Summary
Objectives
As individuals may be colonized with multiple strains of Staphylococcus aureus at different body sites, the objectives of this study were to determine whether S. aureus polyclonal colonization exists within one body niche and the optimal sampling sites and culture methodology to capture the diversity of S. aureus strains in community-dwelling individuals.
Methods
Swabs were collected from the nares, axillae, and inguinal folds of 3 children with community-associated S. aureus infections and 11 household contacts, all with known S. aureus colonization. S. aureus isolates were recovered from each body niche using 4 culture methods and evaluated for polyclonality using phenotypic and genotypic strain characterization methodologies.
Results
Within individuals, the mean (range) number of phenotypes and genotypes was 2.4 (1–4) and 3.1 (1–6), respectively. Six (43%) and 10 (71%) participants exhibited phenotypic and genotypic polyclonality within one body niche, respectively. Broth enrichment yielded the highest analytical sensitivity for S. aureus recovery, while direct plating to blood agar yielded the highest genotypic strain diversity.
Conclusions
This study revealed S. aureus polyclonality within a single body niche. Culture methodology and sampling sites influenced the analytical sensitivity of S. aureus colonization detection and the robustness of phenotypic and genotypic strain recovery.
Keywords: Staphylococcus aureus, polyclonal colonization, strain diversity, broth enrichment
Introduction
Staphylococcus aureus is a commensal bacterium with the potential to cause community-associated and nosocomial infections.1,2 S. aureus often resides in the anterior nares, oropharynx, inguinal folds, axillae, and rectum of otherwise healthy people.3–6 Colonization with S. aureus is a known risk factor for skin and soft tissue infections; in extreme cases, S. aureus may cause invasive life-threatening infections such as osteomyelitis, bacteremia, and pneumonia.7,8
Within an individual, multiple colonizing strain types of S. aureus have been recovered from different anatomical niches.9 This finding raises the inquiry of polyclonality within an anatomic niche. S. aureus polyclonality is an important consideration from a biological and clinical standpoint. The simultaneous carriage of more than one subtype of S. aureus at one body site opens the opportunity for horizontal gene transfer; the transfer of antibiotic resistance and virulence factors between co-colonizing strains could contribute to the pathogenicity of the particular isolate.10
When studying the carriage and transmission of S. aureus, it is important to not only consider the prevalence of polyclonal colonization within one niche, but also the culture methods resulting in the recovery of the highest strain diversity. The majority of studies rely on the selection and analysis of a single colony from a positive culture plate, not accounting for polyclonality despite its potential importance to molecular epidemiology investigations.11 Polyclonality has primarily been studied in healthcare settings or immunocompromised patients. These studies have demonstrated that 7–30% of individuals colonized with S. aureus possess multiple strain types within one body niche.10,12 While important, these findings within populations with healthcare exposure may be limited in their application to individuals in community settings.
In the current study, we sought to address the issue of intra-individual S. aureus strain diversity in pediatric index patients diagnosed with community-associated methicillin-resistant S. aureus (CA-MRSA) infections and their household contacts. Using genotypic and phenotypic analysis, the objective of this study was to determine whether multiple strain types of S. aureus coexist in one body niche within an individual. Additionally, we sought to determine the optimal sampling sites and culture methodology to capture the greatest diversity of S. aureus strains within a population.
Methods
Participant Selection
This study population was derived from a longitudinal study and consisted of 14 participants, all of whom were colonized with S. aureus: 3 pediatric index patients with cutaneous or invasive CA-MRSA infections and 11 healthy household contacts of these patients. The cohort was recruited from Saint Louis Children’s Hospital and community pediatric practices as described previously.13 At enrollment, swabs (Eswab, Becton Dickinson [BD], Franklin Lakes, NJ) were collected for culture from the anterior nares, axillae, and inguinal folds of each participant to determine S. aureus colonization status. In the present study, S. aureus isolates recovered from the same niche (i.e. the axillae, inguinal folds, or anterior nares) of 14 participants (designated A-N) were evaluated for polyclonality as described below. This study was approved by the Washington University School of Medicine Human Research Protection Office. Written informed consent was obtained from all participants.
