Abstract
Gene deletion and protein expression are cornerstone procedures for studying metabolism in any organism, including methane-producing archaea (methanogens). Methanogens produce coenzymes and cofactors not found in most bacteria, therefore it is sometimes necessary to express and purify methanogen proteins from the natural host. Protein expression in the native organism is also useful when studying post-translational modifications and their effect on gene expression or enzyme activity. We have created several new suicide plasmids to complement existing genetic tools for use in the methanogen, Methanosarcina acetivorans. The new plasmids are derived from the commercially available E. coli plasmid, pNEB193, and cannot replicate autonomously in methanogens. The designed plasmids facilitate markerless gene deletion, gene transcription, protein expression, and purification of proteins with cleavable affinity tags from the methanogen, Methanosarcina acetivorans.
1. Introduction
Genetic methods for Methanosarcina species are well developed, and making mutations on the chromosome is a routine procedure in several laboratories [1–5]. We sought to increase the ease-of-use for these tools to facilitate cloning, protein expression, and molecular biology experiments. The plasmid tools we have created complement existing methods and expand the repertoire of in vivo experiments possible in methanogens. Of particular need is the ability to express tagged proteins in methanogens to facilitate protein purification from the native host.
Methanogens survive by reducing carbon substrates to methane gas in a process called methanogenesis [6]. They employ unique enzymes and cofactors to activate carbon for reduction, and simultaneously generate a transmembrane ion gradient that is used for ATP synthesis [7, 8]. Unusual cofactors used in methanogens include coenzyme M, coenzyme B, methanopterins, methanophenazine, dimethylbenzimidazolyl cobamide, and deazaflavin F420 [9–25]. Because of these unusual cofactors, it may be difficult or impossible to express some methanogen proteins in heterologous hosts that do not produce these cofactors. If cofactor binding is essential for proper protein folding, the absence of the cofactor may result in misfolded and/or insoluble protein. If the protein does fold properly in a heterologous host, it is possible that host cofactors may bind in the active site in place of the native cofactor. For instance, dimethylbenzimidazolyl cobamide is structurally similar, though not identical to cobalamin [26–28]. The methanogen methanol:corrinoid methyltransferase, MtaB, and the corrinoid protein, MtaC, from Methanosarcina barkeri, have been purified from E. coli and biochemically characterized [29–31]. MtaB and MtaC expressed in E. coli are insoluble, and must be refolded in vitro after purification. As a result, MtaC is devoid of cofactor and must be reconstituted with the non-native corrin cofactor, hydroxycobalamin. While heterologously expressed, refolded, and reconstituted protein can be suitable for biochemical characterization, these treatments introduce the possibility of producing structural artefacts that can inhibit crystal formation. As such, overexpression of proteins in the native organism can be desirable to purify soluble protein populated with the biologically relevant cofactor. The crystal structure of the MtaBC complex was successfully obtained using protein purified from M. barkeri [32].
Dimethylbenzimidazolyl cobamide is not the only exotic cofactor found in methanogens. Coenzyme F420 is a deazaflavin, and structurally similar to flavin mononucleotide (FMN) in E. coli [19, 33–36]. To our knowledge, no predicted flavin-binding proteins from methanogens has been heterologously expressed or crystallized to date. One reason for the paucity of methanogen flavoprotein structures could perhaps be because annotated flavin adenine dinucleotide (FAD) or FMN binding sites may in fact be F420 binding sites. Therefore E. coli flavins may not be able to bind correctly in F420 binding site, resulting in unstable or misfolded protein. Methanogens also express many proteins with catalytic or structural iron/sulfur clusters, which do not have homologs in E. coli [37–45]. Therefore, expression of iron/sulfur cluster proteins in E. coli runs the risk of producing insoluble or misfolded protein, which may or may not be able to be reconstituted in vitro with Fe2+ and S2− [46].
Several methanogen proteins which do not require cofactors have been successfully expressed from E. coli, such as histone-like proteins, glutamine synthetase GlnK, and CRISPR Cas6 [47–50]. However, in some circumstances though proteins are not anticipated to require a cofactor or iron/sulfur cluster, expression of methanogen proteins in E. coli can still be challenging due to differences in codon usage between the two organisms [51–54]. Codon usage is significantly different between E. coli and methanogens. Translation of methanogen proteins can be accomplished using E. coli expression strains engineered to produce rare codons, however the yields can be low [55–57]. The yield of heterologously expressed methanogen protein can be increased by codon optimizing the nucleotide sequence for E. coli [55, 57]. Synthesis of codon-optimized genes is more expensive than traditional cloning involving PCR amplification of the gene of interest. Taking into account the high proportion of methanogen proteins with iron/sulfur clusters and unique cofactors, we perceived a need for a wider array of molecular tools for protein expression and purification in methanogens.
To address the need for plasmids that can be used to express and purify protein from methanogens, we designed new suicide plasmids based on the features of pMP44 and pJK026A [58, 59]. pMP44 is useful for markerless deletion of genes using homologous recombination [58]. However, pMP44 replicates in the E. coli host at a relatively low-copy number and must be propagated in a pir+ strain [60, 61]. Plasmid pJK026A and its derivatives can be used for inserting DNA at a ϕC31 phage att site which has been added to the chromosome [59]. It is useful for expressing protein in Methanosarcina, or for studying transcription and translational fusions [2]. pJK026A family plasmids are 11.7 Kb, and must be purified from a trfA+ E. coli strain [62]. The plasmid sizes, low copy number, and need for separate E. coli host strains, are attributes that can present technical challenges during cloning. We wanted to determine if the features of pMP44 and pJK026A could be used to create smaller plasmids that are suitable for high-copy replication in DH5α or DH10β E. coli hosts. The new suicide plasmids are designed to 1) use conventional, commercially-available E. coli hosts, 2) simplify and speed up the cloning process, and 3) combine features in a multifunctional plasmid that can stably integrate onto the M. acetivorans chromosome and be used for in vivo protein expression and purification via Strep-Tag II and histidine affinity tags [63–65].
2. Materials and Methods
2.1 Growth of cultures
E. coli was grown in Lysis Broth (LB) at 37°C with shaking [66]. M. acetivorans strains were grown at 35°C in HS medium as described [67]. Table 1 lists all the E. coli and M. acetivorans strains used in this study. The following additions were added as required (final concentration): ampicillin (100 µg ml−1), kanamycin (50 µg ml−1), chloramphenicol (8 or 35 µg ml−1), rhamnose (10 mM), histidine (0.1 mM), puromycin (2 µg ml−1), 8-aza-diaminopurine (8-ADP) (20 µg ml−1), trimethylamine (50 mM), methanol (125 mM), and acetate (40 or 120 mM).
Table 1.
Strains used in this study
| NB# | Genotype | Purpose | Reference |
|---|---|---|---|
| E. coli strains | |||
| NB3 | 5α F' lacIq | parent | New England Biolabs |
| NB4 | 10β | parent | New England Biolabs |
| NB100 | 10β/pNB721 | Promotorless pac (opt) for conditional essentiality test of promoters in M. acetivorans | This study |
| NB101 | 10β/pNB722 | pac (opt) vector for homologous recombination repair of mutants, gene deletion by homologous recombination (marked) in M. acetivorans | This study |
| NB104 | 5α F' lacIq/pNB723 | Gene deletion by homologous recombination (markerless) in M. acetivorans | This study |
| NB128 | 5α F' lacIq/pNB724 | pac (opt) vector for homologous recombination repair of mutants, gene deletion by homologous recombination (marked) (unique SpeI restriction site) in M. acetivorans | This study |
| NB131 | 5α F' lacIq/pNB727 | operon insertion into ϕC31 attP site on the chromosome in M. acetivorans | This study |
| NB133 | 5α F' lacIq/pNB729 | Expression of native or tagged protein in M. acetivorans | This study |
| NB134 | 5α F' lacIq/pNB730 | Expression of native or tagged protein (unique BamHI restriction site) in M. acetivorans | This study |
| NB161 | 5α F' lacIq/pALD1 | MA4421 deletion by homologous recombination (markerless) in M. acetivorans | This study |
| NB224 | 5α F' lacIq/pSK1 | Expresses native UidA in M. acetivorans | This study |
| NB225 | 5α F' lacIq/pSK2 | Expresses strep-his-UidA-his-strep protein in M. acetivorans | This study |
| NB238 | 10β / pNB735 | Expresses native or tagged protein in M. acetivorans. Amino-terminal strep-his tag is cleavable with thrombin protease. | This study |
| NB239 | 10β/ pNB737 | Expresses native or tagged protein in M. acetivorans. Amino-terminal his tag is cleavable with thrombin protease. | This study |
| M. acetivorans strains | |||
| NB34 | Δhpt::ϕC31 int, attP | parent | [59] |
| NB218 | Δhpt::ϕC31 int, attP, ΔMA4421 | ΔMA4421 mutant | This study |
| NB231 | Δhpt::ϕC31 int, att:pSK1 | Expresses native UidA protein | This study |
| NB232 | Δhpt::ϕC31 int, att:pSK2 | Expresses strep-his-UidA-his-strep tagged protein | This study |
2.2 DNA techniques and cloning procedures
PCR Primers and DNA sequences in Table 2 were designed using Vector NTI software (Life Technologies Corporation, Grand Island, NY). Genes, oligos, and multiple cloning sites were synthesized commercially by Integrated DNA Technologies (IDT, Coralville, IA) and Life Technologies Corporation (Grand Island, NY). Various PCR techniques were employed during the course of this work, including overlap extension and site-directed mutagenesis [68, 69]. For all PCR amplifications, Phusion Flash PCR Master Mix was used as a source of proofreading DNA polymerase (Life Technologies Corporation (Grand Island, NY)). DNA purification was carried out using Wizard kits from Promega (Madison, WI). DNA fragments were joined using T4 DNA ligase (New England Biolabs, Ipswich, MA) or GeneArt kits (Life Technologies Corporation (Grand Island, NY)). Restriction enzymes (AscI, BamHI, NdeI, NcoI, EcoRI, SphI, XbaI) were purchased from New England Biolabs (Ipswich, MA). All plasmids were sequenced by Eurofins Operon MWG (Huntsville, AL).
