Abstract
Much progress has been made in elucidating the molecular networks required for specifying retinal cells, including photoreceptors, but the downstream mechanisms that maintain identity and regulate differentiation remain poorly understood. Here, we report that the transcription factor Glass has a dual role in establishing a functional Drosophila eye. Utilizing conditional rescue approaches, we confirm that persistent defects in ommatidium patterning combined with cell death correlate with the overall disruption of eye morphology in glass mutants. In addition, we reveal that Glass exhibits a separable role in regulating photoreceptor differentiation. In particular, we demonstrate the apparent loss of glass mutant photoreceptors is not only due to cell death but also a failure of the surviving photoreceptors to complete differentiation. Moreover, the late reintroduction of Glass in these developmentally stalled photoreceptors is capable of restoring differentiation in the absence of correct ommatidium patterning. Mechanistically, transcription profiling at the time of differentiation reveals that Glass is necessary for the expression of many genes implicated in differentiation, i.e. rhabdomere morphogenesis, phototransduction, and synaptogenesis. Specifically, we show Glass directly regulates the expression of Pph13, which encodes a transcription factor necessary for opsin expression and rhabdomere morphogenesis. Finally, we demonstrate the ability of Glass to choreograph photoreceptor differentiation is conserved between Drosophila and Tribolium, two holometabolous insects. Altogether, our work identifies a fundamental regulatory mechanism to generate the full complement of cells required for a functional rhabdomeric visual system and provides a critical framework to investigate the basis of differentiation and maintenance of photoreceptor identity.
Keywords: Glass, Pph13, prominin, photoreceptor, differentiation
INTRODUCTION
The generation of a functional visual system requires coordinated development and integration of many neuronal and non-neuronal cell types. In Drosophila, each ommatidium, the functional visual unit of the compound eye, consists of eight neuronal photoreceptors and twelve accessory cells. During early development, the specification and recruitment of these twenty cells are initiated in a sequential wave as the morphogenetic furrow moves across the eye imaginal disc (Ready et al., 1976; Wolff and Ready, 1993). To date, the transcriptional and cell signaling pathways that govern photoreceptor cell specification, establishment of the repetitive array of ommatidia and the numerous genetic networks required for ommatidial subtype specification, i.e. the mosaic patterning of opsin expression within each ommatidium, are well characterized (reviewed in (Bao, 2010; Charlton-Perkins and Cook, 2010; Pichaud, 2014; Rister and Desplan, 2011; Treisman, 2013; Wernet and Desplan, 2004)). Nonetheless, the molecular and cellular events that link ommatidium patterning and specification with the terminal differentiation and maintenance of cell fate to form a functional unit remain undefined.
With respect to terminal differentiation, the homeodomain transcription factors Orthodenticle (Otd) (Ranade et al., 2008; Tahayato et al., 2003; Vandendries et al., 1996) and Pph13 (Goriely et al., 1999; Zelhof et al., 2003) are necessary for distinct and critical aspects of rhabdomere morphogenesis, phototransduction and synaptogenesis. The loss-of-function mutations in either otd or Pph13 leads to severe defects in photoreceptor terminal differentiation, morphogenesis and function (Mishra et al., 2010; Vandendries et al., 1996; Zelhof et al., 2003), and the combinatorial activity of both factors is essential for the proper transformation of the photoreceptor apical membrane into the rhabdomere, the phototransduction organelle (Mishra et al., 2010). With respect to function, otd mutants result in the absence of photosensitivity to ultraviolet light, while disruption of Pph13 leads to a complete loss of photosensitivity (Tahayato et al., 2003; Vandendries et al., 1996; Zelhof et al., 2003). Also, Otd is required for the proper innervation of photoreceptors into the lamina and medulla (Mencarelli and Pichaud, 2015). Lastly, the function and relationship between Otd and Pph13 with respect to differentiation of photoreceptors is not limited to Drosophila, but conserved among Pancrustaceans (Mahato et al., 2014).
In contrast, a third transcription factor, Glass (Gl), appears to be regulating multiple aspects eye development. glass encodes a the zinc-finger transcription factor (Moses et al., 1989) and it was first identified by Hermann Muller in 1913 as a mutation that resulted in a glassy-eyed appearance (Bridges and Morgan, 1923). Subsequent characterization revealed that the loss of glass results in irregular facet organization and a complete lack of definable rhabdomere-forming photoreceptor cells in adults (Moses et al., 1989; Treisman and Rubin, 1996). In fact, glass is essential for the development of photoreceptor cells in all three organs in which photoreceptors occur: the larval Bolwig’s organ, the adult compound eye, and the adult ocelli (Moses et al., 1989); gl mutant photoreceptors initiate neuronal development but are reported to die (Moses et al., 1989; Ready et al., 1986). Also, Glass expression is not limited to photoreceptor neurons. Glass is initially expressed in all cells posterior of the morphogenetic furrow before being restricted to the developing photoreceptor and cone cells. Later, Glass expression is detected in both photoreceptors and pigment cells (Ellis et al., 1993; Moses et al., 1989; Moses and Rubin, 1991), suggesting that the disrupted eye morphology in adults is the result of developmental defects associated with all retinal cell types.
To date genetic approaches have identified Glass transcriptional targets necessary for retinal cell specification and ommatidium patterning that directly correlate with the overall deformation of gl mutant eyes. For example, Lozenge, Prospero, and Boss are all Glass targets (Hayashi et al., 2008; Treisman and Rubin, 1996; Yan et al., 2003). Interestingly, all are required for the proper specification of the R7 equivalence group, photoreceptors R1, R6, R7 and cone cells. The loss of Boss converts the R7 photoreceptor into a cone cell (Reinke and Zipursky, 1988), whereas in prospero mutants the R7 photoreceptor now takes on characteristics of the R8 photoreceptor (Cook et al., 2003; Kauffmann et al., 1996; Morey et al., 2008). Moreover, Lozenge is not only required in for correct specification of the R7 equivalence group but also is a direct regulator of prospero transcription (Daga et al., 1996; Flores et al., 1998; Hayashi and Saigo, 2001; Hayashi et al., 2008; Xu et al., 2000); Lozenge appears to represents a feed forward loop in promoting eye development with Glass (Potier et al., 2014; Treisman, 2013; Yan et al., 2003) and thus is a critical read-out of Glass activity and downstream patterning of ommatidium. Lastly, Glass targets are not limited to molecules necessary for cell type specification. chaoptin is a Glass target (Moses et al., 1989; Naval-Sanchez et al., 2013) and is essential for the formation of photoreceptor rhabdomeres (Reinke et al., 1988; Van Vactor et al., 1988). Moreover, recent genomic analyses (Naval-Sanchez et al., 2013; Potier et al., 2014) indicate Glass could have a broad role in the development of the entire eye structure contributing to the specification, differentiation, and possibly maintenance of all retinal cell fates.
In this report, we address the paradoxical observation of the absence of recognizable terminally differentiated photoreceptors in adults given the initial presence of specified photoreceptor neurons in gl mutants (Moses et al., 1989; Ready et al., 1986); is the absence of terminal differentiated photoreceptors a direct consequence of earlier photoreceptor developmental defects in specification and ommatidium patterning, cell death, or a manifestation of a defect in transcriptional program for differentiation? In order to investigate this question we have further examined the glass mutant phenotypes. Our observations indicate the disruption of external eye morphology mainly reflects the earlier patterning defects of both neuronal and non-neuronal cells while excess retinal cell death, including photoreceptors, plays only a subtle role in contributing to the overall defects observed. Whereas we find glass mutant photoreceptors are dying, many persist throughout pupal development but fail to complete differentiation. However, photoreceptor differentiation is partially restored by introducing Glass expression after ommatidial patterning is complete, suggesting separable functions in patterning and differentiation. In agreement, transcriptome analysis reveals that Glass choreographs the expression of numerous genes essential for differentiation, including the homeodoamin protein Pph13. Lastly, genetic analyses in the red flour beetle Tribolium demonstrate that Glass function is conserved between these rhabdomeric visual systems. Overall, our work identifies Glass as a significant fundamental molecular link between rhabdomeric retinal cell type patterning and photoreceptor differentiation necessary to produce a functional visual unit.
MATERIALS AND METHODS
Drosophila strains and genetics
All stocks and crosses were maintained and staged at 23°C unless otherwise indicated. The following stocks were utilized in this study: w1118, gl3, gl60j, UAS-LacZ.NZ, UAS-GFP::lacZ.nls, UAS-p35, UAS-mCD8::GFP, heat-shock-GAL4, elav-GAL4, GMR-GAL4, ey-Flp; FRT82B GMR-myr.GFP, FRT82B gl60j. hs-Pph13 and hs-Glass were generated by inserting cDNAs into pCaSpeR-hs. so-GAL4 was obtained from Dr. Volker Hartenstein (Chang et al., 2003b) The gl-BAC construct (Slattery et al., 2014) was a gift from Dr. K. White. The BAC utilized was CH321-23N19 (P[acman] Resources) and AV007 (GenBank: KF411445.1) represents the sequence of the epitope tags; the epitope tag includes both GFP and FLAG epitopes. All constructs were either randomly integrated into the genome or integrated into chromosome position 65B2 (Rainbow Transgenic Flies, Inc). The heat shock protocol for expression of Glass, Pph13 and p35 consisted of three one-hour heat shocks at 37°C equally spaced per day from 24hrs APF until eclosion.
Enhancer analysis
All DNA sequences representing wild type or the mutated enhancer region of Drosophila Pph13 were cloned into pLacZattB and integrated into genomic position 22A3. Candidate Glass binding sites were mined by creating a weighted matrix derived from published validated Glass binding sites using Target Explorer (Hayashi et al., 2008; Naval-Sanchez et al., 2013; Sosinsky et al., 2003). Mutagenesis was performed using QuikChange Multi Site-Directed Mutagenesis Kit (Agilent Technologies). See Table S6 for mutated base pairs. The DNA sequence representing the enhancer region of Tribolium Pph13 was cloned into the pChs-Gal4 vector and integrated into random genomic positions.
Scanning and transmission electron microscopy and immunofluorescence staining
All procedures were performed as previously described in (Nie et al., 2012; Nie et al., 2014). The primary antibodies used in this study were: mouse anti-elav (9F8A9, 1:200, Developmental Studies Hybridoma Bank); rat anti-elav (7E8A10, 1:200, Developmental Studies Hybridoma Bank); mouse anti-cut (2B10, 1:200, Developmental Studies Hybridoma Bank); mouse anti-Dlg (1:50, Developmental Studies Hybridoma Bank); mouse anti-Lz (1:200, Developmental Studies Hybridoma Bank); mouse anti-Boss (1:200, Dr. S.L. Zipursky); chicken anti- βgal (1:500); rabbit anti-GFP (1:500, Life Technologies); rabbit anti-Cleaved Drosophila Dcp-1 (1:100, Cell Signaling); mouse anti-FLAG (1:500, Sigma); rabbit anti-dPph13 (1:50) (Zelhof et al., 2003); guinea pig anti-Otd (1:500, Dr. T. Cook) (Charlton-Perkins et al., 2011); rabbit anti-Prom (1:100) (Zelhof et al., 2006); mouse anti-Chp (24B10, 1:50; Developmental Studies Hybridoma Bank); DAPI (1:500 of 0.5ug/ml) Secondary antibodies (1:200, Jackson ImmunoResearch Laboratories); Rhodamine, Alexa Fluor 647 or fluorescein conjugated phalloidin (1:200, Life Technologies). TUNEL staining was performed as described (Lin et al., 2004) using the In Situ Cell Death Detection Kit (Fluorescein) from Roche Diagnostics. Confocal images were captured on a Leica TCS SP5 or a Nikon A1 microscope. TEM and SEM imaging was conducted with a JOEL 1010 and JSM 5800LV microscopes, respectively. All pictures were processed in Adobe Photoshop.