Laboratory Procedures
Culture methods
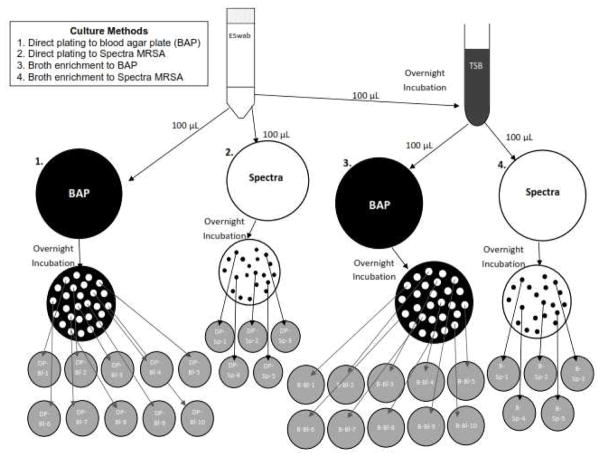
From each culture swab, 100 μL of Eswab eluate was inoculated onto each of 1) trypticase soy agar with 5% sheep blood agar plate (BAP) (BBL, BD), 2) Spectra MRSA (Remel, Lenexa, KS) (a chromogenic agar selective and differential for MRSA), and 3) tryptic soy broth with 6.5% NaCl (BBL, BD), and then incubated overnight at 35°C. From the broth cultures, 100 μL aliquots were plated to BAP and Spectra MRSA and incubated at 35°C overnight. For each BAP, up to 10 colonies consistent with S. aureus (large, cream to golden colored and β-hemolytic) were selected by an experienced microbiologist; for each Spectra MRSA, up to 5 colonies consistent with S. aureus (denim blue in color) were selected. Each colony selected from BAP and Spectra MRSA were subsequently subcultured to individual BAPs and incubated overnight at 35°C ( Figure 1).
Figure 1.
Four separate culture methods were compared: 1) direct plating to blood agar plate (BAP), 2) direct plating to Spectra MRSA, 3) broth enrichment with subculture to BAP, and 4) broth enrichment with subculture to Spectra MRSA.
In all, 4 separate culture methods were compared: 1) direct plating to BAP, 2) direct plating to Spectra MRSA, 3) broth enrichment with subculture to BAP, and 4) broth enrichment with subculture to Spectra MRSA (Figure 1). Results from a rapid latex agglutination test for S. aureus identification (Staphaurex, Remel), catalase activity, and Gram staining confirmed the identity of S. aureus isolates. Variations exist in the number of isolates recovered from each individual in the study due to instances where fewer colonies consistent with S. aureus were present or isolates selected were subsequently identified to be an organism other than S. aureus.
Phenotypic analysis
Antibiotic susceptibility testing for each clone was performed on Mueller-Hinton agar (BBL, BD) using the Kirby-Bauer disk-diffusion method and interpreted in accordance with Clinical and Laboratory Standards Institute 2012 Guidelines.14 Susceptibility to cefoxitin (as a surrogate for methicillin resistance), clindamycin (including inducible clindamycin resistance as determined by the double disk diffusion D-test), erythromycin, trimethoprim-sulfamethoxazole (TMP-SXT), rifampin, tetracycline, ciprofloxacin, linezolid, and ceftaroline were recorded; for the purpose of analysis, isolates with a positive D-test were classified as clindamycin resistant.15 Unique antibiotic susceptibility patterns were assigned a designation (AA, BB, etc.), followed by an –R or –S annotation, reflecting resistance or susceptibility to methicillin, respectively (Table 1).
Table 1.
Phenotypic Staphylococcus aureus strain characterization by antibiotic susceptibility patterns
| Antibiotic susceptibility patterna | Methicillinb | Clindamycinc | Erythromycin | Ciprofloxacin |
|---|---|---|---|---|
| AA-R | Res | Susc | Res | Susc |
| BB-R | Res | Susc | Susc | Susc |
| CC-R | Res | Susc | Susc | Inter |
| DD-R | Res | Susc | Res | Res |
| EE-S | Susc | Susc | Susc | Susc |
| FF-S | Susc | Res | Res | Susc |
Abbreviations: Res, resistant; Susc, susceptible; Inter, intermediate
Note: All isolates were susceptible to trimethoprim-sulfamethoxazole, rifampin, tetracycline, linezolid, and ceftaroline.
The –R or –S designation to the strain classification reflects resistance (-R) or susceptibility (-S) to methicillin, respectively.
As predicted by cefoxitin testing.
One isolate was both clindamycin susceptible and D-test positive and thus considered clindamycin resistant.
Genotypic analysis
DNA extraction for each clone was performed using the BiOstic Bacteremia DNA kit (MoBio Laboratories, Carlsbad, CA). Strain typing by repetitive-sequence PCR (repPCR) was performed as described previously.13,16 For the purpose of our analyses, distinct repPCR strains were assigned a designation (RS1, RS2, etc).