Table 2.
Sequences of DNA synthesized in this study
| Purpose | Name | Sequence (5’-3’) | Reference |
|---|---|---|---|
| Amplify pac (opt) gene from pMS86 | oNB115 | TCTAGAGTGATTCTCATGACCGAATATAAAC | This study |
| oNB116 | GCATGCACTAGTTCATGCTCCAGGTTTCCTG | This study | |
| Amplify PmcrB(M. voltae) from pMP44 | oNB128 | ACTAGTCGGTTTGCGTATTGGCG | This study |
| oNB129 | ACTAGTTCCTATTTTTTTGATATATACATCATAACA | This study | |
| Amplify hpt (opt) from pMS66 | oNB106 | TCTAGATCACTGATTTCCAAAAACATCTTTAATCTCAACTCC | This study |
| oNB127 | TCTAGACATGGTTGAAAGGCTTAAAGATTCCC | This study | |
| Removes SpeI restriction site from pNB722 | oNB151 | GCATGCAAGCTTGGCGTAATCATG | This study |
| oNB152 | GCATGCTCATGCTCCAGGTTTCC | This study | |
| Amplify ϕC31 attB from pJK026A | oNB117 | TCTAGAATGAATCAACAACTCTCCTGGCGCA | This study |
| oNB118 | TCTAGACGCTGGCGATTCAGGTTCATCATG | This study | |
| Amplify entire pNB724 | oNB110 | ACTAGTCTTGTCTGCTCCCGGCATCCG | This study |
| oNB111 | ACTAGTCCCGTCAGGGCGCGTCA | This study | |
| Amplify entire pNB727 | oNB130 | ACTAGTGGTGTGAAATACCGCACAGATGCGTAA | This study |
| oNB131 | ACTAGTTGACGCGCCCTGACGGG | This study | |
| Amplify expression cassette from pNB716 (inserts in pIDTSmart-Amp) | oNB183, M13 forward - 20 | GTGTAAAACGACGGCCAGTTTATCTAGTCA | Integrated DNA Technolog ies (IDT) |
| oNB184, M13 reverse -27 | CCTCAGGAAACAGCTATGACATCAAGCT | Integrated DNA Technolog ies (IDT) | |
| Removes second BamHI site from pNB729 | oNB185 | GGGGGCGCGCCGGATCTTAATTAAGTCTAG | This study |
| oNB186 | CTAGACTTAATTAAGATCCGGCGCGCCCCC | This study | |
| Creation of MA4421 deletion fusion fragment | oNB250 | AAAAAAAAAAAAGGCGCGCCCTGGATTTTTTACAGATTCTAATGATTCCAGG | This study |
| oNB251 | GCTCTGCATATATCTTGGATCTTATACCCCATGCTGAACTACAGAACGTT | This study | |
| oNB252 | AACGTTCTGTAGTTCAGCATGGGGTATAAGATCCAAGATATATGCAGAGC | This study | |
| oNB253 | AAAAAAAAAAAAGGCGCGCCCCTGCCCCTCACATAATCGTGC | This study | |
| oNB311 | GCCCTGGTTTGGTTCCCGGTTTACCAGAGAATGGAGGTATAAGATCCAAGAT | This study | |
| oNB312 | ATCTTGGATCTTATACCTCCATTCTCTGGTAAACCGGGAACCAAACCAGGGC | This study | |
| Amplifies MA4421 chromosomal region | oNB274 | GACCTTCTGGTGGATTGTTG | This study |
| oNB318 | GCAAAGCTTGTATACAGGGCAG | This study | |
| oNB319 | CTCGGAAGCATGGTCTATCC | This study | |
| Amplifies uidA from pJK026A | oNB369 | CATATGTTACGTCCTGTAGAAACCCCAACCCG | This study |
| oNB370 | CCATGGTACGTCCTGTAGAAACCCCAACCCG | This study | |
| oNB371 | GGATCCTCATTGTTTGCCTCCCTGCTGCGG | This study | |
| oNB372 | GGATCCTTTTGTTTGCCTCCCTGCTGCGGTT | This study | |
| pNB723 insert sequencing | oNB301 | GGCTGGCTTAACTATGCGGCATC | This study |
| oNB302 | GCACCGTGGGTTTATATTCGGTCATGAGAATC | This study | |
| pNB730 MCS insert sequencing | oNB303 | CGTCAGGGCGCGTCATTAACTACT | This study |
| att integration of pNB730 at the hpt locus | ϕC31 screenall#1 | GAAGCTTCCCCTTGACCAAT | [59] |
| ϕC31 screen-C2A#1 | TTGATTCGGATACCCTGAGC | [59] | |
| ϕC31 screen- pJK200#1 | GCAAAGAAAAGCCAGTATGGA | [59] | |
| oNB317 | GATGAGTGGCAGGGCGGGGCGTAAT | This study | |
| Thrombin-cleavable tagged protein | Strep-His-Thrombin | ATTAAGGAGGAAATTCATATG TGGAGCCACCCTCAGTTCGAGA AACA TCACCATCACCATCATCA CCATCTGGTGCCGCGTGGCTCT TCCATGGAAGGCGCGCCGGA TC CAAGCTTGGGCCCTCG |
This study |
| His-Thrombin | ATTAAGGAGGAAATTCATATGC ATCACCATCACCATCATCACCA TCT GGTGCCGCGTGGCTCTTCC ATGGAAGGCGCGCCGGATCCA AGCTTGGGCCCTCG |
This study |
2.3 Transformation
Plasmids used and created in this study are listed in Table 3. E. coli was transformed by electroporation and plated onto LB agar plates (1.5% w/v agar) containing the appropriate antibiotic [70]. M. acetivorans was transformed using the liposome-mediated transformation method [71]. After transformation and recovery, M. acetivorans cells were plated on HS medium with 50 mM trimethylamine as carbon source solidified with 1.4% agar and incubated in Wolfe incubators (Coy Laboratory Products, Grass Lake, MI) under premixed 20% CO2/79.9% N2/0.1% H2S atmosphere (Matheson).
Table 3.