RNA-Seq
For each condition, triplicate sets of 48hrs APF w1118 and gl3 pupal heads were collected for total RNA extraction. Library construction and sequencing were performed as described (Mahato et al., 2014). HISeq read sequences were cleaned using Cutadapt version 1.2.1 (Rehm et al., 2011) to remove adapter sequences and perform quality trimming. Cutadapt was run with the following parameters, “-a AGATCGGAAGAGC – m 40 –q 30”. The resulting reads were mapped against the Drosophila release FB2014_06 gene models using TopHat2 version 2.0.13 (Kim et al., 2013) with the parameters “–b2-very-sensitive–read-edit-dist 2 –maxmultihits 100 –library-type fr-firststrand”. Read counting was done for each gene using htseq-count from the HTSeq package version 0.5.4p3 (Anders et al., 2015)with the “–stranded = reverse” parameter. Read counts were normalized across samples using the DESeq2 package (version 1.4.5) in R/Bioconductor (Gentleman et al., 2004). DESeq2 (Love et al., 2014)was further used to detect statistically significant differences in expression between two conditions using the binomial test with a .05 adjusted p-value cutoff. Gene expression values for adult eye were downloaded from FlyBase and are based on microarray expression data from the FlyAtlas project (http://flyatlas.org/) (Chintapalli et al., 2007). The cutoffs for high (>=500) and moderate (>=100) expression are as defined by FlyBase. The complete data set will be available at the Sequence Read Archive at NCBI.
Prediction of Glass Binding Sites
We retrieved the gl_SOLEXA_5_FBgn0004618 binding site motif from Fly Factor Survey (Beckstette et al., 2009; Zhu et al., 2011). We used the motif to search the FlyBase r6.03 assembly for binding sites using PoSSumSearch2 with a p-value cutoff of 1E-04. We then used custom scripts to filter potential binding sites based on the following criteria: (1) binding sites not located within 5000bp upstream or downstream of a gene or within the first intron of a transcript (according to FlyBase r6.03 annotation) were removed. (2) binding sites overlapping with any CDS were removed. (3) if multiple binding sites overlapped, the one with the lowest p-value was retained and others removed. (4) motifs without a perfect match in the first five positions (“GAAGC”) were removed.
RT-PCR reactions
RNA-seq data were validated using semi-quantitative end-point reverse-transcription PCR (RT-PCR) reactions. RNA extraction was as described above. First-strand synthesis was performed using SuperScript III First-Strand kit (Invitrogen) and PCR amplification was done with a S1000 Thermal Cycler (bio-rad). Cycle number and primers were individually optimized for each gene. List of all primer pairs was provided in Table S6.
Tribolium RNAi injections
To generate RNAi knockdown animals, 1 ug/ul of total probe, DsRNA was injected into m26- contains a 3XP3-RFP reporter, or vw late stage larvae (Lorenzen et al., 2007; Tomoyasu and Denell, 2004). Two independent DsRNAs were tested and eyes from at least five different subjects were examined to confirm phenotypes. The regions listed for each gene are listed in Table S6.
CHIP-qPCR
Chromatin samples were generated from 48 hrs APF pupal retinas of gl60j, gl-BAC/gl60j, gl-BAC. As a negative control, chromatin samples generated from flies expressing nuclear GFP::lacZ.nls expressed with GMR-GAL4. Procedures were based upon (Slattery et al., 2014; Zhang et al., 2014) but a detailed methodology is provided in Supplemental Material. Primers for PCR are listed in Table S6.
RESULTS
Loss of Glass results in disorganization of the entire eye structure
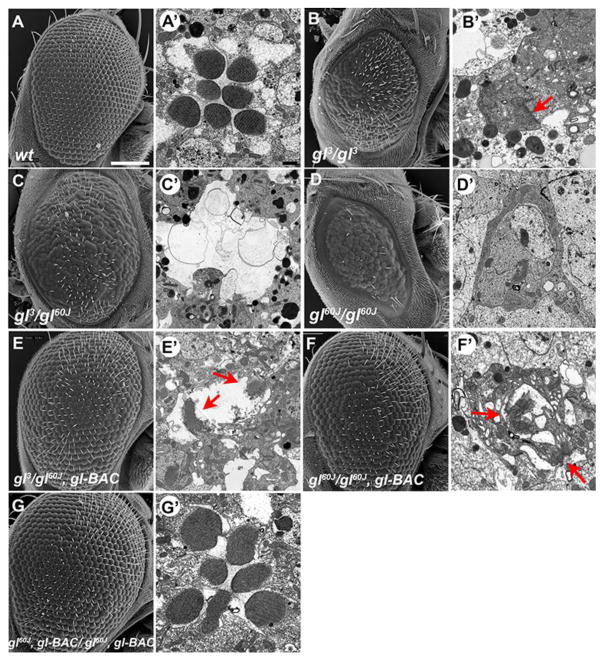
To investigate and reconfirm the phenotypes associated with the failure of eye development in gl mutants, we first examined the gl mutant phenotypes observed in both a hypomorphic allele, gl3, and null allele, gl60j, of glass (Moses et al., 1989) utilizing both scanning (SEM) and transmission (TEM) electron microscopy. As previously described (Moses et al., 1989; Ready et al., 1986), the SEM results reconfirmed that gl mutants possessed fewer and fused ommatidia as compared to wild type (Fig. 1A–D). With TEM analysis, we observed some, but few differentiated photoreceptors limited only to gl3 retinas (Fig. 1B′) but not in gl60j retinas (Fig. 1C′,D′). These “differentiated” photoreceptors were distinguished by the presence of a rhabdomere, but the rhabdomeres were morphologically disrupted, smaller and malformed compared to wild type (Fig. 1A′). Individual cell types were impossible to distinguish based on morphology and the boundaries between ommatidia were obscured in all allelic combinations (Fig. S1A–C).
Fig. 1. The loss of Glass results in defects in eye morphology and absence of differentiated photoreceptors.
(A–G) Transmission and Scanning electron microscopy of (A) wild type, (B–D) gl mutant retinas, and (E–G) gl mutant retinas containing a glass genomic rescue construct (gl-BAC). In gl mutant retinas the detection of differentiated photoreceptors was based on the presence of a rhabdomere, red arrows. The TEM images in B′ – F′ putatively highlight photoreceptor cells in the gl mutant alleles. Samples are from newly emerged adult Drosophila. SEM Scale Bar-100 um TEM Scale Bar-1um
To ensure that the phenotypic disruptions observed were due to the absence of Glass activity, we examined eye development in the presence of a genomic construct (gl-BAC), in which epitope tags were added to the carboxy terminus of the glass transcript (Slattery et al., 2014). The presence of one copy of the genomic construct rescued the overall external morphology of the eyes in all glass mutant allelic combinations (Fig. 1E,F). In contrast, TEM analysis demonstrated there was not a corresponding rescue of photoreceptor differentiation (Fig. 1E′,F′). Any detection of differentiated photoreceptors was similar to gl3 homozygotes, in both frequency of occurrence and shape. Only in the presence of two copies of the genomic construct did we observe complete rescue of all defects, eye patterning and photoreceptor differentiation (Fig. 1G). These results suggested that our tagged version of Glass did not have complete wild-type activity; for example glass mutant phenotypes were rescued by a considerable smaller genomic construct in which the protein was not tagged (Moses et al., 1989). Nonetheless, this result and the lack of identifiable differentiated photoreceptors in a gl hypomorphic allele, despite the less severe defects in external morphology (Moses et al., 1989), suggests that photoreceptor differentiation may require higher levels of Glass activity and that Glass’ functions in ommatidium patterning and photoreceptor differentiation may be mechanistically separable.
glass defects in ommatidium patterning persist through retina development
What are the biological events that contribute to the defects both in eye morphology and in photoreceptor differentiation? As noted, the role of Glass in early retinal specification has been well documented (reviewed in (Treisman, 2013)). First, as described previously, disruption of neuronal patterning was observed immediately as cells were specified in the third instar eye imaginal disc and was cell autonomous (Fig. S2A–D) (Moses et al., 1989; Treisman and Rubin, 1996). Likewise, and as expected, the expression of Lozenge (Lz) and Bride of Sevenless (Boss), two Glass targets necessary for retinal cell patterning, were reduced or missing in glass mutants, respectively (Fig. 2A–B,D–E). Nevertheless, whether these early defects are sustained and persist through retina development has not been described. To determine this, we examined the consequence of the loss of glass on the subsequent maintenance of neuronal and non-neuronal cell types at an intermediate time point in development, 48 hrs APF. First and foremost, we examined gl mutants with markers for photoreceptor cells (ELAV), cone cells (Cut), and primary pigment cells (sine oculis –lacZ). In gl60j and gl3 alleles we observed that all cell types, photoreceptors, cone, and pigment cells, were present (Fig. 3). However, as compared to wild type, no ommatidium in gl mutants contained the correct number of each retinal cell type. For example, gl mutant ommatidia did not have the full complement of eight photoreceptors. In addition, cone and primary pigment cells were either too few or too many in number (Fig. 3, Fig. S3A,B). Furthermore, there was a complete disruption of organization of retinal cells into distinct identifiable ommatidia as observed with Disc-large (Dlg) immunoflouresence (Fig. 5G,H); Dlg marks the cell membranes of various retinal cells highlighting the invariant hexagonal array of each ommatidium (Fan et al., 2010; Martin-Bermudo et al., 2015). Overall, these results suggested Glass is not essential for the presence of any general class of retinal cell, photoreceptor, cone or pigment cell. However, as previously described Glass is critical for the correct specification and patterning of each cell type in each ommatidium (Hayashi et al., 2008; Treisman and Rubin, 1996; Yan et al., 2003) and the disorganization of the adult eye structure reflects the persistence of these earlier defects in patterning.
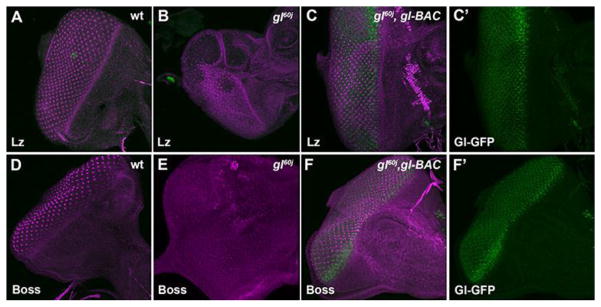
Fig. 2. The rescue of eye morphology with one copy of gl-BAC corresponds with the restoration of Lozenge and Boss expression.
