Abstract
Osteoclasts are resorptive cells important for homeostatic bone remodeling and pathological bone resorption. Emerging evidence suggests an important role for epigenetic mechanisms in osteoclastogenesis. A recent study showed that epigenetic silencing of the negative regulator of osteoclastogenesis Irf8 by DNA methylation is required for osteoclast differentiation. In this study, we investigated the role of EZH2, which epigenetically silences gene expression by histone methylation, in osteoclastogenesis. Inhibition of EZH2 by the small molecule GSK126, or decreasing its expression using antisense oligonucleotides, impeded osteoclast differentiation. Mechanistically, EZH2 was recruited to the IRF8 promoter after RANKL stimulation to deposit the negative histone mark H3K27me3 and downregulate IRF8 expression. GSK126 attenuated bone loss in the ovariectomy mouse model of postmenopausal osteoporosis. Our findings provide evidence for an additional mechanism of epigenetic IRF8 silencing during osteoclastogenesis that likely works cooperatively with DNA methylation, further emphasizing the importance of IRF8 as a negative regulator of osteoclastogenesis.
Introduction
Osteoclasts are large, multi-nucleated cells that are the primary effectors of bone resorption, making them indispensible for bone remodeling and repair and maintaining mineral homeostasis. Two key cytokines that promote osteoclast differentiation from myeloid precursors are M-CSF and receptor activator of NF-κB ligand (RANKL) (1). M-CSF induces expression of RANK, the receptor for RANKL, which activates signaling by MAPK, NF-κB and AP-1 pathways. These pathways work in concert with calcium signals provided by costimulatory receptors to induce expression of osteoclast-associated genes, including the transcription factor NFATc1 that coordinates the osteoclast differentiation program (2, 3). More recently, it has become clear that osteoclast differentiation is opposed by negative regulators including transcription factors IRF8, MAFB and BCL6 that need to be repressed after RANKL stimulation for osteoclastogenesis to proceed (4). IRF8 plays a key role in restraining osteoclastogenesis and bone resorption in vivo, and works in part by suppressing NFATc1 expression (5).
While RANKL-induced signaling pathways have been intensively studied, epigenetic regulatory mechanisms of the osteoclast differentiation program are not as well elucidated (6). In the context of cell differentiation, epigenetic regulation typically refers to DNA or histone/chromatin modifications that regulate gene expression. Previous reports of RANKL-induced histone modifications at the NFATc1 promoter (7) and suppression of osteoclastogenesis by inhibitors of the epigenetic regulators HDACs and BET proteins suggest that regulation of osteoclast differentiation likely has a strong epigenetic component (8, 9). Recently, Nishikawa et al. described epigenetic repression of Irf8 by Dnmt3a-mediated DNA hypermethylation, which elevated IRF8 expression and suppressed osteoclastogenesis and bone resorption in disease models. Interestingly, DNA methylation of Irf8 occurred subsequent to prior downregulation of Irf8 expression by an as yet unidentified mechanism and served to stabilize gene silencing. As suppressive chromatin modifications often precede and are associated with DNA methylation, these findings suggest that the initial phase of IRF8 repression may be mediated by chromatin-based mechanisms (6, 10).
One key mechanism of chromatin-based stable gene silencing is deposition of H3K27me3 by the Polycomb repressive complex 2 (PRC2). The negative H3K27me3 histone mark silences genes by recruiting corepressors and PRC1, which leads to DNA methylation (11). Trimethylation of H3K27 is catalyzed by PRC2 component EZH2. EZH2 typically functions to promote proliferation and repress differentiation genes and is associated with aggressive malignancies (12), and pharmacological targeting of EZH2 has shown promise in preclinical tumor models (13, 14).
We are interested in identifying epigenetic regulators of osteoclastogenesis with potential to become novel therapeutic targets of pathological bone resorption. In this study, we show that EZH2 is required for osteoclast differentiation and that inhibition of EZH2 attenuates bone loss in an ovariectomy model of osteoporosis. EZH2 was recruited to the IRF8 promoter, where deposition of H3K27me3 was associated with IRF8 repression, thus allowing osteoclastogenesis to proceed. These results identify a new mechanism of EZH2 action in osteoclast precursors that shows promise for therapeutic targeting in bone resorptive disease.