Statistical Analysis
Data were analyzed with descriptive statistics using SPSS 22 for Windows (IBM SPSS, Chicago, IL). The numbers of colonies needed to be selected in order to detect all strain types within each body niche, as well as the strain diversity recovered by sampling multiple body sites, were calculated. T-tests were used to compare mean colonization burden across levels of genotypic strain diversity. The analytical sensitivity of each method was calculated using the following formula:
Methods of characterization were compared by the discriminatory index (D), calculated using the standard formula for this metric:17
where N is the total number of strains in the sample population, S is the total number of subtypes described, and nj is the number of strains belonging to each of the subtypes. A value of 1 is considered to be highly discriminatory and a value of 0 is not discriminatory.17
Results
Of 14 participants, 8 (57%) were male, 10 (71%) were African-American, and the median age was 8.5 years (range 0.5–37 years). Colonization cultures collected from the anterior nares, axillae, and inguinal folds by all culture methods yielded 336 S. aureus isolates. By niche, 178 (53%) isolates were from the anterior nares, 74 (22%) were from the axillae and 84 (25%) were from the inguinal folds. Six subjects were colonized at 1 body site (most commonly the nares) and 8 were colonized at multiple body sites. Fifteen repPCR strain types (genotypes) were recovered; 1 strain type (designated RS7) comprised 39% of all isolates (Table 2). Based on patterns of antibiotic resistance (phenotypes), a total of 2 methicillin-susceptible S. aureus (MSSA) phenotypes and 4 MRSA phenotypes were recovered (Table 1). All isolates recovered in this investigation were susceptible to TMP-SXT, rifampin, tetracycline, linezolid, and ceftaroline.
Table 2.
Genotypic and phenotypic Staphylococcus aureus strain characterization by body site and culture method
| Number of isolates collected by body niche and culture method
| |||||||
|---|---|---|---|---|---|---|---|
| repPCR strain type | Phenotype | Anterior Nares | Axillae | Inguinal folds | |||
|
| |||||||
| Broth enrichment | Direct plating | Broth enrichment | Direct plating | Broth enrichment | Direct plating | ||
| RS1, n=1 (%) | EE-S | 33 | 33 | 1 (100) | 33 | ||
|
| |||||||
| RS2, n=61 (%) | EE-S | 19 (31) | 12 (20) | 17 (28) | 1 (2) | 4 (6) | 8 (13) |
|
| |||||||
| RS3, n=28 (%) | AA-R | 1 (3.5) | |||||
| EE-S | 6 (21) | 5 (18) | 3 (11) | 3 (11) | 9 (32) | 1 (3.5) | |
|
| |||||||
| RS4, n=29 (%) | AA-R | 2 (7) | |||||
| EE-S | 17 (59) | 10 (34) | 3 | ||||
|
| |||||||
| RS5, n=1 (%) | EE-S | 33 | 1 (100) | ||||
|
| |||||||
| RS6, n=3 (%) | EE-S | 3 (100) | |||||
|
| |||||||
| RS7, n=130 (%) | AA-R | 36 (28) 3 | 15 (12) | 16 (12) | 3 (2) | 11 (8) 3 | 16 (12) |
| BB-R | 6 (4) | 2 (2) | 33 | ||||
| CC-R | 1 (1) | 33 | |||||
| DD-R | 15 (12) | 33 | |||||
| EE-S | 2 (2) | 7 (5) | 33 | ||||
|
| |||||||
| RS8, n=38 (%) | AA-R | 2 (5) | 1 (3) | ||||
| EE-S | 3 (8) | 1 (3) | 7 (18) 3 | 6 (16) | 13 (34) | 4 (10) | |
| FF-S | 1 (3) | ||||||
|
| |||||||
| RS9, n=1 (%) | EE-S | 33 | 33 | 1 (100) | |||
|
| |||||||
| RS10, n=35 (%) | EE-S | 10 (28) | 16 (46) | 1 (3) | 8 (23) | ||
|
| |||||||
| RS11, n=1 (%) | AA-R | 33 | 1 (100) | 3 | 33 | ||
|
| |||||||
| RS12, n=1 (%) | AA-R | 1 (100) | |||||
|
| |||||||
| RS13, n=1 (%) | EE-S | 1 (100) 3 | |||||
|
| |||||||
| RS14, n=5 (%) | EE-S | 2 (40) | 3 (60) | ||||
|
| |||||||
| RS15, n=1 (%) | EE-S | 33 | 33 | 1 (100) | 33 | ||
Abbreviations: repPCR, repetitive-sequence PCR
Note: Phenotype descriptions are listed in Table 1. Percentages represent proportion of isolates within each repPCR strain type.