Plasmids used in this study
| Name | Features | Purpose | Reference |
|---|---|---|---|
| pNEB193 | pUC19 ori, bla +, lacZ+ | Parent vector | New England Biolabs |
| pMP44 | oriR6K, bla+, Pmcr (M. barkeri) hpt, Pmcr (M. voltae) pac | Gene deletion by homologous recombination (markerless) | [58] |
| pJK026A | oriV, repE, sopABC, cat, ϕC31 attB, Pmcr (M. voltae) pac-hpt, PmcrB uidA | Gene insertion into ϕC31 attP site on the chromosome | [59] |
| pMS86 | pac (opt) | synthesized | This study |
| pMS66 | hpt (opt) | synthesized | This study |
| pNB716 | strep-his MCS his-strep expression cassette | synthesized | This study |
| pNB721 | pUC19 ori, bla +, lacZ+, pac (opt) | Promotorless pac (opt) for conditional essentiality test of promoters in M. acetivorans | This study |
| pNB722 | pUC19 ori, bla +, lacZ+, PmcrB(M. voltae) pac (opt) | pac (opt) vector for homologous recombination repair of mutants, KO by homologous recombination (marked) in M. acetivorans | This study |
| pNB723 | pUC19 ori, bla +, lacZ+, Pmcr (M. voltae) pac- hpt | Gene deletion by homologous recombination (markerless) in M. acetivorans | This study |
| pNB724 | pUC19 ori, bla +, lacZ+, PmcrB(M. voltae) pac (opt) | pac (opt) vector for homologous recombination repair of mutants, gene deletion by homologous recombination (marked) (unique SpeI restriction site) in M. acetivorans | This study |
| pNB727 | pUC19 ori, bla +, lacZ+, ϕC31 attB, PmcrB(M. voltae) pac (opt) | operon insertion into ϕC31 attP site on the chromosome | This study |
| pNB729 | pUC19 ori, bla +, lacZ+, ϕC31 attB, PmcrB(M. voltae) pac (opt), strep-his MCS his-strep expression cassette | Expression of native or tagged protein in M. acetivorans | This study |
| pNB730 | pUC19 ori, bla +, lacZ+, ϕC31 attB, PmcrB(M. voltae) pac (opt), strep-his MCS his-strep expression cassette | Expression of native or tagged protein (unique BamHI restriction site) in M. acetivorans | This study |
| pALD1 | pUC19 ori, bla +, lacZ+, PmcrB(M. voltae) pac- hpt, ΔMA4421 deletion fusion | MA4421 deletion by homologous recombination (markerless) in M. acetivorans | This study |
| pSK1 | pUC19 ori, bla +, lacZ+, ϕC31 attB, PmcrB(M. voltae) pac (opt), uidA | Expresses native UidA in M. acetivorans | This study |
| pSK2 | pUC19 ori, bla +, lacZ+, ϕC31 attB, PmcrB(M. voltae) pac (opt), strep-his- uidA-his-strep | Expresses strep-his-UidA-his-strep protein in M. acetivorans | This study |
| pNB735 | pUC19 ori, bla +, lacZ+, ϕC31 attB, PmcrB(M. voltae) pac (opt), strep-his (thrombin) MCS his-strep expression cassette | Expression of native or tagged protein in M. acetivorans. Amino-terminal strep-his tag is cleavable with thrombin protease. | This study |
| pNB737 | pUC19 ori, bla +, lacZ+, ϕC31 attB, PmcrB(M. voltae) pac (opt), his (thrombin) MCS his-strep expression cassette | Expression of native or tagged protein in M. acetivorans. Amino-terminal his tag is cleavable with thrombin protease. | This study |
2.4 Western Blot Analysis
Sample protein concentration was determined using the Coomassie Plus Protein Assay Reagent (Life Technologies Corporation (Grand Island, NY)). Samples were diluted with 6X Cracking Buffer (348 mM Tris pH 6.8, 349 mM SDS, 600 mM DTT, 4.1 mM glycerol, 180 µM bromophenol blue), boiled for 10 minutes, and 2 µg each sample were loaded per lane on a 12% sodium dodecyl sulfate-polyacrylamide gel (Bio-Rad, Hercules, CA). Three microliters of Precision Plus Protein Dual Color Standards (Bio-Rad, Hercules, CA) and 1.5 µL of Precision Plus Protein WesternC Standards (Bio-Rad, Hercules, CA) were used as markers. Proteins were separated at 15 mA per gel for 30 minutes and 30 mA per gel for 45 minutes. Proteins were transferred to a polyvinylidene difluoride membrane (PVDF) (Bio-Rad, Hercules, CA) for 1 hour at 100 V in transfer buffer (25 mM Tris, 192 mM glycine, 15% methanol). The membrane was blocked with 25 mL 5% nonfat dry milk in Tris-buffered saline (137 mM NaCl, 20 mM Tris pH 7.6) with 0.1% TWEEN 20 (TBST) overnight and probed with a 1:4000 dilution of Strep-Tag II Antibody, HRP Conjugate (Novagen, EMD Millipore, Temecula, CA) in 20 mL of Blocking Solution. Strep-tagged protein was detected with Pierce ECL Western Blotting Substrate (Life Technologies Corporation (Grand Island, NY)).
2.5 β-glucuronidase enzyme assays
Cell extract was assayed for β-glucuronidase activity as described [59]. Briefly, 10 ml exponential phase cultures of strains listed in Table 1 were harvested by centrifugation in a TX-750 Swinging Bucket Rotor with 15 mL conical tube adapters at 4031 × g for 3 minutes at room temperature. Cells were resuspended in 200 µl of 50 mM Tris-Cl, 1 mM DTT pH 8.0 buffer, followed by the addition of 1 u of DNaseI (Life Technologies Corporation (Grand Island, NY)) and Halt Protease Inhibitor Cocktail, EDTA-Free (Life Technologies Corporation (Grand Island, NY)) to a final concentration of 1X. Cells were lysed on ice for 10 minutes, and insoluble cell debris was removed by centrifugation in F21–48×1.5/2.0 rotor at 14000 × g for 10 minutes at room temperature. Cleavage of p-nitrophenyl glucuronide to p-nitrophenol was detected by increased absorbance at 415 nm in a Tecan Sunrise plate spectrophotometer (Tecan US, Inc., Morrisville NC). The extinction coefficient of p-nitrophenol was determined in Solid 96 Well Plates (Fisher catalog #21-377-205) with a path length of 0.5 cm at 415 nm. Protein concentration was measured using the Coomassie Plus Protein Assay Reagent (Life Technologies Corporation (Grand Island, NY)).
3. Results
3.1 pNB723 plasmid design
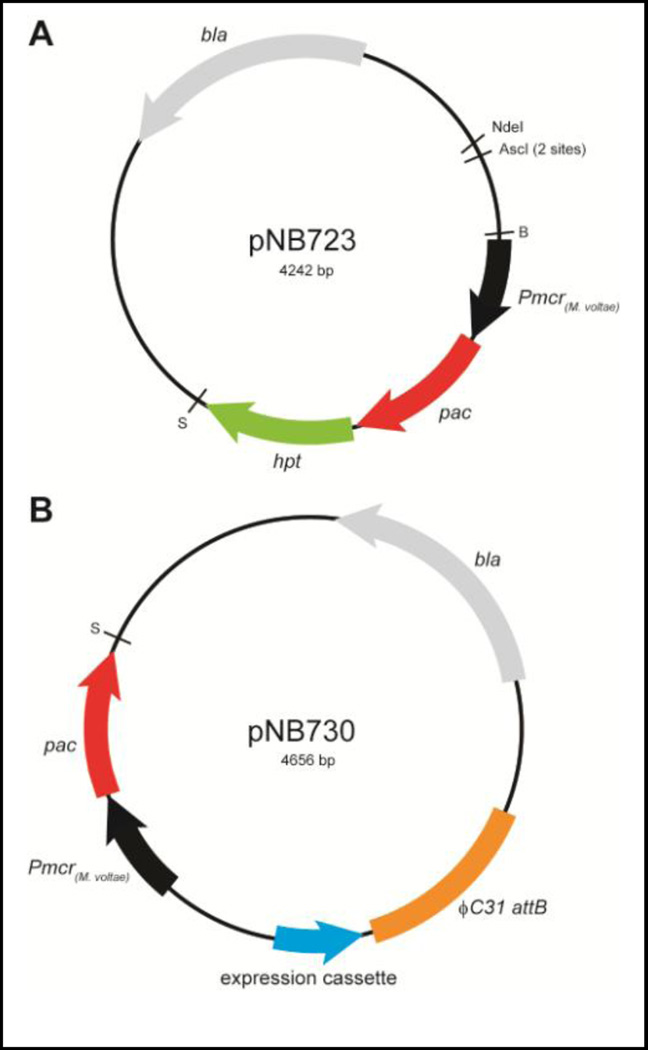
We created a high-copy plasmid, pNB723, for markerless deletion of Methanosarcina genes to circumvent the need to use pir+ E. coli hosts (Figures 1 and S1). To construct pNB723, we used pNEB193 as the E. coli plasmid scaffold (New England Biolabs, Ipswich, MA). pNEB193 is a small, high-copy, commercially available vector with a pUC19 origin of replication, a bleomycin (ampicillin) resistance cassette, and a PT7 lacZ cassette for blue-white selection of plasmids containing inserts at the multiple cloning site. To select Methanosarcina strains which have recombined the plasmid onto the chromosome, we added the pac (puromycin acetyltransferase) gene at the unique XbaI and SphI restriction sites on pNEB193. We optimized the pac codons for expression in Methanosarcina, thereby lowering the %GC content from 73.1% to 48.5%, and eliminating interference in sequencing reactions that can occur when plasmids contain high %GC stretches (Figure S2). The optimized pac gene was amplified from pMS86 using oligos oNB115 and oNB116, which added a XbaI restriction site at the 5’ end of the gene, and tandem SpeI and SphI restriction sites at the 3’ end of the gene. The resulting plasmid carrying a promotorless optimized pac gene is pNB721.
Figure 1.
pNB723 and pNB730 plasmid maps. Genes encoding puromycin acetyltransferase (pac, red) and hypoxanthine phosphoribosyltransferase (hpt, green) are codon-optimized for expression in Methanosarcina.
The Methanococcus voltae PmcrB promoter from pMP44 was cloned upstream of the pac gene at the XbaI restriction site, creating plasmid pNB722. The PmcrB(M. voltae) promoter will constitutively express the pac gene in Methanosarcinales. PmcrB(M. voltae) was cloned from pMP44 using oligos oNB128 and oNB129, which added XbaI sites at each end of the gene. Orientation of the promoter was verified by DNA sequencing to ensure that the pac gene will be expressed in the host strain.