Materials and Methods
In vitro osteoclastogenesis assays
Osteoclast precursors (OCPs) derived from human blood monocytes were stimulated in triplicate wells with RANKL (40 ng/ml; Peprotech 310-01) for 5 days and stained for TRAP as previously described (8).
In vivo mouse assay
All animal experiments were approved by the Hospital for Special Surgery Institutional Animal Care and Use Committee. 11-week-old sham operated or ovariectomized C3H mice (n = 6/group) from Charles River Laboratory (Wilmington, MA) were randomized and treated intraperitoneally with GSK126 (3 mg per 20 g body weight) or vehicle control 20% Captisol 5 days per week for 5 weeks beginning 3 weeks after ovariectomy. μ-CT analysis was performed as previously described (8).
Locked nucleic acid (LNA)-mediated knockdown
Human CD14+ monocytes were transfected with an LNA oligonucleotide targeting EZH2 (Exiqon 300600) or a random sequence negative control oligonucleotide (Exiqon 300610) with the Amaxa Monocyte Nucleofector kit (Lonza VPA-1007) and program Y-001 (Amaxa Biosystems, Cologne, Germany).
ChIP-qPCR
ChIP-qPCR experiments were performed as described in Qiao et al (15). Antibodies for H3K27me3 and EZH2 were from Millipore (17-622) and Active Motif (39875) respectively. Primer sequences are available on request.
Gene expression analysis
Real time quantitative PCR was performed as previously described (15). Primer sequences are available on request.
Western Blot
Western blots were performed using the following antibodies: NFATc1 from Millipore (MABS409), EZH2 (3147S), NF-κB p65 (4764), NF-κB p50 (3035), NF-κB p52 (3017), AKT (9272) from Cell Signaling Technology, and c-Fos, IRF8, TBP, and p38α from Santa Cruz Biotechnology (sc-52, sc-6058, sc-535).
Dynamic histomorphometry
To measure bone mineralization in vivo OVX model, mice were intraperitoneally injected with Calcein (green) at 10 μg/g (body weight) twice, 7 days apart. Two days after the second injection, femurs were collected, fixed in 4% paraformaldehyde, embedded in OCT compound (Thermo Fisher Scientific, Whaltham, MA), and then cut into 7 μm sections, as described previously (16). Histomorphometry analysis using Osteometric software (Osteomeasure) was performed on trabecular bone within the femur metaphysis. Mineral apposition rate (MAR) was determined by measuring the distance between two fluorochrome-labeled mineralization fronts. The mineralizing surface was determined by measuring the double labeled surface and one-half of single labeled surface, and by then expressing this value as a percentage of total bone surface. The Bone Formation Rate (BFR) was then expressed as MAR x mineralizing surface/total bone surface, using a surface referent.
Osteoblast differentiation analysis
Primary osteoblast precursors were isolated from calvaria of wild type mice using digestion solution containing type I collagenase and dispase II (Invitrogen). Cells were cultured with α-MEM containing 10% FBS, 50 μg/ml of ascorbic acid, and 8 mM beta-glycerophosphate as described previously (8). For osteoblast marker gene expression, total mRNA was purified from osteoblast cultures at day 10. Osteoblasts were stained with Alizarin Red after 21 days of culture and Alizarin Red staining was quantified by colorimetric analysis at OD405.
Statistics
Statistical testing was performed on fold changes between conditions using pooled data from independent experiments. All statistical analyses were performed with Graphpad Prism 5.0 software using the 2-tailed, unpaired t-test (two conditions), one-way ANOVA for multiple comparisons (more than two conditions) with post hoc Tukey test. p<0.05 was taken as statistically significant.
