediated suppression of RICTOR expression led to a reduction of mTOR activity this does not reverse the inhibition of autophagy. Red colours represent autophagy-reducing effects and, on the other side, blue colours are used for autophagy-promoting effects and dashed lines indicate no predicted relationships.
In accordance with our findings, Holla et al. described an inhibitory role of miR155 on interferon-γ-induced autophagy in macrophages 42. However, another study found that miR-155 enhanced hypoxia-induced autophagy in human cancer cell lines by inhibiting the mTOR pathway 43. Although we also found a reduction of mTOR activity in response to mimic miR-155 transfection, this was not sufficient to reverse the strong inhibitory effect of miR155 on autophagy flux. Thus it is likely that the final effect of miR-155 on autophagy is stimulus and cell-specific and may be dependent on other factors implicated in the post-transcriptional and post-translational regulation of autophagy-related genes.
MiR-155 has been implicated in other aspects linked to OA pathophysiology, in particular in regulating inflammatory responses 44–46. For example, CD14+ monocytes and macrophages overexpressing miR-155 exhibited increased production of proinflammatory cytokines while these were reduced in miR-155-deficient mice 47. In line with these data, Li et al. showed previously that miR-155 upregulated TNF-α and IL-1β in peripheral blood macrophages 48.
The involvement of miRs across human diseases has been widely reported and motivated the development of miRs-based therapies. miRs are promising targets as a single miR is able to regulate multiple genes in dysregulated pathways in disease. Thus, the identification of a single miR, involved simultaneously in several disease-related pathways, discloses a potent therapeutic target.
Our study reveals that miR-155 can profoundly impact the autophagic cascade. We showed an inverse correlation between the miR-155 levels and autophagic flux in human chondrocytes. Furthermore, we demonstrated that increased levels of miR-155 inhibit GABARAPL1, ATG3, ATG5, ATG14, FOXO3, MAP1LC3, and ULK1 expression, and drastically suppress autophagy independently of its regulation on RICTOR expression and on mTOR signalling. In light of this study we can hypothesize that miR-155 is responsible, at least in part, to autophagy failure associated with the pathophysiology of OA.
Although further investigations on the role of miR-155 in dysfunction of autophagy and in other processes involved in the development of OA are required, the present study proposes an attractive opportunity to develop a future strategy based on miR-155 blockage for OA treatment.
Conclusions
These findings demonstrate that miR-155 is a potent suppressor of autophagy in human chondrocytes by reducing the expression of key autophagic proteins. As miR-155 is increased in OA, this represents a mechanism that might contribute to the autophagy defects in OA cartilage.
Supplementary Material
MiR-155 suppresses autophagy-related genes in primary human chondrocytes. Cells were transfected for 24h with LNA miR-155 and LNA negative control (LNA nc), mimic miR-155 and mimic negative control (mimic nc). (A) qRT-PCR analysis of mRNA expression levels of direct and indirect targets of miR-155 in human primary chondrocytes following transfection with LNA or (B) mimic (n=4 independent experiments separate with chondrocyte preparations). Values are expressed as mean ± SD, *P<0.05, **P<0.01, ***P<0.001.
Acknowledgments
The authors acknowledge Merissa Olmer and Stuart Duffy for technical assistance.
Grant Funding
This study was supported by National Institutes of Health (AG007996) and the Sam and Rose Stein Endowment Fund. Drs. D’Adamo and Flamigni’s work was supported by grants from University of Bologna (Marco Polo, RFO).
Footnotes
Authors’ Contributions
ML had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Study design: ML, SD, FF. Acquisition of data: SD, YM, OA. Analysis and interpretation of data: SD, OA, ML. Manuscript preparation and approval: SD, OA, YM, FF, ML.
Conflict of interest
None.