A discriminatory index was calculated for each method of characterization: genotypic, phenotypic, and a combination of the genotypic and phenotypic strain characterization methods (aggregate). RepPCR was more discriminatory than antibiotic susceptibility patterns, but the aggregate resulted in the highest discriminatory power (Table 3).
Table 3.
Discriminatory indices of strain characterization methods within participants
| Participant ID | Strain characterization method
|
|||||
|---|---|---|---|---|---|---|
| Genotypic | Phenotypic | Aggregatea | ||||
|
| ||||||
| # Distinct strains | Discriminatory index | # Distinct strains | Discriminatory index | # Distinct strains | Discriminatory index | |
| A | 6 | 0.60 | 1 | n/a | 6 | 0.60 |
| B | 5 | 0.40 | 4 | 0.51 | 9 | 0.61 |
| C | 6 | 0.78 | 3 | 0.43 | 7 | 0.81 |
| D | 1 | n/a | 1 | n/a | 1 | n/a |
| E | 6 | 0.71 | 2 | 0.47 | 8 | 0.83 |
| F | 3 | 0.29 | 1 | n/a | 3 | 0.29 |
| G | 2 | 0.60 | 2 | 0.60 | 2 | 0.60 |
| H | 2 | 0.34 | 1 | n/a | 2 | 0.34 |
| I | 5 | 0.76 | 2 | 0.29 | 5 | 0.76 |
| J | 3 | 0.59 | 3 | 0.33 | 5 | 0.64 |
| K | 1 | n/a | 1 | n/a | 1 | n/a |
| L | 2 | 0.54 | 1 | n/a | 2 | 0.54 |
| M | 1 | n/a | 1 | n/a | 1 | n/a |
| N | 1 | n/a | 1 | n/a | 1 | n/a |
Abbreviations: n/a, not applicable
Aggregate represents the combination of genotypic and phenotypic strain characterization methods.
Of note, some S. aureus repPCR strain types appeared to be commonly detected together, i.e. they were only recovered when a certain other strain type was present in the same body niche of the same participant. This pattern was observed for repPCR strain type RS8 (correlated with RS7), RS10 (correlated with RS2) and RS4 (correlated with RS7). Polyclonality with these strains was not specific to a particular niche.
Strain Diversity by Culture Method
Within each body niche, the analytical sensitivity of each culture method to recover S. aureus was determined. Broth enrichment to BAP was the most sensitive of the four culture methods with 100% recovery of S. aureus and 75% recovery of MRSA from both the anterior nares and axillae and 100% recovery of both S. aureus and MRSA from the inguinal folds. The direct plating to BAP culture method was the second most sensitive recovering 42%, 57%, and 63% of S. aureus isolates from the anterior nares, axillae, and inguinal folds, respectively (Table 4). In all circumstances in which direct plating techniques yielded S. aureus isolates, broth enrichment techniques also recovered S. aureus isolates.
Table 4.
Analytical sensitivity for recovering Staphylococcus aureus by different culture methods within a body niche
| Anterior Nares | Axillae | Inguinal Folds | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| All S. aureus n=12 (%) | MRSA only n=8 (%) | All S. aureus n=7 (%) | MRSA only n=4 (%) | All S. aureus n=8 (%) | MRSA only n=3 (%) | |
| Direct plating to BAP | 5 (42%) | 1 (13%) | 4 (57%) | 1 (25%) | 5 (63%) | 3 (100%) |
| Direct plating to Spectra MRSAa | 3 (25%) | 3 (38%) | n/a | n/a | 2 (25%) | 2 (67%) |
| Broth enrichment to BAP | 12 (100%) | 6 (75%) | 7 (100%) | 3 (75%) | 8 (100%) | 3 (100%) |
| Broth enrichment to Spectra MRSAa | 4 (33%) | 4 (50%) | 1 (14%) | 1 (25%) | 1 (13%) | 1 (33%) |
Abbreviations: MRSA, methicillin-resistant S. aureus; BAP, blood agar plate
Spectra MRSA is selective for the growth of MRSA.