Finally, we cloned a hypoxanthine phosphoribosyltransferase gene (hpt) at the SpeI and SphI restriction sites in pNB722 so that the resulting plasmid, pNB723, expresses the hpt gene in an operon with the pac gene. The hpt gene is a counterselection marker that can be used to create a markerless gene deletion when transformants are plated on the purine analog, 8-aza-diaminopurine (8-ADP) [58]. The hpt gene was also codon-optimized for expression in Methanosarcina, which resulted in lowering the %GC content from 47.6% to 36.7% (Figure S3). The optimized hpt gene was amplified from pMS66 using oligos oNB106 and oNB127, which added a XbaI site at the 5’ and at the 3’ ends of the gene. Directionality of the hpt gene was verified by DNA sequencing. The resulting pNB723 plasmid has unique NdeI and BamHI sites, and two AscI restriction sites, which can be used to clone DNA sequences for deletion of genes in Methanosarcina.
3.2 Deletion of MA4421 using pNB723
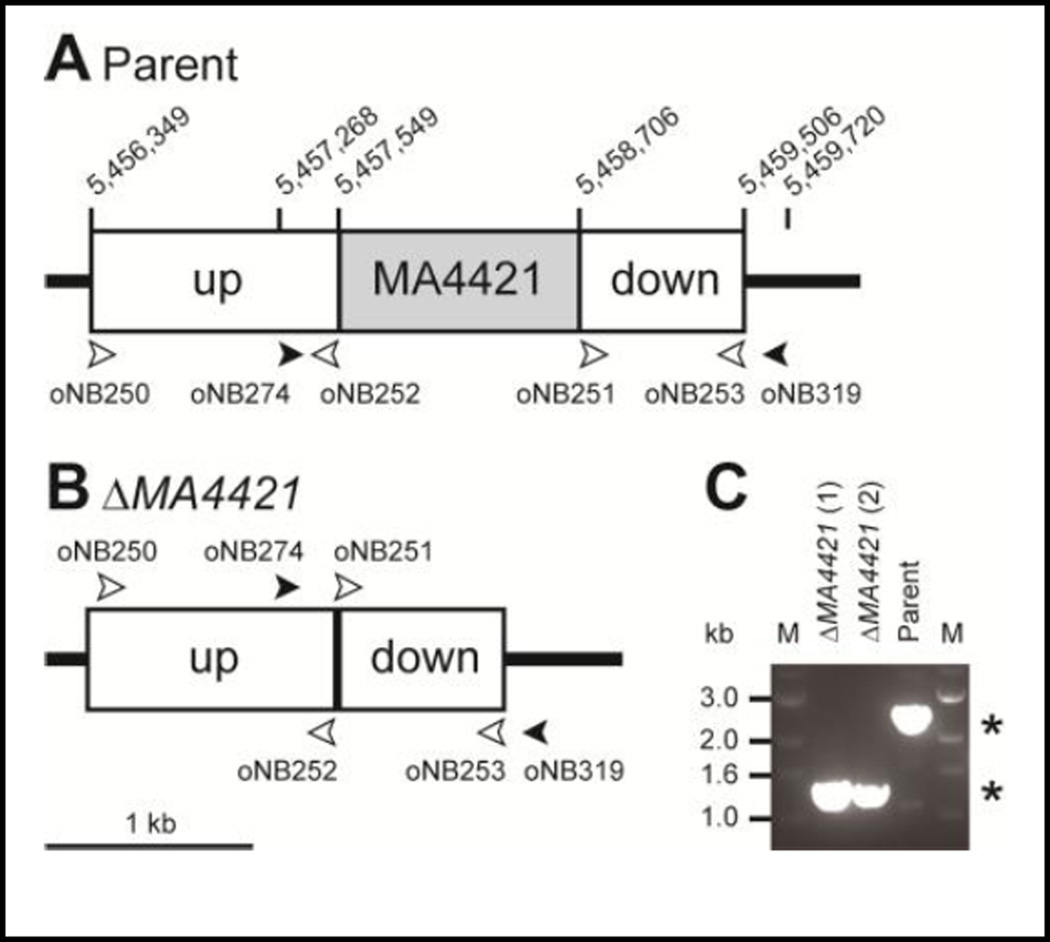
To demonstrate that pNB723 functions as designed, we used it to delete the MA4421 prenyl reductase gene from the M. acetivorans chromosome (Figure 2). For the plasmid validation purposes here, the gene to be deleted was expected to be nonessential. The DNA sequence 5’ upstream of the MA4421 gene was amplified using primers oNB250 and oNB252. The 3’ downstream DNA sequence was amplified using primers oNB251 and oNB253. The 5’ and 3’ sequences were fused using oligos oNB311 and oNB312, and cloned into the pNB723 plasmid at the AscI restriction site, resulting in plasmid pALD1 (Figure S4). Plasmid pALD1 was transformed into M. acetivorans strain NB34 using liposomes, and puromycin as a selection agent. The puromycin-resistant colonies were streaked for isolation, grown in liquid medium without puromycin, and plated onto agar containing 8-ADP to counterselect for the hpt gene. The resulting 8-ADP-resistant colonies were streaked for isolation, and grown in liquid medium without puromycin or 8-ADP. Markerless deletion of the MA4421 gene was confirmed by PCR amplifying the MA4421 deletion junctions from genomic DNA using the oNB274, oNB318 and oNB319 oligos (Figure 2). In this PCR strategy, the screening oligos do not anneal to the 5’ upstream or 3’ downstream DNA sequences that were used to construct the pALD1 deletion plasmid. Methanosarcina cells can carry several copies of the chromosome, and PCR amplification for the deleted gene is essential to ensure that all copies of the gene have been deleted [72]. In addition, plasmids may integrate in unpredictable ways if there is a region of low complexity or if the gene is essential. Surviving cells may, for instance, create large deletions, amplifications, or insertions to preserve essential gene function while also generating a false-positive in a PCR screen. As additional measures to confirm strain identity, genome resequencing and/or Southern blots using probes specific for the deleted gene, for flanking genomic regions, or for pac or bla (to verify plasmid insertion at the expected location) is also indicated (Figure S5).
Figure 2.
Deletion of MA4421 from the chromosome using pALD1. Schematics of the MA4421 genomic locus in the parental strain (A), and in the deletion mutant (B), are shown. (C) PCR results with oligos oNB274 and oNB319 showing deletion of MA4421 in two isogenic isolates. Gray box= MA4421 coding sequence. White boxes= DNA sequences upstream (“up”) and downstream (“down”) of the MA4421 gene that were used to create plasmid pALD1. Open arrowheads= annealing site of PCR oligos used to construct pALD1. Solid arrowheads= annealing site of PCR oligos used to screen for deletion of MA4421 on the chromosome. M= DNA size marker. Kb= kilobasepairs. The asterisks denote the expected amplicon sizes.
3.3 pNB730 plasmid design
We created a pNEB193-derived plasmid for expression of tagged protein in Methanosarcina. Using oligos oNB151 and oNB152, we removed the SpeI restriction site, creating plasmid pNB724. pNB724 was amplified using oligos oNB110 and oNB111, which creates SpeI restriction sites at the 5’ and 3’ ends of the linear amplification product. To insert the ϕC31 phage attB attachment site that allows the plasmid to recombine with the ϕC31 attP site on the M. acetivorans NB34 chromosome, we amplified the ϕC31 phage attB site from pJK026A using oligos oNB117 and oNB118, which creates XbaI restriction sites at the 5’ and 3’ ends of the amplification product. The XbaI-digested attB fragment was ligated into the SpeI-digested pNB724 amplicon to create pNB727. pNB727 was verified by DNA sequencing.
Next, we designed an expression cassette with multiple restriction sites to facilitate cloning (Figure 3). The cassette, encoded on plasmid pNB716, contains the PmcrB promoter from pJK026A and a multiple cloning site (MCS) flanked by sequences encoding the Strep-Tag II peptide (strep, WSHPQFEK) and histidine tags (his, HHHHHHHH). The Strep-Tag II peptide was codon optimized for expression in M. acetivorans (Figure 3, orange shaded sequences). The 5’ and 3’ tag sequences were not identical so as to prevent homologous recombination that would result in loss of the MCS or of the gene to be expressed. The expression cassette was designed such that cloning a gene into the NdeI site results in expression of native protein. Cloning the gene into the NcoI site results in protein with an amino-terminal his-strep tag. Carboxy-tagged protein can be expressed by removing the stop codon from the gene and cloning into the BamHI, ApaI, or NruI restriction sites. Therefore this expression cassette can be used to express native, amino-tagged, carboxy-tagged, or dual-tagged protein depending on the restriction sites used. A strong translational stop signal was added after the 3’ his-strep tag sequence by introducing four stop codons within a 20 bp region. The expression cassette was amplified from pNB716 using oligos oNB183 and oNB184 and digested with XbaI restriction enzyme.