Ethics approval
This study was conducted with the approval of the Human Subjects Committee and the Institutional Animal Care and Use Committee at The Scripps Research Institute.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Control CfD, Prevention. Prevalence of disabilities and associated health conditions among adults--United States, 1999. MMWR Morb Mortal Wkly Rep. 2001;50:120–5. [PubMed] [Google Scholar]
- 2.Loeser RF. Aging and osteoarthritis: the role of chondrocyte senescence and aging changes in the cartilage matrix. Osteoarthritis Cartilage. 2009;17:971–9. doi: 10.1016/j.joca.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldring MB, Goldring SR. Osteoarthritis. J Cell Physiol. 2007;213:626–34. doi: 10.1002/jcp.21258. [DOI] [PubMed] [Google Scholar]
- 4.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 6.Caramés B, Taniguchi N, Otsuki S, Blanco FJ, Lotz M. Autophagy is a protective mechanism in normal cartilage, and its aging-related loss is linked with cell death and osteoarthritis. Arthritis Rheum. 2010;62:791–801. doi: 10.1002/art.27305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Vasheghani F, Li YH, Blati M, Simeone K, Fahmi H, et al. Cartilage-specific deletion of mTOR upregulates autophagy and protects mice from osteoarthritis. Ann Rheum Dis. 2014 doi: 10.1136/annrheumdis-2013-204599. [DOI] [PubMed] [Google Scholar]
- 8.Caramés B, Kiosses WB, Akasaki Y, Brinson DC, Eap W, Koziol J, et al. Glucosamine activates autophagy in vitro and in vivo. Arthritis Rheum. 2013;65:1843–52. doi: 10.1002/art.37977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuzaki T, Matsushita T, Tabata Y, Saito T, Matsumoto T, Nagai K, et al. Intra-articular administration of gelatin hydrogels incorporating rapamycin-micelles reduces the development of experimental osteoarthritis in a murine model. Biomaterials. 2014;35:9904–11. doi: 10.1016/j.biomaterials.2014.08.041. [DOI] [PubMed] [Google Scholar]
- 10.Borzì RM, Guidotti S, Minguzzi M, Facchini A, Platano D, Trisolino G, et al. Polyamine delivery as a tool to modulate stem cell differentiation in skeletal tissue engineering. Amino Acids. 2014;46:717–28. doi: 10.1007/s00726-013-1607-9. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki H, Takayama K, Matsushita T, Ishida K, Kubo S, Matsumoto T, et al. Autophagy modulates osteoarthritis-related gene expression in human chondrocytes. Arthritis Rheum. 2012;64:1920–8. doi: 10.1002/art.34323. [DOI] [PubMed] [Google Scholar]
- 12.Takayama K, Kawakami Y, Kobayashi M, Greco N, Cummins JH, Matsushita T, et al. Local intra-articular injection of rapamycin delays articular cartilage degeneration in a murine model of osteoarthritis. Arthritis Res Ther. 2014;16:482. doi: 10.1186/s13075-014-0482-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barter MJ, Bui C, Young DA. Epigenetic mechanisms in cartilage and osteoarthritis: DNA methylation, histone modifications and microRNAs. Osteoarthritis Cartilage. 2012;20:339–49. doi: 10.1016/j.joca.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 16.Zhu H, Wu H, Liu X, Li B, Chen Y, Ren X, et al. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy. 2009;5:816–23. doi: 10.4161/auto.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Bell LN, Okamura J, Kim MS, Mohney RP, Guerrero-Preston R, et al. Phospho-ΔNp63α/SREBF1 protein interactions: bridging cell metabolism and cisplatin chemoresistance. Cell Cycle. 2012;11:3810–27. doi: 10.4161/cc.22022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korkmaz G, le Sage C, Tekirdag KA, Agami R, Gozuacik D. miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy. 2012;8:165–76. doi: 10.4161/auto.8.2.18351. [DOI] [PubMed] [Google Scholar]
- 19.Morgado AL, Xavier JM, Dionísio PA, Ribeiro MF, Dias RB, Sebastião AM, et al. MicroRNA-34a Modulates Neural Stem Cell Differentiation by Regulating Expression of Synaptic and Autophagic Proteins. Mol Neurobiol. 2014 doi: 10.1007/s12035-014-8794-6. [DOI] [PubMed] [Google Scholar]
- 20.Fang Y, Xue JL, Shen Q, Chen J, Tian L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology. 2012;55:1852–62. doi: 10.1002/hep.25576. [DOI] [PubMed] [Google Scholar]
- 21.Seca H, Lima RT, Lopes-Rodrigues V, Guimaraes JE, Almeida GM, Vasconcelos MH. Targeting miR-21 induces autophagy and chemosensitivity of leukemia cells. Curr Drug Targets. 2013;14:1135–43. doi: 10.2174/13894501113149990185. [DOI] [PubMed] [Google Scholar]
- 22.Blanco FJ, Ochs RL, Schwarz H, Lotz M. Chondrocyte apoptosis induced by nitric oxide. Am J Pathol. 1995;146:75–85. [PMC free article] [PubMed] [Google Scholar]