Examination of early patterning retinal gene expression Lozenge (A–C) and Boss (D–F) in wild-type (A,D), gl60j (B,E), and gl60j/gl60j, gl-BAC 3rd instar eye imaginal discs (C′,F′). (C′,F′) gl60j/gl60j, gl-BAC discs stained for GFP to highlight the Gl-GFP fusion protein expression pattern.
Fig. 3. Patterning defects include both neuronal and non-neuronal cells.
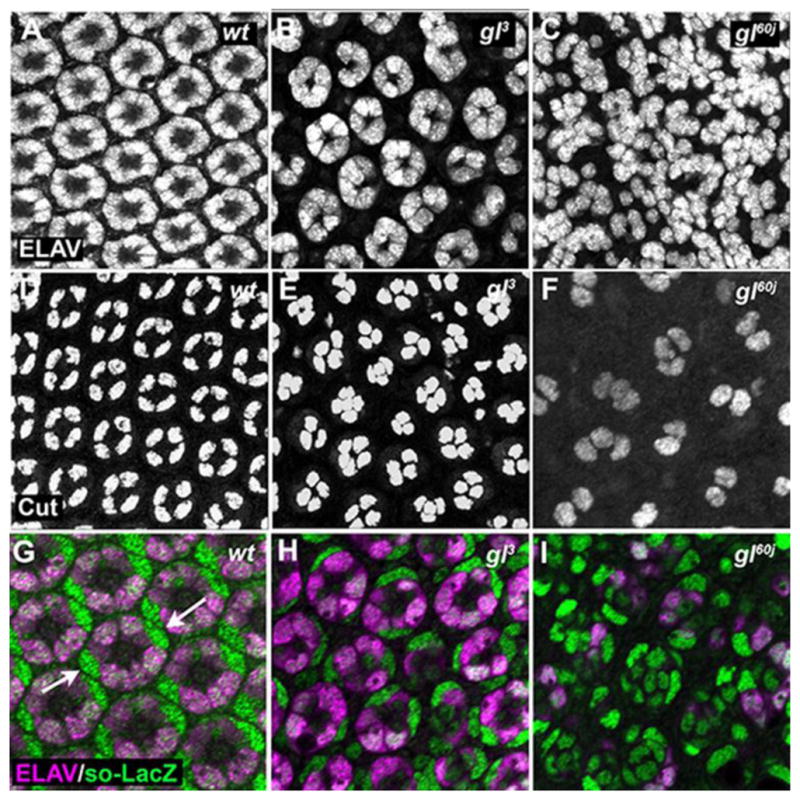
Analysis of ELAV (A–C), Cut (D–F), and so-LacZ (G–I) expression (green) in wild-type (A,D,G) and gl3 (B,E, H) and gl60j (C,F,I) 48 hrs APF eyes. (G–I) Retinas are countered stain ELAV (magenta). The primary pigment cells were distinguished by their unique oblong nuclei (arrows).
Fig. 5. One copy of gl-BAC restores retinal cell types and numbers within each ommatidium.
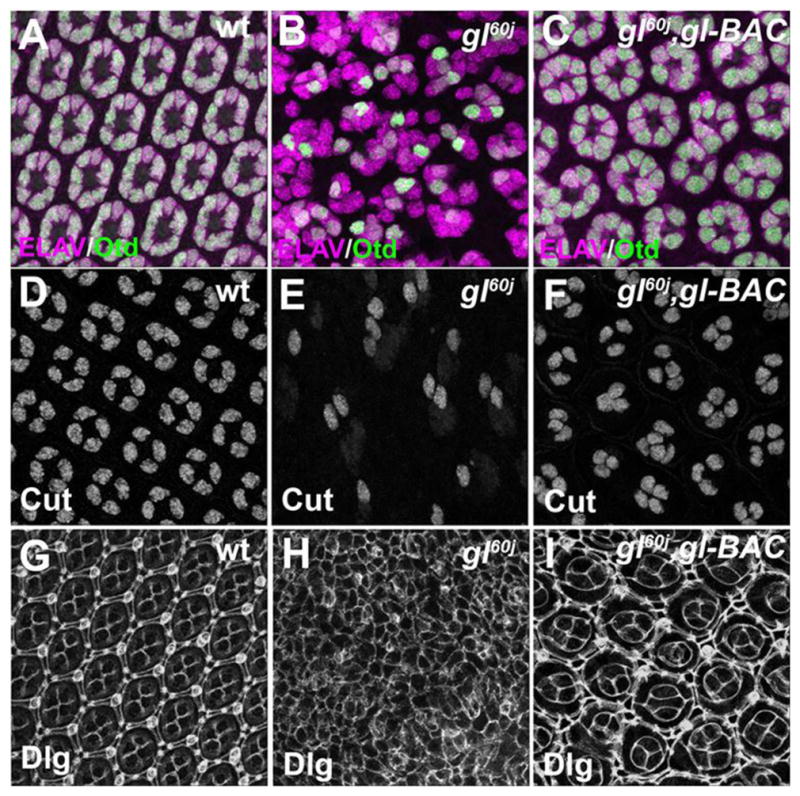
Analysis of ELAV and Otd (A–C), Cut (D–F), and Dlg (G–I) expression in wild-type (A,D,G) and gl60j (B,E, H) and gl60j/gl60j, gl-BAC (C,F,I) 48 hrs APF eyes.
gl mutant photoreceptors are capable of persisting through pupal development
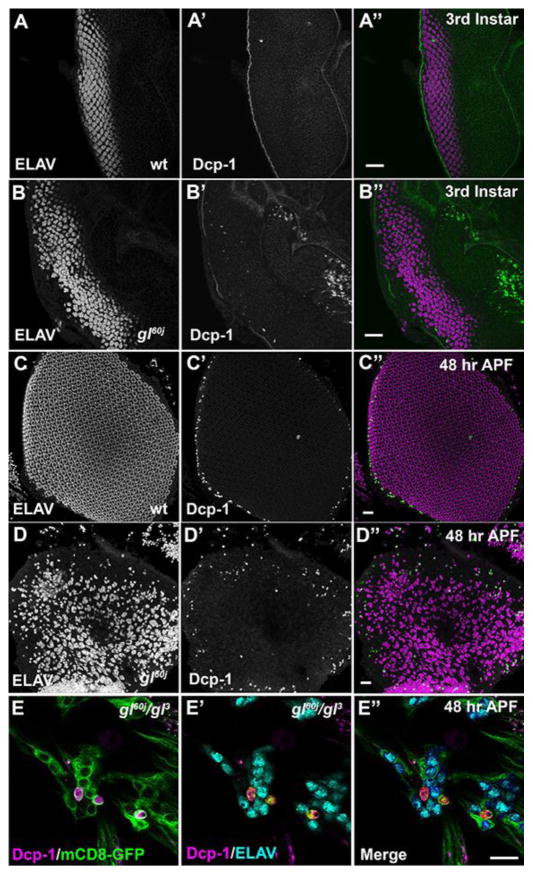
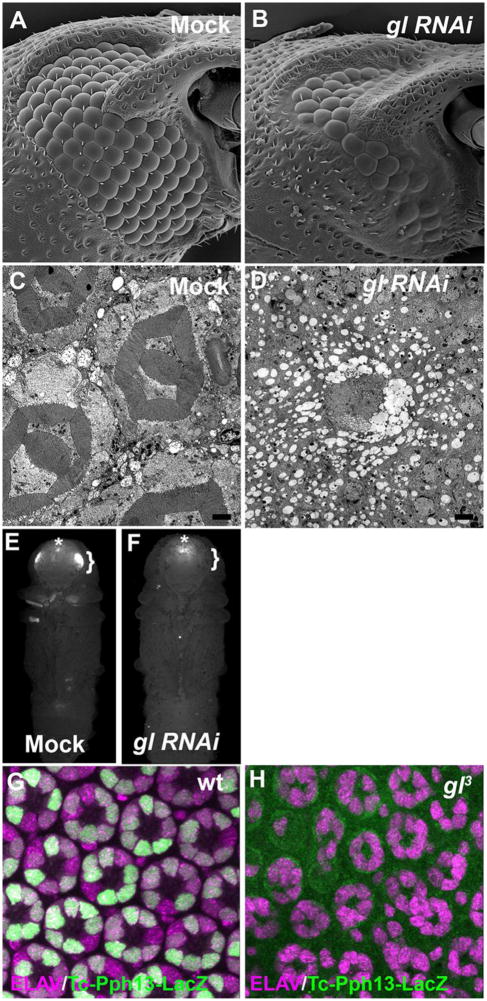
Besides defects in specification and patterning contributing to the disruption of eye morphology, a second potential causative factor responsible for gl mutant phenotypes, including the apparent lack of photoreceptors in the adult retina, was cell death (Moses et al., 1989; Ready et al., 1986). To explore this possibility, we examined the expression pattern of cleaved Death caspase-1 (Dcp-1) (Kondo et al., 2006) and we utilized TUNEL labeling to identify dying cells. We did not observe any aberrant or excessive cell death in the retina fields of third instar larvae as compared to wild type suggesting that the defects in ommatidium patterning were not the result of cell death. (Fig. 4A,B and Fig. S4 A,B). By 48 hrs after puparium formation (APF) besides the characteristic presence of cell death around the periphery of the retinal field (Lin et al., 2004; Tomlinson, 2003; Wolff and Ready, 1993; Wolff and Ready, 1991) we now observed the co-localization of Dcp-1 with ELAV positive in gl mutant neurons but the dying photoreceptors were limited in number (Fig. 4D,E and S4). At 72 hrs APF in gl60j null mutants, there was extensive cell death in the retinal field and again associated with ELAV positive neurons but not all neurons (Fig. S5). Due to the complete loss of retinal tissue integrity of gl60j retinas, we were unable to perform dissections at 96 hrs APF to determine the extent of cell death. Nonetheless, in gl60j/gl3 mutant retinas there were populations of both ELAV positive, Dcp-1 negative neurons as well as ELAV positive, Dcp-1 positive neurons (Fig. S5E,F). To further explore the contribution of cell death to the disruption of eye development in gl mutants, we attempted to inhibit or lessen apoptosis by the overexpression of p35 (Hay et al., 1994) either by driving expression with so-GAL4 or heat shock-GAL4. In either case, even though we could detect a decrease in Dcp-1 positive cells (Fig. S6A–H) there was no rescue of overall morphology or the appearance of recognizable photoreceptors in newly eclosed adults (Fig. S6D,H). Altogether, these results suggested the initial defects in specification and ommatidium patterning are the major contributors to the disruption of eye development in gl mutants and cell death may be a secondary effect of a discontinued developmental program. With respect to the fate of the photoreceptors, photoreceptors are dying throughout pupal development but many putative specified photoreceptors are capable of surviving to the point of eclosion.
Fig. 4. Loss of Glass results in death of some photoreceptors.