Results and Discussion
EZH2 is required for osteoclast differentiation and NFATc1 expression
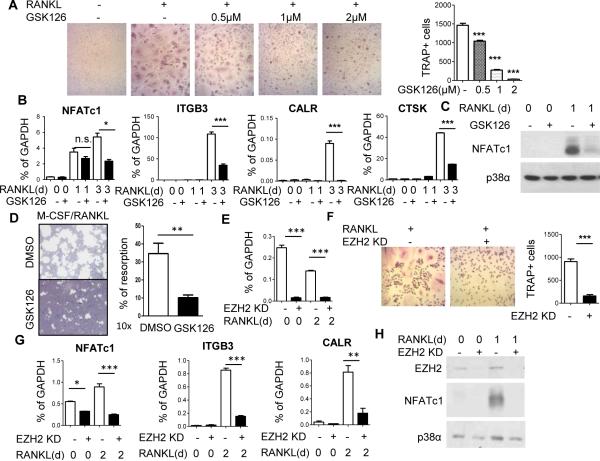
We tested the effects of the selective EZH2 inhibitor GSK126 on osteoclast differentiation using human blood-derived OCPs, which are post-mitotic cells thought to correspond to circulating quiescent OCPs that migrate to sites of bone erosion and differentiate into osteoclasts (17). The human system offers the advantage of testing the effects of EZH2 inhibition on osteoclastogenesis separately from effects on proliferation and survival of progenitors that can occur in mouse bone marrow precursors. Previously GSK126 has been described to bind EZH2 and inhibit its methyltransferase activity without affecting total histone H3 or PRC2 amounts (13). A dose response experiment showed that inhibitory effects of GSK126 on RANKL-induced differentiation of TRAP+ multinucleated osteoclasts became apparent at 0.5 μM and were essentially complete at 2 μM GSK126 (Fig. 1A). Inhibition of the osteoclast differentiation program by GSK126 was corroborated by inhibition of RANKL-induced expression of genes encoding β3 integrin (ITGB3), calcitonin receptor (CALCR) and cathepsin K (CTSK), which are markers of intermediate and late stages of osteoclast differentiation and important for osteoclast function. RANKL induction of NFATc1, which is required for osteoclast differentiation and promotes the expression of many osteoclast genes such as ITGB3, was also strongly suppressed by GSK126 (Fig. 1B and 1C). Accordingly, osteoclast-mediated resorption on osteoassay substrate was markedly suppressed in GSK126-treated osteoclast cultures compared to vehicle-treated controls. (Fig 1D). As a specificity control, we found that GSK126 did not affect basal expression or induction of various immune response genes (unpublished observations).
Figure 1. EZH2 is required for osteoclast differentiation and NFATc1 expression.
A. Monocytes pre-treated with DMSO or indicated concentrations of GSK126 were stimulated for 5 days with RANKL and stained for TRAP. Data are shown as mean ± SEM of triplicates from a representative experiment; N = 3.
B. RT-qPCR analysis of mRNA from cells pre-treated with GSK126 or DMSO for 1 day and stimulated with RANKL for 0, 1 or 3 days. Data are shown as mean ± SEM of triplicates from one representative experiment. Percent inhibition by GSK126 at day 3 was significant (p < 0.05, one tailed t-test) for pooled data where N = 4.
C. Immunoblot of NFATc1 expression in cells treated with GSK126. Blots shown are representative; N = 5.
D. Bone resorption assay. Left panel: Representative images are shown from four experiments. Right panel: % of resorption area relative to total area of osteoassay plate. Statistical analysis was performed using two-tailed, unpaired t-test. **P =0.0065
E. Effective knockdown (KD) of EZH2 gene expression using EZH2-specific LNAs relative to control LNAs.
F. Representative photomicrographs (left) and cell counts (right) of TRAP staining of OCPs transfected with control or EZH2-specific LNAs. Percent suppression was significant for pooled data; N = 5. Statistical analysis was performed using two-tailed, unpaired t-test. ***P =0.0003
G. RT-qPCR analysis of osteoclast gene expression in cells transfected with control or EZH2-specific LNAs. Data are shown as mean ± SEM of triplicates from one of four experiments; p < 0.05 for percent suppression in pooled data where N = 4.
H. Immunoblot of EZH2 and NFATc1 expression in cells transfected with control or EZH2-specific LNAs. Blots shown are representative; N = 5.
Statistical testing was then applied to pooled data (one-way ANOVA with post hoc Tukey test for all data except D and F). The results of statistical tests: *P <0.05; ** P <0.01; *** P <0.005, n.s=not significant for pooled data are shown in the respective figure panels.