Figure 3.
Multiple cloning sites of pNEB193-derived plasmids used for protein expression in Methanosarcina. The ribosome binding site is in bold font. Unique restriction sites are underlined. Green triangle= translation start site. Red square= stop codon. Orange box= Strep-Tag II sequence. Blue box= histidine tag sequence. Gray box= thrombin recognition sequence.
pNB727 was amplified using oligos oNB130 and oNB131, resulting in a linearized amplicon containing SpeI restriction sites at the 5’ and 3’ termini. The pNB727 amplicon was digested with SpeI, then ligated with the XbaI-digested expression cassette from pNB716, to create the plasmid pNB729. Finally, oligos oNB185 and oNB186 were used to amplify pNB729 and remove the BamHI restriction site upstream of the pac expression cassette. The resulting plasmid, pNB730, contains unique NdeI, NcoI, BamHI, ApaI, and NruI restriction sites for cloning genes into the expression cassette multiple cloning site. pNB730 was verified by DNA sequencing (Figure S6).
3.4 Native and tagged expression of uidA using pNB730
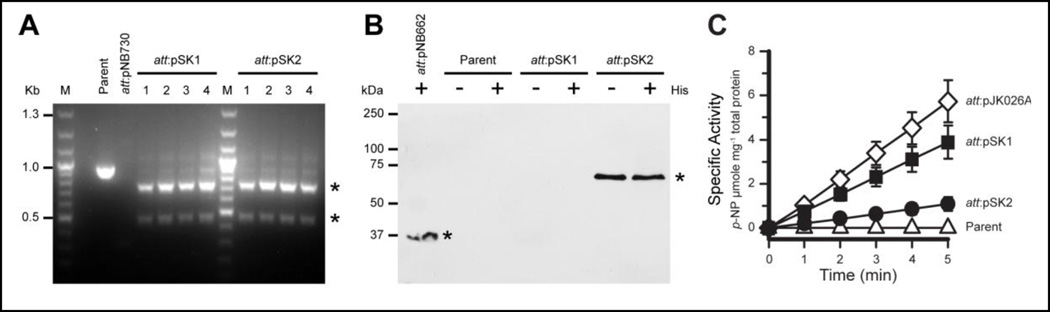
We used the β-glucuronidase (uidA) gene to measure tagged and untagged protein expression in M. acetivorans from the expression cassette we created (Figure 4). The uidA gene was amplified from pJK026A using oligos oNB369 and oNB371 and cloned into the NdeI and BamHI sites of pNB730 to create plasmid pSK1 (expresses native UidA) (Figure S7). The uidA gene was also amplified from pJK026A using oligos oNB370 and oNB372, and cloned into the NcoI and BamHI sites to create plasmid pSK2 (expresses dual-tagged UidA) (Figure S8). Plasmids pSK1 and pSK2 were transformed into M. acetivorans strain NB34. Cells which had recombined the plasmid onto the chromosome at the ϕC31 attP site were selected using puromycin. Puromycin-resistant colonies were streaked for isolation, grown in liquid medium without antibiotic, and screened by PCR.
Figure 4.
Expression of tagged UidA protein in M. acetivorans. (A) Verification of integration of pSK1 and pSK2 on the chromosome. Genomic DNA from four isolates of each transformation were screened by PCR. M= marker. (B) Western blot of strep-his-UidA-his-strep protein expressed in M. acetivorans. Two micrograms of total protein from each strain were separated by denaturing PAGE. Western blots were probed with anti-strep antibodies. His= cultures were supplemented with 0.1 mM histidine. (C) Triplicate cultures of each strain were assayed in triplicate (N=9). Specific activity reported in nmoles min−1 mg−1 lysate.
To screen for integration at the attP site, oligos “C31 screen-all#1”, “C31 screen C2A #1”, “C31 screen pJK200#1”, and oNB317 were used in a four-oligo PCR reaction with genomic DNA [59]. In this four-oligo PCR amplification, genomic DNA from strains which have integrated a single copy of pNB730-derived plasmids will produce amplicons of 740 and 471 bp. A 301bp band is amplified by plasmid alone or if multiple copies of the pNB730-derived plasmid has integrated at the ϕC31 attP site. Parental genomic DNA template will result in amplification of a 910 bp fragment. Using this screen, we verified the creation of strains NB231 (Δhpt::ϕC31 int, att:pSK1) and NB232 (Δhpt::ϕC31 int, att:pSK2), which had recombined the respective plasmid at the ϕC31 attP site on the chromosome. Plasmid integration was also confirmed by Southern blot using the uidA gene as a probe (Figure S9).
Next, we verified expression of UidA enzyme from the integrated pSK1 or pSK2 plasmids (Figure 4). To verify expression of tagged protein, we analyzed cell extract from strains NB231 (att:pSK1), NB232 (att:pSK2) by Western blot using anti-strep-tag antibodies. The parent extract was used as a negative control, and the positive control was cell extract from strain NB75, which expresses strepHdrD2 (37 kDa) from an integrated copy of the pJK026A-derived plasmid, pNB665. The cell extract from strains that expressed tagged protein had a single strep-tagged protein band at the expected size of 71 kDa, whereas the parent extract and NB231 (att:pSK1, which expresses untagged UidA protein), had no visible bands. We also added 0.1 mM histidine to cultures to determine if adding exogenous histidine to the medium could increase expression of his-tagged protein. It would be reasonable to hypothesize that methanogens may produce limiting quantities histidine to synthesize large quantities of a his-tagged protein. Histidine supplementation, assuming it could be transported into the cell and used to charge histinyl tRNAs, may alleviate this limitation and result in higher expression levels. However, addition of histidine had no measurable effect on protein expression. Finally, we verified that the UidA protein expressed from pNB730 was properly folded and active. As expected, cell extract from strains NB231 (att:pSK1, expresses untagged protein) and NB232 (att:pSK2, expresses dual-tagged protein) had detectable β-glucuronidase activity, whereas extract from the parent strain had no detectable β-glucuronidase activity (Figure 4). We noted that activity of untagged β-glucuronidase (pSK1) is higher than the dual-tagged β-glucuronidase (pSK2), demonstrating that amino and/or carboxy-terminal peptide tags can affect enzyme function and may not reflect differences in translation efficiency from identical promoters.
3.5 Creation of plasmids for expression of protein with cleavable affinity tags
In some circumstances (i.e., if it interferes with enzyme activity, or for protein crystallography) it may be preferable to have the ability to cleave affinity tags from expressed protein. Therefore we designed two additional plasmids based on pNB730, which include thrombin cleavage sites (Figure 2). The thrombin recognition site (LVPRGS) was optimized for expression in M. acetivorans [73, 74]. Plasmid pNB735 has the thrombin site immediately downstream of the 5’ strep-tag and histidine tag, before the NcoI site where a gene of interest can be cloned. The complementary plasmid, pNB737, was created to express proteins with a thrombin-cleavable amino-terminal his tag. The sequences of both plasmids, pNB735 (Figure S10) and pNB737 (Figure S11), have been verified by DNA sequencing.
4. Discussion
We have succeeded in creating a suite of easy-to-use plasmids for gene deletion and expression of affinity tagged protein in Methanosarcina acetivorans. We have also demonstrated the utility of these plasmids in deleting genes from the chromosome (MA4421), and in expressing active enzymes in vivo. Depending on the restriction sites used for insertion of the gene of interest, the expressed protein either contains Strep-Tag II and histidine affinity tags or is untagged. The small, high-copy plasmids are compatible with ligation-independent cloning methods such as GeneArt Seamless Cloning and Assembly Kits (Invitrogen). The ease of propagation in E. coli, and the ease of cloning make the pNB723 and pNB730 family plasmids compatible with modern synthetic biology experiments. Though not demonstrated in this work,, plasmid pNB730 can be used for a wide array of experiments including mutant complementation, purification of proteins to study post-translational modification, and metabolic engineering applications in addition to expression of foreign proteins in the cell [75–77].
pNB735 and pNB737 plasmids will also make it easier to express protein in Methanosarcina for purification and crystallography purposes. To advance methanogen structure/function studies, we anticipate plasmid tools designed specifically for protein purification in methanogens, such as pNB735 and pNB737, may make it possible to obtain large quantities of pure, correctly folded protein from the native organism. Purification of protein from the native host may enable correct protein folding and population of the active site with the physiological cofactor. After purification, affinity tags can be removed by digestion with thrombin protease. The pNEB193-derived plasmids we created add to the expanding repertoire of genetic and protein expression tools in M. acetivorans and other Methanosarcina species [5, 58, 59, 78, 79].