Analysis of Dcp-1 expression (green) in wild-type (A,C) and gl60j developing eyes (B,D). (A,B) 3rd instar eye imaginal discs. (C,D) 48 hrs APF retina. In all cases the tissue is countered stain with ELAV (magenta) to mark the putative photoreceptors. Scale Bars - 20um. (E) Analysis of Dcp-1 and ELAV expression in gl60j/gl3 48 hrs APF retinas expressing ELAV-GAL4 driving UAS-mCD8-GFP. GFP labels the membranes of the photoreceptors and note the two distinct populations of Dcp-1 positive and negative neurons. Scale Bars - 10um.
Rescue of adult eye morphology is dependent on the restoration of correct patterning
Our previous results demonstrated that one copy of gl-BAC was capable of rescuing overall eye morphology in gl mutants. As Glass was required for ommatidium patterning, then based upon our SEM results (Fig. 1E,F) we would predict that the presence of a single copy of gl-BAC would restore the wild-type pattern of each ommatidial cell type. In wild type, photoreceptor, cone, and pigment cells are present, have unique cell shapes and occupy distinct positions within each ommatdium at 48 hrs APF. As expected, the presence of gl-BAC in a gl mutant background significantly increased the number of ommatidia with the correct numbers of photoreceptor (Fig. 5A–C, Fig. S3A) and cone cells (Fig. 5D–F, Fig. S3B). In addition, the restoration of patterning was exemplified by immunofluoresence staining against Disc-large (Dlg) (Fig. 5I). This rescue was associated with the reestablishment of genes required for patterning retinal cell fates. By monitoring GFP expression, we first observed that one copy of gl-BAC was sufficient to recapitulate the broad gl expression pattern posterior of the morphogenetic furrow and then reduction of expression into a subset of cells (Fig. 2C′,F′). More importantly, both the temporal and spatial patterns of Lz (Fig. 2C) and Boss (Fig. 2F) were rescued. Combined with the restoration of adult eye morphology, these observations reconfirmed Glass is a critical molecule in generating the correct population of diverse cell types in each ommatidium and suggested that the level of Glass activity provided by one copy of gl-BAC was sufficient for improving ommatidium patterning and overall eye morphology and size.
gl mutant photoreceptors neurons fail to differentiate
The apparent absence of photoreceptors in gl mutants (Fig. 1B′–D′) was consistent with previous reports suggesting that photoreceptors were dying (Moses et al., 1989; Ready et al., 1986). However, we observed two contrasting results: the lack of identifiable photoreceptors in adult gl mutant retinas and the presence of ELAV positive neurons in gl mutant retinas extending to the point of eclosion. These observations suggested an alternate fate for the photoreceptors in gl mutants. To examine the fate of specified gl mutant photoreceptors, we investigated ELAV expression in gl mutant retinas at several developmental time points in addition to noting their differentiation status, i.e. rhabdomere morphogenesis. As noted, ELAV retinal positive cells were detected at all stages for gl3 and gl60j retinas through 72 hrs APF (Fig. 3, S2, 4, S5) and in 96 hrs APF gl60j/gl3 retinas (Fig. S5).
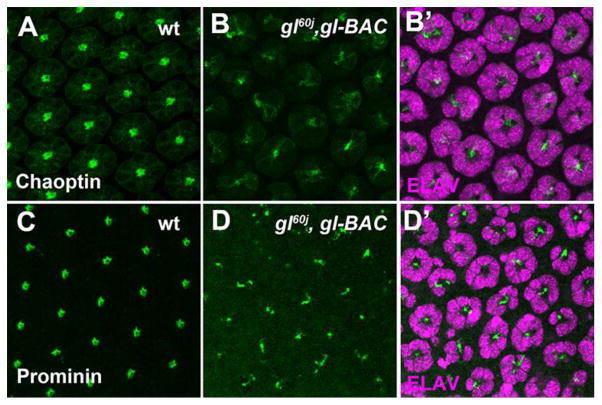
Second, even though ELAV positive neurons were present, we noted the complete lack of rhabdomere morphogenesis initiation in both gl60j and gl3 mutant photoreceptors. At 48 hrs APF, the transformation of the apical membrane is a critical marker of photoreceptor differentiation. This transformation is observed by the accumulation and organization of F-Actin, Prominin, and Chaoptin on the apical surface (Fig. 6). In gl3 retinas, we did not detect the expression of Chaoptin (Fig. 6B,E), a target of Glass (Moses et al., 1989; Naval-Sanchez et al., 2013). In contrast, F-Actin was accumulating in the center of some neuronal clusters but the localization was not organized as in wild type and there was substantial accumulation around the periphery of the cells (Fig. 6A,D). Furthermore, we did not detect or observe any accumulation of Prominin (Nie et al., 2012; Zelhof et al., 2006) on the apical surface of mutant photoreceptors (Fig. 6C,F). Additionally, we examined expression of Otd and Pph13, factors responsible for transcriptional control of differentiation. We noted that the temporal initiation of Otd expression was normal (Fig. S7A–C). However, Otd expression was not detected in every ELAV positive neuron and this mosaic expression of Otd was maintained at 48 hrs APF, at the time of rhabdomere morphogenesis (Fig. 5B). In contrast, Pph13 expression was absent in gl mutants (Fig. S8A,B). Overall, our data suggested that although photoreceptors were being specified in the absence of Glass activity, gl mutations led to a complete lack of rhabdomere morphogenesis and the inability to form a functional photoreceptor among the surviving neurons.
Fig. 6. glass mutant photoreceptors do not initiate differentiation.
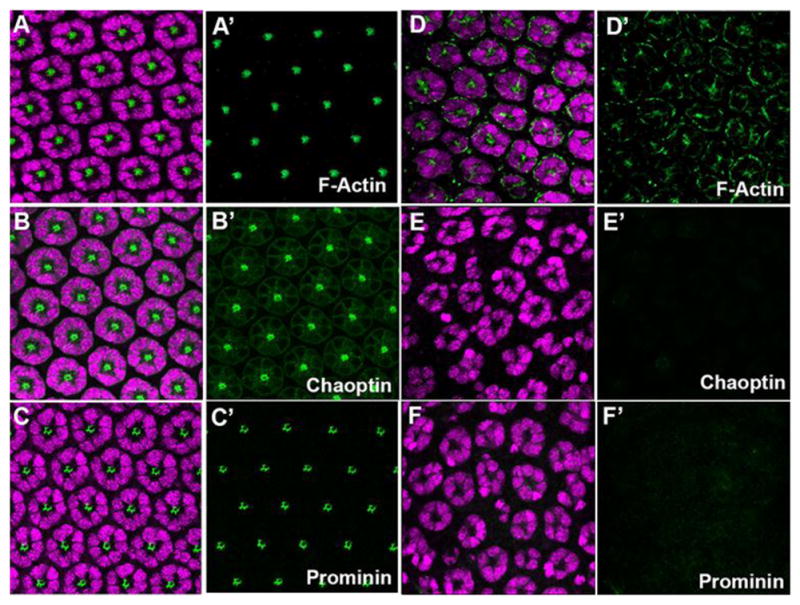
Immunofluoresence staining of markers of rhabdomere morphogenesis in wild-type (AC) and gl3 retinas (D–F). (A, D) ELAV (magenta) and F-Actin (green). (B,E) ELAV (magenta) and Chaoptin (green). (C,F) ELAV (magenta) and Prominin (green). All panels represent a projection of a confocal stack.
Rescuing photoreceptor differentiation with Glass
Our ability to disconnect the defects in retina patterning from the failure in photoreceptor differentiation in gl mutant retinas suggested that Glass activity was required at two phases of eye development to generate a functional ommatdium. In addition, successful Glass mediated ommatidium patterning did not guarantee photoreceptor differentiation. In agreement, we observed that with one copy of gl-BAC, Chaoptin and Prominin could be detected in some photoreceptors at 48 hrs APF, but there was no organized or distinct patterning representative of rhabdomere morphogenesis as observed in wild type (Fig. 7). In addition, while gl-BAC was capable of rescuing the mosaic pattern of Otd expression but this in itself was not sufficient to rescue differentiation (Fig. 5C, Fig. S3C). Intriguingly, whereas Otd expression was rescued, Pph13 was absent in this genetic background (Fig. S8C); one copy of gl-BAC was not capable of restoring Pph13 expression even though transgenic Glass, i.e. Glass:GFP, was detected (Fig. S8D–F).
Fig. 7. Photoreceptor differentiation is not restored with one copy of gl-BAC.
Immunofluoresence staining of markers of rhabdomere morphogenesis in wild-type (A,C) and gl60j/gl60,gl-BAC retinas (B,D). (A, B) Chaoptin (green). (C,D) Prominin (green). Panels B′,D′ are counterstained with ELAV(magenta) to highlight the photoreceptor clusters. All panels represent a projection of a confocal stack.
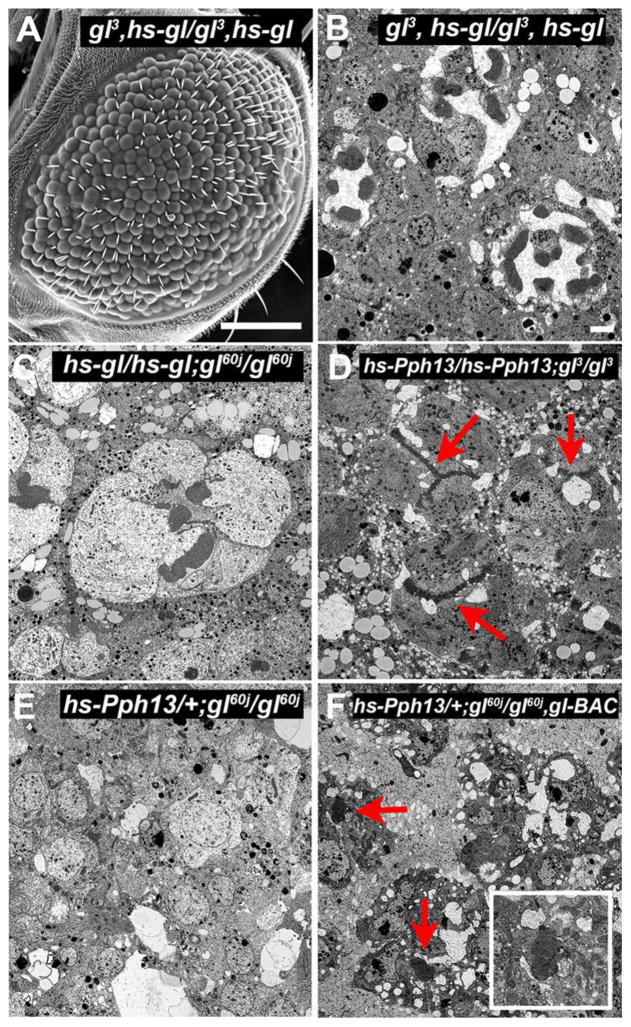
If these developmentally dependent Glass’ functions were separable, we should be able to reinitiate photoreceptor development in the disrupted patterning in gl mutant retinas. The patterning of the retinal field and the movement of the morphogenetic furrow are completed by 12 hrs APF (Wolff and Ready, 1993). Therefore, we reasoned that pulses of Glass activity after this time point should not rescue the overall morphology of the eye but should rescue photoreceptor differentiation. Consistent with this hypothesis, we observed that periodic pulses of Glass after 24 hrs APF neither rescued nor induced additional defects with respect to eye morphology (Fig. 8A). However, the reintroduction of Glass was capable of restoring differentiation in gl mutant retinas, as indicated by the appearance of rhabdomeres, which were separated by a distinct inter-rhabdomeral space (Fig. 8B,C). Many of the “rescued” ommatidia did not have the full complement of eight photoreceptors in agreement with the presence of defects in ommatidium patterning and consistent with our hypothesis of two temporal and separable requirements of Glass activity for eye development.