To confirm that GSK126-induced suppression of osteoclastogenesis was mediated by EZH2 inhibition, we knocked down EZH2 expression using locked nucleic acids (LNAs) and analyzed osteoclast differentiation and gene expression. EZH2 knock down (Fig. 1E) effectively suppressed osteoclast differentiation (Fig. 1F) and accordingly suppressed RANKL-induced expression of ITGB3 and CALCR (Fig. 1G). These results support a role for EZH2 in osteoclast differentiation and that GSK126 suppresses osteoclastogenesis by inhibiting EZH2 catalytic activity. Strikingly, LNA-mediated knockdown of EZH2 expression also strongly suppressed RANKL-induced expression of NFATc1 (Fig. 1G and 1H). These results show that EZH2 promotes osteoclast differentiation by enabling RANKL induction of NFATc1 to drive downstream osteoclast differentiation.
As EZH2 is a negative regulator of gene expression, its positive role in NFATc1 induction suggests an indirect mechanism of action. One possible indirect mechanism would be regulation of proteins or signaling molecules in pathways linking RANKL stimulation with induction of NFATc1 expression. We found that either inhibition of EZH2 or EZH2 knockdown had minimal effects on RANK expression (Supp. Fig 1A and 1B) and RANKL-mediated NFκB activation (Supp. Fig. 1C). However, GSK126 attenuated RANKL-mediated induction of Fos protein (Supp. Fig. 1D), which is required for NFATC1 expression, and inhibited induction of Fos mRNA at the 1 day time point (Supp. Fig. 1E). Thus, EZH2 promotes osteoclast differentiation in part by affecting Fos that is required to activate NFATc1.
EZH2 directly silences IRF8 expression
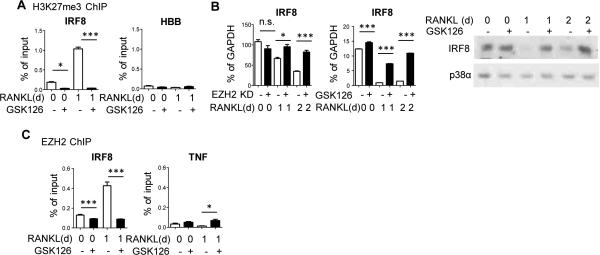
In addition to signals that promote NFATc1 expression, effective osteoclastogenesis requires downregulation of transcriptional repressors, such as IRF8, BCL6 or MAFB, that oppose the differentiation program (4). Perhaps the most established of these is IRF8, which works at least in part by suppressing NFATc1 expression (5). Thus, we tested the hypothesis that EZH2 directly represses IRF8, BCL6 or MAFB by depositing H3K27me3 at these gene loci, and that inhibition of EZH2 would restore expression of these negative regulators, thereby suppressing osteoclastogenesis. First, we found using chromatin immunoprecipitation (ChIP) assays that H3K27me3 accumulated at the IRF8 promoter 1 day after RANKL stimulation (Fig. 2A), and was associated with the expected decrease in IRF8 expression after RANKL stimulation (Fig. 2B, white bars). Strikingly, GSK126 abrogated the negative H3K27me3 histone mark at the IRF8 promoter (Fig. 2A), which was associated with a reversal of the RANKL-induced downregulation of IRF8 mRNA and protein (Fig. 2B). We found that EZH2 was recruited to the IRF8 promoter concomitant with deposition of H3K27me3 (Fig. 2C), which supports a direct mechanism of action. EZH2 recruitment to IRF8 was suppressed by GSK126, which is consistent with a previously described positive feedback loop where H3K27me3 marks reinforce EZH2 recruitment (11). In contrast to IRF8, neither H3K27me3 accumulation nor EZH2 recruitment was detected at the BCL6 or MAFB promoters after RANKL stimulation (Supp. Fig 1F and 1G), although GSK126 had a modest and likely indirect effect on BCL6 expression one day after RANKL stimulation (Supp. Fig 1H). Collectively, these results show that EZH2 is recruited to IRF8 during the early stages of RANKL-induced osteoclast differentiation, and that EZH2-mediated H3K27me3 deposition represses IRF8 to facilitate osteoclastogenesis. The elevation in IRF8 expression that occurs when EZH2 is inhibited or knocked down would suppress osteoclast differentiation, and helps explain diminished osteoclastogenesis that was observed when cells were treated with GSK126 or EZH2-specific LNAs.
Figure 2. EZH2 directly silences IRF8 expression.