The broth enrichment to BAP culture method recovered 201 isolates (60% of all recovered isolates), yielding 9 distinct genotypes. Of note, 4 genotypes were uniquely recovered by this method (RS1, RS4, RS9, RS13). The direct plating to BAP culture method recovered 88 isolates (26%), yielding 11 distinct genotypes; 5 genotypes were only recovered by this method (RS5, RS6, RS12, RS14, RS15). The broth enrichment to Spectra MRSA culture method recovered 28 isolates (8%), yielding 1 distinct genotype. The direct plating to Spectra MRSA method recovered 19 isolates (6%), yielding 2 distinct genotypes; genotype RS11 was solely recovered by this method. Overall, broth enrichment culture methods recovered more phenotypes (6) than direct plating methods (3). The mean genotypic strain diversity recovered by the direct plating to BAP culture method was higher than the broth enrichment to BAP culture methods (Table 5).
Table 5.
Strain diversity by culture methods and body niche
| Sites | Overall | METHODS
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Direct plating to BAP | Broth enrichment to BAP | Direct plating to Spectra MRSA | Broth enrichment to Spectra MRSA | |||||||||||||||||
|
| ||||||||||||||||||||
| Genotypic | Phenotypic | Genotypic | Phenotypic | Genotypic | Phenotypic | Genotypic | Phenotypic | Genotypic | Phenotypic | |||||||||||
|
| ||||||||||||||||||||
| Mean | Median (Range) |
Mean | Median (Range) |
Mean | Median (Range) |
Mean | Median (Range) |
Mean | Median (Range) |
Mean | Median (Range) |
Mean | Median (Range) |
Mean | Median (Range) |
Mean | Median (Range) |
Mean | Median (Range) |
|
| All sites | 3.1 | 2.5 (1–6) | 1.7 | 1.0 (1–4) | 2.1 | 2.0 (1–4) | 1.3 | 1.0 (1–3) | 2.3 | 2.0 (1–4) | 1.5 | 1.0 (1–4) | 1.3 | 1.0 (1–2) | 1.0 | 1.0 (1–1) | 1.0 | 1.0 (1–1) | 1.3 | 1.0 (1–2) |
| Anterior Nares | 2.8 | 2.0 (1–6) | 1.6 | 1.0 (1–4) | 2.6 | 2.0 (2–4) | 1.4 | 1.0 (1–3) | 1.8 | 1.5 (1–3) | 1.3 | 1.0 (1–3) | 1.3 | 1.0 (1–2) | 1.0 | 1.0 (1–1) | 1.0 | 1.0 (1–1) | 1.3 | 1.0 (1–2) |
| Axillae | 2.0 | 2.0 (1–4) | 1.4 | 1.0 (1–2) | 2.3 | 2.0 (2–3) | 1.3 | 1.0 (1–2) | 1.6 | 1.0 (1–3) | 1.7 | 2.0 (1–2) | n/a | n/a | n/a | n/a | 1.0 | 1.0 (1–1) | 1.0 | 1.0 (1–1) |
| Inguinal Folds | 1.9 | 2.0 (1–3) | 1.3 | 1.0 (1–2) | 1.4 | 1.0 (1–2) | 1.2 | 1.0 (1–2) | 1.9 | 2.0 (1–3) | 1.3 | 1.0 (1–2) | 1.0 | 1.0 (1–1) | 1.0 | 1.0 (1–1) | 1.0 | 1.0 (1–1) | 1.0 | 1.0 (1–1) |
Abbreviations: BAP, blood agar plate; MRSA, methicillin-resistant S. aureus
Note: n/a – no isolates recovered using direct plating to Spectra MRSA from axillae cultures. Data represented as the mean and median (range) number of genotypic and phenotypic strain types by culture method at each body site. Range represents the minimum and maximum number of strain types recovered.
Strain Diversity within Individuals
The mean number of isolates recovered per participant (by all culture methods) was 24 (range 5–53). Five participants were colonized exclusively with MSSA (36%), 3 exclusively with MRSA (21%), and 6 with both MRSA and MSSA (43%). Both MRSA and MSSA phenotypes were recovered within a single body niche from all 6 individuals in the latter group. The mean number of phenotypes detected within an individual was 2.4 (range 1–4). The mean number of genotypes recovered from an individual was 3.1 (range 1–6).
Using the number of isolates recovered per participant as a surrogate for burden of S. aureus colonization, patients more heavily colonized with S. aureus had greater genotypic strain diversity. On average, 12 isolates were recovered from participants with 1 S. aureus genotype, while 29 isolates were recovered from participants with multiple genotypes (p=0.049). Employing the median number of genotypes to generate groups for comparison, 36 isolates were recovered from participants with more than 2 genotypes, compared to 12 isolates in participants with 2 or less genotypes (p<0.001).