Fig. 8. Rescue of photoreceptor differentiation with Glass and Pph13.
Transmission and Scanning electron microscopy of (A,B) gl3, heat-shock(hs)-gl/gl3, heat-shock-gl (C) heat-shock-gl/heat-shock-gl; gl60j/gl60j, (D) heat-shock-Pph13/heat-shock-Pph13; gl3/gl3, (E) heat shock-Pph13/+; gl60j/gl60j. (F) heat shock-Pph13/+; gl60j/gl60j, gl-BAC. Inset represents a higher magnification of the rhabdomeres present. Samples were from 96hrs APF. In all cases, white pupae were collected and aged for 24 hrs before subjected to three 1 hr heat shocks/day. SEM Scale Bar-100um, TEM Scale Bar-1um.
Targets of Glass include critical factors for phototransduction and rhabdomere morphogenesis
To further explore how Glass promotes photoreceptor differentiation, we identified potential transcriptional targets of Glass. To date, a combination of transcriptome analyses, binding site analyses, and comparisons among Drosophila species have identified 173 putative Glass targets (Naval-Sanchez et al., 2013; Potier et al., 2014). However, these transcriptome analyses were limited to the third instar eye imaginal disc. In contrast, our results demonstrated at least two critical phases of Glass function, hinting at the existence of unique targets at different developmental times. Therefore, we reasoned that examination of the changes in the Glass transcriptome at 48 hrs APF would provide a more accurate representation of the variations in gene expression required for differentiation. In addition, by utilizing the gl3 allele we could bypass the severity of the organizational defects observed in gl60j alleles and potentially eliminate or reduce the changes in transcription due to the earlier loss of Glass function. Thus, we performed RNA-seq analysis of control and gl3 mutant retinas prepared from 48 hrs APF pupae. From this analysis, we identified 2781 candidates with a ranked DEseq2 adjusted P-value of less than or equal to .05 and of those genes, 1254 targets demonstrated at least a log-2-fold decrease in expression upon removal of Glass (the top fifty candidates are listed in Table S1). Intriguingly, a third of these potential targets were either eye-enriched (Xu et al., 2004) or known to participate in photoreceptor differentiation.
In order to parse our complete data set and identify candidate genes responsible for mediating Glass directing differentiation we filtered our data set to previously generated expression analyses to identify candidate genes that had moderate or high expression in the eye (see methods). We found thirty-two and twenty-one candidates that demonstrated moderate or high expression in the adult retina, respectively (Table S2, S3). However, this analysis was limited because our comparisons were at two different developmental stages (48 hrs APF vs. adult), not all of the candidates have been assayed for expression in the eye, and some genes may be retina specific but only expressed at low levels. Nonetheless, of the twenty-one candidates that demonstrate high expression, ten of these genes have known roles in photoreceptor differentiation (Table S3). In addition, we examined for the presence of a Glass binding sites (See Methods and Table S1,S7) to highlight potential direct transcriptional targets and utilized RT-PCR of a subset of putative targets in gl60j null retinas to confirm the decrease in transcript levels detected by RNA-seq in the gl3 hypomorph (Fig. S9).
Interestingly, our transcriptome data identified Prominin as a putative transcriptional target of Glass; Prominin is essential for the correct morphogenesis and organization of the rhabdomeres within each ommatidium (Nie et al., 2012; Zelhof et al., 2006). This assessment was consistent with the lack of Prominin immunofluorescence staining during the initiation of rhabdomere morphogenesis in gl mutants (Fig. 6F) and the decrease in transcripts at 48 hrs APF as compared to wild type in gl60j null mutant retinas (Figure S9). Furthermore, the identification of Prominin supported our notion for the existence of unique Glass targets at different developmental stages. Prominin is neither expressed in the 3rd instar eye imaginal disc nor required until 40 hrs APF (Nie et al., 2014; Zelhof et al., 2006), and as such, could not have been identified in the earlier screens (Naval-Sanchez et al., 2013; Potier et al., 2014). Overall, our transcriptome data support the model that Glass represents a global upstream regulator that choreographs a network of genes required for generating the rhabdomere (e.g. chaoptin and prominin), genes required for phototransduction (e.g trp, arr2, gβ76C, ninaA), organizing the phototransduction molecules into functional signaling complexes (inaD, inaC), and synaptogenesis (CG9935/ekar).
Glass directs the transcription of Pph13
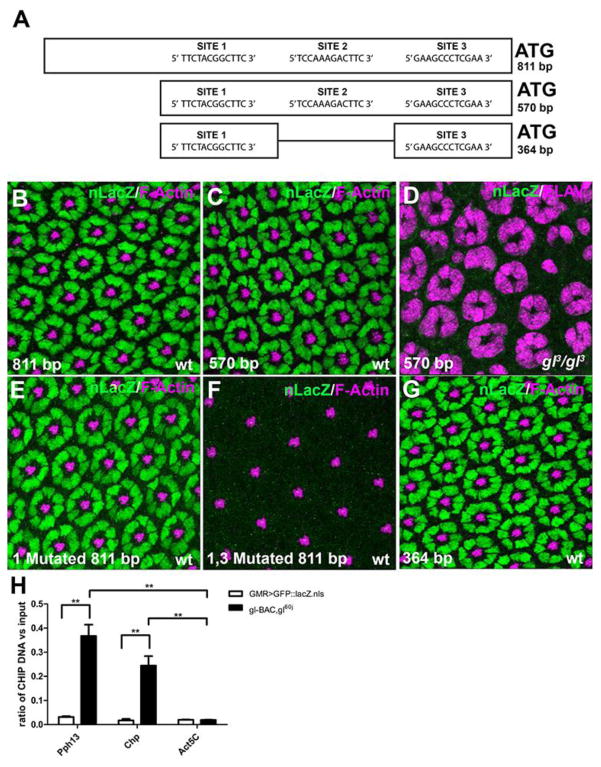
Our transcriptome data highlighted a second notable down-regulated target, Pph13, which was likely integral in mediating Glass directed differentiation. For example, the loss of Pph13 could explain the down-regulation of previously identified Pph13 transcriptional targets: Def, CG30900, arr2, trp, gβ76C, and PIP82 (Mishra et al., 2010) (Table S4). As noted, Pph13 expression was not detected at all in gl mutants, suggesting Pph13 was a direct transcriptional target (Fig. S8B). To test this hypothesis, we identified a genomic fragment from the Pph13 locus that reproduced the endogenous Pph13 expression pattern when linked to a reporter (Fig. 9A). We found that a minimum fragment of 811 base pairs (bp) contained all the elements that activated and limited Pph13 expression to photoreceptors (Fig. 9A,B). The fragment could be reduced to 570 bp (Fig. 9A,C), but any further 5′ or 3′ deletion abolished expression. Consistent with the absence of Pph13 protein, all transcriptional activity was lost from these fragments in gl3 retinas (Fig. 9D). Bioinformatics analysis detected three potential Glass binding sites within this fragment (Fig. 9A). Mutation of any individual site did not eliminate expression (Fig. 9E and data not shown), but reporter gene activity was completely eliminated when sites 1 and 3 were mutated in tandem (Fig. 9F). In addition, a fragment containing only these two sites was sufficient to reproduce Pph13 reporter expression (Fig. 9G). Lastly, in vivo binding analysis utilizing chromatin immunoprecipitation followed by quantitative PCR analysis indicated that Glass occupied these sites at 48 hr APF (Fig. 9H). Overall, these findings demonstrated Glass was essential for Pph13 transcription and suggested that Pph13 mediates transcriptional instruction by Glass during photoreceptor differentiation.
Fig. 9. Pph13 is a direct transcriptional target of Glass.
(A) Schematic of genomic fragments (811, 570, and 364 bp) fused to nuclear LacZ (nLacZ) and sequence of potential Glass binding sites within the fragments. The ATG represents the first codon of Pph13. Immunofluorescence staining of 48 hr wild-type (B–C,E–G) or gl3 retinas (D). (B–C) nLacZ expression (green) counterstained with F-actin (magenta) of the 811 bp (B) and 570 bp (C) reporter constructs. (D) nLacZ expression (green) in gl3 retinas counterstained with ELAV (magenta) of the 570 bp fusion construct. (E) Immunofluoresence staining of nLacZ expression (green) in 48 hr APF wild-type retinas counterstained with F-actin (magenta) of the 811 bp construct with binding Site 1 mutated. (F) nLacZ expression (green) in 48 hr APF wild-type retinas counterstained with F-actin (magenta) of the 811 bp construct with binding Site 1 and Site 3 mutated. (G) nLacZ expression (green) in 48 hr APF wild-type retinas counterstained with F-actin (magenta) of the 364 bp construct. (H) ChIP-qPCR of 48 hrs APF gl60j, gl-BAC/gl60j, gl-BAC (black bars) and GMR-GAL4/+; UAS-GFP::nlacZ/+ (white bars) against Pph13, chaoptin, and actin5C. The Y-axis represents the ratio of the qPCR of pellet DNA normalized to input. Error bars represent standard error of measurement for three independent experiments. **p<0.01.
Our findings that Pph13 is a direct transcriptional target and that late expression of Glass can re-initiate differentiation presented an opportunity to assay how Pph13 contributes to photoreceptor differentiation in gl mutant retinas. Can the presence of Pph13 in gl mutant photoreceptors rescue the differentiation of photoreceptors? First, pulses of Pph13 expression subsequent to the completion of ommatidium patterning did not further disrupt or rescue overall gl mutant eye morphology (data not shown). Second, we found the ability of Pph13 to re-initiate differentiation was dependent upon the amount of Glass activity present. In the gl3 hypomorph, Pph13 could rescue differentiation as observed by the presence of definable rhabdomeres (Fig. 8D). However, such rescue was not observed in the gl60j null retinas but restored in the presence of a single copy of our gl-BAC genomic rescue construct in the gl60j mutant background (Fig. 8E,F). These observations indicated that Glass regulated photoreceptor differentiation was partially directed through Pph13 but the degree of rescue relies on Glass-dependent, Pph13-independent targets, e.g. chaoptin and prominin.