A. ChIP assays for H3K27me3 at the IRF8 or negative control HBB promoter in cells pretreated with GSK126 or DMSO and stimulated with RANKL. Data are shown as mean ± SEM of triplicates from one representative experiment. Pooled data (N = 4) was analyzed using repeated measures one-way ANOVA with post hoc Tukey test. *P <0.05; *** P <0.005.
B. IRF8 mRNA expression in RANKL-stimulated EZH2 knockdown and control cells (left panel) or GSK126 treated cells (center panel) and IRF8 protein expression in GSK126 treated cells stimulated with RANKL (right panel). PCR data is shown as mean ± SEM of triplicates from one experiment; N = 3. This figure shows data from representative experiments. Statistical testing was then applied to the representative data (one-way ANOVA with post hoc Tukey test). *P <0.05; ** P <0.01; *** P <0.005, n.s=not significant.
Blots shown are representative; N = 3.
C. ChIP assays for EZH2 at the IRF8 or negative control TNF promoter in cells pretreated with GSK126 or DMSO and stimulated with RANKL. Data shown as mean ± SEM of triplicates from one representative experiment; N=4. Statistics were performed as in B. *P <0.05; *** P <0.005.
GSK126 increases bone mass in ovariectomized mice
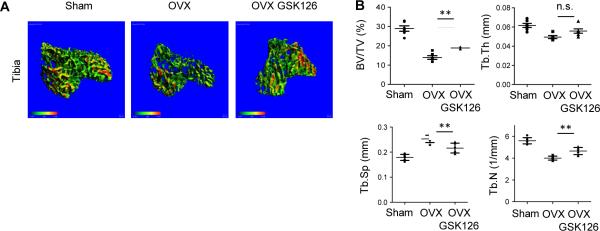
We then tested whether GSK126 could suppress pathological bone resorption in vivo using an ovariectomy (OVX) model of osteoporosis. GSK126 treatment was started 3 weeks after OVX and continued for 5 weeks. Microcomputed tomography (μCT) analysis revealed that compared to mice that underwent sham surgery, vehicle-treated OVX mice exhibited a nearly 60% reduction in trabecular bone volume in tibiae (Fig. 3A and 3B). GSK126 treatment significantly increased tibial bone mass relative to vehicle-treated mice (Fig. 3A and 3B). GSK126 treatment also significantly increased trabecular number and decreased trabecular spacing in tibiae (Fig. 3B). However, an effect of GSK126 was not observed in femurs. We also tested the effect of GSK126 on bone formation of ovariectomized mice. GSK126 treatment showed minimal effects on mineral apposition rate (MAR) and bone formation rate (BFR) in OVX mice compared to vehicle-treated OVX mice (Supplementary Fig. 2A and 2B). Of note, EZH2 has been implicated in osteoblast differentiation and it has been shown that short-term treatment (three days) with GSK126 promotes in vitro osteoblast differentiation (18). Consistently, long-term treatment (two weeks) with GSK-126 showed a trend towards increased in vitro osteoblast differentiation (Supplementary Fig. 2C-E). Taken together, our results suggest that GSK126 has beneficial effects on pathologic bone resorption.
Figure 3. GSK126 increases bone mass in ovariectomized mice.
A. μ-CT images (N = 6) of trabecular bone from tibiae of sham, ovariectomized (OVX), and GSK126 treated OVX mice.
B. Measurements of bone volume to total volume (BV/TV), trabecular thickness (Tb.Th), trabecular spacing (Tb.Sp), and trabecular number (Tb.N) for the indicated mice (N = 6). Statistical significance of differences was assessed using one-way ANOVA with post hoc Tukey test. **<0.01; n.s=not significant.
The importance of epigenetic mechanisms in regulating osteoclast differentiation is becoming increasingly appreciated and represents an emerging research area. This research has therapeutic implications, as inhibitors of the epigenetic regulators HDACs, BET proteins, and Dnmt3a were previously shown to effectively suppress pathological bone loss in several disease models, including osteoporosis, arthritis, inflammatory osteolysis, and bony tumor metastasis (5, 6, 8, 9, 19). This study adds to this body of work by identifying a key role for EZH2 and associated H3K27me3 in promoting RANKL-induced osteoclast differentiation. Our work supports a model whereby RANKL induces EZH2 recruitment and H3K27me3-mediated repression of IRF8, thereby releasing the IRF8-mediated ‘brakes’ on osteoclastogenesis. In addition, EZH2 regulates RANKL-induced expression of Fos, an essential transcription factor in osteoclast differentiation, suggesting that EZH2 controls both positive and negative regulators of osteoclastogenesis. Inhibition of EZH2 did not affect known mechanisms that regulate Fos expression in OCPs, such as IFN signaling, and did not affect expression of microRNAs that regulate Fos expression (20, 21). Understanding mechanisms by which EZH2 regulates Fos expression needs further investigation.