Strain Diversity by Body Niche
The mean number of isolates recovered per body niche (by all culture methods) was 12 (range 3–28). Participants were colonized most frequently in the anterior nares (12, 86%), followed by the inguinal folds (8, 57%), and axillae (7, 50%). MRSA constituted 46% (82 of 178) of the isolates recovered from the anterior nares, 24% (18 of 74) of the axillae isolates, and 33% (28 of 84) of the inguinal folds isolates. Eight of 14 individuals were colonized with S. aureus in multiple body niches: 5 were colonized in all three body niches, 2 were colonized in the nares and inguinal folds, and 1 was colonized in the nares and axillae. The remaining 6 individuals were colonized exclusively in one body site: 4 in the nares, 1 in the axillae, and 1 in the inguinal folds.
Six (43%) participants exhibited phenotypic polyclonality within a single body niche. Two of these participants (33%) exhibited polyclonality at all body niches in which S. aureus was recovered. The body niches most frequently colonized with multiple phenotypes were the anterior nares and axillae (3 of 6 participants each, 50%), while 2 participants (33%) had different phenotypes recovered from the inguinal folds. The most predominant phenotypes were AA-R and EE-S; both were recovered from all three body sites. Phenotype FF-S was only recovered from the inguinal folds while BB-R, CC-R, and DD-R were only recovered from the anterior nares. The median number of colonies needed to be selected per plate to detect all the phenotypic strain diversity within a body niche (by all 4 culture methodologies) was 1.
Ten (71%) participants exhibited genotypic polyclonality within one body niche. Of these, 8 (80%) exhibited polyclonality in each body niche in which S. aureus was recovered. The body niche most frequently colonized with multiple S. aureus genotypes was the axillae (5 of 7 participants, 71%) followed by the anterior nares (8 of 12 participants, 67%) and inguinal folds (5 of 8 participants, 63%). Five repPCR strain types were recovered from all three body sites (RS2, RS3, RS7, RS8, RS10), with RS7 being the predominant strain type in all three sites. Strain types RS5, RS6, RS11, RS12 and RS13 were unique to the anterior nares, RS1 and RS15 were unique to the axillae, and RS9 was unique to the inguinal folds. The median number of colonies needed to be selected to detect all the genotypic strain diversity within a body niche was 3 (by direct to BAP culture methodology), 2 (by broth enrichment to BAP), and 1 (by both direct and broth enrichment to Spectra).
Selecting one colony from a sample derived from each of the three body niches within a participant (anterior nares, axillae, and inguinal folds) recovered a majority of the genotypic and phenotypic strain diversity. Nine of the 15 genotypes and 4 of the 6 phenotypes were recovered. The remaining 6 genotypes and 2 phenotypes not recovered by selecting just one colony from each body niche were each recovered only once in the study population.
Discussion
While S. aureus colonization is a symbiotic state for many individuals, for some, this colonization poses risk for subsequent infection or transmission of the organism to others.4,5 Given this risk, active surveillance is frequently conducted in healthcare settings to identify carriers of MRSA and subsequently implement precautions to prevent transmission to other patients. Given the varying burden of organism at various body niches, the sensitivity of the culture method to detect colonized individuals is an important consideration. Several investigators have demonstrated the advantage of broth enrichment in the recovery of S. aureus. Mernelius et al, determined that the use of broth enrichment increased recovery of S. aureus isolates by 46%.18,19 In the present study, participants were swabbed at 3 body sites (anterior nares, axillae, and inguinal folds) and 4 culture methods were employed, including broth enrichment, to detect S. aureus colonization. This study confirmed that inclusion of a broth enrichment step increases the recovery of S. aureus isolates. The omission of broth enrichment would have led to falsely negative cultures in 6 of 14 participants (43%), while the omission of the direct plating culture method would not have led to any falsely negative cultures for S. aureus recovery. An important consideration is the finding that, in the present study, broth enrichment resulted in slightly decreased genotypic strain diversity than direct plating to BAP, likely due to the propagation of a predominant clone which out-competes other strains. Investigators should consider overall study goals when selecting optimization of S. aureus strain type diversity vs. recovery. As expected, Spectra MRSA culture methods yielded fewer colonies given the lower number of colonies chosen (5, rather than 10) and the MRSA selectivity of Spectra MRSA.