Conservation of Glass function between holometabolous insects
The Drosophila adult visual system is one example of the two fundamental visual systems we find in nature, rhabdomeric and ciliary. Our studies in Drosophila have now described a key regulatory link between rhabdomeric cell type patterning and differentiation mediated by Glass and Pph13. Both of these molecules are conserved in Pancrustaceans (Liu and Friedrich, 2004; Rivera et al., 2010) and homologs can be found in other protostomes including Apylsia californica ((Mahato et al., 2014) and NCBI Ref. Seq.: XM_005091098.1). As such, whether the Glass-Pph13 regulatory interaction is conserved among Protostomes rhabdomeric visual systems remains to be explored. As a first step in deciphering the potential evolutionary role of Glass, we investigated the function of Glass in Tribolium castaneum. Previous reports have demonstrated that both Glass and Pph13 are expressed in the developing photoreceptors of the Tribolium castaneum adult visual system (Liu and Friedrich, 2004; Mahato et al., 2014). Utilizing RNAi, SEM analysis of Glass RNAi knockdowns revealed an identical phenotype as compared to Drosophila; there was an overall reduction of eye size, including a reduction in ommatidium number, as well as the presence of fused ommatidia (Fig. 10A,B). Additionally, TEM analysis confirmed the complete absence of any recognizable ommatidium structure (Fig. 10C,D). To test whether the regulatory interaction between Glass and Pph13 was conserved we performed two sets of complementary experiments. First, we have previously demonstrated that in Drosophila and Tribolium Pph13 was sufficient and necessary for 3XP3-reporter expression, a photoreceptor specific reporter (Mahato et al., 2014; Mishra et al., 2010). If Glass was regulating the transcription of Pph13, we would predict 3XP3 expression to be abolished upon knockdown of Glass function. Indeed, the introduction of glass dsRNA prior to pupariation was sufficient to eliminate 3XP3-RFP expression (Fig. 10E,F). Second we have identified a 1163 bp fragment immediately upstream of the first codon of Tribolium Pph13 that when placed in Drosophila drives reporter expression that phenocopies the endogenous expression of Drosophila Pph13 (Fig. 10G). The fragment not only contains several putative Glass binding sites (data not shown) but transcriptional activity was eliminated upon the removal of Glass (Fig. 10H). Altogether, our data demonstrated that the function of Glass and its regulatory interaction with Pph13 is conserved between these two visual systems and may represent an ancestral regulatory cassette to create a functional rhabdomeric visual system.
Fig. 10. Evolutionary conservation of Glass function in rhabdomeric visual systems.
(A–D) Scanning and Transmission electron microscopy of mock-injected Tribolium (A,C) and Tribolium injected with dsRNA directed against Tc glass (B,D). TEM Scale Bar 2um. (E,F) Fluorescence imaging of 3XP3-RFP in mock-injected Tribolium (E) and Tribolium injected with dsRNA directed against Tc glass (F). Brackets outline eye specific expression of 3XP3-RFP and an asterisk marks the non Pph13-dependent enhancer staining in the brain. (G,H) Merged image of immunofluoresence staining of ELAV (magenta) and nLacZ (green) of the 1163 bp reporter fusion construct in wild-type (G) and gl3 mutant (H) Drosophila retinas.
DISCUSSION
Two temporal roles for Glass function
The requirement for Glass in eye development has been well described (Ellis et al., 1993; Moses et al., 1989; Moses and Rubin, 1991; Ready et al., 1986; Treisman and Rubin, 1996), but the molecular mechanisms leading to the observed phenotypes have remained elusive (Potier et al., 2014). Our re-evaluation of glass mutant phenotypes and our investigation of potential molecular mechanisms have revealed a critical aspect of Glass biology. First, our data confirms and is in agreement with previous reports that the loss of Glass function resulted in early defects in ommatidium patterning and cell type specification and that the loss of identified downstream targets of Glass correlate with the overall defects in the disruption of eye morphology (Hayashi et al., 2008; Naval-Sanchez et al., 2013; Potier et al., 2014; Treisman and Rubin, 1996; Yan et al., 2003). Moreover, our results demonstrated that these defects persist through eye development. Interestingly, staining with Dlg, and confirmed in our TEM analyses, suggest that the patterning defects or potentially maintenance of cell fate is furthered hampered or abolished in glass mutants as development proceeds. For example, even though we detect markers for pigment and cone cells, the Dlg staining clearly reveals that the morphological cell shape changes associated with these cell types is not occurring; the cone and pigment cells have not adopted their correct morphology or position within an ommatdium. In addition, we also examined the role of cell death. Initially, we did not detect any difference between wild type and mutant retina fields but by 48 hrs APF, we observed an increase in the number of apoptotic cells. By 72 hrs in gl60j null mutation the cell death was extensive, especially in the periphery of gl60j null retinas. Nonetheless, the induced expression of the caspase inhibitor p35 failed to rescue any aspect of glass mutant phenotype: the reduction of eye size, the presence of the naked cuticle surrounding the ommatidium field, the disorder of facet patterning and the failure in photoreceptor differentiation. These results suggest that cell death may be a secondary consequence of a failed developmental program and this highlights the importance of Glass for the establishment of the correct patterning of each ommatidium and the subsequent execution of the developmental program of retinal cells.
The execution of a developmental program, and second temporal role of Glass in eye development, is clearly observed with respect to photoreceptors. Our results indicate the apparent loss of photoreceptors in gl mutants was not solely a consequence of cell death (Moses et al., 1989; Ready et al., 1986) but rather a combined outcome of photoreceptors dying and the inability of the remaining mutant photoreceptors to differentiate. Our examination of different developmental stages indicated that there are always two populations of ELAV positive neurons, Dcp-1 positive and negative. There is the possibility that all surviving neurons may eventually die during adult life, but in either case these neurons do not have any characteristics of or potentially function as photoreceptors. Our data indicate that the surviving photoreceptors fail to differentiate and this failure was independent of ommatidium patterning. Our conclusions were supported by two complementary rescue experiments and examination of gl mutant photoreceptors during metamorphosis. glass is a recessive mutation and as such, one copy of our genomic rescue construct (gl-BAC) should have rescued all phenotypes. Instead, we only observed a rescue of eye morphology and the subsequent restoration of spatial and temporal expression of genes critical for ommatdium patterning. Yet, despite the clear presence of photoreceptors and proper patterning, there was a failure to detect photoreceptors in TEM sections based on morphology. In addition, proteins critical for initiating and executing photoreceptor differentiation were absent in gl mutant photoreceptors and those that were detectable were not sufficient for differentiation to occur. Nonetheless, the photoreceptors still maintained the capacity to differentiate, as the reintroduction of Glass function was capable of producing morphologically differentiated photoreceptors. Significantly, photoreceptor differentiation was occurring after patterning of the ommatidium was completed, demonstrating that the role of Glass in patterning of the ommatidium was separable from the role of Glass in photoreceptor differentiation.
What is the molecular basis of the two separable functions of Glass in eye development? Our data and previously published data (Ellis et al., 1993; Moses et al., 1989; Treisman and Rubin, 1996) suggested two non-mutually exclusive scenarios. The first is the level of Glass activity, namely, the failure of photoreceptor differentiation was due to the reduction of active Glass protein. Our study reinforced the previous assessment that gl3 represents a hypomorphic allele; the coding sequence is not effected (Moses et al., 1989). Likewise, we believe we have a complementary scenario with our gl-BAC construct. The sequences required for expression are unaffected. However, by placing epitope tags at the 3′ end of the protein in close proximity to the DNA binding zinc-fingers, we may have inadvertently compromised the ability of Glass to bind DNA or interact with co-factors for proper activation of transcription. Our data supports the idea that the amount of active protein was not sufficient for photoreceptor differentiation but adequate for proper patterning and differentiation of the non-neuronal cone and pigment cells.
The second mechanism, supported by our data, was that the Glass transcriptional targetome changes over developmental time. For example, if we compare the previous 173 identified third instar eye imaginal disc targets (Naval-Sanchez et al., 2013; Potier et al., 2014) to our 1254 identified candidates, there is an overlap of forty-three targets and only seven of those demonstrated at least a log-2-fold decrease in our data set (Table S5). This difference could be attributed to the restriction of Glass to only certain retina cell types by 48 hrs APF. However, this would not explain why Glass targets Pph13 and prominin were expressed at 48 hrs APF but not in the photoreceptors of the eye imaginal disc. Moreover, this change in transcriptional targets can be related to a change in Glass activity and/or a differential requirement of Glass activity for transcriptional activation over developmental time. Ellis et al. had previously demonstrated that non-photoreceptor cells are developmentally restricted in their response to ectopic expression of Glass. In addition, the proximal sequence to the Glass binding site, located in the Rh1 promoter, was required to restrict expression to only photoreceptors (Ellis et al., 1993). A similar scenario can be applied to the transcription of Pph13 and prominin. In addition to limiting spatial expression, cis-regulatory sequences could also be directing temporal expression.
Glass mediated photoreceptor differentiation requires a combination of targets
Besides revealing separable functions of Glass, we found that the specified neurons, as assayed by ELAV staining, were present at all stages in both alleles tested up to the point of eclosion. More importantly, it is not that specified gl mutant, ELAV positive neurons, were transformed to another retinal fate but rather they were stalled in the differentiation pathway leading to a functional photoreceptor. As such, by utilizing the gl3 hypomorph allele and performing our transcriptome analysis at 48 hrs APF, we maximized the ability to identify the factors critical for Glass mediated differentiation. Our analysis has identified numerous characterized factors responsible for every key aspect of photoreceptor differentiation in addition to new factors that may reveal more avenues through which Glass mediates differentiation. Moreover, our data highlight the importance of the interplay between the entire Glass transcriptome for generating a functional photoreceptor. For example, we have demonstrated that Pph13 is a direct transcriptional target but we found that reintroduction of Pph13 was not sufficient to rescue rhabdomere morphogenesis in gl null retinas compared to gl3 retinas. We attribute this difference not to the inability of Pph13 to activate downstream components but rather to the dependency on other Glass targets, i.e. Chaoptin and Prominin, for the presence of rhabdomeres. The generation of a rhabdomere is dependent upon the interplay between Chaoptin, Prominin, EYS, Crumbs (Gurudev et al., 2014; Nie et al., 2014; Zelhof et al., 2006). In a gl60j retina, regardless of the presence of Pph13 a rhabdomere cannot form due to the absence of Chaoptin. In a gl3 or gl60j/gl60j,gl-BAC retina, there was residual Chaoptin and Prominin activity and in combination with Pph13 generated malformed rhabdomeres. Nevertheless, the rhabdomeres had a distinct prominin phenotype; there were pockets of inter-rhabdomeral space and the rhabdomeres were fused together (Zelhof et al., 2006). We believe the addition of Pph13 increased the likelihood of generating rhabdomeres on all photoreceptors present.