A recent report highlighted the importance of stable epigenetic silencing of Irf8 at a later stage of osteoclastogenesis by Dnmt3a-mediated DNA methylation, but did not clarify the mechanism that initially repressed IRF8 expression. Our findings indicate that the earlier stages of IRF8 repression are mediated by a chromatin-based H3K27me3-mediated mechanism. Interestingly, EZH2-mediated chromatin regulation and Dnmt3a-mediated DNA methylation have been shown to cooperate to silence genes in other systems (11, 22), and EZH2 occupancy and H3K27me3 can serve as a recruitment platform for Dnmt3a (23). Thus, our work suggests that EZH2 and Dnmt3a may cooperate to silence IRF8 expression during osteoclast differentiation.
In conclusion, this study provides evidence of a novel mechanism of epigenetic regulation of osteoclastogenesis by EZH2. The repression of IRF8 by EZH2 further highlights IRF8 as an important negative regulator of NFATc1 expression that must be silenced for RANKL-induced osteoclastogenesis. Effective inhibition of EZH2 by GSK126 in human osteoclast precursors provides a proof of concept supporting potential clinical applications of similar inhibitors and molecules that target chromatin regulators for bone resorptive diseases and perhaps also bone metastases.
Supplementary Material
Footnotes
This work was supported by NIH grants DE019420, AR046713, AR061430, AR062047, and AI044938
References
- 1.Takayanagi H. Osteoimmunology: shared mechanisms and crosstalk between the immune and bone systems. Nature reviews. Immunology. 2007;7:292–304. doi: 10.1038/nri2062. [DOI] [PubMed] [Google Scholar]
- 2.Lorenzo J, Horowitz M, Choi Y. Osteoimmunology: interactions of the bone and immune system. Endocrine reviews. 2008;29:403–440. doi: 10.1210/er.2007-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakashima T, Hayashi M, Takayanagi H. New insights into osteoclastogenic signaling mechanisms. Trends in endocrinology and metabolism: TEM. 2012;23:582–590. doi: 10.1016/j.tem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Zhao B, Ivashkiv LB. Negative regulation of osteoclastogenesis and bone resorption by cytokines and transcriptional repressors. Arthritis research & therapy. 2011;13:234. doi: 10.1186/ar3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao B, Takami M, Yamada A, Wang X, Koga T, Hu X, Tamura T, Ozato K, Choi Y, Ivashkiv LB, Takayanagi H, Kamijo R. Interferon regulatory factor-8 regulates bone metabolism by suppressing osteoclastogenesis. Nature medicine. 2009;15:1066–1071. doi: 10.1038/nm.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivashkiv LB. Metabolic-epigenetic coupling in osteoclast differentiation. Nature medicine. 2015;21:212–213. doi: 10.1038/nm.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yasui T, Hirose J, Tsutsumi S, Nakamura K, Aburatani H, Tanaka S. Epigenetic regulation of osteoclast differentiation: possible involvement of Jmjd3 in the histone demethylation of Nfatc1. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2011;26:2665–2671. doi: 10.1002/jbmr.464. [DOI] [PubMed] [Google Scholar]
- 8.Park-Min KH, Lim E, Lee MJ, Park SH, Giannopoulou E, Yarilina A, van der Meulen M, Zhao B, Smithers N, Witherington J, Lee K, Tak PP, Prinjha RK, Ivashkiv LB. Inhibition of osteoclastogenesis and inflammatory bone resorption by targeting BET proteins and epigenetic regulation. Nature communications. 2014;5:5418. doi: 10.1038/ncomms6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamoureux F, Baud'huin M, Rodriguez Calleja L, Jacques C, Berreur M, Redini F, Lecanda F, Bradner JE, Heymann D, Ory B. Selective inhibition of BET bromodomain epigenetic signalling interferes with the bone-associated tumour vicious cycle. Nature communications. 2014;5:3511. doi: 10.1038/ncomms4511. [DOI] [PubMed] [Google Scholar]