Consistent with prior investigations, sampling multiple body niches in the present cohort also increased the recovery of S. aureus isolates and strain diversity.4–6,20,21 In a community-based study of children presenting with S. aureus skin infections and their household contacts, participants were swabbed in the anterior nares, axillae, and inguinal folds to detect S. aureus colonization. Obtaining extra-nasal cultures resulted in identification of 32% of S. aureus- colonized individuals, and specifically 41% of MRSA-colonized individuals, who would not have been detected if only the anterior nares had been sampled.5 In the present study, had the anterior nares been the only body site sampled, S. aureus colonization would not have been detected in 14% of individuals; 47% of isolates and 3 genotypes would not have been recovered.
The present study of community-dwelling participants also aimed to determine whether multiple strain types of S. aureus colonize an individual and whether polyclonal colonization exists within one body niche within an individual. Several prior studies have addressed this question in populations either associated with a healthcare setting or considered immunocompromised. Mongkolrattanothai and colleagues analyzed nasal and perianal samples of children undergoing elective surgery for polyclonal colonization and found that 30% of positive S. aureus swabs contained multiple genotypic strains as determined by multiple-locus variable-number tandem-repeat fingerprinting (MLVF).12 In a study of nasal colonization in current and past intravenous drug users, Cespedes et al. employed pulsed-field gel electrophoresis (PFGE) as well as multilocus sequence (MLST), S. aureus protein A (spa), and accessory gene regulator (agr) typing to develop a mathematical prediction model of polyclonality among colonized individuals. This model predicted 7% of S. aureus-colonized individuals would carry more than one strain.10 Using genotypic and phenotypic analysis, we determined that 71% of our study population was colonized with more than one strain of S. aureus overall, and more specifically, within the same body niche. Of note, polyclonality was consistent across body sites.
This study has several limitations. First, although our study population was limited, we sampled multiple body sites and interrogated a large number of isolates with multiple culture methods and strain characterization methods. Additionally, due to low colonization abundance for some subjects, the inability to consistently collect 10 colonies for analysis from BAP and 5 colonies from Spectra MRSA precluded normalization of the results.
This study revealed the presence of S. aureus polyclonality within a single body niche. Further, selection of culture method and sampling sites influenced the analytical sensitivity of S. aureus colonization detection, as well as the robustness of genotypic and phenotypic strain recovery. The analysis of co-colonizing strains may contribute to a better understanding of horizontal gene transfer. Additionally, these analyses may inform the mechanisms by which co-colonizing strains interact and influence the progression to or protection from infection and disease states. Prospective studies following individuals with polyclonal colonization will address these pertinent questions.
Highlights.
S. aureus polyclonality was assessed by phenotypic and genotypic methods.
Polyclonality existed within a single body niche in community-dwelling individuals.
Inclusion of broth enrichment increased recovery of S. aureus isolates.
Sampling multiple body niches increased recovery and diversity of S. aureus isolates.
Acknowledgments
We thank David Hunstad, MD, for project inspiration. Funding for this project was provided by the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital; National Institutes of Health grants K23-AI091690 and UL1-TR000448; and grant R01-HS021736 from the Agency for Healthcare Research and Quality. These funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Agency for Healthcare Research and Quality.