- 23.Goldring MB, Birkhead JR, Suen LF, Yamin R, Mizuno S, Glowacki J, et al. Interleukin-1 beta-modulated gene expression in immortalized human chondrocytes. J Clin Invest. 1994;94:2307–16. doi: 10.1172/JCI117595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shikhman AR, Brinson DC, Valbracht J, Lotz MK. Cytokine regulation of facilitated glucose transport in human articular chondrocytes. J Immunol. 2001;167:7001–8. doi: 10.4049/jimmunol.167.12.7001. [DOI] [PubMed] [Google Scholar]
- 25.Carames B, Kiosses WB, Akasaki Y, Brinson DC, Eap W, Koziol J, et al. Glucosamine activates autophagy in vitro and in vivo. Arthritis Rheum. 2013;65:1843–52. doi: 10.1002/art.37977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akasaki Y, Alvarez-Garcia O, Saito M, Carames B, Iwamoto Y, Lotz MK. FoxO transcription factors support oxidative stress resistance in human chondrocytes. Arthritis Rheumatol. 2014;66:3349–58. doi: 10.1002/art.38868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oeste CL, Seco E, Patton WF, Boya P, Pérez-Sala D. Interactions between autophagic and endo-lysosomal markers in endothelial cells. Histochem Cell Biol. 2013;139:659–70. doi: 10.1007/s00418-012-1057-6. [DOI] [PubMed] [Google Scholar]
- 28.Fisch KM, Akagi R, Alvarez-Garcia O, Teramura T, Muramatsu Y, Saito M, et al. Integrative omics profiling reveals dysregulated novel pathways mediated by microRNAs and DNA methylation in osteoarthritis. Arthritis Rheum; ACR/ARHP Annual Meeting; Boston, MA. 2014. p. Abstract 1007.p. 11. [Google Scholar]
- 29.Lotz MK, Carames B. Autophagy and cartilage homeostasis mechanisms in joint health, aging and OA. Nat Rev Rheumatol. 2011;7:579–87. doi: 10.1038/nrrheum.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–75. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–41. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–95. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–6. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 36.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 37.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 38.Caramés B, Olmer M, Kiosses WB, Lotz M. The relationship of autophagy defects and cartilage damage during joint aging in a mouse model. Arthritis Rheumatol. 2015;67:1568–76. doi: 10.1002/art.39073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J, Guan KL. Regulation of the autophagy initiating kinase ULK1 by nutrients: roles of mTORC1 and AMPK. Cell Cycle. 2011;10:1337–8. doi: 10.4161/cc.10.9.15291. [DOI] [PubMed] [Google Scholar]
- 40.Füllgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol. 2014;15:65–74. doi: 10.1038/nrm3716. [DOI] [PubMed] [Google Scholar]
- 41.Xiong X, Tao R, DePinho RA, Dong XC. The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J Biol Chem. 2012;287:39107–14. doi: 10.1074/jbc.M112.412569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holla S, Kurowska-Stolarska M, Bayry J, Balaji KN. Selective inhibition of IFNG-induced autophagy by Mir155- and Mir31-responsive WNT5A and SHH signaling. Autophagy. 2014;10:311–30. doi: 10.4161/auto.27225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wan G, Xie W, Liu Z, Xu W, Lao Y, Huang N, et al. Hypoxia-induced MIR155 is a potent autophagy inducer by targeting multiple players in the MTOR pathway. Autophagy. 2014;10:70–9. doi: 10.4161/auto.26534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–9. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 45.Nettelbladt E, Sundblad L. Protein patterns in synovial fluid and serum in rheumatoid arthritis and osteoarthritis. Arthritis Rheum. 1959;2:144–51. doi: 10.1002/1529-0131(195904)2:2<144::aid-art1780020206>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 46.Du F, Yu F, Wang Y, Hui Y, Carnevale K, Fu M, et al. MicroRNA-155 deficiency results in decreased macrophage inflammation and attenuated atherogenesis in apolipoprotein E-deficient mice. Arterioscler Thromb Vasc Biol. 2014;34:759–67. doi: 10.1161/ATVBAHA.113.302701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurowska-Stolarska M, Alivernini S, Ballantine LE, Asquith DL, Millar NL, Gilchrist DS, et al. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc Natl Acad Sci U S A. 2011;108:11193–8. doi: 10.1073/pnas.1019536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X, Tian F, Wang F. Rheumatoid arthritis-associated microRNA-155 targets SOCS1 and upregulates TNF-α and IL-1β in PBMCs. Int J Mol Sci. 2013;14:23910–21. doi: 10.3390/ijms141223910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MiR-155 suppresses autophagy-related genes in primary human chondrocytes. Cells were transfected for 24h with LNA miR-155 and LNA negative control (LNA nc), mimic miR-155 and mimic negative control (mimic nc). (A) qRT-PCR analysis of mRNA expression levels of direct and indirect targets of miR-155 in human primary chondrocytes following transfection with LNA or (B) mimic (n=4 independent experiments separate with chondrocyte preparations). Values are expressed as mean ± SD, *P<0.05, **P<0.01, ***P<0.001.