- 10.Nishikawa K, Iwamoto Y, Kobayashi Y, Katsuoka F, Kawaguchi S, Tsujita T, Nakamura T, Kato S, Yamamoto M, Takayanagi H, Ishii M. DNA methyltransferase 3a regulates osteoclast differentiation by coupling to an S-adenosylmethionine-producing metabolic pathway. Nature medicine. 2015;21:281–287. doi: 10.1038/nm.3774. [DOI] [PubMed] [Google Scholar]
- 11.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lund K, Adams PD, Copland M. EZH2 in normal and malignant hematopoiesis. Leukemia. 2014;28:44–49. doi: 10.1038/leu.2013.288. [DOI] [PubMed] [Google Scholar]
- 13.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, 3rd, Diaz E, LaFrance LV, Mellinger M, Duquenne C, Tian X, Kruger RG, McHugh CF, Brandt M, Miller WH, Dhanak D, Verma SK, Tummino PJ, Creasy CL. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 14.Zhao X, Lwin T, Zhang X, Huang A, Wang J, Marquez VE, Chen-Kiang S, Dalton WS, Sotomayor E, Tao J. Disruption of the MYC-miRNA-EZH2 loop to suppress aggressive B-cell lymphoma survival and clonogenicity. Leukemia. 2013;27:2341–2350. doi: 10.1038/leu.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiao Y, Giannopoulou EG, Chan CH, Park SH, Gong S, Chen J, Hu X, Elemento O, Ivashkiv LB. Synergistic activation of inflammatory cytokine genes by interferon-gamma-induced chromatin remodeling and toll-like receptor signaling. Immunity. 2013;39:454–469. doi: 10.1016/j.immuni.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Won HY, Mun SH, Shin B, Lee SK. Contradictory role of CD97 in basal and tumor necrosis factor alpha-induced osteoclastogenesis in vivo. Arthritis & rheumatology. 2015 doi: 10.1002/art.39538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sorensen MG, Henriksen K, Schaller S, Henriksen DB, Nielsen FC, Dziegiel MH, Karsdal MA. Characterization of osteoclasts derived from CD14+ monocytes isolated from peripheral blood. Journal of bone and mineral metabolism. 2007;25:36–45. doi: 10.1007/s00774-006-0725-9. [DOI] [PubMed] [Google Scholar]
- 18.Dudakovic A, Camilleri ET, Xu F, Riester SM, McGee-Lawrence ME, Bradley EW, Paradise CR, Lewallen EA, Thaler R, Deyle DR, Larson AN, Lewallen DG, Dietz AB, Stein GS, Montecino MA, Westendorf JJ, van Wijnen AJ. Epigenetic Control of Skeletal Development by the Histone Methyltransferase Ezh2. The Journal of biological chemistry. 2015;290:27604–27617. doi: 10.1074/jbc.M115.672345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon JA, Stein JL, Westendorf JJ, van Wijnen AJ. Chromatin modifiers and histone modifications in bone formation, regeneration, and therapeutic intervention for bone-related disease. Bone. 2015 doi: 10.1016/j.bone.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takayanagi H, Kim S, Matsuo K, Suzuki H, Suzuki T, Sato K, Yokochi T, Oda H, Nakamura K, Ida N, Wagner EF, Taniguchi T. RANKL maintains bone homeostasis through c-Fos-dependent induction of interferon-beta. Nature. 2002;416:744–749. doi: 10.1038/416744a. [DOI] [PubMed] [Google Scholar]
- 21.Dunand-Sauthier I, Santiago-Raber ML, Capponi L, Vejnar CE, Schaad O, Irla M, Seguin-Estevez Q, Descombes P, Zdobnov EM, Acha-Orbea H, Reith W. Silencing of c-Fos expression by microRNA-155 is critical for dendritic cell maturation and function. Blood. 2011;117:4490–4500. doi: 10.1182/blood-2010-09-308064. [DOI] [PubMed] [Google Scholar]
- 22.Yoo KH, Hennighausen L. EZH2 methyltransferase and H3K27 methylation in breast cancer. International journal of biological sciences. 2012;8:59–65. doi: 10.7150/ijbs.8.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.