The multiple cloning site we designed for pNB730 and derivative plasmids contained a UAG stop codon to terminate translation of the carboxy-terminal his-strep affinity tag. In Methanosarcina, UAG can either be translated as a pyrrolysine residue, or will be recognized as a termination signal, depending on whether a PYLIS element is encoded in the 3’ untranslated region of the RNA. When a PYLIS element is absent, approximately 70% of the translated polypeptides will stop at the UAG, while 30% of the time pyrrolysine will be incorporated into the growing protein chain, and translation continues until a second UAA or UGA stop codon is encountered. In the pNB730 multiple cloning site, the next in-frame stop codon is 336 bp downstream. If pyrrolysine had been incorporated in the uidA translation product, we would expect to detect two bands, one at 71 kDa, and the pyrrolysine read-through product at 83 kDa. In anti-strep immunoblots we only detected a single band at 71 kDa, indicating that translation was terminated at the first UAG codon. Kryzcki and coworkers noted that in highly expressed monomethylamine methyltransferase genes, the +1 and +2 nucleotides after the pyrrolysine-coding UAG codon are often GG [80]. Others have observed that the efficiency of pyl incorporation at UAG codons in heterologous systems can vary with the gene context [81, 82]. This contextual dependence on translational termination has been described in eukarya [83]. Our data suggests that the +1 and +2 nucleotides after the stop codon, TT, may disfavor pyrrolysine incorporation and instead results in translation termination in methanogens.
5. Conclusions
Methanogenic archaea produce several unusual coenzymes and cofactors that are not synthesized by E. coli, thereby constraining the ability to use E. coli as a heterologous host for overexpression and purification of a subset of methanogen proteins. To address this limitation, we have created a suite of plasmids for gene deletion and protein overexpression in Methanosarcina species. The new plasmids are derived from the small, high-copy E. coli plasmid, pNEB193, and can be propagated in standard E. coli cloning strains. We have successfully used the new plasmids to overexpress a native or his-strep tagged β-glucuronidase and to delete the gene MA4421 from the chromosome. These plasmids complement the growing list of genetic tools available for studying methanogen biology, and will be especially useful for identifying post-translational modifications in methanogen proteins, and for expressing proteins with amino- or carboxy-terminal affinity tags that can be cleaved with thrombin.
Supplementary Material
Highlights.
We have created a suite of user-friendly plasmids for methanogens.
The new plasmids are now compatible with ligation-independent cloning.
We validated plasmids for markerless gene deletion
Plasmids were used to express a native and his-tagged reporter gene.
Acknowledgments
The authors wish to thank William Metcalf for providing plasmids (pMP44, pJK026A) and M. acetivorans strain WWM82 (Δhpt::ϕC31 int, attP). This publication was made possible by National Science Foundation grant number IOS-1449525, National Institutes of Health Grant Number P20 RR-17675 from the National Center for Research Resources, and by the Nebraska Tobacco Settlement Biomedical Research Development Funds. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the funding agencies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ehlers C, Jager D, Schmitz RA. Establishing a markerless genetic exchange system for Methanosarcina mazei strain Go1 for constructing chromosomal mutants of small RNA genes. Archaea. 2011;2011:439608. doi: 10.1155/2011/439608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buan N, Kulkarni G, Metcalf W. Genetic methods for methanosarcina species. Methods Enzymol. 2011;494:23–42. doi: 10.1016/B978-0-12-385112-3.00002-0. [DOI] [PubMed] [Google Scholar]
- 3.Whitman WB, Tumbula DL, Yu JP, Kim W. Development of genetic approaches for the methane-producing archaebacterium Methanococcus maripaludis. Biofactors. 1997;6:37–46. doi: 10.1002/biof.5520060105. [DOI] [PubMed] [Google Scholar]
- 4.Bertani G, Baresi L. Genetic transformation in the methanogen Methanococcus voltae PS. J Bacteriol. 1987;169:2730–2738. doi: 10.1128/jb.169.6.2730-2738.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mondorf S, Deppenmeier U, Welte C. A novel inducible protein production system and neomycin resistance as selection marker for Methanosarcina mazei. Archaea. 2012;2012:973743. doi: 10.1155/2012/973743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thauer RK. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. 1998 Marjory Stephenson Prize Lecture. Microbiology. 1998;144(Pt 9):2377–2406. doi: 10.1099/00221287-144-9-2377. [DOI] [PubMed] [Google Scholar]
- 7.Deppenmeier U. The membrane-bound electron transport system of Methanosarcina species. J Bioenerg Biomembr. 2004;36:55–64. doi: 10.1023/b:jobb.0000019598.64642.97. [DOI] [PubMed] [Google Scholar]
- 8.Kulkarni G, Kridelbaugh DM, Guss AM, Metcalf WW. Hydrogen is a preferred intermediate in the energy-conserving electron transport chain of Methanosarcina barkeri. Proc Natl Acad Sci U S A. 2009;106:15915–15920. doi: 10.1073/pnas.0905914106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bobik TA, Wolfe RS. Physiological importance of the heterodisulfide of coenzyme M and 7-mercaptoheptanoylthreonine phosphate in the reduction of carbon dioxide to methane in Methanobacterium. Proc Natl Acad Sci U S A. 1988;85:60–63. doi: 10.1073/pnas.85.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ankel-Fuchs D, Bocher R, Thauer RK, Noll KM, Wolfe RS. 7-Mercaptoheptanoylthreonine phosphate functions as component B in ATP-independent methane formation from methyl-CoM with reduced cobalamin as electron donor. FEBS Lett. 1987;213:123–127. doi: 10.1016/0014-5793(87)81476-x. [DOI] [PubMed] [Google Scholar]
- 11.Leigh JA, Rinehart KL, Jr, Wolfe RS. Methanofuran (carbon dioxide reduction factor), a formyl carrier in methane production from carbon dioxide in Methanobacterium. Biochemistry. 1985;24:995–999. doi: 10.1021/bi00325a028. [DOI] [PubMed] [Google Scholar]
- 12.Gunsalus RP, Wolfe RS. Stimulation of CO2 reduction to methane by methylcoenzyme M in extracts Methanobacterium. Biochem Biophys Res Commun. 1977;76:790–795. doi: 10.1016/0006-291x(77)91570-4. [DOI] [PubMed] [Google Scholar]
- 13.Wood JM, Wolfe RS. Propylation and purification of a B12 enzyme involved in methane formation. Biochemistry. 1966;5:3598–3603. doi: 10.1021/bi00875a031. [DOI] [PubMed] [Google Scholar]
- 14.Mayr S, Latkoczy C, Kruger M, Gunther D, Shima S, Thauer RK, Widdel F, Jaun B. Structure of an F430 variant from archaea associated with anaerobic oxidation of methane. J Am Chem Soc. 2008;130:10758–10767. doi: 10.1021/ja802929z. [DOI] [PubMed] [Google Scholar]
- 15.Hinderberger D, Piskorski RP, Goenrich M, Thauer RK, Schweiger A, Harmer J, Jaun B. A nickel-alkyl bond in an inactivated state of the enzyme catalyzing methane formation. Angew Chem Int Ed Engl. 2006;45:3602–3607. doi: 10.1002/anie.200600366. [DOI] [PubMed] [Google Scholar]
- 16.Kruger M, Meyerdierks A, Glockner FO, Amann R, Widdel F, Kube M, Reinhardt R, Kahnt J, Bocher R, Thauer RK, Shima S. A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature. 2003;426:878–881. doi: 10.1038/nature02207. [DOI] [PubMed] [Google Scholar]
- 17.Fischer R, Thauer RK. Ferredoxin-dependent methane formation from acetate in cell extracts of Methanosarcina barkeri (strain MS) FEBS Lett. 1990;269:368–372. doi: 10.1016/0014-5793(90)81195-t. [DOI] [PubMed] [Google Scholar]
- 18.Ankel-Fuchs D, Thauer RK. Methane formation from methyl-coenzyme M in a system containing methyl-coenzyme M reductase, component B and reduced cobalamin. Eur J Biochem. 1986;156:171–177. doi: 10.1111/j.1432-1033.1986.tb09563.x. [DOI] [PubMed] [Google Scholar]
- 19.Eirich LD, Vogels GD, Wolfe RS. Proposed structure for coenzyme F420 from Methanobacterium. Biochemistry. 1978;17:4583–4593. doi: 10.1021/bi00615a002. [DOI] [PubMed] [Google Scholar]