Conservation of Glass regulatory pathways for neuronal differentiation
Much progress has been made in elucidating the molecular networks required for specifying retinal cells, including photoreceptors, but the downstream mechanisms that maintain identity and regulate differentiation remain poorly understood. Our description of the conservation of Glass’s function in eye/photoreceptor development and the crosstalk between Glass and Pph13 in two insects suggest that this transcriptional interaction may be a key regulatory link between Pancrustaceans compound eye patterning and rhabdomeric photoreceptor differentiation. Moreover, our studies can serve as the entry point to answer a further question: does this pathway have broader conservation for rhabdomeric differentiation? It is indeed the case that the presence of homologs of Glass and Pph13 were discovered not only in Pancrustaceans, but also in other protostomes such as Aplysia californica, which have a different structural organization of eye in contrast to the compound eye but contain rhabdomeric photoreceptors (Jacklet, 1969; Jacklet et al., 1972; Liu and Friedrich, 2004; Mahato et al., 2014; Rivera et al., 2010). Thus further research on these protostomes will reveal whether Glass-Pph13 interplay acts as an evolutionary ancient transcriptional motif between the morphogenesis of various eye structures and the ensuing differentiation of rhabdomeric photoreceptor neurons. Second, it has been postulated that Otd, another shared transcriptional regulator for rhabdomere differentiation among Pancrustaceans, represents the ancestral state for development and function of the prototype photoreceptor and Pph13 joined the regulatory network during the acquisition of a more sophisticated rhabdomere organization at a later stage (Friedrich et al., 2016). This hypothesis is supported by the fact that Otx/Otd homologs are instrumental in directing both rhabdomeric and ciliary photoreceptor differentiation and deuterostomes appear to lack Pph13 homologs (Friedrich et al., 2016; Mahato et al., 2014). As Pph13 expression requires Glass, and if it is true that the Glass-Pph13 interaction represented an ancestral regulatory mechanism during the evolution of rhabdomeric photoreceptors, the lack of Pph13 orthologs outside protostomes can be partially explained by the absence of glass orthologs. In fact, the distribution of glass orthologs is not found throughout the entire Bilateria. Using phylogenetic analysis, Rivera et al. found a putative glass homolog in the chordate Branchistoma floridae and suggested a loss of glass gene family early in chordate evolution (Rivera et al., 2010). Although it has been reported that vertebrate ZNF500 and ZFP64 are either phylogenetically distantly related to glass or actually glass orthologs (Etchberger et al., 2007; Mack et al., 1997), our limited analysis with reciprocal-BLAST best-hit does not support this view. Therefore, phylogenetically dating the glass and Pph13 interaction will be helpful to uncover whether the emergence of these two genes coincided within certain phylum and photoreceptor architecture and thus address the hypothesis that Glass-Pph13- regulated rhabdomere differentiation diverged from the ancestral mechanisms driven by Otx proteins during evolution.
Interestingly, our observation that the surviving glass mutant photoreceptors failed to complete photoreceptor differentiation suggests that Glass exerts a broader role in regulating neuronal development and identity maintenance. This role is similar to the role described for the C. elegans homolog of Glass che (chemotaxis-defective)-1 (Uchida et al., 2003). In C. elegans the development and function of ASE gustatory neurons requires che-1 (Chang et al., 2003; Etchberger et al., 2009; Etchberger et al., 2007; Uchida et al., 2003). Che-1 is exclusively expressed in C. elegans ASE gustatory neurons (Etchberger et al., 2007; Uchida et al., 2003). In contrast, Glass expression is not merely restricted in photoreceptor neurons, but is also detected in retinal non-neuronal cells, indicating other roles of Glass in the development of visual and nervous system (Ellis et al., 1993; Moses et al., 1989; Moses and Rubin, 1991). With respect to photoreceptors, the loss of glass can lead to cell death but of those photoreceptors that survive they do not complete differentiation, exhibiting loss of photoreceptor specific gene expression and severe morphological defects. In comparison, che-1 deficient neurons fail to sense taste cues and lose the expression of ASE specific markers; however, ASE neurons are still present in correct number and anatomical localization and the morphological features of sensory neurons are still preserved (Chang et al., 2003; Etchberger et al., 2009; Etchberger et al., 2007; Lewis and Hodgkin, 1977; Uchida et al., 2003). Thus che-1 is only required for determining the terminal ASE features of differentiated sensory neurons (Etchberger et al., 2009). Therefore, even though each appear to act at the top of the regulatory hierarchy to direct the transcription of a large battery of sensory specific gene expression, the specific functions of each homolog has diverged with respect to sensory neuron specification, differentiation, and maintenance of neuronal identity.
Overall, our study has now generated a paradigm in which the contribution of specific Glass targets or combinations thereof can be addressed to better understand both the transition of neuronal specification to differentiation and the mechanisms of photoreceptor differentiation. Furthermore, the conserved transcriptional link between Glass and Pph13 will aid as a key hallmark to unravel the evolutionary developmental history of rhabdomeric visual systems.
Supplementary Material
Highlights.
Glass represents a transcriptional link between photoreceptor specification and patterning and terminal differentiation.
glass mutant photoreceptors are capable of surviving pupal development but fail to differentiate.
The role of Glass in ommatidium patterning and photoreceptor differentiation are separable.
Pph13 is a direct transcriptional target of Glass.
Acknowledgments
Funding
This work was supported by the NIH (R21 EY024125) and the NSF (IOS-1353267) to A.C.Z.
We thank Dr. T. Cook, the Bloomington Drosophila Stock Center and Developmental Studies Hybridoma Bank for reagents. We thank the assistance of Dr. J. Powers and the Indiana University Light Microscopy Imaging Center for image generation. We thank A. Hurlbert, Drs. M. Friedrich, N. Sokol, J. Tennessen and anonymous reviewers for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S. Two themes on the assembly of the Drosophila eye. Current topics in developmental biology. 2010;93:85–127. doi: 10.1016/B978-0-12-385044-7.00004-7. [DOI] [PubMed] [Google Scholar]
- Beckstette M, Homann R, Giegerich R, Kurtz S. Significant speedup of database searches with HMMs by search space reduction with PSSM family models. Bioinformatics. 2009;25:3251–3258. doi: 10.1093/bioinformatics/btp593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges CB, Morgan TH. The third-chromosome group of mutant characters of Drosophila melanogaster. Carnegie Institution of Washington; Washington: 1923. [Google Scholar]
- Chang S, Johnston RJ, Jr, Hobert O. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes Dev. 2003;17:2123–2137. doi: 10.1101/gad.1117903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton-Perkins M, Cook TA. Building a fly eye: terminal differentiation events of the retina, corneal lens, and pigmented epithelia. Current topics in developmental biology. 2010;93:129–173. doi: 10.1016/B978-0-12-385044-7.00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton-Perkins M, Whitaker SL, Fei Y, Xie B, Li-Kroeger D, Gebelein B, Cook T. Prospero and Pax2 combinatorially control neural cell fate decisions by modulating Ras- and Notch-dependent signaling. Neural development. 2011;6:20. doi: 10.1186/1749-8104-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Cook T, Pichaud F, Sonneville R, Papatsenko D, Desplan C. Distinction between color photoreceptor cell fates is controlled by Prospero in Drosophila. Dev Cell. 2003;4:853–864. doi: 10.1016/s1534-5807(03)00156-4. [DOI] [PubMed] [Google Scholar]
- Daga A, Karlovich CA, Dumstrei K, Banerjee U. Patterning of cells in the Drosophila eye by Lozenge, which shares homologous domains with AML1. Genes Dev. 1996;10:1194–1205. doi: 10.1101/gad.10.10.1194. [DOI] [PubMed] [Google Scholar]
- Ellis MC, O’Neill EM, Rubin GM. Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development. 1993;119:855–865. doi: 10.1242/dev.119.3.855. [DOI] [PubMed] [Google Scholar]
- Etchberger JF, Flowers EB, Poole RJ, Bashllari E, Hobert O. Cis-regulatory mechanisms of left/right asymmetric neuron-subtype specification in C. elegans. Development. 2009;136:147–160. doi: 10.1242/dev.030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchberger JF, Lorch A, Sleumer MC, Zapf R, Jones SJ, Marra MA, Holt RA, Moerman DG, Hobert O. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 2007;21:1653–1674. doi: 10.1101/gad.1560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Lee TV, Xu D, Chen Z, Lamblin AF, Steller H, Bergmann A. Dual roles of Drosophila p53 in cell death and cell differentiation. Cell death and differentiation. 2010;17:912–921. doi: 10.1038/cdd.2009.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores GV, Daga A, Kalhor HR, Banerjee U. Lozenge is expressed in pluripotent precursor cells and patterns multiple cell types in the Drosophila eye through the control of cell-specific transcription factors. Development. 1998;125:3681–3687. doi: 10.1242/dev.125.18.3681. [DOI] [PubMed] [Google Scholar]
- Friedrich M, Cook T, Zelhof AC. Ancient default activators of terminal photoreceptor differentiation in the pancrustacean compound eye: the homeodomain transcription factors Otd and Pph13. Current Opinion in Insect Science. 2016;13:33–42. doi: 10.1016/j.cois.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goriely A, Mollereau B, Coffinier C, Desplan C. Munster, a novel paired-class homeobox gene specifically expressed in the Drosophila larval eye. Mech Dev. 1999;88:107–110. doi: 10.1016/s0925-4773(99)00168-9. [DOI] [PubMed] [Google Scholar]
- Gurudev N, Yuan M, Knust E. chaoptin, prominin, eyes shut and crumbs form a genetic network controlling the apical compartment of Drosophila photoreceptor cells. Biology open. 2014;3:332–341. doi: 10.1242/bio.20147310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Saigo K. Diversification of cell types in the Drosophila eye by differential expression of prepattern genes. Mech Dev. 2001;108:13–27. doi: 10.1016/s0925-4773(01)00466-x. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Xu C, Carthew RW. Cell-type-specific transcription of prospero is controlled by combinatorial signaling in the Drosophila eye. Development. 2008;135:2787–2796. doi: 10.1242/dev.006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacklet JW. Electrophysiological organization of the eye of Aplysia. J Gen Physiol. 1969;53:21–42. doi: 10.1085/jgp.53.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacklet JW, Alvarez R, Bernstein B. Ultrastructure of the eye of Aplysia. J Ultrastruct Res. 1972;38:246–261. doi: 10.1016/s0022-5320(72)90002-0. [DOI] [PubMed] [Google Scholar]