Footnotes
Conflicts of interest
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hulten KG, Kaplan SL, Gonzalez BE, Hammerman WA, Lamberth LB, Versalovic J, et al. Three-year surveillance of community onset health care-associated Staphylococcus aureus infections in children. Pediatr Infect Dis J. 2006;25(4):349–53. doi: 10.1097/01.inf.0000207404.50143.1e. [DOI] [PubMed] [Google Scholar]
- 2.Naimi TS, LeDell KH, Como-Sabetti K, Borchardt SM, Boxrud DJ, Etienne J, et al. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA. 2003;290(22):2976–84. doi: 10.1001/jama.290.22.2976. [DOI] [PubMed] [Google Scholar]
- 3.Faden H, Lesse AJ, Trask J, Hill JA, Hess DJ, Dryja D, et al. Importance of colonization site in the current epidemic of staphylococcal skin abscesses. Pediatrics. 2010;125(3):e618–624. doi: 10.1542/peds.2009-1523. [DOI] [PubMed] [Google Scholar]
- 4.Fritz SA, Camins BC, Eisenstein KA, Fritz JM, Epplin EK, Burnham CA, et al. Effectiveness of measures to eradicate Staphylococcus aureus carriage in patients with community-associated skin and soft-tissue infections: a randomized trial. Infect Control Hosp Epidemiol. 2011;32(9):872–80. doi: 10.1086/661285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fritz SA, Hogan PG, Hayek G, Eisenstein KA, Rodriguez M, Krauss M, et al. Staphylococcus aureus colonization in children with community-associated Staphylococcus aureus skin infections and their household contacts. Arch Pediatr Adolesc Med. 2012;166(6):551–7. doi: 10.1001/archpediatrics.2011.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller LG, Eells SJ, Taylor AR, David MZ, Ortiz N, Zychowski D, et al. Staphylococcus aureus colonization among household contacts of patients with skin infections: risk factors, strain discordance, and complex ecology. Clin Infect Dis. 2012;54(11):1523–35. doi: 10.1093/cid/cis213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lina G, Piemont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29(5):1128–32. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 8.Pannaraj PS, Hulten KG, Gonzalez BE, Mason EO, Jr, Kaplan SL. Infective pyomyositis and myositis in children in the era of community-acquired, methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis. 2006;43(8):953–60. doi: 10.1086/507637. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez M, Hogan PG, Burnham CA, Fritz SA. Molecular epidemiology of Staphylococcus aureus in households of children with community-associated S. aureus skin and soft tissue infections. J Pediatr. 2014;164(1):105–111. doi: 10.1016/j.jpeds.2013.08.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cespedes C, Said-Salim B, Miller M, Lo SH, Kreiswirth BN, Gordon RJ, et al. The clonality of Staphylococcus aureus nasal carriage. J Infect Dis. 2005;191(3):444–52. doi: 10.1086/427240. [DOI] [PubMed] [Google Scholar]
- 11.Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev. 1997;10(3):505–520. doi: 10.1128/cmr.10.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mongkolrattanothai K, Gray BM, Mankin P, Stanfill AB, Pearl RH, Wallace LJ, et al. Simultaneous carriage of multiple genotypes of Staphylococcus aureus in children. J Med Microbiol. 2011;60(Pt 3):317–22. doi: 10.1099/jmm.0.025841-0. [DOI] [PubMed] [Google Scholar]
- 13.Fritz SA, Hogan PG, Singh LN, Thompson RM, Wallace MA, Whitney K, et al. Contamination of environmental surfaces with Staphylococcus aureus in households with children infected with methicillin-resistant S. aureus. JAMA Pediatr. 2014 doi: 10.1001/jamapediatrics.2014.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; twenty-second informational supplement. Clinical and Laboratory Standards Institute; Wayne, PA: 2012. [Google Scholar]
- 15.Lewis JS, 2nd, Jorgensen JH. Inducible clindamycin resistance in Staphylococci: should clinicians and microbiologists be concerned? Clin Infect Dis. 2005;40(2):280–5. doi: 10.1086/426894. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez M, Hogan PG, Satola SW, Crispell E, Wylie T, Gao H, et al. Discriminatory indices of typing methods for epidemiologic analysis of contemporary Staphylococcus aureus strains. Medicine (Baltimore) 2015;94(37):e1534. doi: 10.1097/MD.0000000000001534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26(11):2465–6. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mernelius S, Lofgren S, Lindgren PE, Matussek A. The role of broth enrichment in Staphylococcus aureus cultivation and transmission from the throat to newborn infants: results from the Swedish hygiene intervention and transmission of S. aureus study. Eur J Clin Microbiol Infect Dis. 2013;32(12):1593–8. doi: 10.1007/s10096-013-1917-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Ogtrop ML. Effect of broth enrichment cultures on ability to detect carriage of Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39(9):2169. doi: 10.1128/aac.39.9.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McAllister SK, Albrecht VS, Fosheim GE, Lowery HK, Peters PJ, Gorwitz R, et al. Evaluation of the impact of direct plating, broth enrichment, and specimen source on recovery and diversity of methicillin-resistant Staphylococcus aureus isolates among HIV-infected outpatients. J Clin Microbiol. 2011;49(12):4126–30. doi: 10.1128/JCM.05323-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lauderdale TL, Wang JT, Lee WS, Huang JH, McDonald LC, Huang IW, et al. Carriage rates of methicillin-resistant Staphylococcus aureus (MRSA) depend on anatomic location, the number of sites cultured, culture methods, and the distribution of clonotypes. Eur J Clin Microbiol Infect Dis. 2010;29(12):1553–9. doi: 10.1007/s10096-010-1042-8. [DOI] [PubMed] [Google Scholar]