- 20.Keltjens JT, Vogels GD. Methanopterin and methanogenic bacteria. Biofactors. 1988;1:95–103. [PubMed] [Google Scholar]
- 21.van Beelen P, Labro JF, Keltjens JT, Geerts WJ, Vogels GD, Laarhoven WH, Guijt W, Haasnoot CA. Derivatives of methanopterin, a coenzyme involved in methanogenesis. Eur J Biochem. 1984;139:359–365. doi: 10.1111/j.1432-1033.1984.tb08014.x. [DOI] [PubMed] [Google Scholar]
- 22.Keltjens JT, Daniels L, Jannsen HG, Borm PJ, Vogels GD. A novel one-carbon carrier (carboxy-5,6,7,8-tetrahydromethanopterin) isolated from Methanobacterium thermoautotrophicum and derived from methanopterin. Eur J Biochem. 1983;130:545–552. doi: 10.1111/j.1432-1033.1983.tb07184.x. [DOI] [PubMed] [Google Scholar]
- 23.Keltjens JT, Huberts MJ, Laarhoven WH, Vogels GD. Structural elements of methanopterin, a novel pterin present in Methanobacterium thermoautotrophicum. Eur J Biochem. 1983;130:537–544. doi: 10.1111/j.1432-1033.1983.tb07183.x. [DOI] [PubMed] [Google Scholar]
- 24.Abken HJ, Tietze M, Brodersen J, Baumer S, Beifuss U, Deppenmeier U. Isolation and characterization of methanophenazine and function of phenazines in membrane-bound electron transport of Methanosarcina mazei Go1. J Bacteriol. 1998;180:2027–2032. doi: 10.1128/jb.180.8.2027-2032.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pol A, van der Drift C, Vogels GD. Corrinoids from Methanosarcina barkeri: structure of the alpha-ligand. Biochem Biophys Res Commun. 1982;108:731–737. doi: 10.1016/0006-291x(82)90890-7. [DOI] [PubMed] [Google Scholar]
- 26.Harms U, Thauer RK. The corrinoid-containing 23-kDa subunit MtrA of the energy-conserving N5-methyltetrahydromethanopterin:coenzyme M methyltransferase complex from Methanobacterium thermoautotrophicum. EPR spectroscopic evidence for a histidine residue as a cobalt ligand of the cobamide. Eur J Biochem. 1996;241:149–154. doi: 10.1111/j.1432-1033.1996.0149t.x. [DOI] [PubMed] [Google Scholar]
- 27.Kengen SW, Daas PJ, Duits EF, Keltjens JT, van der Drift C, Vogels GD. Isolation of a 5-hydroxybenzimidazolyl cobamide-containing enzyme involved in the methyltetrahydromethanopterin: coenzyme M methyltransferase reaction in Methanobacterium thermoautotrophicum. Biochim Biophys Acta. 1992;1118:249–260. doi: 10.1016/0167-4838(92)90282-i. [DOI] [PubMed] [Google Scholar]
- 28.Krautler B, Moll J, Thauer RK. The corrinoid from Methanobacterium thermoautotrophicum (Marburg strain). Spectroscopic structure analysis and identification as Co beta-cyano-5'-hydroxybenzimidazolyl-cobamide (factor III) Eur J Biochem. 1987;162:275–278. doi: 10.1111/j.1432-1033.1987.tb10596.x. [DOI] [PubMed] [Google Scholar]
- 29.Sauer K, Thauer RK. Methanol:coenzyme M methyltransferase from Methanosarcina barkeri -- substitution of the corrinoid harbouring subunit MtaC by free cob(I)alamin. Eur J Biochem. 1999;261:674–681. doi: 10.1046/j.1432-1327.1999.00355.x. [DOI] [PubMed] [Google Scholar]
- 30.Sauer K, Thauer RK. Methanol:coenzyme M methyltransferase from Methanosarcina barkeri--identification of the active-site histidine in the corrinoid-harboring subunit MtaC by site-directed mutagenesis. Eur J Biochem. 1998;253:698–705. doi: 10.1046/j.1432-1327.1998.2530698.x. [DOI] [PubMed] [Google Scholar]
- 31.Sauer K, Thauer RK. Methanol:coenzyme M methyltransferase from Methanosarcina barkeri. Zinc dependence and thermodynamics of the methanol:cob(I)alamin methyltransferase reaction. Eur J Biochem. 1997;249:280–285. doi: 10.1111/j.1432-1033.1997.t01-1-00280.x. [DOI] [PubMed] [Google Scholar]
- 32.Hagemeier CH, Krer M, Thauer RK, Warkentin E, Ermler U. Insight into the mechanism of biological methanol activation based on the crystal structure of the methanol-cobalamin methyltransferase complex. Proc Natl Acad Sci U S A. 2006;103:18917–18922. doi: 10.1073/pnas.0603650103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klein AR, Thauer RK. Re-face specificity at C14a of methylenetetrahydromethanopterin and Si-face specificity at C5 of coenzyme F420 for coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase from methanogenic Archaea. Eur J Biochem. 1995;227:169–174. doi: 10.1111/j.1432-1033.1995.tb20373.x. [DOI] [PubMed] [Google Scholar]
- 34.Kunow J, Schworer B, Setzke E, Thauer RK. Si-face stereospecificity at C5 of coenzyme F420 for F420-dependent N5,N10-methylenetetrahydromethanopterin dehydrogenase, F420- dependent N5,N10-methylenetetrahydromethanopterin reductase and F420H2:dimethylnaphthoquinone oxidoreductase. Eur J Biochem. 1993;214:641–646. doi: 10.1111/j.1432-1033.1993.tb17964.x. [DOI] [PubMed] [Google Scholar]
- 35.Jaenchen R, Schonheit P, Thauer RK. Studies on the biosynthesis of coenzyme F420 in methanogenic bacteria. Arch Microbiol. 1984;137:362–365. doi: 10.1007/BF00410735. [DOI] [PubMed] [Google Scholar]
- 36.Coenzyme F420: another example of the diversity in structure and function of natural flavins. Nutr Rev. 1980;38:88–90. doi: 10.1111/j.1753-4887.1980.tb05849.x. [DOI] [PubMed] [Google Scholar]
- 37.Fielding AJ, Parey K, Ermler U, Scheller S, Jaun B, Bennati M. Advanced electron paramagnetic resonance on the catalytic iron-sulfur cluster bound to the CCG domain of heterodisulfide reductase and succinate: quinone reductase. J Biol Inorg Chem. 2013;18:905–915. doi: 10.1007/s00775-013-1037-x. [DOI] [PubMed] [Google Scholar]
- 38.Wang S, Tiongson J, Rasche ME. Discovery and Characterization of the First Archaeal Dihydromethanopterin Reductase, an Iron-Sulfur Flavoprotein from Methanosarcina mazei. J Bacteriol. 2014;196:203–209. doi: 10.1128/JB.00457-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lessner DJ, Ferry JG. The archaeon Methanosarcina acetivorans contains a protein disulfide reductase with an iron-sulfur cluster. J Bacteriol. 2007;189:7475–7484. doi: 10.1128/JB.00891-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korbas M, Vogt S, Meyer-Klaucke W, Bill E, Lyon EJ, Thauer RK, Shima S. The iron-sulfur cluster-free hydrogenase (Hmd) is a metalloenzyme with a novel iron binding motif. J Biol Chem. 2006;281:30804–30813. doi: 10.1074/jbc.M605306200. [DOI] [PubMed] [Google Scholar]
- 41.Hedderich R, Hamann N, Bennati M. Heterodisulfide reductase from methanogenic archaea: a new catalytic role for an iron-sulfur cluster. Biol Chem. 2005;386:961–970. doi: 10.1515/BC.2005.112. [DOI] [PubMed] [Google Scholar]
- 42.Lyon EJ, Shima S, Boecher R, Thauer RK, Grevels FW, Bill E, Roseboom W, Albracht SP. Carbon monoxide as an intrinsic ligand to iron in the active site of the iron-sulfur-cluster-free hydrogenase H2-forming methylenetetrahydromethanopterin dehydrogenase as revealed by infrared spectroscopy. J Am Chem Soc. 2004;126:14239–14248. doi: 10.1021/ja046818s. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi Y, Tokumoto U. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J Biol Chem. 2002;277:28380–28383. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- 44.Zhao T, Cruz F, Ferry JG. Iron-sulfur flavoprotein (Isf) from Methanosarcina thermophila is the prototype of a widely distributed family. J Bacteriol. 2001;183:6225–6233. doi: 10.1128/JB.183.21.6225-6233.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertram PA, Karrasch M, Schmitz RA, Bocher R, Albracht SP, Thauer RK. Formylmethanofuran dehydrogenases from methanogenic Archaea. Substrate specificity, EPR properties and reversible inactivation by cyanide of the molybdenum or tungsten iron-sulfur proteins. Eur J Biochem. 1994;220:477–484. doi: 10.1111/j.1432-1033.1994.tb18646.x. [DOI] [PubMed] [Google Scholar]
- 46.Kulzer R, Pils T, Kappl R, Huttermann J, Knappe J. Reconstitution and characterization of the polynuclear iron-sulfur cluster in pyruvate formate-lyase-activating enzyme. Molecular properties of the holoenzyme form. J Biol Chem. 1998;273:4897–4903. doi: 10.1074/jbc.273.9.4897. [DOI] [PubMed] [Google Scholar]
- 47.Grayling RA, Becktel WJ, Reeve JN. Structure and stability of histone HMf from the hyperthermophilic archaeon Methanothermus fervidus. Biochemistry. 1995;34:8441–8448. doi: 10.1021/bi00026a027. [DOI] [PubMed] [Google Scholar]