- Kauffmann RC, Li S, Gallagher PA, Zhang J, Carthew RW. Ras1 signaling and transcriptional competence in the R7 cell of Drosophila. Genes Dev. 1996;10:2167–2178. doi: 10.1101/gad.10.17.2167. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Senoo-Matsuda N, Hiromi Y, Miura M. DRONC coordinates cell death and compensatory proliferation. Mol Cell Biol. 2006;26:7258–7268. doi: 10.1128/MCB.00183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JA, Hodgkin JA. Specific neuroanatomical changes in chemosensory mutants of the nematode Caenorhabditis elegans. J Comp Neurol. 1977;172:489–510. doi: 10.1002/cne.901720306. [DOI] [PubMed] [Google Scholar]
- Lin HV, Rogulja A, Cadigan KM. Wingless eliminates ommatidia from the edge of the developing eye through activation of apoptosis. Development. 2004;131:2409–2418. doi: 10.1242/dev.01104. [DOI] [PubMed] [Google Scholar]
- Liu Z, Friedrich M. The Tribolium homologue of glass and the evolution of insect larval eyes. Developmental biology. 2004;269:36–54. doi: 10.1016/j.ydbio.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Lorenzen MD, Kimzey T, Shippy TD, Brown SJ, Denell RE, Beeman RW. piggyBac-based insertional mutagenesis in Tribolium castaneum using donor/helper hybrids. Insect Mol Biol. 2007;16:265–275. doi: 10.1111/j.1365-2583.2007.00727.x. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack HG, Beck F, Bowtell DD. A search for a mammalian homologue of the Drosophila photoreceptor development gene glass yields Zfp64, a zinc finger encoding gene which maps to the distal end of mouse chromosome 2. Gene. 1997;185:11–17. doi: 10.1016/s0378-1119(96)00607-5. [DOI] [PubMed] [Google Scholar]
- Mahato S, Morita S, Tucker AE, Liang X, Jackowska M, Friedrich M, Shiga Y, Zelhof AC. Common transcriptional mechanisms for visual photoreceptor cell differentiation among Pancrustaceans. PLoS Genet. 2014;10:e1004484. doi: 10.1371/journal.pgen.1004484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Bermudo MD, Bardet PL, Bellaiche Y, Malartre M. The vav oncogene antagonises EGFR signalling and regulates adherens junction dynamics during Drosophila eye development. Development. 2015;142:1492–1501. doi: 10.1242/dev.110585. [DOI] [PubMed] [Google Scholar]
- Mencarelli C, Pichaud F. Orthodenticle Is Required for the Expression of Principal Recognition Molecules That Control Axon Targeting in the Drosophila Retina. PLoS Genet. 2015;11:e1005303. doi: 10.1371/journal.pgen.1005303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M, Oke A, Lebel C, McDonald EC, Plummer Z, Cook TA, Zelhof AC. Pph13 and orthodenticle define a dual regulatory pathway for photoreceptor cell morphogenesis and function. Development. 2010;137:2895–2904. doi: 10.1242/dev.051722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey M, Yee SK, Herman T, Nern A, Blanco E, Zipursky SL. Coordinate control of synaptic-layer specificity and rhodopsins in photoreceptor neurons. Nature. 2008;456:795–799. doi: 10.1038/nature07419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses K, Ellis MC, Rubin GM. The glass gene encodes a zinc-finger protein required by Drosophila photoreceptor cells. Nature. 1989;340:531–536. doi: 10.1038/340531a0. [DOI] [PubMed] [Google Scholar]
- Moses K, Rubin GM. Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev. 1991;5:583–593. doi: 10.1101/gad.5.4.583. [DOI] [PubMed] [Google Scholar]
- Naval-Sanchez M, Potier D, Haagen L, Sanchez M, Munck S, Van de Sande B, Casares F, Christiaens V, Aerts S. Comparative motif discovery combined with comparative transcriptomics yields accurate targetome and enhancer predictions. Genome research. 2013;23:74–88. doi: 10.1101/gr.140426.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J, Mahato S, Mustill W, Tipping C, Bhattacharya SS, Zelhof AC. Cross species analysis of Prominin reveals a conserved cellular role in invertebrate and vertebrate photoreceptor cells. Developmental biology. 2012;371:312–320. doi: 10.1016/j.ydbio.2012.08.024. [DOI] [PubMed] [Google Scholar]
- Nie J, Mahato S, Zelhof AC. The actomyosin machinery is required for Drosophila retinal lumen formation. PLoS Genet. 2014;10:e1004608. doi: 10.1371/journal.pgen.1004608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichaud F. Transcriptional regulation of tissue organization and cell morphogenesis: the fly retina as a case study. Dev Biol. 2014;385:168–178. doi: 10.1016/j.ydbio.2013.09.031. [DOI] [PubMed] [Google Scholar]
- Potier D, Davie K, Hulselmans G, Naval Sanchez M, Haagen L, Huynh-Thu VA, Koldere D, Celik A, Geurts P, Christiaens V, Aerts S. Mapping gene regulatory networks in Drosophila eye development by large-scale transcriptome perturbations and motif inference. Cell reports. 2014;9:2290–2303. doi: 10.1016/j.celrep.2014.11.038. [DOI] [PubMed] [Google Scholar]
- Ranade SS, Yang-Zhou D, Kong SW, McDonald EC, Cook TA, Pignoni F. Analysis of the Otd-dependent transcriptome supports the evolutionary conservation of CRX/OTX/OTD functions in flies and vertebrates. Dev Biol. 2008;315:521–534. doi: 10.1016/j.ydbio.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Ready DF, Tomlinson A, Lebovitz RM. Building an Ommatidium: Geometry and Genes. In: Hilfer SR, Sheffield JB, editors. Development of Order in the Visual System. Springer-Verlag; New York: 1986. pp. 97–125. [Google Scholar]
- Rehm P, Borner J, Meusemann K, von Reumont BM, Simon S, Hadrys H, Misof B, Burmester T. Dating the arthropod tree based on large-scale transcriptome data. Mol Phylogenet Evol. 2011;61:880–887. doi: 10.1016/j.ympev.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Reinke R, Krantz DE, Yen D, Zipursky SL. Chaoptin, a cell surface glycoprotein required for Drosophila photoreceptor cell morphogenesis, contains a repeat motif found in yeast and human. Cell. 1988;52:291–301. doi: 10.1016/0092-8674(88)90518-1. [DOI] [PubMed] [Google Scholar]
- Reinke R, Zipursky SL. Cell-cell interaction in the Drosophila retina: the bride of sevenless gene is required in photoreceptor cell R8 for R7 cell development. Cell. 1988;55:321–330. doi: 10.1016/0092-8674(88)90055-4. [DOI] [PubMed] [Google Scholar]
- Rister J, Desplan C. The retinal mosaics of opsin expression in invertebrates and vertebrates. Dev Neurobiol. 2011;71:1212–1226. doi: 10.1002/dneu.20905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera AS, Pankey MS, Plachetzki DC, Villacorta C, Syme AE, Serb JM, Omilian AR, Oakley TH. Gene duplication and the origins of morphological complexity in pancrustacean eyes, a genomic approach. BMC evolutionary biology. 2010;10:123. doi: 10.1186/1471-2148-10-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M, Ma L, Spokony RF, Arthur RK, Kheradpour P, Kundaje A, Negre N, Crofts A, Ptashkin R, Zieba J, Ostapenko A, Suchy S, Victorsen A, Jameel N, Grundstad AJ, Gao W, Moran JR, Rehm EJ, Grossman RL, Kellis M, White KP. Diverse patterns of genomic targeting by transcriptional regulators in Drosophila melanogaster. Genome research. 2014;24:1224–1235. doi: 10.1101/gr.168807.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosinsky A, Bonin CP, Mann RS, Honig B. Target Explorer: An automated tool for the identification of new target genes for a specified set of transcription factors. Nucleic Acids Res. 2003;31:3589–3592. doi: 10.1093/nar/gkg544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahayato A, Sonneville R, Pichaud F, Wernet MF, Papatsenko D, Beaufils P, Cook T, Desplan C. Otd/Crx, a dual regulator for the specification of ommatidia subtypes in the Drosophila retina. Dev Cell. 2003;5:391–402. doi: 10.1016/s1534-5807(03)00239-9. [DOI] [PubMed] [Google Scholar]
- Tomlinson A. Patterning the peripheral retina of the fly: decoding a gradient. Dev Cell. 2003;5:799–809. doi: 10.1016/s1534-5807(03)00326-5. [DOI] [PubMed] [Google Scholar]
- Tomoyasu Y, Denell RE. Larval RNAi in Tribolium (Coleoptera) for analyzing adult development. Dev Genes Evol. 2004;214:575–578. doi: 10.1007/s00427-004-0434-0. [DOI] [PubMed] [Google Scholar]
- Treisman JE. Retinal differentiation in Drosophila. Wiley interdisciplinary reviews. Developmental biology. 2013;2:545–557. doi: 10.1002/wdev.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman JE, Rubin GM. Targets of glass regulation in the Drosophila eye disc. Mech Dev. 1996;56:17–24. doi: 10.1016/0925-4773(96)00508-4. [DOI] [PubMed] [Google Scholar]
- Uchida O, Nakano H, Koga M, Ohshima Y. The C. elegans che-1 gene encodes a zinc finger transcription factor required for specification of the ASE chemosensory neurons. Development. 2003;130:1215–1224. doi: 10.1242/dev.00341. [DOI] [PubMed] [Google Scholar]
- Van Vactor D, Jr, Krantz DE, Reinke R, Zipursky SL. Analysis of mutants in chaoptin, a photoreceptor cell-specific glycoprotein in Drosophila, reveals its role in cellular morphogenesis. Cell. 1988;52:281–290. doi: 10.1016/0092-8674(88)90517-x. [DOI] [PubMed] [Google Scholar]
- Vandendries ER, Johnson D, Reinke R. orthodenticle is required for photoreceptor cell development in the Drosophila eye. Dev Biol. 1996;173:243–255. doi: 10.1006/dbio.1996.0020. [DOI] [PubMed] [Google Scholar]
- Wernet MF, Desplan C. Building a retinal mosaic: cell-fate decision in the fly eye. Trends in cell biology. 2004;14:576–584. doi: 10.1016/j.tcb.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready D. Pattern formation in the Drosophila retina. In: Bate M, Arias AM, editors. The Development of Drosophila melanogaster. Cold Spring Harbor Press; Cold Spring Harbor: 1993. pp. 1277–1325. [Google Scholar]
- Wolff T, Ready DF. Cell death in normal and rough eye mutants of Drosophila. Development. 1991;113:825–839. doi: 10.1242/dev.113.3.825. [DOI] [PubMed] [Google Scholar]
- Xu C, Kauffmann RC, Zhang J, Kladny S, Carthew RW. Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye. Cell. 2000;103:87–97. doi: 10.1016/s0092-8674(00)00107-0. [DOI] [PubMed] [Google Scholar]
- Xu H, Lee SJ, Suzuki E, Dugan KD, Stoddard A, Li HS, Chodosh LA, Montell C. A lysosomal tetraspanin associated with retinal degeneration identified via a genome-wide screen. EMBO J. 2004;23:811–822. doi: 10.1038/sj.emboj.7600112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Canon J, Banerjee U. A transcriptional chain linking eye specification to terminal determination of cone cells in the Drosophila eye. Dev Biol. 2003;263:323–329. doi: 10.1016/j.ydbio.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Zelhof AC, Hardy RW, Becker A, Zuker CS. Transforming the architecture of compound eyes. Nature. 2006;443:696–699. doi: 10.1038/nature05128. [DOI] [PubMed] [Google Scholar]
- Zelhof AC, Koundakjian E, Scully AL, Hardy RW, Pounds L. Mutation of the photoreceptor specific homeodomain gene Pph13 results in defects in phototransduction and rhabdomere morphogenesis. Development. 2003;130:4383–4392. doi: 10.1242/dev.00651. [DOI] [PubMed] [Google Scholar]
- Zhang B, Mehrotra S, Ng WL, Calvi BR. Low levels of p53 protein and chromatin silencing of p53 target genes repress apoptosis in Drosophila endocycling cells. PLoS genetics. 2014;10:e1004581. doi: 10.1371/journal.pgen.1004581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LJ, Christensen RG, Kazemian M, Hull CJ, Enuameh MS, Basciotta MD, Brasefield JA, Zhu C, Asriyan Y, Lapointe DS, Sinha S, Wolfe SA, Brodsky MH. FlyFactorSurvey: a database of Drosophila transcription factor binding specificities determined using the bacterial one-hybrid system. Nucleic Acids Res. 2011;39:D111–117. doi: 10.1093/nar/gkq858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.