- 48.Ehlers C, Grabbe R, Veit K, Schmitz RA. Characterization of GlnK1 from Methanosarcina mazei strain Go1: complementation of an Escherichia coli glnK mutant strain by GlnK1. J Bacteriol. 2002;184:1028–1040. doi: 10.1128/jb.184.4.1028-1040.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richter H, Zoephel J, Schermuly J, Maticzka D, Backofen R, Randau L. Characterization of CRISPR RNA processing in Clostridium thermocellum and Methanococcus maripaludis. Nucleic Acids Res. 2012;40:9887–9896. doi: 10.1093/nar/gks737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ehlers C, Weidenbach K, Veit K, Forchhammer K, Schmitz RA. Unique mechanistic features of post-translational regulation of glutamine synthetase activity in Methanosarcina mazei strain Go1 in response to nitrogen availability. Mol Microbiol. 2005;55:1841–1854. doi: 10.1111/j.1365-2958.2005.04511.x. [DOI] [PubMed] [Google Scholar]
- 51.Torarinsson E, Klenk HP, Garrett RA. Divergent transcriptional and translational signals in Archaea. Environ Microbiol. 2005;7:47–54. doi: 10.1111/j.1462-2920.2004.00674.x. [DOI] [PubMed] [Google Scholar]
- 52.Karlin S, Brocchieri L, Trent J, Blaisdell BE, Mrazek J. Heterogeneity of genome and proteome content in bacteria, archaea, and eukaryotes. Theor Popul Biol. 2002;61:367–390. doi: 10.1006/tpbi.2002.1606. [DOI] [PubMed] [Google Scholar]
- 53.Bell SD, Jackson SP. Transcription and translation in Archaea: a mosaic of eukaryal and bacterial features. Trends Microbiol. 1998;6:222–228. doi: 10.1016/s0966-842x(98)01281-5. [DOI] [PubMed] [Google Scholar]
- 54.Koonin EV, Mushegian AR, Galperin MY, Walker DR. Comparison of archaeal and bacterial genomes: computer analysis of protein sequences predicts novel functions and suggests a chimeric origin for the archaea. Mol Microbiol. 1997;25:619–637. doi: 10.1046/j.1365-2958.1997.4821861.x. [DOI] [PubMed] [Google Scholar]
- 55.Makrides SC. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol Rev. 1996;60:512–538. doi: 10.1128/mr.60.3.512-538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saxena P, Walker JR. Expression of argU, the Escherichia coli gene coding for a rare arginine tRNA. J Bacteriol. 1992;174:1956–1964. doi: 10.1128/jb.174.6.1956-1964.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Makoff AJ, Oxer MD, Romanos MA, Fairweather NF, Ballantine S. Expression of tetanus toxin fragment C in E. coli: high level expression by removing rare codons. Nucleic Acids Res. 1989;17:10191–10202. doi: 10.1093/nar/17.24.10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pritchett MA, Zhang JK, Metcalf WW. Development of a markerless genetic exchange method for Methanosarcina acetivorans C2A and its use in construction of new genetic tools for methanogenic archaea. Appl Environ Microbiol. 2004;70:1425–1433. doi: 10.1128/AEM.70.3.1425-1433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guss AM, Rother M, Zhang JK, Kulkarni G, Metcalf WW. New methods for tightly regulated gene expression and highly efficient chromosomal integration of cloned genes for Methanosarcina species. Archaea. 2008;2:193–203. doi: 10.1155/2008/534081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Filutowicz M, McEachern MJ, Helinski DR. Positive and negative roles of an initiator protein at an origin of replication. Proc Natl Acad Sci U S A. 1986;83:9645–9649. doi: 10.1073/pnas.83.24.9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stalker DM, Filutowicz M, Helinski DR. Release of initiation control by a mutational alteration in the R6K pi protein required for plasmid DNA replication. Proc Natl Acad Sci U S A. 1983;80:5500–5504. doi: 10.1073/pnas.80.18.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas CM, Smith CA, Shingler V, Cross MA, Hussain AA, Pinkney M. Regulation of replication and maintenance functions of broad host-range plasmid RK2. Basic Life Sci. 1985;30:261–276. doi: 10.1007/978-1-4613-2447-8_21. [DOI] [PubMed] [Google Scholar]
- 63.Ko JH, Chung WJ, Koh S, Park BC, Kwon ST, Kim CH, Lee DS. Metal affinity engineering of proinsulin carrying genetically attached (His)10-X-Met affinity tail and removal of the tag by cyanogen bromide. Biosci Biotechnol Biochem. 1994;58:1694–1699. doi: 10.1271/bbb.58.1694. [DOI] [PubMed] [Google Scholar]
- 64.Voss S, Skerra A. Mutagenesis of a flexible loop in streptavidin leads to higher affinity for the Strep-tag II peptide and improved performance in recombinant protein purification. Protein Eng. 1997;10:975–982. doi: 10.1093/protein/10.8.975. [DOI] [PubMed] [Google Scholar]
- 65.Schmidt TG, Skerra A. One-step affinity purification of bacterially produced proteins by means of the "Strep tag" and immobilized recombinant core streptavidin. J Chromatogr A. 1994;676:337–345. doi: 10.1016/0021-9673(94)80434-6. [DOI] [PubMed] [Google Scholar]
- 66.Uetake H, Luria SE, Burrous JW. Conversion of somatic antigens in Salmonella by phage infection leading to lysis or lysogeny. Virology. 1958;5:68–91. doi: 10.1016/0042-6822(58)90006-0. [DOI] [PubMed] [Google Scholar]
- 67.Metcalf WW, Zhang JK, Shi X, Wolfe RS. Molecular, genetic, and biochemical characterization of the serC gene of Methanosarcina barkeri Fusaro. J Bacteriol. 1996;178:5797–5802. doi: 10.1128/jb.178.19.5797-5802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 69.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 70.Dower WJ, Miller JF, Ragsdale CW. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Metcalf WW, Zhang JK, Apolinario E, Sowers KR, Wolfe RS. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc Natl Acad Sci U S A. 1997;94:2626–2631. doi: 10.1073/pnas.94.6.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hildenbrand C, Stock T, Lange C, Rother M, Soppa J. Genome copy numbers and gene conversion in methanogenic archaea. J Bacteriol. 2011;193:734–743. doi: 10.1128/JB.01016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Keil B. Specificity of Proteolysis. Berlin Heidelberg: Springer; 1992. Essential Substrate Residues for Action of Endopeptidases; pp. 43–228. [Google Scholar]
- 74.Huntington JA. Molecular recognition mechanisms of thrombin. J Thromb Haemost. 2005;3:1861–1872. doi: 10.1111/j.1538-7836.2005.01363.x. [DOI] [PubMed] [Google Scholar]
- 75.Calo D, Kaminski L, Eichler J. Protein glycosylation in Archaea: sweet and extreme. Glycobiology. 2010;20:1065–1076. doi: 10.1093/glycob/cwq055. [DOI] [PubMed] [Google Scholar]
- 76.Eichler J, Adams MW. Posttranslational protein modification in Archaea. Microbiol Mol Biol Rev. 2005;69:393–425. doi: 10.1128/MMBR.69.3.393-425.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Forbes AJ, Patrie SM, Taylor GK, Kim YB, Jiang L, Kelleher NL. Targeted analysis and discovery of posttranslational modifications in proteins from methanogenic archaea by top-down MS. Proc Natl Acad Sci U S A. 2004;101:2678–2683. doi: 10.1073/pnas.0306575101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Macauley SR, Zimmerman SA, Apolinario EE, Evilia C, Hou YM, Ferry JG, Sowers KR. The archetype gamma-class carbonic anhydrase (Cam) contains iron when synthesized in vivo. Biochemistry. 2009;48:817–819. doi: 10.1021/bi802246s. [DOI] [PubMed] [Google Scholar]
- 79.Demolli S, Geist MM, Weigand JE, Matschiavelli N, Suess B, Rother M. Development of beta -Lactamase as a Tool for Monitoring Conditional Gene Expression by a Tetracycline-Riboswitch in Methanosarcina acetivorans. Archaea. 2014;2014:725610. doi: 10.1155/2014/725610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krzycki JA. Translation of UAG as Pyrrolysine. Nucleic Acids Mol Bi. 2010;24:53–77. [Google Scholar]
- 81.Ou W, Uno T, Chiu HP, Grunewald J, Cellitti SE, Crossgrove T, Hao X, Fan Q, Quinn LL, Patterson P, Okach L, Jones DH, Lesley SA, Brock A, Geierstanger BH. Site-specific protein modifications through pyrroline-carboxy-lysine residues. Proc Natl Acad Sci U S A. 2011;108:10437–10442. doi: 10.1073/pnas.1105197108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Longstaff DG, Blight SK, Zhang L, Green-Church KB, Krzycki JA. In vivo contextual requirements for UAG translation as pyrrolysine. Mol Microbiol. 2007;63:229–241. doi: 10.1111/j.1365-2958.2006.05500.x. [DOI] [PubMed] [Google Scholar]
- 83.Beier H, Grimm M. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res. 2001;29:4767–4782. doi: 10.1093/nar/29.23.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.